Abstract
Rotaviruses are a major cause of acute gastroenteritis in children worldwide. Early stages of rotavirus assembly in infected cells occur in viroplasms. Confocal microscopy demonstrated that viroplasms associate with lipids and proteins (perilipin A, ADRP) characteristic of lipid droplets (LDs). LD-associated proteins were also found to colocalize with viroplasms containing a rotaviral NSP5-enhanced green fluorescent protein (EGFP) fusion protein and with viroplasm-like structures in uninfected cells coexpressing viral NSP2 and NSP5. Close spatial proximity of NSP5-EGFP and cellular perilipin A was confirmed by fluorescence resonance energy transfer. Viroplasms appear to recruit LD components during the time course of rotavirus infection. NSP5-specific siRNA blocked association of perilipin A with NSP5 in viroplasms. Viral double-stranded RNA (dsRNA), NSP5, and perilipin A cosedimented in low-density gradient fractions of rotavirus-infected cell extracts. Chemical compounds interfering with LD formation (isoproterenol plus isobutylmethylxanthine; triacsin C) decreased the number of viroplasms and inhibited dsRNA replication and the production of infectious progeny virus; this effect correlated with significant protection of cells from virus-associated cytopathicity. Rotaviruses represent a genus of another virus family utilizing LD components for replication, pointing at novel therapeutic targets for these pathogens.
Rotaviruses are a major cause of acute gastroenteritis in infants and young children, producing a high burden of disease worldwide and over 600,000 deaths per annum, mainly in developing countries (43). Recently, two live attenuated rotavirus vaccines (49, 58) have been licensed in various countries, and their widespread use in universal mass vaccination programs is being implemented (55).
Rotaviruses form a genus of the Reoviridae family. They contain a genome of 11 segments of double-stranded RNA (dsRNA) encoding six structural proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6). After entry into the host cell the outer layer of the triple-layered particles (TLPs; infectious virions) is removed in endocytic vesicles, and the resulting double-layered particles (DLPs) actively transcribe mRNAs from the 11 RNA segments and release them into the cytoplasm. The mRNAs are translated into proteins but also act as templates for dsRNA synthesis (RNA replication). The early stages of viral morphogenesis and viral RNA replication occur in cytoplasmic inclusion bodies termed “viroplasms.” Partially assembled DLPs are released from viroplasms and receive their outer layer in the rough endoplasmic reticulum (RER), forming TLPs (for details, see Estes and Kapikian [20]).
The rotavirus nonstructural proteins NSP2 and NSP5 are major components of viroplasms (20, 47). These two proteins alone are sufficient to induce the formation of viroplasm-like structures (VLS) (21). Blocking of either NSP2 or NSP5 in rotavirus-infected cells significantly reduces viroplasm formation and the production of infectious viral progeny (11, 54, 57). Until now, host cell proteins involved in viroplasm formation have not been identified.
Morphological similarities between viroplasms and lipid droplets (LDs) prompted us to investigate their relationship. Both structures have phosphoproteins (NSP5 and perilipin A, respectively) inserted on their surface in ringlike shapes (16, 34). LDs are intracellular organelles involved in lipid and carbohydrate metabolism. They consist of a neutral lipid core surrounded by a phospholipid monolayer containing LD-associated proteins; those include proteins of the PAT family consisting of perilipin, adipophilin (adipose differentiation-related protein [ADRP]), and TIP-47 (9, 37). Lipolysis from LDs is regulated by hormones such as catecholamines, which trigger the phosphorylation of hormone-sensitive lipase (HSL) and perilipin A and induce LD fragmentation. Incubating adipocytes with the β-adrenergic agonist isoproterenol and the phosphodiesterase inhibitor isobutylmethylxanthine (IBMX) activates this pathway (27, 34). LD formation can also be blocked by triacsin C, a specific inhibitor of long chain acyl coenzyme A synthetases (30, 39).
We demonstrate here that rotavirus viroplasms colocalize with the LD-associated proteins perilipin A and ADRP and also with the lipids of LDs. These interactions appear to be required for the formation of functional viroplasms and the production of infectious viral progeny, since compounds dispersing LDs or blocking LD formation significantly decrease the number and size of viroplasms and the amount of infectious progeny. Taken together, these findings strongly suggest a critical role of LDs in rotavirus replication.
MATERIALS AND METHODS
Cells.
The cell lines MA104, Caco-2, BSC-1, and Cos-7 were used and maintained in Eagle's minimal essential medium (EMEM), supplemented with 5 to 10% fetal calf serum (Gibco) and antibiotics. An MA104 cell line expressing enhanced green fluorescent protein (EGFP)-tagged NSP5 (termed NSP5-EGFP, tagged at the C terminus) (16) was cultured in EMEM supplemented with 10% fetal calf serum. For confocal microscopy (CM) assays, cells were grown on glass coverslips. Cell viability was measured by the trypan blue exclusion assay. For transfection experiments, MA104 cells were first infected with recombinant vaccinia virus expressing the T7 polymerase (24) for 1 h at a multiplicity of infection (MOI) of 1 and then transfected with 2 to 4 μg of plasmid DNA (see below) mixed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cells were usually fixed at 6 to 8 h posttransfection (unless stated otherwise) and analyzed by CM.
Viruses.
The bovine rotavirus RF strain (G6P6[1]) and the porcine rotavirus OSU strain (G5P9[7]) were used and replicated in MA-104 cells. Rotavirus grown in large amounts was concentrated by ultracentrifugation, treated with Freon, and TLPs and DLPs were purified by repeated CsCl gradient ultracentrifugation as described elsewhere (2). DLPs in CsCl solution were deionized by passage over a G25 Sephadex column. Infectivity of the virus was determined in cell culture and expressed as 50% tissue culture infective doses (TCID50)/ml. The T7 recombinant vaccinia virus (24) was propagated in BSC-1 cells and infectivity expressed in PFU/ml. The MOI of cells was between 0.5 and 5, unless stated otherwise.
Confocal microscopy.
The primary and secondary antibodies used are listed in Tables 1 and 2, respectively. Cells on coverslips were processed for CM as described by Nejmeddine et al. (40) with slight modifications. Cell monolayers were blocked with 1% bovine serum albumin (BSA) plus 0.1% Triton X-100 in phosphate-buffered saline (PBS) and then washed with PBS. Coverslips were incubated with primary antibodies diluted in PBS containing 1% BSA, washed three times, and incubated with appropriate dilutions of secondary antibody (for details, see Tables 1 and 2) for 1 h in the presence of 2 μg of Hoechst 33342 (Sigma-Aldrich)/ml. (There was no homology between the peptide against which the perilipin A antibodies were raised and any of the rotavirus proteins. Furthermore, extensive tests excluded cross-reactivity of the perilipin A-specific, affinity-purified antibodies with rotavirus-specific proteins.) After three washes, the coverslips were mounted on glass slides with Prolong Gold Antifade mounting medium (Molecular Probes). For lipid staining, unfixed Cos-7 cells infected with rotavirus were stained at 4 to 8 h postinfection (p.i.) with Nile Red (100 ng/ml) for 4 h at 4°C and fixed with 2% paraformaldehyde in PBS containing 10 mM CaCl2 for 1 h at 4°C. Alternatively, Cos-7 cells were fixed with 3% paraformaldehyde for 30 min at room temperature and then incubated for 30 min with BODIPY 493/503 (4,4-difluoro-1,3,5,7,8 pentamethyl-4-bora 3a,4a-diaza-s-indacene; 1 μg/ml, diluted from 1-mg/ml stock in dimethyl sulfoxide; Molecular Probes) (34). Coverslips were then washed five times with PBS (10 min per wash) and counterstained with NSP5-, VP2-, and VP6 antibodies for 1 h, followed by incubation with secondary antibody conjugated with Alexa Fluor 488. The coverslips were then mounted as described above. The samples were analyzed by CM by using a Leica DM Libre TCS SP instrument.
TABLE 1.
Primary antibodies used for confocal microscopy
| Proteina | Animal | Dilution | Source (catalog no.) |
|---|---|---|---|
| Perilipin A | Rabbit | 1:500 | Abcam (ab3526) |
| ADRP | Rabbit | 1:500 | Abcam (ab52356) |
| Perilipin A | Goat | 1:5,000 | Abcam (ab60269) |
| NSP2 | Mouse | 1:500 | Oscar Burrone |
| NSP2 | Guinea pig | 1:200 | Oscar Burrone |
| NSP5 | Mouse | 1:500 | Oscar Burrone |
| NSP5 | Guinea pig | 1:1,000 | Oscar Burrone |
| VP1 | Guinea pig | 1:500 | John Patton |
| VP2 | Guinea pig | 1:500 | John Patton |
| VP3 | Guinea pig | 1:500 | John Patton |
| VP6 | Guinea pig | 1:500 | John Patton |
That is, the protein against which the antibody was raised.
TABLE 2.
Secondary antibodies used for confocal microscopy
| Antibody | Conjugate | Dilution | Supplier (catalog no.) |
|---|---|---|---|
| Goat anti-mouse IgG (H+L) | Alexa Fluor 488 | 1:500 | Invitrogen (A11029) |
| Donkey anti-goat IgG (H+L) | Alexa Fluor 633 | 1:500 | Invitrogen (A21082) |
| Goat anti-rabbit IgG (H+L) | Alexa Fluor 633 | 1:500 | Invitrogen (A21070) |
| Goat anti-rabbit IgG (H+L) | FITCa | 1:500 | Southern Biotech (4050-02) |
| Goat anti-guinea pig IgG | Texas Red | 1:300 | Santa Cruz (sc-2996) |
| Goat anti-guinea pig IgG | FITC | 1:500 | Santa Cruz (sc-2441) |
| Polyclonal goat anti-mouse immunoglobulin | FITC | 1:500 | Biostat (F047902) |
| Donkey anti-rabbit IgG (H+L) | Cy3 | 1:500 | Jackson Immunoresearch Europe, Ltd. (711-166-152) |
FITC, fluorescein isothiocyanate.
The numbers of viroplasms per cell were counted in 100 cells, and the average diameter of viroplasms was measured for ∼100 viroplasms (by cophotography of a calibration bar and use of the ImageJ 1.37c program http://rsbweb.nih.gov/ij/).
Iodixanol gradient ultracentrifugation of extracts of rotavirus-infected and uninfected cells.
Discontinuous gradients of iodixanol (Opti-Prep; Axis-Shield, United Kingdom) (37) were preformed, using different concentrations in 60 mM Tris-HCl buffer (pH 7.4) containing 0.25 M sucrose, and incubated for several hours at 4°C according to the manufacturer's instructions. RV-infected or uninfected cells were lysed at 6 h p.i. for 10 min at 4°C using a hypotonic, detergent-free buffer supplemented with protease inhibitor (Roche Diagnostics, according to the manufacturer's instructions) (38). Subsequently, a homogenate was produced with 20 to 30 strokes of a glass Dounce homogenizer using a tight-fitting pestle and layered onto the preformed iodixanol gradients. The tubes were centrifuged at 80,000 × g for 16 h at 10°C using a Beckman SW 50.1 rotor to produce a flotation gradient (38, 42), and 0.4-ml fractions were obtained manually from the top and analyzed for the presence of viral dsRNA, perilipin A, and NSP5. The density of fractions was determined by refractometry.
FRET.
The occurrence of fluorescence resonance energy transfer (FRET) implies that two molecules are in close proximity (<10 nm) (19, 23). FRET was performed with the use of an in-house-developed instrument based on a confocal microscope Olympus FluoView 300 (Olympus UK, Ltd., Watford, United Kingdom) coupled with a supercontinuum laser source (SC450; Fianium, Southampton, United Kingdom) (19). The SC450 provides ∼5-ps pulses at a repetition rate of 40 MHz, suitable for time correlated single photon counting (TCSPC). The system was upgraded with a PMC.100-20 photomultiplier tube and a SPC.830 board for TCSPC, both supplied by Becker & Hickl GmbH (Berlin, Germany). EGFP was excited at 485 nm, and the fluorescence emission was collected over the 513- to 533-nm range. Average count rates were kept at ∼104 cps in order to avoid pulse pile-up. All images were acquired at room temperature over a time period of 90s using a ×60 magnification (oil immersion objective lens) and a large (300-μm) pinhole in order to collect enough photons.
TCSPC data were analyzed with SPCImage (Becker & Hickl GmbH) by fitting a single-exponential decay and the scattered light (5, 17). Typically, 1,000 to 5,000 photons per LD were collected, and no binning was applied for data analysis in order to avoid possible edge artifacts. It was possible to discriminate LDs from the background autofluorescence and scattered light by thresholding pixels exhibiting more than 10 photons of peak intensity. The images in Fig. 7B and D were generated by using a binning procedure for representation purposes (nonbinned data are not shown).
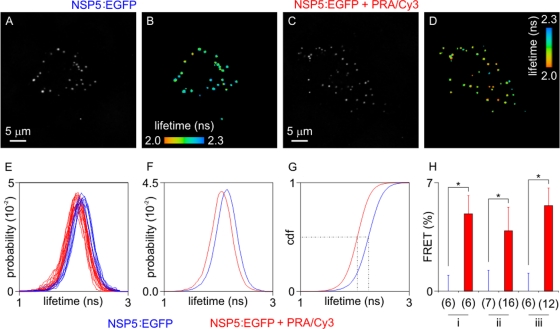
FIG. 7.
FRET between NSP5-EGFP and perilipin A in viroplasms of rotavirus-infected cells (at 6 h p.i.). Cells expressing NSP5-EGFP were imaged by TCSPC. EGFP fused to the C terminus of NSP5 acted as the donor fluorophore, and Cy3-antibody targeted PerA served as acceptor for FRET. Panels A and C show the fluorescence intensities, and panels B and D show the fluorescence lifetimes. Panels A and B represent the autofluorescence controls; panel C and D show cells in which perilipin A is detected by Cy3-labeled antibody. Fluorescence lifetime probabilities are recorded in panels E and F. In panel E, fluorescence lifetime histograms computed on individual cells show a shift toward lower values. The average shift can be clearly resolved in the cumulative histograms (F) and in the cumulative distribution functions (cdf; panel G, derived from panel F). (E to G) Results from experiment ii. Dashed lines in panel G show the median of the fluorescence lifetime for both control cells and cells exhibiting FRET. FRET efficiencies were statistically significant in three independent experiments (panel H: experiments i to iii; *, P < 0.01; numbers in brackets indicate numbers of cells compared, with control cells to the left and cells exhibiting FRET to the right of experiments i to iii).
Fluorescence lifetime distributions were exported from SPCImage and processed with the Matlab (Mathworks, Natick, MA) toolbox ImFluo (18) (see also http://laser.ceb.cam.ac.uk and http://www.quantitative-microscopy.org).
For each cell, FRET efficiencies were computed according to the following equation: E = 1 − (τ/τ0), where τ is the fluorescence lifetime of each cell and τ0 is the fluorescence lifetime of the control. τ was determined by the median of the fluorescence lifetime distributions for each cell. In order to minimize sensitivity to noise and to biases caused by non-normal distributions, medians were computed by finding the fluorescence lifetime value at which cumulative distribution functions (17) equal a probability of 50%. τ0 was estimated by the average of the fluorescence lifetimes (τ) computed on a control sample where no acceptor was present. Statistical testing was performed with a two-tailed Student t test with Origin (OriginLab Corp., Northampton, MA).
RNA extraction and agarose gel electrophoresis.
RNAs of purified virions or DLPs or of totals or fractions of cell lysates were extracted as described previously (22). Separation of genomic RNA segments was by electrophoresis through 1% agarose (Invitrogen) gels using 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.0]) at 60 to 70 V for 1 h, followed by staining with ethidium bromide (1 μg/ml) for 20 min and photography in a UV transilluminator. Negative images of the photographs were densitometrically evaluated by using the ImageJ program (http://rsbweb.nih.gov/ij/).
Analysis of cell extracts or gradient fractions by Western blotting.
Western blotting followed established procedures (2). Full-range protein rainbow markers (Amersham) were used for calibration. Blots were reacted with anti-NSP2 or anti-NSP5 antibodies at a dilution of 1:1,000 each, with anti-perilipin A antibody at a dilution of 1:500 (for details on antibodies, see Table 1) and with mouse anti-alpha actin antibody (Abcam) at a dilution of 1:5,000. Bound antibody was reacted with anti-species (mouse, guinea pig, and rabbit) IgG secondary antibodies conjugated with horseradish peroxidase (HRP; Dako, Ely, United Kingdom), and binding of secondary antibody was visualized by a Pierce Western blotting substrate (Thermo Scientific) or the Amersham ECL Plus Western blotting detection system (GE Healthcare) using X-ray film.
Treatment of cells with chemicals affecting LD homeostasis.
MA104 cells were incubated in 1× EMEM containing 0.5 to 1 mM IBMX (Sigma) and 10 to 20 μM isoproterenol (Sigma). When these two chemicals were used in combination, they were found to disperse and/or fragment LDs in adipocytes (34). The chemicals were added immediately or at 1 h p.i. to RV-infected MA104 cells or uninfected controls. The cells were incubated for 4 to 16 h p.i.
Triacsin C (10 μM; Biomol International) was transfected into uninfected MA104 cells or at 1 h p.i. using Lipofectamine 2000, according to the manufacturer's instructions.
Statistics.
Arithmetic means ± the standard deviations (SD) were calculated and compared by a two-tailed Student t test. Frequencies were compared by using a chi-square test. Differences at P < 0.05 were regarded as significant.
RESULTS
Components of rotavirus inclusion bodies (viroplasms) colocalize with LD-associated proteins in MA104 and Caco-2 cells.
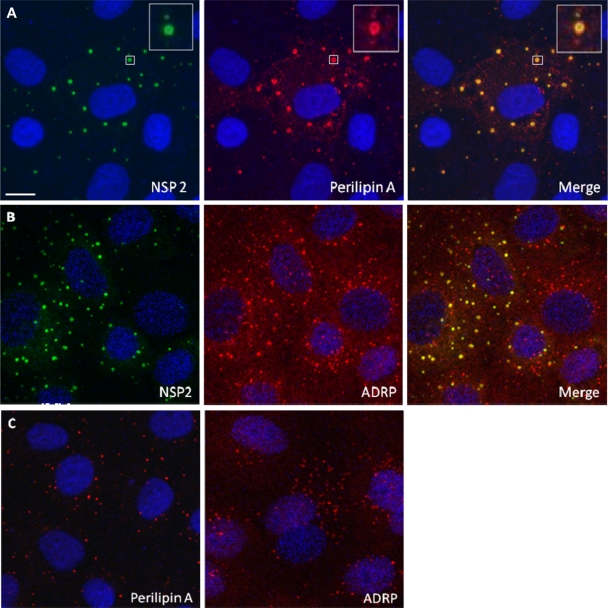
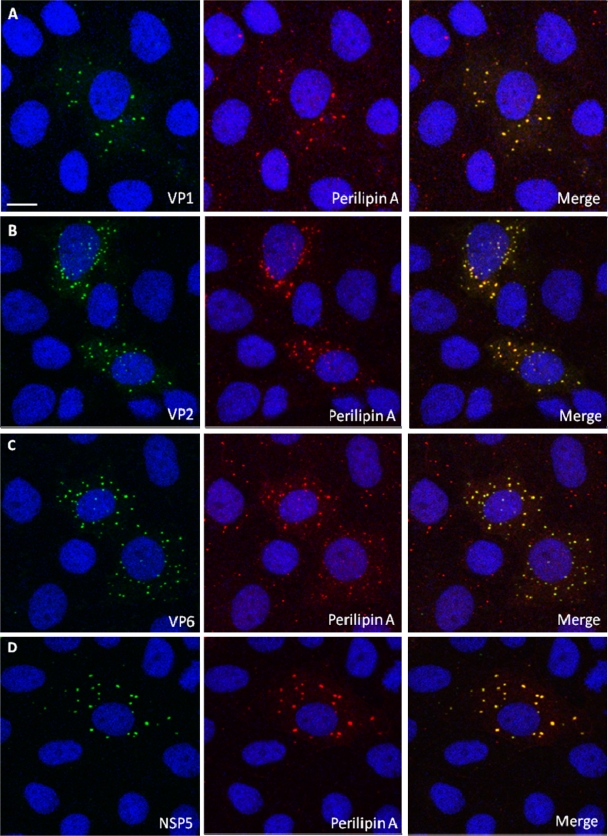
Using CM on rotavirus-infected MA104 cells (21), we found colocalization of NSP2 with both ADRP and perilipin A in viroplasms (Fig. 1 A and B). Identical results were obtained when using a different perilipin A antibody (raised in goats), or Caco-2 cells, a human gut-derived cell line supporting rotavirus replication (data not shown). We also found that perilipin A colocalizes with other rotavirus proteins previously shown to be components of viroplasms (25, 46, 47), VP1, VP2, VP6, and NSP5, in viroplasms (Fig. 2). The data indicate close association of known rotaviral viroplasm components with major LD-associated proteins.
FIG. 1.
NSP2 colocalizes with LD-associated proteins perilipin A and ADRP in viroplasms. Confocal images of rotavirus-infected MA104 cells at 8 h p.i. were obtained. (A and B) Viroplasms were detected with anti-NSP2 antibodies, followed by visualization with Alexa Fluor 488 (green)-labeled secondary antibody, whereas LD-associated proteins were detected with anti-perilipin A (A) and anti-ADRP antibodies (B), followed by reaction with Alexa Fluor 633 (red)-labeled secondary antibody. An individual viroplasm of panel A has been magnified and is shown in the inserts. (C) Confocal images of uninfected MA104 cells stained for perilipin A or ADRP and both for NSP2 according to the procedure used in panels A and B. Uninfected cells only show LDs. Scale bar, 10 μm.
FIG. 2.
Perilipin A colocalizes with VP1, VP2, VP6, and NSP5 in viroplasms. Shown are CM images of rotavirus-infected MA104 cells at 6 h p.i. Viroplasms were detected with anti-perilipin A antibodies visualized by Alexa Fluor 633 (red)-conjugated secondary antibodies, while VP1 (A), VP2 (B), VP6 (C), and NSP5 (D) were detected by monospecific antibodies raised in guinea pigs, followed by visualization with secondary antibodies conjugated with Alexa-Fluor 488 (green). Scale bar, 10 μm.
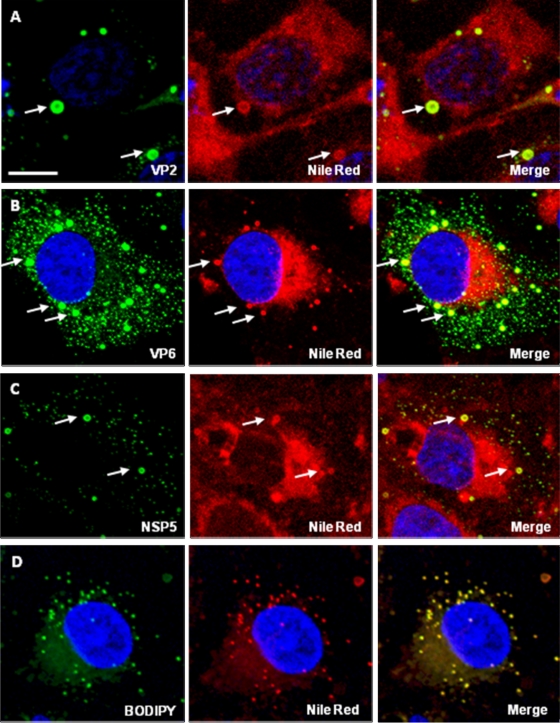
Costaining of Cos-7 cells, an African green monkey kidney derived cell line supporting rotavirus replication, with the fluorophore Nile Red, which stains LDs (7, 26), and antibodies to VP2, VP6, and NSP5, demonstrated colocalization of viral proteins with lipids in viroplasms (Fig. 3 A to C). Some diffuse cytoplasmic staining of lipids was also seen in uninfected cells (Fig. 3D); in rotavirus-infected cells, there was no reactivity with rotavirus-specific proteins in this area. (Figure 3D also demonstrates that Nile Red and BODIPY react with identical lipid structures.)
FIG. 3.
Rotavirus proteins colocalize with lipids in viroplasms. Confocal images of rotavirus-infected Cos-7 cells costained with Nile Red and antibodies to VP2 (A, 8 h p.i.), VP6 (B, 8 h p.i.), and NSP5 (C, 4 h p.i.). (D) uninfected cell controls stained with Bodipy (left) and Nile Red (center). The merged picture is shown on the right. Scale bar, 10 μm.
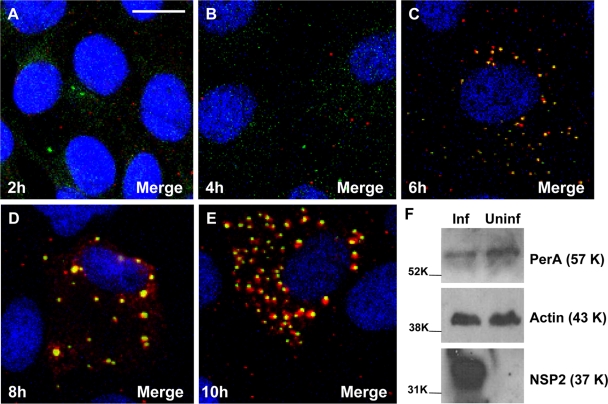
A time course on rotavirus-infected MA104 cells by CM revealed that viroplasms form separately at early times p.i. (2 to 4 h; Fig. 4 A and B) and from 5 to 6 h p.i. onward recruit LD components (Fig. 4C to E). Western blotting of extracts of rotavirus-infected and uninfected cells showed that the overall expression of perilipin A was not increased in the rotavirus-infected cells (Fig. 4F).
FIG. 4.
(A to E) Time course analysis of viroplasm development in rotavirus-infected MA104 cells showing the recruitment of LD-associated proteins. MA104 cells on coverslips were infected with rotavirus RF strain at an MOI of 1, and cells were fixed at 2, 4, 6, 8, and 10 h p.i. as indicated. Viroplasms were detected with anti-perilipin A antibodies visualized by Alexa Fluor 633 (red)-conjugated secondary antibodies, while NSP5 was detected by monospecific antibodies raised in guinea pigs, followed by visualization with secondary antibodies conjugated with Alexa Fluor 488 (green). (F) MA104 cells on six-well plates were infected with rotavirus RF strain at an MOI of 3 for 6 h. A mock-infected control was also produced. Cells were lysed, and the extracts were processed for Western blotting as described in Materials and Methods. Detection of perilipin A, NSP5 and actin was by specific antibodies, followed by reaction with HRP-conjugated secondary antibodies and substrates as described in Methods. The molecular masses (in kilodaltons) of the calibration protein markers are indicated to the left. Scale bar, 10 μm.
Rotavirus NSP2 and NSP5 in VLS colocalize with perilipin A in MA104 cells.
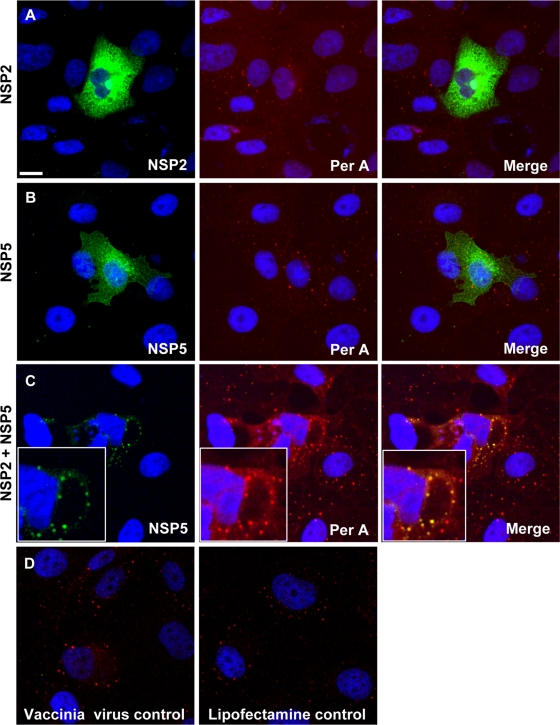
VLS can be produced in uninfected cells by coexpression of NSP2 and NSP5 (21). Perilipin colocalizes with VLS but not with the individually expressed viral proteins (Fig. 5). Thus, coexpression of these two viral proteins alone is sufficient to drive interaction of VLS with LD components, indicating an interplay of viral and cellular components independent of rotavirus replication.
FIG. 5.
NSP5 colocalizes with perilipin A in VLS. MA104 cells were infected with a vaccinia virus recombinant expressing T7 polymerase and transfected after 1 h. (A) Transfection of plasmid expressing NSP2 only. (B) Transfection of plasmid expressing NSP5 only. (C) Cotransfection of NSP2- and NSP5-expressing plasmids, leading to the production of VLS (see magnification in inserts). Perilipin A only colocalizes with VLS and not the individual nonstructural rotavirus proteins. (D) Vaccinia virus-infected and mock-transfected controls. Costaining was by anti-NSP2, anti-NSP5, and anti-perilipin A antibodies, followed by staining with secondary antibodies labeled with Alexa Fluor 488 and Alexa Fluor 633. Scale bar, 10 μm.
FRET demonstrates proximity of NSP5-EGFP and perilipin A.
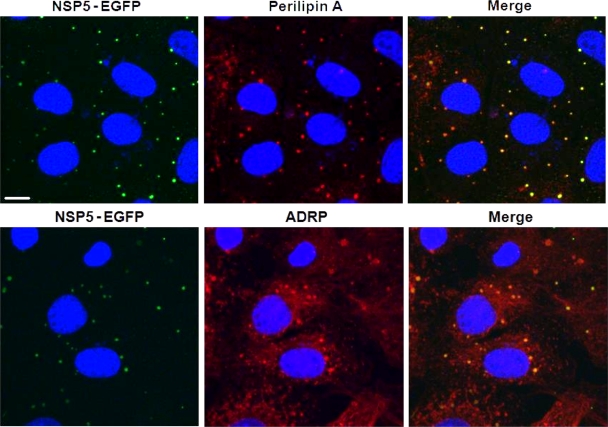
Uninfected MA104 cell lines stably expressing an NSP5-EGFP fusion protein showed diffuse cytoplasmic localization of this protein by autofluorescence. Upon rotavirus infection, NSP5-EGFP colocalized with the other viral components in viroplasms (16) and also with perilipin A and ADRP (Fig. 6). Close spatial proximity of NSP5-EGFP with perilipin A was further demonstrated by Fluorescence Resonance Energy Transfer (FRET) (33, 59). Figure 7 shows the fluorescence intensity (panels A and C) and fluorescence lifetime (panels B and D) of NSP5-EGFP in viroplasms of rotavirus-infected cells measured by time-correlated single photon counting as well as probability and cumulative distribution functions (panels E to G). Cells labeled with donor and acceptor fluorophores exhibited 5.0% ± 0.7% FRET efficiency (mean ± the SD, n = 34) compared to 0.0% ± 0.7% for the cells containing only the donor fluorophore in viroplasms (n = 19). The computation of FRET efficiencies was mostly based on nonbinned data (details not shown).
FIG. 6.
NSP5-EGFP colocalizes with LD-associated proteins perilipin A and ADRP in viroplasms. Confocal images of MA104 cells stably expressing EGFP-NSP5 infected with rotavirus at 8 h p.i. NSP5-EGFP is visualized by its autofluorescence (green), while LD-associated proteins were detected with monospecific antibodies, followed by visualization with Alexa Fluor 633 (red)-labeled secondary antibody. Scale bar, 10 μm.
The redistribution of perilipin A in virus-infected cells depends on viroplasm formation.
Perilipin A was observed to be redistributed in rotavirus-infected cells only when viroplasms are formed (Fig. 1 and 6). MA104 cells transfected with siRNA against the NSP5 mRNA of the porcine OSU rotavirus strain (11) showed a significant reduction in both, the number of cells forming viroplasms and perilipin A redistribution (data not shown). As expected (11), cells infected with the OSU rotavirus and treated with OSU NSP5-specific siRNA replicated significantly less viral RNA (data not shown) and produced 100 times less infectious progeny virus (Table 3).
TABLE 3.
Infectious porcine rotavirus OSU recovered at 16 h p.i. from MA104 cells transfected with OSU NSP5 siRNA and with irrelevant siRNA and from untransfected MA104 cells
| Parameter | Log TCID50 for cell groupa: |
Difference (group B − group C) |
|||
|---|---|---|---|---|---|
| A | B | C | Log | Antilog | |
| Titration 1 | 8.3 | 8.5 | 6.2 | 2.3 | 200 |
| Titration 2 | 8.3 | 8.5 | 6.4 | 2.1 | 126 |
| Mean ± SD | 8.3 ± 0 | 8.5 ± 0 | 6.3 ± 0.1 | 2.2 | 158 |
Cell groups: A, rotavirus infected only; B, rotavirus infected and transfected with SA11 NSP5-specific small interfering RNA (siRNA); C, rotavirus infected and transfected with OSU NSP5-specific siRNA.
Viroplasm components and perilipin A colocalize in low-density fractions of rotavirus-infected cell extracts separated by iodixanol gradient ultracentrifugation.
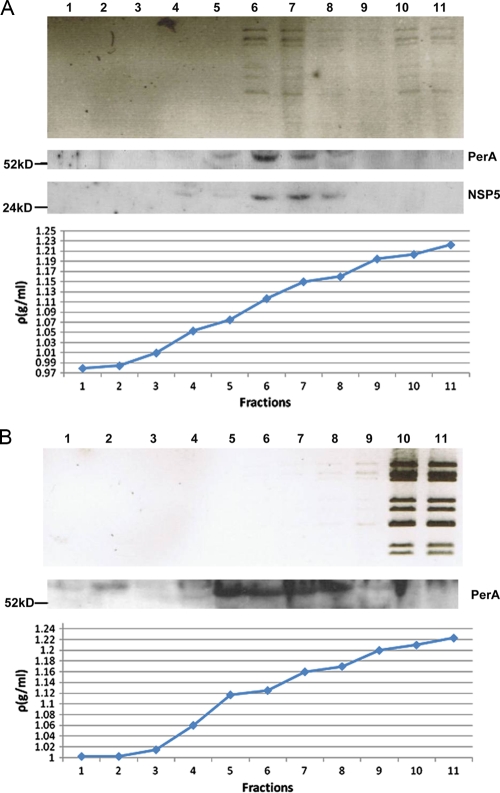
Equilibrium ultracentrifugation of rotavirus-infected cell extracts through iodixanol gradients showed that most of the viral dsRNA was found in the same low-density fractions (1.11 to 1.15 g/ml) as both NSP5 and perilipin A (Fig. 8 A), again implying that nascent subviral particles are in viroplasms and associated with LD components. As a control, noninfected MA104 cell extract, spiked with CsCl gradient-purified and deionized DLPs, was subjected to the same procedure. Free DLPs sedimented to the bottom fractions of the gradient without interaction with low density fractions (Fig. 8B). Perilipin A was found in the same low-density fractions of uninfected and rotavirus-infected cell extracts. In addition, there was a perilipin A band in fraction 2 (density of 1.01) of the gradient of the uninfected cell extract which was not seen in the corresponding fraction of rotavirus-infected cell extract, suggesting that this material was usurped by rotavirus viroplasms as shown above by CM (Fig. 4). (The perilipin A band in fraction 2 of the uninfected cell extract gradient migrates slightly slower than the bands in fractions 5 to 8 and may represent a higher phosphorylated form.) In the rotavirus-infected cell extract gradient (Fig. 8A), viral dsRNA (although not the majority) is also found in the bottom fractions, originating either from free DLPs or DLPs released from viroplasms that are broken during the ultracentrifugation procedure.
FIG. 8.
Fractions of iodixanol gradients in which extracts of rotavirus-infected (A) and uninfected cells spiked with CsCl gradient-purified and deionized DLPs (B) have been separated by ultracentrifugation (for details, see Materials and Methods). Fractions were analyzed for the presence of rotavirus dsRNA, NSP5, and perilipin A. Fractions 6 and 7 of the rotavirus-infected cell extract showed maximal colocalization of all three components at densities of 1.11 to 1.15 g/ml. Fractions of uninfected cell extracts were also probed for the presence of NSP5, which was negative as expected (results not shown). The sizes of marker proteins are indicated in kilodaltons.
Drug-induced fragmentation of LDs decreases viroplasm formation, viral RNA replication and the yield of infectious progeny.
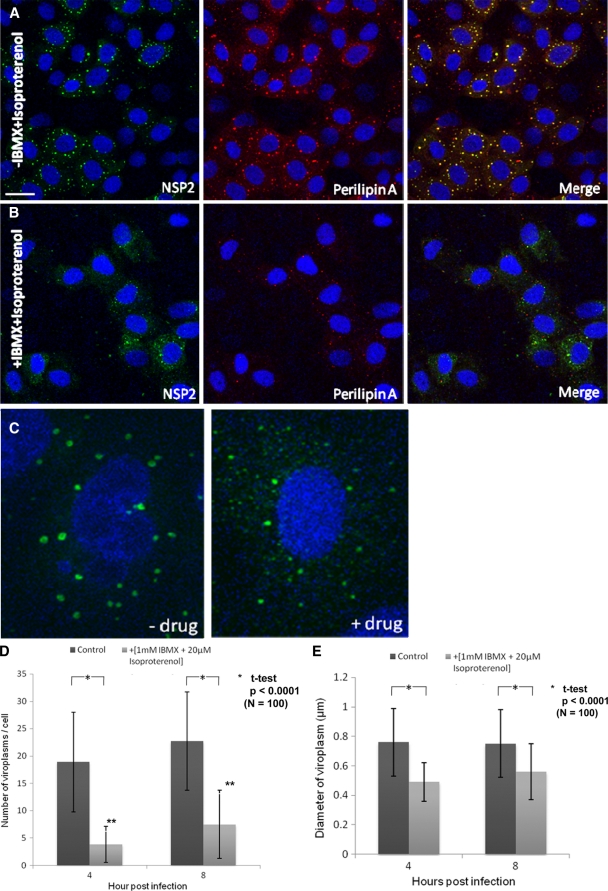
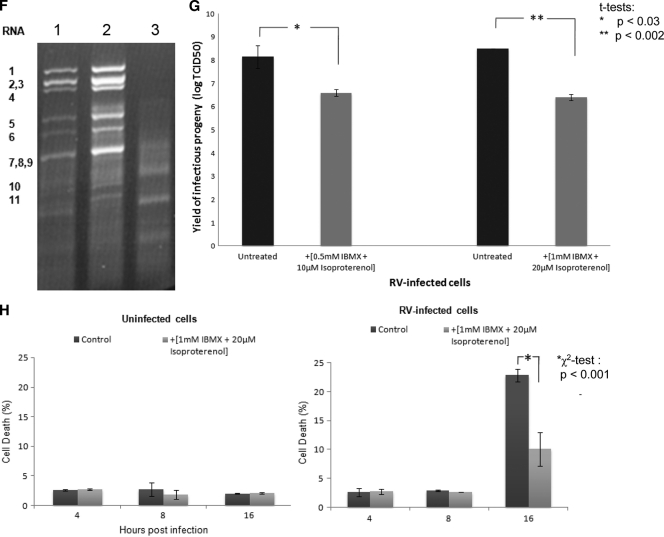
Isoproterenol and IBMX raise cellular cyclic AMP and, in combination, disperse LDs into smaller microdroplets in adipocytes (27, 34). Rotavirus-infected MA104 cells were incubated with 1 mM IBMX and 20 μM isoproterenol for 4 and 8 h. The average numbers and sizes of viroplasms in the treated cells were significantly reduced at both time points (Fig. 9 A, B, D, and E). After 4 h of treatment, NSP2 appeared to be uniformly dispersed in minute foci (Fig. 9C). At 16 h p.i., in the presence of IBMX and isoproterenol, the amount of viral dsRNA was decreased by ∼4-fold (Fig. 9F; details of the densitometric evaluation are not shown), and the titers of infectious viral progeny were 120 to 200 times lower (Fig. 9G). Viability tests indicated that the drugs had no toxic effect on the cells (Fig. 9H). In contrast, at 16 h p.i., the drug-treated cells showed significantly higher viability compared to the untreated control (Fig. 9H). This reduction in cytopathicity correlated well with the decreases in viral RNA replication and in the amount of infectious viral progeny in drug-treated cells (Fig. 9F and G).
FIG. 9.
LD fragmenting chemicals IBMX and isoproterenol disperse viroplasms and decrease their number and size. (A and B) Confocal images of MA104 cells infected with rotavirus in the absence (A) or presence (B) of 1 mM IBMX and 20 μM isoproterenol at 8 h p.i. Viroplasms were reacted with anti-NSP2 antibodies and visualized with Alexa Fluor 488 (green)-labeled secondary antibody. Scale bar, 20 μm. (C) Individual rotavirus-infected cells stained for NSP2 in the absence or presence of drugs. The drugs lead to the fragmentation of viroplasms as shown for LDs by Marcinkiewicz et al. (34). (D and E) Numbers of viroplasms per cell and average diameters of viroplasms at 4 and 8 h p.i. All cells were infected with rotavirus at an MOI of 10. Only typical viroplasms were counted, not the apparently fragmented organelles (see panel C). Significant differences (P values) were determined by two-tailed Student t tests. (F) IBMX and isoproterenol decreases viral RNA replication (by 4-fold; densitometrically evaluated by the ImageJ program). An ethidium bromide-stained agarose gel (1%) shows the dsRNA profiles of rotavirus in MA104 cells infected for 16 h in the presence (lane 1) or absence (lane 2) of IBMX and isoproterenol, as well as uninfected and chemically treated cell controls (lane 3). The 11 RNA segments are denoted to the left. (G) Infectious rotavirus recovered from MA104 cells infected for 16 h in the presence or absence of different concentrations of IBMX and isoproterenol. (H) Viability of uninfected and rotavirus-infected MA104 cells at 4, 8, and 16 h after treatment with 1 mM IBMX and 20 μM isoproterenol.
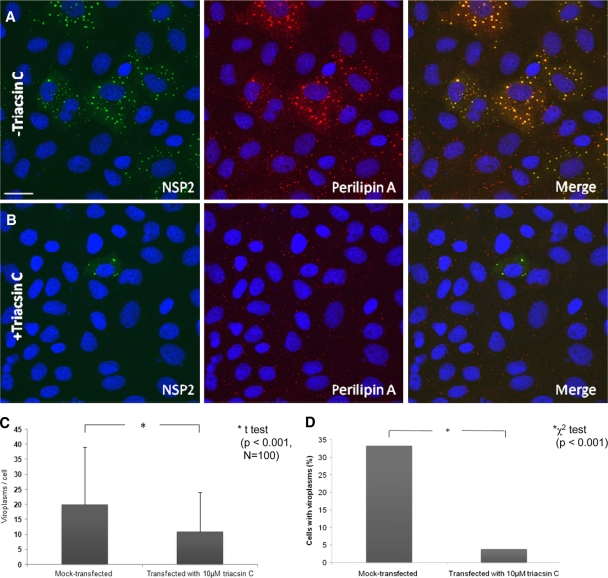
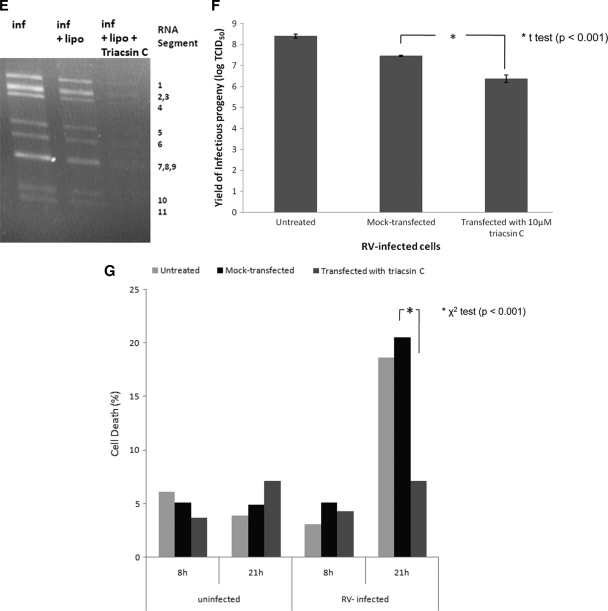
To further investigate the significance of LD components for rotavirus replication, we used the LD-blocking compound triacsin C (10, 30, 39). Transfection of rotavirus-infected cells with 10 μM triacsin C led to a significant decrease in the number of cells containing viroplasms (Fig. 10 A, B, and D), the number of viroplasms/cell (Fig. 10A, B, and C), the amount of viral RNA replication by ∼4-fold (Fig. 10E; details of the densitometric evaluation not shown), and the yield of infectious viral progeny (Fig. 10F). The reduction in viroplasm formation was again associated with an increased cell viability at 21 h p.i. (Fig. 10G).
FIG. 10.
Triacsin C decreases viroplasm number and percentage of cells infected. (A and B) Confocal images of MA104 cells infected with rotavirus in the absence (A) or presence (B) of 10 μM triacsin C which was transfected with Lipofectamine 15 min after infection, which lasted for 6 h. Viroplasms were reacted with anti-NSP2 and anti-perilipin A antibodies and visualized with Alexa Fluor 488 (green)-labeled secondary antibody and Alexa Fluor 633 (red)-labeled secondary antibody, respectively. Scale bar, 20 μm. (C and D) Numbers of viroplasms/cell (C) and percentages of cells (D) carrying viroplasms in RV-infected MA104 cells at 16 h p.i. The numbers were compared by the Student t test (C) and the chi-squared test (D), respectively. (E) MA104 cells were infected with rotavirus at an MOI of 3 for 16 h (inf) in the absence of any other chemical, in the presence of Lipofectamine (inf + lipo), or in the presence of both Lipofectamine and triacsin C (inf + lipo + triacsin C, applied 15 min p.i.). At 16 h p.i., RNA was extracted from the supernatant of both fractions, separated on a 1% agarose gel, and detected by ethidium bromide staining. Under triacsin C treatment, the yield of dsRNA was decreased by 4-fold (densitometric evaluation using the ImageJ program). (F) Yield of infectious rotavirus progeny from mock-transfected or triacsin C-transfected MA104 cells at 16 h p.i. (G) Viability of uninfected and rotavirus-infected MA104 cells at 8 and 21 h after no further treatment, mock transfection with Lipofectamine 2000 and transfection with Lipofectamine 2000 containing 10 μM triacsin C.
DISCUSSION
Rotaviruses begin their assembly in cytoplasmic inclusion bodies termed viroplasms, also described as “factories” of RV particle production (3). We have shown by structural and functional analyses that viroplasms associate with components of, and physically resemble, LDs. Ringlike staining of perilipin A has been observed, particularly on larger viroplasms (Fig. 1A), as has also been seen for NSP5 (16), another phosphoprotein. Second, we demonstrated that chemical compounds affecting the integrity of LDs also disrupt viroplasms, leading to a decrease in both viral RNA replication and the production of infectious viral progeny.
LDs are the main sites of lipid storage in eukaryotic cells and are evolutionarily conserved. They consist of a hydrophobic core containing mainly triacylglycerides surrounded by a phospholipid monolayer containing cholesterol (56) into which numerous cellular proteins are inserted. Best characterized are proteins of the PAT family but, recently, more than 100 proteins have been found to be associated with LDs (4, 28), some of which are recognized as regulators of lipid metabolism, governing lipid homeostasis, intracellular trafficking, and cell signaling, often through phosphorylation pathways (4, 28, 53, 60). However, the functions of many of these proteins remain largely uncharacterized (28, 56). LDs are highly dynamic organelles interacting with various cellular compartments (36).
The CM data demonstrated that perilipin A and ADRP, two LD-associated proteins, colocalize with the rotaviral proteins found in viroplasms, i.e., with the nonstructural proteins NSP2 and NSP5 (Fig. 1 and 2) and the structural proteins VP1, VP2, and VP6 (Fig. 2). This association was further confirmed using perilipin A antibodies raised in a different host species (goat) and infecting Caco-2 cells. Interestingly, perilipin A appears as a ring-shaped structure around the viroplasms (Fig. 1A, inset), as has been observed for perilipin A and other LD-associated proteins on the surface of LDs (34-36, 48). It was confirmed that NSP5 colocalized with NSP2, VP1, VP2, VP6, and perilipin A in viroplasms (Fig. 2). All controls, omitting either primary or secondary antibodies or both, had the expected outcome (data not shown). The close association of rotavirus viroplasms with LD components was also demonstrated by observing colocalization of rotavirus proteins with lipids in viroplasms (Fig. 3A to C). The detection of perilipin A in NSP2/NSP5-induced VLS indicated that these two nonstructural viral proteins suffice to successfully recruit LD components (Fig. 5), i.e., independently of a natural rotavirus infection. A time course analysis of rotavirus infection demonstrated the recruitment of LD components to viroplasms very clearly (Fig. 4). Rotavirus proteins are not normal components of LDs, and their large concentration in viroplasms likely alters the morphology of LD components, particularly at later stages of the rotavirus replication cycle (Fig. 4). Such changes appear to be less prominent in LDs of HCV-infected cells (48).
Fluorescence lifetime imaging (Fig. 7) revealed energy transfer between NSP5-EGFP and perilipin A labeled with a Cy3-conjugated secondary antibody, confirming close spatial proximity between NSP5 and perilipin A within viroplasms.
Analysis of extracts of rotavirus-infected cells on iodixanol gradients showed colocalization of viral dsRNA, NSP5, and perilipin A in low-density fractions (Fig. 8A), also indicating close interaction between viroplasms and LD components. (This experiment was controlled by a gradient of uninfected cells spiked with purified DLPs [Fig. 8B], which sedimented through the gradient to the bottom fractions without interaction with low-density fractions.) Intimate involvement of LD components with viral replication has previously been demonstrated for hepatitis C virus (HCV) (8, 29, 38, 48), for the related GB virus B (29), for dengue virus (50), and to some extent for orthoreovirus (12). Interaction of the 2C protein of human parechovirus 1 with LDs has been observed (31); however, the significance of this interaction for viral replication is not clear. The core and some nonstructural proteins of HCV were found to colocalize with LDs in infected hepatocytes (38). However, HCV is not synthesized in separate “virus factories,” as has been shown for members of the Reoviridae family (3, 41). HCV mutants, in which the core protein cannot associate with LDs, produce less infectious virus (8), demonstrating that LDs are essential for HCV replication (38). Propagation of the obligate intracellular parasite Chlamydia was also shown to depend on functionally intact LDs (32). For dengue virus, interaction of hydrophobic regions of the capsid protein with LDs was shown to occur and to be essential for viral replication, and the fatty acid synthase inhibitor C75 (decreasing the number of LDs in treated cells) was found to reduce dengue virus replication significantly (50).
LD metabolism is regulated by its associated proteins, phosphorylation of some of them leading to lipolysis (27, 34). Chemicals active in promoting this pathway (β-adrenergic stimulants and phosphodiesterase inhibitors) can fragment and disperse LDs (34); these chemicals were shown to interfere with the formation of viroplasms, viral RNA replication, and the production of infectious rotavirus progeny.
NSP5 is phosphorylated and hyperphosphorylated to various degrees (1). It is plausible that NSP5 is anchored into the phospholipid monolayer of LDs by its predicted C-terminal amphipathic helix (51), where it then can have access to the phosphorylation pathways characteristic of LD-associated cellular proteins. This would be consistent with the documented ringlike structures formed by NSP5 in viroplasms (16), as is observed for LD-associated proteins (34, 36).
NSP5-EGFP has a diffuse cytoplasmic distribution in uninfected MA104 cells but becomes associated with viroplasms and LD-specific proteins as soon as the cells are infected with rotavirus. VLS formed in uninfected cells after cotransfection with NSP2- and NSP5-expressing plasmids (21) also associate with LD-specific proteins.
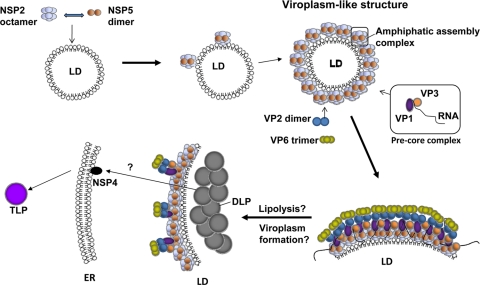
A model of the sequence of events (Fig. 11) envisages that NSP2 and NSP5 recruit LD proteins and lipids and establish an amphipathic assembly complex, which could then interact with VP1 (2) before or after VP1 binds rotavirus single-stranded RNA. The building blocks for rotavirus precores (2, 25, 45) may be assembled on the surface of the phospholipid LD monolayer (Fig. 11). At some stage, these assemblies become viroplasms containing cores and DLPs and may lose some or all of their lipids (Fig. 11). Further details of this transition are not clear (14) and remain to be investigated (Immune electron microscopy work is in progress.) After leaving viroplasms, genome-containing DLPs enter the RER using NSP4 as an intracellular receptor (20), and further maturation into TLPs takes place (Fig. 11). A somewhat analogous model has been proposed for the production of HCV virions for which the presence of LDs is essential (38).
FIG. 11.
Proposed schematic model of early rotavirus morphogenesis. LDs serve as a platform (LD-associated proteins not shown) to which NSP2 and NSP5 attach to form VLS. The VLS in turn associate with pre-core complexes (consisting of VP1, VP3, and segmental plus RNA), VP2 and VP6. Viroplasms then form and fuse (20) by as-yet-unknown mechanisms; this is likely to be accompanied by lipolysis. DLPs assembled in viroplasms make contact with NSP4 inserted into membranes of the ER and are then further processed to become TLPs.
The association of RV- and LD-associated cellular proteins was found to be of functional significance. Chemicals which inhibit LD formation, either by enhancing the phosphorylation of perilipin A, leading to lipolysis (IMBX and isoproterenol) (34), or by inhibiting the formation of LDs (triacsin C) (10), were reproducibly shown to interfere with the RV replication cycle, significantly reducing the number and size of viroplasms formed, viral RNA replication, and the production of infectious progeny (Fig. 9 and 10). Fragmentation of viroplasms by (IBMX plus isoproterenol) may affect their recognized functions of subviral particle assembly and viral RNA replication. The chemicals used are unlikely to affect viral particle adsorption, penetration, and early transcription since their effect was identical when applied to cells at 1 h p.i. (unpublished data). The effect of other inhibitors of LD formation on rotavirus replication is under investigation, and it is likely that more active compounds will be found. The data provided strengthen the proposition that association of rotavirus proteins with LD components is essential for RV replication and that viroplasms at 5 to 10 h p.i. may be considered as rotavirus-modified forms of LDs.
The reduction in the production of infectious rotavirus progeny by the chemicals used was not due to a toxic effect on cell viability (Fig. 9 and 10). On the contrary, at 16 to 21 h p.i., rotavirus-infected cells treated with the chemicals showed significantly less cell death than untreated, rotavirus-infected cells. These results are in line with the fact that the rotavirus replication cycle is completed by 12 h p.i. (20), after which time the cells usually are lysed.
The association of viroplasms with LD components presents another example of a virus hijacking a cellular organelle for its own replication. This finding raises many questions that remain at present unanswered. The number and nature of the interactions of RV proteins with LD-associated proteins need to be studied further. Identification of LD proteins important for rotavirus replication will lead to a better understanding not only of the early stages of rotavirus morphogenesis but also of the functions of LD-associated proteins (36). The rotavirus protein NSP4 has recently been found to associate with autophagosomal marker LC3-positive vesicles which form caplike structures on viroplasms (6). Studies in hepatocytes have indicated an interaction of autophagosomal components with LDs and involvement of autophagy in LD metabolism under stress conditions (52). Rotavirus infection elicits upregulation of a number of stress-related genes (15), and thus it is possible that NSP4 is involved in the lipolysis of viroplasms via “macrolipophagy” (52). Rotavirus NSP4 also interacts with caveolin-1 (44) which has been shown to be involved in the modulation of LD biogenesis and metabolism (13). All of these interactions require further study. The cytoplasmic “virus factories” (3) of other genera of the Reoviridae (Orthoreovirus, Orbivirus, etc.) may also depend on interaction with LD-associated components.
The antiviral effect of LD-fragmenting drugs as described here for rotavirus-infected cells in vitro is promising in terms of the development of novel therapeutics; however, it needs to be confirmed in animal models of rotavirus infection and disease. Such data may open the door for the development of effective therapies of disease after infection with rotaviruses and other viruses whose replication depends on the interaction with LDs.
Acknowledgments
We thank Ray Hicks for technical support of the CM studies and Wilson Li for advice. Discussions with Oscar Burrone were particularly appreciated. Oscar Burrone, Francesca Arnoldi, and Roberta Contin provided the MA104 cell line constitutively expressing NSP5-EGFP and NSP2- and NSP5-specific antisera. Antisera against VP1, VP2, VP3, and VP6 were a gift from John Patton, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD. Simon Schlachter was instrumental in developing the TCSPC system.
N.K. was supported by an Amgen Foundation Vacation Scholarship. The general support by Addenbrooke's Biomedical Research Centre is acknowledged. This study was funded by The Wellcome Trust (grants RG46760 to W.C. and WT082031MA to A.L. and U.D.). W.C. was supported by the Wellcome Trust 4-year Ph.D. program in Infection and Immunity.
W.C. carried out many of the experiments and was involved in writing the manuscript. M.G. contributed to the planning of the experiments. A.E. and C.F.K. planned and carried out the FRET experiments and contributed to the writing of the manuscript. N.K. carried out the time course experiment. N.C., S.C., and G.T. carried out the combined staining for lipids and viroplasm-associated proteins and contributed to the writing of the manuscript. A.L. and U.D. planned and supervised the work and were responsible for the writing of the manuscript. All data indicated as “not shown” are available upon request.
Footnotes
Published ahead of print on 24 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. [DOI] [PubMed] [Google Scholar]
- 2.Arnoldi, F., M. Campagna, C. Eichwald, U. Desselberger, and O. R. Burrone. 2007. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J. Virol. 81:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamford, D. H., and L. Mindich. 2004. Viral molecular machines: replication systems within the inner cores of dsRNA viruses. Virus Res. 101:1. [Google Scholar]
- 4.Bartz, R., J. K. Zehmer, M. Zhu, Y. Chen, G. Serrero, Y. Zhao, and P. Liu. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256-3265. [DOI] [PubMed] [Google Scholar]
- 5.Becker, W., A. Bergmann, M. A. Hink, K. König, K. Benndorf, and C. Biskup. 2004. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc. Res. Tech. 63:58-66. [DOI] [PubMed] [Google Scholar]
- 6.Berkova, Z., S. E. Crawford, G. Trugnan, T. Yoshimori, A. P. Morris, and M. K. Estes. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 80:6061-6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonilla, E., and A. Prelle. 1987. Application of nile blue and nile red, two fluorescent probes, for detection of lipid droplets in human skeletal muscle. J. Histochem. Cytochem. 35:619-621. [DOI] [PubMed] [Google Scholar]
- 8.Boulant, S., P. Targett-Adams, and J. McLauchlan. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204-2213. [DOI] [PubMed] [Google Scholar]
- 9.Brasaemle, D. L., T. Barber, N. E. Wolins, G. Serrero, E. J. Blanchette-Mackie, and C. Londos. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38:2249-2263. [PubMed] [Google Scholar]
- 10.Brasaemle, D. L., B. Rubin, I. A. Harten, J. Gruia-Gray, A. R. Kimmel, and C. Londos. 2000. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 275:38486-38493. [DOI] [PubMed] [Google Scholar]
- 11.Campagna, M., C. Eichwald, F. Vascotto, and O. R. Burrone. 2005. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J. Gen. Virol. 86:1481-1487. [DOI] [PubMed] [Google Scholar]
- 12.Coffey, C. M., A. Sheh, I. S. Kim, K. Chandran, M. L. Nibert, and J. S. Parker. 2006. Reovirus outer capsid protein micro1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J. Virol. 80:8422-8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen, A. W., B. Razani, W. Schubert, T. M. Williams, X. B. Wang, P. Iyengar, D. L. Brasaemle, P. E. Scherer, and M. P. Lisanti. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes 53:1261-1270. [DOI] [PubMed] [Google Scholar]
- 14.Cuadras, M. A., B. B. Bordier, J. L. Zambrano, J. E. Ludert, and H. B. Greenberg. 2006. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J. Virol. 80:3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 76:4467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichwald, C., J. F. Rodriguez, and O. R. Burrone. 2004. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 85:625-634. [DOI] [PubMed] [Google Scholar]
- 17.Esposito, A., C. P. Dohm, P. Kermer, M. Bähr, and F. S. Wouters. 2007. Alpha-synuclein and its disease-related mutants interact differentially with the microtubule protein tau and associate with the actin cytoskeleton. Neurobiol. Dis. 26:521-531. [DOI] [PubMed] [Google Scholar]
- 18.Esposito, A., H. C. Gerritsen, and F. S. Wouters. 2005. Fluorescence lifetime heterogeneity resolution in the frequency domain by lifetime moments analysis. Biophys. J. 89:4286-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esposito, A., H. C. Gerritsen, F. S. Wouters, and U. Resch-Genger. 2008. Fluorescence lifetime imaging microscopy: quality assessment and standards, p. 117-142. In O. S. Wolfbeis (ed.), Standardization and quality assurance in fluorescence measurements II. Springer, Berlin, Germany.
- 20.Estes, M. K., and A. Z. Kapikian. 2007. Rotaviruses, p. 1917-1974. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, PA.
- 21.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two nonstructural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 22.Follett, E. A., and U. Desselberger. 1983. Cocirculation of different rotavirus strains in a local outbreak of infantile gastroenteritis: monitoring by rapid and sensitive nucleic acid analysis. J. Med. Virol. 11:39-52. [DOI] [PubMed] [Google Scholar]
- 23.Frank, J. H., A. D. Elder, J. Swartling, A. R. Venkitaraman, A. D. Jeyasekharan, and C. F. Kaminski. 2007. A white light confocal microscope for spectrally resolved multidimensional imaging. J. Microsc. 227:203-215. [DOI] [PubMed] [Google Scholar]
- 24.Fuerst, T. R., and B. Moss. 1989. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells. Importance of the 5′ untranslated leader. J. Mol. Biol. 206:333-348. [DOI] [PubMed] [Google Scholar]
- 25.Gallegos, C. O., and J. T. Patton. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616-627. [DOI] [PubMed] [Google Scholar]
- 26.Genicot, G., J. L. Leroy, A. V. Soom, and I. Donnay. 2005. The use of a fluorescent dye, Nile red, to evaluate the lipid content of single mammalian oocytes. Theriogenology 63:1181-1194. [DOI] [PubMed] [Google Scholar]
- 27.Gross, D. N., H. Miyoshi, T. Hosaka, H. H. Zhang, E. C. Pino, S. Souza, M. Obin, A. S. Greenberg, and P. F. Pilch. 2006. Dynamics of lipid droplet-associated proteins during hormonally stimulated lipolysis in engineered adipocytes: stabilization and lipid droplet binding of adipocyte differentiation-related protein/adipophilin. Mol. Endocrinol. 20:459-466. [DOI] [PubMed] [Google Scholar]
- 28.Guo, Y., T. C. Walther, M. Rao, N. Stuurman, G. Goshima, K. Terayama, J. S. Wong, R. D. Vale, P. Walter, and R. V. Farese. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 30.Igal, R. A., P. Wang, and R. A. Coleman. 1997. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem. J. 324:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogerus, C., O. Samuilova, T. Poeyry, E. Jokitalo, and T. Hyypiae. 2007. Intracellular localization and effects of individually expressed human parechovirus 1 non-structural proteins. J. Gen. Virol. 88:831-841. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, Y., J. Cocchiaro, and R. H. Valdivia. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646-1651. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz, J. R. 1999. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum Publishers, New York, NY.
- 34.Marcinkiewicz, A., D. Gauthier, A. Garcia, and D. L. Brasaemle. 2006. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281:11901-11909. [DOI] [PubMed] [Google Scholar]
- 35.Martin, S., K. Driessen, S. J. Nixon, M. Zerial, and R. G. Parton. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280:42325-42335. [DOI] [PubMed] [Google Scholar]
- 36.Martin, S., and R. G. Parton. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell. Biol. 7:373-378. [DOI] [PubMed] [Google Scholar]
- 37.Miura, S., J. W. Gan, J. Brzostowski, M. J. Parisi, C. J. Schultz, C. Londos, B. Oliver, and A. R. Kimmel. 2002. Functional conservation for lipid storage droplet association among perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277:32253-32257. [DOI] [PubMed] [Google Scholar]
- 38.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089-1097. (Erratum, 9:1216.) [DOI] [PubMed] [Google Scholar]
- 39.Namatame, I., H. Tomoda, H. Arai, K. Inoue, and S. Omura. 1999. Complete inhibition of mouse macrophage-derived foam cell formation by triacsin C. J. Biochem. 125:319-327. [DOI] [PubMed] [Google Scholar]
- 40.Nejmeddine, M., G. Trugnan, C. Sapin, E. Kohli, L. Svensson, S. Lopez, and J. Cohen. 2000. Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol. 74:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 42.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parr, R. D., S. M. Storey, D. M. Mitchell, A. L. McIntosh, M. Zhou, K. D. Mir, and J. M. Ball. 2006. The rotavirus enterotoxin NSP4 directly interacts with the caveolar structural protein caveolin-1. J. Virol. 80:2842-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pesavento, J. B., S. E. Crawford, M. K. Estes, and B. V. Prasad. 2006. Rotavirus proteins: structure and assembly. Curr. Top. Microbiol. Immunol. 309:189-219. [DOI] [PubMed] [Google Scholar]
- 46.Petrie, B. L., D. Y. Graham, H. Hanssen, and M. K. Estes. 1982. Localization of rotavirus antigens in infected cells by ultrastructural immunocytochemistry. J. Gen. Virol. 63:457-467. [DOI] [PubMed] [Google Scholar]
- 47.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 48.Rouillé, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruiz-Palacios, G. M., I. Pérez-Schael, F. R. Velázquez, H. Abate, T. Breuer, S. C. Clemens, B. Cheuvart, F. Espinoza, P. Gillard, B. L. Innis, Y. Cervantes, A. C. Linhares, P. López, M. Macías-Parra, E. Ortega-Barría, V. Richardson, D. M. Rivera-Medina, L. Rivera, B. Salinas, N. Pavía-Ruz, J. Salmerón, R. Rüttimann, J. C. Tinoco, P. Rubio, E. Nuñez, M. L. Guerrero, J. P. Yarzábal, S. Damaso, N. Tornieporth, X. Sáez-Llorens, R. F. Vergara, T. Vesikari, A. Bouckenooghe, R. Clemens, B. de Vos, M. O'Ryan, et al. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 354:11-22. [DOI] [PubMed] [Google Scholar]
- 50.Samsa, M. M., J. A. Mondotte, N. G. Iglesias, I. Assunção-Miranda, G. Barbosa-Lima, A. T. Da Poian, P. T. Bozza, and A. V. Gamarnik. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sen, A., N. Sen, and E. R. Mackow. 2007. The formation of viroplasm-like structures by the rotavirus NSP5 protein is calcium regulated and directed by a C-terminal helical domain. J. Virol. 81:11758-11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh, R., S. Kaushik, Y. Wang, Y. Xiang, I. Novak, M. Komatsu, K. Tanaka, A. M. Cuervo, and M. J. Czaja. 2009. Autophagy regulates lipid metabolism. Nature 458:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tansey, J. T., A. M. Huml, R. Vogt, K. E. Davis, J. M. Jones, K. A. Fraser, D. L. Brasaemle, A. R. Kimmel, and C. Londos. 2003. Functional studies on native and mutated forms of perilipins: a role in protein kinase A-mediated lipolysis of triacylglycerols. J. Biol. Chem. 278:8401-8406. [DOI] [PubMed] [Google Scholar]
- 54.Taraporewala, Z. F., P. Schuck, R. F. Ramig, L. Silvestri, and J. T. Patton. 2002. Analysis of a temperature-sensitive mutant rotavirus indicates that NSP2 octamers are the functional form of the protein. J. Virol. 76:7082-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tate, J. E., C. A. Panozzo, D. C. Payne, M. M. Patel, M. M. Cortese, A. L. Fowlkes, and U. D. Parashar. 2009. Decline and change in seasonality of US rotavirus activity after the introduction of rotavirus vaccine. Pediatrics 124:465-471. [DOI] [PubMed] [Google Scholar]
- 56.Tauchi-Sato, K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507-44512. [DOI] [PubMed] [Google Scholar]
- 57.Vascotto, F., M. Campagna, M. Visintin, A. Cattaneo, and O. R. Burrone. 2004. Effects of intrabodies specific for rotavirus NSP5 during the virus replicative cycle. J. Gen. Virol. 85:3285-3290. [DOI] [PubMed] [Google Scholar]
- 58.Vesikari, T., D. O. Matson, P. Dennehy, P. van Damme, M. Santosham, Z. Rodriguez, M. J. Dallas, J. F. Heyse, M. G. Goveia, S. B. Black, H. R. Shinefield, C. D. Christie, S. Ylitalo, R. F. Itzler, M. L. Coia, M. T. Onorato, B. A. Adeyi, G. S. Marshall, L. Gothefors, D. Campens, A. Karvonen, J. P. Watt, K. L. O'Brien, M. J. DiNubile, H. F. Clark, J. W. Boslego, P. A. Offit, P. M. Heaton, et al. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 354:23-33. [DOI] [PubMed] [Google Scholar]
- 59.Wouters, F. S., P. J. Verveer, and P. I. Bastiaens. 2001. Imaging biochemistry inside cells. Trends Cell. Biol. 11:203-221. [DOI] [PubMed] [Google Scholar]
- 60.Zehmer, J. K., Y. Huang, G. Peng, J. Pu, R. G. Anderson, and P. Liu. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics 9:914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]