Abstract
The murine coronavirus mouse hepatitis virus (MHV) induced the expression of type I interferon (alpha/beta interferon [IFN-α/β]) in mouse oligodendrocytic N20.1 cells. This induction is completely dependent on virus replication, since infection with UV light-inactivated virus could no longer induce IFN-α/β. We show that MHV infection activated both transcription factors, the IFN regulatory factor 3 (IRF-3) and nuclear factor κB (NF-κB), as evidenced by phosphorylation and nuclear translocation of IRF-3 and an increased promoter binding activity for IRF-3 and NF-κB. Furthermore, the cytoplasmic pattern recognition receptor retinoic acid-inducible gene I (RIG-I) was induced by MHV infection. Knockdown of RIG-I by small interfering RNAs blocked the activation of IRF-3 and subsequent IFN-α/β production induced by MHV infection. Knockdown of another cytoplasmic receptor, the melanoma-differentiation-associated gene 5 (MDA5), by small interfering RNAs also blocked IFN-β induction. These results demonstrate that MHV is recognized by both RIG-I and MDA5 and induces IFN-α/β through the activation of the IRF-3 signaling pathway. However, knockdown of RIG-I only partially blocked NF-κB activity induced by MHV infection and inhibition of NF-κB activity by a decoy peptide inhibitor had little effect on IFN-α/β production. These data suggest that activation of the NF-κB pathway might not play a critical role in IFN-α/β induction by MHV infection in oligodendrocytes.
Innate immune response is the first line of host defense against invading microorganisms and ultimately regulates the adaptive immune response generated by both T and B cells (27, 33). The innate immune system recognizes conserved molecular structures expressed by diverse groups of pathogens, known as pathogen-associated molecular patterns (PAMPs), through specific pathogen recognition receptor (PRR) molecules, which initiate signaling pathways that ultimately lead to the expression of antiviral genes (1, 4, 5, 11, 16, 17, 30, 49). Type I interferons (alpha/beta interferon [IFN-α/β]) represent the most critical innate antiviral genes. For RNA viruses, double-stranded RNAs (dsRNAs) and single-stranded RNAs (ssRNAs), which are either present in viral genomes or generated during viral replication, are two major PAMPs and can be detected by most cell types through Toll-like receptor (TLR)-dependent and TLR-independent molecules (1, 4). TLR-3 is the first identified PRR that specifically recognizes dsRNA (2). It is present on the cell surface or in the lumen of the endosomes. TLR-3 interacts with incoming viral dsRNA through its N-terminal leucine-rich repeat domain, resulting in the recruitment of adaptor molecules, such as TRIF (Toll/interleukin-1 [IL-1] receptor domain containing adaptor inducing IFN-β) (28, 29, 32, 46). TRIF subsequently interacts with several kinases, leading to the activation of transcription factors nuclear factor κB (NF-κB) and IFN regulatory factor 3 (IRF-3) (1, 4, 17, 25, 26). Retinoic acid-inducible gene I (RIG-I) and melanoma-differentiation-associated gene 5 (MDA5) are two related cytoplasmic RNA helicases that have been shown to serve as cytosolic cellular sensors for RNA viruses, independently of TLR (42, 49, 50). RIG-I contains two caspase recruitment domain (CARD)-like motifs near its N terminus and a downstream DExD/H-box helicase domain. Binding of cytoplasmic viral dsRNA or 5′-triphosphate RNA (3p-RNA) with the RIG-I helicase domain results in a conformational change of RIG-I, allowing its CARD to recruit a CARD-containing adaptor, eventually leading to the activation and nuclear translocation of IRF-3 and NF-κB (4, 7, 8, 10, 18, 39, 45). Along with ATF-2-c-Jun, these transcription factors bind to different regions on the promoter of the IFN-β gene in the nucleus and lead to expression of the antiviral IFN-β and/or proinflammatory cytokine genes (24, 47). Once IFN-β is produced and secreted, it interacts with the heterodimeric IFN receptor (IFNR) on neighboring cells, leading to further amplification of IFN-β and induction of IFN-α and more than 100 IFN-stimulated genes (ISGs), which ultimately leads to the establishment of an “antiviral state” (9, 34, 38).
The murine coronavirus mouse hepatitis virus (MHV) is an enveloped, single-stranded RNA virus. The viral genome is a positive-sense RNA, capped at the 5′ end and polyadenylated at the 3′ end, and contains multiple open reading frames (20). During cell entry, the viral genome is delivered to the cytoplasm either at the cell surface through fusion between viral envelope and plasma membrane or from the endosome following endocytosis (31). Once in the cytoplasm, the genomic RNA is transported to the endoplasmic reticulum (ER) (53) where the 5′-end open reading frame of the viral genome is translated into a polymerase polyprotein, which is then cleaved into 16 nonstructural proteins (nsp's) (40). These nsp's form replication complexes that are associated with double-membranous vesicles derived from modified ER membranes (12, 19). The RNA polymerase complex synthesizes a nested set of genomic and subgenomic mRNAs and negative-strand counterparts as well as their corresponding dsRNA replication intermediates that can potentially be the source for recognition by the cytoplasmic PRRs.
MHV can infect rodents and cause diseases of the central nervous system (CNS) that range from acute fulminant fatal encephalitis to chronic demyelination. However, the severity of CNS disease is significantly influenced by both viral genetics and host factors. While the roles of an adaptive immune response in the pathogenesis of MHV-caused CNS diseases have been relatively well studied in the past, information about the contribution of antiviral innate immunity is rather limited (3, 35). Previous studies have shown that MHV infection in fibroblast cells does not induce type I IFNs (37, 44, 48, 52). It has been suggested that the inability of MHV to induce type I IFN is likely due to masking of viral dsRNAs in double-membranous vesicles during replication and preventing them from being recognized by cellular PRRs (44, 52). This is a logical assumption since MHV infection in the same fibroblast cells did not inhibit IFN-α/β production induced by double-stranded RNAs or Sendai virus (52). It has also been reported that MHV replication is insensitive to treatment with type I interferon in fibroblast cells and that this insensitivity is partially attributable to the expression of a viral IFN antagonist (48). In contrast, MHV replication was significantly enhanced in the CNS of IFNR-knockout mice compared to the wild-type mice, suggesting that virus replication is sensitive to type I IFN response in vivo (13, 36). Further, it has been recently shown that MHV infection induces type I IFNs in primary brain macrophages and microglia but not in primary neurons or astrocytes, a finding which correlates with virus replication and disease severity (36). These results suggest that type I IFNs play an important role in MHV replication and in pathogenesis of the CNS diseases. Taken together, these published data indicate that the ability of MHV to induce type I IFNs is cell type specific.
During the course of studying MHV infection in oligodendrocytes, we noticed that MHV replication is severely restricted. We then searched for potential antiviral factors that might be induced in MHV-infected oligodendrocytes. We discovered that MHV infection indeed induced type I IFNs. We further found that RIG-I was upregulated by MHV infection and that viral recognition by RIG-I and MDA5 is the driving force for the activation of IRF-3 and the ensuing induction of IFN-α/β. Production of IFN-α/β is likely an important factor in restricting subsequent MHV replication in oligodendrocytes.
MATERIALS AND METHODS
Virus, cells, and reagents.
The N20.1 cell line is a cell line clone derived from mouse primary cultures of oligodendrocytes conditionally immortalized by transformation with a temperature-sensitive mutant of the simian virus 40 large-T antigen (43). It was kindly provided by Anthony Compadre (University of California Los Angeles School of Medicine). The N20.1 cell line represents a mature oligodendrocyte line because it expresses the myelin basic protein (MBP) and myelin proteolipid protein (PLP) but a line that is at a very early stage of maturity based on the expression of the MBP and PLP alternatively spliced mRNA population (43). In addition, the N20.1 cell line can be recognized by the A2B5 antibody, which recognizes a surface galactolipid expressed by oligodendrocyte progenitor cells; the A007 antibody, which is specific for sulfatide and found on cells committed to the oligodendrocyte lineage as well as mature oligodendrocytes; and the anti-GC antibody, which recognizes the protein expressed specifically by mature oligodendrocytes (43). The N20.1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium with 10% fetal bovine serum (FBS) and G418 (100 μg/ml) at the permissive temperature of 34°C. Cells were grown either in T25 flasks or on glass coverslips coated with poly-d-lysine. Mouse fibroblast 17Cl-1 cells (kindly provided by Susan Baker, Loyola University of Chicago) were cultured in DMEM. Mouse astrocytoma DBT cells were cultured in Eagle minimal essential medium (EMEM) and were used for virus propagation and plaque assay as described previously (21). Either the wild-type mouse hepatitis virus A59 (MHV-A59) or recombinant MHV-A59/green fluorescent protein (GFP) was used for infection throughout the study. MHV-A59/GFP expresses the GFP that replaced the viral nonstructural protein 4 (NS4) (kindly provided by Susan Weiss, University of Pennsylvania School of Medicine). The virus was propagated in DBT cells. The virus was semipurified from the cultured medium through a 30% (wt/vol) sucrose cushion by ultracentrifugation at 27,000 rpm at 4°C for 4 h in the SW28 rotor (Beckman Coulter). Virus pellets were resuspended in serum-free medium, and the titer was determined by plaque assay on DBT cells as described previously (21). Purified virus was used for infection of N20.1 cells at a multiplicity of infection (MOI) of 10 throughout this study. The following antibodies were used in this study: rabbit anti-IRF-3 polyclonal antibody (catalog no. 4962), rabbit anti-phospho-IRF-3 (Ser396) monoclonal antibody (MAb) (4D4G) (catalog no. 4947), rabbit anti-NF-κB p65 polyclonal antibody (catalog no. 3034), and rabbit anti-RIG-1 polyclonal antibody (catalog no. 4520) were purchased from Cell Signaling Inc. while mouse anti-β-actin MAb (catalog no. A2228) was obtained from Sigma-Aldrich. IFN-α and IFN-β were purchased from Thermo Scientific. The NF-κB p65 decoy peptide (PTD-p65-p6) and the control peptide (PTD) were purchased from Imgenex, and their stock solutions of 5 mM were prepared in distilled water and stored at −20°C. Oligonucleotide poly(I:C) was purchased from Amersham-Pharmacia, while small interfering RNAs (siRNAs) against mouse RIG-I (catalog no. sc-61481) and its control siRNA (catalog no. sc-37007), mouse MDA5 (catalog no. sc-61011), and the enhanced GFP (EGFP) siRNA (catalog no. sc-36869) were obtained from Santa Cruz Biotechnology, Inc. These oligonucleotides were transfected into N20.1 cells at 50 nM [for poly(I:C)] or at 100 nM (for siRNAs) with the Lipofectamine (Invitrogen) or Neuroporter (Fisher Scientific) reagent according to the manufacturer's instructions.
RNA isolation and conventional or real-time quantitative RT-PCR (qRT-PCR).
Intracellular total RNAs were isolated from N20.1 cells using the Qiagen RNeasy Plus minikit (catalog no. 74134) according to the manufacturer's protocol and treated with RNase-free DNase I (Qiagen). The concentration of the RNA samples was determined with a spectrophotometer (model U2001; Hitachi). One nanogram of the total RNA sample was used in a reverse transcription (RT) reaction (25 μl) using the iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer's instructions. The reaction was carried out in an Applied Biosystems 9800 ThermaCycler in a 96-well plate for 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C, and the reaction mixture was then held at 4°C. Real-time PCR was carried out in a 50-μl reaction mixture containing 1 μl RT product, 1× iQ SYBR green Supermix PCR master mix, and 100 nM primers using an iCycler IQ Multicolor real-time PCR detection system (Bio-Rad). PCR was performed in a 96-well optical plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 57° for 1 min. The primer pairs used in the PCR were 5′-IFN-α (5′-ACA AGG CTG CCC CGA CTA C-3′, forward) and 3′-IFN-α (5′-TGG AAG ACT CCT CCC AGG TAT ATG-3′, reverse) for IFN-α and 5′-IFN-β (5′-GGC TTC CAT CAT GAA CAA CAG GT-3′, forward) and 3′-IFN-β (5′-AGG TGA GGT TGA TCT TTC CAT TCA-3′, reverse) for IFN-β. The primer pair for β-actin was 5′-GGC TAT GCT CTC CCT CAC G-3′ (forward) and 5′-CGC TCG GTC AGG ATC TTC AT-3′ (reverse). Results were analyzed using the iCycler IQ Multicolor real-time PCR detection system (Bio-Rad Laboratories). Gene expression levels in various experimental groups were calculated after normalizing cycle thresholds (CT) against the housekeeping gene β-actin and are presented as picogram values. The cycle threshold is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. Relative gene expression values were determined using the 2−ΔΔCT method of Livak and Schmittgen (23).
For detection of murine RIG-I mRNA, conventional RT-PCR was carried out as described previously (22). The forward and reverse primers were 5′-RIG-I (5′-GCC AGA GTG TGA GAA TCT CAG TCA G-3′) and 3′-RIG-I (5′-GAG AAC ACA GTT GCC TGC TGC TCA T-3′), respectively. The resultant RT-PCR product is 285 bp in length. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control in the RT-PCR with the forward primer 5′-CCA TGA CAA CTT TGG CAT TG-3′ and the reverse primer 5′-CCT GAA GTC GCA GGA GAC AAC C-3′. The PCR products were analyzed by agarose gel electrophoresis.
For detection of murine MDA5 mRNA, a two-step nested RT-PCR was carried out according to the manufacturer's protocol (Santa Cruz Biotechnology). Briefly, the first-step PCR was carried out in 30 cycles with a pair of outside primers and the second-step PCR was done in an additional 30 cycles with a pair of inside primers. Both primer pairs were purchased from Santa Cruz Biotechnology (catalog no. sc-61011-PR). A pair of primers specific for β-actin were used in the same PCR as a control for normalization.
ELISA.
The enzyme-linked immunosorbent assay (ELISA) system (PBL Biomedical Laboratories) was used for detection and quantification of IFN-α/β according to the manufacturer's instructions. Briefly, N20.1 cells were infected with MHV-A59/GFP at an MOI of 10. Culture supernatants were collected at various time points postinfection (p.i.) for determining the protein levels of IFN-α/β. ELISA was performed in 96-well plates that were precoated with an antibody specific to mouse IFN-α or IFN-β. The plate was incubated with the culture supernatants and controls for 2 h at room temperature and then washed with a wash buffer five times. The mouse IFN-α/β conjugate was added to each well and incubated for an additional 2 h followed by 5 washes. The substrate solution was added to each well and incubated for 30 min. The reaction was stopped by the addition of a stop solution. The plate was read with a microplate reader (BioTek Instruments, Inc.) at 450 nm, and 540 nm was used as a correction. The IFN protein concentration was obtained based on a standard curve generated by performing parallel assays using known amounts of IFN-α or IFN-β included in the kits.
UV inactivation of viruses.
To inactivate the virus by UV light, purified virus was diluted to a concentration of 107 PFU per ml in serum-free medium. Aliquots of 1 ml were added to each well of the 6-well tissue culture plate. The plates were placed on ice at a distance of 14 cm from the UV light in a UV cross-linker (Fisher Scientific) and exposed to UV light at an energy level of 1,200 μW/ms for 30 min. Viral inactivation was confirmed by titration of samples before and after UV exposure and by the absence of cytopathic effect (CPE) after inoculation of DBT cell monolayers with the UV-irradiated virus samples. Preliminary experiments indicated that a 10-min exposure to such UV light completely inactivated MHV infectivity.
Western blot analysis.
For Western blot analysis, whole-cell lysates were prepared using cell lysis buffer (catalog no. 9038; Cell Signaling Inc.) according to the manufacturer's protocol. The protein concentration was measured by using the Bio-Rad protein assay kit. Equal amounts of total proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a nitrocellulose membrane (Bio-Rad) for 7 h at 40 V in the transfer buffer. Membranes were blocked for 1 h at room temperature with 5% powdered skim milk in Tris-buffered saline (10 mM Tris-Cl [pH 7.5], 150 mM NaCl) with 0.05% Tween 20 (TBST). The membranes were incubated with a primary antibody at 4°C overnight, followed by incubation with a secondary horseradish peroxidase (HRP)-conjugated antibody for 1 h at room temperature. Blots were developed with an enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia) and exposed to X-ray film (Fuji). After the membranes were incubated in stripping buffer (10 mM β-mercaptoethanol, 2% [wt/vol] SDS, 62.5 mM Tris, pH 6.7) at 55°C for 30 min, they were washed in TBST, blocked, and incubated with anti-β-actin MAb (as an internal control) at room temperature for 1 h. Detection of β-actin was carried out following incubation with the secondary anti-mouse IgG antibody conjugated with HRP and ECL.
Immunofluorescence assay (IFA).
N20.1 cells were seeded onto 12-mm-diameter coverslips coated with poly-d-lysine, and 48 h later cells were either mock infected or infected with MHV at an MOI of 10. Transfection of poly(I:C) was performed using Lipofectin transfection reagent (Invitrogen) at a concentration of 8 μg/ml, according to the manufacturer's instructions. Coverslips were removed at the indicated times, fixed in 4% formaldehyde for 10 min at 25°C, washed three times in phosphate-buffered saline (PBS), and permeabilized with 0.1% Triton X-100 for 1 min at 25°C. The coverslips were then washed two times in PBST (1× PBS with 0.04% Tween 20), incubated in blocking solution (5-mg/ml bovine serum albumin [BSA] in PBST) for 1 h at 25°C, and incubated overnight at 4°C with primary antibody against IRF-3 (sc-9083; Santa Cruz Biotechnology) or rabbit anti-NF-κB p65 monoclonal antibody (catalog no. 3035; Cell Signaling Inc.). After being washed three times with PBST, cells were incubated with secondary antibody (fluorescein isothiocyanate [FITC]-conjugated goat anti-rabbit IgG) (Sigma-Aldrich) for 1 h. Coverslips were then washed three times in PBS. Finally, the coverslips were mounted on glass slides with a mounting medium. Cells were viewed on an Olympus fluorescent microscope at ×40 magnification (Olympus IX70), and images were taken with a digital camera (MagnaFire).
Assay for transcription factor activity.
For determining the activity of the transcription factors, nuclear extracts were isolated with a nuclear extraction kit (Active Motif), quantified by using a Quick Start Bradford protein assay kit (Bio-Rad Laboratories), and stored at −80°C for further use. The promoter binding activity of the transcription factors (IRF-3 and NF-κB p65) in the nuclear extracts was determined with TransAM ELISA-based kits (Active Motif). Briefly, nuclear extracts were added to the wells of a 96-well plate with an immobilized oligonucleotide that contains the consensus binding site for the given transcription factor (IRF-3 or NF-κB p65). Following incubation and extensive washes, the quantity of bound transcription factor was then determined by use of a primary antibody and a secondary antibody conjugated to horseradish peroxidase in an ELISA-based format. Wild-type and mutated consensus oligonucleotides were used for competitive binding with the transcription factors to monitor the specificity of the assay. The wild-type oligonucleotide competes for IRF-3 or NF-κB p65 binding to the oligonucleotide immobilized on the plate, while the mutated oligonucleotide does not. These controls were included in each assay.
Statistical analysis.
The results are expressed as means ± standard deviations (SD), and the mean values were compared using the Student t test. Values of P < 0.05, P < 0.01, and P < 0.001 were considered statistically significant.
RESULTS
MHV replication is severely restricted in oligodendrocytes and is sensitive to treatment with type I IFNs.
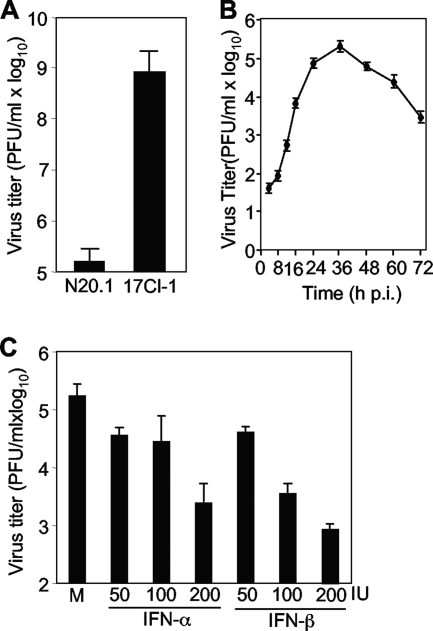
During the course of studying MHV infection in oligodendrocytes, we observed that MHV replication was severely restricted. When mouse oligodendrocytic N20.1 cells were infected with MHV-A59/GFP at an MOI of 10 for 24 h, the virus titer reached approximately 5 log10 PFU/ml. In contrast, the same virus grew to a significantly higher titer (≈9 log10 PFU/ml) in fibroblast 17Cl-1 cells for the same period of time (Fig. 1A). The one-step growth curve of MHV-A59/GFP in N20.1 cells was further assessed. It was found that the virus titer reached the plateau of approximately 5 log10 at 36 h p.i. and then gradually and continually declined (Fig. 1B). This result led us to hypothesize that antiviral factors might be present or induced in oligodendrocytes, which in turn restrict viral replication. We thus tested whether type I IFNs, which are considered to be universal antiviral factors, contribute to this restriction. N20.1 cells grown in a 24-well plate were treated with either IFN-α or IFN-β for 24 h prior to and throughout the infection. Cells were infected with MHV-A59/GFP at an MOI of 10. At 24 h p.i., virus titers in the culture medium were determined. The results showed that virus titers were reduced by approximately 2 log10 after treatment with either IFN-α or IFN-β at 200 IU and that the inhibitory effect was dose dependent (Fig. 1C). The data also indicate that IFN-β is a more potent inhibitor of virus replication than is IFN-α (Fig. 1C). These findings suggest that type I IFNs might be induced in oligodendrocytic N20.1 cells by MHV infection.
FIG. 1.
MHV replication is severely restricted and sensitive to type I interferon treatment in oligodendrocytes. (A) Restriction of MHV growth in oligodendrocytes. Oligodendrocyte N20.1 cells or fibroblast 17Cl-1 cells were infected with MHV-A59/GFP at an MOI of 10. Virus titer in culture medium was determined at 24 h p.i. by plaque assay. (B) One-step growth curve of MHV-A59/GFP in N20.1 cells. (C) Sensitivity of MHV replication to IFN treatment. N20.1 cells were treated with either IFN-α or IFN-β at 50, 100, and 200 international units (IU) for 24 h and infected with MHV-A59/GFP at an MOI of 10 in the presence of the respective IFN. At 24 h p.i., virus titer in culture medium was determined by plaque assay. The results are expressed as the mean PFU per ml for three independent experiments. Error bars indicate standard deviations of the means. M, mock treatment.
MHV infection induced type I interferon response in oligodendrocytes.
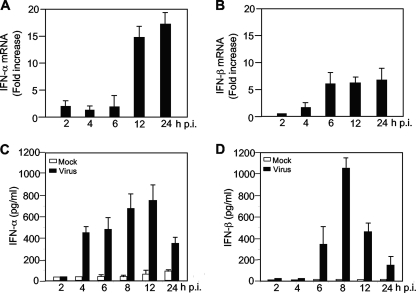
Recent studies suggest that the antiviral innate immune response to MHV infection varies widely among different cell types (36, 37, 48, 52). MHV induces a type I IFN response in primary macrophages and microglia but not in several fibroblast cell lines, primary hepatocytes, neurons, or astrocytes. Since oligodendrocytes are some of the major target cells for MHV infection and persistence in the central nervous system and since MHV replication is restricted and sensitive to IFN treatment in oligodendrocytes (Fig. 1), we wondered whether MHV infection could induce type I IFN in oligodendrocytes. N20.1 cells were infected with MHV-A59/GFP at an MOI of 10. At various time points p.i., intracellular RNAs were isolated and the mRNAs for IFN-α/β were quantified with real-time quantitative RT-PCR. As shown in Fig. 2A and B, both IFN-α/β mRNAs were induced at 2 h p.i. and continued to increase throughout the 24-h period, indicating that IFN-α/β transcription is activated by MHV infection. It has been shown previously that although MHV infection induced transcription of IFN-β in fibroblast cells, IFN-β protein was not detected (37). Thus, we also determined the protein level for IFN-α/β in culture medium at various times p.i. with ELISA. The results showed that both IFN-α/β proteins were produced and secreted into the medium (Fig. 2C and D). IFN-α protein was first detected at 4 h p.i. at approximately 400 pg/ml and reached a plateau (≈800 pg/ml) at 12 h p.i. IFN-β protein was first detected at 6 h p.i. and rapidly reached a plateau (≈1,000 pg/ml) by 8 h p.i. In contrast to the mRNAs, which continuously increased throughout the 24-h period, IFN-α/β protein levels rapidly decreased thereafter and reached <400 and <200 pg/ml, respectively, at 24 h p.i. This result suggests a potential translational inhibition during the late stage of virus infection. Nevertheless, these findings clearly demonstrate that IFN-α and IFN-β are induced in oligodendrocytes by MHV infection.
FIG. 2.
Induction of type I interferon in oligodendrocytes by MHV infection. Oligodendrocyte N20.1 cells were infected with MHV-A59/GFP at an MOI of 10. Cells and culture medium were separately collected at 0, 2, 4, 6, 12, and 24 h p.i. for mRNA (A and B) and protein (C and D) detection, respectively. (A and B) Kinetics of IFN-α and IFN-β mRNA induction. Intracellular RNAs were isolated, and the amounts of IFN-α/β mRNAs were determined by real-time quantitative RT-PCR. The results are expressed as fold increase over those at 0 h p.i. for three independent experiments and are normalized to β-actin for each time point. Error bars indicate standard deviations of the means. (C and D) Kinetics of IFN-α/β protein induction. The amounts of IFN-α/β protein in the supernatants were determined with a respective ELISA kit. The results are expressed as the mean picograms per ml for three independent experiments. Error bars indicate standard deviations of the means.
Virus replication is required for IFN-α/β induction.
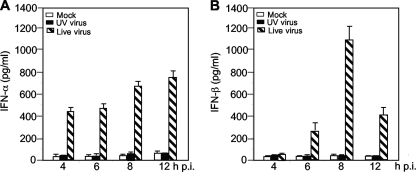
For most RNA viruses, induction of IFN-α/β can be triggered at the cell surface or in the endosome by TLR-3 or in the cytoplasm by cytoplasmic RNA helicases. One way to discriminate these two pathways is to determine whether virus entry induces IFN-α/β or whether virus replication (dsRNA) is required for IFN-α/β induction. Thus, N20.1 cells were infected with live MHV-A59/GFP at an MOI of 10 or with UV-inactivated MHV-A59/GFP at an equivalent MOI of 10 of preinactivated virus. Mock-infected cells were used as a control. At various time points p.i. as indicated, culture medium was collected for determining the amount of IFN-α/β proteins by ELISA. As shown in Fig. 3, the amounts of both IFN-α and IFN-β were at the background level in cells infected with the UV-inactivated virus, similar to those from mock-infected controls, indicating that virus replication is absolutely required for IFN-α/β induction in oligodendrocytes. This result suggests that MHV is most likely recognized by cytoplasmic RNA helicases.
FIG. 3.
Virus replication is required for IFN-α/β induction. N20.1 cells were infected with UV-inactivated MHV-A59/GFP or live MHV-A59/GFP at an MOI of 10 or mock infected as a control. At various times p.i. as indicated, culture medium was collected for determination of the IFN-α/β protein levels with respective ELISA kits. The results are expressed as the mean picograms per ml for three independent experiments. Error bars indicate standard deviations of the means.
Activation of the transcription factors IRF-3 and NF-κB in oligodendrocytes by MHV infection.
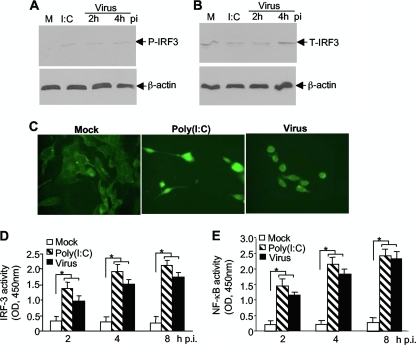
Since the initial induction of IFN-α/β transcription involves activation of transcription factors IRF-3 and NF-κB, their activation by MHV infection was then assessed in N20.1 cells with three complementary approaches. Cells were infected with MHV-A59/GFP at an MOI of 10 or transfected with poly(I:C) as a positive control. Mock-infected cells were used as a negative control. At 2 and 4 h p.i., cell lysates were extracted and assayed for the phosphorylation status of IRF-3 using a phosphor-specific antibody in Western blotting. β-Actin was used as an internal control for protein normalization. The results show that while total IRF-3 was detected in all samples, phosphorylated IRF-3 was detected only in virus-infected or poly(I:C)-transfected cells, not in mock-infected cells, indicating that IRF-3 was activated by MHV infection (Fig. 4A and B). It is noted that the low level of phosphorylated IRF-3 protein is most likely the result of overall IRF-3 protein levels in this type of cell (compare Fig. 4A with 4B). We then determined the nuclear translocation of IRF-3 following virus infection or poly(I:C) transfection (Fig. 4C). Consistent with the phosphorylation status of IRF-3, IRF-3 was translocated to the nucleus in virus-infected or poly(I:C)-transfected N20.1 cells but not in mock-infected N20.1 cells, confirming the activation of IRF-3 by MHV infection. Finally we used a commercial assay kit to determine the activity of IRF-3 in the nuclear extracts. The results show that IRF-3 activities were significantly increased in virus-infected and poly(I:C)-transfected nuclear extracts at 2, 4, and 8 h p.i. (or posttransfection [p.t.]) compared to those in mock-infected controls (P < 0.01) (Fig. 4D). In a parallel experiment, we determined the activity of NF-κB in the nuclear extracts and found that the activities of NF-κB were significantly increased in virus-infected and poly(I:C)-transfected cells over those in the mock-infected controls (P < 0.01) (Fig. 4E). These results demonstrate that both IRF-3 and NF-κB are activated in oligodendrocytes by MHV infection.
FIG. 4.
Activation of the transcription factor IRF-3 in oligodendrocytes by MHV infection. N20.1 cells were infected with MHV-A59/GFP at an MOI of 10 (Virus), mock infected as a negative control (Mock), or transfected with poly(I:C) as a positive control [I:C or Poly(I:C)]. (A and B) IRF-3 is phosphorylated by MHV infection. At 2 and 4 h p.i. or 4 h posttransfection, cell lysates were isolated for detection of phosphorylated and total IRF-3 (P-IRF-3 and T-IRF-3, respectively) by Western blot analysis. β-Actin was used as an internal control. Lanes M, mock infection. (C) IRF-3 is translocated to the nucleus during MHV infection. At 4 h p.i. or posttransfection, cells were fixed and the intracellular localization of IRF-3 was visualized by IFA staining with an antibody to IRF-3. (D and E) Activation of IRF-3 and NF-κB by MHV infection. At 2, 4, and 8 h p.i. or posttransfection, nuclear extracts were prepared and the IRF-3 or NF-κB activity was determined with the respective ELISA-based assay, in which the oligonucleotide containing the IRF-3 or NF-κB consensus binding site is immobilized on the microplate for capturing the activated transcription factor. The amount of the bound complexes was determined with a microplate reader as optical density at 450 nm (OD450). The values represent the means of the readings from three samples, and error bars are the standard deviations of the means. The asterisks indicate statistical significance (P < 0.05) between the comparison groups.
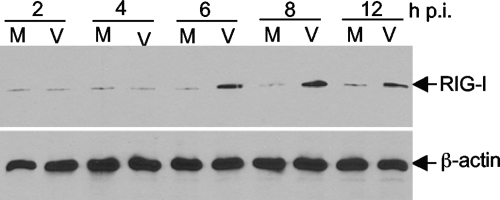
RIG-I is upregulated by MHV infection.
The results from UV-inactivated virus suggest that MHV is most likely recognized by cytoplasmic RNA helicases. One of the three known cytoplasmic RNA helicases is RIG-I. Since RIG-I is also an IFN-stimulated gene (ISG) product, its expression can be upregulated by IFN-α/β through the so-called positive feedback mechanism (17). We wondered whether RIG-I is also upregulated by MHV infection in N20.1 cells. Cells were infected with MHV-A59 at an MOI of 10 or mock infected as a control. At various time points p.i. as indicated, cell lysates were isolated and RIG-I protein levels were determined by Western blot analysis. As shown in Fig. 5, the expression of RIG-I was indeed upregulated by MHV infection, compared to mock-infected controls. The upregulation was detected beginning at 6 h p.i. and continued until the end of the experiment (12 h p.i.). This result thus establishes that RIG-I is upregulated by MHV infection in N20.1 cells.
FIG. 5.
RIG-I is upregulated in oligodendrocytes by MHV infection. N20.1 cells were infected with MHV-A59/GFP at an MOI of 10 (V) or mock infected as a control (M). At various time points p.i. as indicated, cell lysates were isolated for determining the RIG-I protein level by Western blot analysis. β-Actin was used as an internal control.
Recognition of MHV by RIG-I is responsible for triggering IFN-α/β production in oligodendrocytes.
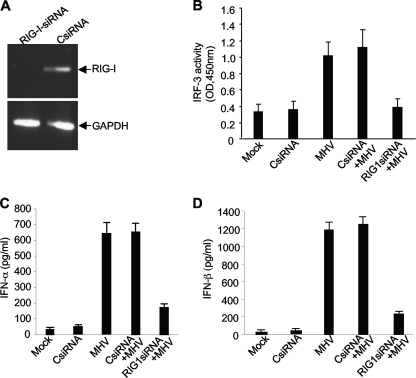
To establish that the activation of the transcription factors and the ensuing production of IFN-α/β are initiated through the recognition by RIG-I, we performed an experiment in which RIG-I mRNA was knocked down by specific siRNAs. Cells were transfected with a set of siRNAs specifically targeting the mRNA of RIG-I. At 24 h posttransfection (p.t.), the presence of RIG-I mRNA was detected by RT-PCR. It was found that RIG-I mRNA was completely knocked down when transfected with the RIG-I siRNA compared to that when transfected with a nonspecific control siRNA (CsiRNA) (Fig. 6A). We then determined whether RIG-I is required for the activation of the transcription factor IRF-3. At 24 h following transfection with RIG-I siRNAs or control siRNAs, cells were infected with MHV. Mock-infected and mock-transfected cells were used as controls. At 8 h p.i., nuclear extracts were isolated for determining the IRF-3 activity using the commercial assay kit. Figure 6B shows that knockdown of RIG-I mRNA completely blocked the activation of IRF-3 by MHV infection. This blockage is specific for RIG-I, since transfection with nonspecific siRNAs did not inhibit IRF-3 activity induced by MHV infection. This result indicates that activation of the IRF-3 signaling pathway is mediated through RIG-I during MHV infection. To further determine whether RIG-I is responsible for the induction of IFN-α/β by MHV infection, medium collected at 8 h p.i. from the same experiment was used for determining the production of IFN-α/β. In general agreement with the inhibitory effect on IRF-3 activity, knockdown of RIG-I drastically but incompletely blocked the production of IFN-α/β, while IFN-α/β induction was not affected by transfection with nonspecific control siRNAs (Fig. 6C and D). Taken together, these data demonstrate that production of IFN-α/β in oligodendrocytes induced by MHV infection is largely mediated through the activation of the IRF-3 signaling pathway that is triggered by the recognition of viral RNAs by RIG-I.
FIG. 6.
RIG-I is responsible for the activation of IRF-3 and production of IFN-α/β in oligodendrocytes induced by MHV infection. (A) Knockdown of RIG-I by siRNAs. N20.1 cells were transfected with a pool of siRNAs specific for RIG-I (RIG-I-siRNA) or the nonspecific control siRNA (CsiRNA). At 48 h posttransfection, intracellular RNAs were isolated and the mRNAs for RIG-I were detected by RT-PCR using primers specific to the mouse RIG-I gene. The housekeeping gene GAPDH was used as an internal control. (B) MHV-induced IRF-3 activation was blocked by knockdown of RIG-I. N20.1 cells were transfected with the RIG-I siRNAs or CsiRNA or mock transfected. At 24 h posttransfection, cells were infected with MHV-A59/GFP at an MOI of 10 or mock infected. At 8 h p.i., nuclear extracts were isolated for determining the IRF-3 activity as described in the legend to Fig. 4. (C and D) Viral recognition by RIG-I is critical for the IFN-α/β induction. Cells were treated as described for panel B. At 8 h p.i. culture medium was collected for determination of IFN-α/β proteins by ELISA. The results are expressed as the mean picograms per ml for three independent experiments. Error bars indicate standard deviations of the means.
Activation of the NF-κB signaling pathway is not critical for the production of IFN-α/β induced by MHV infection in oligodendrocytes.
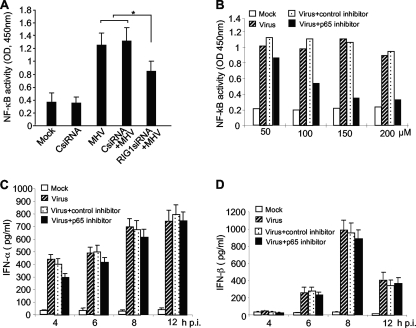
Since the transcription factor NF-κB is activated in oligodendrocytes by MHV infection (Fig. 4E), we wondered whether the activation of NF-κB is also mediated through RIG-I, as in the case for IRF-3. N20.1 cells were transfected with RIG-I siRNAs or nonspecific control siRNAs or mock transfected as a control. Cells were then infected with MHV-A59/GFP at an MOI of 10 or mock infected at 24 h p.t. At 8 h p.i., nuclear extracts were prepared for assaying the NF-κB activity. Unlike IRF-3, NF-κB activity was only partially blocked by knockdown of RIG-I, although the reduction of NF-κB activity was statistically significant (P < 0.05) compared to that for cells treated with CsiRNA (Fig. 7A). This result suggests that additional signaling pathways other than RIG-I might have contributed to the activation of NF-κB in MHV-infected N20.1 cells.
FIG. 7.
Role of NF-κB in MHV-induced IFN-α/β production in oligodendrocytes. (A) Activation of NF-κB is partially mediated through RIG-I. N20.1 cells were transfected with the RIG-I siRNAs or nonspecific control siRNA (CsiRNA) or mock transfected. At 24 h posttransfection, cells were infected with MHV-A59/GFP at an MOI of 10 or mock infected. At 8 h p.i., nuclear extracts were isolated for determining the NF-κB activity with an ELISA-based assay kit as described in Materials and Methods. The asterisk indicates statistical significance (P < 0.05) between the comparison groups. (B) Optimization of NF-κB inhibitor. N20.1 cells were treated with NF-κB p65 decoy peptide inhibitor or the nonspecific control peptide inhibitor at 50 to 200 μM or mock treated for 1 h prior to infection. Cells were then infected with MHV-A59/GFP at an MOI of 10 or mock infected for 2 h in the presence of the inhibitors, and nuclear extracts were then prepared for determining the NF-κB activity as described for panel A. “Mock” indicates mock treatment and mock infection; “Virus” indicates virus infection only. (C and D) Effect of RIG-I knockdown on IFN-α/β induction. N20.1 cells were pretreated with NF-κB p65 inhibitor or control inhibitor at 150 μM and infected with the virus as described for panel B. At various time points as indicated, culture medium was harvested for determining IFN-α/β protein levels by ELISA. The results are expressed as the mean picograms per ml for three independent experiments. Error bars indicate standard deviations of the means.
We then determined whether NF-κB plays a role in transcription of IFN-α/β. N20.1 cells were pretreated with the NF-κB decoy p65 peptide inhibitor or a nonspecific control peptide inhibitor for 12 h and then infected with MHV-A59/GFP at an MOI of 10 or mock infected as a control. Since the 50% inhibitory concentration (IC50) for the decoy p65 inhibitor can vary in different types of cells, we first determined the IC50 in N20.1 cells even though the manufacturer's recommended concentration is 50 μM. The results show that the IC50 for the p65 inhibitor was approximately 100 μM in N20.1 cells (Fig. 7B). Thus, a higher concentration (150 μM) of the p65 inhibitor was chosen in subsequent experiments to ensure effective inhibition of NF-κB. We then determined the IFN-α/β production at various time points p.i. as indicated and found that neither IFN-α nor IFN-β was significantly affected by the treatment with the decoy p65 peptide inhibitor (Fig. 7C and D). These results demonstrate that inhibition of NF-κB activation failed to block IFN-α/β production, suggesting that NF-κB does not play a critical role in the induction of IFN-α/β in oligodendrocytes by MHV infection.
Recognition by MDA5 is also involved in MHV-induced IFN-β signaling in oligodendrocytes.
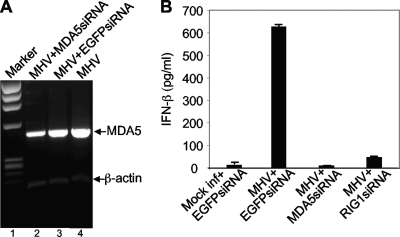
To determine if MDA5 can recognize MHV and mediate type I IFN induction, we initially used an antibody to MDA5 to determine its presence in virus- and mock-infected N20.1 cells by Western blot analysis. Unfortunately, no MDA5 protein was detected in virus-infected cells at various times p.i. or in mock-infected cells (data not shown). Thus, it was not clear whether MDA5 protein was absent in N20.1 cells or whether the antibody lacked reactivity. We then used RT-PCR to detect its mRNA. While the amount of MDA5 mRNA was extremely low in N20.1 cells, the nested RT-PCR using a total of 60 cycles was able to amply MDA5 mRNA in MHV-infected N20.1 cells (Fig. 8A, lanes 2 to 4). To determine whether MDA5 plays a role in MHV-induced IFN-β signaling, cells were transfected with MDA5 siRNAs or EGFP siRNA as a negative control for 24 h. The effectiveness of siRNAs in knocking down MDA5 mRNA was assessed by RT-PCR. It was found that the amount of MDA5 mRNA decreased by approximately 40% compared to that following transfection with the control EGFP siRNA while the amount of β-actin mRNA remained unchanged (compare lane 3 with lane 2 in Fig. 8A). Importantly, knockdown of MDA5 mRNA by its siRNAs drastically blocked the induction of IFN-β by MHV infection compared to that by the control EGFP siRNA (Fig. 8B). As a positive control for this experiment, knockdown of RIG-I mRNA by siRNAs also drastically blocked IFN-β induction (Fig. 8B). These results demonstrate that recognition by MDA5 is also involved in MHV-induced IFN-β signaling.
FIG. 8.
Role of MDA5 in MHV-induced IFN-β signaling. (A) Knockdown of MDA5 mRNA by siRNA. N20.1 cells were transfected with siRNAs specific to MDA5 or EGFP or mock transfected. At 24 h posttransfection, cells were infected with MHV-A59/GFP at an MOI of 10. At 24 h p.i., cellular mRNAs were isolated and amplified by RT-PCR with primers specific to mouse MDA5 or β-actin (see Materials and Methods). The molecular weight marker is indicated on the left (lane 1), and the bands representing MDA5 and β-actin are indicated with arrows. (B) Effect of MDA5 knockdown on IFN-β induction. The experiment was identical to that described for panel A but with additional controls that include mock infection and transfection with EGFP siRNA as a negative control for the inability to induce IFN-β and MHV infection and transfection with RIG-I siRNA as a positive control for the inhibition of IFN-β induction. The amount of IFN-β protein was quantified with the ELISA kit and expressed as the mean picograms per ml for three independent experiments. Error bars indicate standard deviations of the means.
Type I IFN induction contributes at least in part to the restriction of MHV replication in oligodendrocytes.
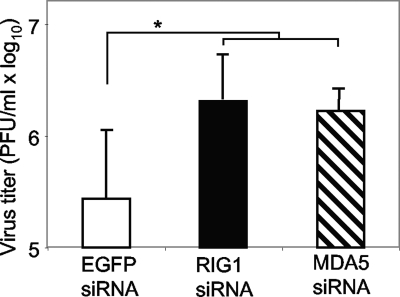
Results presented in Fig. 1 suggest that type I IFN may play a role in restricting MHV production in oligodendrocytes. To directly test this hypothesis, induction of IFN-β in oligodendrocyte N20.1 cells by virus infection was blocked by knockdown of RIG-I or MDA5 by transfection with the respective siRNAs. Transfection with siRNA to EGFP was used as a negative control. The effectiveness of gene knockdown and of IFN-β inhibition was assessed with RT-PCR and ELISA, respectively, as shown in Fig. 8. The effect of IFN-β inhibition on infectious virus production was determined at 24 h p.i. by plaque assay. As shown in Fig. 9, virus titer increased slightly (approximately 1 log10) but consistently and statistically significantly following knockdown of RIG-I or MDA5 compared to that in the EGFP siRNA control. These results thus demonstrate that the innate antiviral IFN response to MHV infection contributes as least in part to restriction of subsequent virus production in oligodendrocytes.
FIG. 9.
Effect of knockdown of RIG-I or MDA5 on virus production. N20.1 cells were transfected with siRNAs specific to RIG-I, MDA5, or EGFP for 24 h and then infected with MHV-A59/GFP at an MOI of 10. At 24 h p.i., virus titer in the culture medium was determined by plaque assay. The results are expressed as the mean PFU per ml for three independent experiments. Error bars indicate standard deviations of the means. The asterisk indicates statistical significance (P < 0.05) between the comparison groups.
DISCUSSION
In this study we have shown that MHV replication is severely restricted in oligodendrocyte N20.1 cells but is robust in fibroblast 17Cl-1 cells (≈4-log10 difference in virus titer) (Fig. 1A). This difference cannot be explained solely by the differential susceptibility of the two types of cells, even though only up to 30% of the N20.1 cells are susceptible to MHV infection compared to >90% for 17Cl-1 cells (data not shown). The lower virus titer in N20.1 cells at 24 h p.i. (Fig. 1A) is not due to slower growth, since the one-step growth curve shows that the virus titer decreased after 36 h and never reached more than 5.5 log10 during the 72 h p.i. (Fig. 1B). Thus, this finding suggests that the N20.1 cell line may contain antiviral factors that are either present endogenously or induced by virus infection. Interestingly, several recent studies have reported on the inability of MHV to induce antiviral type I IFNs in fibroblast cells, including studies of 17Cl-1 cells and their insensitivity to treatment with type I IFNs (37, 44, 48, 52). These findings prompted us to further examine whether the N20.1 oligodendrocyte line is unique in its type I IFN response to MHV infection. Indeed, we found that MHV infection of the N20.1 cell line induced IFN-α/β (>800 pg/ml) within 12 h after infection (Fig. 2). Furthermore, MHV replication was severely inhibited when cells were pretreated with either IFN-α or IFN-β (Fig. 1C). These findings suggest that the type I IFN response to initial MHV infection may play a role in restricting subsequent MHV production in N20.1 cells (Fig. 1A). In support of this hypothesis is the direct evidence from our results showing that inhibition of IFN-β induction by knockdown of RIG-I or MDA5 mRNA with specific siRNAs increased virus titer in N20.1 cells (Fig. 8 and 9). Our findings further support the notion that different types of cells respond differently to MHV infection with respect to type I IFN induction (36).
The signaling pathways that lead to the induction of IFN-α/β have been established in the current study. We show that virus particles alone are not sufficient to induce IFN-α/β during cell entry (Fig. 3), suggesting that recognition of virions by TLR-3 either at the plasma membrane or in the endosome does not play a role in IFN-α/β induction in oligodendrocytes. Similarly, the viral genomic RNA prior to release into the cytoplasm (uncoating) is likely not recognized by TLR-3, although we have not specifically examined the activation of the TLR-3 signaling pathway during entry. Instead, virus replication is absolutely required for IFN-α/β induction (Fig. 3). This suggests that dsRNAs generated during virus replication are most likely recognized by cytoplasmic PRRs and induce IFN-α/β. In support of this notion is the evidence that knockdown of the cytoplasmic PRR RIG-I or MDA5 almost completely blocked the induction of IFN-α/β by MHV infection (Fig. 6 and 8). Consistent with the well-documented signaling pathway initiated by RIG-I, both downstream transcription factors, IRF-3 and NF-κB, were activated in oligodendrocytes during MHV infection (Fig. 4). Interestingly, knockdown of RIG-I by siRNAs completely blocked the activation of IRF-3 but only partially inhibited NF-κB activation (Fig. 6 and 7). Does this finding suggest that other upstream signals that might be triggered by virus infection also partially contribute to the activation of NF-κB? Recently we have shown that MHV infection in primary mouse astrocytes activates NF-κB through the activation of a mitogen-activated protein kinase, Jun N-terminal protein kinase (JNK), which is also dependent on virus replication (51). Thus, it is possible that MHV replication in oligodendrocytes may activate the JNK signaling pathway, which in turn activates NF-κB. Further studies are necessary to determine this possibility.
Most intriguing, however, is the finding that blocking NF-κB binding to the promoter of IFN-α/β genes with the decoy p65 peptide inhibitor was unable to inhibit IFN-α/β induction by MHV infection (Fig. 7). This result is reminiscent of what we recently found for the regulation of tumor necrosis factor alpha (TNF-α) in MHV-infected primary mouse astrocytes (51). In that study, inhibition of the JNK signaling pathway blocked the activation of NF-κB and the induction of TNF-α while inhibition of NF-κB with the decoy p65 peptide did not block TNF-α induction (51). Such a coincidence prompted us to reexamine whether the decoy p65 peptide was also effective at the recommended concentration of 50 μM in oligodendrocyte N20.1 cells. Interestingly, we found that the decoy p65 peptide inhibitor at 50 μM had little effect on NF-κB activity in oligodendrocytes even though there was a clear dose-dependent inhibition (Fig. 7). This result suggests that the inhibitory effect of the decoy peptide is specific to NF-κB but that it requires a much higher concentration for an effective inhibition in oligodendrocytes. Thus, a concentration of 150 μM for the decoy peptide, which almost completely blocked the activity of NF-κB, was used in subsequent experiments. However, the decoy peptide even at such a high concentration did not have any significant effect on NF-κB activity (Fig. 7). Although it is well known that the p50 subunit of NF-κB sometimes has an inhibitory effect on target gene transcription, to date there is no evidence indicating an inhibitory effect for the p65 subunit of NF-κB (6). Since the detection kit specifically detects the p65 activity in the nuclear extracts and since the NF-κB activity was inhibited specifically by the decoy p65 peptide in the controls (Fig. 7A and B), the inability of decoy p65 peptide to block IFN-α/β induction demonstrates that in the presence of an activated IRF-3 pathway, NF-κB is dispensable in MHV-induced IFN-α/β expression in oligodendrocytes. However, the present study does not address whether and to what extent the NF-κB pathway contributes to MHV-induced IFN-α/β expression in the absence of a functional IRF-3 pathway.
A recent study from Weiss's laboratory shows that MHV induces IFN-β in the macrophages and microglia in the brains of wild-type mice (36). Furthermore, MHV appears to be recognized specifically by MDA5 since IFN-β is still induced in bone marrow-derived macrophages generated from mice lacking MyD88, RIG-I, and TLR-3 but not MDA5 (36). Our study shows that knockdown of either RIG-I mRNA or MDA5 mRNA in N20.1 cells by the respective siRNAs significantly blocked the induction of IFN-β upon MHV infection (Fig. 8), indicating that MHV can be recognized by both RIG-I and MDA5 in N20.1 cells. However, our results do not distinguish the relative roles played by RIG-I and MDA5 in sensing MHV and signaling IFN induction. Nevertheless, combined data from this study and those from the work of Roth-Cross et al. (36) clearly show that recognition of MHV by RIG-I appears critical for IFN-β induction in oligodendrocytes but dispensable in bone marrow-derived macrophages. The difference in recognition by different PRRs is most likely due to cell type specificity, since the same MHV cannot be recognized by either RIG-I or MDA5 in fibroblast cells (52). Cell-type-specific involvement of RIG-I and differential recognition of RNA viruses by RIG-I and MDA5 have been previously reported (14, 15).
Our analysis of the kinetics of IFN-α/β gene expression (Fig. 2) has identified an interesting regulatory pattern. The IFN-α/β mRNAs appeared to continually increase throughout the 24-h period after virus infection. However, their protein levels peaked at 12 and 8 h p.i., respectively, and rapidly declined thereafter (Fig. 2). This gene expression pattern suggests that the dsRNAs generated during viral replication within the first 24 h or perhaps longer can be continuously recognized by the intracellular PRRs, which induce the transcription of IFN-α/β, while their expression is likely inhibited at late times of virus infection at the posttranscriptional level. It is not clear why the kinetics of mRNA and protein levels at early times of virus infection are different for IFN-α and IFN-β. The inhibitory effect on IFN-α/β protein expression at late times of virus infection is likely exerted by secreted IFN-α/β proteins themselves since IFN-α and IFN-β are well-known inducers of multiple ISGs, many of which have the ability to inhibit translation of viral and cellular capped mRNAs, such as phosphorylating eukaryotic initiation factor 2α (eIF-2α) by protein kinase R (PKR) or degrading RNA by 2′,5′-oligoadenylate synthetase/RNase L (38, 41). The reduction of virus titer at late times of infection or the induction of one of the ISGs, RIG-I, is consistent with this interpretation (Fig. 1 and 5). However, it remains to be seen what ISGs are induced during MHV infection and whether they are responsible for the inhibition of IFN-α/β or other cellular and viral protein expressions at late times during virus replication.
Acknowledgments
We thank Anthony Compadre (University of California at Los Angeles School of Medicine, Los Angeles, CA) for kindly providing the N20.1 cells and Susan Weiss (University of Pennsylvania School of Medicine, Philadelphia, PA) for the recombinant MHV-A59/GFP.
This work was supported by Public Health Service grant NS047499 and in part by P30 core grant NS047546 and grant AI061204 from the National Institutes of Health. J.L. is partially supported by the China Scholarship Council, China.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, C. C., T. E. Lane, and S. A. Stohlman. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutler, B., C. Eidenschenk, K. Crozat, J. Imler, O. Takeuchi, J. A. Hoffmann, and S. Akira. 2007. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 7:753-766. [DOI] [PubMed] [Google Scholar]
- 5.Boehme, K. W., and T. Compton. 2004. Innate sensing of viruses by toll-like receptors. J. Virol. 78:7867-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brasier, A. R. 2006. The NF-kappaB regulatory network. Cardiovasc. Toxicol. 6:111-130. [DOI] [PubMed] [Google Scholar]
- 7.Cayley, P. J., J. A. Davies, K. G. McCullagh, and I. M. Kerr. 1984. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur. J. Biochem. 143:165-174. [DOI] [PubMed] [Google Scholar]
- 8.Cui, S., K. Eisenächer, A. Kirchhofer, K. Brzózka, A. Lammens, K. Lammens, T. Fujita, K. K. Conzelmann, A. Krug, and K. P. Hopfner. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, S. 2002. Pattern recognition receptors: doubling up for the innate immune response. Cell 111:927-930. [DOI] [PubMed] [Google Scholar]
- 12.Gosert, R., R. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ireland, D. D., S. A. Stohlman, D. R. Hinton, R. Atkinson, and C. C. Bergmann. 2008. Type 1 interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J. Virol. 82:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 15.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, and M. M. Yamamoto. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 16.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Dato, J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I andMDA5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 19.Knoops, K., M. Kikkert, S. H. E. van den Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. E. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:1957-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Y., Y. Cai, and X. Zhang. 2003. Induction of caspase-dependent apoptosis in cultured rat oligodendrocytes by murine coronavirus is mediated during cell entry and does not require virus replication. J. Virol. 77:11952-11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., and X. Zhang. 2005. Expression of cellular oncogene Bcl-xL prevents coronavirus-induced cell death and converts acute to persistent infection in progenitor rat oligodendrocytes. J. Virol. 79:47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 25.Martinand, C., C. Montavon, T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 73:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinand, C., T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1998. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus (EMCV)-infected cells. Eur. J. Biochem. 254:248-255. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov, R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819-826. [DOI] [PubMed] [Google Scholar]
- 28.Meylan, E., K. Burns, K. Hofmann, V. Blancheteau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503-507. [DOI] [PubMed] [Google Scholar]
- 29.Meylan, E., J. Curren, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 30.Meylan, E., J. Tschopp, and M. Karin. 2006. Intracellular pattern recognition receptors in the host response. Nature 442:39-44. [DOI] [PubMed] [Google Scholar]
- 31.Nash, T. C., and M. J. Buchmeier. 1997. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology 233:1-8. [DOI] [PubMed] [Google Scholar]
- 32.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 156:4746-4756. [DOI] [PubMed] [Google Scholar]
- 33.Pichlmair, A., and C. Reise-Sousa. 2007. Innate recognition of viruses. Immunity 27:370-383. [DOI] [PubMed] [Google Scholar]
- 34.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 35.Rempel, J. D., S. J. Murray, J. Meisner, and M. J. Buchmeier. 2004. Differential regulation of innate and adaptive immune responses in viral encephalitis. Virology 318:381-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth-Cross, J. K., S. J. Bender, and S. R. Weiss. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82:9829-9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth-Cross, J. K., L. Martínez-Sobrido, E. P. Scott, A. García-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81:7189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadler, A. J., and B. R. G. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 40.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanow, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 42.Takahasi, K., M. Yoneyama, T. Nishihori, R. Hirai, H. Kumeta, R. Narita, M. Gale, Jr., F. Inagaki, and T. Fujita. 2008. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 29:428-440. [DOI] [PubMed] [Google Scholar]
- 43.Verity, A. N., D. Bredesen, C. Vonderscher, V. W. Handley, and A. T. Campagnoni. 1993. Expression of myelin protein genes and other myelin components in an oligodendrocytic cell line conditionally immortalized with a temperature-sensitive retrovirus. J. Neurochem. 60:577-587. [DOI] [PubMed] [Google Scholar]
- 44.Versteeg, G. A., P. J. Bredenbeek, S. H. van den Worm, and W. J. Spaan. 2007. Group coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll-IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 47.Yang, H., G. Ma, C. H. Lin, M. Orr, and M. G. Wathelet. 2004. Mechanism for transcriptional synergy between interferon regulatory factor (IRF)-3 and IRF-7 in activation of the interferon-beta gene promoter. Eur. J. Biochem. 271:3693-3703. [DOI] [PubMed] [Google Scholar]
- 48.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoneyama, M., and T. Fujita. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54-65. [DOI] [PubMed] [Google Scholar]
- 50.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 51.Yu, D., H. Zhu, Y. Liu, J. Cao, and X. Zhang. 2009. Regulation of proinflammatory cytokine expression in primary mouse astrocytes by coronavirus infection. J. Virol. 83:12204-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, H., and S. Perlman. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, H., D. Yu, and X. Zhang. 2009. The spike protein of murine coronavirus regulates viral genome transport from the cell surface to the endoplasmic reticulum during infection. J. Virol. 83:10653-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]