Abstract
The efficient release of newly assembled retrovirus particles from the plasma membrane requires the recruitment of a network of cellular proteins (ESCRT machinery) normally involved in the biogenesis of multivesicular bodies and in cytokinesis. Retroviruses and other enveloped viruses recruit the ESCRT machinery through three classes of short amino acid consensus sequences termed late domains: PT/SAP, PPXY, and LYPXnL. The major late domain of Rous sarcoma virus (RSV) has been mapped to a PPPY motif in Gag that binds members of the Nedd4 family of ubiquitin ligases. RSV Gag also contains a second putative late domain motif, LYPSL, positioned 5 amino acids downstream of PPPY. LYPXnL motifs have been shown to support budding in other retroviruses by binding the ESCRT adaptor protein Alix. To investigate a possible role of the LYPSL motif in RSV budding, we constructed PPPY and LYPSL mutants in the context of an infectious virus and then analyzed the budding rates, spreading profiles, and budding morphology. The data imply that the LYPSL motif acts as a secondary late domain and that its role in budding is amplified in the absence of a fully functional PPPY motif. The LYPXL motif proved to be a stronger late domain when an aspartic acid was substituted for the native serine, recapitulating the properties of the LYPDL late domain of equine infectious anemia virus. The overexpression of human Alix in the absence of a fully functional PPPY late domain partially rescued both the viral budding rate and viral replication, supporting a model in which the RSV LYPSL motif mediates budding through an interaction with the ESCRT adaptor protein Alix.
Retroviruses acquire their lipid envelopes from the plasma membrane as they bud from the cell. Although the structural protein Gag is both necessary and sufficient for the assembly of virus-like particles (VLPs), the membrane scission step of virus egress requires the recruitment of a network of cellular proteins normally involved in two analogous cellular membrane fission events, the budding of cargo-containing vesicles into multivesicular bodies (MVBs) (for review, see references 1, 5, 11, and 50) and the separation of two daughter cells during cytokinesis (3, 4). This cellular network of proteins, collectively called the ESCRT (endosomal sorting complex required for transport) machinery, includes four sequentially recruited high-molecular-weight protein complexes (ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III) and is essential for the transport of transmembrane cargo proteins to the lysosome for degradation via an MVB intermediate.
In addition to the multiprotein ESCRT complexes, other proteins are required to promote the budding of vesicles into the MVB. Ubiquitin ligases (such as Nedd4) monoubiquitinate both ESCRT components and transmembrane cargo proteins, tagging them for the MVB pathway. Adaptor proteins connect cargo proteins to ESCRT complexes or ESCRT complexes to each other. Ultimately, the final membrane fission event of vesicle budding is mediated by an AAA ATPase (Vps4).
Retroviruses as well as other enveloped viruses use three amino acid consensus sequences, PPXY, PT/SAP, and LYPXnL, as docking sites for the components of the cellular ESCRT machinery. The deletion or mutation of these sequences, termed late domains, results in the failure of the virus to recruit the budding machinery to the site of assembly and thereby results in a block at the late stage of virus release in which fully assembled but immature virus particles remain attached to the plasma membrane. The PPXY late domain interacts with the WW domains of the Nedd4 family of ubiquitin ligases. Multiple ESCRT components bind to monoubiquitin tags on both cargo and ESCRT proteins. The PT/SAP late domain binds the ESCRT-I complex component, Tsg101 (tumor susceptibility gene 101). The LYPXnL late domain interacts with an adaptor protein of the ESCRT pathway, Alix (ALG-2-interacting protein X; also called AIP1) (reviewed in reference 12). Alix interacts with both Tsg101 of the ESCRT-I complex and CMHP4 of the ESCRT-III complex. A possible fourth class of late domains for the paramyxovirus SV5 was reported previously (47). The late domain function in this case has been mapped to an FPIV sequence in the M (matrix) protein. To date, this motif has yet to be shown to be important for the budding of any other virus, and an FPIV-interacting cellular protein has yet to be identified.
Often, retroviruses rely on multiple late domains for efficient budding (2, 13, 16, 29, 30). For example, in addition to its PT/SAP motif in human immunodeficiency virus type 1 (HIV-1) p6, which binds Tsg101 (6, 14, 34, 52), HIV-1 also harbors an Alix-binding LYPXnL motif that functions in budding (13, 33, 34, 48, 52). Mutation of this LYPXnL motif results in only a modest reduction in HIV-1 budding (10). However, the effects of mutations in the LYPXnL motif become more obvious in the context of a minimal Gag in which the globular domain of MA and the N-terminal domain of CA are absent (48). Furthermore, the role of this motif also seems to vary among cell types. For example, the deletion of this motif decreases HIV-1 particle production 2- to 3-fold in COS-7 cells (15) but has no consequence for HeLa cells (7). The relationship of the two viral late domains to each other is unknown. It is possible that they are partially redundant, are cooperative (since they act at slightly different steps in the ESCRT pathway), or are cell type specific. It has been observed that the mutation of one late domain has a larger effect on budding than the mutation of the other, implying a hierarchy of function. For example, in HIV-1, PTAP acts as the dominant late domain and LYPXnL acts as a secondary late domain. Equine infectious anemia virus (EIAV) seems to be an exception in that it relies only on a single LYPDL motif for late domain function.
Like other retroviruses, the avian alpharetrovirus Rous sarcoma virus (RSV) requires the ESCRT pathway for release, as evidenced by the observation that a dominant-negative mutant of the ATPase Vps4, which is required for the final step of the ESCRT pathway that releases the ESCRT-III complex, inhibits RSV budding in a dose-dependent manner (37). Mutational analysis mapped the RSV late domain to the PPPY motif in the small spacer peptide p2b of Gag (41, 54, 56). This PPPY motif was previously shown to interact with chicken members of the Nedd4 family of ubiquitin ligases (21, 51). RSV Gag also harbors an LYPSL late domain consensus motif 5 amino acids downstream from PPPY in the p10 domain, which could potentially promote budding via an interaction with Alix.
Alix, a 97-kDa adaptor protein with diverse functions, is composed of an N-terminal Bro1 domain, a central V domain, and a C-terminal proline-rich region (10, 22, 26, 58). The proline-rich region is assumed to be unstructured and binds Tsg101 and endophilins. The Bro1 domain, which binds CHMP4, is curved and resembles a banana shape. CHMP4 binding is functionally important for promoting HIV-1 budding (10). It was suggested previously that its convex face may allow Alix to sense negative curvatures in membranes (17, 22). At least for HIV-1, the Alix Bro1 domain also interacts with the Gag NC domain (42, 43). The central V domain of Alix, which is named for its shape, has a conserved hydrophobic pocket on the second arm near the apex of the V that is responsible for the binding of the LYPXnL late domains of HIV-1 and EIAV (10, 26, 58).
In the present study, we investigated the role of the LYPSL motif in RSV budding and replication. We report here that not only the PPPY motif but also the LYPSL motif act as late domains. The contribution of the LYPSL motif to the budding rate and spreading rate is secondary to that of the PPPY motif but increases in the absence of a fully functional PPPY motif. The Alix overexpression-mediated rescue of PPPY mutants supports a model in which the LYPSL late domain functions through an interaction with Alix.
MATERIALS AND METHODS
Plasmids.
The RSV proviral constructs were derived from the RCAS vector system (9) and contained a green fluorescent protein (GFP) marker expressed as a separate message in place of v-src. Mutations in p2 and p10 of Gag were introduced by two-step PCR. The PCR product was digested with SacI and SacII, resulting in a 1,547-bp fragment that was inserted between a unique SacI site (nucleotide 261 of RCAS) located 110 nucleotides upstream of the start of the gag gene and a unique SacII site (nucleotide 1807 of RCAS) located in the sequence that encodes the C terminus of the CA domain of Gag. Sequences of all PCR-derived regions of the constructs were verified by DNA sequencing (Cornell University Life Sciences Core Laboratories Center).
Cell culture.
The ev-0-derived chicken fibroblast cell line DF1 (ATCC CRL 12203) was grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 5% fetal bovine serum (Gemini Bio-Products), 5% NuSerum (BD Biosciences), 2% heat-inactivated chicken serum (Invitrogen), 1% glutamine (Invitrogen), and 1% vitamins (Invitrogen). Transfection was achieved with Fugene 6 or Fugene HD (Roche) according to the manufacturer's recommendations. Infection was initiated by filtering medium from cells transfected with proviral DNA through 0.45-μm syringe filters (Pall) and applying it to virgin cells.
Measurement of virus budding kinetics.
Measurement of virus budding kinetics was carried out by the transfection of DF1 cells with either wild-type (WT) or mutant proviral RSV DNA and then allowing the viral infection to spread. The spread of infection was confirmed by the detection of GFP by fluorescence microscopy or flow cytometry. Infected DF1 cells growing on 60-mm plates were labeled with 100 to 300 μCi [35S]methionine for 30 min (pulse). The radioactive medium was then replaced by unlabeled medium (chase). Virus-containing medium was collected serially at various time points, with prewarmed medium being added to the cells after each collection. Debris in the virus-containing medium was cleared by low-speed centrifugation at 5,000 rpm for 5 min, and the virus was then collected by centrifugation for 20 min at 70,000 rpm in a TLA 100.4 rotor (Beckman). Virus pellets were dissolved in SDS sample buffer, and the viral proteins were resolved by SDS-PAGE. Virus release over time was measured by quantitating the band corresponding to CA at each time point using ImageQuant software (Molecular Dynamics). Budding half-time, or the time at which half of the final amount of labeled virus had been released, was calculated by using the equation Y = A × [1 − exp(−B × x)], where Y equals the CA signal from ImageQuant units and x equals time. The budding rate was ultimately expressed as the rate relative to that of the wild type.
Generation of an Alix-overexpressing DF1 cell line.
The HEK293-based packaging cell line Phoenix-ampho was cotransfected with the pQCXIP (Clontech) retroviral vector expressing human Alix (hAlix) and with a plasmid expressing the G protein of vesicular stomatitis virus. The medium was collected 1 to 2 days later and used to infect DF1 cells. A population of transduced hAlix-expressing DF1 cells was selected in the presence of 2 μg/ml puromycin. Control DF1 cells were established by transduction with the empty pQCXIP vector and also selected with puromycin.
Measurement of viral spread.
DF1 cells were transfected with wild-type or mutant proviral RSV DNA carrying a GFP marker. For most experiments, 1 day posttransfection and at approximately 80 to 100% confluence, the cells were trypsinized, diluted 1:10 with nontransfected cells, and reseeded onto fresh plates at a density to give approximately 30% confluence. For Alix rescue experiments, the nontransfected cells were either DF1 cells transduced to express human Alix or control DF1 cells transduced with the empty pQCXIP vector and thus expressing only the puromycin resistance gene. Trypsinized and washed cells were fixed for flow cytometry by incubation in 1% formaldehyde in phosphate-buffered saline (PBS) for 15 min.
SEM.
Scanning electron microscopy (SEM) analyses were performed as described previously (23). DF1 cells were plated onto glass coverslips containing a gold finder grid for cell identification and then transfected by using Fugene 6 (Roche) with wild-type or mutant RSV proviral DNA. Nineteen hours after transfection, cells were fixed in 4% paraformaldehyde and imaged by fluorescence microscopy to identify the transfected cells and assess relative transfection levels. The cells were then fixed in 2.5% glutaraldehyde, dehydrated, critical-point dried, coated with a thin layer of platinum, and imaged at the University of Missouri Electron Microscopy core facility with a Hitachi S4700 cold-cathode scanning electron microscope.
RESULTS
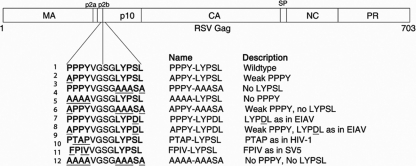
The PPPY motif in the p2b segment of RSV Gag was previously shown to play a critical role in budding (54) by engaging members of the Nedd4 family of E3 ubiquitin ligases (21). RSV Gag also possesses a putative, but as-yet-uncharacterized, LYPSL late domain motif in p10, 5 amino acids downstream from PPPY (Fig. 1). Since late domains often reside in close proximity to each other (2, 16, 18, 28, 34), and since the LYPXnL consensus sequences have been shown to be important for the release of other retroviruses (45, 48), we sought to determine the role of this LYPXnL sequence in RSV budding. Preliminary data from our laboratory (not shown) and another laboratory (21) failed to show a major role for this motif in RSV release. We hypothesized that this result is due to the presence of the strong primary late domain PPPY. To uncover the contribution of the LYPSL motif to budding, we weakened the primary PPPY late domain, as was done originally for murine leukemia virus (MLV) (31) and then also for HIV-1 (10, 49). To measure budding in the most physiologically relevant context and to avoid the expression inconsistency associated with transfection, various PPPY and LYPSL mutants were constructed in the context of an infectious proviral RSV DNA that carries a GFP marker (Fig. 1). All mutants proved to be infectious and able to spread at least slowly in DF1 chicken fibroblasts, as evidenced by Western blotting and by the detection of the GFP marker by fluorescence microscopy. Even a double knockout of the PPPY and LYPSL motifs (AAAA-AAASA) was weakly infectious.
FIG. 1.
Overview of late domain mutations within RSV Gag. Established and putative late domain positions are in boldface type, and mutated amino acids are underlined. The PPPY and LYPSL sequences are located in the p2b and p10 domains of the Gag polyprotein, respectively. Mutants were constructed in the context of infectious RSV proviral DNA carrying a GFP reporter gene. LYPDL, PTAP, and FPIV are exogenous late domains used by EIAV, HIV-1, and the paramyxovirus SV5, respectively. MA, matrix; CA, capsid; SP, spacer; NC, nucleocapsid; PR, protease.
The LYPSL motif functions in virus release and virus spread.
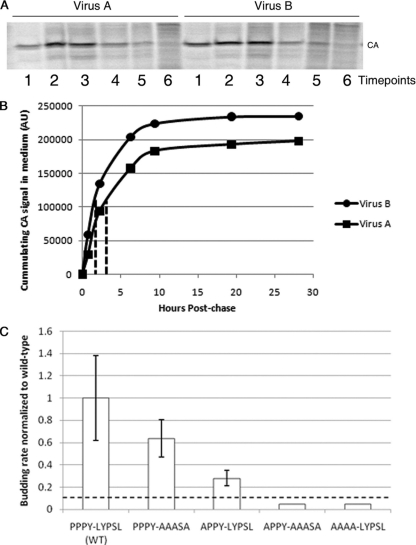
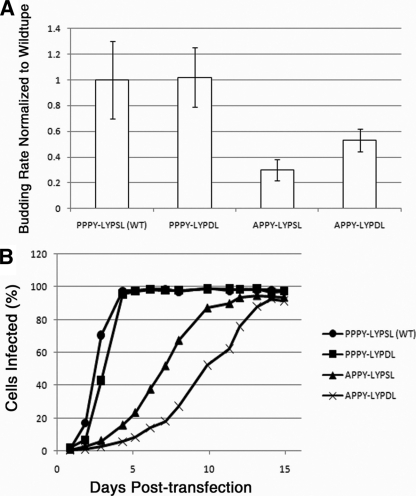
To examine the role of the LYPSL motif in RSV budding, we first mutated the LYPXL consensus motif to alanines, yielding the amino acid sequence AAASA in place of LYPSL (Fig. 1, line 3). Although YPXL is often considered the core consensus sequence of this motif, we chose to also mutate the preceding leucine residue because it was shown to contribute to Alix binding and is present just upstream of the YPXL motifs implicated in budding in other viruses (24, 38, 48). To measure the budding rate of this and other late domain mutants, DF1 chicken fibroblasts were transfected with proviral RSV DNA, and the infection was allowed to spread for at least 1 week. Infection of the majority of cells was confirmed by the detection of the GFP marker by flow cytometry or fluorescence microscopy. The infected cells were pulse-labeled with [35S]methionine, and virus-containing medium was collected serially approximately every hour. We chose this method, i.e., serial collection at several time points from the same plate of cells, to avoid inconsistencies among different plates of cells and to obtain several time points to increase the reliability of the data. However, a disadvantage of this method is that the amount of Gag left in the cells at each time point cannot be determined. The virus was collected by centrifugation, the viral proteins were resolved by SDS-PAGE, and the band corresponding to CA was quantitated by PhosphorImager analysis (typical examples are shown in Fig. 2A). Plotting of the cumulative amount of virus released over time (Fig. 2B) allowed us to calculate the time at which half of the total amount of labeled virus had been released from the cell (Fig. 2B), and this budding half-time was normalized to that of wild-type RSV.
FIG. 2.
Effects of mutation of the LYPSL motif on the budding rate. (A and B) Overview of the method used to measure budding rates of late domain mutants. (A) Example of SDS-PAGE PhosphorImager scans of the CA bands representing different time points for one of the mutants (virus A) (left) and for the wild type (virus B) (right). Each lane represents the virus collected from the medium at one time point, starting with the first collection after the pulse (lanes 1). (B) Example plots of cumulative virus release. The CA bands from the experiment shown in A were quantified, and the cumulative intensity was plotted as a function of the chase time. The half-time of virus release for these two curves is shown by the dotted lines. AU, arbitrary units. (C) Budding rates of late domain mutants. Budding half-times, or the time at which half of the final amount of labeled virus was released into the medium, were calculated based on curves like those in B and then normalized to the budding half-time of the wild type in the same experiment. The normalized budding rate on the y axis is the reciprocal of the budding half-time. The budding rates of the APPY-AAASA and AAAA-LYPSL mutants were lower than the detection level of this assay, represented by the dashed line, and thus remain undetermined. Error bars represent standard deviations from the means. The P values obtained from a Student's t test for the budding rates for the wild type and the PPPY-AAASA mutant and for the wild type and the APPY-LYPSL mutant were both significant (P < 0.05).
Mutation of the entire LYPSL consensus motif to alanines (PPPY-AAASA) led to less than a 2-fold reduction in the budding rate, which was only marginally statistically significant (Fig. 2C, compare bars 1 and 2 with error bars). In contrast, the mutation of only the first proline of the PPPY late domain, APPY-LYPSL, reduced the budding rate about 3-fold (Fig. 2C, compare bars 1 and 3). In the context of this partially weakened PPPY motif (APPY-LYPSL), budding became sensitive to mutations in LYPSL (Fig. 2C, compare bars 3 and 4), with a 10-fold reduction in the budding rate for the APPY-AAASA mutant compared with the wild type (Fig. 2C, compare bars 1 and 4). Thus, we conclude that the LYPSL sequence functions in RSV release, although this contribution to budding rate, as measured by pulse-chase, is fully exposed only when the primary PPPY late domain is compromised.
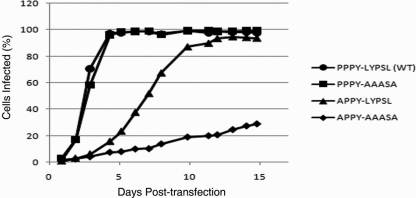
Presumably, a reduction in the rate of particle release should translate into a reduction in the rate of virus spread. As another way to assess viral release, we monitored the spread of the wild-type and mutant viruses using GFP fluorescence detection by flow cytometry. DF1 cells were transfected with proviral RSV DNA containing either wild-type or mutant PPPY and LYPSL motifs and carrying a GFP reporter. One day after transfection the cells were diluted with a 10-fold excess of nontransfected cells so that only about 1% of the cell population expressed GFP at that time. Sample populations of cells were then taken at daily intervals and fixed for analysis by flow cytometry. A representative set of spreading profiles is shown in Fig. 3. We found that the spreading rate of the mutant PPPY-AAASA was indistinguishable from that of the wild type (Fig. 3). Thus, the observed marginal reduction in the budding rate (Fig. 2C, bar 2), if it is indeed real, has no appreciable effect on the rate of virus spread under these conditions. However, the significant budding defect measured for the APPY-LYPSL mutant did translate into slower spreading (Fig. 3). Likewise, the APPY-AAASA mutant, which budded too slowly to be reliably measurable by metabolic labeling, spread more slowly still (Fig. 3). Although the per-particle infectivity of the late domain mutants in principle might be decreased due to incorrect or incomplete Gag processing or RNA splicing (see below), we found that, overall, the spreading profiles of the mutants described above reproducibly correlated with the measured budding rates. In summary, these findings illustrate a definite contribution of the LYPSL motif to RSV budding and spreading when the PPPY motif is weakened.
FIG. 3.
Effect of LYPSL mutations on the spread of infection. DF1 cells were transfected with proviral DNA harboring late domain mutations. At 1 day posttransfection, cells were diluted 1:10 with fresh cells, and infection was monitored at daily intervals by GFP detection via flow cytometry.
Overexpression of Alix rescues budding of PPPY mutants.
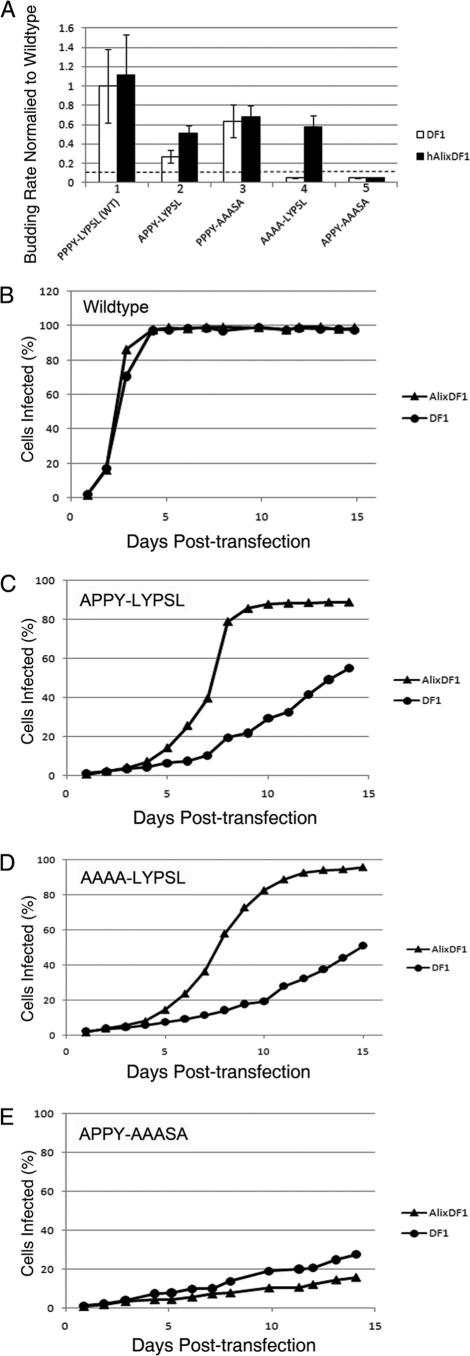
Since the LYPSL motif does contribute to RSV budding, we wanted to know if it acts by binding the protein Alix. If so, high-level expression of Alix might be able to rescue the budding and spreading rates of PPPY mutants, as was shown previously for HIV-1 (10, 49). To this end, we established a DF1 cell line that expresses hAlix at a level about 20-fold above that of endogenous Alix, as evidenced by Western blotting using an anti-hAlix rabbit serum (data not shown). Since the processes of transduction and selection might alter cell properties, control DF1 cells were transduced with an empty vector and thus express only the puromycin resistance gene.
Alix overexpression appeared to marginally increase the budding rate of the WT and APPY-LYPSL mutants, but this increase was not statistically significant (Fig. 4A, bar sets 1 and 3). However, Alix overexpression did significantly increase the budding rate of the PPPY mutant AAAA-LYPSL in a statistically significant manner (Fig. 4A, bar set 4). In contrast, an increase in the budding rate in Alix-overexpressing cells was not observed for the APPY-AAASA mutant in which the presumed Alix-binding LYPSL motif was absent (Fig. 4A, bar set 5). The budding rates of the APPY-AAASA mutant in both DF1 cells transduced with hAlix and in DF1 cells transduced with the control puromycin resistance gene were at least ∼20-fold lower than those of the wild type, which was below the level of detection in this budding assay. (Fig. 4A, bar set 5). This finding agrees with data from experiments showing that Alix overexpression failed to rescue the budding of HIV-1 ΔPTAP mutants if the Y and P residues of the LYPXnL motif were mutated (10, 49).
FIG. 4.
Partial rescue of the budding and spreading rates of the PPPY mutants by the overexpression of human Alix. (A) Comparison of the budding rates of the wild type and late domain mutants in DF1 control cells (white bars) or DF1 cells overexpressing human Alix (black bars). The P value obtained from a Student's t test for the budding rates of the APPY-LYPSL mutant in DF1 cells and AlixDF1 cells was significant (P < 0.05). The P value for the AAAA-LYPSL mutant was unable to be calculated since the budding rate in DF1 cells is below the sensitivity of the assay, indicated by the dashed line. (B to E) Spreading profiles of the wild type (B) and the APPY-LYPSL (C), AAAA-LYPSL (D), and APPY-AAASA (E) mutants were generated by flow cytometry.
The rescue of the budding rate by the overexpression of Alix correlated with an increase in the rate of spread in cultured cells (Fig. 4B and C). In replicate experiments, wild-type RSV spread at a similar or slightly higher rate in cells expressing Alix (Fig. 4B). The PPPY mutants APPY-LYPSL and AAAA-LYPSL, which retained the presumed Alix-binding site, showed a higher rate of spread in hAlix cells than in control DF1 cells (Fig. 4C and D). The overexpression of Alix failed to rescue the spreading rate of the PPPY mutant APPY-AAASA, which lacks the Alix-binding site (Fig. 4E). This mutant spread equally slowly in control cells and Alix cells. Taken together, these data show that high levels of Alix lead to an LYPSL-dependent rescue of the budding and spreading of PPPY mutants.
Heterologous late domains support RSV budding to various degrees.
Since late domains are modular protein-binding motifs and often retain their ability to promote virus budding when placed in different regions of Gag or in different viruses (27, 32, 40, 41, 56, 57), we investigated how well late domain motifs from other viruses could satisfy the requirements for RSV budding. Although others have examined the effects of exogenous late domains on RSV budding using assays involving the transient transfection of Gag constructs into COS-1 cells (41), we did so using infectious RSV in the more relevant DF1 chicken fibroblast cell line.
According to isothermal calorimetry assays, Alix binds the LYPDL-containing p9 sequence of EIAV more tightly than the LYPLTSL-containing p6 sequence of HIV-1 (10, 38, 58). To examine the possibility that the LYPDL sequence represents the optimal Alix-binding site, we inserted the LYPDL motif of EIAV in place of LYPSL. Although no difference in the budding rate was observed when the PPPY motif remained intact, when PPPY was handicapped by mutating it to APPY, the LYPDL motif rescued the budding rate better than did the native LYPSL (Fig. 5, compare the third and fourth bars). This finding correlates well with its reported higher Alix-binding affinities in vitro. Nevertheless, the wild-type RSV LYPSL motif allowed RSV to spread faster in cell culture in the context of the APPY than did the EIAV LYPDL motif (Fig. 5B). We interpret this result to mean that the wild-type sequence gives RSV a replication advantage, either because the lower-affinity binding of Alix is preferred or because some other aspect of this sequence, at the protein level or at the RNA level, is preferred.
FIG. 5.
Partial rescue of the budding and spreading rates of the PPPY mutant by the EIAV LYPDL motif. Shown are budding rates (A) and spreading profiles (B) of PPPY mutants in context of the native LYPSL motif or the EIAV LYPDL late domain. The P value obtained from a Student's t test for the APPY-LYPSL and the APPY-LYPDL mutants was significant (P < 0.05).
Although the absolute spreading rates of wild-type or a given mutant virus varied from experiment to experiment, depending on factors such as cell density and the frequency of passage, the relative spreading rates of the various viruses were invariably consistent over many experiments. Table 1 summarizes the budding rate rankings compared to the spreading rate rankings of the various late domain mutants. For all but one mutant, a lower budding rate translated into a lower spreading rate. The one exception was the APPY-LYPDL mutant. While the APPY-LYPDL mutant budded more quickly than the APPY-LYPSL mutant, it consistently spread more slowly than the APPY-LYPSL mutant as well as the AAAA-LYPSL mutant (Fig. 5B). Thus, this modest decrease in viral fitness must be due to factors other than the budding rate.
TABLE 1.
Comparison of budding and spreading rates in DF1 cells
| Mutant | Rank order of mutant |
|
|---|---|---|
| Buddinga | Spreadingb | |
| PPPY-LYPSL (WT) | 1 | 1 |
| PPPY-LYPDL | 2 | 2 |
| PPPY-AAASA | 3 | 3 |
| APPY-LYPDL | 4 | 6 |
| APPY-LYPSL | 5 | 4 |
| AAAA-LYPSL | NA | 5 |
| APPY-AASA | NA | 7 |
| AAAA-AAASA | NA | 8 |
The number represents the rank order for the rate of budding as measured by pulse-chase experiments like that shown in Fig. 2C. Although the raw data for the budding rate varied among experiments, the rank order was invariant for all experiments that included some or all of these mutants. NA, not applicable because the rate was too low to be measured by pulse-chase labeling.
The number represents the rank order for the rate of spreading as measured by flow cytometry in experiments like that shown in Fig. 3. Although the actual rate of spread varied among experiments, the rank order was invariant for all experiments that included some or all of these mutants.
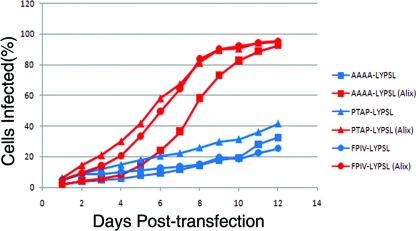
To further characterize the late domain requirements of RSV, we replaced the primary PPPY late domain with the heterologous late domain PTAP or FPIV. The PTAP sequence recruits Tsg101 and helps mediate budding for a variety of viruses, including Ebola virus (34), human T-cell leukemia virus type 1 (HTLV-1) (2, 53), and simian immunodeficiency virus (SIV) in addition to HIV-1. Earlier work with Gag expression vectors transiently transfected into COS-1 cells had shown that replacing the 12-amino-acid residues of RSV p2b (which includes PPPY but not LYPSL) with a 12-amino-acid sequence, including the HIV-1 PTAP but not the HIV-1 LYPXnL sequence, leads to a 3-fold reduction in budding (41). FPIV is a less-characterized late domain responsible for the release of the paramyxovirus SV5 (47). The cellular FPIV-binding factor remains to be identified.
To test whether RSV can rely on PTAP or FPIV in place of PPPY, we measured the budding and spreading rates of these mutants (Fig. 1, lines 8 and 9). Both 4-amino-acid motifs (without their native adjoining sequences) supported only an extremely low budding rate, below the sensitivity of pulse-chase analysis (data not shown). Both PTAP-LYPSL and FPIV-LYPSL mutants were infectious and able to spread through DF1 cell culture but at a rate that was indistinguishable from that of the AAAA-LYPSL mutant (Fig. 6). Furthermore, the rate of spread of the PTAP-LYPSL and FPIV-LYPSL mutants was higher in cells overexpressing hAlix than in control DF1 cells. The rescue observed was comparable to that observed with the AAAA-LYPSL mutant in Alix-overexpressing cells. These data suggest that in the context of RSV p2b, the minimal 4-residue PTAP and FPIV late domain motifs do not engage the ESCRT machinery in a way that promotes RSV budding, as also found for some other minimal late domain swaps (40).
FIG. 6.
Heterologous PTAP and FPIV late domains do not support budding in place of the PPPY late domain. Shown are spreading profiles comparing mutants in which the PPPY was replaced with AAAA, the HIV late domain PTAP, or the SV5 late domain FPIV.
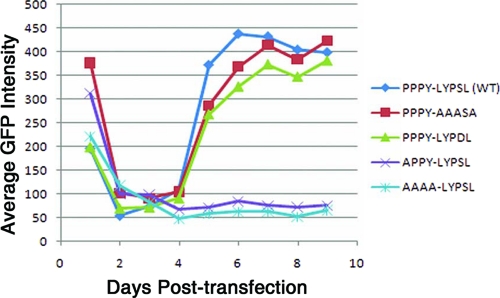
Low GFP expression is selected for cells infected with severe late domain mutants.
In spreading infections of wild-type as well as of mutant viruses, the average steady-state fluorescence of the GFP-positive cell population varied depending on the severity of the late domain mutation. At the beginning of the spreading assay, all transfected cells, both those expressing wild-type DNA and those expressing mutant DNAs, showed a similar high level of fluorescence (day 1) (Fig. 7), presumably reflecting the uptake of multiple DNA molecules. The fluorescence intensity then dropped for both the wild type and mutants (days 2 to 4) (Fig. 7). After this initial drop, the fluorescence gradually increased over the next several days in cells infected with wild-type virus or with mutants having minimal effects on budding rate, such as PPPY-AAASA and PPPY-LYPDL (days 4 to 7) (Fig. 7). In these cells the average fluorescence leveled off on day 7, about 2 days after most of the cells had been infected, as gauged by eye with an epifluorescence microscope. In contrast, in the cells infected with the mutants exhibiting significant reductions in the budding rate, such as APPY-LYPSL and AAAA-LYPSL, the average fluorescence remained low even as the infection spread through the culture (days 4 to 9) (Fig. 7).
FIG. 7.
The GFP intensity of infected cells is lower in cells infected with severe late domain mutants. The average fluorescence intensity of GFP-positive cells was determined as the virus spread through the population of DF1 cells.
Since GFP is expressed from the same promoter as Gag, these data suggest that cells expressing high levels of GFP, and, therefore, also presumably high levels of Gag, are selected against in the seriously crippling late domain mutants. We speculate that this selection is due to the cytotoxic effects associated with the accumulation of masses of virus particles on the cell surface.
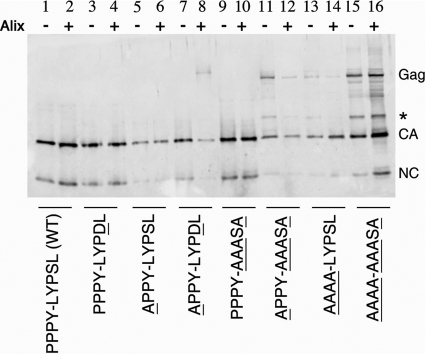
Late domain mutants have cleavage defects.
RSV particle maturation occurs when the viral protease (PR) dimerizes and liberates itself from the Gag polyprotein and cleaves Gag into its individual domains, causing a gross morphological change inside the particle. Viral maturation occurs during or shortly after budding from the cell membrane and is required for infectivity. For HIV-1, late domain mutants were reported to have Gag proteolytic cleavage abnormalities (14, 15). In RSV, both the PPPY and LYPSL motifs are in immediate proximity of a cleavage site that lies between them. To address if the observed spreading may reflect cleavage deficiencies in addition to slowed budding, we examined the cleavage profiles of the late domain mutants produced in either control DF1 cells or hAlix-overexpressing DF1 cells. Filtered medium was obtained from cells infected with the wild type or the indicated mutant, and the virus was collected by centrifugation. The virus pellets were resolved by SDS-PAGE and subjected to immunoblotting with an antiserum against the CA and NC proteins (Fig. 8). We found that the more severe the budding defect, the less complete the proteolysis of Gag. Wild-type virions contained Gag that was fully processed into CA and NC (Fig. 8, lane 1). The production of virus in cells that overexpress hAlix had no effect on wild-type Gag processing (Fig. 8, lane 2). The processing profiles of Gag from the PPPY-LYPDL and PPPY-AAASA virions, whether produced in control or hAlixDF1 cells, were also indistinguishable from that of the wild type (Fig. 8, lanes 3, 4, 9, and 10). In contrast, the viral cleavage profiles of the severe late domain mutants APPY-AAASA, AAAA-LYPSL, and AAAA-AAASA contained not only fully processed CA and NC but also various levels of unprocessed Gag and intermediate cleavage products (Fig. 8, lanes 11 to 16).
FIG. 8.
Gag polyprotein cleavage in late domain virions. Gag proteins in wild-type and mutant virus pellets from control DF1 cells (−) or hAlix-DF1 cells were analyzed by immunoblotting with anti-CA rabbit serum.
Since the budding of PPPY mutants is more rapid in hAlixDF1 cells, we wanted to investigate whether producing virus in these cells would result in more complete Gag cleavage. The production of virions in hAlixDF1 cells had no significant effect on Gag processing in wild-type, PPPY-LYPDL, APPY-LYPSL, and PPPY-AAASA virions. However, the ratio of CA to Gag in the AAAA-LYPSL virions, which exhibited a dramatic budding and spreading rescue in hAlixDF1 cells, appeared to be somewhat higher in the virions produced by hAlixDF1 cells than in control DF1 cells (Fig. 8, compare lane 13 with lane 14). Thus, improved budding and virus spread may correlate with a lesser cleavage defect.
Virus budding defects are visualized by scanning electron microscopy.
The slowed budding and spreading for late domain mutants do not exclude the possibility that the mutations also perturb other steps in retrovirus replication, such as trafficking or assembly. To confirm that the observed defects are due to a block at the membrane scission step of budding, we performed SEM on DF1 cells transiently transfected with wild-type or mutant proviral DNA carrying the GFP marker gene. Due to the rapid budding rate of RSV compared with that of other retroviruses, wild-type RSV particles are very rarely observed in the act of budding by thin-section transmission electron microscopy (TEM). SEM allows a much larger portion of the cell surface to be visualized, making this the preferred technique for the examination of budding. The fluorescence level of a number of typical individual cells was recorded to allow viral gene expression to be gauged, and the same cells were then visualized by SEM.
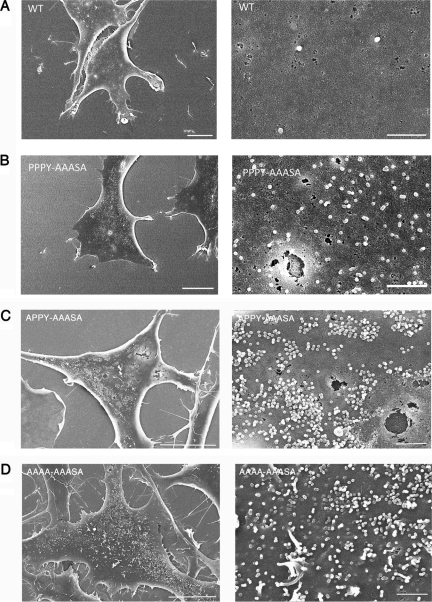
Compared with the surfaces of cells transfected with wild-type RSV (Fig. 9A), the surfaces of cells transfected with the PPPY-AAASA mutant were decorated with larger numbers of particles (Fig. 9B), which is indicative of a late budding defect. To underpin this visual conclusion in a semiquantitative manner, we estimated the budded or budding virus-like particles on several dozen cells with roughly equivalent GFP fluorescence and, thus, presumably equivalent levels of Gag expression. Although the particle numbers on individual cells varied over a wide range, the PPPY-AAASA mutant on average showed approximately a 5-fold increase over the wild type, as exemplified in Fig. 9A and B. This signature late domain phenotype implicates the LYPSL motif in budding, even in the context of a fully functional primary PPPY late domain.
FIG. 9.
Budding morphology of late domain mutants. Shown are representative scanning electron micrographs of DF1 cells transiently transfected with the wild type (A) and the PPPY-AAASA (B), APPY-AAASA (C), and AAAA-AAASA (D) mutants. The cells shown had similar levels of GFP fluorescence. The scale bars are 10 μm in the left images and 1 μm in the right images.
Cells transfected with the double mutant APPY-AAASA (Fig. 9C) or AAAA-AAASA (Fig. 9D) exhibited much more densely arrayed particles on the surface, with particle number estimates ranging to several thousand. The surface of cells transfected with the full double knockout not only was crowded with fully assembled particles but also displayed particles that appeared to be arrested at various earlier stages of budding. Thin-section TEM showed these particles to be immature (data not shown). In summary, for the severe late domain mutants, the results of the SEM analysis are fully consistent with their phenotypes measured by other assays and imply that the primary defect in the virus life cycle can be ascribed to defective budding.
DISCUSSION
In most previous studies of late domains, budding defects were evaluated in transiently transfected cells, commonly by quantifying Gag shed into the medium. Transient transfection typically leads to overexpression, and since virus assembly presumably is concentration dependent, the observed budding phenotypes under these conditions may not accurately or quantitatively reflect the physiological function of the late domain. For this reason, we chose to examine putative late domain mutants primarily in the context of infected cells, which were derived by virus spread after transfection with an infectious RSV DNA carrying a GFP marker gene.
We have shown by three techniques that the RSV LYPSL motif, which is located several residues downstream of the well-known PPPY sequence in Gag, is a secondary late domain. The function of the LYPSL motif became most obvious when the PPPY motif was ablated or partially compromised by mutation. By measuring the half-time of Gag release with pulse-chase labeling, the rate of viral spread with flow cytometry, and the number of budding structures on the surface of cells with SEM, we were able to evaluate the relative contributions of PPPY and LYPSL to efficient budding. The three techniques proved to be complementary and consistent. The pulse-chase measurements were better able to distinguish among the moderately compromised budding mutants, while the flow cytometry assays were better able to distinguish and rank the severely compromised budding mutants. SEM was the most sensitive technique, and unlike the other assays, SEM analysis revealed a delay in budding when the LYPSL motif alone was mutated (PPPY-AAASA). The presumptive explanation for this greater sensitivity is that SEM focuses on the assembly and budding steps, while pulse-chase kinetics measure the product of several steps, including not only assembly and budding but also Gag trafficking. For example, in the HIV-1 system, the time required for the formation of a virus particle is only 5 to 10 min, as visualized in real time by total-internal-reflection microscopy (20). A similar measurement is not available for RSV. In contrast, in our hands, in preliminary studies the half-time for HIV-1 VLP release by pulse-chase labeling in DF1 cells is about 2 h (data not shown), about twice as long as that for RSV.
The contribution of the LYPSL sequence to RSV budding was also apparent upon the overexpression of its presumed ESCRT-binding partner, Alix. Like the LYPSL mutation itself (PPPY-AAASA), Alix overexpression had different effects on the budding and spreading of RSV depending on the wild-type or mutational status of the primary late domain, but its role could be readily observed both when the PPPY motif was handicapped (APPY-LYPSL) and when it was absent (AAAA-LYPSL).
In RSV budding, the secondary role of the LYPSL motif compared to the role of the dominant PPPY motif does not necessarily imply a secondary role of the LYPSL-binding protein (presumably Alix) compared to the role of the PPPY-binding protein (Nedd4 or a related E3 ubiquitin ligase). For example, mutation of the LYPSL motif might have a weaker effect on budding because Alix has alternative means of interacting with RSV Gag, while Nedd4 does not. In the HIV-1 system, Alix binds not only the LYPXL motif in the p6 domain in Gag but also the upstream NC domain, and the latter interaction apparently is functionally important (8, 42, 43). The possibility that RSV NC may have a late domain-like function is consistent with a previous report that certain RSV NC mutations lead to budding-arrest phenotypes (25). In our system, it was not possible to examine the contribution of an NC-Alix interaction to RSV budding because infectivity requires an intact NC domain, and we used infected cells for these assays.
Efficient retrovirus replication requires a balance of unspliced and spliced RNA. This balance serves to provide sufficient quantities of full-length RNA for packaging and for the correct expression of Gag and GagPol on the one hand and of mRNA for Env on the other hand. In simple retroviruses, several regulatory mechanisms contribute to maintaining an optimal ratio of the two types of RNA. In RSV, a cis-acting element termed the negative regulator of splicing (NRS) reduces splicing and thereby helps maintain an optimal level of full-length viral RNA in the cytoplasm (36). The NRS also promotes polyadenylation (55). Defined broadly, the NRS is a 230-nucleotide sequence that overlaps both the PPPY and LYPSL motifs at the RNA level (35, 36, 39). Thus, mutations in the late domain motifs unavoidably alter the sequence of the NRS. We cannot entirely exclude the possibility that the phenotypes observed here are due in part to changes in splicing. For example, mutationally compromising the NRS would decrease Gag synthesis, thereby decreasing the production of infectious particles and, thus, the rate of virus spread to neighboring cells. However, the effects of characterized NRS mutants on unspliced RNA levels are generally in the range of only 2-fold. Since the budding rates for the various late domain mutants described here correlate almost perfectly with their spreading rates, we assume that any replication disadvantage caused by changes in the NRS is minimal. In addition, SEM analysis illustrates that the mutant viruses exhibit late domain phenotypes qualitatively proportional to the severity of the mutation, consistent with a replication block occurring at the stage of virus release and not at the level of viral RNA splicing or protein synthesis.
For mutants with severe late domain phenotypes, such as AAAA-LYPSL in the absence of Alix overexpression, the steady-state level of GFP fluorescence in infected cells remained low, at about 20% of the level of that in cells expressing wild-type virus. Presumably, Gag expression mirrors GFP fluorescence. The most likely explanation of this downregulation of viral gene expression is that the accumulation of Gag and virus particles at the plasma membrane is toxic and that cells are therefore selected for low expression. The rapidity of this downregulation seems surprising, and the mechanism underlying selection is uncertain. For uncompromised virus that spreads rapidly, we hypothesize that the rise in fluorescence between days 3 and 6 (Fig. 7) is due to several virus particles infecting each cell before superinfection resistance is established. According to this model, for severely compromised viruses that spread slowly, in the initially infected cell, more time is available to establish superinfection resistance before potentially superinfecting viruses are encountered. According to another model that is not mutually exclusive, the proviral integration sites may be selected for low expression. The distribution of fluorescence intensity and therefore, presumably, Gag expression for wild-type-infected cells varies by about 10-fold, most likely due to the properties of integration sites. Sites leading to high-level expression may be inconsistent with cell survival or cell division. However, this type of selection is unlikely to account entirely for the observed low fluorescence of mutant viruses, since we did not observe a massive reduction in cell numbers in the 3- to 6-day time frame, which would be predicted by this model.
A complication of the experimental logic used in these experiments is that the GFP marker gene may be lost during passage of the virus. This well-known phenomenon is due to a replication disadvantage of alpharetroviruses that express a marker gene like GFP from a spliced RNA or, indeed, that carry the v-src gene in the original Rous sarcoma virus (44). In our hands, this loss was sporadic and unpredictable, although it seemed to occur more frequently in the severely compromised late domain mutants. Thus, in some experiments, a subpopulation of cells never became fluorescent, even after long times, although they were infected as evidenced by immunofluorescence staining. While the conclusions that we have drawn from the experiments presented here are not affected by the sporadic loss of the GFP gene, for other experiments, interpretations may be complicated, requiring the monitoring of infection by other methods.
One surprising finding from our study is that RSV infection is able to spread in the absence of both late domains. While this spread is slow and the resulting infected cells appear unhealthy, clearly, the virus can gain access to cells without a functional PPPY or LYPSL sequence (46). Several precedents for late domain-independent replication have been reported both for HIV-1 (7, 15) and for MLV (46). For example, in the MLV system, all known late domain-like sequences were ablated without an abrogation of infectivity. One model that accounts for this observation is that RSV Gag or GagPol has one or more additional motifs that can act as late domains to recruit the ESCRT machinery. Depending on the degeneracy allowed in the consensus sequences, one can find additional motifs that might act as late domains. An alternative model is that cell-to-cell spread by the virus, which is known to be much more efficient than spread via infectious virus particles released into the medium (19), does not have an absolute requirement for the ESCRT machinery. Further work will be required to test these models.
Acknowledgments
We thank Wesley Sundquist for the human Alix DNA clone and for antiserum to the protein and A. Devrim Güçlü for help in computing budding half-times.
This work was supported by USPHS grants CA20081 to V.M.V. and AI073098 to M.C.J.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlton, J. G., M. Agromayor, and J. Martin-Serrano. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U. S. A. 105:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908-1912. [DOI] [PubMed] [Google Scholar]
- 5.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dussupt, V., M. P. Javid, G. Abou-Jaoude, J. A. Jadwin, J. de La Cruz, K. Nagashima, and F. Bouamr. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 5:e1000339. [DOI] [PMC free article] [PubMed]
- 9.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 10.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841-852. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O., and A. J. Mouland. 2006. The cell biology of HIV-1 and other retroviruses. Retrovirology 3:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, K., J. H. Hurley, and E. O. Freed. 2007. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat. Rev. Microbiol. 5:912-916. [DOI] [PubMed] [Google Scholar]
- 13.Fujii, K., U. M. Munshi, S. D. Ablan, D. G. Demirov, F. Soheilian, K. Nagashima, A. G. Stephen, R. J. Fisher, and E. O. Freed. 2009. Functional role of Alix in HIV-1 replication. Virology 391:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317-323. [DOI] [PubMed] [Google Scholar]
- 18.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, J., N. M. Sherer, G. Heidecker, D. Derse, and W. Mothes. 2009. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 7:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouvenet, N., P. D. Bieniasz, and S. M. Simon. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., S. Sitaraman, A. Hierro, B. M. Beach, G. Odorizzi, and J. H. Hurley. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson, D. R., M. C. Johnson, W. W. Webb, and V. M. Vogt. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 102:15453-15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazert, C., N. Chazal, L. Briant, D. Gerlier, and J. C. Cortay. 2008. Refined study of the interaction between HIV-1 p6 late domain and ALIX. Retrovirology 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, E. G., and M. L. Linial. 2006. Deletion of a Cys-His motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology 347:226-233. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S., A. Joshi, K. Nagashima, E. O. Freed, and J. H. Hurley. 2007. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14:194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcucci, K. T., Y. Martina, F. Harrison, C. A. Wilson, and D. R. Salomon. 2008. Functional hierarchy of two L domains in porcine endogenous retrovirus (PERV) that influence release and infectivity. Virology 375:637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., and P. D. Bieniasz. 2003. A bipartite late-budding domain in human immunodeficiency virus type 1. J. Virol. 77:12373-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 35.McNally, M. T., and K. Beemon. 1992. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J. Virol. 66:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally, M. T., R. R. Gontarek, and K. Beemon. 1991. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology 185:99-108. [DOI] [PubMed] [Google Scholar]
- 37.Medina, G., Y. Zhang, Y. Tang, E. Gottwein, M. L. Vana, F. Bouamr, J. Leis, and C. A. Carter. 2005. The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic 6:880-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munshi, U. M., J. Kim, K. Nagashima, J. H. Hurley, and E. O. Freed. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 282:3847-3855. [DOI] [PubMed] [Google Scholar]
- 39.O'Sullivan, C. T., T. S. Polony, R. E. Paca, and K. L. Beemon. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses SRC mRNA splicing and promotes polyadenylation. Virology 302:405-412. [DOI] [PubMed] [Google Scholar]
- 40.Ott, D. E., L. V. Coren, T. D. Gagliardi, and K. Nagashima. 2005. Heterologous late-domain sequences have various abilities to promote budding of human immunodeficiency virus type 1. J. Virol. 79:9038-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through nucleocapsid. J. Virol. 82:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2009. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J. Virol. 83:7185-7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portsmouth, D., D. Deitermann, B. Salmons, W. H. Gunzburg, and M. Renner. 2008. Transgene expression facilitated by the v-src splice acceptor can impair replication kinetics and lead to genomic instability of Rous sarcoma virus-based vectors. J. Virol. 82:1610-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabo, Y., N. Laham-Karam, and E. Bacharach. 2008. Basal budding and replication of the murine leukemia virus are independent of the gag L domains. J. Virol. 82:9770-9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 49.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Usami, Y., S. Popov, E. Popova, M. Inoue, W. Weissenhorn, and H. G. Gottlinger. 2009. The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 37:181-184. [DOI] [PubMed] [Google Scholar]
- 51.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 78:13943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H., N. J. Machesky, and L. M. Mansky. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilusz, J. E., and K. L. Beemon. 2006. The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J. Virol. 80:9634-9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhai, Q., R. D. Fisher, H. Y. Chung, D. G. Myszka, W. I. Sundquist, and C. P. Hill. 2008. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43-49. [DOI] [PubMed] [Google Scholar]