Abstract
Although most inbred mouse strains are highly susceptible to mouse hepatitis virus (MHV) infection, the inbred SJL line of mice is highly resistant to its infection. The principal receptor for MHV is murine CEACAM1 (mCEACAM1). Susceptible strains of mice are homozygous for the 1a allele of mCeacam1, while SJL mice are homozygous for the 1b allele. mCEACAM1a (1a) has a 10- to 100-fold-higher receptor activity than does mCEACAM1b (1b). To explore the hypothesis that MHV susceptibility is due to the different MHV receptor activities of 1a and 1b, we established a chimeric C57BL/6 mouse (cB61ba) in which a part of the N-terminal immunoglobulin (Ig)-like domain of the mCeacam1a (1a) gene, which is responsible for MHV receptor function, is replaced by the corresponding region of mCeacam1b (1b). We compared the MHV susceptibility of these chimeric mice to that of SJL and B6 mice. B6 mice that are homozygous for 1a are highly susceptible to MHV-A59 infection, with a 50% lethal dose (LD50) of 102.5 PFU, while chimeric cB61ba mice and SJL mice homozygous for 1ba and 1b, respectively, survived following inoculation with 105 PFU. Unexpectedly, cB61ba mice were more resistant to MHV-A59 infection than SJL mice as measured by virus replication in target organs, including liver and brain. No infectious virus or viral RNA was detected in the organs of cB61ba mice, while viral RNA and infectious virus were detected in target organs of SJL mice. Furthermore, SJL mice produced antiviral antibodies after MHV-A59 inoculation with 105 PFU, but cB61ba mice did not. Thus, cB61ba mice are apparently completely resistant to MHV-A59 infection, while SJL mice permit low levels of MHV-A59 virus replication during self-limited, asymptomatic infection. When expressed on cultured BHK cells, the mCEACAM1b and mCEACAM1ba proteins had similar levels of MHV-A59 receptor activity. These results strongly support the hypothesis that although alleles of mCEACAM1 are the principal determinants of mouse susceptibility to MHV-A59, other as-yet-unidentified murine genes may also play a role in susceptibility to MHV.
Differences in susceptibility to a number of viral infections have been documented among inbred mouse strains (20). These differences have been studied as models for the various degrees of susceptibility of individual humans to some viral infections. Numerous host factors have been found to be involved in such differences (2, 15). For example, allelic variations in the virus receptor and coreceptor for HIV-1 are important host factors influencing susceptibility to HIV-1 infection (36).
A virus receptor is a molecule with which the virus interacts at an initial step of infection. Therefore, receptors are crucial host determinants of virus susceptibility (15, 16). A variety of receptor proteins has been identified for many different viruses, including the murine coronavirus mouse hepatitis virus (MHV) (12, 50). The principal receptor for MHV is murine carcinoembryonic antigen-related cell adhesion molecule 1 (mCEACAM1; previously called Bgp or MHVR [3]), which is in the immunoglobulin (Ig) superfamily (12, 50). Four isoforms of mCEACAM1a (1a) are expressed on the plasma membranes of a variety of murine cells and tissues (14). The two mCEACAM1 isoforms with a molecular mass of 100 to 120 kDa are composed of four Ig-like ectodomains, a transmembrane (TM) domain, and either a long or a short cytoplasmic tail (Cy) (3, 22). Two other isoforms consist of two Ig-like domains, with either long or short Cy (3, 22). The N-terminal (N) domain is responsible for virus binding (10, 24), the induction of conformational changes in the viral spike protein (S), and membrane fusion during virus entry and syncytium formation (13, 24). The replacement of the N-terminal domain of mCEACAM1a with that of the murine homolog of the poliovirus receptor (PVR) yields a functional receptor for MHV (10), and Ceacam1a-knockout mice are completely resistant to infection with the hepatotropic A59 strain of MHV (17, 25).
Wild mice have two alleles of the mCeacam1 gene, called mCeacam1a and mCeacam1b. Inbred mouse strains that are homozygous for mCeacam1a, including BALB/c, C57BL/6 (B6), C3H, and A/J mice, etc., are highly susceptible to infection with strains of MHV. In contrast, the SJL line of inbred mice, which is resistant to death from MHV infection, is homozygous for the mCeacam1b allele (5, 11, 50). The most extensive differences in amino acid sequence between mCEACAM1a and mCEACAM1b are found in the N-terminal domain, where the virus-binding region is located (21, 22, 32). It was initially reported by Boyle et al. that mCEACAM1a proteins had MHV-A59 virus-binding activity in a virus overlay protein blot, while mCEACAM1b did not (5). Those authors speculated that the different viral affinities of these mCEACAM1 proteins may account for the various MHV-A59 susceptibilities of BALB/c mice compared to those of SJL mice (49). However, Yokomori and Lai (53) and Dveksler et al. (11) previously showed that when recombinant CEACAM1a and CEACAM1b proteins are expressed at high levels on cultured cells, both proteins have MHV-A59 receptor activity. Yokomori and Lai suggested that the difference in MHV susceptibility between BALB/c and SJL mice does not depend solely upon the interaction of the virus with mCEACAM1 proteins (52, 53). Dveksler et al. suggested that small differences in MHV-A59 receptor activity between mCEACAM1a and mCEACAM1b could result in very large biological differences during multiple cycles of infection in in vivo infection (11). We then quantitatively showed that recombinant mCEACAM1a expressed in BHK cells has 10- to 30-times-higher MHV-binding activity than mCEACAM1b (31). Similar results were observed in other laboratories (7, 32). Because the mCeacam1 gene is located on chromosome 7 (34) and the gene controlling MHV-A59 susceptibility and the resistance of BALB/c mice versus SJL mice is also located on chromosome 7 close to the mCeacam1 gene (40), we speculated that the mCeacam1 gene is identical to the gene that determines the susceptibility and/or resistance of mice to MHV-A59 and MHV-JHM infection.
To examine the above-described hypothesis, we used progeny mice produced by crossing BALB/c and SJL mice. F2 mice and F1 mice backcrossed to SJL mice were examined for the mCeacam1 genotype and for MHV-JHM susceptibility (30). Mice homozygous for mCeacam1a (1a/1a) and heterozygous mice (1a/1b) were susceptible to lethal MHV-JHM infection, while mice homozygous for mCeacam1b (1b/1b) were not killed by inoculation with MHV-JHM. These data are consistent with the hypothesis that the susceptibility of mice to MHV is determined by the mCeacam1a allele (30). However, this classical genetic analysis could not prove that mCeacam1 alone determines the susceptibility or resistance of mice to MHV-JHM infection, because this methodology cannot rule out the possibility that a different unknown host gene located close to mCeacam1 on chromosome 7 could also affect MHV-JHM susceptibility. Therefore, we used gene replacement in B6 embryonic stem (ES) cells to create a mouse strain in which the exon encoding the N-terminal part of the N-terminal Ig domain of mCeacam1a was replaced with the corresponding region of mCeacam1b from SLJ mice. We bred the chimeric mCeacam1 gene on the B6 background (called B6 chimeric mCeacam1ba, or cB61ba). We compared these mice, wild-type B6 mice, and SJL mice for their susceptibilities to MHV-A59 infection. We confirmed that the expression of mCEACAM1a makes mice susceptible to lethal infection with MHV-A59. However, surprisingly, we found that cB61ba mice were profoundly resistant to MHV-A59 infection, while the virus could replicate at low levels in SJL mice in a self-limited, unapparent infection. Our results suggest that one or more as-yet-unidentified murine genes may also contribute to murine susceptibility and/or resistance to MHV-A59 infection.
MATERIALS AND METHODS
Generation of the targeting vector.
To generate chimeric mCeacam1ba mice, a fragment of DNA encoding amino acids (aa) 1 to 70 of the ∼107-aa-long N-terminal domain of mCeacam1a was replaced by homologous recombination with the corresponding fragment of mCeacam1b from SJL mice (Fig. 1Aa) (11, 48). The DNA fragment corresponding to the sequence from the promoter of mCeacam1a to just upstream of the N domain, from −917 bp upstream to nucleotide (nt) 84 in the 1a open reading frame (ORF), was amplified from a wild-type B6 genomic clone by using primers 84R and 5′F, which has an additional KpnI site on its 5′ end. The second DNA fragment containing the N domain, nt 65 to 417 in the 1b ORF (encoding amino acids 22 to 139), was amplified from a plasmid clone of 1b from SJL mice by using primers 65F and 5′R with a BamHI site. By using these two fragments and primers 5′F and 5′R, we further amplified the DNA fragment containing the promoter of 1a (28) and encoding aa 1 to 70 in the N domain of 1b, which includes the virus-binding region in 1a (28, 48). This fragment was digested with KpnI and BamHI to isolate cDNA that encodes the chimeric N domain and then inserted into the KpnI and BamHI sites of pBluescript (Stratagene, TX). The DNA fragment encoding amino acid 70 to exon 5 of mCeacam1a was digested with BamHI from a C57BL/6 genomic clone and then inserted into the BamHI site of the plasmid described above, containing the chimeric N domain of mCeacam1ba. To insert the neomycin (neo) selection cassette with loxP sequences on both the 5′ and 3′ ends into the targeting vector, a neo selection cassette was digested with XhoI and KpnI from pPNT (46) (kindly provided by R. C. Mulligan, Harvard University) and then cloned into the EcoRV site of pBS246 (Gibco BRL, CA) to place the cassette between two loxP sequences. This loxP-neo-loxP cassette was cloned into the XhoI site in intron 2 of the chimeric mCeacam1ba fragment. The resulting targeting vector was digested with NotI to linearize it for ES cell injection. Primers used in the present study are shown in Table 1.
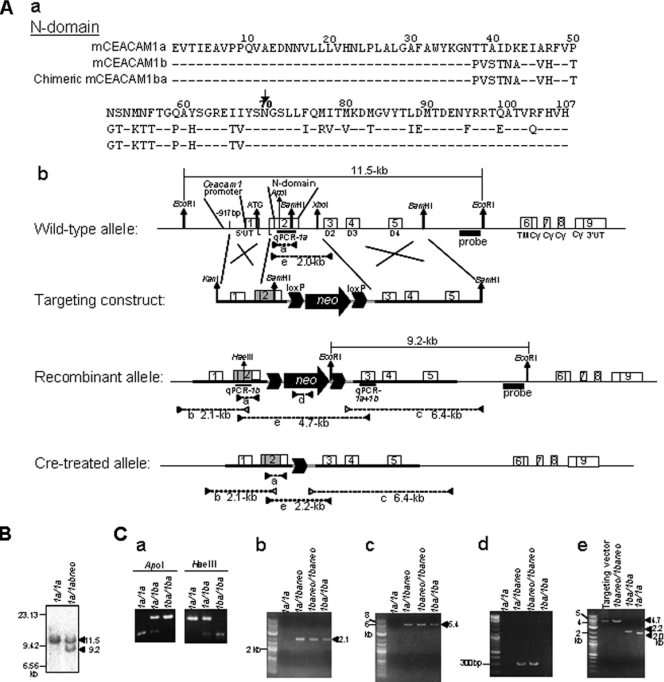
FIG. 1.
Replacement in the mCeacam1a gene of B6 mice with the portion of exon 2 that encodes amino acids 1 to 70 of domain 1 of mCeacam1b of SJL mice. (A) (a) Comparison of the amino acid sequences of the N-terminal domains of the mCEACAM1a, mCEACAM1b, and chimeric mCEACAM1ba proteins of engineered cB61ba mice. For 1b and chimeric 1ba, only amino acids that differ from 1a are shown. Data are from data reported under GenBank accession no. M77196 (CEACAM1a) and M96934 (CEACAM1b). (b) The mCeacam1a gene is composed of nine exons (numbered boxes) preceded by the mCeacam1 promoter (28). Exon 1 consists of the 5′ untranslated region (5′UT), an initiator ATG codon, and the 5′ half of the leader (L) sequence. Exon 2 encompasses the 3′ half of L and the first Ig domain, the N-terminal (N) domain (D1), which has MHV-binding activity (28, 48). Exons 3, 4, and 5 encode three constant-type Ig domains (D2, D3, and D4, respectively). Exon 6 encodes the transmembrane (TM) domain and a part of the cytoplasmic tail (Cy). The rest of Cy is encoded in exons 7, 8, and 9, and alternative splicing generates long and short Cy tails. The 3′ end of the substituted region in mCeacam1a is at a BamHI site that encodes amino acids 1 to 70. The remainder of the chimeric mCeacam1ba gene is identical to mCeacam1a. The targeting construct that leads to recombination events in ES cells has the loxP-neo-loxP selection cassette (arrow and two large arrowheads) inserted into the unique XhoI site in intron 2 (targeting construct). The replaced allele contains a chimeric exon 2 that encodes a chimeric N-terminal domain (D1) consisting of amino acids 1 to 70 of mCeacam1b (gray box) and amino acids 71 to 107 of D1 and the remainder of mCeacam1a (white box) that was replaced by homologous recombination (recombinant and Cre-treated alleles). (B) Southern blot analysis of genomic DNAs from recombinant ES cell clones. By using a probe (black box), indicated in A, we replaced 9.2-kb and detected 11.5-kb wild-type EcoRI fragments in DNA isolated from ES cells. (C) Genotyping of mice using PCR and restriction digestion. a to e correspond to fragments a to e depicted in A. (a) PCR fragments amplified with primers 104F and 431R that correspond to conserved sequences of 1a and 1b were digested with 1a-specific ApoI and 1b-specific HaeIII. (b) The 2.1-kb 5′ border region in the replaced 1baneo allele was PCR amplified by using a primer 5′ upstream of the targeting construct (−997F) and a 1b-specific primer (239R). (c) The 6.4-kb border region in the replaced 1baneo allele was amplified by nested PCR by using plasmid vector-specific primers and primers downstream of the 3′ end of the targeting construct (BS1F and 2204F for first-round PCR and BS2F and 2134R for second-round PCR). (d) To confirm the deletion of the loxP-neo-loxP cassette in the 1ba allele resulting from Cre treatment, an approximately 300-bp neo gene-specific DNA fragment was PCR amplified with primers (neoF and neoR). (e) The 4.7-kb DNA fragment from exons 2 to 3 containing the loxP-neo-loxP cassette in the recombinant 1baneo allele was altered in the 1ba allele by the removal of the loxP-neo-loxP cassette via the Cre-loxP system to yield the 2.2-kb fragment. PCR was done with a pair of primers (104F and 579R).
TABLE 1.
Primers and probes used in this study
| Primer or probe | Sequence (5′→3′)a | Positions in GenBank accession no. or vectorb |
|---|---|---|
| 5′F | CTCCGGTACCCTGGATGACCAACGAAAGAATAC | 1479-1501 in AF287911 |
| 5′R | ATGAAATCGCACAGTCGCCTGAGTA | 3574-3550 in AF287911 |
| 65F | CCTCACTTTTAGCCTCCTGGAG | 134-155 in M77196 |
| 84R | CCAGGAGGCTAAAAGTGAGG | 153-134 in M77196 |
| 104F | GAAGTCACCATTGAGGCTGT | 172-191 in M77196 |
| 239R | ACAATTTCAGCGTTTGTAGACACA | 256-233 in M96934 |
| 431R | ATGAAATCGCACAGTCGCCT | 486-467 in M77196 |
| 579R | GGCTTTCACCATTTCTGCTC | 628-609 in M77196 |
| 2134R | GGCAGAAGAGACATAGGTGGG | 10267-10247 in AF287911 |
| 2204R | GACATGGTTGCCCCAGAGACC | 10337-10317 in AF287911 |
| −997F | GGCGAAAGCTCACAGTCAAC | 1331-1350 in AF287911 |
| 5intF | ATTTTGCTGGCTTTACTAATTTAGT | 11498-11522 in AF287911 |
| 5intR | CTCAATAGTTATATGCTAGCAGCTG | 11979-11955 in AF287911 |
| BS1F | TTAGGTCCCTCGAAGAGGTTC | 259-279 in pBS246 |
| BS2F | CACTAGTACTGGCCATTGCGGC | 279-299 in pBS246 |
| neoF | GGAGAGGCTATTCGGCTATGAC | 656-677 in pPNT |
| neoR | TACTTTCTCGGCAGGAGCAAGG | 947-926 in pPNT |
| mMCEACAM1ab-P | AGAAGGTGACAGGCTGA | 633-649 in M77196 |
| mMCEACAM1ab-F | TGGAGCAGAAATGGTGAAAGC | 607-627 in M77196 |
| mMCEACAM1ab-R | GAGTCCTGTTGCCCTCAGACA | 673-653 in M77196 |
| mMCEACAM1a-P | CAGCAATGGATCCC | 375-388 in M77196 |
| mMCEACAM1a-F | GCATACAGCGGCAGAGAGATAA | 349-370 in M77196 |
| mMCEACAM1a-R | CCTTCATGGTGATCATTTGGAA | 415-394 in M77196 |
| mMCEACAM1b-P | ACAGGCACTAATAAGAC | 267-283 in M96934 |
| mMCEACAM1b-F | GTCTACAAACGCTGAAATTGTACATTTT | 236-263 in M96934 |
| mMCEACAM1b-R | TGTGTGCAGGCCCTGTTG | 303-286 in M96934 |
The restriction site is underlined.
Position according to the sequence of the GenBank accession no. or vector.
Generation of homozygous chimeric B6 mCeacam1ba mice.
The targeting vector was injected into the B6 ES cell line MS12 (19), and cell clones resistant to G418 were selected. Homologous recombination in G418-resistant MS12 ES cell clones was confirmed by Southern blot analysis as follows (3, 17). First, 5 μg of genomic DNA isolated from ES cells was digested with EcoRI, and fragments were separated on 0.75% agarose gels. The DNA fragments were transferred onto a positively charged nylon membrane (GE Healthcare, Buckinghamshire, England) and hybridized with a probe located in the fifth intron (Fig. 1). The DNA probe was generated by PCR from a B6 genomic clone by using primers 5intF and 5intR (Table 1) and labeled with [α-32P]CTP by using a Megaprime kit (GE Healthcare). The selected clones were injected into BALB/c blastocysts, and the blastocysts were implanted into the uteri of ICR (CD1) mice. Chimeric mice born to the implanted mice were mated with wild-type B6 mice homozygous for 1a, and mice heterozygous for 1a and chimeric mCeacam1baneo were obtained. The heterozygous mice were crossed to generate mice homozygous for chimeric mCeacam1baneo. To remove the loxP-neo-loxP cassette in intron 2, these mice were crossed with B6.FVB-TgN(EIIa-Cre)C5379Lmgd mice, which express Cre recombinase (37). Because the original strain of B6.FVB-TgN(EIIa-Cre)C5379Lmgd mice was established in the FVB/N background, we backcrossed them with B6 mice for more than 11 generations to ensure that the chimeric mCecacam1ba gene was in the B6 genetic background (37). The resultant mice, heterozygous for 1a and chimeric 1ba, were then crossed to obtain mice homozygous for the chimeric 1ba genotype. Littermates from this mating that were found to be homozygous for 1a were used as controls (B6) for the genotyping of homozygous chimeric mCeacam1ba mice.
Genotyping of progeny mice.
Genotyping of each progeny mouse was performed on genomic DNA prepared from the tail according to standard procedures (23). DNA targeting the replaced region by using primers 104F and 431R (fragment a) (Fig. 1Ab) was amplified by PCR and then digested with 1a-specific ApoI or with HaeIII, which is on 1b as well as the chimeric 1ba. To confirm that the replacement was inserted correctly, we performed PCR to detect the 5′-and 3′-flanking sequences of the replaced allele. To detect the 5′-flanking region, we used primer pair 239R and −997F, which is located upstream of primer 5′F (fragment b) (Fig. 1Ab). To detect the 3′ crossover region of the replaced chimeric mCeacam1ba allele, we performed nested PCR with two sets of primers, BS1F and 2204R for first-round PCR and BS2F and 2134R for subsequent rounds of PCR (fragment c) (Fig. 1Ab). To confirm that the loxP-neo-loxP cassette was removed, PCR was performed to detect the fragments of the neo gene and the loxP-neo-loxP cassette by using two sets of primers, neoF and neoR and 104F and 579R, respectively (fragments d and e) (Fig. 1Ab).
Real-time PCR analysis for mCeacam1 mRNA.
To determine the levels of mRNA specific for 1a and 1b in tissues from 6- to 8-week-old male B6, SJL, and cB61ba mice, total RNA was extracted by using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. cDNAs were synthesized from these RNAs by using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Takara Bio, Shiga, Japan) and an oligo(dT) primer (Invitrogen, CA) according to the manufacturers' instructions. A series of reactions was performed by using LightCycler 480 Probes Master (Roche Diagnostics, Mannheim, Germany). To quantitate both 1a and 1b mRNAs, the target sequence conserved in both alleles at exon 3 of mCeacam1 was amplified with primers mCeacam1ab-F and mCeacam1ab-R and with the hybridization probe mCeacam1ab-P, labeled with the 5′-fluorescein 6-carboxyfluorescein (FAM). To quantify the level of expression of mCeacam1a mRNA specifically, we used a target sequence found within exon 2 of 1a but not 1b. To amplify 1a and detect the amplified fragments, we used primers mCeacam1a-F and mCeacam1a-R and hybridization probe mCeacam1a-P labeled with the 5′-fluorescein FAM. To quantitate the expression of mRNA encoding both mCeacam1b and chimeric mCeacam1ba but not mCeacam1a, we used a target sequence in exon 2 of 1b and the chimera, and to amplify 1b and detect the amplified fragments, we used primers mCeacam1b-F and mCeacam1b-R and hybridization probe mCeacam1b-P, labeled with the 5′-fluorescein FAM. These fragments were amplified under the following conditions: 95°C for 5 min and 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 1 s). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was also quantified by using the TaqMan rodent GAPDH control reagent VIC probe (Applied Biosystems, CA) as an internal control for RNA quality.
Western blot analysis of mCEACAM1 glycoproteins.
To detect the sizes and relative amounts of mCEACAM1a, mCEACAM1b, and chimeric mCEACAM1ba that were expressed in various tissues of homozygous male mice, tissues were taken from 6- to 8-week-old male B6, SJL, and cB61ba mice; snap-frozen; and stored at −80°C. Tissues were homogenized on ice with glass homogenizers in lysis buffer (0.65% NP-40 in phosphate-buffered saline [PBS]) containing Complete protease inhibitor (Roche Diagnostics). After centrifugation at 3,000 rpm for 5 min at 4°C, the supernatant was used as the total cell protein sample. The protein concentrations of the samples were determined by using a BCA protein assay kit (Pierce, IL). Next, 10 μg of total protein from the liver and spleen and 5 μg of total protein from the ileum of the three genotypes of mice were separated by SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto Clear Blot Membrane-p (Atta, Tokyo, Japan). The expression of both the mCEACAM1a and mCEACAM1b proteins was detected with polyclonal rabbit anti-mCEACAM1-specific antibody 655, while 1a but not 1b or 1ba was detected with mouse anti-mCEACAM1a-specific monoclonal antibody (MAb) CC1 (11, 39, 49). The membranes were subsequently reacted with anti-rabbit mouse or anti-mouse rabbit immunoglobulin G (IgG) antibody labeled with horseradish peroxidase, respectively. The immune complexes were visualized by using an ECL Plus Western blotting detection system (GE Healthcare) with LAS-3000 (Fujifilm, Tokyo, Japan). GAPDH detected with anti-GAPDH-specific mouse MAb (Imgenex, CA) was used as an internal control.
Immunohistochemical detection of mCEACAM1 proteins in tissues.
To visualize the mCEACAM1a, mCEACAM1b, and chimeric mCEACAM1ba proteins expressed in tissues of mice, the liver and ileum collected from 6- to 8-week-old male wild-type B6, SJL, and cB61ba mice were fixed in 10% neutral buffered formalin (pH 7.4) or 4% paraformaldehyde and subjected to routine histological examination. Formalin-fixed tissues were paraffin embedded and used to detect the expression of mCEACAM1 (1a, 1b, and chimeric 1ba) with polyclonal rabbit anti-mCEACAM1-specific antibody 655, while paraformaldehyde-fixed tissues were used to detect mCEACAM1a with anti-1a-specific MAb CC1 with streptavidin-biotin complex methods (Dako, Glostrup) according to the manufacturer's instructions (12, 48).
Propagation and plaque assay of MHV-A59.
To analyze the differences in the susceptibilities of different mouse strains to MHV-A59 versus neurotropic MHV-JHM, we previously used intracerebral inoculation (30). Envelopes of MHV-JHM virions contain both the spike (S) protein, which binds to CEACAM1 proteins, as well as the hemagglutinin-esterase (HE) protein, which binds to oligosaccharide moieties on the cell surface, while MHV-A59 envelopes contain only the S protein (38). In addition, MHV-JHM can infect the brains of mCeacam1a-knockout mice (25) in a receptor-independent manner, while MHV-A59 cannot. Therefore, in the present study, we analyzed mouse strain differences in MHV susceptibility by intraperitoneal (i.p.) inoculation with MHV-A59. MHV-A59 was propagated in DBT cells as previously described for MHV-JHM (45). Aliquots of the virus were stored at −80°C until use. Viral infectivity in cell cultures or murine tissues was determined by using a previously described plaque assay with delayed brain tumor (DBT) cells in 24-well plates (Becton Dickinson, NJ) (43, 45). Liver, spleen, brain, and blood collected from MHV-A59-inoculated male mice homozygous for different mCEACAM1 alleles were snap-frozen on dry ice, homogenized in PBS (pH 7.2) to make a 10% (wt/vol) homogenate, and centrifuged at 3,000 rpm at 4°C for 10 min. The supernatants were collected and used for virus titration.
Virus inoculation of mice.
To determine the 50% lethal dose (LD50) of MHV-A59 in B6 and SJL mice versus cB61ba mice, we inoculated homozygous 6- to 8-week-old male mice i.p. with 100 μl of 10-fold serial dilutions of virus containing 105 to 102 PFU. Animals were checked daily for mortality for 2 weeks after virus inoculation. The LD50 was calculated by the method of Reed and Muench (33). To detect the multiplication of virus in mice, we sacrificed animals inoculated i.p. with 105 PFU on days 2, 4, 7, and 10 postinfection (p.i.). Tissues were homogenized, and the titers of infectious virus were determined by plaque assay as described above. These tissue homogenates were also utilized to determine the levels of viral mRNAs plus viral genomic RNA by real-time quantitative PCR (qPCR), as described below.
Real-time qPCR analysis of levels of viral RNA expressed in mouse tissues.
We mixed 200 μl of the supernatants of the 10% tissue homogenate obtained as described above with 1 ml Isogen (Nippon Gene) and extracted total RNA according to the manufacturer's instructions. Real-time reverse transcription-qPCR was performed to quantitate the relative amounts of viral mRNA and genomic RNA by using 500 ng of total RNA and LightCycler RNA master mix (Roche Diagnostics) as previously described (47).
Quantitation of antivirus antibody in virus-inoculated mice.
Chimeric cB61ba and SJL mice inoculated i.p. with 105 PFU of MHV-A59 were euthanized on day 10 or 14, and their sera were collected. The relative levels of murine anti-MHV antibody in the mouse sera were assayed by using an enzyme-linked immunosorbent assay (ELISA) as reported previously (26, 51), with a lysate of MHV-JHM-infected DBT cells as an immobilized antigen.
Histopathology and immunohistochemistry.
B6, SJL, and cB61ba mice inoculated i.p. with 105 PFU of MHV-A59 were euthanized on day 2 or 4 after inoculation. The livers and other tissues were removed, fixed in 10% neutral buffered formalin (pH 7.4), and subjected to routine pathological examination. To detect virus antigens, we used polyclonal rabbit anti-MHV-JHM serum with a streptavidin-biotin complex (Dako) according to the manufacturer's instructions.
Preparation of PMs.
Peritoneal macrophages (PMs) were cultured as described previously by Taguchi et al. (42), with a slight modification. Briefly, 8-week-old male mice homozygous for either mCeacam1a (B6), mCeacam1b (SJL), or chimeric mCeacam1ba (cB61ba) were inoculated i.p. with 2 ml of sterile thioglycolate medium (Millipore, Billerica, MA). Four days later, the mice were euthanized, and 6 to 8 ml of chilled PBS (pH 7.2) containing kanamycin (100 μg/ml) and heparin (5 units/ml) was then injected into the peritoneal cavity. After massaging the abdomen, we withdrew peritoneal exudates, which were centrifuged at 1,000 rpm for 5 min at 4°C. The cells were resuspended in 0.2% NaCl and incubated for 2 min on ice to induce hemolysis. An equal volume of 1.6% NaCl was immediately added to make the suspension isotonic. After three washes with PBS, cells were cultured in Dulbecco's minimal essential medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS) (Tissue Culture Biologicals, Los Alamitos, CA) at 37°C for 1 h. Nonadherent cells were then removed, and adherent cells were subsequently cultured for 1 day. The levels of mCeacam1a, mCeacam1b, or chimeric mCeacam1ba mRNA expression were determined by quantitative reverse transcription-PCR (qRT-PCR), and the susceptibility of the PMs to MHV-A59 infection was determined 1 day after plating of the PMs.
MHV-A59 susceptibility of peritoneal macrophages.
To quantitatively compare the MHV-A59 susceptibilities of PMs from different mouse strains, PMs were inoculated with MHV-A59 at a multiplicity of infection (MOI) of 1 and incubated for 1 h at 37°C. After three washes, the cells were further cultured for 12 h and 24 h. Infectious virus in culture supernatants was titrated by plaque assay, as described above. PMs were then fixed with acetone-methanol (1:1) for 2 min at room temperature, and intracellular viral antigens were detected by immunofluorescence assay (IFA) as described below.
Expression of the mCEACAM1b protein in BHK cell.
BHK cells grown on 24-well plates were transfected with the expression plasmid pKS336 (35), encoding the four domains with short-tail isoforms of mCEACAM1b or chimeric mCEACAM1ba, by using FuGENE 6 reagent (Roche Diagnostics) according to the manufacturer's instructions. To compare the levels of expression of the mCEACAM1 proteins in transiently transfected cells, we lysed the cells with 200 μl/well of 1× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 2.5% 2-mercaptoethanol, 2.5% SDS, 10% sucrose, 0.004% bromophenol blue) at 34 h posttransfection and compared the levels of the mCEACAM1 proteins by Western blotting, for which we used rabbit anti-mCEACAM1 polyclonal antibody 665 as described above. To analyze the virus receptor activities of the mCEACAM1 proteins, we cultured the cells for 34 h after transfection with plasmids and then inoculated them with MHV-A59 at an MOI of 1 for 1 h at 37°C. After three washes, the cells were further cultured for 12 h and 24 h. The infectious virus in the supernatant medium was then collected and titrated by a plaque assay, and the cells were fixed with acetone-methanol (1:1) for 2 min. The expression of intracellular viral antigens was detected by an indirect IFA.
Indirect IFA.
To detect intracellular viral antigens, cells inoculated with MHV-A59 were washed three times with PBS, fixed as described above, and incubated with polyclonal rabbit anti-MHV-JHM serum for 1 h at room temperature. After three washes with PBS, the cells were further incubated with fluorescein isothiocyanate-conjugated anti-rabbit IgG (Zymed Laboratories, CA).
RESULTS
Generation of chimeric mCeacam1ba mice.
To generate B6 mice expressing 1b, we replaced the gene segment encoding amino acids 1 to 70 in the N domain of 1a of wild-type B6 mice (1a/1a) with that of 1b of SJL mice (1b/1b) (Fig. 1A). The MHV receptor function in CEACAM1 proteins resides in this region of the N domain (32, 48). The targeting plasmid was transfected into the ES cell line M12, derived from B6 mice (19), and neomycin-resistant cell clones were selected. Recombinant G418-resistant ES cell clones were identified by Southern blot analysis. The 9.2-kb recombinant mCeacam1-specific EcoRI fragment and the 11.5-kb wild-type allele-specific EcoRI fragment were present in recombinant ES cells (Fig. 1A and B). Four independent ES cell clones carrying the expected recombination event were injected into BALB/c blastocysts and implanted into the uteri of ICR mice. Hair-color-chimeric mice born after implantation were mated with B6 mice (1a/1a), and the mCeacam1 genotypes of the resulting progeny mice were examined by PCR to select mice with a 1baneo genotype (Fig. 1Ca). Out of a total 815 hair-color-chimeric mice obtained, female 2 exhibited the 1baneo genotype. To generate heterozygous 1baneo mice, one of these two chimeras was mated with B6 (1a/1a) mice, and F1 progeny were genotyped by PCR and restriction digestion by mCeacam1a-specific ApoI and mCeacam1b-specific HaeIII to detect heterozygotes with both the 1a and 1baneo genes. The heterozygous mice were intercrossed to generate homozygous 1baneo/1baneo mice (Fig. 1Ca). To remove the loxP-neo-loxP cassette in intron 2, the 1baneo/1abneo mice were crossed with B6 background EIIA-Cre mice that express Cre recombinase (37). The resulting heterozygous 1a/1ba mice were intercrossed to generate homozygous 1ba/1ba mice, for which the genotype was identified by PCR and restriction digestion (Fig. 1Ca). Those replacement events were confirmed by PCR analysis to detect both the 5′ and 3′ border regions of the recombination site. The 2.1-kb 5′ recombination-specific fragment was detected in 1a/1baneo, 1baneo/1baneo, and 1ba/1ba mice but was not found in 1a/1a mice (Fig. 1Cb). The 6.4-kb 3′ recombination-specific fragment was present in 1a/1baneo, 1baneo/1baneo, and 1ba/1ba mice but was not found in 1a/1a mice (Fig. 1Cc). To confirm the deletion of the loxP-neo-loxP cassette, we performed PCR to detect the fragments of the neo gene and the loxP-neo-loxP cassette. An approximately 300-bp, neo-specific fragment, present in 1baneo/1baneo, was not detected in 1ba/1ba (Fig. 1Cd). The 4.7-kb fragment from exons 2 to 3 containing the loxP-neo-loxP cassette found in 1baneo/1baneo mice was not present in homozygous 1ba/1ba mice, which instead had a 2.2-kb fragment due to the removal of the loxP-neo-loxP cassette (Fig. 1Ce). The replacement event was also confirmed by sequencing of the mCeacam1 cDNA synthesized from mRNA extracted from the liver of 1ba/1ba mice.
General health states of the progeny.
The mice with the 1ba/1ba genotype (cB61ba) were viable and healthy under pathogen-free conditions. We have maintained a colony of these mice for 5 years and have not noticed any reduction in fertility, bone or cartilage abnormalities, tumors, or abnormal behavior.
Expression of the 1b protein in gene-replaced cB61ba mice.
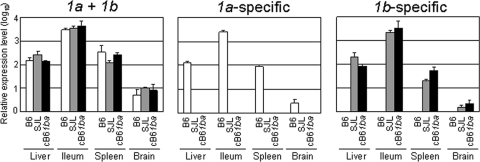
We have examined the levels of mRNA to mCeacam1, 1a or 1b, independently, with real-time PCR using mRNA-specific primers in cB61ba, B6, and SJL mouse tissues. As shown in Fig. 2, comparable levels of mRNA to mCeacam1 were detected in various organs of cB61ba, B6, and SJL mice, and there was no significant difference in the levels of expression of mCeacam1 mRNAs among those three strains of mice. With primers specific for the region of the mCeacam1a allele encoding aa 1 to 70, mRNA was detected only in the organs of B6 but not cB61ba or SJL mice (Fig. 2). As expected, PCR with primers specific for the corresponding region of the mCeacam1b allele detected mRNA only in cB61ba and SJL mouse organs and not in B6 organs (Fig. 2).
FIG. 2.
Expression of mCeacam1 mRNA in the liver, ileum, spleen, and brain in homozygous gene-replaced B6 (1a/1a), SJL (1b/1b), and cB61ba (1ba/1ba) mice. cDNA was synthesized from the RNA isolated from these tissues of four mice in each mouse strain by using Isogen, as described in Materials and Methods, and subjected to real-time qPCR to detect both 1a and 1b (1a + 1b), only 1a (1a-specific), or only 1b (1b-specific). Regions detected by qPCR are indicated by fine black lines in Fig. 1Ab. qPCR of GAPDH cDNA was used as an internal control. Error bars represent the standard deviations of the results from four independent samples.
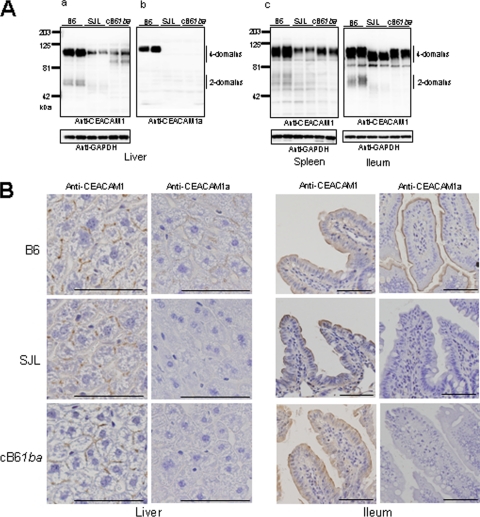
We also examined the levels of the mCEACAM1ba protein expressed in homozygous cB61ba mice and compared them with the levels of expression of mCEACAM1a in B6 mice and mCEACAM1b in SJL mice (Fig. 3). The mouse mCeacam1 gene produces four major splice variants of mRNA that encode either four or two Ig domains that are linked through a TM domain to either a short or a long cytoplasmic domain (Cy), producing four isoforms of mCEACAM1 proteins (22, 28). The four-domain mCEACAM1 isoforms were suggested to be the principal MHV receptors in vivo (4). In the livers of B6, cB61ba, and SJL mice, Western blotting showed the expression of the 100- to 120-kDa mCEACAM1 proteins containing four Ig domains. The expression level of the four-Ig isoforms of 1a in B6 mouse liver was apparently higher than that of four-Ig isoforms of 1ba or 1b in livers of cB61ba or SJL mice. However, there was no significant difference between the levels of four-Ig 1ba protein compared to those of the 1b protein expressed in livers of cB61ba and SJL mice (Fig. 3Aa). The 50- to 60-kDa isoforms of mCEACAM1 with two Ig-like domains were marginally detectable in extracts of B6 mouse livers and were not detected in livers of cB61ba or SJL mice (Fig. 3Aa). Clearly, cB61ba mice expressed the 1ba protein but not 1a, because mCEACAM1 of cB61ba reacted with antibody that recognized both 1a and 1b but did not react with 1a-specific antibody, as shown in Fig. 3Ab. In the spleen, B6 mice apparently expressed larger amounts of the 1a protein than cB61ba and SJL mice expressed the 1b protein (Fig. 3Ac). These findings were similar to those seen for the expression pattern in the liver (Fig. 3Aa). However, there was no striking difference in the levels of 1b expressed in spleens of cB61ba and SJL mice (Fig. 3Ac). In contrast, similar levels of the mCEACAM1 1a, 1b, and 1ba proteins were expressed in the intestine (ileum) of B6, SJL, and cB61ba mice, respectively (Fig. 3Ac). These results collectively indicated that gene-replaced cB61ba mice expressed the chimeric 1ba protein, but not 1a, at the same level as that of the 1b protein expressed in SJL mice. Low levels of expression of 1b in the liver and spleen of cB61ba and SJL mice, compared with that of 1a in B6 mice, might be attributable to an instability of 1b-specific mRNA in those organs, but not in the intestine, since comparable levels of Ceacam1 mRNA were detected in each organ of all three strains of mice.
FIG. 3.
Expression of mCEACAM1 proteins in the liver, spleen, and ileum of homozygous wild-type B6 (1a/1a), SJL (1b/1b), and cB61ba (1ba/1ba) mice. (A) Proteins were extracted from these tissues from two mice of each strain, as described in Materials and Methods, and analyzed for 1a and 1b by Western blotting. (a and c) To detect both 1a and 1b mCEACAM1 proteins, we used rabbit polyclonal antibody 655 against mCEACAM1a. (b) To detect mCEACAM1a only, MAb CC1, which is specific for 1a, was used. GAPDH was used as a loading control. (B) The expression of mCEACAM1 proteins in murine tissues was studied by immunohistochemistry with polyclonal goat antibody that detects both mCEACAM1a and mCEACAM1b and with MAb CC1, which detects only mCEACAM1a. mCEACAM1 proteins were expressed in bile canaliculi and cell contacts in the liver and on apical surfaces of intestinal epithelia. In the ileum of cB61ba mice, the chimeric mCEACAM1ba protein localized by polyclonal antibody was similar to that found in wild-type B6 and SJL mice. Bar, 100 μm. Magnifications, ×200 (liver) and ×100 (ileum).
We have also examined the expression patterns of mCEACAM1 proteins in a variety of organs by immunohistochemistry. As shown in Fig. 3B, mCEACAM1 proteins were localized similarly in the liver in bile canaliculi and at cell-cell contacts and on the apical membranes of epithelial cells in the ileum of cB61ba, B6, and SJL mice. Because mCEACAM1 proteins of cB61ba and SJL mice were detected by a polyclonal antibody that detects both mCEACAM1a and mCEACAM1b but not by 1a-specific monoclonal antibody CC1, these two mouse strains expressed a 1b epitope(s) in domain 1.
Susceptibility of gene-replaced chimeric B61ba mice.
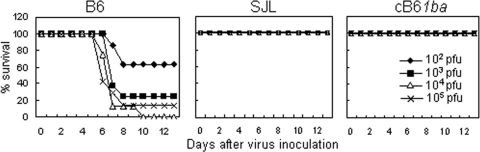
To see the differences in the susceptibilities of different mouse strains to MHV, we previously inoculated neurotropic MHV-JHM intracerebrally (30). In the present study, we investigated the susceptibilities of different mouse strains to intraperitoneal infection with hepatotropic MHV-A59. MHV-A59 was previously utilized to examine mouse strain differences in susceptibility to MHV infection (42). To study the effect of mCEACAM1 alleles on susceptibility to MHV-A59, we compared the LD50 values of MHV-A59 in B6 (1a/1a), SJL(1b/1b), and cB61ba (1ba/1ba) mice inoculated i.p. with 10-fold serial dilutions of virus, from 102 to 105 PFU/100 μl, and recorded mortality daily for 2 weeks after virus inoculation. As shown in Fig. 4, all SJL and cB61ba mice, with 1b/1b and 1ba/1ba genotypes, respectively, survived for more than 2 weeks, even after inoculation with 105 PFU, while B6 mice with the 1a/1a genotype died by 5 to 10 days postinfection after inoculation with more than 102 PFU. The LD50 calculated by the method of Reed and Muench for B6 mice was 102.5 PFU, while that for cB61ba and SJL mice was more than 105 PFU. These results showed that cB61ba mice were as resistant to MHV as SJL mice and led us to suggest that the mouse strain differences in MHV susceptibility are determined by the 1a and 1b alleles.
FIG. 4.
Susceptibility of cB61ba, wild-type B6, and SJL mice to death from MHV-A59. MHV-A59, from 102, 103, 104, and 105 PFU/100 μl, was inoculated i.p. into seven to eight mice in each group, and mortality was checked daily for 2 weeks after virus inoculation.
Virus growth and antibody production in cB61b/1a mice.
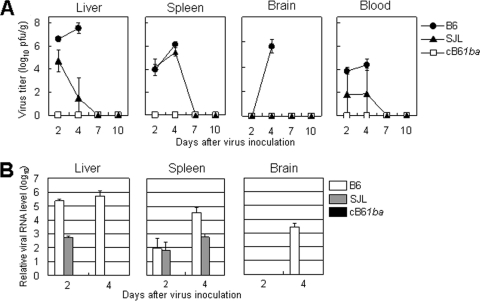
We then assayed virus replication in the major target organs of chimeric B61ba mice compared to B6 and SJL mice. Mice were inoculated i.p. with 105 PFU of MHV-A59, and virus titers in the liver, spleen, brain, and blood were determined at intervals after inoculation. Two different methods were employed to quantify virus replication: a plaque assay to determine the infectious-virus titers and real-time PCR to monitor the level of MHV-specific mRNA. As shown in Fig. 5A, high titers of MHV-A59 were found in the livers of B6 mice as early as day 2, and these titers increased on day 4. In B6 mice, infectious virus was also detected on days 2 and 4 in spleen and blood and on day 4 in brain. In contrast, no infectious virus was detected in any organs of cB61ba mice throughout the experimental period, while virus replication was detected in SJL mice in all organs except for the brain although at much lower titers than in B6 mouse organs. Real-time PCR showed similar results, with high and moderate levels of viral mRNA and genomic RNA in B6 and SJL mouse organs but no viral RNA in cB61ba mice (Fig. 5B). These results demonstrated that cB61ba mice were more profoundly resistant to infection with MHV-A59 than SJL mice, even though these two mouse strains had similar LD50 values.
FIG. 5.
Virus growth and viral mRNA in B6, SJL, and cB61ba mice. Mice were inoculated i.p. with MHV-A59 (105 PFU/100 μl), and tissues shown were collected on days 2, 4, and 7. Virus titers (A) and the relative levels of viral mRNA and genomic RNA (B) in those tissues were measured by plaque assay and real-time PCR, respectively. All B6 mice died 5 to 7 days after virus inoculation. Error bars represent standard deviations of the results from four independent samples.
Since no virus replication was detected in cB61ba mice, we inoculated 105 PFU of MHV-A59 into cB61ba and SJL mice and tested their sera for the development of antiviral antibodies. Under certain conditions, virus infection was confirmed by the presence of antibody, even if no infectious virus was detected in the target organs of MHV. As shown in Table 2, by 10 days after infection, all SJL mice had developed antibodies to MHV-A59, while by day 14, none of the cB61ba mice had developed any antiviral antibody. These results suggested that cB61ba mice did not permit MHV-A59 replication and were fully resistant to MHV-A59 infection.
TABLE 2.
Antivirus antibody production in cB61ba and SJL mice after inoculation with MHV-A59a
| Mouse | dpi | No. of mice with antibody/no. of mice tested | Avg antibody titer (range) |
|---|---|---|---|
| SJL | 10 | 4/4 | 450 (200-800) |
| cB61ba | 10 | 0/4 | <50 |
| cB61ba | 14 | 0/4 | <50 |
Mice were inoculated i.p. with 105 PFU of MHV-A59, and serum antibody was examined on day 10 or 14 postinfection (dpi) by an ELISA.
Histopathology and immunohistochemistry.
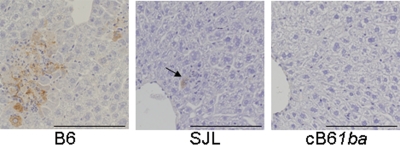
In B6 mice on days 2 and 4 after inoculation with 105 PFU of MHV-A59, the liver contained multiple foci of necrosis and infiltration of inflammatory cells. Abundant viral antigens were observed in liver lesions. In contrast, at day 2, the livers of SJL mice appeared mostly normal, with rare, tiny foci that consisted of a few degenerating hepatocytes expressing viral antigens and inflammatory cells (Fig. 6). However, in cB61ba mice, no histological changes and no viral antigens were detected in the liver or other organs throughout the experimental period. These observations are consistent with the results of virus replication in the three different mouse strains, showing that cB61ba mice are fully resistant to MHV infection.
FIG. 6.
Immunohistochemical studies of representative viral liver lesions in B6, SJL, and cB61ba mice on day 2 after i.p. inoculation with MHV-A59 (105 PFU). A large focus of necrosis and inflammation and abundant viral antigens were observed in livers of wild-type B6 mice. A tiny focal lesion containing viral antigens (arrow) was detected in SJL liver. No histological changes or viral antigens were detected in the organs of MHV-A59-inoculated cB61ba mice. Antigens were detected with polyclonal rabbit anti-MHV serum by using diaminobenzidine for visualization and counterstaining with hematoxylin. Bar, 200 μm. Magnification, ×200.
MHV-A59 susceptibility of peritoneal macrophages from gene-replaced cB61ba mice.
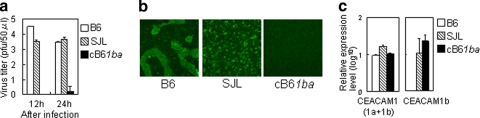
It is generally accepted that mouse susceptibility to MHV is reflected by the susceptibility of macrophages from the peritoneal cavity to MHV infection (2, 40, 42). Therefore, we compared the MHV susceptibilities of PMs isolated from B6, cB61ba, and SJL mice as described in Materials and Methods. MHV-A59 replicated very efficiently in macrophages derived from B6 (1a/1a) mice, slightly less efficiently in those from SJL (1b/1b) mice, but not at all in macrophages from cB61ba (1ba/1ba) mice, as revealed by virus yield (Fig. 7a). MHV-A59 induced the formation of large syncytia in PMs from B6 mice and tiny syncytia in PMs from SJL mice. No syncytium formation or viral antigens were seen in PMs from cB61ba mice (Fig. 7b). The amounts of mCEACAM1 mRNAs in PMs were not significantly different among the three mouse lines as shown by real-time PCR (Fig. 7c). These results indicated that the profound resistance of cB61ba mice to MHV-A59 infection in vivo is also detectable in macrophages isolated from these animals.
FIG. 7.
Susceptibility of PMs of wild-type B6, SJL, and cB61ba mice to MHV-A59 infection. (a) PMs were inoculated with MHV-A59 at an MOI of 1 and incubated for 12 or 24 h. Virus titers in the culture medium were measured by plaque assay. Error bars represent standard deviations of the results from three independent samples in one experiment. (b) MHV-A59-inoculated PMs at 24 h after inoculation were fixed with acetone-methanol, and viral antigens in the cells were detected by IFA with anti-MHV-JHM rabbit polyclonal antibody. (c) To determine the expression levels of mCEACAM1 mRNA in the PMs 1 day after plating of the cells, cDNA was synthesized from RNA from cultured PMs, and real-time qPCR was done to detect either a region of mCeacam1 outside domain 1 that is common to both 1a and 1b (1a + 1b) or a region in domain 1 of Ceacam1b that is specific for 1b as well as 1ba.
MHV receptor activity of recombinant chimeric mCEACAM1ba proteins in transfected hamster cells.
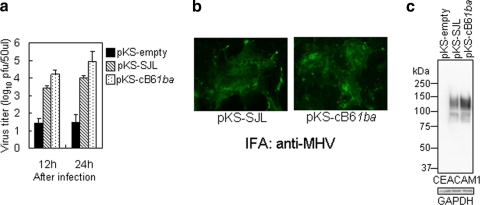
Since cB61ba mice expressing chimeric 1ba did not permit virus replication in target organs or PMs, we examined whether the chimeric mCEACAM1ba protein transiently expressed in hamster cells is a functional MHV-A59 receptor. We and others have previously shown that BHK cells expressing recombinant mCEACAM1b proteins with four Ig domains have MHV receptor activities, although they were 10- to 30-fold lower than those of mCEACAM1a (11, 31, 32). Before engineering mice to express the mCeacam1ba gene, we confirmed that the chimeric mCEACAM1ba protein transiently expressed in BHK cells from a plasmid had MHV receptor activity similar to that of mCEACAM1b, as previously reported (48). To see whether the chimeric mCEACAM1ba protein had MHV-A59 receptor activity when transiently expressed in transfected hamster cells, we cloned the cDNAs of 1b from SJL liver and chimeric 1ba from cB61ba liver, expressed the mCEACAM1b or mCEACAM1ba protein in BHK cells, and examined susceptibility to MHV-A59 infection as described in Materials and Methods. BHK cells expressing either recombinant mCEACAM1b or chimeric mCEACAM1ba showed almost equal MHV-A59 receptor activities (Fig. 8a and b). The slightly higher MHV receptor activity of recombinant chimeric 1ba than that of 1b (Fig. 8a) could be due to the slightly higher level of expression of 1ba than that of chimeric 1b in these cells, as shown in Fig. 8c. These results are in good agreement with previously reported data showing that the MHV-A59-binding activity of this chimeric mCEACAM1ba receptor protein was not significantly different from that of mCEACAM1b (48).
FIG. 8.
Comparison of MHV-A59 receptor activities of the recombinant mCEACAM1b protein and chimeric mCEACAM1ba protein. BHK cells were transiently transfected with pKS336 containing cDNAs encoding four domains with short-tail isoforms of mCEACAM1b or chimeric mCEACAM1ba from cB61ba mice or no insert (empty). (a) The cells were inoculated with MHV-A59 at an MOI of 1 and incubated for 12 or 24 h. Virus titers in the culture were measured by a plaque assay. Error bars represent standard deviations of the results from three independent samples from one experiment. (b) Those cells were fixed with acetone-methanol, and viral antigens in the cytoplasm were examined by IFA with anti-MHV rabbit polyclonal antibody. (c) At 34 h posttransfection, cell lysates prepared as described in Materials and Methods were analyzed by Western blotting using rabbit polyclonal antibody 655, which detects the mCEACAM1a, mCEACAM1b, and mCEACAM1ba proteins. GAPDH was used as an internal control.
DISCUSSION
In the present study, we analyzed whether or not different alleles of the mCeacam1 gene are responsible for the susceptibility and resistance of mice to MHV infection. We established B6 mice in which part of the N-terminal domain of the mCeacam1a gene was replaced by mCeacam1b derived from SJL mice by genetic engineering technology. Genotyping of these mice indicated that the 1a gene of B6 was successfully replaced with a chimeric 1ba gene. Gene-replaced B61ba mice expressed the chimeric mCEACAM1ba protein to the same level as that of mCEACAM1b expressed in SJL (1b/1b) mice, and its distribution in organs was the same as that in SJL mice. Homozygous cB61ba (1ba/1ba) mice did not show any gross abnormalities by visual inspection, were as healthy as B6 (1a/1a) mice, and had a similar life span. These findings led us to suggest that in cB61ba mice, the chimeric mCEACAM1ba protein can perform the normal cellular functions of mCEACAM1a. By using B6 mice, SJL mice, and the genetically engineered cB61ba mice, we showed that the mCeacam1a allele was responsible for the high degree of susceptibility of B6 mice to MHV-A59 infection and that mCeacam1b was not sufficient to account for the lesser susceptibility of SJL mice, since cB61ba mice were revealed not to be less susceptible than SJL mice but fully resistant to MHV-A59 infection.
What is the possible mechanism responsible for the differences in susceptibility found between SJL and cB61ba mice? Both of those strains have the 1b/1b genotype in terms of the virus-binding region of domain 1 of the mCEACAM1 protein, and both strains express comparable levels of CEACAM1, but SJL mice permit limited, asymptomatic MHV-A59 infection, while cB61ba mice are fully resistant to MHV-A59 replication. There was no apparent difference in MHV-A59 receptor activity between the 1b and 1ba proteins that were transiently expressed at high levels on BHK cells. Thus, it might be possible to speculate that 1b works as a less functional receptor in SJL mice, while it does not work at all in B6 mice, even if the molecule is expressed indistinguishably from that in SJL mice. However, this possibility would be unlikely, since mCEACAM1ba is a fully functional protein in genetically engineered cB61ba mice, which lack any detectable abnormalities. mCeacam1-knockout mice on the B6 background differ from wild-type B6 mice in showing impaired insulin clearance, abnormal weight gain, and reduced fertility (8). Insofar as we have determined, there is no significant difference between cB61ba and B6 mice in insulin clearance, suggesting that the mCEACAM1ba protein in cB61ba mice functions in insulin clearance like the wild-type mCEACAM1a protein (data not shown). Thus, the chimeric mCEACAM1ba protein is assumed to have less MHV-A59 receptor activity than mCEACAM1a, as revealed with transiently transfected BHK cells. These findings led us to hypothesize that an as-yet-unidentified molecule present in SJL mice but not in B6 mice might be critical to permit limited, asymptomatic MHV-A59 replication in mCeacam1b SJL mice. This hypothesis suggests that MHV resistance and/or susceptibility may be governed by several murine genes. This idea is in good agreement with an initial description of differences in susceptibility to MHV among mouse strains. Stohlman and Frelinger previously reported that two different genes are involved in susceptibility and/or resistance to MHV infection (41). Others also reported the involvement of other genes of mice in MHV susceptibility (1, 9). To see the existence of a hypothetical molecule in SJL but not in B6 mice, we are currently comparing the levels of MHV-A59 susceptibility of 1b-expressing B6 cells versus 1b-expressing SJL cells by using embryo fibroblasts.
Alternatively, it is possible that other murine proteins with weaker MHV receptor activities may be involved in the observed differences in MHV-A59 susceptibility between SJL and cB61ba mice. Two murine proteins, in addition to mCEACAM1, have thus far been reported to serve as weak MHV receptors. One protein is mCEACAM2, and the other is a murine pregnancy-specific glycoprotein (PSG), both of which are members of the Ig superfamily (6, 27). CEACAM2 has less MHV-A59 receptor activity in vitro than mCEACAM1a, and it is expressed in different organs than mCEACAM1a (27). PSG works as a functional receptor for some strains of MHV, including MHV-A59, but not for certain other strains (6). Although these two murine proteins were shown to be functional receptors for some MHV strains in cultured cells, it is likely that these are not functional receptors for MHV-A59 in B6 mice because mCeacam1a-knockout B6 mice are fully resistant to MHV-A59 infection by the intranasal route, even though these mice supposedly express the CEACAM2 and PSG proteins (17, 25). However, if alleles or isoforms of these proteins were expressed in SJL mice that were different from those in B6 mice, it is possible that these proteins in SJL mice could serve as MHV receptors. Therefore, we analyzed mCEACAM2 and PSG sequences of SJL and B6 mice. The predicted amino acid sequence of mCEACAM2 from SJL mouse liver (GenBank accession no. AB500065) showed only one amino acid substitution in the signal sequence compared with that of B6 mouse liver (accession no. X760875), and this mutation appears unlikely to influence the virus receptor activity of mCEACAM2 in SJL mice. The predicted amino acid sequences of PSG from SJL and B6 mice (accession no. AB500066) were identical. Therefore, those two possible alternative MHV receptors would probably not explain the differing MHV-A59 susceptibilities of SJL and cB61ba mice.
It has been well documented that the susceptibility of mice to MHV was reflected by the susceptibilities of macrophages isolated from the peritoneal cavity; macrophages from resistant or susceptible mice were resistant or susceptible to MHV infection, respectively (2, 42, 44). In this study we examined the susceptibilities of macrophages from B6, SJL, and cB61ba mice and showed that macrophages from B6 and SJL mice supported MHV-A59 infection, while those from cB61ba mice failed to do so. Our macrophage data and the data on the susceptibilities of these three mouse lines to MHV-A59 are in good agreement. Thus, peritoneal macrophages are an ideal tool for the detection of as-yet-unknown factors that may affect the susceptibility of SJL mice to MHV-A59 infection.
MHV is one of the most prevalent infectious diseases in laboratory mouse colonies and presumably in wild mouse populations as well (18). Although most of the MHV isolates from mouse colonies are of low virulence or avirulent in immunocompetent adult mice and are pathogenic only for immunodeficient mice, such as suckling or nude mice, some isolates of these viruses may establish persistent infection even in immunocompetent mice without clinically apparent disease, which continues for years (18). The resistance of wild mice to lethal MHV infection would provide a selective advantage in an adverse environment. Although among inbred laboratory mouse strains, the mCeacam1b allele is retained in only SJL mice, we have previously shown that all wild mouse subspecies distributed throughout the world contain both the mCeacam1a and mCeacam1b alleles (29). Thus, each murine subspecies is maintained by susceptible and also less susceptible mouse populations. Furthermore, the present study showed that some mice with 1b/1b could be fully resistant to MHV infection when those mice have the same genetic background as that of B6 mice. A similar role for an allele of the HIV coreceptor in mediating resistance to HIV infection was reported previously (36).
In the present study, we showed that homozygosity for the mCeacam1a allele is likely responsible for the high susceptibility of most lines of inbred mice to lethal infection with MHV-A59. In contrast, homozygosity for the mCeacam1b allele in the SJL background does not fully explain why SJL mice can allow limited virus replication in an asymptomatic, nonlethal infection following i.p. inoculation with MHV-A59, since B6 mice with homozygous mCeacam1ba, functionally equivalent to mCeacam1b, showed no asymptomatic, limited infection but showed full resistance. We postulate that an as-yet-unidentified murine gene in SJL mice but not in B6 mice may account for why SJL mice are susceptible to limited MHV-A59 replication. Accordingly, it would be valuable to identify the postulated modifying gene in progeny mice obtained by crossing SJL and cB61ba mice and then backcrossing with SJL versus cB61ba mice. Genetic analysis and the study of the susceptibility of progeny of each offspring to MHV-A59 replication will identify the chromosome and locus in the SJL genome that encode the postulated gene modifying susceptibility to MHV-A59 infection.
Acknowledgments
We thank members of the Department of Animal Models for Human Diseases of the National Institute of Neuroscience, where this project started, especially Tateki Kikuchi for valuable discussions and encouragement and Hisae Kikuchi and Sachiko Tateishi for the breeding of gene-replaced mice. We also thank Kazuya Shirato for the real-time PCR for mCeacam1 and the Meiji Dairies Corporation for the MS12 ES cell line.
This work was financially supported by grants-in-aid from the Ministry of Education, Sports, Culture, and Technology of Japan (to F.T.) and grant AI25231 from the National Institutes of Health (to K.V.H.).
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Asanaka, M., and M. M. C. Lai. 1993. Cell fusion studies identified multiple cellular factors involved in mouse hepatitis virus entry. Virology 197:732-741. [DOI] [PubMed] [Google Scholar]
- 2.Bang, F. B., and A. Warwick. 1960. Mouse macrophages as host cells for the mouse hepatitis virus and the genetic basis of their susceptibility. Proc. Natl. Acad. Sci. U. S. A. 46:1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauchemin, N., P. Draber, G. Dveksler, P. Gold, S. Gray-Owen, F. Grunert, S. Hammarström, K. V. Holmes, A. Karlsson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von Kleist, C. Wagener, S. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252:243-249. [DOI] [PubMed] [Google Scholar]
- 4.Blau, D. M., C. Turbide, M. Tremblay, M. Olson, S. Létourneau, E. Michaliszyn, S. Jothy, K. V. Holmes, and N. Beauchemin. 2001. Targeted disruption of the Ceacam1 (MHVR) gene leads to reduced susceptibility of mice to mouse hepatitis virus infection. J. Virol. 75:8173-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, J. F., D. G. Weismiller, and K. V. Holmes. 1987. Genetic resistance to mouse hepatitis virus correlates with absence of virus-binding activity on target tissues. J. Virol. 61:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D. S., M. Asanaka, K. Yokomori, F. Wang, S. B. Hwang, H. Li, and M. M. C. Lai. 1995. A pregnancy-specific glycoprotein is expressed in the brain and serves as a receptor for mouse hepatitis virus. Proc. Natl. Acad. Sci. U. S. A. 92:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, W., and R. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeAngelis, A. M., G. Heinrich, T. Dai, T. A. Bowman, P. R. Patel, S. J. Lee, E. G. Hong, D. Y. Jung, A. Assmann, R. N. Kulkarni, J. K. Kim, and S. M. Najjar. 2008. Carcinoembryonic antigen-related cell adhesion molecule 1: a link between insulin and lipid metabolism. Diabetes 57:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dindzans, V. J., E. Skamene, and G. A. Levy. 1986. Susceptibility/resistance to mouse hepatitis virus strain 3 and macrophage procoagulant activity are genetically linked and controlled by two non-H2-linked genes. J. Immunol. 137:2355-2360. [PubMed] [Google Scholar]
- 10.Dveksler, G. S., A. A. Basile, C. B. Cardellichio, and K. V. Holmes. 1995. Mouse hepatitis virus receptor activities of an MHVR/mph chimera and MHVR mutants lacking N-linked glycosylation of the N-terminal domain. J. Virol. 69:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dveksler, G. S., C. W. Dieffenbach, C. B. Cardellichio, K. McCuaig, M. N. Pensiero, G. S. Jiang, N. Beauchemin, and K. V. Holmes. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors of the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G. S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-68891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dveksler, G. S., M. N. Pensiero, C. W. Dieffenbach, C. B. Cargellichio, A. A. Bacile, P. E. Elia, and K. Holmes. 1993. Mouse hepatitis virus strain A59 and blocking antireceptor monoclonal antibody bind to the N-terminal domain of cellular receptor. Proc. Natl. Acad. Sci. U. S. A. 90:1716-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfraind, C., S. G. Langreth, C. B. Cardellichio, R. Knobler, J. P. Coutelier, M. Dubois-Dalcq, and K. V. Holmes. 1995. Tissue and cellular distribution of an adhesion molecule in the carcinoembryonic antigen family that serves as a receptor for mouse hepatitis virus. Lab. Invest. 73:615-627. [PubMed] [Google Scholar]
- 15.Haller, O. 1981. Inborn resistance of mice to orthomyxoviruses. Curr. Top. Microbiol. Immunol. 95:25-52. [DOI] [PubMed] [Google Scholar]
- 16.Haywood, A. M. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J. Virol. 68:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemmila, E., C. Turbide, M. Olson, S. Jothy, K. V. Holmes, and N. Beauchemin. 2004. Ceacam1a−/− mice are completely resistant to infection by murine coronavirus mouse hepatitis virus A59. J. Virol. 78:10156-10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homberger, F. R. 1977. Enterotropic mouse hepatitis virus. Lab. Anim. 31:97-115. [DOI] [PubMed] [Google Scholar]
- 19.Kawase, E., H. Suemori, N. Takahashi, K. Okazaki, K. Hashimoto, and N. Nakatsuji. 1994. Strain difference in establishment of mouse embryonic stem (ES) cell lines. Int. J. Dev. Biol. 3:385-390. [PubMed] [Google Scholar]
- 20.Malo, D., and E. Skamene. 1994. Genetic control of host resistance to infection. Trends Genet. 10:365-371. [DOI] [PubMed] [Google Scholar]
- 21.McCuaig, K., M. Rosenberg, P. Nédellec, C. Turbide, and N. Beauchemin. 1993. Expression of the Bgp gene and characterization of mouse colon biliary glycoprotein isoforms. Gene 127:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCuaig, K., C. Turbide, and N. Beauchemin. 1992. mmCGM1a: a mouse carcinoembryonic antigen gene family member, generated by alternative splicing, functions as an adhesion molecule. Cell Growth Differ. 3:165-174. [PubMed] [Google Scholar]
- 23.Miller, S. A., D. D. Dykes, and H. F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura, S. H., K. Nakagaki, and F. Taguchi. 2004. N terminus domain of murine coronavirus receptor CEACAM1 is responsible for fusogenic activation and conformational changes of the spike protein. J. Virol. 78:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura, T. A., E. A. Travanty, L. Oko, H. Bielefeldt-Ohmann, S. R. Weiss, N. Beauchemin, and K. V. Holmes. 2008. The spike glycoprotein of murine coronavirus MHV-JHM mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a−/− mice. J. Virol. 82:755-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Research Council. 1991. Infectious diseases of mice and rats. National Academies Press, Washington, DC.
- 27.Nédellec, P., G. S. Dveksler, E. Daniels, C. Turbide, B. Chow, A. A. Basile, K. V. Holmes, and N. Beauchemin. 1994. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J. Virol. 68:4525-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nédellec, P., C. Turbide, and N. Beauchemin. 1995. Characterization and transcriptional activity of the mouse biliary glycoprotein 1 gene, a carcinoembryonic antigen-related gene. Eur. J. Biochem. 231:104-114. [DOI] [PubMed] [Google Scholar]
- 29.Ohtsuka, N., K. Tsuchiya, E. Honda, and F. Taguchi. 2001. A study on mouse hepatitis virus receptor genotype in the wild mouse. Adv. Exp. Med. Biol. 494:237-240. [DOI] [PubMed] [Google Scholar]
- 30.Ohtsuka, N., and F. Taguchi. 1997. Mouse susceptibility to mouse hepatitis virus infection is linked to viral receptor genotype. J. Virol. 71:8860-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtsuka, N., Y. K. Yamada, and F. Taguchi. 1996. Difference in virus-binding activity of two distinct receptor proteins for mouse hepatitis virus. J. Gen. Virol. 77:1683-1692. [DOI] [PubMed] [Google Scholar]
- 32.Rao, P. V., S. Kumari, and T. M. Gallagher. 1997. Identification of a contiguous 6-residue determinant in the MHV receptor that controls the level of virion binding to cells. Virology 229:336-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 34.Robbins, J., P. Robbins, C. A. Kozak, and R. Callahan. 1991. The mouse biliary glycoprotein gene (Bgp): partial nucleotide sequence, expression, and chromosomal assignment. Genomics 10:583-587. [DOI] [PubMed] [Google Scholar]
- 35.Saijo, M., T. Qing, M. Niikura, A. Maeda, T. Ikegami, W. Sakai, C. Prehaud, I. Kurane, and S. Morikawa. 2002. Immunofluorescence technique using HeLa cells expressing recombinant nucleoprotein for detection of immunoglobulin G antibodies to Crimean-Congo hemorrhagic fever virus. J. Clin. Microbiol. 40:372-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 37.Satoh, Y., S. Endo, T. Ikeda, K. Yamada, M. Ito, M. Kuroki, T. Hiramoto, O. Imamura, Y. Kobayashi, Y. Watanabe, S. Itohara, and K. Takishima. 2007. Extracellular signal-regulated kinase 2 (ERK2) knockdown mice show deficits in long-term memory; ERK2 has a specific function in learning and memory. J. Neurosci. 27:10765-10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shieh, C. K., H. J. Lee, K. Yokomori, N. La Monica, S. Makino, and M. M. Lai. 1989. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J. Virol. 63:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, A. L., C. B. Cardellichio, D. F. Winograd, S. S. de Souza, S. W. Barthold, and K. V. Holmes. 1991. Monoclonal antibody to the receptor for murine coronavirus MHV-A59 inhibits viral replication in vivo. J. Infect. Dis. 163:879-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, M. S., R. E. Click, and P. G. W. Plagemann. 1984. Control of mouse hepatitis virus replication in macrophages by a recessive gene on chromosome 7. J. Immunol. 134:428-432. [PubMed] [Google Scholar]
- 41.Stohlman, S. A., and J. A. Frelinger. 1978. Resistance to fatal central nervous system disease by mouse hepatitis virus, strain JHM. 1. Genetic analysis. Immunogenetics 6:277-281. [Google Scholar]
- 42.Taguchi, F., N. Hirano, Y. Kiuchi, and K. Fujiwara. 1976. Difference in response of mouse hepatitis virus among susceptible mouse strains. Jpn. J. Microbiol. 20:293-302. [DOI] [PubMed] [Google Scholar]
- 43.Taguchi, F., A. Yamada, and K. Fujiwara. 1980. Resistance to highly virulent mouse hepatitis virus acquired by mice after low-virulence infection: enhanced antiviral activity of macrophages. Infect. Immun. 29:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taguchi, F., R. Yamaguchi, S. Makino, and K. Fujiwara. 1981. Correlation between growth potential of mouse hepatitis viruses in macrophages and their virulence for mice. Infect. Immun. 34:1059-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taguchi, F., and S. Matsuyama. 2002. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J. Virol. 76:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe, R., S. Matsuyama, and F. Taguchi. 2006. Receptor-independent infection of murine coronavirus: analysis by spinoculation. J. Virol. 80:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wessner, D. R., P. C. Shick, J. H. Lu, C. B. Cardellichio, S. E. Gagneten, N. Beauchemin, K. V. Holmes, and G. S. Dveksler. 1998. Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 72:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, R. K., G. S. Jiang, S. W. Snyder, M. F. Frana, and K. V. Holmes. 1990. Purification of the 110-kilodalton glycoprotein receptor for mouse hepatitis virus (MHV)-A59 from mouse liver and identification of a nonfunctional, homologous protein in MHV-resistant SJL/J. mice. J. Virol. 64:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, R. K., G. S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada, Y. K., M. Yabe, K. Takimoto, K. Nakayama, and M. Saitoh. 1998. Application of nested polymerase chain reaction to detection of mouse hepatitis virus in fecal specimens during a natural outbreak in an immunodeficient mouse colony. Exp. Anim. 47:261-264. [DOI] [PubMed] [Google Scholar]
- 52.Yokomori, K., and M. M. C. Lai. 1992. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J. Virol. 66:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokomori, K., and M. M. C. Lai. 1992. The receptor for mouse hepatitis virus in the resistant mouse strain SJL is functional: implications for the requirement of a second factor for viral infection. J. Virol. 66:6931-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]