Abstract
The products of numerous open reading frames (ORFs) present in the genome of human cytomegalovirus (CMV) have not been characterized. Here, we describe the identification of a new CMV protein localizing to the nuclear envelope and in cytoplasmic vesicles at late times postinfection. Based on this distinctive localization pattern, we called this new protein nuclear rim-associated cytomegaloviral protein, or RASCAL. Two RASCAL isoforms exist, a short version of 97 amino acids encoded by the majority of CMV strains and a longer version of 176 amino acids encoded by the Towne, Toledo, HAN20, and HAN38 strains. Both isoforms colocalize with lamin B in deep intranuclear invaginations of the inner nuclear membrane (INM) and in novel cytoplasmic vesicular structures possibly derived from the nuclear envelope. INM infoldings have been previously described as sites of nucleocapsid egress, which is mediated by the localized disruption of the nuclear lamina, promoted by the activities of viral and cellular kinases recruited by the lamina-associated proteins UL50 and UL53. RASCAL accumulation at the nuclear membrane required the presence of UL50 but not of UL53. RASCAL and UL50 also appeared to specifically interact, suggesting that RASCAL is a new component of the nuclear egress complex (NEC) and possibly involved in mediating nucleocapsid egress from the nucleus. Finally, the presence of RASCAL within cytoplasmic vesicles raises the intriguing possibility that this protein might participate in additional steps of virion maturation occurring after capsid release from the nucleus.
Cytomegalovirus (CMV) is a highly prevalent betaherpesvirus that can cause severe multiorgan disease in immunocompromised individuals (45). The ability of this virus to infect an exceptionally wide variety of different cell types substantially contributes to pathogenesis (5, 68). CMV tropism is largely determined by a finely tuned interplay between cellular and viral factors, many of which act at the earliest stages of infection (30, 68). We recently showed that the cellular protein vimentin is required for efficient onset of infection in primary human foreskin fibroblasts (HF). Interestingly, the degree of reliance on the presence and integrity of vimentin intermediate filaments is dependent on the virus strain, with the broadly tropic strain TB40/E being more negatively affected than the HF-adapted, attenuated strain AD169 (44).
Serial passage of clinical isolates in HF or in endothelial cells (EC) has produced strains with different tropisms. The attenuated strains AD169 and Towne were developed as vaccine candidates by propagation in HF for more than 50 (AD169) and 125 (Towne) serial passages (19, 53, 61). During this process, both strains, compared to clinical isolates, accumulated multiple mutations and genomic deletions, resulting in the loss of more than 19 open reading frames (ORFs) (8). The number of serial passages in HF of another commonly used strain, Toledo, has been more moderate (19, 54, 58). This, however, did not prevent the emergence of numerous genomic mutations, including the inversion of an ∼15-kb fragment (8, 16, 56). As a consequence of these changes, productive infections by AD169, Towne, and Toledo are largely restricted to HF. In contrast, propagation of clinical isolates in EC has yielded a series of strains with more-intact genomes and broader tropisms, such as TB40/E, VHL/E, and FIX (VR1814) (25, 60, 71). These strains retain the ability to grow in a wider variety of cell types, including EC, epithelial cells, and dendritic cells (DC), in addition to HF (23, 28, 59, 60, 68).
The UL128, UL130, and UL131A gene products were recently identified as essential mediators of CMV infection of EC and epithelial cells (26, 72, 73) and of virus transfer from infected EC to monocyte-derived DC (23). Each of these proteins is independently required for the broader tropisms of EC-propagated CMV isolates (63, 64), and the presence of mutations affecting their functionality has been directly linked to the inability of AD169, Towne, and Toledo to initiate productive infections in EC and epithelial cells (26, 72, 73).
We have shown that mature Langerhans-type DC differentiated in vitro from CD34+ hematopoietic progenitor cells are highly permissive to direct infection with TB40/E or VHL/E, with 48 to 72% of cells in culture expressing the viral immediate-early genes IE1 and IE2 at 48 h postinfection (hpi) (28). In contrast, only 2 to 5% and 0% of mature Langerhans cells were IE1/IE2 positive after exposure to Towne and Toledo, respectively. However, productive infection was detected in 12 to 17% of cells infected with AD169, despite the fact that this strain lacks expression of the UL131A gene as a consequence of a frameshift mutation (26). These results suggested the existence of additional viral genes with products involved in mediating tropisms for specific cell types, such as DC. To identify possible candidates, we compared the amino acid sequence of each ORF found in the genome of TB40-BAC4, a sequenced clone of the TB40/E strain in a bacterial artificial chromosome (BAC) (GenBank accession number EF999921) (69), to the sequence of each ORF found in AD169 and AD169-BAC (accession numbers X17403 and AC146999) (10, 49), Towne and Towne-BAC (accession numbers FJ616285, AC146851, and AY315197) (17, 18, 49), and Toledo-BAC (accession number AC146905) (49). The product of a putative ORF, originally identified by Murphy et al. and named conserved ORF 29 (c-ORF29) (49), was considered of particular interest because the amino acid sequence of the putative protein encoded by Toledo and Towne was extended by 79 residues compared to the putative protein encoded by TB40/E and AD169. This led to our speculation that that the extended version might result in a nonfunctional version of the c-ORF29-encoded protein. We thus focused our studies on the products of this ORF.
Here, we show for the first time that CMV c-ORF29 encodes a protein expressed at early to late times postinfection (p.i.) and localizes to the nuclear rim in peculiar invaginations of the nuclear lamina and in cytoplasmic vesicular structures at late times p.i. Based on this localization pattern, we named this gene product nuclear rim-associated cytomegaloviral protein, or RASCAL. Surprisingly, no difference was observed in the distributions of RASCAL during infection of HF with TB40/E or Towne, suggesting that the intracellular trafficking of this protein is not affected by the presence of the additional residues at the C terminus of RASCAL from strain Towne (RASCALTowne). Ectopic expression of RASCAL in human embryo kidney 293T (HEK293T) cells further revealed that this protein requires the presence of the nuclear egress complex (NEC) member UL50 to reach the nuclear rim, while coimmunoprecipitation (co-IP) assays provided evidence for the existence of an interaction between RASCAL and UL50. These findings suggest that RASCAL may be a new component of the NEC with possible roles in remodeling the nuclear lamina during nucleocapsid egress from the nucleus.
MATERIALS AND METHODS
In silico analysis.
RASCAL amino acid sequences were analyzed using the following programs: ClustalW2 (32), NetPhosK (4), NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/), SignalP3.0 (2), Kyte-Doolittle hydropathy plot (31), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), TopPred II (11), dense alignment surface (DAS) (13), ESLpred (3), HSLpred (22), TargetP1.1 (20), and SubLoc v1.0 (29).
Cells and viruses.
HF and HEK293T cells were gifts of E. S. Mocarski, Emory University, Atlanta, GA, and were propagated in Dulbecco's modified Eagle medium supplemented with 10% fetal clone serum III (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin, 4 mM HEPES, and 1 mM sodium pyruvate (all from Gibco Invitrogen Corp.). HF were used between passages 17 and 27 postisolation. Human CMV strains AD169varATCC and TB40/E and the green fluorescent protein (GFP)-tagged derivative of TownevarRIT3, Towne/GFP-IE2 (J. Xu, D. Formankova, and E. S. Mocarski, unpublished), were originally obtained from the American Type Culture Collection, from C. Sinzger (Tübingen, Germany), and from E. S. Mocarski (Emory University, Atlanta, GA), respectively. Propagation and purification of all strains were performed as previously described (28).
HF infection and cell transfection.
HF were plated at a density of 5 × 104 cells/cm2 3 days prior to exposure to AD169, TB40/E, or Towne/GFP-IE2 at a multiplicity of infection (MOI) of 3 or 5. Mock-infected samples were exposed to culture medium alone. After virus adsorption at 37°C in 5% CO2 for 1 h, the inoculum was removed, and cells were washed three times with medium prior to incubation in fresh medium until they were harvested at different times p.i. For phosphonoformic acid (PFA) treatment, HF were exposed to Towne/GFP-IE2 (MOI of 3) for 1 h at 37°C, washed, and further incubated in fresh culture medium containing 300 μg/ml of PFA (Sigma, St. Louis, MO). HEK293T cell transfection was performed using PolyFect (Qiagen) as per the manufacturer's guidelines.
mRNA isolation and RT-PCR.
mRNA was isolated from mock-, TB40/E-, or Towne/GFP-IE2-infected HF using the μMACS mRNA isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). First-strand cDNA synthesis was performed using SuperScript III reverse transcriptase according to the manufacturer's guidelines (Gibco Invitrogen Corp.). c-ORF29TB40/E was amplified with the forward primer GCGGATCCTAATGGGGGAACGCC (labeled a > in Fig. 2A, top) and the reverse primer GTGGATCCAGAGATGCGGAAAAGCC (labeled < b in Fig. 2A, top), c-ORF29Towne was amplified with the forward primer GCGGATCCTAATGGGGGAACGCC (labeled a > in Fig. 2A, top) and the reverse primer CCTCGAGAAAAGCACGCAAGC (labeled < c in Fig. 2A, top), vimentin was amplified with the forward primer CGGATCCATGTCCACCAG and the reverse primer CGAATTCTTCAAGGTCAT, UL99 was amplified with the forward primer CGGATCCATGGGTGGCGAACTCT and the reverse primer GGATATCTGAAAGGACAAGGGGGCG, and β-actin cDNA was amplified with the forward primer GGTCATCACCATTGGCAATGAGCGG and the reverse primer GGACTCGTCATACTCCTGCTTGCTG.
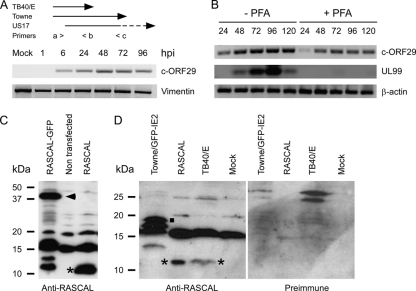
FIG. 2.
RASCAL expression in infected and transfected cells. (A, top) Schematic depiction of the primers used to amplify c-ORF29 by RT-PCR from TB40/E- or Towne/GFP-IE2-infected HF. The > and < symbols depict the direction of polymerization. The sequence of primer a > is complementary to nucleotides 1 to 13 of c-ORF29TB40/E and of c-ORF29Towne, the sequence of primer < b is complementary to nucleotides 277 to 291 of c-ORF29TB40/E and of c-ORF29Towne and to nucleotides 189 to 203 of US17, while the sequence of primer < c is complementary to nucleotides 515 to 528 of c-ORF29Towne and nucleotides 427 to 440 of US17. (Bottom) RT-PCR analysis of c-ORF29 transcription in mock- or TB40/E-infected HF at the indicated times p.i., using primers a > and < b. Amplification of vimentin's cDNA was used as the PCR control. (B) RT-PCR analysis of c-ORF29 and of UL99 transcription in HF infected with TB40/E in the presence (+) or absence (−) of PFA (300 μg/ml). Amplification of β-actin's cDNA was used as the PCR control. (C) Immunoblot analysis results of RASCALTB40/E expression in protein extracts from HEK293T cells nontransfected or transfected with expression plasmids encoding RASCALTB40/E-GFP or RASCALTB40/E. The blot was incubated with an anti-RASCAL polyclonal Ab as described in Materials and Methods. Expected molecular masses were 10.6 kDa for RASCALTB40/E and 37.6 kDa for RASCALTB40/E-GFP. Asterisk, RASCALTB40/E; arrowhead, RASCALTB40/E-GFP. (D) Immunoblot analysis of RASCAL expression in protein extracts from mock-, TB40/E-, or Towne/GFP-IE2-infected HF and in HEK293T cells expressing RASCALTB40/E. The same membrane was incubated first with an anti-RASCAL polyclonal Ab (left) and subsequently with the preimmune serum (right) as described in Materials and Methods. Expected molecular masses were 10.6 kDa for RASCALTB40/E and 19.4 kDa for RASCALTowne. Asterisk, RASCALTB40/E; square, RASCALTowne.
Plasmid construction.
L-RASCALTB40/E (where the prefix “L” stands for LNCX) was generated by excising GFP from LNCX-GFP (LGFP) (36) using EcoRI and EcoRV and by replacing it with the RASCALTB40/E sequence amplified by PCR using a forward primer containing the EcoRI restriction site (GGAATTCATGGGGGAACGCCGTGTG) and a reverse primer containing the RASCALTB40/E stop codon and the EcoRV restriction site (GCGATATCTTAAGATGCGGAAAAGCCA). L-RASCALTB40/E-GFP was generated by cloning RASCALTB40/E, lacking its stop codon, in frame with the N terminus of GFP using the EcoRI and BamHI restriction sites present in LGFP. The forward primer used to make L-RASCALTB40/E-GFP was the same as that used to make L-RASCALTB40/E. The reverse primer (GTGGATCCAGAGATGCGGAAAAGCC) contained a BamHI restriction site and was designed to remove the RASCAL stop codon. Hemagglutinin-tagged LNCX UL50 (LNCX UL50-HA) was generated by amplifying the UL50 sequence from cDNA populations derived from AD169-infected HF using the forward primer AGAATTCATGGAGATGAACAAGGTT, containing an EcoRI restriction site, and the reverse primer TTTTGTCGACTCAAGCGTAATCTGGAACATCGTATGGGTAGTCGCGGTGTGCGGAG, containing the HA tag nucleotide sequence. This PCR product was cloned into the pSC-B vector (Stratagene, La Jolla, CA) and subsequently excised using EcoRI before being ligated into the EcoRI restriction site of LNCX (43). FLAG-tagged L-UL53 (L-UL53-FLAG) was created by amplifying the UL53 sequence from cDNA populations derived from TB40/E-infected HF using the forward primer AGAATTCATGTCTAGCGTGAGCG, containing the EcoRI restriction site, and the reverse primer TTTTGTCGACTCACTTGTCATCGTCGTCCTTGTAGTCAGGCGCACGAATGCTGTTGA, containing the FLAG tag nucleotide sequence and the SalI restriction site. The UL53-FLAG PCR product was ligated into the EcoRI and SalI sites of LGFP after excision of the GFP coding sequence.
Generation of anti-RASCAL Abs.
Polyclonal rabbit antibodies (Abs) were raised by ProSci Inc. (Poway, CA) against the RASCAL peptide YAPFDSHRRHVSELRGHRD conjugated to the keyhole limpet hemocyanin. Abs were affinity purified and were provided at a final concentration of 1.7 mg/ml.
Immunoblot analysis.
Cell pellets from mock-, TB40/E-, or Towne/GFP-IE2-infected HF and from HEK293T cells nontransfected or transiently transfected with L-RASCALTB40/E-GFP or with L-RASCALTB40/E were resuspended in 3% sodium dodecyl sulfate (SDS) lysis buffer containing 125 mM Tris-HCl (pH 6.8), 3% SDS, 10 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride, and complete EDTA-free protease inhibitor cocktail (Roche). Cell lysates were boiled at 100°C for 5 min, and cell debris were eliminated by centrifugation at 16,100 × g for 3 min. Protein concentrations were determined with the DC protein assay kit (Bio-Rad). Protein extracts were then separated by electrophoresis on a 15% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes. Membranes were blocked overnight at 4°C in blocking buffer containing 10 mM Tris-Cl (pH 7.5), 100 mM NaCl, 5% milk powder, 0.1% Tween 20 prior to incubation with rabbit anti-RASCAL Abs (1:1,000) or with mouse anti-HA Abs (1:1,000) for 1 h at room temperature (RT). Membranes were rinsed in wash buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, and 0.05% Tween 20) and were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:4,000; Vector Laboratories) or goat anti-mouse IgG (1:5,000; Vector Laboratories) for 1 h at RT. For reprobing, membranes were incubated in 62.5 mM Tris-Cl (pH 6.8), 100 mM β-mercaptoethanol, and 2% SDS at 50°C for 30 min, rinsed with phosphate-buffered saline (PBS) containing 0.1% Tween 20, blocked overnight at 4°C, and incubated with preimmune rabbit serum (1:1,000) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (1:4,000), both for 1 h at RT. Signal was developed by enhanced chemiluminescence (ECL plus kit; GE Healthcare).
Coimmunoprecipitation assays.
HEK293T cells were transfected with LNCX (control) or were cotransfected with L-RASCALTB40/E and LNCX UL50-HA at a 1:3 molar ratio or with LNCX UL50-HA and L-UL53-FLAG at a 1:1 molar ratio in T25 flasks. At 24 h posttransfection, cells were harvested in 500 μl of co-IP buffer containing 50 mM Tris-HCl (pH 8), 100 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, and complete EDTA-free protease inhibitor cocktail (Roche) prior to the addition of 5 μg of mouse monoclonal anti-HA Abs (Invitrogen) or of polyclonal anti-RASCAL Abs for 2 h at 4°C under rotation. Extracts were incubated with 100 μl of protein A-Sepharose bead slurry (Sigma) for 2 h at 4°C, pelleted, washed in lysis buffer containing 200 mM NaCl, and subjected to immunoblot analysis as described above.
Immunofluorescence staining analysis.
Cells were fixed in 3.7% paraformaldehyde (Fisher Chemicals, Fairlawn, NJ) for 30 min at RT, permeabilized in 0.2% Triton X-100 (USB Corporation, Cleveland, OH) for 20 min on ice, and blocked in 100% horse serum (HS; PML Microbiologicals, Wilsonville, OR) for 30 min at RT. Cells were then incubated with rabbit polyclonal anti-RASCAL Abs (1:500) alone or in combination with monoclonal Abs directed against the FLAG tag (1:1,000; clone M2; Sigma, St. Louis, MO), the HA tag (1:500; Invitrogen), IE1/IE2 (1:500; fluorescein isothiocyanate [FITC]-conjugated MAb810F; Chemicon, Temecula, CA), lamin B (1:50; Santa Cruz Biotechnology, Santa Cruz, CA), or lamin A/C (1:50; Santa Cruz Biotechnology, Santa Cruz, CA). Samples were washed in PBS-0.05% Tween 20 prior to incubation with Alexa Fluor 594-conjugated goat anti-rabbit IgG Abs (1:500; Molecular Probes, Eugene, OR) alone or in combination with FITC-conjugated goat anti-mouse IgG Abs (1:100; Invitrogen, Carlsbad, CA). For control stainings, cells were incubated with preimmune sera diluted 1:500 in 100% HS. For dual staining of cells expressing UL50-HA and UL53-FLAG, samples were incubated with mouse monoclonal anti-FLAG Abs and rabbit polyclonal anti-HA Abs (1:500; Zymed), followed by FITC-conjugated goat anti-mouse IgG Abs and Alexa Fluor 594-conjugated goat anti-rabbit IgG Abs. All Abs were diluted in 100% HS and were incubated on samples for 1 h at RT. Nuclei were labeled with Hoechst 33342 (0.2 mg/ml; Molecular Probes, Eugene, OR) for 3 min at RT and were mounted in 90% glycerol-10% PBS containing 2.5 g/liter of 1,4-diazabicyclo(2,2,2)octane (DABCO; Alfa Aesar, Pelham, NH). Samples were analyzed on a Zeiss Axioskop 2 magneto-optical trap fluorescence microscope equipped with a QImaging Retiga 1300-coded monochrome 12-bit camera. Images were captured and pseudocolored using Northern Eclipse version 7.0 software. Confocal images were acquired on a Zeiss LSM 510 META ConfoCor2 confocal laser scanning microscope equipped with Zeiss LSM 510 META image processing software.
RESULTS
Identification of the ORF encoding RASCAL and in silico analysis of the predicted amino acid sequence.
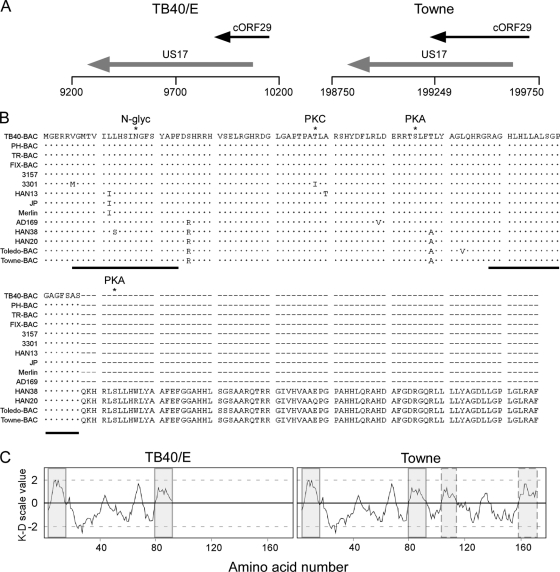
The genomic region of TB40-BAC4 corresponding to nucleotides 9100 to 11200 was screened for the presence of ORFs with the potential to encode proteins of at least 80 amino acids (aa) and containing a 5′ ATG codon. One ORF of 294 nucleotides encoding a putative protein of 97 aa was found. As described for c-ORF29 (48, 49), the sequence of this ORF was located on the negative strand of the genome (nucleotides 9855 to 10148 in TB40-BAC4) and partially overlapped (206 nucleotides) the 5′ end of the US17 gene (Fig. 1A, left). An ortholog of this ORF was found in the Towne-BAC genome, spanning nucleotides 199167 to 199697 (in GenBank accession no. AY315197) and overlapping the 5′ end of the US17 gene by 442 nucleotides (Fig. 1A, right). In contrast to what was previously reported for c-ORF29 (48, 49), however, searches of the nonredundant nucleotide collection databases using the BLASTN algorithm failed to reveal the presence of a full-length c-ORF29 ortholog in the genome of chimpanzee CMV. A ClustalW2 global alignment of the predicted amino acid sequence of c-ORF29 from strain TB40-BAC4 (RASCALTB40/E) with sequences from 13 other human CMV strains showed that RASCAL is highly conserved and that not only strains Towne and Toledo, but also two new CMV strains, HAN20 and HAN38, carry a longer version of this protein, containing 79 additional amino acids (Fig. 1B). No significant homology between either isoform of RASCAL and proteins of cellular or viral origin was detected in extensive searches of multiple amino acid sequence databases.
FIG. 1.
Genomic location of the c-ORF29 gene and in silico analysis of the RASCAL amino acid sequence. (A) Schematic map of the TB40-BAC4 (GenBank accession no. EF999921) and of the Towne-BAC (accession no. AY315197) genomic regions corresponding to nucleotides 9200 to 10200 (TB40/E) and 198750 to 199750 (Towne). The black horizontal line depicts the viral genome, with vertical lines positioned every 500 nucleotides. ORFs are represented by arrows pointing in the direction of transcription. (B) ClustalW2 alignment of RASCAL amino acid sequences from 14 human CMV strains. Viral genome accession numbers are as follows: TB40-BAC, EF999921; PH-BAC, AC146904; TR-BAC, AC146906; FIX-BAC, AC146907; 3157, GQ221974; 3301, GQ466044; HAN13, GQ221973; JP, GQ221975; Merlin, AY44689; AD169, X17403; HAN38, GQ396662; HAN20, GQ396663; Toledo-BAC, AC146905; and Towne-BAC, AC146851. Dots and dashes indicate identical and absent amino acids, respectively. The asterisks mark the specific amino acid predicted to be phosphorylated by PKA or PKC or to be N glycosylated (N-glyc). The black lines underscore the putative TMDs. (C) Kyte-Doolittle (K-D) hydropathy plot of RASCALTB40/E and RASCALTowne amino acid sequences, performed using a window size of 9 aa. The shaded boxes include the stretch of amino acids corresponding to the putative TMDs, while the boxes with the dashed borders include the two additional hydrophobic regions of more than 17 aa detected in RASCALTowne.
Two potential phosphorylation sites with scores greater than 80% were identified in RASCALTB40/E by the NetPhosK software, one of which was specific for protein kinase C (PKC) at Thr 48 and the other for protein kinase A (PKA) at Ser 65. One additional site for PKA was found in the longer version of the protein at Ser 103 (Fig. 1B). A single potential N-glycosylation site was detected by the NetNGlyc software at residue Asn 17 (Fig. 1B). However, the SignalP3.0 algorithm failed to predict the presence of a signal sequence at the N terminus of RASCAL, suggesting that this protein may not be recognized by the glycosylation machinery in vivo.
A Kyte-Doolittle hydropathy plot of RASCALTB40/E revealed the presence of two regions of at least 17 aa with overall hydrophobicity values greater than 1, one located at the N terminus (aa 6 to 24) and the other at the C terminus (aa 79 to 97) (Fig. 1C, left, shaded boxes). Both were predicted to contain a transmembrane domain (TMD) by three protein topology prediction algorithms, TMpred, TopPred II, and DAS. The amino acid sequences of these putative TMDs are highly conserved in all strains (Fig. 1B, black lines). Although strains Merlin, JP, and HAN13 carry a single amino acid replacement of Leu 12 with Ile, HAN38 carries a Leu 13-to-Ser substitution, and 3301 carries a Val 6-to-Met substitution, none of these changes altered the outcome of the predictions. Two additional hydrophobic regions consisting of more than 17 aa were detected in RASCALTowne (Fig. 1C, right, boxes with dashed borders), but neither was predicted to be a TMD by all three software programs.
RASCAL expression in infected and transfected cells.
To determine if c-ORF29 was expressed during infection, we performed reverse transcriptase-PCR (RT-PCR) analyses of mRNA extracts from mock-, TB40/E-, or Towne/GFP-IE2-infected HF using the primers schematically depicted in the top panel of Fig. 2A. A single product of approximately 300 nucleotides was detected in extracts from TB40/E-infected cells amplified with primers a > and < b (Fig. 2A, bottom). Mock-infected samples and control reaction mixtures lacking the cDNA template were negative (Fig. 2A, bottom, and data not shown). Although the sequence of primer < c is complementary to nucleotides 9621 to 9624 of the TB40/E genome, no product was obtained when the same extracts were amplified with primers a > and < c (not shown), suggesting that no transcript spanning the region between these primers was produced in cells infected with TB40/E. In contrast, a single product of about 550 nucleotides was observed in extracts from Towne/GFP-IE2-infected cells amplified using primers a > and < c (not shown). Sequence analysis of this PCR product confirmed the presence of a T-to-C mutation at position 292, resulting in the conversion of a TAA stop codon into a CAA codon coding for the amino acid Gln. RT-PCR analyses of mRNA extracts from HF infected with TB40/E in the presence of PFA revealed that c-ORF29 expression was reduced, but not completely abrogated, in the absence of viral DNA replication (Fig. 2B). In contrast, no expression of the true late gene UL99 was detected in PFA-treated cells (Fig. 2B). Together, these data indicate that c-ORF29 is transcribed with early-late kinetics during infection and confirm that the Towne strain carries a longer version of this ORF.
To determine if c-ORF29 did encode a protein, affinity-purified, polyclonal rabbit Abs were raised against residues 21 to 39 of the predicted RASCALTB40/E amino acid sequence. The specificities of these Abs were tested using Western blot analyses of protein extracts from HEK293T cells transiently transfected with expression plasmids encoding RASCALTB40/E only or RASCALTB40/E fused to the N terminus of the GFP amino acid sequence (RASCALTB40/E-GFP). Two proteins with expected molecular masses of approximately 11 kDa (Fig. 2C, asterisk) and 38 kDa (Fig. 2C, arrowhead) were detected in extracts from cells transfected with expression plasmids encoding RASCALTB40/E and RASCALTB40/E-GFP, respectively. These proteins were not recognized by the anti-RASCAL Abs in extracts from nontransfected cells (Fig. 2C) or by the preimmune serum (not shown). Moreover, the 38-kDa protein was also specifically detected in membranes reprobed with anti-GFP monoclonal Abs (not shown), indicating that this polypeptide did contain both the RASCAL and the GFP epitopes. Two additional bands of 15 and 20 kDa were observed in extracts from both transfected and nontransfected cells and were considered to be cellular proteins nonspecifically recognized by the anti-RASCAL Abs. Finally, two products with molecular masses of less than 15 kDa were observed exclusively in cells expressing RASCALTB40/E-GFP, suggesting that they might correspond to degradation products of the fusion protein.
To assess if RASCAL was expressed in CMV-infected cells, protein extracts from mock-, TB40/E-, and Towne/GFP-IE2-infected HF were separated by SDS-polyacrylamide gel electrophoresis (PAGE). The same membrane was probed with anti-RASCAL Abs and, after being stripped, with preimmune serum. A single band of approximately 11 kDa was detected in extracts from TB40/E-infected HF and in extracts from HEK293T cells expressing RASCALTB40/E, used as the control (Fig. 2D, left, asterisks). This band was not detected in samples from mock- and Towne/GFP-IE2-infected cells. In addition, two proteins with molecular masses larger than 15 kDa but smaller than 20 kDa were observed exclusively in extracts from Towne/GFP-IE2-infected HF (Fig. 2D, left). While the larger protein (Fig. 2D, left, square) is likely to correspond to RASCALTowne (predicted molecular mass, 19.4 kDa), the smaller protein remains uncharacterized. None of these bands, including the nonspecific protein of approximately 15 kDa recognized by the anti-RASCAL Abs in HF and HEK293T cell extracts, was detected by the preimmune serum (Fig. 2D, right).
RASCAL localizes at the nuclear envelope and in cytoplasmic vesicular structures during infection.
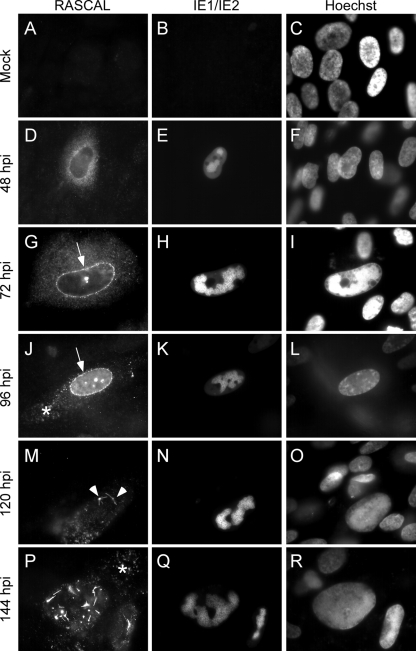
To establish the subcellular localization of RASCAL during infection, mock-, AD169-, TB40/E-, and Towne/GFP-IE2-infected HF (MOI of 5) were harvested at 24, 48, 72, 96, 120, and 144 hpi. Cells were stained with the preimmune serum or with the affinity-purified anti-RASCAL Abs and with FITC-conjugated anti-IE1/IE2 Abs. Nuclei were highlighted with Hoechst 33342. No signal was observed in mock-infected cells (Fig. 3A to C), indicating that no cellular proteins were recognized nonspecifically by the anti-RASCAL Abs in immunofluorescence staining assays. Similarly, staining of infected cells with the preimmune serum did not yield any specific signal (not shown). At 24 hpi, RASCAL fluorescence was barely detectable (not shown). By 48 hpi, a prominent cytoplasmic signal was observed in infected cells, with a punctuate pattern more densely concentrated around the nucleus (Fig. 3D to F). At 72 hpi, RASCAL accumulation in a perinuclear ring was clearly detectable in a large proportion of cells (Fig. 3G to I, arrow), and by 96 hpi most of the RASCAL signal emanated from the nuclear rim, with very little diffuse fluorescence remaining in the cytoplasm (Fig. 3J to L). In some cells, RASCAL appeared to also concentrate in cytoplasmic vesicular compartments distributed from the nuclear envelope to the cell surface (Fig. 3J to L, asterisk). At 96 hpi and, more prominently, at 120 hpi, RASCAL fluorescence appeared to originate almost exclusively from peculiar structures likely located on the nuclear envelope and characterized by a central “knot” with rod-like extensions (Fig. 3M to O, arrowheads). These nuclear envelopes and the cytoplasmic structures became clearly visible at 144 hpi (Fig. 3P to R), at a time when few or no perinuclear rings were still detectable. The same staining pattern was observed in AD169- and TB40/E-infected cells (not shown), indicating that the presence of the 79 additional amino acids in RASCALTowne did not interfere with its localization.
FIG. 3.
RASCAL intracellular localization. Mock-infected (A to C) or Towne/GFP-IE2-infected HF (MOI of 5) (D to R) were harvested at the indicated times p.i. and were stained with affinity-purified rabbit anti-RASCAL polyclonal Abs followed by Alexa Fluor 594-conjugated goat anti-rabbit Abs. The signal emitted from the GFP-IE2 protein was further amplified with FITC-conjugated anti-IE1/IE2 Abs, and nuclear DNA was stained with Hoechst 33342. The arrows point at RASCAL accumulation at the nuclear rim, the arrowheads indicate the peculiar structures observed on the nuclear surface at late times p.i., and the asterisks mark the locations of the cytoplasmic RASCAL-positive vesicles. Original magnification, ×400.
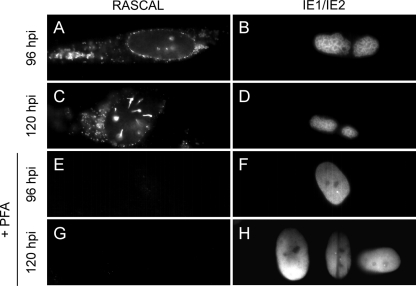
To determine if the RASCAL protein was produced in the absence of viral DNA replication, HF were infected with Towne/GFP-IE2 (MOI of 3) in the presence or absence of PFA. Cells were harvested at 24, 48, 72, 96, 120, and 144 hpi and stained for RASCAL and for IE1/IE2. As expected, intranuclear accumulation of IE1/IE2 at the sites of viral genome replication (52) was observed exclusively in untreated HF (Fig. 4B and D). In these cells, RASCAL expression was detected starting from 24 hpi, and signal accumulated at the nuclear rim, on the nuclear envelope, and in cytoplasmic vesicles at late times p.i. (Fig. 4A and C). In contrast, no RASCAL-specific signal was detected in PFA-treated cells at each time p.i. (Fig. 4E and G and data not shown), suggesting that expression of the RASCAL protein was reduced to levels below detection in the absence of viral DNA replication.
FIG. 4.
RASCAL expression in the absence of viral DNA synthesis. HF infected with Towne/GFP-IE2 (MOI of 3) in the presence or absence of PFA (300 μg/ml) were harvested at the indicated times p.i. and stained with affinity-purified rabbit anti-RASCAL polyclonal Abs followed by Alexa Fluor 594-conjugated goat anti-rabbit Abs and FITC-conjugated anti IE1/IE2 Abs. Original magnification, ×400.
RASCAL and lamin B colocalize at the nuclear lamina and in cytoplasmic vesicular structures.
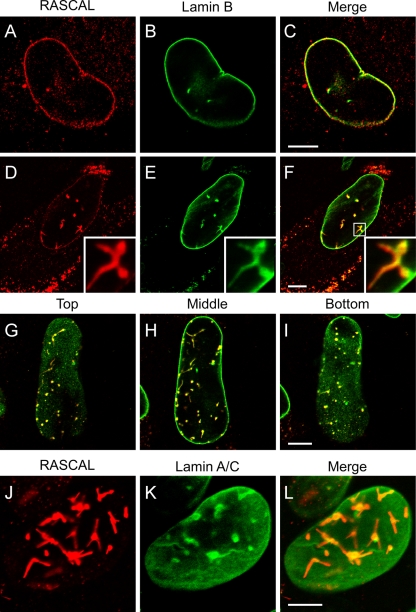
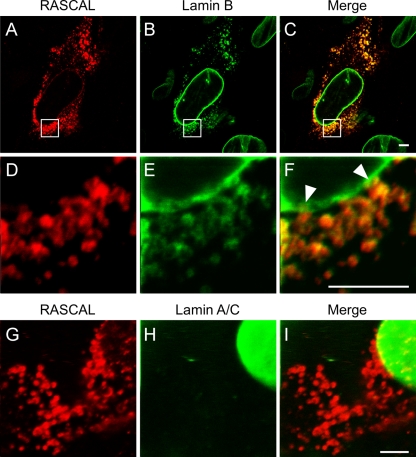
To establish if RASCAL was located at the nuclear lamina, confocal microscopy images were acquired of TB40/E-infected HF harvested at 72 and 96 hpi and stained for RASCAL and lamin B or for RASCAL and lamin A/C. TB40/E was used in place of Towne/GFP-IE2 in these experiments to avoid interference between the GFP-IE2 nuclear fluorescence and the signal from Alexa Fluor 488-conjugated secondary Abs. At 72 hpi, a clear overlay of the RASCAL and lamin B signals was observed at the nuclear envelopes of infected cells (Fig. 5A to C). While the lamin B staining was uniformly distributed at the nuclear lamina (Fig. 5B), RASCAL appeared to accumulate in a more punctuate pattern (Fig. 5A). RASCAL and lamin B colocalization was also clearly detected in the structures observed in the nuclei of infected cells at late times p.i. (Fig. 5D to F). These structures were also observed in confocal images corresponding to the midsections of nuclei, suggesting that they might correspond to the extensive invaginations of the inner nuclear membrane (INM) described in ultrastructural studies and in immunofluorescence staining analyses of human- and mouse-CMV-infected cells at late times p.i. (6, 7, 14, 24, 35, 50, 62, 67). To determine if this was the case, images were acquired at three different focal planes of a single nucleus, corresponding to the top (Fig. 5G), middle (Fig. 5H), and bottom (Fig. 5I) sections of the nucleus. Several of the RASCAL- and lamin B-positive structures appeared to stretch from the top to the bottom of the nuclear volume (Fig. 5G to I), indicating that they did indeed represent deep, tunnel-like folds of the nuclear lamina that penetrate through the entire volume of the nucleus. Extensive overlap was also detected between the RASCAL and lamin A/C signals, although the majority of the lamin A/C signal appeared to emanate from the central “knot” of the invaginations and to be less intense along the rod-like extensions (Fig. 5J to L). Confocal images of infected cells at 96 hpi also revealed that both RASCAL and lamin B (Fig. 6A to C), but not lamin A/C (Fig. 6D to F), were present in the cytoplasmic vesicle-like compartments appearing at late times p.i., suggesting that these proteins might remain associated even when not localizing at the nuclear lamina. Higher-magnification images of the nuclear rim uncovered the existence of close connections between the vesicles and the nuclear envelope, suggesting that these cytoplasmic structures may originate from the nuclear membrane (Fig. 6G to I, arrowheads).
FIG. 5.
RASCAL colocalization with lamin B and with lamin A/C at the nuclear envelopes of infected cells. Confocal images of TB40/E-infected HF (MOI of 5) harvested at 72 hpi (A to C) or 96 hpi (D to L) and stained with affinity-purified rabbit anti-RASCAL polyclonal Abs followed by Alexa Fluor 594-conjugated goat anti-rabbit Abs (red), with monoclonal anti-lamin B Abs followed by FITC-conjugated anti-mouse Abs (green), or with monoclonal anti-lamin A/C Abs followed by FITC-conjugated anti-mouse Abs (green). Bar size, 10 μm.
FIG. 6.
RASCAL colocalization with lamin B but not lamin A/C in cytoplasmic vesicles. Confocal images of TB40/E-infected HF (MOI of 5) harvested at 96 hpi and stained with affinity-purified rabbit anti-RASCAL polyclonal Abs followed by Alexa Fluor 594-conjugated goat anti-rabbit Abs (red), with monoclonal anti-lamin B Abs followed by FITC-conjugated anti-mouse Abs (green) (A to F), or with monoclonal anti-lamin A/C Abs followed by FITC-conjugated anti-mouse Abs (green) (G to I). The area magnified in panels D to F is framed by a square box in panels A to C. Arrowheads indicate the points of close contact between the cytoplasmic vesicles and the nuclear lamina. Bar size, 5 μm.
Identical RASCAL- and lamin B- or lamin A/C-positive structures were also observed in HF infected with Towne/GFP-IE2 or with AD169 (not shown).
RASCAL interacts with UL50 and requires its presence to gather at the nuclear lamina.
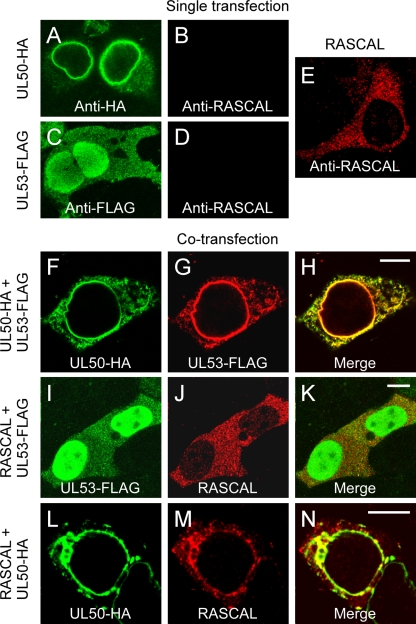
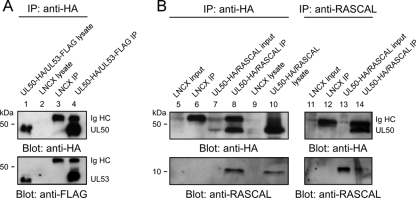
At early times p.i. (48 hpi), RASCAL localizes mainly in a cytoplasmic punctuate pattern (Fig. 3D to F). Relocalization to the nuclear rim occurs only later in infection (72 hpi [Fig. 3G to I]), at a time when both UL50 and UL53 are abundantly expressed and gathered at the NEC. This suggested that RASCAL might require the presence of other NEC components to reach the nuclear lamina. To test if, akin to UL53, RASCAL relocalization to the nuclear lamina was mediated by UL50, three expression plasmids were constructed, one containing RASCALTB40/E, one containing a C-terminal FLAG-tagged version of UL53 (UL53-FLAG), and one containing a C-terminal HA-tagged version of UL50 (UL50-HA), as described previously (14, 40). HEK293T cells transfected with one of these constructs were stained with rabbit anti-RASCAL Abs and with mouse anti-HA or mouse anti-FLAG Abs. No signal was detected in cells expressing UL50-HA or UL53-FLAG after being stained with anti-RASCAL Abs (Fig. 7B and D), indicating that these Abs did not cross-react with either protein. As previously shown (7, 41, 42), UL50-HA localized at the nuclear lamina and in cytoplasmic vesicular structures (Fig. 7A), while UL53-FLAG was predominantly nuclear, with some cytoplasmic dotted staining (Fig. 7C). When expressed in the absence of CMV infection, RASCALTB40/E did not accumulate at the nuclear rim but displayed a punctuate cytoplasmic staining similar, although not completely identical, to that observed in infected HF at 48 and 72 hpi (compare Fig. 7E to Fig. 3D and G). Coexpression of UL50-HA and UL53-FLAG induced the relocalization of UL53-FLAG to the endoplasmic reticulum and to the nuclear rim (Fig. 7F to H), as expected (7, 40). Expression of RASCALTB40/E in the presence of UL53-FLAG did not modify the localization of either protein (Fig. 7I to K), while coexpression of RASCALTB40/E and UL50-HA triggered the accumulation of RASCALTB40/E in a UL50-positive, perinuclear region (Fig. 7L to N). These data indicate that the presence of UL50, but not of UL53, is required to mediate RASCALTB40/E tethering to the nuclear rim and suggest that RASCALTB40/E may interact with UL50. Expression of RASCALTB40/E in the presence of both UL50-HA and UL53-FLAG did not change the localization pattern of either RASCALTB40/E or UL53, and both proteins still accumulated at the nuclear lamina together with UL50 (not shown), suggesting that UL50 may bind to RASCALTB40/E and UL53 via two different interaction domains. To investigate if RASCAL did indeed interact with UL50, protein extracts from HEK293T cells transfected with LNCX or cotransfected with L-RASCALTB40/E and LNCX UL50-HA were subjected to co-IP using anti-HA or anti-RASCAL Abs. As a control, extracts from HEK293T cells transfected with LNCX or cotransfected with LNCX UL50-HA and L-UL53-FLAG were subjected to co-IP using anti-HA Abs. As expected (7, 40, 41, 65), UL53-FLAG did coimmunoprecipitate with UL50-HA (Fig. 8A, lane 4), while no signal was observed in immunoprecipitates from control cells (Fig. 8A, lane 3). Specific bands of about 44 kDa (UL50-HA) and 10 kDa (RASCAL) were detected in cell lysates (Fig. 8B, left, lane 10) and in immunoprecipitates (Fig. 8B, left, lane 8, and right, lane 14) from L-RASCALTB40/E- and LNCX UL50-HA-cotransfected cells, irrespective of whether anti-HA or anti-RASCAL Abs were used as immunoprecipitating reagents. No bands were detected in extracts from control cells (Fig. 8B, left, lanes 5, 6, and 9, and right, lanes 11 and 12). Together, these data indicate that RASCAL and UL50 can be found in the same complex and suggest that RASCAL is likely to be a new NEC member.
FIG. 7.
Localization of UL50-HA, UL53-FLAG, and RASCALTB40/E in transfected cells. (A to D) Confocal images of HEK293T transiently transfected with expression plasmids encoding UL50-HA (A and B) or UL53-FLAG (C and D) and stained with mouse anti-HA Ab (A), mouse anti-FLAG Abs (C), or affinity-purified rabbit anti-RASCAL polyclonal Abs (B and D) followed by FITC-conjugated goat anti-mouse Abs (green) or Alexa Fluor 594-conjugated goat anti-rabbit Abs. (E to M) Confocal images of HEK293T coexpressing UL50-HA and UL53-FLAG (E to F), UL53-FLAG and RASCALTB40/E (H to J), or UL50-HA and RASCALTB40/E (K to M). Cells were stained with rabbit anti-HA and mouse anti-FLAG Abs followed by Alexa Fluor 594 goat anti-rabbit Abs (green) and FITC-conjugated goat anti-mouse Abs (red) (E to G), with mouse anti-FLAG and rabbit anti-RASCAL Abs followed by FITC-conjugated goat anti-mouse Abs (green) and Alexa Fluor 594-conjugated goat anti-rabbit Ab (red) (H to J), or with mouse anti-HA and rabbit anti-RASCAL Abs followed by FITC-conjugated goat anti-mouse Abs (green) and Alexa Fluor 594-conjugated goat anti-rabbit Abs (red) (K to M). Bar size, 10 μm.
FIG. 8.
Determination of RASCAL's interaction with UL50 by co-IP. (A) Immunoblot analysis of protein extracts from HEK293T cells transiently transfected with LNCX or cotransfected with L-RASCALTB40/E and LNCX UL50-HA. Proteins were denatured in 3% SDS lysis buffer (lysate) or were subjected to co-IP with anti-HA Abs (left) or with anti-RASCAL Abs (right) prior to separation on 10% (UL50-HA) or 15% (RASCALTB40/E) SDS-PAGE gels. An aliquot corresponding to 2% of the original co-IP buffer volume was loaded onto the gels as the input control (input). Membranes were probed with anti-HA (1:500) or anti-RASCAL (1:1,000) Abs. (B) Immunoblot analysis of protein extracts from HEK293T cells transiently transfected with LNCX or cotransfected with LNCX UL50-HA and L-UL53-FLAG. Proteins were denatured in 3% SDS lysis buffer (lysate) or were subjected to co-IP with anti-HA Abs, prior to separation on 10% SDS-PAGE gels. Membranes were probed with anti-HA (1:500) or anti-FLAG (1:500) Abs. Expected molecular masses were 43.9 kDa for UL50-HA, 10.6 kDa for RASCALTB40/E, and 43.3 kDa for UL53-FLAG.
DISCUSSION
In this report, we show that the putative CMV gene c-ORF29 encodes a protein, RASCAL, displaying a dual localization to the nuclear lamina and to cytoplasmic vesicles at late times during infection. Accumulation of RASCAL at the nuclear lamina requires the presence of UL50, and both proteins engage in direct or indirect interactions, indicating that RASCAL is likely to be a new component of the NEC. The occurrence of lamin B-positive vesicular structures in the cytoplasm of infected cells at late times p.i. has been previously reported (42, 47, 66), but the nature and function of these vesicles has not yet been determined. The presence of RASCAL and lamin B within these structures suggests that they may originate from the nuclear envelope. We thus speculate that RASCAL may play a role in facilitating the transition of nucleocapsids across the nuclear envelope and may have additional functions in promoting virion maturation and trafficking toward the plasma membrane.
Akin to all herpesviruses, CMV genome replication and encapsidation occur in the nuclei of host cells (45). Release of newly assembled nucleocapsids into the cytoplasm necessarily entails crossing of the nuclear envelope, in a process presumed to involve an initial envelopment of nucleocapsids by budding through the INM, followed by a de-envelopment step as virions traverse the outer nuclear membrane (ONM) (34, 37, 38, 50, 67). The INM is structurally supported by the nuclear lamina, a dense fibrillar network composed of A- and B-type lamins and their partners (57). Localized destabilization of this structure is required for virions to gain access to the INM and is mediated by components of the NEC, a multiprotein complex composed of the viral proteins UL50, UL53, and UL97 and of the cellular proteins p32, lamin B receptor (LBR), and protein kinase C (PKC) (41). UL50 is a key mediator of the NEC formation and directly interacts with UL53, p32, and PKC (41). Recruitment of UL53 and PKC to the nuclear lamina is strictly dependent on the presence of UL50, while p32 can reach this location also via interactions with LBR (7, 40, 41). UL50 also acts to enhance the accumulation at the NEC of the viral kinase UL97, although UL97 recruitment is largely mediated by binding to p32 (35).
Similar to UL53 and PKC, RASCAL localization to the nuclear rim is dependent on UL50 (Fig. 7L to N). RASCAL and UL50 could also be coimmunoprecipitated (Fig. 8B), indicating the existence of direct or indirect interactions between the two proteins. Thus, RASCAL may either be a new UL50 binding partner or may be recruited to the NEC via interactions with PKC and/or p32.
Both RASCAL and UL53 were able to reach the nuclear lamina when coexpressed in the presence of UL50 (not shown), indicating a lack of competition between these two proteins for binding to UL50. Thus, if the RASCAL-to-UL50 link is direct, two distinct binding domains must exist on the surface of UL50, one for RASCAL and one for UL53. Although our data do not exclude the possibility of an interaction between RASCAL and UL53, complete overlap of signals emanating from each of these proteins was not observed in the cytoplasm of cotransfected cells, and expression of UL53-FLAG did not result in the relocation of RASCAL to the nucleus (Fig. 7I to K). We thus believe that the formation of RASCAL-UL53 complexes is unlikely to occur.
Quite interestingly, no differences were observed in the intracellular localization of RASCALTB40/E and RASCALTowne during infection, despite the fact that the RASCAL version encoded by Towne is substantially longer than that encoded by TB40/E (Fig. 1 and 2). As both proteins accumulate at the NEC, the domains required for this localization must be located within the first 97 aa of RASCAL, while the presence of the 79 additional amino acids in RASCALTowne does not contribute to, or interfere with, the recruitment process. We thus speculate that the two proteins, although similarly localized, will prove to be functionally different and that these differences will be dependent on their ability to interact with specific cellular and/or viral proteins in addition to the components of the NEC. Studies are currently in progress to identify the exact domains required for the tethering of RASCALTB40/E and RASCALTowne to the nuclear lamina and to isolate the potential binding partners of each protein.
Extensive overlap of the RASCAL and lamin B signals was observed at the nuclear rim and in intranuclear invaginations of the INM in TB40/E-, Towne/GFP-IE2-, or AD169-infected HF (Fig. 5A to I and data not shown). Substantial infoldings of the INM have been described in a number of ultrastructural studies of CMV-infected cells, and the presence of UL50, UL53, and UL97 at these sites was documented by immunoelectron and immunofluorescence microscopy (6, 7, 14, 24, 35, 50, 62, 67). These invaginations were shown to be sites of nucleocapsid budding across the INM and were proposed to enhance the efficiency of virion egress from the nucleus by increasing the surface area of the INM and by acting as channels for the unhindered transport of primary enveloped virions toward the ONM (6). Confocal microscopy imaging of cells stained for RASCAL and lamin B clearly showed that both these proteins are located at sites found deep within the nuclear volume (Fig. 5D to I). In contrast, lamin A/C colocalization with RASCAL was less prominently in the nuclear invaginations and more evident at the openings of the INM channels (Fig. 5J to L). Although these staining pattern differences could potentially be due to some variation in the binding efficiency of each Ab, it is also conceivable that they might reflect functional differences between the two types of lamins (57). Together, these results strongly suggest that RASCAL is associated with the INM and provide further support to the hypothesis that RASCAL is a new component of the NEC.
Although expression of UL50 and UL53 was reported to be sufficient for the remodeling of the nuclear lamina and for the formation of INM invaginations in COS7 cells (7), the presence of nuclear foldings was not observed in HeLa (40) or in HEK293T (Fig. 7F to H) cells coexpressing these proteins. Expression of RASCALTB40/E in the presence of UL53 (Fig. 7I to K), UL50 (Fig. 7L to N), or both (not shown) also did not induce the formation of INM folds, indicating that RASCAL expression on its own, or in conjunction with UL50 and UL53, is not sufficient to trigger the development of these structures. These data suggest that the generation of INM invaginations in the absence of infection may be cell type dependent and may be affected by the intracellular content and availability of PKC, a kinase whose activity is required to enhance the disassembly of the nuclear lamina during mitosis (12) and during egress of herpes simplex virus type 1 (HSV-1) and of murine and human CMV (33, 40, 46, 47, 51). In addition to PKC, the viral kinase UL97 is recruited to the nuclear lamina (35) and substantially contributes to its phosphorylation-mediated dissolution (27, 35, 41, 42, 55). The presence of these two kinases at the NEC fostered the speculation that they might phosphorylate specific NEC components. Although no significant posttranslational modification was detected for UL53 in infected HF (14), phosphorylation of both UL50 and UL53 was reported to occur after in vivo labeling of transfected cells with 33P (40), and phosphorylation of UL50 was shown to be mediated by PKC (40).
We did not observe substantial changes in the SDS-PAGE mobility of RASCALTB40/E from either infected or transfected cells (Fig. 2C and D), despite the predicted presence of potential phosphorylation sites for both PKC and PKA (Fig. 1B). Protein phosphorylation, however, has been reported to increase, decrease, or leave unaffected the apparent molecular weights of proteins separated on SDS-PAGE gels. It is thus conceivable that both RASCALTB40/E and RASCALTowne might indeed be phosphorylated. The potential presence of this modification might explain the appearance of a faster-migrating band in protein extracts from Towne-infected cells (Fig. 2D), particularly considering that the amino acid sequence of RASCALTowne is predicted to contain two additional phosphorylation sites compared to that of RASCALTB40/E. Further analyses are in progress to clarify this issue.
While expression of UL53 was reported to occur with late kinetics (14), c-ORF29 transcription was observed starting from 6 hpi (Fig. 2A), and its protein product was detected at low levels starting from 24 hpi, with signal intensities markedly increasing at later times (Fig. 3). These data suggest that RASCAL accumulation in infected cells may begin before synthesis of UL53 and, possibly, of UL50, while RASCAL gathering at the nuclear rim occurs exclusively at late times p.i., when both UL50 and UL53 are present within the NEC. c-ORF29 transcription was reduced, but not completely abolished, in the absence of viral DNA replication (Fig. 2B), while signal from the protein product of c-ORF29 became completely undetectable by immunofluorescence staining analysis of PFA-treated cells (Fig. 4). These results suggest that, as with the synthesis of another NEC component, UL97, RASCAL is expressed with early-late kinetics, requiring viral DNA replication for maximum expression (39).
As previously mentioned, the c-ORF29 and US17 nucleotide sequences partially overlap. Although the kinetics of US17 gene expression have not been studied in detail, the presence of early transcripts hybridizing to the US17 gene region was detected in microarray studies of viral gene expression in Towne-infected HF (9). Interestingly, however, synthesis of the US17 protein was observed starting from 72 hpi and was entirely inhibited by treatment with PFA (15). These data suggest that, although both the US17 and c-ORF29 genes are located on the negative strand of the genome and have partially overlapping sequences, their transcription may be controlled by different promoters.
While expression of both UL50 and UL53 is absolutely necessary for the production of viral progeny from infected cells (18, 74), growth of two distinct US17 deletion mutant viruses in HF was not impaired (18, 74). As the c-ORF29 and US17 nucleotide sequences extensively overlap, it is likely that deletion of c-ORF29 will also not lead to dramatic reductions in mutant virus yields, at least in HF. Although the absence of RASCAL expression might still affect the efficiency of virion egress in HF, we expect that more dramatic effects may be observed in cell types other than HF.
Substantial overlap of the RASCAL and lamin B, but not lamin A/C, signals was observed in cytoplasmic vesicles distributed from the nucleus to the cytoplasm of infected HF at late times p.i. (Fig. 3J, M, and P, 4A and C, and 6A to F). Although the existence of similar vesicles has been mentioned in previous analyses of human- or murine-CMV-infected cells stained for lamin B (47, 66), their origin and nature has not yet been established and is the subject of ongoing investigations. Our confocal microscopy images indicate that they are likely to originate from the nuclear envelope (Fig. 6), an assumption further reinforced by the fact that they do appear to contain lamin B. It is currently unclear, however, whether they consist of a single or of a double membrane and if they are derived from the INM, the ONM, or both. As these structures have been noticed before in cells expressing exogenous UL50 (7), it is possible that they may contain additional NEC components and possibly even nucleocapsids. If so, they may constitute a completely novel, alternative route of virion maturation and transport from the nucleus to the cell surface. Alternatively, they may represent sites of viral particle degradation, similar to the nuclear envelope-derived, four-layered structures described in the cytoplasm of HSV-1-infected HF (1), which were recently shown to be autophagosomes (21).
The presence of RASCAL at these sites raises the intriguing possibility that RASCAL might remain associated with virions during egress, possibly becoming a tegument component. UL50 was reported to be an envelope glycoprotein in proteomic analyses of the CMV particles (70), and UL53 was shown by immunoelectron microscopy to be a tegument protein (14), although this localization was not confirmed by the proteomic study (70). It is thus conceivable that RASCAL may accompany newly formed nucleocapsids during their journey from the nucleus to the periphery, perhaps by triggering the formation of nucleus-derived vesicles. If so, RASCAL may indeed function as a new determinant of viral tropism by enhancing virion egress from specific cell types or, if included in the tegument, by promoting entry of viral particles into uninfected cells.
Acknowledgments
This investigation was supported by operating grants to L.H. from the Canadian Institutes of Health Research and from the Natural Sciences and Engineering Research Council.
We are grateful to J. S. Mymryk and G. A. Dekaban for critical reading of the manuscript.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Alexander, D. E., S. L. Ward, N. Mizushima, B. Levine, and D. A. Leib. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81:12128-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bhasin, M., and G. P. S. Raghava. 2004. ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 32:W414-W419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom, N., T. Sicheritz-Ponten, R. Gupta, S. Gammeltoft, and S. Brunak. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633-1649. [DOI] [PubMed] [Google Scholar]
- 5.Britt, W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417-470. [DOI] [PubMed] [Google Scholar]
- 6.Buser, C., P. Walther, T. Mertens, and D. Michel. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camozzi, D., S. Pignatelli, C. Valvo, G. Lattanzi, C. Capanni, P. Dal Monte, and M. P. Landini. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89:731-740. [DOI] [PubMed] [Google Scholar]
- 8.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 11.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 12.Collas, P., L. Thompson, A. P. Fields, D. L. Poccia, and J. C. Courvalin. 1997. Protein kinase C-mediated interphase lamin B phosphorylation and solubilization. J. Biol. Chem. 272:21274-21280. [DOI] [PubMed] [Google Scholar]
- 13.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 14.Dal Monte, P., S. Pignatelli, N. Zini, N. M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005-1012. [DOI] [PubMed] [Google Scholar]
- 15.Das, S., Y. Skomorovska-Prokvolit, F.-Z. Wang, and P. E. Pellett. 2006. Infection-dependent nuclear localization of US17, a member of the US12 family of human cytomegalovirus-encoded seven-transmembrane proteins. J. Virol. 80:1191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 17.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 18.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elek, S. D., and H. Stern. 1974. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet 1:1-5. [DOI] [PubMed] [Google Scholar]
- 20.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 21.English, L., M. Chemali, and M. Desjardins. 2009. Nuclear membrane-derived autophagy, a novel process that participates in the presentation of endogenous viral antigens during HSV-1 infection. Autophagy 5:1026-1029. [DOI] [PubMed] [Google Scholar]
- 22.Garg, A., M. Bhasin, and G. P. S. Raghava. 2005. Support vector machine-based method for subcellular localization of human proteins using amino acid compositions, their order, and similarity search. J. Biol. Chem. 280:14427-14432. [DOI] [PubMed] [Google Scholar]
- 23.Gerna, G., E. Percivalle, D. Lilleri, L. Lozza, C. Fornara, G. Hahn, F. Baldanti, and M. G. Revello. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86:275-284. [DOI] [PubMed] [Google Scholar]
- 24.Gilloteaux, J., and M. R. Nassiri. 2000. Human bone marrow fibroblasts infected by cytomegalovirus: ultrastructural observations. J. Submicrosc. Cytol. Pathol. 32:17-45. [PubMed] [Google Scholar]
- 25.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamirally, S., J. P. Kamil, Y. M. Ndassa-Colday, A. J. Lin, W. J. Jahng, M. C. Baek, S. Noton, L. A. Silva, M. Simpson-Holley, D. M. Knipe, D. E. Golan, J. A. Marto, and D. M. Coen. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 77:7563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua, S. J., and Z. R. Sun. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721-728. [DOI] [PubMed] [Google Scholar]
- 30.Isaacson, M. K., L. K. Juckem, and T. Compton. 2008. Virus entry and innate immune activation. Curr. Top. Microbiol. Immunol. 325:85-100. [DOI] [PubMed] [Google Scholar]
- 31.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 32.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 33.Leach, N., S. L. Bjerke, D. K. Christensen, J. M. Bouchard, F. Mou, R. Park, J. Baines, T. Haraguchi, and R. J. Roller. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81:10792-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, C. P., and M. R. Chen. 2010. Escape of herpesviruses from the nucleus. Rev. Med. Virol. [Epub ahead of print.] doi: 10.1002/rmv.643. [DOI] [PubMed]
- 35.Marschall, M., A. Marzi, P. aus dem Siepen, R. Jochmann, M. Kalmer, S. Auerochs, P. Lischka, M. Leis, and T. Stamminger. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357-33367. [DOI] [PubMed] [Google Scholar]
- 36.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2009. Herpesvirus assembly: an update. Virus Res. 143:222-234. [DOI] [PubMed] [Google Scholar]
- 39.Michel, D., I. Pavic, A. Zimmermann, E. Haupt, K. Wunderlich, M. Heuschmid, and T. Mertens. 1996. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J. Virol. 70:6340-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milbradt, J., S. Auerochs, and M. Marschall. 2007. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 88:2642-2650. [DOI] [PubMed] [Google Scholar]
- 41.Milbradt, J., S. Auerochs, H. Sticht, and M. Marschall. 2009. Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90:579-590. [DOI] [PubMed] [Google Scholar]
- 42.Milbradt, J., R. Webel, S. Auerochs, H. Sticht, and M. Marschall. 2010. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. [Epub ahead of print.] doi: 10.1074/jbc.M109.063628. [DOI] [PMC free article] [PubMed]
- 43.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7:980-990. [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, M., and L. Hertel. 2009. Onset of human cytomegalovirus replication in fibroblasts requires the presence of an intact vimentin cytoskeleton. J. Virol. 83:7015-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In A. P. M. Howley and D. M. Knipe (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 46.Morris, J. B., H. Hofemeister, and P. O'Hare. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 48.Murphy, E., and T. Shenk. 2008. Human cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 325:1-19. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadimitriou, J. M., G. R. Shellam, and T. A. Robertson. 1984. An ultrastructural investigation of cytomegalovirus replication in murine hepatocytes. J. Gen. Virol. 65(Pt. 11):1979-1990. [DOI] [PubMed] [Google Scholar]
- 51.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 53.Plotkin, S. A., T. Furukawa, N. Zygraich, and C. Huygelen. 1975. Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 12:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plotkin, S. A., S. E. Starr, H. M. Friedman, E. Gonczol, and R. E. Weibel. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860-865. [DOI] [PubMed] [Google Scholar]
- 55.Prichard, M. N. 2009. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 19:215-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 57.Prokocimer, M., M. Davidovich, M. Nissim-Rafinia, N. Wiesel-Motiuk, D. Bar, R. Barkan, E. Meshorer, and Y. Gruenbaum. 2009. Nuclear lamins: key regulators of nuclear structure and activities. J. Cell. Mol. Med. [Epub ahead of print.] doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed]
- 58.Quinnan, G. V., Jr., M. Delery, A. H. Rook, W. R. Frederick, J. S. Epstein, J. F. Manischewitz, L. Jackson, K. M. Ramsey, K. Mittal, S. A. Plotkin, et al. 1984. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann. Intern. Med. 101:478-483. [DOI] [PubMed] [Google Scholar]
- 59.Revello, M. G., D. Lilleri, M. Zavattoni, M. Stronati, L. Bollani, J. M. Middeldorp, and G. Gerna. 2001. Human cytomegalovirus immediate-early messenger RNA in blood of pregnant women with primary infection and of congenitally infected newborns. J. Infect. Dis. 184:1078-1081. [DOI] [PubMed] [Google Scholar]
- 60.Riegler, S., H. Hebart, H. Einsele, P. Brossart, G. Jahn, and C. Sinzger. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81:393-399. [DOI] [PubMed] [Google Scholar]
- 61.Rowe, W. P., J. W. Hartley, S. Waterman, H. C. Turner, and R. J. Huebner. 1956. Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc. Soc. Exp. Biol. Med. 92:418-424. [PubMed] [Google Scholar]
- 62.Ruebner, B. H., K. Miyai, R. J. Slusser, P. Wedemeyer, and D. N. Medearis, Jr. 1964. Mouse cytomegalovirus infection. An electron microscopic study of hepatic parenchymal cells. Am. J. Pathol. 44:799-821. [PMC free article] [PubMed] [Google Scholar]
- 63.Ryckman, B. J., M. A. Jarvis, D. D. Drummond, J. A. Nelson, and D. C. Johnson. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryckman, B. J., B. L. Rainish, M. C. Chase, J. A. Borton, J. A. Nelson, M. A. Jarvis, and D. C. Johnson. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sam, M. D., B. T. Evans, D. M. Coen, and J. M. Hogle. 2009. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez, V., P. C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 68.Sinzger, C., M. Digel, and G. Jahn. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325:63-83. [DOI] [PubMed] [Google Scholar]
- 69.Sinzger, C., G. Hahn, M. Digel, R. Katona, K. L. Sampaio, M. Messerle, H. Hengel, U. Koszinowski, W. Brune, and B. Adler. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359-368. [DOI] [PubMed] [Google Scholar]
- 70.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waldman, W. J., W. H. Roberts, D. H. Davis, M. V. Williams, D. D. Sedmak, and R. E. Stephens. 1991. Preservation of natural endothelial cytopathogenicity of cytomegalovirus by propagation in endothelial cells. Arch. Virol. 117:143-164. [DOI] [PubMed] [Google Scholar]
- 72.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]