Abstract
We previously reported on a panel of HIV-1 clade B envelope (Env) proteins isolated from a patient treated with the CCR5 antagonist aplaviroc (APL) that were drug resistant. These Envs used the APL-bound conformation of CCR5, were cross resistant to other small-molecule CCR5 antagonists, and were isolated from the patient's pretreatment viral quasispecies as well as after therapy. We analyzed viral and host determinants of resistance and their effects on viral tropism on primary CD4+ T cells. The V3 loop contained residues essential for viral resistance to APL, while additional mutations in gp120 and gp41 modulated the magnitude of drug resistance. However, these mutations were context dependent, being unable to confer resistance when introduced into a heterologous virus. The resistant virus displayed altered binding between gp120 and CCR5 such that the virus became critically dependent on the N′ terminus of CCR5 in the presence of APL. In addition, the drug-resistant Envs studied here utilized CCR5 very efficiently: robust virus infection occurred even when very low levels of CCR5 were expressed. However, recognition of drug-bound CCR5 was less efficient, resulting in a tropism shift toward effector memory cells upon infection of primary CD4+ T cells in the presence of APL, with relative sparing of the central memory CD4+ T cell subset. If such a tropism shift proves to be a common feature of CCR5-antagonist-resistant viruses, then continued use of CCR5 antagonists even in the face of virologic failure could provide a relative degree of protection to the TCM subset of CD4+ T cells and result in improved T cell homeostasis and immune function.
Entry of human immunodeficiency virus (HIV) into target cells is a complex, multistep process that is initiated by interactions between the viral envelope (Env) protein gp120 and the host cell receptor CD4, which trigger conformational changes in gp120 that form and orient the coreceptor binding site (9, 24). Upon binding to coreceptor, which is either CCR5 or CXCR4 for primary HIV isolates, Env undergoes further conformational changes resulting in insertion of the gp41 fusion peptide into the host cell membrane and gp41-mediated membrane fusion (8, 15, 26). Targeting stages of the HIV entry process with antiretroviral drugs is a productive method of inhibiting HIV replication, as demonstrated by the potent antiviral effects of small-molecule CCR5 antagonists and fusion inhibitors (23, 35, 49). As with other antiretroviral drugs, HIV can develop resistance to entry inhibitors, and a detailed understanding of viral and host determinants of resistance will be critical to the optimal clinical use of these agents.
The coreceptor binding site that is induced by CD4 engagement consists of noncontiguous regions in the bridging sheet and V3 loop of gp120 (4, 18, 42, 43, 50). Interactions between gp120 and CCR5 occur in at least two distinct areas: (i) the bridging sheet and the stem of the V3 loop interact with sulfated tyrosine residues in the N′ terminus of CCR5, and (ii) the crown of the V3 loop is thought to engage the extracellular loops (ECLs), particularly ECL2, of CCR5 (10-12, 14, 18, 28). Small-molecule CCR5 antagonists bind to a hydrophobic pocket in the transmembrane helices of CCR5 and exert their effects on HIV by altering the position of the ECLs, making them allosteric inhibitors of HIV infection (13, 31, 32, 46, 52). The conformational changes in CCR5 that are induced by CCR5 antagonists vary to some degree with different drugs, as evidenced by differential binding of antibodies and chemokines to various drug-bound forms of CCR5 (47, 54).
CCR5 antagonists are unusual among antiretroviral agents in that they bind to a host protein rather than a viral target, and therefore the virus cannot directly mutate the drug binding site to evade pharmacologic pressure. Nevertheless, HIV can escape susceptibility to CCR5 antagonists. One mechanism by which this occurs is the use of the alternative HIV coreceptor, CXCR4. In vivo, this has most often been manifest as the outgrowth of R5/X4-tropic HIV isolates that were present in the patient's circulating viral swarm prior to therapy (17, 27, 55). A second mechanism of HIV resistance to CCR5 antagonists is the use of drug-bound CCR5 as a coreceptor for entry. Resistant viruses that utilize drug-bound CCR5 have been identified following in vitro passaging with multiple CCR5 antagonists (1, 2, 22, 33, 36, 51, 56). Recently, we identified a panel of viral Envs able to use aplaviroc (APL)-bound CCR5 that were isolated from a patient (21, 48). The Envs from this patient were cross resistant to the CCR5 antagonists AD101, TAK779, SCH-C, and maraviroc. Surprisingly, this antiretroviral-naïve patient harbored Envs resistant to aplaviroc prior to the initiation of therapy. In the present study, we have examined viral and host factors that contribute to aplaviroc resistance and examined the consequences of resistance for viral tropism. Aplaviroc resistance determinants were located within the V3 loop of gp120, although additional residues diffusely spread throughout the gp120 and gp41 proteins modulated the magnitude of drug resistance. The resistant virus displayed altered interactions between gp120 and CCR5 such that the virus became critically dependent upon the N′ terminus of drug-bound CCR5. This differential recognition of CCR5 in the presence of aplaviroc was also associated with increased dependence on a higher CCR5 receptor density for efficient virus infection and a tropism shift toward effector memory cells on primary CD4+ T cells.
MATERIALS AND METHODS
Creation of chimeric Envs.
We previously described a panel of Envs isolated from a patient (P5) that were resistant to complete inhibition by aplaviroc and other CCR5 antagonists (48). Two representative Envs from this panel, pre5.2 and post 5.1, were chosen for the present study. Chimeric envs between pre5.2 and JRFL and between pre5.2 and post5.1 were created by overlap PCR. Junctions between envs were constructed at the end of the V1/V2 loop, at the end of the V3 loop, and at the end of gp120. Chimeric viruses were confirmed by sequencing.
Virus infection assays.
Pseudotyped viruses were produced by digestion of patient envs and chimeric constructs with KpnI and XbaI and subcloning into a pCI expression construct containing hepatitis B virus PRE to enable high-level, rev-independent Env expression. The Env-pCI-PRE plasmids were cotransfected into 293T/17 cells with either a luciferase-based reporter HIV core (pNL4.3-Vpr+-Env−-Luc+) or an enhanced green fluorescent protein (EGFP)-based HIV core (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3-deltaE-EGFP [catalog no. 11100] from Haili Zhang, Yan Zhou, and Robert Siliciano). Supernatants were harvested after incubation for 3 days and filtered through a 0.45-μm filter. GFP pseudotypes were concentrated by ultracentrifugation through a 20% sucrose cushion and resuspended in phosphate-buffered saline (PBS). Pseudotyped viruses were normalized for p24 content and were tested for Env incorporation by Western blot analysis and to ensure that Env incorporation did not differ greatly (>20%) between preparations. In all cases, the amount of virus utilized was empirically determined to be within the linear range of the infection assay. At 3 days postinfection, cells were lysed and luciferase activity analyzed on a luminometer. For infection assays in the presence of aplaviroc, cells were maintained in the presence of 10 μM drug from 30 min prior to infection until 3 days postinfection.
Mutant CCR5 receptor assays.
293T cells stably transfected with human CD4 were transfected with the with wild-type or mutant CCR5 genes using Lipofectamine (Invitrogen). The CCR5s transfected included wild-type, Y3A, Q4A, Y10A, D11A, Y14A, Y15A, N24A, Q27A, H88A, Q93A, N98A, S179A, H181A, F182A, P183A, Q186A, Y187A, F189A, W190A, E262A, N267A, N268A, C269A, and N273A CCR5s. Expression of CD4 and CCR5 was quantified by flow cytometry at 24 h posttransfection using anti-CD4 (BD) and anti-CCR5 (CTC5 [R&D Systems] and 2D7 [BD]) antibodies. Cells were infected with luciferase-based pseudoviruses at 24 h posttransfection via spinoculation (450 × g, 2 h), and infection was quantified after 3 days as detailed above.
Affinofile cell assays.
Infection of the HEK293-based CD4/CCR5 dual-inducible cell line (293-Affinofile) with luciferase-based pseudoviruses was performed as previously described (19). Briefly, 96-well plates were seeded with 1.0 × 104 inducible cells 2 days prior to CD4 and CCR5 induction. Cells were induced using 2-fold serial dilutions from 5 to 0.156 ng/ml of minocycline (resulting in six induction conditions for CD4) and from 2 to 0.0156 μM ponasterone A (resulting in eight induction conditions for CCR5). Cells were incubated for 18 h at 37°C, after which induction medium was removed and cells were infected with pseudoviruses normalized for p24 content. Infection was quantified after 3 days as described above.
Flow cytometry.
Stimulation plates were prepared by treating 48-well plates with 10 μl/ml of anti-CD3, 10 μl/ml anti-CD28, and 0.1% bovine serum albumin (BSA) in PBS for 12 h at 4°C. Plates were washed and seeded with CD4+ T cells isolated from leukapheresis (RosetteSep CD4+ T cell kit; Stemcell Technologies) at a concentration of 4 × 106 cells/ml in the presence of 20 U/ml interleukin-2 (IL-2) (Sigma) for 3 days. Cells were then transferred to V-bottom plates in the presence of medium containing IL-2 and infected with 50 ng p24 equivalent of GFP-pseudotyped virus by spinoculation (1,200 g, 2 h). Cells were resuspended, transferred back to flat-bottom plates, and incubated for an additional 3 days at 37°C. Cells were stained with anti-CCR7 (unlabeled IgM; BD) antibody at 37°C for 30 min, followed by staining with anti-mouse IgM (phycoerythrin [PE] conjugated; Invitrogen), anti-CD45RO (PE-TR conjugated; Beckman Coulter), anti-CD3 (Qdot 655 conjugated; Invitrogen), and anti-CD4 (Alexa 700 conjugated; BD) for 4°C for 30 min. The percentage of CD4+ T cells in uninfected controls was analyzed for each donor and was >95% for all samples. Cells were then washed and analyzed on an LSR II flow cytometer equipped with a green laser (BD). Between 2.5 × 105 and 1.0 × 106 events were collected on an LSR II flow cytometer (BD) and analyzed with FlowJo software (TreeStar).
Statistical analysis.
Data analyses were performed using the Prism software package (GraphPad Software) and validated by the University of Pennsylvania CFAR Biostatistics Core. All infection, Affinofile, and mutant CCR5 assays were performed at least three times with three, two, and six replicates per experimental condition, respectively. In the figures, error bars represent the standard errors of the means.
RESULTS
Envelope determinants responsible for CCR5 antagonist resistance.
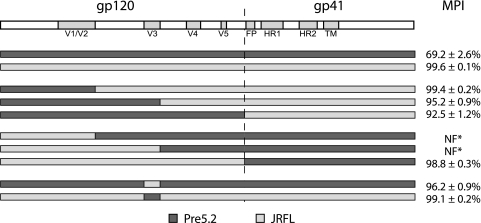
We previously described Env proteins derived from a patient treated with the CCR5 antagonist aplaviroc that exhibited resistance to all CCR5 antagonists tested (48). This patient (P5) harbored resistant Envs both prior to and after treatment with aplaviroc, designated pre5 and post5 clones, respectively. We concluded that these Envs were able to utilize the drug-bound conformation of CCR5 to mediate entry and infection. In addition, none of these Envs were able to use CXCR4. Because all Env clones isolated from this patient were resistant to complete suppression by aplaviroc, we performed a protein BLAST search on the Env clone demonstrating the greatest resistance to aplaviroc (clone pre5.2) and identified JRFL as a closely related, commonly studied HIV strain that is sensitive to inhibition by aplaviroc and other CCR5 antagonists. Since mutations in the V3 loop are commonly associated with resistance to CCR5 antagonists (22, 33, 53, 56), we constructed chimeric viruses in which the V3 loops of pre5.2 and JRFL were exchanged. Pseudotype infection assays were performed on NP2/CD4/CCR5 cells in the presence or absence of 10 μM aplaviroc, a level empirically determined to be 3 log units higher than required to saturate CCR5 receptors on this cell line. Viral infection was quantified by measuring luciferase activity, and maximal percent inhibition (MPI) by aplaviroc was calculated. The MPI by aplaviroc for virions bearing the pre5.2 Env was 64%, and that for JRFL was >99% (Fig. 1). When the V3 loop from JRFL was introduced into the pre5.2 Env, the MPI increased to 96% (P = 0.001). In contrast, introducing the V3 loop from pre5.2 into the JRFL Env did not impart any resistance to drug (MPI = 99%). These results suggest that there are residues within the V3 loop of pre5.2 that are critical for resistance to aplaviroc but that these residues depend on the overall context of Env.
FIG. 1.
The V3 loop of pre5.2 contains residues required for APL resistance. Chimeric constructs between the APL-resistant pre5.2 Env and the sensitive JRFL Env were created. Pseudotyped viruses bearing the parental or chimeric Envs were used to infect NP2/CD4/CCR5 cells in the presence or absence of 10 μM APL, and maximal percent inhibition (MPI) with standard errors of the means (SEM) were calculated. The V3 loop of pre5.2 contains residues required for APL resistance, but these mutations are dependent on the Env context and did not transfer resistance to the JRFL gp160. NF*, viruses that did not infect NP2/CD4/CCR5 cells in the absence of drug (nonfunctional).
To identify determinants outside the V3 loop that contributed to aplaviroc resistance, we constructed a series of chimeric Envs between clone pre5.2 and JRFL and used these to produce HIV-1 pseudoviruses. Introduction of the N′ terminus through V1/V2 of pre5.2 into the JRFL Env did not confer resistance to aplaviroc (MPI = 99%), though transferring the N′ terminus through V3 or all of gp120 from pre5.2 into the JRFL background resulted in small but significant reductions in inhibition by aplaviroc (MPI = 95 and 92%; P = 0.002 and P = 0.001, respectively). The modest reduction in aplaviroc sensitivity in the chimeric Envs containing all of gp120 from pre5.2 suggested that the gp41 region contained determinants important for resistance to aplaviroc. However, the reciprocal chimera containing all of gp120 from JRFL with gp41 from pre5.2 was nearly completely inhibited by aplaviroc (MPI = 99%). The remaining two chimeras, with the N′ terminus through V1/V2 or through V3 from JRFL in the pre5.2 Env background, were not functional in the pseudotype assay and could not be evaluated.
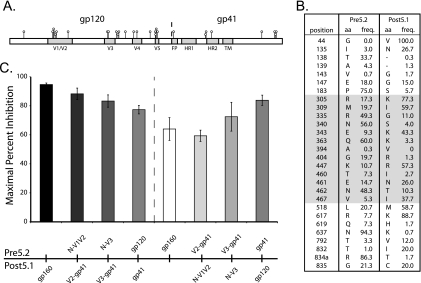
Since several of the pre5.2-JRFL chimeric Envs were nonfunctional and the analysis of the functional clones indicated that determinants might be sensitive to the Env context, we reasoned that construction of chimeras with a more closely related Env might help elucidate the determinants of aplaviroc resistance. While none of the Envs from P5 were completely sensitive to aplaviroc, we identified one Env (post5.1) that exhibited an average MPI of 92%, compared to an MPI of 64% for pre5.2. The post5.1 and pre5.2 Envs differ at 28 amino acids, 20 within gp120 and 8 in gp41 (Fig. 2 A and B), including changes at positions 305 and 309 in the V3 loop and a mutation in the gp41 fusion peptide (numbering is based on the HXB2 strain). None of these changes have previously been linked to CCR5 antagonist resistance. Pre5.2-post5.1 chimeras were constructed using overlap PCR, and pseudoviruses bearing the parental and chimeric Envs were created.
FIG. 2.
Mutations outside the V3 loop can modulate the magnitude of APL resistance. (A) Schematic showing the locations of the amino acid changes between the highly APL-resistant pre5.2 Env and the more sensitive post5.1 Env. (B) Locations of the amino acids residues that differ between pre5.2 and post5.1 and the frequencies of their respective residues in 582 clade B Env sequences from the Los Alamos National Laboratory HIV sequence database. Amino acids (aa) are numbered according to their positions in the HXB2 reference strain. Residues within and between the V3 and V5 regions of gp120 are indicated by the shaded region. (C) Pseudotyped viruses bearing pre5.2, post5.1, or chimeric viruses were used to infect NP2/CD4/CCR5 cells in the presence or absence of 10 μM APL. The V1/V2 loop was not found to contribute to APL resistance, but gp120 residues outside V3 and gp41 residues modulated the extent of drug resistance. Error bars indicate SEM.
The introduction of the N′ terminus through V1/V2, the N′ terminus through V3, and all of gp120 from the highly aplaviroc-resistant pre5.2 Env into the more sensitive post5.1 Env background resulted in progressively higher resistance to aplaviroc (MPI = 88%, 83%, and 77%, respectively) (Fig. 2C), while the reciprocal chimeras had the opposite effect, with a progressive reduction in aplaviroc resistance (MPI = 59%, 72%, and 83%, respectively). These data are consistent with those from the pre5.2-JRFL chimeras in several regards. First, the V1/V2 region from the pre5.2 Env did not contribute significant resistance to aplaviroc in either the JRFL or post5.1 background. Thus, the 13 amino acid changes between the highly resistant pre5.2 and more sensitive post5.1 Envs located within and between the V3 and V5 regions are largely responsible for the role that gp120 plays in modulating aplaviroc resistance (shaded residues in Fig. 2B). Second, gp120 by itself was not sufficient to confer full aplaviroc resistance, suggesting a role for gp41. However, the ability of gp41 to contribute to aplaviroc resistance appears to be dependent on the context of the gp120 domain: when matched with the more diverse JRFL virus, gp41 from pre5.2 did not impart any substantial degree of aplaviroc resistance, whereas when matched with the post5.1 virus, it contributed to a modest but significant increase in resistance (Fig. 2C).
Modeling of mutations between pre5.2 and post5.1 Envs.
To gain insight into how the mutations in the highly aplaviroc-resistant pre5.2 Env might function, the 13 gp120 differences within and between the V3 to V5 regions were modeled onto the structure of JRFL gp120. Only the two mutations in the V3 loop were located in a region thought to be involved in interactions with the CCR5 coreceptor, though our studies indicate that these mutations play a role that is dependent upon Env context. The remaining 11 mutations cluster on the outer face of gp120 in an area close to the region that is thought to interact with gp41. No mutations were present in the inner domain of gp120 or in buried residues in the outer domain of gp120. Since these mutations did not localize to the CD4 binding site or to the bridging sheet or β19 loop, which interact with CCR5, we hypothesized that these mutations might act by altering the fusogenicity of the virus, that is, by increasing the chances of a productive entry event upon engagement of CD4 and CCR5. Therefore, we tested the ability of these Envs to utilize low levels of CD4 and CCR5 for infection in the absence or presence of aplaviroc.
Contribution of CD4 and CCR5 levels on target cells to the magnitude of aplaviroc resistance.
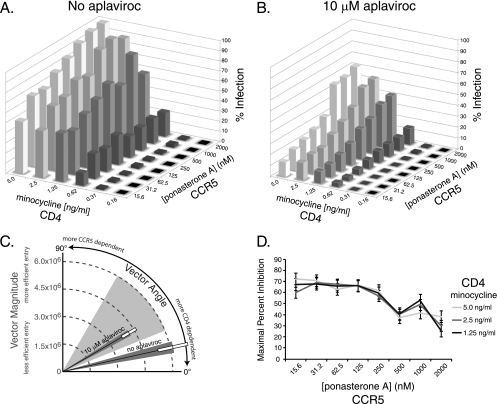
CCR5 levels can influence the magnitude of viral resistance to the coreceptor antagonist vicriviroc (40), and we reasoned that differential expression of CD4 or CCR5 on the cell surface of target cells might affect the efficiency with which Envs from P5 could utilize drug-bound receptor. To test this, we utilized the Affinofile cell line, an HEK293 cell line in which CD4 and CCR5 are under the control of separate, inducible promoters, making it possible to independently vary the surface expression of CD4 and CCR5 over a physiological concentration range (19). The relative dependence of viral Envs on the surface expression levels of CD4 and CCR5 can be mathematically modeled using the VERSA (Viral Entry Receptor Sensitivity Analysis) computational platform (http://versa.biomath.ucla.edu). Using this model, viral infectivity can be described using a single vector: the magnitude reflects the efficiency of entry of the virus, and the angle represents the relative dependence on CD4 or CCR5. Viruses that are completely sensitive to changes in CD4 surface expression but are not affected by varying concentrations of CCR5 have a vector angle of 0°, while viruses that are independent of CD4 concentrations but sensitive to CCR5 levels have a vector angle of 90°.
Infection with pseudotyped virus bearing the pre5.2 Env on the Affinofile cells in the absence of aplaviroc demonstrated that this viral Env was highly sensitive to low concentrations of CD4 on the cell surface. In contrast, pre5.2 was able to utilize very low concentrations of CCR5 on the cell surface efficiently, with robust infection under all conditions (Fig. 3 A). The sensitivity of pre5.2 to CD4 but not CCR5 levels was reflected in the VERSA vector angle of 12 ± 3°. The post5.1 Env in the absence of aplaviroc was slightly more sensitive to lower concentrations of CCR5 and was calculated to have a vector angle of 17 ± 1° (data not shown). These are unusually low vector angles (19) and indicate that these Envs utilize CCR5 very efficiently. In comparison, most primary Envs tested using this system thus far exhibit vector angles of ≥30° (19).
FIG. 3.
The pre5.2 Env becomes increasingly sensitive to CCR5 density in the presence of APL. (A) Infection of pseudotyped viruses bearing the pre5.2 Env on the Affinofile cell line in the absence of drug. Pre5.2 is sensitive to low CD4 levels but is able to use very low CCR5 levels efficiently. Infection is normalized to 100% at the highest CD4 and CCR5 concentration. (B) Infection of pre5.2 pseudotypes on the Affinofile cell line in the presence of 10 μM APL. The pre5.2 Env is still sensitive to low CD4 levels but also displays increasing sensitivity to CCR5 concentrations. Infection levels are proportionate to infection in the absence of drug. (C) Graphical representation of the VERSA vectors of the pre5.2 Env in the presence or absence of drug, showing efficiency of entry and relative dependence on CD4 and CCR5. Dark gray shaded areas represent the SEM of the vector angle; white boxes represent the SEM of the vector magnitude. Vector angles of typical primary HIV-1 isolates are represented by the light gray shaded wedge. (D) Maximal percent inhibition (MPI) of pre5.2 at different CCR5 densities was calculated from the data in panels A and B. APL is more effective at inhibiting viral infection at low CCR5 concentrations. Error bars indicate SEM.
In the presence of 10 μM aplaviroc, pseudoviruses bearing the pre5.2 Env demonstrated a greater sensitivity to the levels of CCR5 in the presence of aplaviroc than was observed in the absence of drug (Fig. 3B and C), resulting in a vector angle of 30 ± 2°. The post5.1 Env in the presence of aplaviroc demonstrated a profile similar to that of the pre5.2 Env, with a vector angle of 30 ± 3° (data not shown). The MPI of pre5.2 by aplaviroc on the Affinofile cells was calculated for all CD4 and CCR5 conditions that supported infection and demonstrated that the MPI decreased with higher levels of CCR5 expression, consistent with previous data (40) (Fig. 3D). These results indicate that the resistance of pre5.2 to aplaviroc can be modulated by CCR5 levels, with decreased inhibition at higher concentrations of coreceptor. Additionally, these Envs are unusually efficient at infecting cells with low levels of CCR5.
Regions of CCR5 required for the ability of pre5.2 to infect in the presence of aplaviroc.
The ability of the pre5.2 and other Env proteins derived from this patient to utilize the drug-bound conformation of CCR5 suggests that these Envs utilize the receptor differently than most other HIV-1 strains. To determine whether pre5.2 utilized the CCR5 receptor differently in the absence or presence of aplaviroc, pseudotype infection assays were performed on 293T cells stably expressing human CD4 and transiently expressing either wild-type CCR5 or one of a panel of CCR5 mutants. Previous studies have indicated that R5-tropic viruses are highly dependent upon both the N′ terminus and the extracellular loops of the CCR5 receptor (10-12, 14, 18, 28), and mutants with alanine substitution mutants in these regions were selected to analyze the interactions between the pre5.2 Env and CCR5 in the absence or presence of aplaviroc. The N′-terminal mutants consisted of Y3A, Q4A, Y10A, D11A, Y14A, Y15A, N24A, and Q27A; the ECL1 mutants were H88A, Q93A, and N98A; the ECL2 mutants were S179A, H181A, F182A, P183A, Q186A, Y187A, F189A, and W190A; and the ECL3 mutants consisted of E262A, N267A, N268A, C269A, and N273A.
On 293T/CD4 cells transiently expressing wild-type CCR5, 10 μM aplaviroc resulted in MPIs of 48.6% ± 5.5% and >99% for pre5.2 and JRFL, respectively (Table 1). The greater resistance of pre5.2 to aplaviroc on this cell line than on the NP2/CD4/CCR5 cells is likely a result of the higher CCR5 expression levels obtained during transient transfections. The N′-terminal tyrosine mutants CCR5 Y3A, Y10A, Y14A, and Y15A were all essential for the resistance phenotype, as pre5.2 was unable to utilize these receptors for entry in the presence of drug (MPI = 93.8, 98.7, 98.2, and 98.1, respectively). Other mutants with mutations within the N′ terminus also inhibited use of drug-bound receptor, but to a lesser extent. Mutations in the ECL regions of CCR5 had more modest effects upon aplaviroc resistance, with MPIs ranging from 35.4 to 87.2%. These data suggest that the pre5.2 Env is critically dependent on residues within the N′ terminus of CCR5, particularly Y3, Y10, Y14, and Y15, for its ability to infect cells in the presence of aplaviroc. In contrast, nearly all changes within the ECL regions of CCR5 did not inhibit infection, indicating that these residues are not essential for the use of drug-bound receptor by the pre5.2 Env. These findings are consistent with recently described CCR5 antagonist-resistant Envs that are dependent on the N′ terminus of CCR5 for entry (5, 25, 36). Similar results were found using maraviroc to block infection, suggesting that pre5.2 relies upon the N′ terminus of CCR5 for entry in the presence of CCR5 antagonists (data not shown).
TABLE 1.
Sensitivity of pre5.2 to inhibition by aplaviroc on mutant CCR5 receptorsa
| CCR5 | MPI (mean ± SEM) | % of wild-type CCR5 resistance (mean ± SEM)b |
|---|---|---|
| Wild type | 48.6 ± 5.5 | 100 |
| N′-terminal mutants | ||
| Y3A | 93.8 ± 1.6 | 11.9 ± 2.5 |
| Q4A | 36.4 ± 4.4 | 125.0 ± 6.3 |
| Y10A | 98.7 ± 0.5 | 2.5 ± 0.9 |
| D11A | 87.3 ± 9.1 | 29.5 ± 22.9 |
| Y14A | 98.2 ± 0.6 | 3.6 ± 1.1 |
| Y15A | 98.1 ± 0.8 | 3.4 ± 1.2 |
| N24A | 60.6 ± 3.3 | 77.3 ± 4.2 |
| Q27A | 54.2 ± 6.0 | 89.1 ± 5.8 |
| ECL1 mutants | ||
| H88A | 67.7 ± 3.2 | 63.2 ± 2.6 |
| Q93A | 54.8 ± 11.2 | 86.1 ± 16.9 |
| N98A | 36.3 ± 8.8 | 124.4 ± 11.6 |
| ECL2 mutants | ||
| S179A | 55.0 ± 1.8 | 90.5 ± 13.6 |
| H181A | 56.8 ± 1.7 | 87.0 ± 13.8 |
| F182A | 68.9 ± 3.4 | 61.3 ± 6.4 |
| P183A | 55.2 ± 3.1 | 89.6 ± 11.9 |
| Q186A | 41.8 ± 6.5 | 113.2 ± 2.6 |
| Y187A | 64.3 ± 5.2 | 71.1 ± 12.6 |
| F189A | 47.3 ± 2.0 | 104.5 ± 9.0 |
| W190A | 35.4 ± 7.6 | 125.7 ± 6.1 |
| ECL3 mutants | ||
| E262A | 54.8 ± 6.5 | 87.6 ± 5.7 |
| N267A | 57.7 ± 9.1 | 82.0 ± 12.7 |
| N268A | 39.0 ± 8.7 | 125.6 ± 33.3 |
| C269A | 87.2 ± 8.2 | 29.4 ± 21.1 |
| N273A | 55.5 ± 1.1 | 89.3 ± 12.7 |
Pseudotyped viruses bearing the pre5.2 Env were used to infect 293 cells expressing CD4 and wild-type CCR5 or one of a panel of CCR5 mutants.
The percentage of wild-type CCR5 resistance is a calculated metric allowing for comparison between mutant receptors: 100% of wild-type CCR5 resistance indicates that a mutant has no effect on drug resistance, while 0% of wild-type CCR5 resistance indicates that a mutant completely abrogates resistance. Lower numbers indicate CCR5 residues essential for the use of drug-bound receptor.
Aplaviroc induces a tropism shift of pseudoviruses bearing the pre5.2 Env, resulting in preferential protection of central memory CD4+ T cells.
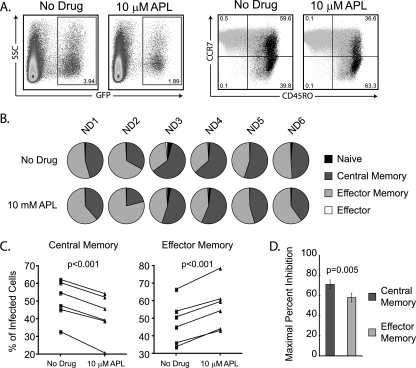
The studies with the CCR5 mutants and Affinofile cells demonstrated that in the presence of aplaviroc, pre5.2 is critically dependent on the N′ terminal tyrosines Y3, Y10, Y14, and Y15 and is more sensitive to CCR5 density. To determine whether this differential usage of CCR5 in the presence of drug translated into differences in the tropism of the virus, we developed a multicolor fluorescence-activated cell sorter (FACS)-based assay for detection of virally infected cells in isolated primary CD4+ T cell subsets. Briefly, peripheral blood mononuclear cells were obtained from six normal donors, and CD4+ T cells were purified by rosette-bead depletion. Cells were stimulated with IL-2, anti-CD3, and anti-CD28 for 3 days and then infected with pre5.2 Env-pseudotyped viruses carrying an EGFP reporter gene in the absence or presence of 10 μM aplaviroc. Following 3 days of incubation, cells were stained for the surface markers CD3, CD4, CD45RO, and CCR7. Subsets of CD4+ T cells were defined using the combination of CCR7 and CD45RO staining: naïve (CCR7+ CD45RO−), central memory (CCR7+ CD45RO+), and effector memory (CCR7− CD45RO+) (45) (Fig. 4 A). Effector cells (CCR7− CD45RO−) (3) accounted for <1% of the CD4+ T cell population. CCR5 expression on each subset was measured independently of viral infection, as CCR5 staining was strongly diminished in the presence of virus, presumably due to either virally induced downregulation or masking of the CCR5 receptor. CCR5 expression levels were highest on effector memory cells, intermediate on central memory cells, and lowest on naïve cells, consistent with previously published results (7, 16, 29, 34). CD4 expression levels were similar between all subsets. HIV-infected cells were defined as CD3+ GFP+ cells, since the pseudotyped viruses downregulated CD4 upon infection and the percentage of CD4+ T cells under uninfected conditions was >95% for all donors. Using a conservative CD3+ GFP+ gate, the background signal from this assay was reproducibly less than 0.01%.
FIG. 4.
APL preferentially protects central memory T cells from infection by pre5.2. (A) Representative flow cytometry data from infection of primary human CD4+ T cells with EGFP viruses pseudotyped with the pre5.2 Env in the absence or presence of 10 μM APL. Virally infected cells were detected by expression of EGFP at 3 days postinfection. The CD4+ T cell subsets infected by virus were determined by staining for CCR7 and CD45RO antigens, which define naïve (CCR7+ CD45RO−), central memory (TCM) (CCR7+ CD45RO+), effector memory (TEM) (CCR7− CD45RO−), and effector (TEMRA) (CCR7− CD45RO−) populations. Infected cells (black) are primarily in the TCM and TEM populations. (B) Pie charts showing the percentage of infected cells within CD4+ T cell subsets in six normal donors in the absence or presence of APL. (C) Line graph representation of the data from panel B, demonstrating that in all patients, APL decreased the percentage of infected cells with the TCM phenotype and increased the percentage of infected cells with the TEM phenotype. (D) Maximal percent inhibition (MPI) on central memory and effector memory T cell subsets. APL inhibited infection of both subsets by pre5.2 but was significantly more effective at preventing infection of central memory cells than at preventing infection of effector memory cells. This preferential protection of TCM cells results in a tropism shift toward TEM cells in the presence of APL. Error bars indicate SEM.
The presence of aplaviroc in the absence of virus infection resulted in a modest, but significant, decrease in the percentage of total CD4+ T cells with the central memory phenotype (TCM) (36.4% ± 3.1% without drug versus 34.7% ± 2.9% with aplaviroc; P = 0.01) (Fig. 4B). Concurrently, the percentage of CD4+ T cells with the effector memory phenotype (TEM) increased in frequency, but this did not reach statistical significance (16.1% ± 1.9% without drug versus 19.7% ± 2.0% with aplaviroc; P = 0.09). These relatively modest alterations in the total CD4+ T cell subsets in the presence of aplaviroc may be due to differential survival of TEM cells, as it has been reported that chemokine antagonists can protect cells from apoptosis (6).
In the absence of aplaviroc, 2.3% ± 0.5% of primary CD4+ T cells were infected by GFP reporter-pseudotyped viruses bearing the pre5.2 Env. Previous experiments have demonstrated that the pre5.2 Env exclusively utilizes the CCR5 receptor for entry and does not infect primary CD4+ T cells from Δ32ccr5-homozygous patients (48). Consistent with a dependence on CCR5 for entry, treatment of cells with 62.5 μM of the CXCR4 antagonist AMD3100 did not significantly decrease infection of primary CD4+ T cells (2.1% ± 0.5%; P = 0.48). Of the infected cells, 50.4% ± 4.5% were of the TCM phenotype, while 47.6% ± 5.0% were of the TEM phenotype (Fig. 4B). In the presence of 10 μM aplaviroc, the percentage of virally infected cells was significantly reduced (to 0.92% ± 0.27%; P = 0.003), resulting in an average MPI for the six donors of 62.1% ± 5.5% (range, 47.8% to 80.7%). However, aplaviroc treatment resulted in a strong tropism shift, with a decrease in the percentage of infected TCM cells (50.4% to 41.8%) and a concomitant increase in the TEM phenotype (47.6% to 56.6%). Aplaviroc was significantly more effective at preventing infection of central memory cells than of effector memory cells, with an average MPI for TCM of 70.5% versus an average MPI for TEM of 57.5%. The differential ability of aplaviroc to protect TCM cells from virus infection relative to TEM cells, which was statistically significant, is consistent with our Affinofile assay data and with results by Pugach and colleagues that have shown that the magnitude of resistance to CCR5 antagonists can be greater on cells with higher expression levels of CCR5 (40). The results reported here show that CCR5 antagonist-resistant Envs that show increasing resistance with higher CCR5 levels can exhibit tropism changes on primary CD4+ T cells in the presence of drug, resulting in preferential protection of the TCM subset.
DISCUSSION
Binding of CD4 to HIV-1 Env induces structural alterations in the gp120 subunit that enable it to interact with at least two extracellular domains of CCR5 (9, 24). The amino-terminal domain of CCR5 interacts with the base of the V3 loop and the bridging sheet region of gp120, with several sulfated tyrosine residues in CCR5 playing important roles (11, 14, 18, 20). The more distal regions of the V3 loop and bridging sheet are thought to interact with the ECLs of CCR5, with ECL2 representing a key determinant (10, 12, 20, 28). Given the sequence variability inherent in the gp120 subunit, it is perhaps not surprising that there is some degree of plasticity in these interactions, as evidenced by the fact that mutations introduced into CCR5 can have markedly different effects on virus infection depending on the virus strain being examined (20, 44). Thus, there is some variability in the relative importance of the CCR5 N′-terminal domain and ECL2 in supporting infection by different HIV-1 strains (41). This underlying plasticity can lead to rare, unexpected outcomes: we identified a patient whose virus exhibited baseline resistance to all CCR5 antagonists tested, which likely contributed to this individual's subsequent virologic failure on an antiviral regimen that included aplaviroc (21, 48). This finding poses two central questions: by what mechanism(s) does resistance to CCR5 antagonists occur, and what are the implications of drug resistance for viral tropism? Addressing the mechanism of resistance can provide basic information on Env-coreceptor interactions and might make it possible to develop tests that can more effectively predict whether a patient will benefit from CCR5 antagonist therapy. Addressing the implications of resistance should make it possible to determine if virologic failure in the face of CCR5 antagonist therapy will affect viral tropism and disease progression in any meaningful way.
Passaging HIV-1 in the presence of increasing concentrations of a CCR5 antagonist in vitro can result in the development of mutations in Env that enable it to recognize the drug-bound conformation of CCR5 (22, 33, 51, 56). A similar resistance pathway can occur in vivo or, as we have shown, can even occur in the absence of drug selection (48, 53). This resistance pathway often, but not always, results in a virus that is resistant to multiple CCR5 antagonists (22, 33, 39, 53, 56). The CCR5 antagonists that have been used clinically all seem to bind to a hydrophobic region in the transmembrane helices of CCR5 and induce conformational alterations in the extracellular domains of the coreceptor that inhibit recognition by gp120 (13, 31, 32, 46, 52). These conformational alterations are similar though not identical as judged by the ability of these compounds to affect the binding of various conformation-dependent monoclonal antibodies to CCR5. Thus, resistance mutations in Env must enable it to recognize the specific conformation of CCR5 that is induced by the specific drug being used or enable it to utilize regions of CCR5 that are not perturbed by drug binding.
Viral determinants critical for CCR5 antagonist resistance are often localized to the V3 loop, consistent with the role of this domain in mediating coreceptor interactions (22, 33, 53, 56). However, while V3 loop mutations appear to be a common though not universal theme underlying resistance to CCR5 antagonists, the specific changes can vary and are often context dependent in that mutations important for CCR5 antagonist resistance often fail to impart complete resistance when introduced into a heterologous background (22, 37). The viruses studied here clearly fall into this category: several mutations in the V3 region were critical for drug resistance, but these did not impart resistance to a heterologous Env protein. In addition, changes outside the V3 region were important for both the presence and the magnitude of drug resistance. Other groups have shown that changes in the C4 region of gp120 and the fusion peptide of gp41 can also play important roles in CCR5 antagonist resistance (1, 36, 37). Precisely how this occurs is not known. Conceivably, changes in Env outside the receptor binding domains can influence the conformation of these domains indirectly or could alter the response of Env to receptor binding, perhaps making induction of the conformational changes needed for membrane fusion occur efficiently even if binding to drug-bound receptor is relatively inefficient. Indeed, nearly all drug-resistant Envs described to date utilize the drug-bound form of CCR5 less efficiently than the drug-free conformation. As a result, the MPI is greatest when CCR5 expression levels are low and tends to fall as CCR5 expression levels increase.
Are there any features of the Env proteins derived from this patient that might predict or at least predispose these viruses to exhibit baseline resistance to CCR5 inhibitors? The development of the Affinofile system by Lee and colleagues provides a way to accurately assess the ability of HIV-1 strains to mediate membrane fusion and virus infection under a variety of CD4 and CCR5 expression levels. Compared to a large panel of primary Env proteins (19; B. Lee, personal communication), the aplaviroc (APL)-resistant Envs studied here were unusually efficient at mediating infection of cells when CCR5 expression levels were low. These viruses were more sensitive to variation in CD4 expression levels, but in vivo this is likely to be less important: CD4 expression is relatively high across different T cell subsets, while CCR5 expression is both lower and more variable. Since the use of drug-bound CCR5 typically occurs with reduced efficiency, Envs that can readily utilize low levels of CCR5 for infection might tolerate mutations associated with drug resistance more readily, making the evolution of drug resistance more efficient.
Further studies with CCR5 antagonist-resistant viruses will clarify the mechanisms by which Env can engage drug-bound CCR5. Perhaps a more interesting question concerns the implications of drug resistance for viral tropism. Since Env is responsible for mediating virus entry into cells, mutations that arise in response to selective pressures imposed by entry inhibitors could potentially alter not only the way in which Env binds to CD4 and coreceptor but also how efficiently membrane fusion-inducing conformational changes are triggered by receptor binding and, as a result, the ability of virus to infect different host cells. It is evident that the mere presence of CD4 and an appropriate coreceptor does not necessarily mean that all viruses capable of using those receptors can enter a given cell; host factors that likely include receptor expression levels and perhaps coreceptor conformation can influence tropism in ways that are not fully understood. We developed a multiparametric FACS-based assay to monitor infection of TCM, TEM, naïve, and effector T cells by virus in the presence or absence of aplaviroc. While each of these subsets expresses similar levels of CD4, their CCR5 expression profiles vary greatly, with TEM cells expressing higher levels of CCR5 than TCM cells and with naïve T cells expressing very low CCR5 levels (7, 16, 29, 34). The viruses studied here infected TCM and TEM cells with similar efficiencies. However, in the presence of APL, TCM CD4+ T cells were preferentially protected compared with TEM CD4+ T cells. This result is consistent with our findings that while the viruses studied here used CCR5 remarkably well, utilization of CCR5 was less efficient in the presence of APL. As TCM cells have lower levels of CCR5 than TEM cells, it follows that in the presence of APL, virus infection of TCM cells will be affected to a greater extent.
The relative tropism shift seen in the presence of APL may have implications for disease progression based on recent studies from simian immunodeficiency virus (SIV) models of HIV infection. A vaccination study of rhesus macaques using a DNA prime/recombinant adenovirus boost regimen resulted in prolonged survival compared with control animals, and survival was associated with preservation of the TCM CD4+ subset (30). More recently, the proliferation of TCM cells has been demonstrated to be critical to the ability to maintain TEM CD4+ cells at extralymphatic sites such as the intestine and lung, where they act as an initial defense against opportunistic pathogens (38). Loss of the TCM subset correlated with a subsequent loss of TEM cells and progression to AIDS. Moreover, among animals with rapid progression to AIDS, infection of the TCM subset was more than three times higher than in normal progressors, indicating that direct virus-mediated killing of TCM cells likely contributes to the decline in the TCM population (38). Although speculative, it is possible that continued treatment with a CCR5 antagonist, even in the presence of resistant viruses, may protect the TCM subset of CD4+ T cells and result in improved T cell homeostasis and immune function. Further analysis of patient populations will be required to evaluate whether there is indeed a clinical benefit for leaving patients on CCR5 antagonists despite the presence of resistant viruses.
Acknowledgments
This work was supported by NIH grants R01 AI40880 and F32 AI077370 and by the University of Pennsylvania Center for AIDS Research.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Anastassopoulou, C. G., T. J. Ketas, P. J. Klasse, and J. P. Moore. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, M., H. Miyake, X. Wang, M. Okamoto, and K. Takashima. 2007. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob. Agents Chemother. 51:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balla-Jhagjhoorsingh, S. S., G. Koopman, P. Mooij, W. Koornstra, S. McCormack, J. Weber, G. Pantaleo, and J. L. Heeney. 2004. Long-term persistence of HIV-1 vaccine-induced CD4+CD45RA-CD62L-CCR7- memory T-helper cells. AIDS 18:837-848. [DOI] [PubMed] [Google Scholar]
- 4.Basmaciogullari, S., G. J. Babcock, D. Van Ryk, W. Wojtowicz, and J. Sodroski. 2002. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J. Virol. 76:10791-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berro, R., R. W. Sanders, M. Lu, P. J. Klasse, and J. P. Moore. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco, J., J. Barretina, G. Henson, G. Bridger, E. De Clercq, B. Clotet, and J. A. Este. 2000. The CXCR4 antagonist AMD3100 efficiently inhibits cell-surface-expressed human immunodeficiency virus type 1 envelope-induced apoptosis. Antimicrob. Agents Chemother. 44:51-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 9.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 10.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accavitti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragic, T., A. Trkola, D. A. Thompson, E. G. Cormier, F. A. Kajumo, E. Maxwell, S. W. Lin, W. Ying, S. O. Smith, T. P. Sakmar, and J. P. Moore. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U. S. A. 97:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 16.Groot, F., T. M. van Capel, J. Schuitemaker, B. Berkhout, and E. C. de Jong. 2006. Differential susceptibility of naive, central memory and effector memory T cells to dendritic cell-mediated HIV-1 transmission. Retrovirology 3:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulick, R. M., Z. Su, C. Flexner, M. D. Hughes, P. R. Skolnik, T. J. Wilkin, R. Gross, A. Krambrink, E. Coakley, W. L. Greaves, A. Zolopa, R. Reichman, C. Godfrey, M. Hirsch, and D. R. Kuritzkes. 2007. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-infected, treatment-experienced patients: AIDS clinical trials group 5211. J. Infect. Dis. 196:304-312. [DOI] [PubMed] [Google Scholar]
- 18.Huang, C. C., S. N. Lam, P. Acharya, M. Tang, S. H. Xiang, S. S. Hussan, R. L. Stanfield, J. Robinson, J. Sodroski, I. A. Wilson, R. Wyatt, C. A. Bewley, and P. D. Kwong. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston, S. H., M. A. Lobritz, S. Nguyen, K. Lassen, S. Delair, F. Posta, Y. J. Bryson, E. J. Arts, T. Chou, and B. Lee. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:11016-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajumo, F., D. A. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 21.Kitrinos, K. M., H. Amrine-Madsen, D. M. Irlbeck, J. M. Word, and J. F. Demarest. 2009. Virologic failure in therapy-naive subjects on aplaviroc plus lopinavir-ritonavir: detection of aplaviroc resistance requires clonal analysis of envelope. Antimicrob. Agents Chemother. 53:1124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuritzkes, D. R. 2009. HIV-1 entry inhibitors: an overview. Curr. Opin. HIV AIDS 4:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBranche, C. C., G. Galasso, J. P. Moore, D. P. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antiviral Res. 50:95-115. [DOI] [PubMed] [Google Scholar]
- 27.Lalezari, J., M. Thompson, P. Kumar, P. Piliero, R. Davey, K. Patterson, A. Shachoy-Clark, K. Adkison, J. Demarest, Y. Lou, M. Berrey, and S. Piscitelli. 2005. Antiviral activity and safety of 873140, a novel CCR5 antagonist, during short-term monotherapy in HIV-infected adults. AIDS 19:1443-1448. [DOI] [PubMed] [Google Scholar]
- 28.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 29.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letvin, N. L., J. R. Mascola, Y. Sun, D. A. Gorgone, A. P. Buzby, L. Xu, Z. Y. Yang, B. Chakrabarti, S. S. Rao, J. E. Schmitz, D. C. Montefiori, B. R. Barker, F. L. Bookstein, and G. J. Nabel. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda, K., D. Das, H. Ogata-Aoki, H. Nakata, T. Miyakawa, Y. Tojo, R. Norman, Y. Takaoka, J. Ding, G. F. Arnold, E. Arnold, and H. Mitsuya. 2006. Structural and molecular interactions of CCR5 inhibitors with CCR5. J. Biol. Chem. 281:12688-12698. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, K., D. Das, P. D. Yin, K. Tsuchiya, H. Ogata-Aoki, H. Nakata, R. B. Norman, L. A. Hackney, Y. Takaoka, and H. Mitsuya. 2008. Involvement of the second extracellular loop and transmembrane residues of CCR5 in inhibitor binding and HIV-1 fusion: insights into the mechanism of allosteric inhibition. J. Mol. Biol. 381:956-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182-199. [DOI] [PubMed] [Google Scholar]
- 34.Mo, H., S. Monard, H. Pollack, J. Ip, G. Rochford, L. Wu, J. Hoxie, W. Borkowsky, D. D. Ho, and J. P. Moore. 1998. Expression patterns of the HIV type 1 coreceptors CCR5 and CXCR4 on CD4+ T cells and monocytes from cord and adult blood. AIDS Res. Hum. Retroviruses 14:607-617. [DOI] [PubMed] [Google Scholar]
- 35.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. U. S. A. 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogert, R. A., L. Ba, Y. Hou, C. Buontempo, P. Qiu, J. Duca, N. Murgolo, P. Buontempo, R. Ralston, and J. A. Howe. 2009. Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc. J. Virol. 83:12151-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogert, R. A., L. Wojcik, C. Buontempo, L. Ba, P. Buontempo, R. Ralston, J. Strizki, and J. A. Howe. 2008. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology 373:387-399. [DOI] [PubMed] [Google Scholar]
- 38.Okoye, A., M. Meier-Schellersheim, J. M. Brenchley, S. I. Hagen, J. M. Walker, M. Rohankhedkar, R. Lum, J. B. Edgar, S. L. Planer, A. Legasse, A. W. Sylwester, M. Piatak, Jr., J. D. Lifson, V. C. Maino, D. L. Sodora, D. C. Douek, M. K. Axthelm, Z. Grossman, and L. J. Picker. 2007. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J. Exp. Med. 204:2171-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugach, P., N. Ray, P. J. Klasse, T. J. Ketas, E. Michael, R. W. Doms, B. Lee, and J. P. Moore. 2009. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor-CCR5 complex at low cell surface CCR5 densities. Virology 387:296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabut, G. E., J. A. Konner, F. Kajumo, J. P. Moore, and T. Dragic. 1998. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J. Virol. 72:3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 16:741-749. [DOI] [PubMed] [Google Scholar]
- 43.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 44.Rucker, J., M. Samson, B. J. Doranz, F. Libert, J. F. Berson, Y. Yi, R. J. Smyth, R. G. Collman, C. C. Broder, G. Vassart, R. W. Doms, and M. Parmentier. 1996. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437-446. [DOI] [PubMed] [Google Scholar]
- 45.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 46.Seibert, C., W. Ying, S. Gavrilov, F. Tsamis, S. E. Kuhmann, A. Palani, J. R. Tagat, J. W. Clader, S. W. McCombie, B. M. Baroudy, S. O. Smith, T. Dragic, J. P. Moore, and T. P. Sakmar. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41-54. [DOI] [PubMed] [Google Scholar]
- 47.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tilton, J. C., H. Amrine-Madsen, J. L. Miamidian, K. M. Kitrinos, J. Pfaff, J. F. Demarest, N. Ray, J. L. Jeffrey, C. C. Labranche, and R. W. Doms. 2010. HIV type 1 from a patient with baseline resistance to CCR5 antagonists uses drug-bound receptor for entry. AIDS Res. Hum. Retroviruses. 26:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilton, J. C., and R. W. Doms. 2010. Entry inhibitors in the treatment of HIV-1 infection. Antiviral Res. 85:91-100. [DOI] [PubMed] [Google Scholar]
- 50.Trkola, A., T. Dragic, J. Arthos, J. M. Binlay, W. C. Olson, G. P. Allaway, C. Cheng-Meyer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 51.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. U. S. A. 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsamis, F., S. Gavrilov, F. Kajumo, C. Seibert, S. Kuhmann, T. Ketas, A. Trkola, A. Palani, J. W. Clader, J. R. Tagat, S. McCombie, B. Baroudy, J. P. Moore, T. P. Sakmar, and T. Dragic. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsibris, A. M., M. Sagar, R. M. Gulick, Z. Su, M. Hughes, W. Greaves, M. Subramanian, C. Flexner, F. Giguel, K. E. Leopold, E. Coakley, and D. R. Kuritzkes. 2008. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 82:8210-8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson, C., S. Jenkinson, W. Kazmierski, and T. Kenakin. 2005. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 67:1268-1282. [DOI] [PubMed] [Google Scholar]
- 55.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]