Abstract
Accidental insertional activation of proto-oncogenes and potential vector mobilization pose serious challenges for human gene therapy using retroviral vectors. Comparative analyses of integration sites of different retroviral vectors have elucidated distinct target site preferences, highlighting vectors based on the alpharetrovirus Rous sarcoma virus (RSV) as those with the most neutral integration spectrum. To date, alpharetroviral vector systems are based mainly on single constructs containing viral coding sequences and intact long terminal repeats (LTR). Even though they are considered to be replication incompetent in mammalian cells, the transfer of intact viral genomes is unacceptable for clinical applications, due to the risk of vector mobilization and the potentially immunogenic expression of viral proteins, which we minimized by setting up a split-packaging system expressing the necessary viral proteins in trans. Moreover, intact LTRs containing transcriptional elements are capable of activating cellular genes. By removing most of these transcriptional elements, we were able to generate a self-inactivating (SIN) alpharetroviral vector, whose LTR transcriptional activity is strongly reduced and whose transgene expression can be driven by an internal promoter of choice. Codon optimization of the alpharetroviral Gag/Pol expression construct and further optimization steps allowed the production of high-titer self-inactivating vector particles in human cells. We demonstrate proof of principle for the versatility of alpharetroviral SIN vectors for the genetic modification of murine and human hematopoietic cells at a low multiplicity of infection.
Mainly due to their ability to efficiently anchor transgenes in cellular chromosomes, retrovirus-based vectors have become one of the most widely used gene transfer systems in basic research and gene therapy. Major obstacles to the development of retrovirus-based vectors are the potential generation of replication-competent retroviruses (RCR), the inadvertent activation or disruption of cellular genes by the integrated vector genome (insertional mutagenesis) (8, 9), and nonphysiological transgene expression levels under the control of retroviral cis-acting sequences. Over recent years, important progress has been made in the development of retrovirus-based vectors, with promising principles developed on the basis of 3 of the 7 genera of the Retroviridae family: Gammaretrovirus (33), Lentivirus (5), and Spumavirus (39). Of these, gammaretroviral vectors have the most advanced split-packaging design, in which cis-acting sequences, required for vector packaging, reverse transcription, and integration, can be entirely separated from the sequences encoding the structural and enzymatically active retroviral proteins. Formation of RCR and potential recombination with retroviral sequences of the target cells thus become very unlikely. Lentiviral vectors are of major interest for their ability to transduce nondividing cells, such as neuronal cells and hepatocytes. Furthermore, these were the first vectors for which high-titer production of vectors with self-inactivating (SIN) long terminal repeats (LTR) was established (5), allowing the expression of the genes of interest under the control of more physiological “internal” promoters. High-titer SIN vectors have subsequently also been established in the gammaretroviral and spumaviral contexts. However, both spumaviral and lentiviral vectors still contain larger portions of residual viral coding sequences in order to achieve efficient packaging.
Alpharetroviruses, with the paradigmatic Rous sarcoma virus (RSV), have attracted increasing interest as an alternative retroviral vector system since the integration patterns of different retroviruses and their derived vectors have been analyzed in greater detail. Murine leukemia virus (MLV) and its derived gammaretroviral vectors have a tendency to integrate close to the transcriptional start sites (TSS), into promoter-proximal and CpG-rich regions (43), increasing the risk of activating cellular proto-oncogenes by enhancer- or promoter-mediated mechanisms. In contrast, human immunodeficiency virus type 1 (HIV-1) and related lentiviral vectors preferentially integrate within active transcription units (34), which may activate growth-promoting genes by both enhancer- and truncation-mediated mechanisms (3, 23, 24). Vectors based on RSV, belonging to the genus Alpharetrovirus, have been shown to have the most neutral, and therefore the most favorable, integration pattern (21). They do not have a strong preference for promoter regions or transcription units. In a rhesus model of hematopoietic stem cell (HSC) gene transfer, alpharetroviral integration was nonclustered and favored neither gene-rich regions, TSS, nor CpG islands (10, 11).
Of the available alpharetroviral vector systems, the most widely used is the RCAS (replication-competent avian leukosis virus [ALV] LTR with a splice acceptor) (13). It is based on RSV, in which the src oncogene is replaced by a transgene of interest. This vector, although very useful for basic nonclinical applications, has disadvantages that preclude its application in advanced forms of genetic cell modification: the presence of gag/pol and env in the vector genome drastically reduces the packaging capacity and poses a major safety concern. Furthermore, the LTRs have not been developed to an advanced SIN design, thus retaining retroviral enhancer-promoter sequences that can perturb both transgene expression and transcriptional control of neighboring genes, and RCAS-derived particles are commonly produced from replication-competent vectors in avian cells. These features pose a high risk of vector recombination with poorly foreseeable consequences.
In the present work, we have developed alpharetroviral vectors that combine the following features: SIN design of the LTRs; split-packaging design with complete elimination of viral coding sequences from the packaging signal; the shortest leader region of all known retroviral vectors, creating space for large gene cassettes; and high-titer production with the potential of pseudotyping for important target populations, such as murine and human hematopoietic cells. Besides introducing a promising tool for genetic cell modification, this work also sheds new light on alpharetroviral replication mechanisms.
MATERIALS AND METHODS
Alpharetroviral vectors and plasmids.
For biosafety reasons, the vector system described was constructed in a split-packaging design, consisting of the alpharetroviral vector (pAlpha.SF.EGFP.wPRE and derivatives; no overlap with gag/pol or envelope plasmids), the alpharetroviral gag/pol helper plasmid (pcDNA3.alpha.gag/pol and derivatives), and the respective envelope plasmid (ecotropic, amphotropic, RD114/TR, and VSVg). VSVg (glycoprotein of vesicular stomatitis virus) (44), RD114/TR (RD114 envelope with the amphotropic cytoplasmic tail) (29), and the ecotropic (25) and amphotropic (strain 4070A) (2) MLV envelopes have been described elsewhere.
The alpharetroviral vector was derived from multiple fragments (5′ LTR and leader; expression unit; 3′ LTR) and was assembled in a series of subcloning steps. To construct the 5′ LTR and the leader region, part of the RSV U3 (from the SphI site on) was amplified using pSRS11.SF.EGFP.pre (33) as a template and primers 5′ RSV sph2 (5′-GCACCGTGCATGCCGATTGGTG-3′) (restriction sites are underlined) and 3′ RSVASLV OL (5′-AATGTGGTGAATGGTCAAATGGCGTTTATTGTATCGAGCTAGGCAC-3′). In a second PCR, the RSV R and U5 regions and the leader region (including the packaging signal ψ; excluding any gag components) were amplified from RCASBP-Y DV (Addgene plasmid 11478) (12-14, 17) using primers 5′ RSVASLV OL (5′-GTGCCTAGCTCGATACAATAAACGCCATTTGACCATTCACCACATTGG-3) and 3′ ASLV nhe (5′-CAGCTAGCGGCCGCTGCTTGATCCGCAGGCCGACCAA-3′) (NheI and NotI sites underlined). The latter two fragments were fused using an overlap-PCR strategy with flanking primers 5′ RSV sph2 and 3′ ASLV nhe. The 3′LTR was amplified from RCASBP-Y DV using 5′ ASLV 3′LTR hind (5′-GAAAGCTTTTAAATATCGATGCGAT-3′) (the HindIII site is underlined) and 3′ ASLV SIN xho (5′-CTCTCGAGAATGAAGCCTTCTGCTTCATGCA-3′) (the XhoI site is underlined). To construct a SIN design and to insert a unique SnaBI site (for the insertion of fragments into the ΔU3 deletion), the overlap primers 3′ ASLVsin OL (5′-AATACAATATCTCTACGTACAAGAGTATTGCATAAGACTACATTTCC-3′) and 5′ ASLVsin OL (5′-TATGCAATACTCTTGTACGTAGAGATATTGTATTTAAGTGCCTAGC-3′) were used together with the two primers mentioned above. The 5′ LTR and leader fragment (SphI/NheI) and the 3′ LTR fragment (HindIII/XhoI) were inserted into a gammaretroviral SIN vector (pSERS11.SF.GFPpre*) backbone harboring a spleen focus-forming virus (SFFV)-driven enhanced green fluorescent protein (EGFP) expression cassette with the safety-optimized posttranscriptional regulatory element of woodchuck hepatitis virus (wPRE) (30, 33). This resulted in the alpharetroviral plasmids pAlpha.SF.EGFP.wPRE (with an intact 3′ U3 region) and pAlpha.SIN.SF.EGFP.wPRE (with a 3′ SIN deletion). In these constructs, the transcription of the alpharetroviral genomic RNA is mediated by the RSV promoter fused to the simian virus 40 (SV40) enhancer.
To design a helper construct for the expression of alpharetroviral Gag/Pol (4,817 bp), we amplified the RSV gag/pol cDNA with primers 5′ ASLVgp eco (5′-CTGAATTCGCCACCAT GGAAGCCGTCATTAAGGTGATT-3′) (the EcoRI site is underlined, and the Kozak consensus sequence is italicized) and 3′ ASLVgp eco (5′-CAGAATTCTTAACTCTCGTTGGCAGCAAGGGT-3′) (the EcoRI site is underlined). This alpharetroviral gag/pol cDNA was inserted into an expression construct harboring the cytomegalovirus (CMV) promoter, the β-globin intron, and the bovine growth hormone poly(A) signal for optimal expression.
In a second approach, we constructed a codon-optimized gag/pol cDNA (4,817 bp). This was synthesized (Biomatik Corp., Markham, Canada) according to the optimal codon preference for Homo sapiens, with GC content adjustment and removal of cryptic splice and polyadenylation sites. The 300 bp surrounding the gag/pol transition was left unchanged to maintain the Gag-to-Pol ratio.
All constructs were verified by sequencing. Cloning and construct details are available on request.
Cell lines and production of viral supernatants.
The human embryonic kidney cell line 293T, the human fibroblast line HT1080, and the murine fibroblast line SC1 were cultured in Dulbecco's modified Eagle's medium (DMEM) with stable glutamine (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin-100 μg/ml streptomycin, and 1 mM sodium pyruvate (all from PAA Laboratories, Pasching, Austria). Alpharetroviral supernatants were produced after transient transfection of 293T cells as previously described for gammaretroviral and lentiviral vectors (5, 30, 32, 33) using ecotropic, amphotropic, gibbon ape leukemia virus (GALV), GALV-TR (cytoplasmically modified), VSVg, or RD114-TR envelopes. Briefly, 5 × 106 293T cells were seeded the day before transfection. Five micrograms of the alpharetroviral vector (pAlpha.SF.EGFP.wPRE and derivatives), 10 μg (unless indicated otherwise) of the alpharetroviral gag/pol helper plasmid (pcDNA3.alpha.gag/pol), and either 1.5 μg of pMD.G (for VSVg pseudotyped particles) or 2 μg of another envelope plasmid were cotransfected using standard calcium phosphate transfection. In some experiments, transfection efficiencies were monitored by cotransfecting 1 μg of the pCMV-DsRed-Express expression plasmid. To harvest viral particles, supernatants were collected 36 h after transfection, filtered through 0.22-μm-pore-size filters, and stored at −80°C before further use.
Human and murine primary cells.
Human CD34-positive cells purified from granulocyte colony-stimulating factor (G-CSF)-mobilized peripheral blood from healthy volunteers were kindly provided by Cytonet GmbH (Hannover, Germany) and were cultivated in Stemspan serum-free medium (Stem Cell Technologies, Vancouver, British Columbia, Canada) supplemented with 200 U/ml penicillin-200 μg/ml streptomycin (PAA), 2 mM l-glutamine (Biochrom), 100 ng/ml hFlt-3 ligand, 100 ng/ml murine stem cell factor (mSCF), and 40 ng/ml murine thrombopoietin (mTPO) (all from Peprotech, Rocky Hill, NJ).
Murine bone marrow cells were flushed from the femurs and tibias of C57BL6/J mice (Charles River Laboratories, Wilmington, MA), and lineage-negative (Lin−) cells were isolated by magnetic sorting using lineage-specific antibodies (Lineage cell depletion kit; Miltenyi, Bergisch-Gladbach, Germany). Lin− cells were prestimulated for 2 days in StemSpan medium (Stem Cell Technologies, Vancouver, CA) containing 10 ng/ml human acidic fibroblast growth factor (hFGF-acidic), 10 ng/ml mSCF, 20 ng/ml mTPO, 20 ng/ml murine insulin-like growth factor 2 (mIGF2) (all from Peprotech, Heidelberg, Germany), 1:1,000 heparin (Liquemin; Roche, Mannheim, Germany) (added 1 day after transduction), 200 U/ml penicillin-200 μg/ml streptomycin, and 2 mM l-glutamine.
Transduction.
A total of 7 × 104 HT1080 cells per well of 12-well plates were plated on the day before transduction. Supernatants containing viral particles were added, and the transduction procedure was assisted by the addition of 4 μg/ml protamine sulfate and by centrifugation for 1 h at 400 × g and 32°C. For transduction of the human primary CD34-positive and murine lineage-negative cells, viral supernatants were loaded onto plates precoated with Retronectin (Takara, Shiga, Otsu, Japan), and transduction was assisted by centrifugation for 30 min at 400 × g and 4°C. Cells were added, and after 24 h, they were transferred to uncoated wells.
Flow cytometry.
Cells were harvested for analysis 4 to 21 days after transduction and were washed with phosphate-buffered saline (PBS). For EGFP-encoding cassettes, cells were pelleted, resuspended in PBS containing 4% heat-inactivated FCS (PAA), and analyzed. Cells transduced with the interleukin-2 common gamma chain (IL2RG) were incubated with a phycoerythrin (PE)-labeled anti-human CD132 antibody (BD Pharmingen, Heidelberg, Germany) for 30 min at 4°C, washed, measured on a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany), and analyzed using CellQuest or FlowJo software (Tree Star, Ashland, OR). CD34 expression was monitored using an allophycocyanin (APC)-labeled anti-human CD34 antibody (BD Pharmingen). A homogeneous cell population was gated as determined by scatter characteristics, and ≥15,000 events were monitored.
Luciferase assay.
To assess LTR transcriptional activity, the corresponding LTR fragments of pAlpha.SF.EGFP.wPRE and pAlpha.SIN.SF.EGFP.wPRE were cloned into synthetic Renilla luciferase reporter vectors (phRG derivatives; Promega, Mannheim, Germany). As a negative control, part of the prokaryotic beta-lactamase gene was cloned into the same reporter vector. Five micrograms of different reporter vectors and 1 μg of pCMV-DsRed-Express were cotransfected using the calcium phosphate transfection method. Thirty-six hours after transfection, cells were harvested, and DsRed expression was monitored via fluorescence-activated cell sorter (FACS) analysis, which served as a measure of transfection efficiency. The cells were lysed, and luciferase activities were determined according to the manufacturer's instructions (Renilla luciferase assay system; Promega) in a Lumat LB950 luminometer (Berthold Technologies, Bad Wildbad, Germany). Using the Bio-Rad protein assay, the protein concentrations of the cell lysates were determined (Bio-Rad, Munich, Germany).
RNA isolation and Northern blot analysis.
RNA was isolated (from 3 × 106 to 5 × 106 cells) using RNAzol (WAK Chemicals, Steinbach, Germany) according to the manufacturer's instructions. The RNA pellet was resuspended in ultrapure water supplemented with 1 μl RiboLock RNase inhibitor (Fermentas, St. Leon-Rot, Germany) and was stored at −20°C. To analyze isolated RNA samples, 10 to 15 μg of each RNA was separated in denaturing formaldehyde gels. RNA was blotted to Biodyne-B membranes (Pall Corp., Pensacola, FL) by capillary transfer and was then heat fixed for 2 h at 80°C. Hybridization was performed with probes corresponding to either the EGFP fragment or 18S rRNA, radiolabeled using the DecaLabel DNA labeling kit (Fermentas). Membranes were exposed to X-ray film (Kodak X-Omat-AR; Kodak GmbH, Stuttgart, Germany), and signal intensities were quantified with a phosphorimaging Storm B20 scanner (Amersham Biosciences, Freiburg, Germany).
Reverse transcription and determination of expression levels by qRT-PCR.
RNA was reverse transcribed using the QuantiTect reverse transcription kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Relative expression levels of 5′ LTR-initiated transcripts were determined by quantitative reverse transcription-PCR (qRT-PCR) using wPRE-specific primers (22) to detect total amounts of vector transcripts and leader-specific primers (leader forw, GTTCGATGACCCTGGTGGAGG; leader rev, CTCACTCTCCGTCTTCCGACGA) to quantify 5′ LTR transcriptional acitivity. From PCR efficiencies and by cross-point deviations between the samples to be compared, relative expression levels were determined according to the method of Pfaffl (27).
Isolation of genomic DNA and determination of vector copy numbers by qRT-PCR.
To isolate genomic DNA from cells, the QIAamp DNA blood kit (Qiagen) was used according to the manufacturer's instructions. Vector copy numbers of transduced cells were determined by quantitative PCR on an Applied Biosystems (Darmstadt, Germany) Step One Plus real-time PCR system using the QuantiTect SYBR green kit (Qiagen) with a reference sample known to contain 9 integrations (clone B, internal standard; kindly provided by U. Modlich, Hannover, Germany) and primers for the vector-specific PRE and the PTBP2 intron (evolutionarily conserved PTBP2 intronic sequence [28]) (19). From PCR efficiencies and by cross-point deviations from the reference sample, the vector copy numbers of unknown samples were determined according to the work of Pfaffl (27).
Protein isolation and Western blotting.
293T cells were cotransfected using 5 μg pAlpha.SIN.SF.EGFP.wPRE, either 2.5 or 10 μg pcDNA3.alpha.gag/pol or its codon-optimized derivative (as indicated), and 2 μg of an ecotropic envelope packaging plasmid as described above. Thirty-six hours after transfection, cells were trypsinized, washed twice with PBS, and lysed using lysis buffer supplemented with proteinase inhibitor (Complete Mini protease inhibitor cocktail; Roche, Mannheim, Germany). Cell lysates normalized for protein levels were fractionated on 12.5% polyacrylamide gels containing sodium dodecyl sulfate (SDS). Following electrophoresis, proteins were transferred to a nitrocellulose membrane (pore size, 0.45 μm; Bio-Rad). The membrane was blocked with 3% dry milk in Tris-buffered saline-Tween 20 (TBST) for 1 h at room temperature and was stained with a monoclonal antibody against avian myeloblastosis virus (AMV)-p19 (AMV-3C2; Developmental Studies Hybridoma Bank, Iowa City, IA) diluted 1:1,000 in 5% dry milk in Tris-buffered saline (TBS) overnight at 4°C. After a wash with TBS, the blot was incubated with a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (Santa Cruz Biotechnologies, Heidelberg, Germany) diluted 1:2,000 in 3% dry milk in TBST for 1 h at room temperature. Detection was performed by chemiluminescence (SuperSignal West Pico chemoluminescence substrate; Thermo Fisher Scientific, Bonn, Germany) according to the manufacturer's instructions. Blots were exposed to Kodak X-Omat-AR film.
Statistical analysis.
Data from experiments are expressed as means ± standard errors of the means (SEM). Student's two-tailed t test was performed for comparison of differences between indicated groups. A P value of <0.05 (*) was considered significant; a P value of <0.01 (**), very significant; a P value of <0.001 (***), extremely significant.
Nucleotide sequence accession number.
The codon-optimized alpharetroviral gag/pol expression sequence has been submitted to GenBank under accession no. HM130053.
RESULTS
Alpharetrovirus-based split-packaging design avoiding sequence overlap between its components.
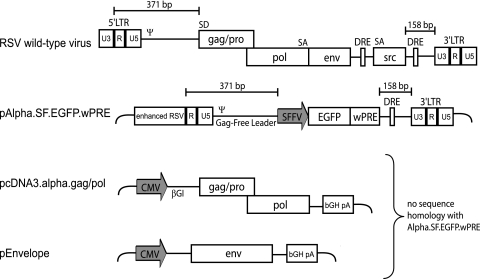
To minimize the risk of RCR formation and to gain maximum space for the insertion of transgene cassettes, we aimed to develop RSV-based vectors with a split-packaging design. As indicated in Fig. 1, we designed a vector in which the RSV coding sequences (gag/pol, env, and src) were completely deleted, leaving only 371 nucleotides in the 5′ untranslated region (5′ UTR), to include R, U5, and the packaging signal (Ψ), and 158 nucleotides in the 3′ UTR, comprising the direct repeat element (DRE) and the polypurine tract (PPT), followed by the 3′ LTR.
FIG. 1.
Alpharetroviral split-packaging design. Shown is a schematic depiction of wild-type RSV and the derived split-packaging system, consisting of the transfer vector pAlpha.SF.EGFP.wPRE (plasmid DNA) and required helper plasmids pcDNA3.alpha.gag/pol and pEnvelope. The long terminal repeats (LTRs) (U3, R, and U5), direct repeat element (DRE), packaging signal (ψ), splice donor and acceptor sites (SD and SA), spleen focus-forming virus promoter (SFFV), enhanced green fluorescent protein (EGFP), woodchuck hepatitis virus posttranscriptional regulatory element (wPRE), cytomegalovirus promoter (CMV), β-globin intron (βGI), and bovine growth hormone polyadenylation signal (bGH pA) are indicated.
In the 5′ U3 region, we introduced a promoter configuration that we have shown to be important for high-titer production of gammaretroviral vectors in transfected human 293T cells. This consists of the simian virus 40 (SV40) enhancer fused to the RSV U3 region (enhanced RSV) (33). The alpharetroviral gag/pol sequences were placed under the control of the human cytomegalovirus (CMV) immediate-early enhancer-promoter, and we took care to remove all of the 5′ UTR sequences that are present in the vector construct and replace these with the β-globin intron. The bovine growth hormone polyadenylation signal served for transcriptional termination, thus removing the alpharetroviral 3′ UTR from the gag/pol expression vector. Envelope proteins derived from RD114 feline leukemia virus, amphotropic or ecotropic MLV, or the glycoprotein of vesicular stomatitis virus (VSVg) were expressed from similarly configured expression vectors, as used in earlier studies (2, 29, 44).
The following design elements are noteworthy. (i) The split design completely avoids sequence overlap between the transfer vector and the packaging construct. (ii) Since the major splice donor of RSV is found in the 5′ region of gag, the vector leader lacks a major splice donor, in contrast to gammaretroviral, spumaviral, and lentiviral vectors. (iii) The remaining alpharetroviral leader sequence providing the packaging signal comprises only 371 nucleotides and is thus 263 and 1,246 nucleotides shorter than the corresponding packaging regions of currently used gammaretroviral and lentiviral vectors, respectively. Considering the sizes of the deleted gag/pol, env, and src genes, the theoretical packaging capacity of the vector is expected to reach 8.4 kb. (iv) The DRE contained in the 3′ UTR of the vector is a cis-acting sequence promoting cytoplasmic expression of full-length RNA (26) and enhancing the efficiency of RNA packaging (37). With this system we could show proof of principle for the transduction of human HT1080 cells (data not shown). However, this configuration still poses a safety risk, residing in the presence of an intact 3′ LTR.
Alpharetroviral SIN design with improved expression.
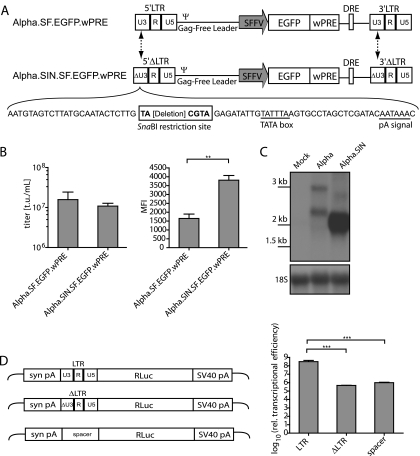
Next, we tested ways to delete the enhancer-promoter sequences from the 3′ U3 region in order to create a SIN design of the vector. Since RSV contains its polyadenylation (pA) signal and related motifs contributing to proper transcriptional termination at the 3′ end of U3, we tested the effects of deletions that preserve ∼40 nucleotides at the 3′ end of U3. The attachment sites required for interaction with the integrase were maintained by preserving 25 nucleotides at the 5′ end of U3. A unique SnaBI restriction site was used to ligate the remaining U3 components, creating opportunities for the introduction of foreign sequences (Fig. 2A). This design left the TATA box intact but deleted upstream promoter and enhancer sequences. As an internal promoter, we used sequences derived from the U3 region of the spleen focus-forming virus (SFFV) (1). A safety-modified version of the woodchuck hepatitis virus posttranscriptional regulatory element (wPRE) (30) was used to further increase the expression of enhanced green fluorescent protein (EGFP), the reporter gene used in our initial studies.
FIG. 2.
Self-inactivating deletion of Alpha.SF.EGFP.wPRE. (A) Schematic depiction of the Alpha.SF.EGFP.wPRE vector (proviral DNA) and its self-inactivating derivative Alpha.SIN.SF.EGFP.wPRE (proviral DNA). The sites of sequence deletions are indicated by dashed arrows. The nucleotide sequences of the self-inactivating U3 region (ΔU3) and the introduced SnaBI restriction site, as well as those of the TATA box and the polyadenylation signal (pA signal), are shown. (B) Comparative titer and mean fluorescence intensity (MFI) analyses as determined on transduced HT1080 cells. Viral particles were generated by cotransfection of pcDNA3.alpha.gag/pol.CO, the VSVg envelope expression plasmid, and either pAlpha.SF.EGFP.wPRE or pAlpha.SIN.SF.EGFP.wPRE. Six days posttransduction, cells were harvested and subjected to flow cytometric analysis. t.u., transducing units. (C) Northern blot analysis of transduced HT1080 cells. A radiolabeled EGFP-specific probe was used to detect all RNA species resulting from EGFP transcription. The expected mRNA sizes for the Alpha.SF.EGFP.wPRE vector are 2,610 bp (LTR transcript) and 1,847 bp (internal transcript). For the Alpha.SIN.SF.EGFP.wPRE vector, the corresponding transcripts are 2,451 bp and 1,688 bp, respectively [sizes without the poly(A) tail are given]. The blot was reprobed with an 18S rRNA-specific probe as a loading control. Molecular size standards are given on the left. (D) Luciferase assay to assess transcriptional activities of the LTRs. 293T cells were transfected, and total protein was lysed as described in Materials and Methods. Relative transcriptional activity was determined as relative light units normalized to the amount of protein and the transfection efficiency. Values were log-transformed, and a Student two-tailed t test was performed for comparison of differences between the indicated groups. A P value of <0.05 (*) was considered significant; a P value of <0.01 (**), very significant; a P value of <0.001 (***), extremely significant.
While the SIN design had only a marginal effect on vector titers, it caused a significant increase in EGFP expression (Fig. 2B), suggesting the presence of either competitive or silencing effects of the intact RSV U3 enhancer-promoter. Next, we addressed the functional consequences of the U3 deletion at the level of the transcript. Polyclonal, unselected cells transduced with vectors at similar multiplicities of infection (MOI) were harvested for preparation of RNA. Northern blot analysis using a radioactively labeled EGFP probe (Fig. 2C) showed that the SIN deletion mediated an increase in the internal message even greater than that indicated by EGFP fluorescence (∼4-fold versus ∼2-fold), which could be explained by the insufficient resolution of flow cytometry under conditions of very high EGFP expression. This analysis also showed that the U3 deletion strongly reduced the expression of the genomic transcript (Fig. 2C). To quantify this effect, we prepared RNA from transduced cells, determined relative expression levels of upstream-initiated and internal transcripts (driven by the internal promoter) by quantitative RT-PCR, and found the upstream-initiated transcription to be approximately 80-fold lower for the self-inactivating vector than for its precursor, Alpha.SF.EGFP.wPRE (data not shown). Furthermore, we assessed the potential residual promoter activity of the deleted U3 region in transient reporter gene assays. Luciferase was cloned under the control of the intact U3 region, the deleted U3 region, or a putative neutral spacer region derived from the prokaryotic beta-lactamase gene. In transiently transfected 293T cells, a reduction of transcriptional activity to background levels was observed with the deleted U3 region (Fig. 2D).
Codon optimization of gag/pol strongly facilitates vector production in human 293T cells.
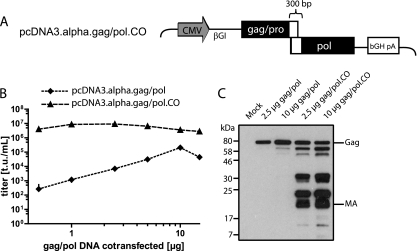
The experiments described above were performed using gag/pol expression vectors in which extensive codon optimization of gag and pol was introduced to match the preferred codon usage found in human cells. Our rationale for codon-optimizing the alpharetroviral gag/pol sequences was based on observations made with lentiviral gag/pol, where codon optimization was shown to overcome nuclear transcript retention (7). We reasoned that similar posttranscriptional limitations may explain the failure of wild-type RSV to replicate in human cells and thus may largely explain the difficulty of producing alpharetroviral vectors in widely used packaging cells such as 293T. When codon-optimizing alpharetroviral gag/pol, we left ∼300 nucleotides surrounding the gag/pro-pol transition unchanged, because this region regulates the ratio of gag/pro and pol expression by ribosomal frameshifting (Fig. 3A). These include the “slippery sequence” of seven nucleotides in the 3′ region of pro, as well as the stem-loop structure in the 5′ region of pol (15, 20).
FIG. 3.
Codon optimization of the gag/pol helper plasmid. (A) Schematic depiction of pcDNA3.alpha.gag/pol.CO, showing the cytomegalovirus promoter (CMV), the β-globin intron (βGI), and the bovine growth hormone polyadenylation signal (bGH pA). Solid rectangles represent areas of codon optimization, while braces indicate sequence that has been exempted from the codon optimization of pcDNA3.alpha.gag/pol. (B) Dilution series of gag/pol helper plasmids. Five micrograms of pAlpha.SIN.SF.EGFP.wPRE, 1.5 μg of the VSVg envelope expression plasmid, and different amounts of either pcDNA3.alpha.gag/pol or its codon-optimized counterpart were cotransfected into 293T cells. Viral supernatants were titrated on HT1080 cells. (C) Western blot of 293T cells. Cells were cotransfected with an ecotropic envelope expression plasmid, pAlpha.SIN.SF.EGFP.wPRE, and different amounts of either pcDNA3.alpha.gag/pol or its codon-optimized counterpart (2.5 or 10 μg, as indicated). Viral proteins were detected by using antibodies against the alpharetroviral matrix (MA) protein. Molecular mass standards are given on the left, while Gag and MA are indicated on the right.
The striking effect of the codon optimization became apparent when alpharetroviral vectors were produced by cotransfecting increasing amounts of the gag/pol expression plasmids with constant amounts of the SIN vector plasmids and the env expression plasmids. Despite the relatively constant transfection efficiencies obtained with 0.5 to 15 μg of the gag/pol plasmid (data not shown), we noted an increase in particle formation dependent on codon optimization that was as great as 4 orders of magnitude at the lowest plasmid dose tested (0.5 μg). While a further increase in the plasmid dose had no effect for the codon-optimized construct, it elevated titers to a maximum of ∼2 × 105 infectious particles per ml of unconcentrated supernatant when the wild-type alpharetroviral sequences were used. This was still at least 1 log unit lower than the maximum level achieved with the codon-optimized construct, which, in pseudotyping with VSVg, typically gave titers on the order of 5 × 106 to 2 × 107/ml of unconcentrated supernatant (Fig. 2B and 3B).
To further characterize this effect, we cotransfected 293T cells with pAlpha.SIN.SF.EGFP.wPRE, different amounts of either pcDNA3.alpha.gag/pol or its codon-optimized derivative (as indicated in Fig. 3C), and 2 μg of an ecotropic envelope expression plasmid to avoid reinfection of producer cells. Thirty-six hours after transfection, cells and associated alpharetroviral particles were lysed for Western blot analysis using antibodies against the alpharetroviral matrix (MA) protein. Efficient proteolytic processing of alpharetroviral Gag/Pol occurred only in cells transfected with the codon-optimized alpharetroviral gag/pol packaging plasmid. To summarize all products detected by the anti-MA antibodies, codon optimization drastically enhanced Gag expression, with a concomitant increase in retroviral cleavage products. It remains to be determined whether the strong effect on functional titers (Fig. 3B) depends on increased synthesis of the Gag precursor with subsequent export through budding particles or on enhanced maturation (see Discussion).
Predictable dose dependence of vector copy number and expression.
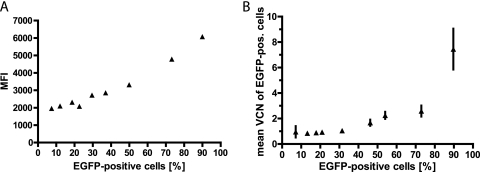
Next, we determined to what extent the transduction level of the target cells can be determined by the dose of vector used. As indicated based on studies performed with gammaretroviral vectors, an increase in the viral particle-to-cell ratio would not only elevate the percentage of transduced cells but also introduce an increasing number of insertions per cell if Poisson statistics apply to the transduction process (6, 16, 41). We thus escalated the MOI to transduce, based on EGFP expression determined by flow cytometry, between 10 and 90% of the target cell population. As indicated in Fig. 4A, the mean fluorescence intensity of transduced cells was relatively constant if <30% of cells were transduced but showed a positive correlation with the percentage of EGFP-positive cells when the transduction rate was increased further. The mean vector copy number of EGFP-positive cells, as determined by real-time PCR detecting the wPRE sequence, confirmed our hypothesis that the increase in EGFP expression depended on the increased likelihood of transferring more than one copy per cell when the MOI was increased (Fig. 4B). Thus, as in gammaretroviral systems, alpharetroviral vectors demonstrate a predictable dose dependence in terms of both the copy number per cell and gene expression per copy, which is an important prerequisite for advanced applications of genetic cell modification.
FIG. 4.
Correlation between the percentage of alpharetrovirally transduced cells and mean fluorescence intensity (MFI) or mean vector copy numbers (VCN). (A) HT1080 cells were transduced with the Alpha.SIN.SF.EGFP.wPRE vector at increasing multiplicities of infection, and 6 days later, the percentage of EGFP-expressing cells and the MFI were determined via flow cytometry. (B) Genomic DNA from transduced cells was harvested, and the mean VCN of EGFP-positive cells were first determined by quantitative PCR and then plotted against the percentage of transduced cells. (The MOIs used were 0.06, 0.13, 0.16, 0.23, 0.39, 0.97, 1.29, 3.24, and 6.47.)
Efficient usage of alpharetroviral vectors in clinically relevant scenarios.
Having established conditions for high-titer production and dose-controlled use of SIN alpharetroviral vectors, we explored their potency in a direct comparison with lentiviral (advanced HIV-1-based SIN design [5, 45]) and gammaretroviral (advanced MLV-based SIN design [31, 33]) vectors. All three SIN vectors contained the same internal expression cassette (SFFV-EGFP-wPRE) (Fig. 5A). We transduced primary hematopoietic cells, a preferred target for retrovirus-based vectors. While a complete analysis of the performance of alpharetroviral vectors in transplanted hematopoietic cells undergoing long-term observation was beyond the scope of these experiments, we set out to explore the potency of SIN alpharetroviral vectors produced in a split-packaging system for the transduction of this cell population.
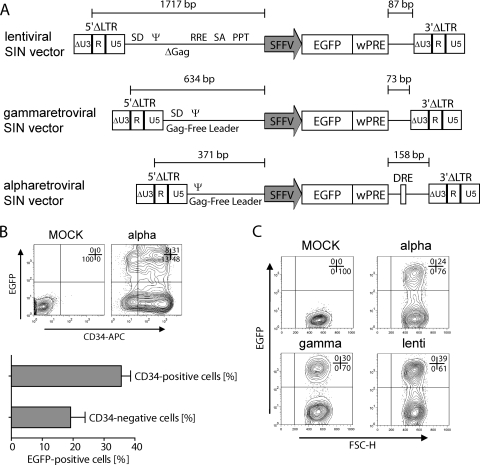
FIG. 5.
Transduction of primary hematopoietic cells. (A) Schematic depiction of lentiviral, gammaretroviral, and alpharetroviral vectors used in this experiment. RRE, Rev-responsive element; PPT, polypurine tract. For other abbreviations, see the legend to Fig. 1. (B) CD34-positive cells were transduced with RD114/TR-pseudpotyped Alpha.SIN.SF.EGFP.wPRE at an MOI of 1. Four to six days after transduction, cells were stained for CD34 expression and were analyzed by flow cytometry. (C) Transduction of murine lineage-negative cells with VSVg-pseudotyped Alpha.SIN.SF.EGFP.wPRE and comparable lentiviral and gammaretroviral SIN vectors at an MOI of 10. Cells were analyzed by FACS 4 days posttransduction.
The choice of the envelope protein is highly important for efficient transduction of hematopoietic cells. We thus tested several options for pseudotyping alpharetroviral vectors. We tested envelopes based on gibbon ape leukemia virus (GALV), feline leukemia virus RD114, the cytoplasmically modified forms of both (GALV/TR and RD114/TR), amphotropic MLV, ecotropic MLV, and VSVg. High titers were achieved with VSVg, RD114/TR, and amphotropic and ecotropic envelopes (data not shown). For the transduction of CD34-positive primary human hematopoietic cells from mobilized peripheral blood, we decided to use RD114/TR particles, based on observations made with lentiviral vectors (29). Of note, a low MOI of 1 (as determined on HT1080 fibroblasts) was sufficient to transduce ∼40% of the target cells with a single exposure on day 3 after stimulation of cells in a cytokine cocktail comprising SCF, TPO, and Flt-3 ligand (4 to 6 days after transduction) (Fig. 5B; confirmed by flow cytometry [n = 3]).
Next, we performed a direct comparison of lentiviral, gammaretroviral, and alpharetroviral vectors in cultured lineage-negative murine hematopoietic cells. We used VSVg pseudotypes in all cases, with an MOI of 10 (as determined on SC1 fibroblasts) for lentiviral, gammaretroviral, and alpharetroviral particles. As determined by flow cytometry on day 4 posttransduction, the transduction rates of alpharetroviral vectors were only slightly lower than those of gammaretroviral vectors, and they were at least half as efficient as lentiviral vectors (Fig. 5C).
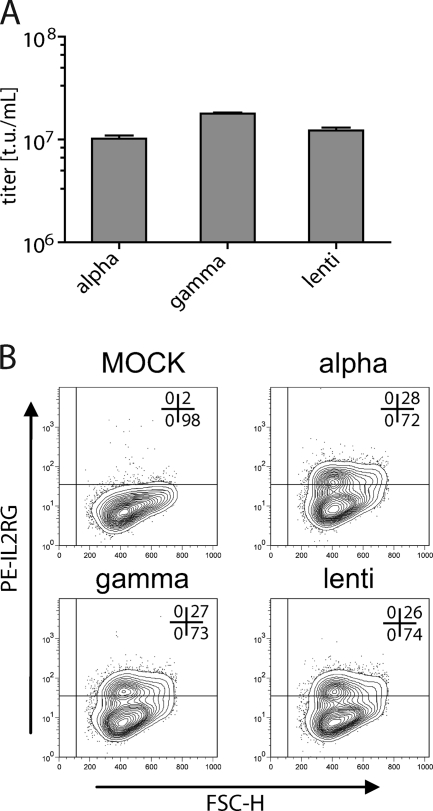
Finally, we performed an intergenus comparison of similarly designed retrovirus-based vectors in the context of a clinically relevant transgene. As we have shown previously (38), a short version of the human elongation factor 1 alpha (EFS) promoter is sufficient to express the interleukin-2 common gamma chain (IL2RG) cDNA to levels that resolve the severe combined immunodeficiency phenotype resulting from mutations in the IL2RG gene (SCID-X1 disease). While previous clinical trials have shown the potential of LTR-driven gammaretroviral vectors for correcting the potentially lethal SCID-X1 phenotype, serious adverse events, caused by the insertional activation of proto-oncogenes from promoter-proximal vector insertions, have been observed in these trials (8, 9). In a sensitive cell system, the internal EFS promoter showed a low risk of insertional proto-oncogene activation after gammaretroviral vector-mediated semirandom integration (46). We thus tested the potential of alpharetroviral vectors for transferring the potentially therapeutic EFS-IL2RG-wPRE cassette into human target cells. As in the experiment described above, lentiviral, gammaretroviral, and alpharetroviral SIN vectors with identical expression cassettes were cloned, produced as VSVg pseudotypes in split-packaging systems, and used to transduce human HT1080 cells at comparable MOI. Vector titers were similar for all three vectors, validating the potency of the newly designed alpharetroviral split-packaging system (Fig. 6A). Furthermore, the IL2RG expression levels were comparable (Fig. 6B). These data illustrate the flexibility of alpharetroviral vectors for incorporating and transferring biologically relevant transgene sequences.
FIG. 6.
Transduction of HT1080 cells with VSVg-pseudotyped lentiviral, gammaretroviral, and alpharetroviral vectors encoding the IL-2 receptor common gamma chain. The vectors were constructed as diagramed in Fig. 5A by replacing SFFV-EGFP with EFS-IL2RG. Transduced cells were stained with a PE-labeled anti-IL2RG antibody and were analyzed by FACS 21 days posttransduction. (A) Comparative titer analyses as determined on transduced HT1080 cells. (B) Cells with similar transduction efficiencies were stained for IL2RG expression and were analyzed by flow cytometry 21 days after transduction.
DISCUSSION
Alpharetroviruses have attracted increasing interest as an alternative retroviral vector system, especially with regard to human gene therapy, due to their favorable integration pattern (10, 11, 21). In the present study, we have developed a versatile and modular self-inactivating alpharetroviral vector system with a split-packaging design, high-titer production in human 293T cells, and the potential of pseudotyping for important target populations, such as murine and human hematopoietic cells.
Alpharetroviral particles are commonly produced in avian cells with vectors based on single constructs containing viral coding sequences and intact LTRs. Even though alpharetroviral vectors have not been reported to be replication competent in human cells, there is a potential risk of vector mobilization, either by mutation or by recombination. The formation of RCR is an even greater threat with the therapeutic use of retrovirus-based gene vectors than insertional mutagenesis (4), and such vectors are therefore unacceptable for clinical applications. To address this safety issue, we have generated an alpharetroviral split-packaging design avoiding sequence overlap between its components (35). The resulting vector backbone not only contains the shortest leader sequence among all retroviral constructs we are aware of (Fig. 5) but also lacks the retroviral major splice donor (in contrast to HIV-1, spumavirus, and MLV vectors), which may prevent some forms of readthrough-dependent insertional gene activation (3, 24). Besides minimizing the risk of RCR formation, the split-packaging design also avoids the potentially immunogenic expression of viral proteins from transduced cells and increases the theoretical packaging capacity (at least 8.4 kb, considering the size of deleted alpharetroviral coding sequences) of the alpharetroviral vector.
To further decrease the risk for insertional mutagenesis, which is caused by activation of neighboring alleles (e.g., proto-oncogenes) via enhancer-mediated mechanisms, we have introduced a SIN deletion into the U3 region, which was complicated by the unusual presence of the alpharetroviral poly(A) signal within U3. Northern blot analysis of transduced polyclonal unselected cells showed that the SIN deletion drastically reduced the expression of the genomic transcript. By quantifying transcript levels of upstream-initiated and internal transcripts, we showed the level of upstream-initiated transcription to be approximately 80-fold lower for the SIN vector than for its precursor containing an intact U3 region. Additionally, we performed transient reporter gene assays, by which we confirmed the strong transcriptional activity of the wild-type U3 region (13, 42) in human cells. While we were still able to detect upstream-initiated transcription in cells transduced with the SIN vector, the transient assay showed a reversion of LTR transcriptional activity to background levels. The residual transcriptional activity detected in transduced cells can be explained by TATA-directed transcription originating from the 5′ LTR, or by readthrough events in cases of intragenic insertions. Further modifications, including a deletion or mutation of the TATA box, need to be evaluated. Similarly, complete elimination of upstream transcription has not been achieved for lentiviral vectors, where the SIN deletion includes the TATA box (18).
Importantly, we were able to produce the alpharetroviral vectors at high titers in human 293T cells, thus reducing the risk of transferring endogenous retroviruses from avian cells. The ability to produce alpharetroviral vectors in 293T cells is somewhat unexpected, because alpharetroviruses are not known to replicate efficiently in mammalian cells (40). Our approach—optimizing gag/pol codon usage for human cells—greatly enhanced alpharetroviral vector production, arguing for the presence of a posttranscriptional alpharetroviral expression block in human cells. In turn, overcoming such an expression block is likely to improve alpharetroviral RNA packaging. According to a competition model, genomic RNA is either translated or packaged in an autogenously regulated manner dependent on the amount of Gag proteins (36). This implies that with increasing Gag concentrations, RNA packaging is eventually favored over translation of genomic RNA, thus contributing to enhanced alpharetroviral particle production. To further address this question, we studied Gag expression in transfected cells. While levels of unprocessed Gag proteins did not increase, major processing of alpharetroviral Gag/Pol occurred only when the codon-optimized plasmid was transfected. Processing may thus be required for proper assembly (40), implying that protease levels are rate-limiting. Since codon optimization is very likely to increase the Gag concentration, packaging is favored over translation, allowing more viral particles to assemble and bud from the membrane. Although we took care to avoid reinfection of the packaging cells by newly produced particles, the increase in the level of Gag subunits detected in Fig. 3C may reflect mature particles that undergo non-receptor-mediated interaction with the transfected cells. Even though the codon optimization drastically increased the titer of alpharetroviral particles, it is unlikely that it is sufficient to circumvent the alpharetroviral replication block in mammalian cells, given the need for tightly balanced splice ratios in retroviral replication.
The newly developed alpharetroviral vector system may facilitate basic molecular analyses of this important retroviral family. Furthermore, our studies suggest that potential species-dependent restrictions of alpharetroviral vector transduction are not profound in murine or human target cells. We showed that alpharetroviral vectors can be pseudotyped with different envelope proteins (RD114/TR, ecotropic and amphotropic, VSVg), and demonstrated a predictable dose dependence of the vector copy number and expression (16, 41). In hematopoietic progenitor cells (murine lineage-negative cells and human CD34-positive cells), the target cell populations for many gene therapy applications in diseases of the blood and immune systems, we showed efficient transduction using a low multiplicity of infection. Finally, we compared HIV-1-based, MLV-based, and alpharetroviral vectors in the setting of SCID-X1, where, due to insertional adverse events (8, 9), vector improvements are urgently needed. In all 3 retroviral vector families, we observed similar expression levels. Of note, both titers and expression levels mediated by the newly developed alpharetroviral SIN vectors are probably sufficient to correct the disease phenotype, since the MLV-based vector has been validated in a SCID-X1 knockout model previously (38).
In summary, the newly developed alpharetroviral split-packaging SIN vector system is an important advancement in the study of retroviral biology and in approaches to the development of safe retrovirus-based vectors for gene therapy.
Acknowledgments
This work was supported by grants from the DAAD and BMBF (Modern Applications in Biotechnology), the Deutsche Forschungsgemeinschaft (REBIRTH Cluster of Excellence and SFB738), the European Union (FP7 project PERSIST, HEALTH-F5-2009-222878), the German Merit Foundation (stipend to T.M.), and the Else-Kröner-Fresenius Stiftung (stipend to A.S.).
We are grateful to Diana Szepe, Girmay Asgedom, Ivonne Fernandez, and Valerie Mordhorst for technical assistance and to Martijn Brugman for consultation on statistics. We thank Steven Hughes (NCI—Frederick, Frederick, MD) and William J. Pavan (NIH, Bethesda, MD) for providing the RCAS-BP vector. The monoclonal antibody against AMV-3C2 developed by David Boettinger was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
The authors declare no competing financial interests.
Footnotes
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Baum, C., S. Hegewisch-Becker, H. G. Eckert, C. Stocking, and W. Ostertag. 1995. Novel retroviral vectors for efficient expression of the multidrug-resistance (mdr-1) gene in early hemopoietic cells. J. Virol. 69:7541-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer, W. R., M. Westphal, W. Ostertag, and D. von Laer. 2002. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J. Virol. 76:1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bokhoven, M., S. L. Stephen, S. Knight, E. F. Gevers, I. C. Robinson, Y. Takeuchi, and M. K. Collins. 2009. Insertional gene activation by lentiviral and gammaretroviral vectors. J. Virol. 83:283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donahue, R. E., S. W. Kessler, D. Bodine, K. McDonagh, C. Dunbar, S. Goodman, B. Agricola, E. Byrne, M. Raffeld, and R. Moen. 1992. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J. Exp. Med. 176:1125-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehse, B., O. S. Kustikova, M. Bubenheim, and C. Baum. 2004. Pois(s)on—it's a question of dose… Gene Ther. 11:879-881. [DOI] [PubMed] [Google Scholar]
- 7.Graf, M., A. Bojak, L. Deml, K. Bieler, H. Wolf, and R. Wagner. 2000. Concerted action of multiple cis-acting sequences is required for Rev. dependence of late human immunodeficiency virus type 1 gene expression. J. Virol. 74:10822-10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina, S., A. Garrigue, G. P. Wang, J. Soulier, A. Lim, E. Morillon, E. Clappier, L. Caccavelli, E. Delabesse, K. Beldjord, V. Asnafi, E. MacIntyre, L. Dal Cortivo, I. Radford, N. Brousse, F. Sigaux, D. Moshous, J. Hauer, A. Borkhardt, B. H. Belohradsky, U. Wintergerst, M. C. Velez, L. Leiva, R. Sorensen, N. Wulffraat, S. Blanche, F. D. Bushman, A. Fischer, and M. Cavazzana-Calvo. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118:3132-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe, S. J., M. R. Mansour, K. Schwarzwaelder, C. Bartholomae, M. Hubank, H. Kempski, M. H. Brugman, K. Pike-Overzet, S. J. Chatters, D. de Ridder, K. C. Gilmour, S. Adams, S. I. Thornhill, K. L. Parsley, F. J. Staal, R. E. Gale, D. C. Linch, J. Bayford, L. Brown, M. Quaye, C. Kinnon, P. Ancliff, D. K. Webb, M. Schmidt, C. von Kalle, H. B. Gaspar, and A. J. Thrasher. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118:3143-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, J., A. Ferris, A. Larochelle, A. E. Krouse, M. E. Metzger, R. E. Donahue, S. H. Hughes, and C. E. Dunbar. 2007. Transduction of rhesus macaque hematopoietic stem and progenitor cells with avian sarcoma and leukosis virus vectors. Hum. Gene Ther. 18:691-700. [DOI] [PubMed] [Google Scholar]
- 11.Hu, J., G. Renaud, T. J. Gomes, A. Ferris, P. C. Hendrie, R. E. Donahue, S. H. Hughes, T. G. Wolfsberg, D. W. Russell, and C. E. Dunbar. 2008. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol. Ther. 16:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, S., and E. Kosik. 1984. Mutagenesis of the region between env and src of the SR-A strain of Rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology 136:89-99. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, S. H. 2004. The RCAS vector system. Folia Biol. (Praha) 50:107-119. [PubMed] [Google Scholar]
- 14.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacks, T., H. D. Madhani, F. R. Masiarz, and H. E. Varmus. 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kustikova, O. S., A. Wahlers, K. Kuehlcke, B. Staehle, A. R. Zander, C. Baum, and B. Fehse. 2003. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood 102:3934-3937. [DOI] [PubMed] [Google Scholar]
- 17.Loftus, S. K., D. M. Larson, D. Watkins-Chow, D. M. Church, and W. J. Pavan. 2001. Generation of RCAS vectors useful for functional genomic analyses. DNA Res. 8:221-226. [DOI] [PubMed] [Google Scholar]
- 18.Logan, A. C., D. L. Haas, T. Kafri, and D. B. Kohn. 2004. Integrated self-inactivating lentiviral vectors produce full-length genomic transcripts competent for encapsidation and integration. J. Virol. 78:8421-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maetzig, T., M. Galla, M. Brugman, R. Loew, C. Baum, and A. Schambach. 2010. Mechanisms controlling titer and expression of bidirectional lentiviral and gammaretroviral vectors. Gene Ther. 17:400-411. [DOI] [PubMed] [Google Scholar]
- 20.Marczinke, B., R. Fisher, M. Vidakovic, A. J. Bloys, and I. Brierley. 1998. Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol. 284:205-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modlich, U., J. Bohne, M. Schmidt, C. von Kalle, S. Knoss, A. Schambach, and C. Baum. 2006. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood 108:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modlich, U., S. Navarro, D. Zychlinski, T. Maetzig, S. Knoess, M. H. Brugman, A. Schambach, S. Charrier, A. Galy, A. J. Thrasher, J. Bueren, and C. Baum. 2009. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 17:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montini, E., D. Cesana, M. Schmidt, F. Sanvito, C. C. Bartholomae, M. Ranzani, F. Benedicenti, L. S. Sergi, A. Ambrosi, M. Ponzoni, C. Doglioni, C. Di Serio, C. von Kalle, and L. Naldini. 2009. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119:964-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063-1070. [DOI] [PubMed] [Google Scholar]
- 26.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 70:3834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahman, L., V. Bliskovski, F. J. Kaye, and M. Zajac-Kaye. 2004. Evolutionary conservation of a 2-kb intronic sequence flanking a tissue-specific alternative exon in the PTBP2 gene. Genomics 83:76-84. [DOI] [PubMed] [Google Scholar]
- 29.Sandrin, V., B. Boson, P. Salmon, W. Gay, D. Negre, R. Le Grand, D. Trono, and F. L. Cosset. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100:823-832. [DOI] [PubMed] [Google Scholar]
- 30.Schambach, A., J. Bohne, C. Baum, F. G. Hermann, L. Egerer, D. von Laer, and T. Giroglou. 2006. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 13:641-645. [DOI] [PubMed] [Google Scholar]
- 31.Schambach, A., J. Bohne, S. Chandra, E. Will, G. P. Margison, D. A. Williams, and C. Baum. 2006. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol. Ther. 13:391-400. [DOI] [PubMed] [Google Scholar]
- 32.Schambach, A., M. Galla, U. Modlich, E. Will, S. Chandra, L. Reeves, M. Colbert, D. A. Williams, C. von Kalle, and C. Baum. 2006. Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp. Hematol. 34:588-592. [DOI] [PubMed] [Google Scholar]
- 33.Schambach, A., D. Mueller, M. Galla, M. M. Verstegen, G. Wagemaker, R. Loew, C. Baum, and J. Bohne. 2006. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 13:1524-1533. [DOI] [PubMed] [Google Scholar]
- 34.Schröder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 35.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonstegard, T. S., and P. B. Hackett. 1996. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 70:6642-6652. [PMC free article] [PubMed] [Google Scholar]
- 37.Sorge, J., W. Ricci, and S. H. Hughes. 1983. cis-acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J. Virol. 48:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornhill, S. I., A. Schambach, S. J. Howe, M. Ulaganathan, E. Grassman, D. Williams, B. Schiedlmeier, N. J. Sebire, H. B. Gaspar, C. Kinnon, C. Baum, and A. J. Thrasher. 2008. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol. Ther. 16:590-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trobridge, G. D. 2009. Foamy virus vectors for gene transfer. Expert Opin. Biol. Ther. 9:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogt, V. M., D. A. Bruckenstein, and A. P. Bell. 1982. Avian sarcoma virus gag precursor polypeptide is not processed in mammalian cells. J. Virol. 44:725-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahlers, A., M. Schwieger, Z. Li, D. Meier-Tackmann, C. Lindemann, H. G. Eckert, D. von Laer, and C. Baum. 2001. Influence of multiplicity of infection and protein stability on retroviral vector-mediated gene expression in hematopoietic cells. Gene Ther. 8:477-486. [DOI] [PubMed] [Google Scholar]
- 42.Weber, F., and W. Schaffner. 1985. Enhancer activity correlates with the oncogenic potential of avian retroviruses. EMBO J. 4:949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 44.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43(Pt. A):99-112. [DOI] [PubMed] [Google Scholar]
- 45.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zychlinski, D., A. Schambach, U. Modlich, T. Maetzig, J. Meyer, E. Grassman, A. Mishra, and C. Baum. 2008. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 16:718-725. [DOI] [PubMed] [Google Scholar]