Abstract
Although the viral Rev protein is necessary for HIV replication, its main function in the viral replication cycle has been controversial. Reinvestigating the effect of Rev on the HIV-1 RNA distribution in various cell lines and primary cells revealed that Rev enhanced cytoplasmic levels of the unspliced HIV-1 RNA, mostly 3- to 12-fold, while encapsidation of the RNA and viral infectivity could be stimulated >1,000-fold. Although this clearly questions the general notion that the nuclear export of viral RNAs is the major function of Rev, mechanistically encapsidation seems to be linked to nuclear export, since the tethering of the nuclear export factor TAP to the HIV-1 RNA also enhanced encapsidation. Interference with the formation of an inhibitory ribonucleoprotein complex in the nucleus could lead to enhanced accessibility of the cytoplasmic HIV-1 RNA for translation and encapsidation. This might explain why Rev and tethered TAP exert the same pattern of pleiotropic effects.
In contrast to simple retroviruses, HIV-1, a lentivirus, utilizes several trans-acting regulatory proteins which fulfill important functions throughout the different phases of the lentiviral replication cycle (recently reviewed in references 16, 30, and 34). One of those regulators is the 16-kDa HIV-1 Rev protein, which is required for the expression of the structural proteins during the late phase of the HIV-1 replication cycle. This effect of Rev has been attributed to its nuclear export activity. After initial transcription of the integrated proviral genome, “early” transcripts are subjected to the cellular splicing machinery. Alternative splicing events caused by the complex genome organization of HIV-1 lead to expression of Rev from a multiply spliced mRNA. Once Rev is translated in the cytoplasm, it is imported into the nucleus, where it binds to a viral RNA element, the Rev response element (RRE), which is present on singly spliced and unspliced transcripts. Rev seems to circumvent further splicing and leads to an increase of the cytoplasmic levels of unspliced and singly spliced viral transcripts by tethering these transcripts to the Crm1 export pathway (13, 28; reviewed in reference 36). The unspliced and singly spliced transcripts serve as templates for translation, and the unspliced RNA is also encapsidated into assembling virus particles.
The magnitude by which Rev enhances lentiviral RNA levels in the cytoplasm has been controversial. Under some experimental conditions, unspliced viral RNA levels in the cytoplasm were detectable only in the presence of Rev, while others observed just a 4-fold enhancement of these cytoplasmic RNA levels by Rev (2, 8, 9, 12, 26, 36, 43, 45). Since Rev was found to stimulate protein levels encoded by the Rev-dependent RNAs to a much larger extent than the cytoplasmic levels of these RNAs, Rev also seems to stimulate translation (6, 25, 35; recently reviewed in reference 17). Consistently, Rev was furthermore shown to enhance the association of the Rev-dependent viral RNA with polysomes (9).
In trying to develop RRE-deficient lentiviral vectors, we previously observed that the deletion of the RRE led to a striking loss of infectivity, despite nearly unchanged levels of unspliced vector RNA in the cytoplasm of the producer cells (27). Further analyses revealed that Rev only moderately enhanced unspliced vector RNA levels in the cytoplasm but increased the encapsidated RNA levels by 2 to 3 orders of magnitude (3). This effect was not due to Rev-mediated stimulation of particle production, since a Rev-independent, codon-optimized Gag-Pol expression plasmid lacking the RRE provided a constant excess of Gag (44).
The lentiviral vector RNA encapsidated in the experiments described above contained large deletions of gag-pol and env also comprising cis-acting regulatory sequences (CRS) and inhibitory sequences (INS) (8, 37-39). Since these sequences have been shown to modulate Rev dependence, the minor effects of Rev on cytoplasmic lentiviral vector RNA levels could simply be due to the absence of such regulatory sequences. Therefore, we have since investigated the influence of Rev on almost-full-length HIV-1 unspliced transcript levels in the cytoplasm and on the encapsidation process. Additionally, we have extended our analyses of the function of Rev to infected T-cell lines and primary peripheral blood mononuclear cells (PBMCs).
MATERIALS AND METHODS
Plasmids.
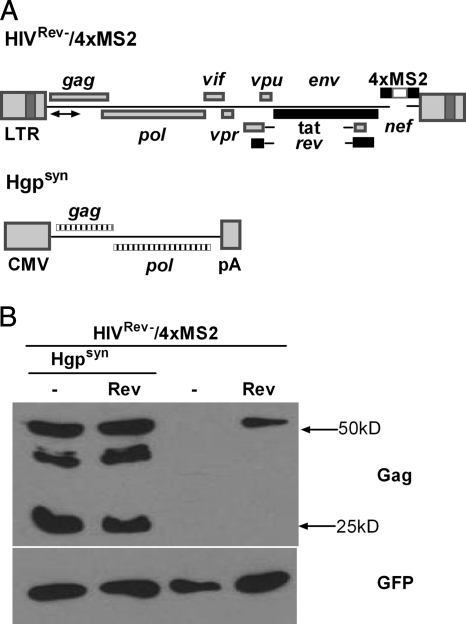
For the cloning of the proviral construct HIVRev−/4xMS2, the env open reading frame (ORF) of pNL4-3Rev−/4xMS2 (47) was inactivated by the deletion of bases 7251 to 7254 (according to the sequence under GenBank accession number AF324493). The rev ORF of the HIVRev−/4xMS2 construct was reactivated by site-directed mutagenesis, leading to HIVRev+/4xMS2. The expression plasmids encoding vesicular stomatitis virus glycoprotein (VSV-G) (pHIT/G), HIV-1 Tat (pcTat), HIV-1 Rev (pcRev), a fusion of the coat protein of phage MS2 and the human nuclear shuttling factor TAP (pMS2-hTAP), and codon-optimized Gag-Pol of HIV-1 (Hgpsyn) were kindly provided by M. Malim, J. Hauber, B. Cullen, and R. Wagner and have been described previously (7, 14, 29, 44).
Cell culture.
Cell cultures of HEK 293T and TZM-bl cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) with 10% fetal calf serum (FCS) and 20 μg/ml ciprofloxacin. Human T-cell lines CEM-SS, HUT78, and Jurkat were cultured in RPMI 1640 (Gibco) with 10% FCS and 20 μg/ml ciprofloxacin. PBMCs were isolated from the blood buffy coats of three healthy donors by Ficoll 400 centrifugation (400 × g, 30 min, 20°C). PBMCs were cultured at 1 × 106 cells/ml in RPMI 1640 with 10% FCS and 20 μg/ml ciprofloxacin and were activated by the addition of 5 μg/ml phytohemagglutinin (PHA) and 100 IU/ml interleukin-2 (IL-2) for 72 h.
Transfections.
Transfections for the production of viral particles were done using the calcium phosphate coprecipitation method. Briefly, 1.5 × 106 293T cells were grown in 25-cm2 flasks for 24 h and were transfected with 2 μg pcTat or 2 μg Hgpsyn, with or without 2 μg pcRev or pMS2-hTAP, respectively. Unless stated otherwise, 500 ng of proviral construct per transfection was used.
A total of 0.1 μg pCMV-GLuc-1 (Targeting Systems, El Cajon, CA) expressing Gaussia luciferase was added to each transfection mix to control for transfection efficiency. The medium was changed after 8 h to remove excessive plasmid DNA.
Transfections for Western blots were done using either calcium phosphate or polyethyleneimine (PEI) as described previously (1). A PEI/DNA (wt/wt) ratio of 1.5 was used. The total amount of DNA transfected was adjusted to 10 μg per transfection by the addition of calf thymus carrier DNA. Transfections were normalized by using Gaussia luciferase as an indicator of transfection efficiency and cell viability. The luciferase activities were used to adjust the RNA copy numbers from the reverse transcription-quantitative PCRs (RT-qPCRs) to determine transfection efficiency.
RNA isolation.
For RNA isolation, cells were detached and washed in 1 ml phosphate-buffered saline (PBS). The pellet was resuspended in 175 μl buffer RLN (50 mM Tris-HCl, pH 8.0; 140 mM NaCl; 1.5 mM MgCl2; 0.5% Nonidet P-40 substitute; 1,000 U/ml RNase inhibitor [Qiagen, Hilden, Germany]; 1 mM dithiothreitol [DTT]) and incubated for 5 min on ice. Debris and nuclei were pelleted (300 × g, 2 min, 4°C), and the cytoplasmic fraction was transferred to 600 μl RLT buffer (Qiagen). For RNA isolation from cell nuclei, the nuclear pellet was washed in 500 μl PBS, again pelleted for 3 min at 300 × g, and then resuspended in 600 μl RLT buffer. The pellet was homogenized using Qiashredder columns (Qiagen) at 13,000 × g for 30 s. The flowthrough was transferred twice to the same column.
RNA isolation was done using an RNeasy Mini kit (Qiagen) according to the manufacturer's instructions, followed by DNase digestion using a Turbo DNA-free kit (Ambion, Austin, TX). The amount of total RNA extracted from the cytoplasmic and nuclear fractions was determined by use of a Quant-iT RiboGreen RNA quantitation kit (Invitrogen, Karlsruhe, Germany).
For RNA isolation from virus particles, supernatants of transfected cells were loaded on top of a 30% sucrose cushion and ultracentrifuged at 150,000 × g for 2 h at 4°C. Virus particles pelleted from 5 ml of cell culture supernatant were resuspended in 150 μl PBS. RNAs were isolated from the resuspended viral particles by use of a QIAamp Viral RNA Mini kit (Qiagen) and were eluted in 45 μl, followed by DNase digestion using a Turbo DNA-free kit.
RT-qPCRs.
HIV-1 unspliced RNA levels were determined using a QuantiTect Probe RT-PCR kit (Qiagen). Primer sequences homologous to a region of gag were taken from the Amplicor HIV-1 Monitor test (31) and are specific for the unspliced HIV-1 RNA. The primers were pSK145 (AGT GGG GGG ACA TCA AGC AGC CAT GCA AAT) and pSKCC1B (TAC TAG TAG TTC CTG CTA TGT CAC TTC C). Serial dilutions of an in vitro transcript were prepared as an RNA standard with known copy numbers. The sensitivity of the assay was below 100 RNA copies/PCR, and the interassay variability was 3%. Cross-reaction of the PCR plasmid with the codon-optimized Hgpsyn plasmid was not detected even after addition of 108 DNA copies of Hgpsyn. This is consistent with the 7 or 8 mismatches between the primer sequences and their target sequence in the codon-optimized gene. HIV-1 unspliced RNA copy numbers in 500 ng cytoplasmic or nuclear RNA or 1 μl of the RNA extracted from the viral particles (corresponding to 6 to 26 ng p24) were determined by direct comparison to serial dilutions of the RNA standard. Elimination of transfected plasmid DNA was routinely confirmed by simultaneously analyzing aliquots of one or two representative samples without adding the reverse transcriptase. Since nonencapsidated extracellular RNA is rather unstable, we did not perform any additional RNase treatment.
Subcellular fractionation was also controlled with a QuantiTect Probe RT-PCR kit (Qiagen), using primers preGAP-DHE6s (CCA CCA ACT GCT TAG CAC C) and preGAP-DHE6a (CTC CCC ACC TTG AAA GGA AAT) (4), homologous to the exon 6-intron 6 junction and to intron 6 of the unprocessed pre-mRNA of the glyceraldehyde-3-phosphate dehydrogenase gene (pre-GAPDH RNA), respectively. Five hundred nanograms of extracted nuclear and cytoplasmic RNAs and serial dilutions of the nuclear RNA were subjected to real-time RT-PCR. This allowed the calculation of the percentage of pre-GAPDH RNA in the cytoplasm relative to the nuclear pre-GAPDH RNA level.
Protein analyses.
For analysis of the purity of cytoplasmic and nuclear fractions, two flasks of 293T cells were transfected as described above. One-half of the pooled cells was used to prepare total cell lysates by the addition of 500 μl of stringent BLP lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 40 mM NaF, 5 mM EDTA, 5 mM EGTA, 1% [vol/vol] Nonidet P-40, 0.1% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS). The other half was fractionated as described for the RNA analyses. Fractions were brought to a final volume of 500 μl, using BLP buffer.
Equal amounts of total protein were subjected to SDS-PAGE prior to staining of the Western blots with antibodies to histone H2B (Epitomics, Burlingame, CA), α-tubulin (Rockland, Gilbertsville, PA), and green fluorescent protein (GFP) (Santa Cruz Biotechnology, Heidelberg, Germany).
For examination of Gag expression levels, transfected cells were lysed in 500 μl BLP buffer. After being adjusted to the same total protein content, lysates were separated by SDS-PAGE, and p24CA was detected using antibody 183-H12-5C (NIH AIDS Research and Reference Reagent Program). The p24CA concentration of viral-particle preparations was determined by an HIV-1 DIY p24 ELISA Kit 2 (Aalto Bio Reagents, Dublin, Ireland).
Infections.
VSV-G-pseudotyped viruses for the infectivity assay were prepared as follows. 293T cells were grown to 75% confluence in a 175-cm2 flask and were transfected with 2 μg pcRev, 5 μg Hgpsyn, 2 μg pcTat, 4 μg pHIT/G, and 10 μg of HIVRev−/4xMS2 or HIVRev+/4xMS2. pEGFP-C1 (1.5 μg) and calf thymus carrier DNA, to a total of 40 μg DNA, were added to each transfection. Transfection medium was changed after 8 h. Supernatants were collected after 48 h and 72 h, filtered through a 0.45-μm filter, and stored at −80°C in aliquots until use. Infectivity was measured on TZM-bl cells by β-galactosidase staining as follows. A total of 5 × 104 TZM-bl cells were seeded in a 24-well plate and grown for 24 h. Cells were treated with 200-μl serial dilutions of the virus-containing supernatant for 4 h. The medium was changed, and cells were grown for 2 days until fixation using 0.5% glutaraldehyde in PBS. After repeated washes in PBS, cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. For infection, T cells or PBMCs were pelleted and resuspended in the virus-containing supernatant, thereby adjusting the multiplicity of infection (MOI) to 0.1. The medium was changed after 4 h. Cells were cultured for a further 36 h until RNA isolation.
Statistical analyses.
To determine whether HIV-1 RNA levels in the cytoplasm and the particles differed significantly in the presence or absence of Rev or MS2-TAP, a two-sided t test was used on the logarithmically transformed values for the RNA copy numbers.
RESULTS
To study the effect of Rev on unspliced HIV-1 RNA levels in the cytoplasm and genomic RNA encapsidation in the context of authentic viral sequences, HIVRev−/4xMS2, a proviral construct with inactivating point mutations in rev, was used (Fig. 1 A). The construct also contained a 4-bp deletion in env preventing virus replication. In addition, a part of the nef open reading frame was replaced by four copies of an RNA stem-loop of the bacteriophage MS2 to allow for complementation of Rev by heterologous export factors (47).
FIG. 1.
Rev-(in)dependent Gag-Pol expression. (A) Map of plasmids HIVRev−/4xMS2 and Hgpsyn. The proviral HIVRev−/4xMS2 construct contains the HIV-1 genome with inactivating point mutations in rev and env. In addition, parts of the nef gene were replaced by 4 repeats of an RNA stem-loop targeted by the MS2 coat protein (marked by the white box). Inactive ORFs are shown as black bars, and active ORFs are shown as gray bars. The arrow indicates the region of the HIV-1 genome detected by RT-qPCR. Hgpsyn encompasses a codon-optimized gag-pol ORF flanked by a strong heterologous promoter (CMV) and a polyadenylation (pA) signal. The codon-optimized ORF is hatched. (B) Western blot analysis with an HIV-1 p24 capsid antibody of total cell lysates transfected with proviral DNA (HIVRev−/4xMS2), with or without the codon-optimized Gag-Pol expression construct (Hgpsyn), in the presence or absence of a Rev expression plasmid (top). A GFP expression plasmid was also cotransfected to control for transfection efficiency by Western blot analysis with an anti-GFP antibody (bottom).
The HIVRev−/4xMS2 construct was transfected into 293T cells in the presence or absence of a Rev expression plasmid. A Tat expression plasmid was always included to enhance expression levels. Since Rev has a strong effect on Gag-Pol expression from proviral constructs, we provided excess levels of Gag-Pol in trans by cotransfection of Hgpsyn. The gag-pol open reading frame of this plasmid has been optimized for mammalian codon usage without changing the amino acid sequence (44). Mutating approximately every fourth nucleotide of gag-pol rendered the Gag-Pol expression completely independent of Rev. As expected, transfection of HIVRev−/4xMS2 led to detectable levels of Gag expression only if it was cotransfected with the Rev expression plasmid (Fig. 1B). The addition of Hgpsyn led to a further increase of the Gag expression level, which was clearly independent of the presence or absence of Rev (Fig. 1B). Thus, any effect of Rev on RNA encapsidation should not be due to the enhancement of Gag-Pol expression.
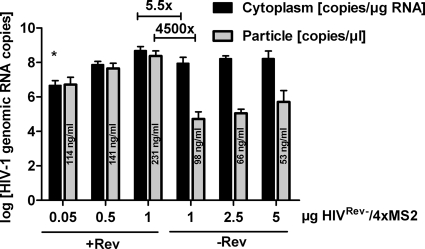
To determine the influence of Rev on the packaging efficiency of proviral RNA, we performed a quantitative RT-PCR-based packaging assay. Briefly, 293T cells were cotransfected with HIVRev−/4xMS2, a Tat expression plasmid, and Hgpsyn in the presence or absence of a Rev expression plasmid. After 48 h, virus particles were pelleted through a 30% sucrose cushion and the particle-associated RNA was extracted. In parallel, cells were lysed under standard conditions to isolate cytoplasmic RNA. Quantitative RT-PCR was performed with primers targeting gag in order to measure the amount of full-length HIV-1 RNA in the cytoplasm and in the viral particles. Hgpsyn-encoded RNA was not detected in this PCR due to its extensive codon optimization. As shown in Fig. 2, Rev increased the cytoplasmic levels of the HIV-1 unspliced RNA after transfection of 1 μg HIVRev−/4xMS2, from 8.9 × 107 to 4.9 × 108 copies/μg extracted RNA. In contrast to this modest increase, Rev enhanced packaged HIV-1 RNA copy numbers approximately 4,500-fold. To exclude a potential nonlinear relationship between the HIV-1 genomic RNA concentration in the cytoplasm and the packaging of this RNA into virus particles, the amount of HIVRev−/4xMS2 DNA transfected was decreased in the presence of Rev and increased in its absence. This led to experimental conditions under which the cytoplasmic HIV-1 unspliced RNA concentration was higher in the absence of Rev than in its presence (Fig. 2). Even under these conditions, encapsidation was at least 350-fold higher in the presence of Rev. The packaging defect in the absence of Rev was not due to inefficient particle production, since the amount of p24 detected in the pelleted particles in the absence of Rev was only 1.2- to 4.3-fold lower than that in the presence of Rev (Fig. 2). This is consistent with previous observations that the budding of virus particles is independent of the amount of viral RNA (15, 24). Thus, the major function of Rev seems to be the enhancement of packaging of the genomic HIV-1 RNA, while there is only a modest effect on cytoplasmic levels of this HIV-1 RNA.
FIG. 2.
Rev enhances packaging of HIV-1 genomic RNA. Unspliced HIV-1 RNA copy numbers in the cytoplasmic RNA fraction and in the RNAs extracted from viral particles are shown. The indicated amounts of HIVRev−/4xMS2 DNA were cotransfected with a constant amount of Hgpsyn in the presence or absence of Rev, as indicated. Values shown are the means and standard errors of the means (SEM) for 3 (* = 2) experiments. The fold enhancement by Rev of full-length HIV-1 RNA levels in the cytoplasm and the particles is given for the 1-μg dose of HIVRev−/4xMS2. Statistical analysis of these results revealed that the difference between the HIV-1 RNA copy numbers was statistically significant (P < 0.01) for the particles but not for the cytoplasm. Numbers inside the bars for the RNA copy numbers in the particles give the means of the p24 concentrations in the particle preparations.
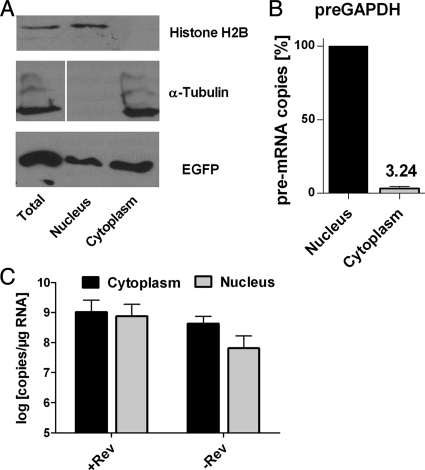
Only modest effects of Rev on cytoplasmic RNA levels of RRE-containing transcripts have been observed before, particularly in the context of subgenomic constructs (9, 11, 26), and they have also been questioned due to concerns of contamination of cytoplasmic fractions with nuclear RNAs (28). We therefore performed various experiments to determine the purity of the cytoplasmic fractions. 293T cells were transfected as before, with an enhanced green fluorescent protein (EGFP) expression plasmid included in the mix, and the cytoplasmic fraction was recovered as described above. In addition, the nuclear fraction was collected after a washing step with the nuclear pellet. Total cell lysates as well as the nuclear and cytoplasmic fractions were then analyzed by Western blotting for EGFP, histone protein H2B, and α-tubulin content (Fig. 3 A). As expected, histone H2B was detected in the nuclear fraction but not in the cytoplasmic fraction, while α-tubulin was present in the cytoplasmic fraction but not in the nuclear fraction. EGFP could be detected in both fractions, confirming proper loading. Histone H2B could be associated more tightly with the nucleus than nuclear RNA and perhaps therefore is not a good marker for contamination of the cytoplasmic fraction with nuclear RNA. We therefore determined the ratio of pre-mRNA levels of the endogenous GAPDH gene (pre-GAPDH) in the cytoplasm and the nucleus. RNAs were extracted from the nuclear and cytoplasmic fractions of transfected 293T cells. The same amounts of extracted nuclear and cytoplasmic RNAs and serial dilutions of the nuclear RNA were subjected to real-time RT-PCR with primers spanning intronic sequences of pre-GAPDH. Similar crossing points of the real-time RT-PCR results for the cytoplasmic RNA samples and a 1:30 dilution of the nuclear RNA preparation indicated that the concentration of the pre-GAPDH RNA in the cytoplasmic RNA was approximately 3% of the concentration of the nuclear RNA (Fig. 3B). Assuming an exclusive nuclear localization of the pre-GAPDH RNA, these results indicate that 3% of the extracted cytoplasmic RNA was actually derived from the nucleus. This nuclear contamination could bias the results regarding the effect of Rev on the cytoplasmic RNA level of the unspliced HIV-1 RNA only if the concentration of this RNA in the nucleus was substantially higher than that in the cytoplasm. Therefore, we also quantified the HIV-1 full-length RNA copy numbers in RNAs extracted from the nuclei and cytoplasm of HIVRev−/4xMS2 DNA-transfected cells. In the presence and absence of Rev, the concentration of the unspliced transcript in the cytoplasmic RNA preparation was higher than that in the nuclear preparation (Fig. 3C). This excludes the possibility that the full magnitude of the enhancing effect of Rev on the cytoplasmic level of full-length HIV-1 RNA is masked by contamination with nuclear RNA.
FIG. 3.
Purity of fractions. (A) Western blot analysis of different fractions of cell lysates. Cell lysates of transfected cells were normalized for total protein content and subjected to SDS-PAGE. Antibodies detecting histone H2B, α-tubulin, and GFP were used. (B) Cellular pre-mRNA of GAPDH was detected via RT-qPCR. The cytoplasmic RNA levels are given as percentages of nuclear RNA levels. Results shown are means plus SEM for 4 experiments. (C) Unspliced HIV-1 RNA in cytoplasmic and nuclear fractions was detected by RT-qPCR. Samples were normalized for RNA content. The results shown are the means plus SEM for 3 experiments.
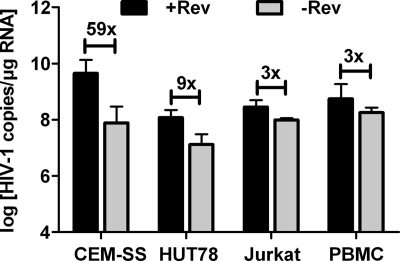
Overexpression by transient transfection of 293T epithelial cells might lead to a spillover of nuclear HIV-1 RNA into the cytoplasm in a Rev-independent manner. Therefore, we also studied the effect of Rev on cytoplasmic RNA levels under rather natural expression levels in lymphoid cell lines, which is considered a more relevant model of HIV infection. The CEM-SS, HUT78, and Jurkat lymphoid T-cell lines were infected with VSV-G-pseudotyped HIV particles transferring either the Rev-deficient HIVRev−/4xMS2 genome or the Rev-proficient HIVRev+/4xMS2 genome. The rev defect of the HIVRev−/4xMS2 construct was complemented in trans during the production of the pseudotypes by cotransfection of a Rev expression plasmid. Vector titers were determined on TZM-bl cells in order to normalize the infectious dose to a multiplicity of infection of 0.1. Forty hours after infection, cytoplasmic RNA was isolated as described above, and the HIV-1 genomic RNA levels were compared between HIVRev−/4xMS2- and HIVRev+/4xMS2-infected T-cell lines. Although all three cell lines were infected with the same stocks of pseudotypes, the effect of Rev on unspliced cytoplasmic HIV-1 RNA levels varied substantially (Fig. 4). In CEM-SS cells, Rev enhanced the level of the genomic transcript in the cytoplasm nearly 60-fold, while the 3- to 9-fold increases observed in Jurkat and HUT78 cells were similar to our observations in transfected 293T cells. Therefore, the HIVRev−/4xMS2 and HIVRev+/4xMS2 pseudotypes were also used to infect activated human PBMCs at a multiplicity of infection of 0.1. Determining the unspliced HIV-1 RNA levels in the cytoplasm of the infected PBMCs revealed that Rev increased these HIV-1 RNA levels just 3-fold (Fig. 4).
FIG. 4.
Effect of Rev on cytoplasmic RNA levels in infected lymphoid cells. T cells were infected at an MOI of 0.1 with VSV-G-pseudotyped HIV-1 constructs differing only in their Rev expression. Unspliced RNA copy numbers in the cytoplasmic fractions were determined by RT-qPCR. Data shown are the means plus SEM for 3 independent experiments. The fold enhancement by Rev is given above the horizontal bars.
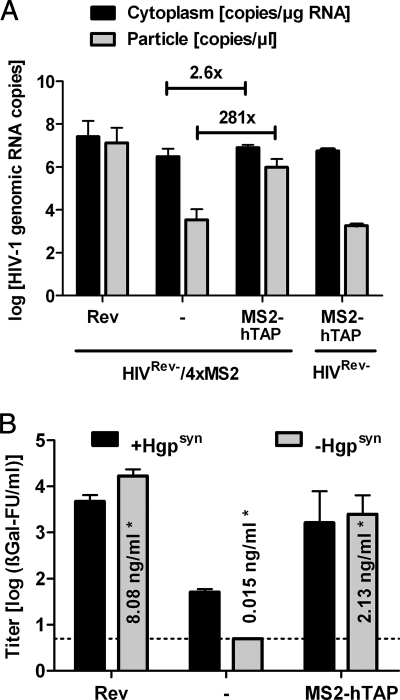
The nuclear export activity of Rev can be replaced functionally by constitutive transport elements (5, 18, 19, 33) or by tethering of heterologous export factors to the HIV-1 RNA (46, 47). To determine whether this is also valid for the packaging effect of Rev, the HIVRev−/4xMS2 DNA was cotransfected with an expression plasmid for a fusion protein consisting of the MS2 coat protein and the human Tip-associated protein (hTAP), a cellular factor known to shuttle mRNA from the nucleus to the cytoplasm. Since the HIVRev−/4xMS2 RNA contains four copies of the MS2 stem-loop in nef, the MS2-hTAP fusion protein can bind to the HIV-1 RNAs. Similar to the case in a previous study (46), the cotransfection of MS2-hTAP with HIVRev−/4xMS2 DNA increased p24 levels approximately 140-fold (Fig. 5 B). However, cotransfection of MS2-hTAP enhanced unspliced HIV-1 RNA levels in the cytoplasm only marginally, while encapsidation of the HIV-1 genomic RNA into particles encoded by cotransfected Hgpsyn was enhanced 280-fold (Fig. 5A). The tethering of hTAP to the MS2 stem-loops was important, since encapsidation of HIVRev− genomic RNA lacking the MS2 stem-loops was strongly impaired even in the presence of MS2-hTAP. Although Rev enhanced encapsidation to an even larger extent, the tethering of hTAP to the HIV-1 genomic RNA can clearly rescue the packaging defect of a Rev-deficient HIV-1. To measure the effect of Rev and MS2-hTAP on HIV-1 infectivity, VSV-G-pseudotyped HIVRev−/4xMS2 particles were produced in the presence or absence of Rev or MS2-hTAP (Fig. 5B). Without Rev and MS2-hTAP, the infectious titer was below the detection limit. Adding Rev or MS2-hTAP increased the infectious titer, at least 3,000- or 500-fold, respectively. Since Rev and MS2-hTAP also increased the secretion of Gag from HIVRev−/4xMS2-transfected cells (533- and 140-fold, respectively), the increase in infectivity could simply be due to enhanced particle production rather than the enhancement of RNA encapsidation. Therefore, we again included Hgpsyn in the cotransfection experiments. Adding Hgpsyn in the presence of Rev or MS2-hTAP did not further increase the infectious titer (Fig. 5B). In the absence of MS2-hTAP and Rev, Hgpsyn allowed the transfer of the HIVRev−/4xMS2 construct, but the infectious titer was approximately 100-fold lower than that in the presence of Rev. Since transfection of Hgpsyn led to higher Gag expression levels than the cotransfection of HIVRev−/4xMS2 and the Rev expression plasmid, infectivity correlates better with encapsidation efficacy than with Gag expression levels.
FIG. 5.
Functional replacement of Rev. (A) Copy numbers of unspliced HIV-1 RNA in the cytoplasmic RNA fraction and in the RNAs extracted from viral particles of cells cotransfected with HIVRev−/4xMS2, HIVRev−, a Rev expression plasmid, and/or the MS2-TAP expression plasmid. The mean values plus SEM for 2 to 4 independent experiments are shown. The fold enhancement by MS2-hTAP of unspliced HIV-1 RNA levels in the cytoplasm and particles is given. Statistical analysis revealed that the difference between the HIV-1 RNA copy numbers in the presence and absence of MS2-TAP was statistically significant (P < 0.05) for the particles but not for the cytoplasm. (B) VSV-G-pseudotyped HIVRev−/4xMS2 particles were prepared by cotransfection with or without Hgpsyn and an expression plasmid for Rev or MS2-TAP. The mean values plus SEM for the infectious titers of 2 or 3 independent experiments are shown. *, mean p24 concentration in the supernatant of cells transfected with HIVRev−/4xMS2 or cotransfected with HIVRev−/4xMS2 and Rev or MS2-TAP.
DISCUSSION
The observation that the tethering of TAP to the HIV-1 RNA enhances the encapsidation of the genomic RNA to a much larger extent than the cytoplasmic RNA level indicates that the packaging function of Rev can be replaced by a heterologous protein. This is consistent with the recent observation that the replacement of the RRE by a constitutive transport element (CTE) leads to efficient genomic RNA encapsidation (32). Since the tethering of TAP to Rev-dependent RNA can also replace the nuclear export function of Rev (46), both activities of Rev seem to be linked mechanistically. However, this raises several questions regarding the precise mechanism by which Rev and TAP enhance the encapsidation process. How can both the Rev-mediated nuclear export via the CRM-1 pathway and the MS2-TAP-triggered nuclear export render the cytoplasmic HIV-1 genomic RNA accessible to packaging, while the same HIV-1 genomic RNA reaching the cytoplasm in the absence of Rev or MS2-TAP is poorly encapsidated? This is particularly puzzling because without a tethered export factor, the unspliced HIV-1 RNA is believed to reach the cytoplasm via the default TAP-mediated mRNA export pathway. One potential explanation could be that the rapid tethering of the unspliced HIV-1 RNA to either of the two nuclear export pathways prevents the formation of a ribonucleoprotein (RNP) complex that is incompetent for translation and encapsidation despite being exported to the cytoplasm. Evidence for the rapid interaction of Rev with the newly synthesized target RNA has been obtained previously (20). In addition to Rev and tethered TAP, CTEs were also shown to enhance the nuclear export and translation of Rev-dependent RNAs (18, 19, 33), as well as the infectivity of lentiviral vector particles (48). Since it is unlikely that different nuclear export factors and constitutive transport elements trigger the formation of RNP complexes mediating the same pattern of pleiotropic effects (enhancement of nuclear export, translation, and packaging), the suppression of the formation of an inhibitory RNP complex by different RNA export factors and constitutive transport elements might be the more plausible mechanism. Whether any of the known cellular cofactors of Rev (recently reviewed in reference 40) is involved in the suppression of the formation of such an inhibitory RNP remains to be determined. The formation of a poorly accessible HIV-1 RNA-protein complex in the absence of Rev might also explain an apparent discrepancy between full-length transcript levels in the cytoplasm, as observed by in situ hybridization and cellular fractionation experiments. In the absence of Rev, there is hardly any cytoplasmic unspliced HIV-1 RNA detectable by in situ hybridization (10, 23). In contrast, we and others have observed substantial levels of unspliced HIV-1 RNA in the cytoplasm by cellular fractionation experiments. Extensive control procedure experiments for the fractionation results argue against the assumption that high levels of full-length transcript detected in the cytoplasmic RNA preparation in the absence of Rev are due to contamination with nuclear RNA. This clearly raises the possibility that the unspliced RNA present in the cytoplasm without Rev escapes efficient detection by in situ hybridization. The mild denaturing conditions during in situ hybridization might not be sufficient to resolve a tightly packed RNP complex that is formed in the absence but not the presence of Rev.
The magnitude of the effect of Rev on genomic HIV-1 RNA levels is influenced not only by the method used to measure the amount of HIV-1 RNA but also by the type of cells analyzed. In infected lymphoid cell lines, Rev enhanced cytoplasmic RNA levels 3- to 60-fold. The latter value was obtained with CEM-SS cells. Interestingly, this cell line was also used in a previous study demonstrating that Rev is absolutely required for nuclear export (28). However, since Rev enhances cytoplasmic RNA levels in primary PBMCs by a factor of only 3, this strong dependence on Rev seems to be a particular property of CEM-SS cells.
Thus, the results of the present study challenge the general notion that the main function of Rev is simply to mediate nuclear export of viral RNAs. Interestingly, the nuclear export pathway of Gag-encoding RNAs has also been observed to modulate the assembly of Gag proteins into particles (21, 22, 41, 42). Therefore, it seems that Rev modulates the RNP complex formed in the nucleus on RRE-containing viral RNAs, thereby either leading to a particular subcytoplasmic compartmentalization or affecting the accessibility of the unspliced HIV-1 RNA for translation and encapsidation.
The detection of substantial levels of unspliced HIV-1 RNA in the cytoplasm in the absence of Rev, and therefore probably also during the early phase of the viral replication cycle, could severely affect HIV-1 spread and persistence. If the structural proteins were indeed produced during the early phase of replication, infected cells could be lysed by antiviral effector mechanisms prior to the release and spread of progeny virus. Therefore, the Rev dependence of nuclear export and translation of the unspliced and singly spliced HIV-1 RNAs might have actually evolved in order to increase the efficiency by which expression of structural proteins during the early phase of the viral replication cycle is prevented. In the late phase of the replication cycle, Rev could then promote coordinated particle production and RNA encapsidation.
Acknowledgments
This work was supported through a grant from the German Research Foundation to K.Ü. (Ue45/11-1). B.G. was supported by the graduate course GRK 1045, funded by the German Research Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to T. Grunwald, N. Ternette, V. V. Temchura, and B. Tippler for experimental help and thank M. Malim, J. Hauber, B. Cullen, and R. Wagner for providing plasmids and D. Cosgrove for a critical reading of the manuscript. TZM-bl cells were obtained from the EVA Centre for AIDS Reagents, NIBSC, United Kingdom, and were donated by J. C. Kappes, X. Wu, and Tranzyme Inc. The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases: HIV-1 p24 hybridoma (183-H12-5C), obtained from Bruce Chesebro and Hardy Chen.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Aricescu, A. R., W. Lu, and E. Y. Jones. 2006. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62:1243-1250. [DOI] [PubMed] [Google Scholar]
- 2.Arrigo, S. J., and I. S. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 5:808-819. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, S., M. Blissenbach, B. Grewe, R. Konietzny, T. Grunwald, and K. Uberla. 2007. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 3:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt, S., T. Grunwald, S. Lucke, A. Stang, and K. Uberla. 2006. Functional replacement of the R region of simian immunodeficiency virus-based vectors by heterologous elements. J. Gen. Virol. 87:2297-2307. [DOI] [PubMed] [Google Scholar]
- 5.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjöld. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. U. S. A. 91:1256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butsch, M., S. Hull, Y. Wang, T. M. Roberts, and K. Boris-Lawrie. 1999. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 73:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coburn, G. A., H. L. Wiegand, Y. Kang, D. N. Ho, M. M. Georgiadis, and B. R. Cullen. 2001. Using viral species specificity to define a critical protein/RNA interaction surface. Genes Dev. 15:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochrane, A. W., K. S. Jones, S. Beidas, P. J. Dillon, A. M. Skalka, and C. A. Rosen. 1991. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65:5305-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Agostino, D. M., B. K. Felber, J. E. Harrison, and G. N. Pavlakis. 1992. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 12:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155-1165. [DOI] [PubMed] [Google Scholar]
- 11.Favaro, J. P., and S. J. Arrigo. 1997. Characterization of Rev function using subgenomic and genomic constructs in T and COS cells. Virology 228:29-38. [DOI] [PubMed] [Google Scholar]
- 12.Favaro, J. P., F. Maldarelli, S. J. Arrigo, and M. G. Schmidt. 1999. Effect of rev on the cytoplasmic localization of intron-containing human immunodeficiency virus type 1 RNA. Virology 255:237-249. [DOI] [PubMed] [Google Scholar]
- 13.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of nondividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. de Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 16.Gramberg, T., N. Sunseri, and N. R. Landau. 2009. Accessories to the crime: recent advances in HIV accessory protein biology. Curr. HIV/AIDS Rep. 6:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groom, H. C. T., E. C. Anderson, and A. M. L. Lever. 2009. Rev: beyond nuclear export. J. Gen. Virol. 90:1303-1318. [DOI] [PubMed] [Google Scholar]
- 18.Grüter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 19.Helga-Maria, C., M. L. Hammarskjöld, and D. Rekosh. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iacampo, S., and A. Cochrane. 1996. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, J., T. Sturgeon, C. Chen, S. C. Watkins, O. A. Weisz, and R. C. Montelaro. 2007. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J. Virol. 81:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, J., T. Sturgeon, O. A. Weisz, W. Mothes, and R. C. Montelaro. 2009. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PLoS One 4:e6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, L., B. W. Guzik, Y.-C. Bor, D. Rekosh, and M.-L. Hammarskjöld. 2003. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 17:3075-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karacostas, V., K. Nagashima, M. A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. U. S. A. 86:8964-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura, T., I. Hashimoto, M. Nishikawa, and J. I. Fujisawa. 1996. A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal beta-actin and viral protein expression. Biochimie 78:1075-1080. [DOI] [PubMed] [Google Scholar]
- 26.Kotsopoulou, E., V. N. Kim, A. J. Kingsman, S. M. Kingsman, and K. A. Mitrophanous. 2000. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74:4839-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucke, S., T. Grunwald, and K. Uberla. 2005. Reduced mobilization of Rev-responsive element-deficient lentiviral vectors. J. Virol. 79:9359-9362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim, M. H., and B. R. Cullen. 1993. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol. Cell. Biol. 13:6180-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 30.Malim, M. H., and M. Emerman. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388-398. [DOI] [PubMed] [Google Scholar]
- 31.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, M. D., O. A. Nikolaitchik, J. Chen, M.-L. Hammarskjöld, D. Rekosh, and W.-S. Hu. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 5:e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nappi, F., R. Schneider, A. Zolotukhin, S. Smulevitch, D. Michalowski, J. Bear, B. K. Felber, and G. N. Pavlakis. 2001. Identification of a novel posttranscriptional regulatory element by using a rev- and RRE-mutated human immunodeficiency virus type 1 DNA proviral clone as a molecular trap. J. Virol. 75:4558-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nekhai, S., and K.-T. Jeang. 2006. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 1:417-426. [DOI] [PubMed] [Google Scholar]
- 35.Perales, C., L. Carrasco, and M. E. Gonzalez. 2005. Regulation of HIV-1 env mRNA translation by Rev protein. Biochim. Biophys. Acta 1743:169-175. [DOI] [PubMed] [Google Scholar]
- 36.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz, S., M. Campbell, G. Nasioulas, J. Harrison, B. K. Felber, and G. N. Pavlakis. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 66:7176-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 66:150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suhasini, M., and T. R. Reddy. 2009. Cellular proteins and HIV-1 Rev function. Curr. HIV Res. 7:91-100. [DOI] [PubMed] [Google Scholar]
- 41.Swanson, C. M., and M. H. Malim. 2006. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic 7:1440-1450. [DOI] [PubMed] [Google Scholar]
- 42.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 9:4155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner, R., M. Graf, K. Bieler, H. Wolf, T. Grunwald, P. Foley, and K. Uberla. 2000. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum. Gene Ther. 11:2403-2413. [DOI] [PubMed] [Google Scholar]
- 45.Ward, A. M., D. Rekosh, and M.-L. Hammarskjold. 2009. Trafficking through the Rev/RRE pathway is essential for efficient inhibition of human immunodeficiency virus type 1 by an antisense RNA derived from the envelope gene. J. Virol. 83:940-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiegand, H. L., G. A. Coburn, Y. Zeng, Y. Kang, H. P. Bogerd, and B. R. Cullen. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi, R., H. P. Bogerd, and B. R. Cullen. 2002. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J. Virol. 76:2036-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]