Abstract
The newly identified retrovirus—the xenotropic murine leukemia virus-related virus (XMRV)—has recently been shown to be strongly associated with familial prostate cancer in humans (A. Urisman et al., PLoS Pathog. 2:e25, 2006). While that study showed evidence of XMRV infection exclusively in the prostatic stromal fibroblasts, a recent study found XMRV protein antigens mainly in malignant prostate epithelial cells (R. Schlaberg et al., Proc. Natl. Acad. Sci. U. S. A. 106:16351-16356, 2009). To help elucidate the mechanisms behind XMRV infection, we show that prostatic fibroblast cells express Xpr1, a known receptor of XMRV, but its expression is absent in other cell lines of the prostate (i.e., epithelial and stromal smooth muscle cells). We also show that certain amino acid residues located within the predicted extracellular loop (ECL3 and ECL4) sequences of Xpr1 are required for efficient XMRV entry. Although we found strong evidence to support XMRV infection of prostatic fibroblast cell lines via Xpr1, we learned that XMRV was indeed capable of infecting cells that did not necessarily express Xpr1, such as those of the prostatic epithelial and smooth muscle origins. Further studies suggest that the expression of Xpr1 and certain genotypes of the RNASEL gene, which could restrict XMRV infection, may play important roles in defining XMRV tropisms in certain cell types. Collectively, our data reveal important cellular determinants required for XMRV entry into different human prostate cells in vitro, which may provide important insights into the possible role of XMRV as an etiologic agent in human prostate cancer.
Prostate cancer is the most common male malignancy in Western countries and the second most common cause of cancer-related deaths in males worldwide (15, 24). The known risk factors for prostate cancer are hormones (i.e., androgens), diet, sex, and race, as well as environmental and genetic factors (27). A recent study suggests that susceptibility to prostate cancer can be influenced by the genetic variations associated with an antagonistic coevolution, which occurs between a specific host locus (RNASEL), known to be involved in antiviral innate immune defense, and a viral pathogen (38). Indeed, several epidemiologic studies have supported the involvement of the RNASEL gene in the prostate cancer etiology (4, 5, 30, 31), whereas other studies do not (9, 22, 34, 43). Some studies have reported that individuals with a single mutated copy of the RNASEL gene have a 50% increased risk for prostate cancer, whereas those with homozygous mutant RNASEL alleles have a 2-fold-increased risk of prostate cancer (5).
The RNASEL gene encodes for the RNase L protein, a constitutively expressed latent endoribonuclease, which mediates the interferon-inducible 2-5A system against viral and/or cellular double-stranded RNAs (8, 16, 20, 23, 49, 50). The RNase L “Q” variant allele (R462Q) shows a 3-fold decrease in catalytic activity compared to the wild-type enzyme (5, 44). The possible association of mutant RNASEL alleles with human prostate cancers suggests an enhanced susceptibility of prostate tissues to a viral agent. This hypothesis has led to the recent identification of a new human retrovirus, xenotropic murine leukemia virus (MuLV)-related virus (XMRV), in 40% of prostate cancer patients with the QQ variant alleles of RNASEL compared to 1.5% among heterozygous (RQ) and wild-type (RR) RNASEL carriers (41). XMRV virus infection appears to be susceptible to inhibition by interferon and its downstream effector RNase L protein (7). However, a recent study has provided some evidence to show that XMRV infection is independent of the RNASEL genotype (34), suggesting that population differences and/or other environmental or genetic factors may influence the impact of RNASEL on prostate cancer development.
The XMRV genome is 8,185 nucleotides in length and shares up to 95% overall nucleotide sequence identity with known xenotropic MuLVs (41). One receptor for xenotropic MuLVs is Xpr1, a 696-amino-acid protein with multiple transmembrane-spanning domains (2). Expression of this protein in Chinese hamster ovary (CHO) cells that are not known to express Xpr1 endogenously confers an enhanced susceptibility of these cells to xenotropic MuLV infection (2). Infection of hamster and mouse cells with XMRV-like virus that is derived from a prostate cancer cell line (22Rv1) also requires Xpr1 as a receptor (18). Earlier studies have demonstrated the importance of certain residues located within the putative third and fourth extracellular loops (ECL3 and ECL4) of Mus dunni's Xpr1 in conferring infection by xenotropic MuLVs (25). Furthermore, it has been shown that the specific and common receptor determinants for xenotropic and polytropic murine retroviruses are simultaneously present in discrete domains of a single Xpr1 protein (42). In the present study, we characterized for the first time the important molecular determinants on Xpr1 required for XMRV infection and investigated the role of RNase L in restricting XMRV infection of various human prostate cancer and noncancerous cell lines.
MATERIALS AND METHODS
Plasmids, cell lines, and viruses.
The pLXSN-Xpr1 plasmid carrying the XPR1 gene was kindly provided by D. Miller (University of Washington). The prostate cancer cell lines DU145, PC-3, and LNCaP were used for XMRV infection. All cells were originally obtained from the American Type Culture Collection and cultured in conditions as recommended by the supplier. The hTERT immortalized fibroblast (Pf179T), epithelial (Ep156T), and smooth muscle (Pm151T) cell lines were provided by V. Rotter (Weizmann Institute of Science, Israel) and cultured in conditions as recommended (19). The human primary (nonimmortalized) prostate fibroblast cells isolated from normal (Pt-N) or cancerous (Pt-C) tissues of prostate cancer patients were kind gifts of L. Chung (Emory University) and were cultured as described previously (39). The primary prostate epithelial cell line (PrEC) was purchased from Lonza (Walkersville, MD) and cultured in conditions as recommended by the supplier. The Chinese hamster ovary cell line (CHO) lacking endogenous Xpr1 expression and showing resistance to XMRV virus infection was used as a negative control in all experiments and grown in F12 medium (Invitrogen).
DU145-C7 cells, which carry integrated copies of XMRV and constitutively produce wild-type XMRV virus, was kindly provided by R. Silverman (Cleveland Clinic) and grown in RPMI medium (7, 41). Supernatant collected from the medium of the virus producing cell line DU145-C7 was centrifuged at 1,500 rpm for 5 min to remove cell debris and filtered through a 0.45-μm-pore-size filter. MuLV pseudotyped viruses carrying the XMRV envelope protein and the green fluorescent protein (GFP)-containing viral genome (referred to here as XMRV-pseudovirus) were prepared by calcium phosphate transfection of 293T cells with a three-vector system as previously reported (26, 29). Briefly, using the calcium phosphate method, 10 million 293T cells per 15-cm2 plates were transfected, in the presence of 25 mM chloroquine, with 14 μg of the pCS2+-mGP plasmid (45) that expresses the MuLV Gag and Pol proteins, 21 μg of pLXSG plasmid that expresses the packageable GFP reporter gene (kindly provided by P. Lewis, Oregon Health Sciences University), and 7 μg of the pDP1-XMRV-env that expresses the XMRV envelope gene. The XMRV-pseudovirus in the transfected supernatant was separated from cellular debris by filtration though a 0.45-μm-pore-size filter.
Transfection and infection.
The CHO cells were seeded into 12-well plates and transfected with wild-type or mutant Xpr1 plasmid using Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen). At 24 h posttransfection, the cells were infected with 200 μl of XMRV-pseudovirus in the presence of 8 μg per ml of Polybrene in F-12 medium without serum and antibiotics. After 4 h of incubation with the pseudovirus, a fresh aliquot of the medium with 1% fetal bovine serum and Pen/Strep (1×) was replaced, and the culture was grown for 3 days. Various immortalized prostate cells were seeded in either six-well plates or 60-mm plates as necessary. The telomerase (hTERT) immortalized epithelial (Ep156T) were transfected with pLXSN-Xpr1 plasmid by using Hyfect transfection reagent (Scientific Denville, Inc.), whereas the immortalized smooth muscle cells (Pm151T) were transfected with the same plasmid by using the Superfect reagent (Qiagen) according to the manufacturers' instructions. Transfection efficiency was monitored by visual scoring of green fluorescent protein expression in parallel transfections of the vector peGFP-N1 (Stratagene) alone and determined to be greater than 30% in each of the transfection reactions. Cells were then infected with either 500 μl of wild-type XMRV virus, which is equivalent to ∼34 U of MuLV RT (New England BioLabs)/ml, or with 350 μl of XMRV-pseudovirus in the presence of Polybrene (8 μg/ml for Ep156T, PrEC, Pf179T, Pt-N, and Pt-C and 1 μg/ml for Pm151T) in their respective media without serum and antibiotics. All other prostate cancer and primary cell lines were infected with XMRV-pseudovirus or wild-type XMRV virus in the presence of 8 μg of Polybrene/ml and in their respective media without serum and antibiotics. After 4 h of incubation with the virus, the medium was replaced with a fresh aliquot of the medium, and the culture was maintained in a tissue culture incubator (for 3 days in the case of XMRV-pseudovirus infections and for 7 days for wild-type XMRV infections). Cells infected with the XMRV-pseudovirus expressing the GFP gene were analyzed under fluorescence microscopy using a Nikon microscope, and images were taken for both GFP-positive cells (using fluorescence microscopy) and total cells in the same area (using bright-field microscopy) at ×20 magnification. The amounts of cells infected with the XMRV-pseudovirus expressing the GFP gene were also analyzed via fluorescence-activated cell sorting (FACS) by using a FACSCalibur (BD Biosciences).
Construction of the plasmid expressing hemagglutinin (HA)-tagged Xpr1 gene and its mutagenesis.
To create the HA tag version of Xpr1, PCR was performed to amplify the Xpr1 coding region using 2 μl of the pLXSN-Xpr1 plasmid as the template in a 50-μl PCR that contains 1 U of high-fidelity Vent DNA polymerase (NEB) and 0.2 μM concentrations of the primers Fxpr1 (5′-ATGCAAGCTTCGGCAGGATGAAGTTCGCCG-3′) and Rxpr1 (5′-ATGCGCGGCCGCTCAAGCGTAATCTGGAACATCGTATGGGTAAGTGTTAGCTTCATCATC-3′). After denaturation at 94°C for 4 min, the reaction proceeded with 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 2 min. The 2.1-kb PCR product was purified with a QIAquick gel extraction kit (Qiagen), cloned into the expression vector pcDNA3.1-Intron A, and sequenced. The QuikChange PCR mutagenesis protocol (Stratagene) was used to generate the Xpr1 mutants ECL3/4 (K500E, Δ582T). The forward and reverse primers used to make the K500E mutant construct were 5′-GCCCTTTACAGCACTCACGAGGAACGAGGTCACTCGG-3′and 5′-CCGAGTGACCTCGTTCCTCGTGAGTGCTGTAAAGGGC-3′, respectively. The primers used to generate a deletion at position 582 were 5′-CCAAATCTCGATTACCTCTACTTTGTTGCCTC-3′ and 5′-GAGGCAACAAAGTAGAGGTAATCGAGATTTGG-3′. The wild-type and mutant Xpr1 constructs were transfected in CHO cell line, which was followed by infection with the XMRV-pseudovirus as described above. XPR1 gene expression was analyzed at 48 h postinfection either by Western blotting with the anti-HA antibody (Santa Cruz) as described in more detail below or by reverse transcription-PCR (RT-PCR). To perform RT-PCR, total RNAs were isolated from the transfected cells and used to synthesize Xpr1 cDNA by using Superscript II (Invitrogen), which was followed by PCR amplification with GoTaq DNA polymerase (Promega) for 4 min at 94°C; followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 45 s at 72°C; and followed finally by 10 min at 72°C in a reaction that contained the Xpr1-F (5′-GGGAGTGAGGGGGAAACGGCAGG-3′) and XPR1-R (5′-AGTGTTAGCTTCATCATCTGTGTCTTCTATC-3′) primers. In addition, a set of universal primers was used to PCR amplify the β-actin gene as a control.
siRNA knockdown of XPR1 and desmoglein-2 genes.
DU145 cells at approximately 50 to 60% confluence were grown in 12-well plates for 24 h before transfection with small interfering RNA (siRNA). Predesigned ON-TARGETplus SMART pool siRNA reagent for XPR1 and control siRNA were purchased from Dharmacon, Inc. The SMART pool siRNAs against XPR1 (catalog no. L-005752-00) were 5′-ACACUAAGGUAUUGAUAGA-3′, 5′-UCGAUUACCUCUACAACUU-3′, 5′-GCUUGCCGCUGUAUUUAAA-3′, and 5′-GGCCUUUCCUCAUUUAGUU-3′, and the control siRNA was ON-TARGETplus nontargeting siRNA 1 (catalog no. D-001810-01-20) of the sequence 5′-UGGUUUACAUGUCGACUAA-3′. siRNA and antibody against desmoglein-2 were kind gifts from A. Nusrat (Emory University). siRNA (20 nM) transfection was performed in triplicate according to the manufacturer's instructions using DharmaFECT transfection reagent 1. Cells were incubated with the transfection medium for 48 h and infected with XMRV pseudovirus. Cells infected with pseudovirus were analyzed by fluorescence microscopy and flow cytometry using the FACSCalibur (BD Biosciences) as described above. The levels of XPR1 gene knockdown were detected by RT-PCR using the primer sets as described above.
RT-PCR and quantitative real-time RT-PCR.
Since none of the commercial polyclonal antibodies (2G8 from Abnova, N-19 from Santa Cruz, and ab13291 from Abcam) proved to be effective at detecting Xpr1 expression by Western blotting or IHC (data not shown), we decided to use RT-PCR and quantitative real-time RT-PCR for this purpose. To this end, total cellular RNAs were isolated from different cell lines, treated with RQ1 DNase (Promega), and extracted with acid phenol-chloroform (5:1, pH 4.5; Omnipur) to remove DNAs. Purified RNAs were used as templates to synthesize Xpr1 cDNAs using Superscript II (Invitrogen), which were then amplified by PCR using the GoTaq DNA polymerase (Promega) at 94°C for 4 min; followed by 30 s at 94°C, 30 s at 58°C, and 45 s at 72°C for 30 cycles; and finally by 10 min at 72°C in a reaction that contained the XPR1out-F (5′-CACTGGTGTTACTACGCTG-3′) and XPR1out-R (5′-GCAACAAAGTTGTAGAGGT-3′) primers as described previously (7). In addition, a set of universal primers was used to PCR amplify the β-actin gene as a control. Quantitative real-time RT-PCR was performed using Sybr green dye mix (Invitrogen) and β-actin as an internal control in a 20-μl reaction mixture containing the primers Xpr1-F (5′-CGATAAGAATGCAGGAGA-3′) and Xpr1-R (5′-GCAACAAAGTTGTAGAGGT-3′) specific for Xpr1. The PCR condition was 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The RNA levels, expressed as threshold cycle (CT) values, were normalized by the β-actin RNA level. RT-PCRs to amplify the XMRV spliced env RNA in cell lysates of infected prostate cancer, immortalized, and primary cell lines were conducted by treatment with GoTaq polymerase (Promega) for 4 min at 94°C; followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 1.5 min at 72°C; and finally by 10 min at 72°C using the forward primer at the expected spliced site at nucleotide (nt) 195 (5′-ACCGTCGGGAG/GCCCTCCAAGCAG-3′) and reverse primer at nt 8185 (5′-TTGCAAACAGCAAAAGGCTTTATTGG-3′).
Western blotting.
Rat monoclonal antibody to spleen focus-forming virus (SFFV) Gag was prepared from R187 cells (American Type Culture Collection) (6). Monoclonal antibodies to α-actin and the HA tag were purchased from Sigma and Santa Cruz Biotech, respectively. Monoclonal antibodies against vimentin, cytokeratin 8, and myosin II LC were kindly provided by A. Nusrat and L. Chung (Emory University). Proteins in cell extracts (20 to 100 μg) were separated in 10 or 12% polyacrylamide-sodium dodecyl sulfate gels, depending on the expected molecular masses of the proteins, and transferred onto polyvinylidene difluoride transfer membrane. Blots were blocked with TBS (Tris-base saline) containing 0.1% Tween (TBS-T) and 5% dried milk for 1 h and then incubated with primary antibodies against Gag, vimentin, cytokeratin 8, myosin II LC, HA tag, or α-actin in 2.5% TBS-T at 4°C overnight with shaking. The blots were washed three times with TBS-T and were further incubated with appropriate secondary antibodies: anti-mouse IgG-HRP for vimentin; cytokeratin, myosin II LC, anti-rabbit for HA; and anti-rat raised in goats for viral Gag protein. After four washes with TBS-T, the protein bands were visualized by using enhanced chemiluminescence detection reagents (Amersham Pharmacia).
Genotype.
All of the telomerase-immortalized primary prostate cell lines used in the present study were genotyped for the R462Q (1385G→A) RNASEL variant using a premade TaqMan genotyping assay (Applied Biosystems). Portions (5 ng) of genomic DNA from each of the cell lines were assayed according to the manufacturer's instructions and analyzed on an Applied Biosystems 7900HT fast real-time PCR system instrument.
RESULTS
Xpr1 as a receptor for XMRV infection.
Previous studies have identified two critical amino acid residues in the extracellular loops (ECLs) of the murine Xpr1 protein, K500 in ECL3 and T582 in ECL4 loop, either of which alone could mediate entry of the xenotropic murine leukemia viruses (XMLVs) (25, 42). Yan et al. defined the specific determinants on Xpr1 protein required for entry of the different types of murine retroviruses (i.e., XMLV, polytropic PMLV, and the wild mouse virus CasE#1) into mouse cells derived from evolutionary divergent species of the Asian mouse species Mus pahari and the laboratory strain NIH 3T3 (46). These authors showed that the K500 residue on the ECL3 is the entry determinant for PMLVs, the T582 residue on the ECL4 is required for CasE#1 entry, but that both residues in the ECL3 and ECL4 are required for XMLV entry into cells (46). Whereas both of these amino acid residues can independently mediate entry of XMLVs, they are, however, not functionally equivalent, since the T582 but not K500 can function as a receptor for the wild mouse CasE#1, while K500 but not T582 mediates efficient entry of the xenotropic AKR6 virus (25, 46). A related study showed that the combination of a substitution mutation in the ECL3 and a single threonine (Thr) deletion at position 582 of the ECL4 of the Xpr1 protein could completely inhibit XMLV infection (25).
Since the molecular determinants on Xpr1 protein required for XMRV infection had never been mapped, we introduced the same mutations at both the ECL3 and ECL4 loops into the Xpr1 expression vector and transfected this mutant construct (Xpr1-M) or the wild-type Xpr1 vector into CHO cell line, which lacks endogenous Xpr1 expression. XMRV-pseudovirus expressing the GFP gene was used to infect these cells. Cells expressing the wild-type XPR1 gene, as detected by RT-PCR and Western blotting (Fig. 1 B and 1C), were expectedly susceptible to XMRV-pseudovirus infection, whereas those expressing the mutant Xpr1 showed no evidence of viral infection (Fig. 1A), demonstrating for the first time the requirement of the ECL3 and ECL4 loops of Xpr1 for XMRV infection. As controls, DU145 prostate cancer cells, which are known to express the Xpr1 protein, were susceptible to XMRV infection, whereas CHO cells with no endogenous expression were not susceptible (Fig. 1A).
FIG. 1.
Validation of Xpr1 requirements for XMRV infection. (A) CHO cells were transfected with wild-type and mutant Xpr1 and examined for GFP expression upon infection with the GFP-tagged XMRV pseudovirus. The fluorescence images for GFP (top panels) and the bright-field images (bottom panels) for the total cell in the same area were taken at ×20 magnification. (B) XPR1 expressions in the transfected CHO cell lines were detected by RT-PCR. (C) XPR1 expressions in the transfected CHO cell lines were detected by Western blotting.
Differential expression of Xpr1 in prostate cancer and immortalized noncancerous cell lines.
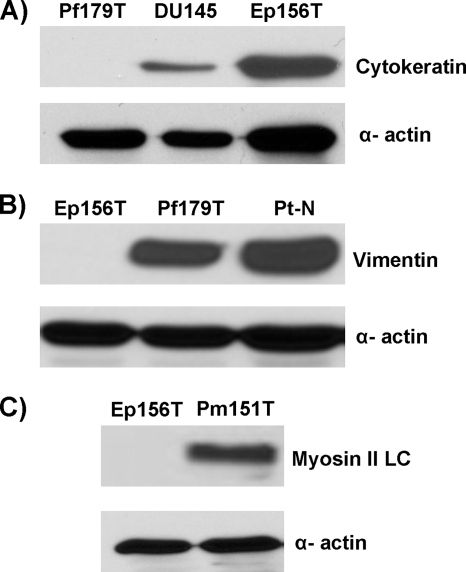
XMRV viral genomic materials and protein antigens have been detected, respectively, by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) in the stromal fibroblast cells of human prostate tissues collected from patients with prostate cancer (7, 41). We reasoned that the susceptibility of cells to XMRV infection must be dependent upon the proper expression of the Xpr1 receptor on the target cells. We therefore attempted to detect XPR1 expression in several prostate cancer and noncancerous cell lines by RT-PCR and/or quantitative real-time RT-PCR. These methods were chosen because none of the commercially available polyclonal antibodies (2G8 from Abnova, N-19 from Santa Cruz, and ab13291 from Abcam) proved to be effective at detecting Xpr1 expression by Western blotting and/or IHC (data not show). We used a series of telomerase-immortalized primary cell lines of the epithelial (Ep156T), smooth muscle (Pm151T), and fibroblast (Pf179T) origins that were previously derived from healthy human prostate glands (19). In addition, we also used nonimmortalized fibroblast cell lines, Pt-N and Pt-C, which were derived from benign and malignant prostate tissues, respectively (39), and the established primary prostatic epithelial PrEC cells (Lonza). Even though most of the telomerase-immortalized cell lines used in the present study, except for the Pf179T, have been characterized (19), we validated the cell-type origins of these and most other primary cell lines used in the present study by determining the expression of the specific protein markers by Western blot analysis with the appropriate antibodies. As expected, the epithelial cells (Ep156T and DU145) expressed cytokeratin 8 (Fig. 2 A), whereas all fibroblast cells (Pf179T, Pt-N, and Pt-C) highly expressed vimentin (Fig. 2B and data not shown), and the smooth muscle cells (Pm151T) expressed myosin II LC (Fig. 2C), correlating with their respective cell-type origins. To detect the expression of Xpr1 transcript, we used several prostate cancer cell lines (DU145, LNCaP, and PC3) as positive controls and CHO cells as a negative control. Xpr1 transcripts were expressed in all prostate cancer cell lines and in all primary fibroblast cell lines tested, but were uniquely absent in the immortalized prostate epithelial (Ep156T) and smooth muscle (Pm151T) cells (Fig. 3 A and B). We also found minimal to no expression of XPR1 in the primary prostatic cell line PrEC as judged by RT-PCR (Fig. 3C). However, we have not determined whether different versions (e.g., isoforms) of Xpr1 are expressed in these cells that can be used by XMRV as its alternative receptors. Overall, these data indicate that XPR1 gene expression is cell type dependent, which may explain why XMRV viral nucleic acid and protein can only be detected in the fibroblast cells of the prostate as reported previously (41).
FIG. 2.
Validation of the cell type origins of the hTERT-immortalized epithelial, smooth muscle, and fibroblast cell lines. (A) Cytokeratin was expressed in the Ep156T cell line but not in the Pf179T cells. (B) Vimentin was expressed in fibroblast cell lines (Pf179T and Pt-N) but not in epithelial cells (Ep156T). (C) Myosin II light-chain protein was expressed in smooth muscle cells (Pm151T) but not in epithelial cells (Ep156T). α-Actin was used as a loading control.
FIG. 3.
Differential expression of XPR1 gene in immortalized cell lines. Xpr1 expression in prostate cancer (A), immortalized prostate cell lines (B), and the primary epithelial cell line PrEC (C) was detected by RT-PCR. β-Actin expression was used as a loading control.
Susceptibility of prostate cancer and noncancerous cell lines to XMRV-pseudovirus infection.
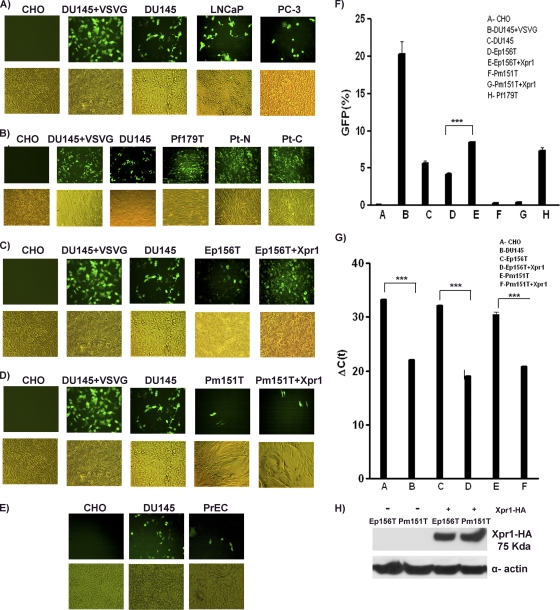
We next sought to determine whether Xpr1 expression correlates with XMRV susceptibility. XMRV-pseudoviruses expressing the GFP gene were used to infect the above-described cell lines that have been examined for Xpr1 expression. At 72 h postinfection, we found that all prostate cancer cell lines tested, which expressed Xpr1, were susceptible to XMRV-pseudovirus infection, whereas CHO cells with no endogenous Xpr1 expression were not (Fig. 4 A and F). Likewise, we showed that all fibroblast cell lines tested, which expressed high levels of XPR1 gene, were also susceptible to XMRV-pseudovirus infection (Fig. 4B and F), a finding consistent with earlier findings on detection of XMRV viral antigens in primary prostatic tissues (41). Surprisingly, we detected low levels of XMRV-pseudovirus infection in immortalized epithelial (Ep156T) and smooth muscle (Pm151T) cell lines, as well as the primary prostatic epithelial (PrEC) cell line (Fig. 4C, D, E, and F), despite the fact that they did not express endogenous Xpr1 as judged by RT-PCR and/or quantitative real-time RT-PCR (Fig. 3B and C and Fig. 4G). Upon Xpr1 overexpression by transient transfection as evidenced by Western blot analysis (Fig. 4H), the epithelial cells (Ep156T) showed marked increase in XMRV-pseudovirus infectivity (P < 0.0001), but the smooth muscle cells (Pm151T) showed no appreciable difference (Fig. 4C, D, and F), the implications for which are discussed below.
FIG. 4.
XMRV pseudovirus infectivity in prostate cancer cell lines (A), immortalized prostate fibroblast cell lines (B), epithelial cell lines with or without Xpr1 transfection (C), smooth muscle cell lines (D) with or without Xpr1 transfection, and primary epithelial cells (PrEC) (E). Representative images in the fluorescence and bright fields were captured at ×20 magnification. The experiments were carried out twice, and each time the experiments were performed in triplicate. (F) The average levels of GFP expression in infected cells were quantified by FACS analysis. Xpr1 expression levels were determined by quantitative real-time RT-PCR (G) and Western blotting (H). Asterisks indicate statistical significance (P < 0.001).
In an effort to confirm prior studies on the requirement of XPR1 expression for XMRV infection (7, 12), we used siRNA technology to reduce the endogenous XPR1 gene expression in DU145 prostate cancer cells (Fig. 5 A), which effectively diminished entry of the XMRV-pseudovirus (P < 0.0002) into these treated cells, whereas those treated with a scrambled siRNA as a negative control did not (Fig. 5C and D), providing strong evidence for Xpr1 as the main receptor of XMRV in these cells. On the contrary, there is no significant difference in the levels of XMRV-pseudovirus infection of the Xpr1 siRNA-treated primary epithelial cells Ep156T compared to those treated with a scrambled siRNA (Fig. 5C and D), suggesting alternative mode(s) of XMRV entry in these cells. As a control for the siRNA knockdown, we similarly transfected the Ep156T cells with siRNA that targets the conserved region of the epithelial cell specific desmoglein-2 gene, which is a member of the cadherin superfamily of genes that mediate adhesion of intercellular junctions (i.e., desmosomes) (36), and showed significant knockdown of this gene (Fig. 5B), implying that the Ep156T primary epithelial cells were indeed susceptible to siRNA transfection and knockdown.
FIG. 5.
Reduction of XPR1 gene expression via siRNA diminished the efficiency of XMRV-pseudovirus entry into DU145 cancer cells. (A) siRNA targeting the XPR1 gene in DU145 effectively reduced XPR1 gene expression, as shown by RT-PCR, whereas a scrambled siRNA molecule did not. (B) siRNA targeting the desmoglein-2 gene effectively reduced its protein expression in Ep156T cells, as shown by Western blot, compared to its expression in the control scrambled siRNA-treated cells. (C and D) Reduced levels of XMRV-pseudovirus infection of DU145 cells when they were treated with Xpr1 siRNA, but similar levels of viral infectivity in similarly treated Ep156T cells. Asterisks indicate statistical significance (P < 0.001).
Susceptibility of prostate cancer and noncancerous cell lines to wild-type XMRV infection.
We next sought to determine whether the cell lines tested above were also susceptible to wild-type XMRV viruses, which were generated from DU145-C7 cells that carry integrated copies of the XMRV viral genome, and hence, constitutively produce wild-type XMRV viral particles. Because XMRV is known to intrinsically replicate very slowly in cell cultures, we have devised an RT-PCR assay to detect XMRV envelope (env) gene expression as a sensitive and specific method to monitor XMRV replication. The viral env transcripts were detected in all infected prostate cancer cell lines and primary cell lines (Fig. 6 A and C), including what appears to be relatively low levels of the env transcript in the epithelial (Ep156T) and smooth muscle (Pm151T) cell lines (Fig. 6E), reflecting the relatively low infectivity of XMRV in these cells due possibly to very low or absent expression of the XPR1 gene (Fig. 3B and 4F). To confirm XMRV viral replication in some of these cells, we carried out Western blot analysis to detect expression of the XMRV Gag protein (Fig. 6B, D, and F) using an anti-SFFV monoclonal antibody that was previously determined to recognize XMRV Gag (41). Taken together, we provide strong evidence to support the role of XPR1 gene expression in enhancing the XMRV infectivity of different cell lines derived from the prostate.
FIG. 6.
XMRV wild-type virus replication in prostate cancer and immortalized prostate epithelial, smooth muscle, and fibroblast cell lines. (A) XMRV env gene expression in prostate cancer cell lines was detected by RT-PCR. (B) Expression of the viral Gag protein in prostate cancer cells was detected by Western blotting. (C) XMRV env detection in all of the prostate fibroblast cell lines by RT-PCR. Detection of viral Gag protein expression was performed by uising Western blots in fibroblast cells (Pf179T) (D) and epithelial (Ep156T) and smooth muscle (Pm151T) cells (F). Five times the amount of Pf179T protein was loaded compared to that of the DU145 cells, since XMRV is known to replicate slowly in the Pf179T cells. (E) XMRV env expression in immortalized epithelial (Ep156T) and smooth muscle (Pm151T) cell lines was detected by RT-PCR.
DISCUSSION
In this report, we show that the receptor (Xpr1) sufficient for XMRV infection is expressed in human prostate stromal fibroblast cell lines but absent in other cell lines derived from the prostate gland, which may help explain previous observations that XMRV viral antigens are present in the stromal fibroblasts of prostate tissues and of peripheral blood collected from cancer patients (41). In addition, we found evidence for XMRV infection of cells that appear not to express detectable levels of XPR1 by RT-PCR assaying, indicating other possible entry pathway(s) for XMRV (34). We also define the molecular determinants of Xpr1 that are required for efficient XMRV infection.
The Xpr1 receptor has been characterized as a transmembrane protein of unknown function, although it shows some homology to yeast genes involved in signal transduction and phosphate transport (2, 40, 48). Previous studies have implicated Xpr1 extracellular loop sequences (ECL3 and ECL4) in mediating entry of the XMLVs. Specifically, two amino acids, K500 in the ECL3 and T582 in ECL4, have been found to be most essential for Xpr1 function as a viral receptor by directly interacting with the viral envelope protein (25, 46). Our study demonstrates for the first time the significance of those two specific amino acids in determining susceptibility to XMRV infection (Fig. 1), validating previous findings that this novel human retrovirus also utilizes Xpr1 as a major receptor for viral entry (46). Recently, Yan et al. screened a panel of polytropic and xenotropic murine retroviruses (X/PMVs), including a novel cytopathic XMV-related virus (Cz524) and XMRV, for infectivity of rodent cells that carried various polymorphic Xpr1 receptors (47). Using a panel of Xpr1 mutants and chimeras, they identified five critical amino acids in ECL3 and ECL4, including E500 and T582, as well as K500, T507, and V508, required to mediate efficient entry of these different viruses (47). It remains to be determined whether these other residues in ECL3 and ECL4 besides K500 and T582 are absolutely required for XMRV entry.
A common mechanism for virus-induced cellular transformation is activation of proto-oncogenes or inactivation of tumor suppressor genes as a consequence of proviral insertions into the host genome (10). Recent studies have provided evidence for integration of XMRV into the genomes of prostate tissues of cancer patients (7, 17), as well as into the 22Rv1 prostate carcinoma cell line (18). However, evidence for common XMRV integration sites within or near proto-oncogenes or tumor suppressor genes in tumor tissues is still lacking (17). FISH and IHC assays have revealed evidence for XMRV infection in the prostate stromal fibroblastic and hematopoietic cells, rather than in other cell types of the prostate (41), arguing that any effect of XMRV on prostate cancer development might involve alterations of the tumor microenvironment.
In search of an explanation for the differential XMRV infectivity of the various prostate cell types, we screened XPR1 gene expression in several prostate cancer and noncancerous cell lines derived from various prostate cell types. By RT-PCR, we found that all of the prostate cancer cell lines, telomerase-immortalized and primary fibroblast cell lines that were isolated from either benign (Pt-N) and malignant (Pt-C) areas of the prostate gland of cancer patients (39) expressed high levels of Xpr1 transcripts, which were completely absent in the benign prostatic epithelial and smooth muscle cell lines (Fig. 3B and 4G). These results are consistent with an earlier observation, which shows XMRV infection in the prostate fibroblasts but not in the epithelial cells of the prostate tissues (41). All cell lines that expressed the XPR1 gene indeed were susceptible to XMRV infection (Fig. 4A, B, and F). In addition, we observed some levels of XMRV infection in the prostate epithelial and smooth muscle cell line even though they appeared to lack Xpr1 expression (Fig. 4C to G). Likewise, we found minimal to no expression of Xpr1 in PrEC cells by the RT-PCR method, which corresponded to relatively low XMRV-pseudovirus infectivity of these cells (Fig. 3C and 4E). This finding is consistent with a recent study, which shows XMRV infection of the benign prostatic epithelial (PrEC) and stromal (PrSC) cells that may consist of fibroblast cells, as well as other cell types in the stroma (12). Related studies have similarly documented XMRV infection of not only prostatic stromal fibroblasts (PrSC) but also human cells of many different tissue types (11, 37), as well as cells from various animal species (37). The authors of these studies have proposed that the in vivo susceptibility to XMRV cannot be determined by the Xpr1 receptor expression alone but that the ability of different cell types to either transactivate the XMRV viral promoter or to restrict virus replication define how efficient the virus can spread in certain tissues (11). Recent studies have also shown that XMRV infectivity can be limited by the presence of known host restriction factors to retroviral replication, such as the human APOBEC3 and tetherin proteins, as well as mouse APOBEC3, tetherin, and Fv1 proteins (11, 28). It is noteworthy that a number of human cells, including breast cells have been shown to be susceptible to infection by the mouse mammary tumor virus (MMTV) (13, 14), in spite of the fact that the human orthologue of the murine transferrin receptor 1 (Trf1), which is the known cellular receptor for MMTV, is not able to support viral entry in human cells (32).
In a recent study, XMRV protein antigens have been detected mainly in malignant prostatic epithelial cells (23%) but only rarely in stromal cells (4%) of prostate cancer tissues collected from one institution (34). The expression of Xpr1 in the XMRV-positive malignant epithelial cells noted in that study is unknown, however. It is important to note that all prostate cancer cell lines used in the current study (DU145, LNCaP, and PC3) are of the epithelial origin, which curiously express high levels of the XPR1 gene, and as such are readily susceptible to XMRV infection (Fig. 3A and 4A). When XPR1 gene expression in DU145 is reduced by siRNA, the efficiency of XMRV entry into these cells is correspondingly reduced (Fig. 5). Taken together, these data suggest that a change in XPR1 gene regulation might have occurred either during the transformation process in vivo or in the process of generating the prostate cancer cell lines that would enhance XMRV infection. Further studies are required to investigate this phenomenon and to determine whether XPR1 gene regulation in different cell types of the prostate gland is influenced by XMRV infection and/or the process of tumorigenesis. Collectively, these results suggest that, in the absence of XPR1 expression, XMRV can use alternative receptor(s) and/or mechanism(s) to gain entry into cells. Additional studies are required in order to elucidate the exact alternative mechanism(s) of XMRV virus entry into cells.
The susceptibility of the telomerase-immortalized epithelial cell line (Ep156T) to XMRV infection appears to be markedly increased upon Xpr1 overexpression (Fig. 4C), demonstrating an important role of Xpr1 in determining the XMRV infectivity. In contrast, the smooth muscle cells (Pm151T) did not appear to support any appreciable increase in XMRV infectivity, even in the setting of Xpr1 overexpression via transfection (Fig. 4D). Since RNase L is required for a complete IFN antiviral response against XMRV (7), we determined whether RNase L might also affect the level of XMRV replication in cell culture. The RNase L “Q” variant allele (R462Q) has been shown to significantly reduce the catalytic activity of the RNase L enzyme (5, 44) and to be associated with XMRV infection in prostate cancer patients. Prostate cancer patients who are homozygous (QQ) for this variant allele of RNASEL show 40% association with XMRV infection compared to 1.5% among those who are either heterozygous (RQ) or wild-type (RR) RNASEL carriers (41). On the contrary, Schlaberg et al. recently analyzed 334 consecutive prostate resection specimens collected from a single institution, which show XMRV infection in 6% of the samples by RT-PCR and in 23% of the samples by IHC (34) but show no association of XMRV infection of the tissues with the RNase L “Q” variant allele (R462Q) (34). Specifically, these researchers did not find any difference in RNASEL allelic distribution between XMRV PCR-positive (50% RR, 43% RQ, and 7% QQ) and PCR-negative (42.7% RR, 47.4% RQ, and 10% QQ) cases, suggesting that more men are probably at risk for XMRV infection than have previously been predicted (34). This and other studies (9, 22, 34, 43), which did not find an association of RNASEL and prostate cancer, suggest that population differences and/or other environmental or genetic factors may impact the development of prostate cancer.
Regardless, we decided to genotype the RNASEL gene in most of the cell lines used in the present study. Genotyping of RNASEL showed that the smooth muscle cell line (Pm151T) was indeed homozygous wild-type (RR) at this locus, which might help explain the relatively low level of XMRV infectivity in these cells, even in the condition of Xpr1 overexpression (Fig. 4D). On the other hand, both the epithelial (Ep156T) and the fibroblast (Pf179T) cells were heterozygous (RQ) for RNASEL, a finding consistent with the fact that these cells were more prone to XMRV infection. Although we cannot formally exclude the possibility that other host factor(s) may also be involved, our results support a role for XPR1 gene expression and RNase L genotype in determining the cellular susceptibility to XMRV infection in vitro. It is noteworthy that the RNASEL locus is found only 1.6 Mb telomeric to the XPR1 gene within the 1q25 HPC1 prostate cancer locus, and evidence of significant association of RNASEL (4, 5, 30, 31) and of nominal association of the XPR1 gene (3) with human prostate cancer has been reported in cases of familial prostate cancer.
In summary, our data reveal important cellular determinants required for XMRV infection of human prostate cells in vitro, which may provide important insights into the possible role of XMRV as an etiologic agent in human prostate cancer. If it can be proven that XMRV infection is indeed widespread and can cause prostate cancer and/or other human diseases (1, 21, 34, 41), it is important to identify effective antivirals against this virus. Toward this end, recent studies have shown that HIV-1 RT inhibitors, such as azidothymidine (AZT) (33, 35), zidovudine (ZDV), and tenofovir disoproxil fumarate (TDF), as well as the integrase inhibitor raltegravir (35), can inhibit XMRV replication in cell culture to various extents, suggesting that they can be used either individually or as a combination therapy to treat XMRV infection (33, 35).
Acknowledgments
This study was supported in part by grants from the American Cancer Society (RSG-06-162-01-GMC), the pilot component of the NIH center grants to the Emory CFAR (P30 AI050409), and the Emory-Winship Cancer Institute (P30 CA138292); by a generous seed grant from D. Liotta to H.L.; and by grant NIH AI-40317 to Y.L. and T. Parslow.
We thank D. Miller for the pLSXN-Xpr1 plasmid; J. Olsen for R187 cells; L. Chung for Pt-C and Pt-N cells; V. Rotter for fibroblast (Pf179T), epithelial (Ep156T), and smooth muscle (Pm151T) cells; R. Silverman for DU145-C7 cells, XMRV, and its molecular clone; A. Nusrat and L. Chung for antibodies against vimentin, cytokerin, and myosin; A. Nusrat and P. Nava for siRNA and antibody against the desmoglein-2 gene; and J. Heimburg-Molinaro for critical reading of the manuscript. We also thank the Emory Biomarker Service Center for genotyping the cell lines.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Arnold, R. S., N. V. Makarova, A. O. Osunkoya, S. Suppiah, T. A. Scott, N. A. Johnson, S. M. Bhosle, D. Liotta, E. Hunter, F. F. Marshall, H. Ly, R. J. Molinaro, J. L. Blackwell, and J. A. Petros. 2010. XMRV infection in patients with prostate cancer: novel serologic assay and correlation with PCR and FISH. Urology 75:755-761. [DOI] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. U. S. A. 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breyer, J. P., K. M. McReynolds, B. L. Yaspan, K. M. Bradley, W. D. Dupont, and J. R. Smith. 2009. Genetic variants and prostate cancer risk: candidate replication and exploration of viral restriction genes. Cancer Epidemiol. Biomarkers Prev. 18:2137-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpten, J., N. Nupponen, S. Isaacs, R. Sood, C. Robbins, J. Xu, M. Faruque, T. Moses, C. Ewing, E. Gillanders, P. Hu, P. Bujnovszky, I. Makalowska, A. Baffoe-Bonnie, D. Faith, J. Smith, D. Stephan, K. Wiley, M. Brownstein, D. Gildea, B. Kelly, R. Jenkins, G. Hostetter, M. Matikainen, J. Schleutker, K. Klinger, T. Connors, Y. Xiang, Z. Wang, A. De Marzo, N. Papadopoulos, O. P. Kallioniemi, R. Burk, D. Meyers, H. Gronberg, P. Meltzer, R. Silverman, J. Bailey-Wilson, P. Walsh, W. Isaacs, and J. Trent. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30:181-184. [DOI] [PubMed] [Google Scholar]
- 5.Casey, G., P. J. Neville, S. J. Plummer, Y. Xiang, L. M. Krumroy, E. A. Klein, W. J. Catalona, N. Nupponen, J. D. Carpten, J. M. Trent, R. H. Silverman, and J. S. Witte. 2002. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 32:581-583. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro, B., W. Britt, L. Evans, K. Wehrly, J. Nishio, and M. Cloyd. 1983. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology 127:134-148. [DOI] [PubMed] [Google Scholar]
- 7.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, B., and R. H. Silverman. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133-4137. [DOI] [PubMed] [Google Scholar]
- 9.Downing, S. R., K. T. Hennessy, M. Abe, J. Manola, D. J. George, and P. W. Kantoff. 2003. Mutations in ribonuclease L gene do not occur at a greater frequency in patients with familial prostate cancer compared with patients with sporadic prostate cancer. Clin. Prostate Cancer 2:177-180. [DOI] [PubMed] [Google Scholar]
- 10.Gray, D. A. 1991. Insertional mutagenesis: neoplasia arising from retroviral integration. Cancer Invest. 9:295-304. [DOI] [PubMed] [Google Scholar]
- 11.Groom, H. C., M. W. Yap, R. P. Galao, S. J. Neil, and K. N. Bishop. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, S., E. A. Klein, J. Das Gupta, K. Hanke, C. J. Weight, C. Nguyen, C. Gaughan, K. A. Kim, N. Bannert, F. Kirchhoff, J. Munch, and R. H. Silverman. 2009. Fibrils of prostatic acid phosphatase fragments boost infections with XMRV (xenotropic murine leukemia virus-related virus), a human retrovirus associated with prostate cancer. J. Virol. 83:6995-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Indik, S., W. H. Gunzburg, P. Kulich, B. Salmons, and F. Rouault. 2007. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology 4:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Indik, S., W. H. Gunzburg, B. Salmons, and F. Rouault. 2005. Mouse mammary tumor virus infects human cells. Cancer Res. 65:6651-6659. [DOI] [PubMed] [Google Scholar]
- 15.Jalava, S. E., K. P. Porkka, H. E. Rauhala, J. Isotalo, T. L. Tammela, and T. Visakorpi. 2009. TCEB1 promotes invasion of prostate cancer cells. Int. J. Cancer 124:95-102. [DOI] [PubMed] [Google Scholar]
- 16.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. U. S. A. 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S., N. Kim, B. Dong, D. Boren, S. A. Lee, J. Das Gupta, C. Gaughan, E. A. Klein, C. Lee, R. H. Silverman, and S. A. Chow. 2008. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 82:9964-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knouf, E. C., M. J. Metzger, P. S. Mitchell, J. D. Arroyo, J. R. Chevillet, M. Tewari, and A. D. Miller. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogan, I., N. Goldfinger, M. Milyavsky, M. Cohen, I. Shats, G. Dobler, H. Klocker, B. Wasylyk, M. Voller, T. Aalders, J. A. Schalken, M. Oren, and V. Rotter. 2006. hTERT-immortalized prostate epithelial and stromal-derived cells: an authentic in vitro model for differentiation and carcinogenesis. Cancer Res. 66:3531-3540. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., and B. C. Tai. 2006. RNASEL gene polymorphisms and the risk of prostate cancer: a meta-analysis. Clin. Cancer Res. 12:5713-5719. [DOI] [PubMed] [Google Scholar]
- 21.Lombardi, V. C., F. W. Ruscetti, J. Das Gupta, M. A. Pfost, K. S. Hagen, D. L. Peterson, S. K. Ruscetti, R. K. Bagni, C. Petrow-Sadowski, B. Gold, M. Dean, R. H. Silverman, and J. A. Mikovits. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585-589. [DOI] [PubMed] [Google Scholar]
- 22.Maier, C., J. Haeusler, K. Herkommer, Z. Vesovic, J. Hoegel, W. Vogel, and T. Paiss. 2005. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br. J. Cancer 92:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malathi, K., J. M. Paranjape, R. Ganapathi, and R. H. Silverman. 2004. HPC1/RNASEL mediates apoptosis of prostate cancer cells treated with 2′,5′-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 64:9144-9151. [DOI] [PubMed] [Google Scholar]
- 24.Marian, C. O., and J. W. Shay. 2009. Prostate tumor-initiating cells: a new target for telomerase inhibition therapy? Biochim. Biophys. Acta 1792:289-296. [DOI] [PubMed] [Google Scholar]
- 25.Marin, M., C. S. Tailor, A. Nouri, S. L. Kozak, and D. Kabat. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 73:9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marino, M. P., M. J. Luce, and J. Reiser. 2003. Small- to large-scale production of lentivirus vectors. Methods Mol. Biol. 229:43-55. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, W. G., A. M. De Marzo, and W. B. Isaacs. 2003. Prostate cancer. N. Engl. J. Med. 349:366-381. [DOI] [PubMed] [Google Scholar]
- 28.Paprotka, T., N. J. Venkatachari, C. Chaipan, R. Burdick, K. A. Delviks-Frankenberry, W. S. Hu, and V. K. Pathak. 2010. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J. Virol. 84:5719-5729. [DOI] [PMC free article] [PubMed]
- 29.Reiser, J. 2000. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 7:910-913. [DOI] [PubMed] [Google Scholar]
- 30.Rennert, H., D. Bercovich, A. Hubert, D. Abeliovich, U. Rozovsky, A. Bar-Shira, S. Soloviov, L. Schreiber, H. Matzkin, G. Rennert, L. Kadouri, T. Peretz, Y. Yaron, and A. Orr-Urtreger. 2002. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am. J. Hum. Genet. 71:981-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rokman, A., T. Ikonen, E. H. Seppala, N. Nupponen, V. Autio, N. Mononen, J. Bailey-Wilson, J. Trent, J. Carpten, M. P. Matikainen, P. A. Koivisto, T. L. Tammela, O. P. Kallioniemi, and J. Schleutker. 2002. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 70:1299-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. U. S. A. 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakuma, R., T. Sakuma, S. Ohmine, R. H. Silverman, and Y. Ikeda. 2010. Xenotropic murine leukemia virus-related virus is susceptible to AZT. Virology 397:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlaberg, R., D. J. Choe, K. R. Brown, H. M. Thaker, and I. R. Singh. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351-16356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Singh, I. R., J. E. Gorzynski, D. Drobysheva, L. Bassit, and R. F. Schinazi. 2010. Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome. PLoS One 5:e9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinberg, M. S., H. Shida, G. J. Giudice, M. Shida, N. H. Patel, and O. W. Blaschuk. 1987. On the molecular organization, diversity and functions of desmosomal proteins. Ciba Found. Symp. 125:3-25. [DOI] [PubMed] [Google Scholar]
- 37.Stieler, K., C. Schulz, M. Lavanya, M. Aepfelbacher, C. Stocking, and N. Fischer. 2010. Host range and cellular tropism of the human exogenous gammaretrovirus XMRV. Virology 399:23-30. [DOI] [PubMed] [Google Scholar]
- 38.Summers, K., and B. Crespi. 2008. Molecular evolution of the prostate cancer susceptibility locus RNASEL: evidence for positive selection. Infect. Genet. Evol. 8:297-301. [DOI] [PubMed] [Google Scholar]
- 39.Sung, S. Y., C. L. Hsieh, A. Law, H. E. Zhau, S. Pathak, A. S. Multani, S. Lim, I. M. Coleman, L. C. Wu, W. D. Figg, W. L. Dahut, P. Nelson, J. K. Lee, M. B. Amin, R. Lyles, P. A. Johnstone, F. F. Marshall, and L. W. Chung. 2008. Coevolution of prostate cancer and bone stroma in three-dimensional coculture: implications for cancer growth and metastasis. Cancer Res. 68:9996-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. U. S. A. 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. DeRisi. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Van Hoeven, N. S., and A. D. Miller. 2005. Use of different but overlapping determinants in a retrovirus receptor accounts for non-reciprocal interference between xenotropic and polytropic murine leukemia viruses. Retrovirology 2:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiklund, F., B. A. Jonsson, A. J. Brookes, L. Stromqvist, J. Adolfsson, M. Emanuelsson, H. O. Adami, K. Augustsson-Balter, and H. Gronberg. 2004. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin. Cancer Res. 10:7150-7156. [DOI] [PubMed] [Google Scholar]
- 44.Xiang, Y., Z. Wang, J. Murakami, S. Plummer, E. A. Klein, J. D. Carpten, J. M. Trent, W. B. Isaacs, G. Casey, and R. H. Silverman. 2003. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 63:6795-6801. [PubMed] [Google Scholar]
- 45.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan, Y., R. C. Knoper, and C. A. Kozak. 2007. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J. Virol. 81:10550-10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan, Y., Q. Liu, and C. A. Kozak. 2009. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, Y. L., L. Guo, S. Xu, C. A. Holland, T. Kitamura, K. Hunter, and J. M. Cunningham. 1999. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21:216-219. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]