Abstract
Vaccines preventing HIV-1 infection will likely elicit antibodies that neutralize diverse strains. However, the capacity for lentiviruses to escape broadly neutralizing antibodies (NAbs) is not completely understood, nor is it known whether NAbs alone can control heterologous infection. Here, we determined that convalescent immune plasma from a horse persistently infected with equine infectious anemia virus (EIAV) neutralized homologous virus and several envelope variants containing heterologous principal neutralizing domains (PND). Plasma was infused into young horses (foals) affected with severe combined immunodeficiency (SCID), followed by challenge with a homologous EIAV stock. Treated SCID foals were protected against clinical disease, with complete prevention of infection occurring in one foal. In three SCID foals, a novel neutralization-resistant variant arose that was found to preexist at a low frequency in the challenge inoculum. In contrast, SCID foals infused with nonimmune plasma developed acute disease associated with high levels of the predominant challenge virus. Following transfer to an immunocompetent horse, the neutralization-resistant variant induced a single febrile episode and was subsequently controlled in the absence of type-specific NAb. Long-term control was associated with the presence of cytotoxic T lymphocytes (CTL). Our results demonstrate that immune plasma with neutralizing activity against heterologous PND variants can prevent lentivirus infection and clinical disease in the complete absence of T cells. Importantly, however, rare neutralization-resistant envelope variants can replicate in vivo under relatively broad selection pressure, highlighting the need for protective lentivirus vaccines to elicit NAb responses with increased breadth and potency and/or CTL that target conserved epitopes.
Development of an effective vaccine will be critical in the efforts to control the human immunodeficiency virus type 1 (HIV-1) pandemic. Unfortunately, vaccines evaluated in completed human efficacy trials have shown moderate to no protective effects, and, clearly, much more work is needed to define the correlates of lentivirus immune protection. Although these correlates are still not entirely known, vaccine strategies that elicit antibodies with broad neutralizing activity are currently of considerable interest, and it is widely believed that HIV-1 envelope glycoproteins that induce broadly neutralizing antibodies (NAbs) will be critical components of a protective vaccine (21, 28, 53, 63).
Equine infectious anemia virus (EIAV) is a macrophage-tropic lentivirus that causes persistent infection in horses worldwide and serves as an important large-animal translational model in which to dissect basic correlates of protective lentiviral immunity (9, 31, 33, 38, 57). EIAV is a naturally occurring lentivirus, and infection results in a predictable course of recurrent episodes of plasma viremia and clinical disease. As with HIV-1 and simian immunodeficiency virus (SIV), EIAV infection is not cleared. However, infected horses eventually control viral replication and clinical disease to remain persistently infected inapparent carriers. Adaptive immune responses, including NAbs, are required for EIAV control since young horses (foals) with severe combined immunodeficiency (SCID), unlike normal foals, fail to eliminate the initial viremia following challenge (46). Equine SCID is caused by a frameshift mutation in the gene encoding the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (55, 60) and has an autosomal recessive mode of inheritance (47). The equine SCID defect is more severe than its murine counterpart in that SCID foals are incapable of forming either coding or signal joints (55). Adoptive transfer of EIAV-specific T and B lymphocytes to a SCID foal results in functional cytotoxic T lymphocytes (CTL) and NAb activity and is protective against homologous EIAV challenge (33).
During acute EIAV infection, each recurrent episode coincides with the emergence of an antigenically distinct EIAV variant as defined by type-specific NAb, which neutralizes virus isolated during early disease episodes but not virus isolated during subsequent disease episodes (2, 20, 22, 43, 52). Amino acid variation primarily occurs within hypervariable regions V1 to V8 of the envelope gp90 surface unit (SU) and particularly within the V3/principal neutralizing domain (PND) region (1, 19, 24, 25, 57). Our work with EIAV-infected SCID foals indicates that significant envelope diversification does not occur in the absence of NAbs but that rapid envelope diversification occurs when adaptive immune responses are reconstituted (35). Thus, adaptive immunity, including NAb, drives selection of EIAV envelope variants during acute infection. Amino acid changes occur primarily within the V3 to V7 hypervariable SU regions, and many changes affect potential N-linked glycosylation sites (PNLGS) (35). Importantly, however, CTL also target the SU, and variants that escape CTL recognizing an EIAV V3/PND epitope have been identified (37, 38). Thus, both NAbs and CTL are capable of contributing to the selection of EIAV SU variants, but the relative contributions of each to such selection are not known.
Recently, SU variation was evaluated in an immunocompetent pony experimentally inoculated with the virulent wild-type Wyoming strain of EIAV (57). Seventy-one distinct V3 variants that partitioned into five major nonoverlapping groups were identified and designated PND1 to PND5. Neutralization assays using chimeric infectious molecular clones containing these PNDs suggested a transition from type-specific NAb responses toward more broadly reactive immune responses during the course of infection and indicated that genetic changes conferring resistance to broadly NAbs lead to recrudescence of clinical disease following a lengthy clinically quiescent period (57). Thus, the NAb response broadens significantly during long-term persistent EIAV infection, and broadly NAbs play a critical role in EIAV immune control.
Studies of nonhuman primates have provided important information regarding the protective effects of NAbs. Passive immunization of macaques with purified immunoglobulin from chimpanzees infected with several different HIV-1 isolates results in complete protection from homologous chimeric simian/human immunodeficiency virus (SHIV) infection when the immunoglobulin is given 24 h prior to challenge (54). Passive transfer of a triple combination of broadly neutralizing human monoclonal antibodies directed against the envelope of a primary HIV-1 isolate results in complete protection against SHIV infection in some macaques while others become infected but exhibit decreased plasma viremia (29). The contribution of T cells to partial protection in these studies is not clear, and the presence or absence of viral escape variants in the unprotected macaques has not been evaluated. In neonatal macaques, various combinations of broadly neutralizing human monoclonal antibodies directed against conserved HIV envelope epitopes administered before and after SHIV challenge result in protection against persistent systemic infection in some animals, but clinical disease develops in others (12-14). Virus-specific T-cell proliferative responses are detected in some of the protected animals, indicating that cellular immune responses occur and likely contribute to protection by eliminating infected cells (13).
Despite the fact that NAbs can block experimental SHIV infection, selection pressure exerted by NAbs plays a critical role in HIV-1 and SIV envelope evolution during infection, and evasion of NAb responses is an important mechanism of HIV-1 and SIV persistence (11, 16, 27, 48, 59). The maturation of a type-specific NAb response in SIV-infected rhesus macaques significantly correlates with diversification in the V1/V2 region of the SIV envelope (50). In HIV-1, NAbs are detectable within the first 2 months postinfection and result in an early and significant selection force on the virus population (49). Escape from NAbs involves many amino acid substitutions with little cross-neutralization between closely related strains, and NAb responses drive the diversification of the HIV-1 envelope during the early stages of infection (16). The early appearance of NAbs in patients with acute HIV-1 infection results in the replacement of neutralization-sensitive virus by successive populations of resistant virus, and virus escape primarily involves changes in N-linked glycosylation (59). Thus, overcoming neutralization escape constitutes a significant barrier to the ultimate efficacy of any NAb-eliciting HIV-1 vaccine.
Because the SCID defect occurs naturally in the horse, it provides a powerful and unique opportunity to finely dissect the protective effects of immune interventions against a naturally occurring lentivirus independent of other de novo adaptive immune responses. This level of dissection is not possible in other lentivirus model systems. The goal of the current study was to determine if broadly NAbs could protect against lentivirus challenge in the complete absence of T lymphocytes and other adaptive immune responses. We hypothesized that convalescent immune plasma from a long-term persistently infected inapparent carrier horse containing antibodies capable of neutralizing homologous and several heterologous EIAV SU PND variants would provide complete protection when infused into SCID foals before experimental virus inoculation. This plasma was administered to four SCID foals 24 h prior to challenge, and four control SCID foals received normal horse plasma. Clinical outcome, plasma viral load, and serum neutralization activity were analyzed in all foals. Although complete protection was achieved in one treated foal, infection occurred in the others. In foals that became viremic, the SU sequence and neutralization phenotype of the breakthrough virus were determined. As part of these experiments, blood containing this virus was inoculated into a naive immunocompetent horse, and the adaptive immune responses associated with its control were further evaluated.
MATERIALS AND METHODS
Horses.
Foals affected with SCID were obtained by selective breeding of Arabian horses and Arabian/mixed-breed pony crosses heterozygous for the SCID trait (10, 33, 46, 47) and included four experimental and four control animals. The experimental animals consisted of three Arabian SCID foals (animals A2239, A2240, and A2241) and one pony-cross SCID foal (animal H703), each of which received infusions of immune plasma prior to virus challenge. The control animals consisted of two Arabian SCID foals (A2245 and A2247) and two pony-cross SCID foals (H707 and H713), each of which received normal horse plasma prior to virus challenge. An additional pony-cross SCID foal (H719) was used as an unchallenged control to confirm that the infused immune plasma did not contain infectious virus. Initial diagnosis of SCID was based on persistent lymphopenia (<1,000 peripheral blood lymphocytes [PBL] per μl) (32, 33) and was confirmed by identification of the homozygous mutation in the DNA-PKcs gene sequence (55). SCID foals were maintained as described previously (33, 36, 45) with some modifications. For the first 24 to 48 h of life, SCID foals were housed in individual box stalls with their dams to allow ingestion of colostrum. After adequate passive transfer of maternal IgG was confirmed, SCID foals were moved into isolation stalls and fed a commercial mare's milk replacer. In an effort to prevent bacterial infection as well as opportunistic lung infection by Pneumocystis carinii, SCID foals were administered systemic antibiotics, which included one or more of the following: trimethoprim-sulfamethoxazole (20 mg/kg of body weight, administered orally [p.o.] every 12 h [q12h]), cefpodoxime (10 mg/kg, p.o. q12h), amikacin (21 mg/kg, p.o. q24h), and chloramphenicol (50 mg/kg, p.o. q8h). SCID foals were 2 weeks of age when used in this experiment.
Immunocompetent horse A2150 was 9 years old and had been infected with EIAV strain WSU5 (EIAVWSU5) for 8 years (38). This horse was a source of immune plasma for infusion into SCID foals. In addition, a 1-year-old naïve immunocompetent horse (A2215) was used as a recipient of a whole-blood transfer that contained a breakthrough virus from one of the experimental SCID foals.
All SCID foals were humanely euthanized at the end of the study. The timing of euthanasia varied and was determined based on the severity of clinical signs associated with EIAV and/or other secondary bacterial/viral infections. All experiments involving horses and foals were approved by the Washington State University Institutional Animal Care and Use Committee.
Neutralization activity of immune plasma.
Long-term EIAV inapparent carrier horse A2150 was used as the immune plasma donor in this study. Virus NAb titers in serum from this horse were determined using a focal reduction assay on equine dermal (ED) cells (3, 57). Briefly, serum was heat inactivated to destroy complement, serially diluted 2-fold in supplemented Dulbecco's modified Eagle's medium (DMEM), and incubated at 37°C for 1 h with 550 focus forming units (FFU) of six EIAV envelope SU variant viruses (see below). Quadruplicate wells of ED cells were inoculated with the virus-serum mixture in the presence of 4 μg of Polybrene, and the culture medium was changed the following day. Cells were incubated for an additional 72 h and fixed with ice-cold methanol, and foci of EIAV-infected cells were detected by immunocytochemistry and enumerated. Results were expressed as the serum neutralization titer, which was defined as the highest serum dilution that yielded a 50% reduction in the FFU count compared with negative-control serum (3).
Viruses used in this assay included the homologous EIAVWSU5 inoculum (43) (GenBank accession number AF247394) and five previously described viruses generated from infectious molecular clones containing heterologous chimeric envelopes (Fig. 1 a) (57). These were derived from a pony experimentally infected with the virulent Wyoming strain of EIAV (EIAVWyo), and each chimeric clone represented a predominant envelope SU genotype corresponding to a different stage of disease. The five clones were previously designated PND1 to PND5 (57) to denote differences in the PND regions among the isolates.
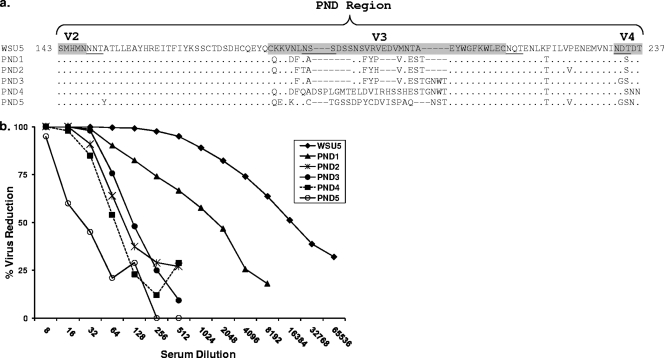
FIG. 1.
(a) Deduced amino acid sequences of the SU PND region of EIAVWSU5 and the five heterologous PND variant molecular clones used to characterize the neutralizing breadth of A2150 immune serum. Hypervariable regions V2 to V4 are labeled and shaded. PNLGS are underlined. (b) Neutralizing activity of A2150 immune serum against homologous EIAVWSU5 and the five heterologous PND variant molecular clones.
Preparation of immune plasma.
Plasma from horse A2150 was collected aseptically and stored at −20°C. Before infusion into SCID foals, the plasma was treated with 1 μM methylene blue and white light (methylene blue-white light) as described previously (15, 40) to inactivate potential infectious virus. Briefly, plasma was thawed, and methylene blue was added, followed by exposure to white light at 2,800 lumens for 1 h. The plasma was then refrozen at −20°C until needed for infusion. The effectiveness of this method for inactivation of infectious EIAV was confirmed by spiking 1 liter of normal horse plasma with 5 × 107 50% tissue culture infective doses (TCID50) of EIAVWSU5, followed by virus titration (3) before and after methylene blue-white light treatment. The pretreatment virus titer was 2 × 103 FFU/ml while the virus titer was zero after methylene blue-white light treatment. Prior to infusion into SCID foals, all plasma treated with methylene blue-white light was confirmed negative for infectious virus by virus titration and real-time reverse-transcription PCR (RT-PCR) (37).
Intravenous infusion of immune plasma.
The volume of immune plasma infused intravenously (i.v.) into each SCID foal was determined by calculating the volume necessary to completely neutralize the challenge virus, based on the neutralization titer of the immune plasma and the plasma volume of the SCID foal recipient. Briefly, previous data indicated that Arabian SCID foals inoculated with 106 TCID50 of EIAVWSU5 develop plasma virus loads between 104 and 105 RNA copies/ml by 2 weeks postinoculation (36). In the current study, SCID foals were 2 weeks of age at inoculation, weighing approximately 40 kg, with a plasma volume of approximately 2 liters. Although a total plasma virus burden of approximately 106 virions would be anticipated in these foals immediately following i.v. challenge with 106 TCID50 of EIAVWSU5, the plasma virus burden anticipated at 2 weeks postinoculation (approximately 5 × 104 RNA copies/ml, or a total of 5 × 107 virions) was used as a neutralization target. This was done to ensure that adequate doses of immune plasma were administered. Since the focal reduction assay described above indicated that a 1:32 dilution of 250 μl of A2150 immune plasma (7.8 μl) was capable of completely neutralizing 550 FFU of input EIAVWSU5 (100% neutralizing titer), 709 ml of A2150 immune plasma would be required to completely neutralize 5 × 107 EIAVWSU5 virions. To be conservative, a volume of 1 liter was chosen for infusion. This was 71 times the volume required to neutralize the actual challenge dose (106 TCID50) at inoculation. Although the experimental pony-cross SCID foal (H703) weighed approximately 40% less than the Arabian SCID foals (with a corresponding lower plasma volume), calculations as above indicated that 1 liter of immune plasma was similarly more than sufficient to completely neutralize an anticipated 2-week-postinoculation total plasma virus burden. Thus, 1 liter of methylene blue-white light-treated A2150 immune plasma was infused i.v. into experimental SCID foals (A2239, A2240, A2241, and H703) at 24 h prior to EIAV challenge and again 7 and 14 days after challenge. Four EIAV-challenged control SCID foals (A2245, A2247, H707, and H713) received 1 liter of methylene blue-white light-treated normal horse plasma at the same time points. One control SCID foal (H719) received 1 liter of methylene blue-white light-treated A2150 immune plasma i.v. without subsequent virus challenge to further confirm that the infused immune plasma as treated did not contain infectious virus.
All SCID foals (except H719) were challenged i.v. with 106 TCID50 of EIAVWSU5 (10, 33, 35, 36, 46). Immunocompetent horse A2215 was inoculated i.v. with 250 ml of whole blood obtained from SCID foal A2239 at 41 days post-EIAV inoculation.
Immunological and virologic monitoring.
Following EIAV challenge in SCID foals (or blood transfer in immunocompetent horse A2215), rectal temperatures and clinical status were recorded daily, and complete blood counts (CBC) with platelet counts were performed on whole blood collected every 2 days. Platelet counts (per μl) at least 3 standard deviations below the prechallenge mean were considered thrombocytopenic. Plasma and serum were collected and stored at −80°C every 2 days. Whole blood was collected from immunocompetent horse A2215 at 404 to 504 days after the infectious blood transfer was received, and peripheral blood mononuclear cells (PBMC) were isolated and either placed directly into stimulation cultures or cryopreserved for later use, as described previously (37).
Virus NAb titers in serum from all challenged foals was determined weekly (or at various other time points in A2215) using the focal reduction assay described above (3, 57). Also in A2215, serum antibodies against EIAV Gag p26 were detected using both enzyme-linked immunosorbent assay (ELISA) and agar gel immunodiffusion (AGID) test kits (VMRD, Inc., Pullman, WA), according to the manufacturer's instructions.
Viral load was determined by extracting plasma viral RNA and performing real-time quantitative RT-PCR as described previously (37). This assay could repeatedly detect dilutions of EIAV RNA containing 10 copies per reaction mixture but not dilutions containing only 1 copy. Thus, the reliable detection limit in assayed plasma was 430 RNA copies/ml when 140 μl of plasma for the RNA extraction and 10 μl of the 60-μl eluate were used per reaction mixture. Assays were repeated independently at least twice to confirm negative results. For peak viremic plasma samples, single-genome amplification (SGA) was performed as described previously (51) to identify potential SU variants. Briefly, cDNA was synthesized using an iScript Select cDNA Synthesis Kit (Bio-Rad, Hercules, CA), and serial dilutions (1:20, 1:60, 1:180, and 1:540) were made. A primary PCR was performed to detect virus by amplification of a 2,353-bp segment immediately 5′ of the envelope gene and including all of gp90 and all but the 3′ end of gp45, with 16 reaction mixtures per dilution in a 96-well format. Primers 5321F (5′-CATGGTCAGCATCGCATTC-3′) and 7674R (5′-GCGAGAGTTCCTTCTTGGGC-3′), along with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), were used in the primary reaction. The primary reaction conditions were as follows: 2 min at 95°C and 35 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min 30 s. A nested PCR was then performed using primers 5366F (5′-CAACCCCTATTACCCAAC-3′) and 7677R (5′-TTCCTTCTTGGGCTTTAATGC-3′) to amplify a 2,311-bp segment of the envelope gene that began 43 bp downstream of the segment amplified in the primary PCR and included most of gp90 and gp45. The nested reaction conditions were 2 min at 95°C and 45 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min 30 s. According to a Poisson distribution, the cDNA dilution that yields PCR products in no more than 30% of wells contains one amplifiable cDNA template per positive PCR more than 80% of the time (44, 51). Thus, positive nested PCR products that met a Poisson distribution were then sequenced.
To confirm the presence or absence of EIAV infection in SCID foals at the end of the study, DNA was extracted from postmortem splenic tissue, and nested PCR was performed to detect provirus by amplification of a 288-bp segment of the gag gene as described previously (33, 38).
An EIAV variant with a divergent SU sequence (designated EIAVWSU5-V55) was identified in this study. To specifically amplify this variant, nested RT-PCR (or PCR) was performed using variant sequence-specific primers. Initial primers 5841F (5′-TCATTGTCAAGAGTATCTATGTAAAGGAC-3′) and 6280R (5′-ATATATCTAGGCGACCCGTCTC-3′) were used to amplify a 399-bp segment of the variant SU that included all of V3 through V6. The primary reaction conditions were 2 min at 95°C and 40 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s. A nested reaction was then performed using primers 5872F (5′-GATCTTAATTCCTCTAACTCTTATTGGG-3′) and 6264R (5′-CGTCTCGCAAACTGTCTC-3′), which amplified a 351-bp segment of the variant SU that began 32 bp downstream of the segment amplified in the primary reaction and included most of V3 through V6. The nested reaction conditions were 2 min at 95°C and 40 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s.
PBMC stimulation and CTL assays.
Freshly isolated or cryopreserved PBMC from immunocompetent horse A2215 were stimulated in vitro for 7 days using Gag peptide pool-pulsed, EIAVWSU5-infected or EIAVWSU5-V55-infected autologous monocytes as described previously (34, 38, 38) with modifications. For peptide stimulation, Gag peptide pool 1, comprising 14 overlapping Gag p15 peptides containing immunodominant CTL epitopes, was used as described previously (34). The EIAVWSU5-V55 variant virus for use in the PBMC stimulation culture and subsequent CTL assay was isolated using methods similar to those described by O'Rourke et al. (42). Very briefly, plasma from SCID foal A2239 (containing EIAVWSU5-V55 exclusively as determined by SGA above) was used to infect equine kidney (EK) cells, which were then expanded and maintained in DMEM with 5% calf serum. Cell culture supernatants were collected to make a virus stock, which was quantitated by titration. Viral RNA was then extracted from this stock and analyzed by SGA to confirm the EIAVWSU5-V55 SU consensus sequence. For stimulation cultures, viruses in 17% fetal bovine serum (FBS) were added to A2215 cryopreserved PBMC in amounts equal to a multiplicity of monocyte infection of 1, assuming that PBMC were 5% monocytes. Virus and PBMC were incubated for 2 h at 37°C with occasional mixing before centrifugation at 250 × g for 10 min. PBMC were then resuspended at 5 × 106/ml of RPMI 1640 medium with 10% FBS, 20 mM HEPES, 10 μg/ml gentamicin, 10 μM 2-mercaptoethanol, and 10 U/ml recombinant human interleukin-2 ([IL-2] Roche Diagnostics, Indianapolis, IN). Tissue culture plates (24 well) were seeded with 2 × 106 PBMC/well and incubated at 37°C with 5% CO2 for 7 days. Virus-specific CTL activity was measured in stimulated PBMC with a 17-h 51Cr release assay (34, 37, 38) using homologous EK target cells infected with either EIAVWSU5 or EIAVWSU5-V55. For Gag peptide pool 1-pulsed target cells, a final concentration of 104 nM for each peptide in the pool was used (34). The following formula for percent specific lysis was used: [(E − S)/(M − S)] × 100, where E is the mean 51Cr beta counts per min of three test wells, S is the mean spontaneous release from three target cell wells without effector cells, and M is the mean maximal release from three target cell wells with 2% Triton X-100 in distilled water. Assays were independently performed at least twice, with effector/target cell ratios from 12:1 to 50:1. For these assays, each well contained ∼30,000 target cells. Only assays with a spontaneous target cell lysis of <15% were used. The standard error (SE) of percent specific lysis was calculated using a formula that accounts for the variability of E, S, and M (56). Specific lysis was considered significant if it exceeded 10% and was at least 2 SEs above that for non-peptide-pulsed (or uninfected) target cells.
Statistical analysis.
A nonparametric Mann-Whitney test (41) was used to compare areas under the curve (AUC) for viral load times the number of days post-EIAV inoculation (dpi) between experimental and challenged control foals using a two-tailed significance level for α of 0.05. For comparisons of the percentages of days that the plasma viral load was >10,000 RNA copies/ml and of the percentages of days of thrombocytopenia between experimental SCID foals and controls, a t test with Welch correction for unequal variances was used with a two-tailed significance level for α of 0.05. A viral load value of >10,000 RNA copies/ml was chosen for comparison because clinically apparent infection with fever and declining platelet counts associated with this viral load are consistently observed in EIAV-infected horses (36-38), and a viral load set point at this level distinguishes between EIAV progressor and nonprogressor horses (24). Statistical analyses were performed using GraphPad InStat, version 3.06, and GraphPad Prism, version 5.01 (GraphPad Software, San Diego, CA).
Nucleotide sequence accession numbers.
Three amplified EIAVWSU5-V55 viral sequences, including a common consensus sequence and two additional sequences with minor amino acid divergence, have been submitted to the GenBank under accession numbers 1238448, 1238466, and 1238469.
RESULTS
A2150 immune serum/plasma had broad neutralizing activity against EIAV.
Serum from long-term EIAV-inapparent carrier horse A2150 contained antibodies capable of neutralizing homologous EIAVWSU5 as well as chimeric heterologous infectious molecular clones PND1, -2, -3, -4, and -5 (Fig. 1b). As expected, the immune serum neutralized EIAVWSU5 more effectively than the five heterologous PND virus clones. Thus, A2150 immune serum was considered to have broad neutralizing activity although titers against heterologous PND viruses were lower than the neutralizing titer against the homologous virus.
Protective effects of broadly neutralizing immune plasma against EIAV infection and clinical disease in SCID foals.
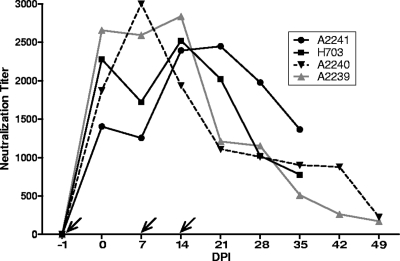
The four experimental SCID foals (A2239, A2240, A2241, and H703) received 1 liter of methylene blue-white light-treated immune plasma i.v., followed by i.v. challenge with 106 TCID50 of EIAVWSU5 24 h later. Then, 1 liter of immune plasma was administered i.v. to each foal 7 and 14 dpi. These passive transfers resulted in peak EIAVWSU5-specific NAb titers between 2,000 and 3,000 in all foals, with similar rates of peak acquisition and subsequent decay. However, the peak titer was reached later and declined more slowly in one foal (A2241) than in the other three (Fig. 2). All four foals maintained a normal platelet count throughout the study (Fig. 3 a to d). Due to the absence of immune-mediated mechanisms, fever (rectal temperature of >101.5°F) can be variable in SCID foal EIAV challenge studies. Fever can also be nonspecific because secondary bacterial, fungal, and other viral (particularly adenovirus) infections can occur. Although fever occurred at various time points in all four foals, (particularly at the end of each experiment), it was not consistently associated with EIAV infection. Foal A2241 did not develop plasma viremia, while animals A2239, A2240, and H703 became viremic at 41, 48, and 33 dpi, respectively. The viral load continued to rise in A2239, with a peak occurring at 55 dpi. These three foals also had detectable proviral DNA in postmortem splenic tissue, confirming infection. In contrast, proviral DNA was not detected in postmortem splenic tissue in foal A2241, confirming that this foal did not become infected.
FIG. 2.
Weekly 50% serum neutralization titers against homologous EIAVWSU5 in experimental SCID foals. Slanted arrows indicate days that immune plasma was infused.
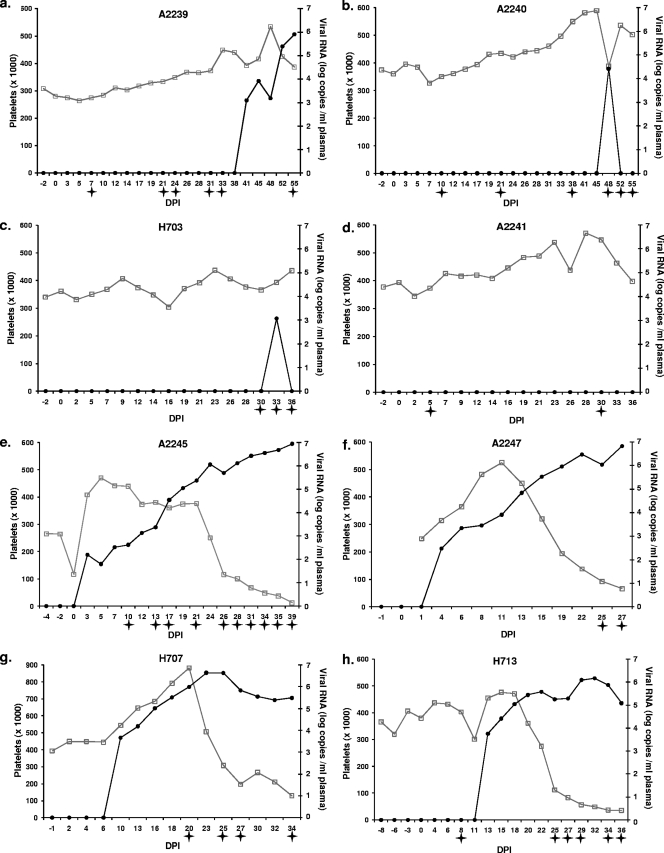
FIG. 3.
Peripheral blood platelet counts (open squares) and plasma viral loads (filled circles) for SCID foals that received A2150 convalescent immune plasma (experimental) (a to d) and normal horse plasma (controls) (e to h). Stars indicate febrile days (rectal temperature of >101.5°F). Platelet counts were not obtained at missing data points.
Four control SCID foals (A2245, A2247, H707, and H713) received methylene blue-white light-treated normal horse plasma prior to EIAV challenge with the same infusion schedules as the experimental SCID foals. No serum neutralization activity was detected at any time point in these control foals (data not shown). All four challenged control SCID foals became viremic by 13 dpi and developed thrombocytopenia and fever (Fig. 3e to h). The unchallenged control SCID foal (H719) that received only methylene blue-white light-treated A2150 immune plasma did not develop thrombocytopenia or plasma viremia, and proviral DNA was not detected in postmortem splenic tissue. In addition to the lack of detectable virus in the treated A2150 immune plasma itself, this result further confirmed that the immune plasma infused into the foals of this study did not contain infectious virus and therefore was not a source of infection in the experimental foals that became viremic.
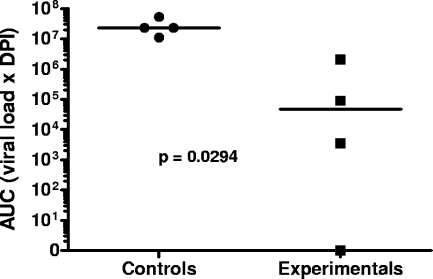
Experimental SCID foals had a significantly lower percentage of days of thrombocytopenia (<1%) than challenged control SCID foals (34%; P = 0.01). Consistent with these findings, experimental SCID foals had a significantly lower percentage of days of plasma viral load of >10,000 copies/ml (<1%) than challenged control SCID foals (61%; P = 0.0002). Lastly, the median AUC value (viral load times the number of days post-EIAV inoculation) for experimental foals (4.7 × 104) was significantly less (P = 0.0294) than that for control foals (2.3 × 107) (Fig. 4).
FIG. 4.
Comparison of AUC values between experimental and control SCID foals challenged with EIAVWSU5. Horizontal bars indicate medians.
Single-genome amplification and sequencing of plasma viral RNA in viremic SCID foals identified a significantly divergent envelope SU variant.
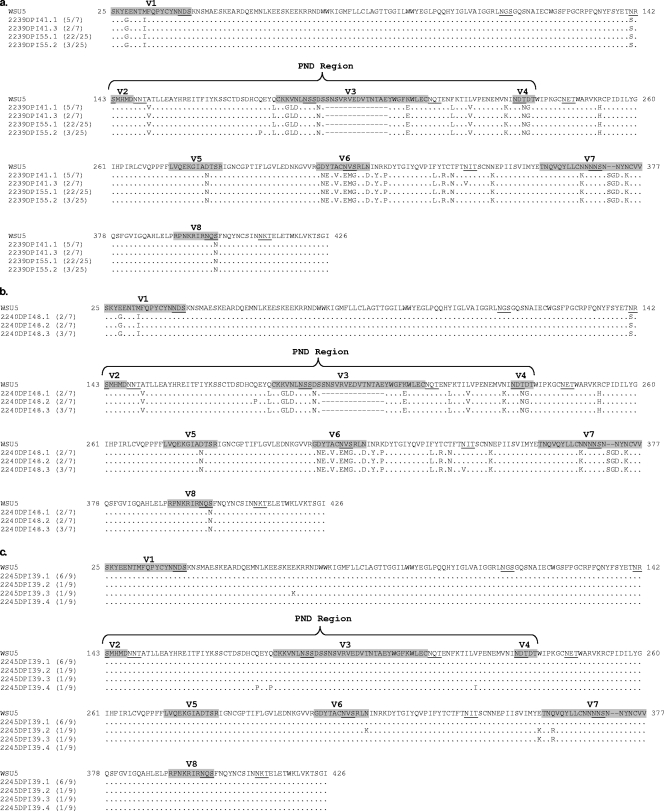
Replicating virus was detected in three of the four SCID foals, and it was of interest to determine the genetic and antigenic properties of the breakthrough virus. Single-genome amplification (SGA) of plasma viral RNA at 41 and 55 dpi in SCID foal A2239 and at 48 dpi in SCID foal A2240 was performed, and the SU sequences of single amplicons were determined. An identical virus genotype, containing the same amino acid substitutions, insertions, and deletions, including a 14-amino-acid deletion in the PND region of SU and a 2-amino-acid insertion in V7, were identified in both foals (Fig. 5 a and b). This variant was designated EIAVWSU5-V55 and was 12.6% divergent from EIAVWSU5 at the amino acid level within SU. Although amino acid changes primarily involved hypervariable regions V1 to V8, changes also occurred at more conserved sites between hypervariable regions. The 14-amino-acid deletion within V3 in the PND spanned two previously mapped neutralization epitopes (ENT and DNT) recognized by murine monoclonal antibodies (1). Importantly, amino acid substitutions in EIAVWSU5-V55 resulted in the creation of new PNLGS and in the shifting or deletion of existing PNLGS. The EIAVWSU5-V55 variant was identified in 32/32 single amplicons from foal A2239 (Fig. 5a) and in 7/7 single amplicons from foal A2240 (Fig. 5b). Although experimental SCID foal H703 developed transient plasma viremia at 33 dpi, single RNA genomes could not be amplified for sequencing. However, the EIAVWSU5-V55 variant provirus was detected by PCR (and confirmed by sequencing) in H703 postmortem splenic DNA at the end of the study using EIAVWSU5-V55-specific primers (data not shown).
FIG. 5.
Deduced amino acid sequences of single plasma viral RNA RT-PCR amplicons from A2239 at 41 and 55 dpi (a), A2240 at 48 dpi (b), and control SCID foal A2245 at 39 dpi (c). The EIAVWSU5 inoculum consensus sequence for comparison is shown at the top. For each unique sequence, the ratio of the number of amplicons with that sequence to the total number of amplicons sequenced is indicated in parentheses. Hypervariable regions V1 to V8 are labeled and shaded, and the PND region is indicated. PNLGS are underlined.
To determine the virus genotype associated with clinical disease in SCID foals receiving normal serum, we performed single-genome amplification and sequencing of plasma viral RNA from control SCID foal A2245 at peak plasma viremia. The EIAVWSU5 inoculum SU sequence was present in nine out of nine amplicons (Fig. 5c), and no variant EIAVWSU5-V55 amplicons were detected. Similarly, SGA of plasma viral RNA from control SCID foals A2247, H707, and H713 at peak viremia identified the same EIAVWSU5 inoculum SU sequence in 9/9, 17/17, and 10/10 amplicons, respectively (data not shown). Although only EIAVWSU5 was identified using SGA of plasma viral RNA from all challenged control SCID foals, nested RT-PCR using EIAVWSU5-V55-specific primers and subsequent sequencing detected the EIAVWSU5-V55 variant in plasma from these control foals (data not shown).
The EIAVWSU5-V55 variant was present as a rare species in the EIAVWSU-5 inoculum.
Identification of the same EIAV SU variant in more than one experimental SCID foal suggested that EIAVWSU5-V55 preexisted in the immune plasma or virus inoculation. Therefore, EIAVWSU5-V55-specific nested RT-PCR was used as above to detect the possible presence of EIAVWSU5-V55 RNA in the methylene blue-white light-treated A2150 immune plasma and in the EIAVWSU5 inoculum. For these experiments, viral RNA was extracted from 1.5 ml of plasma (or virus stock) to enhance the probability of detection. EIAVWSU5-V55 RNA was not detected in the treated A2150 plasma; however, EIAVWSU5-V55 RNA was identified in the EIAVWSU5 virus stock used for inoculation. To determine the relative frequency of the EIAVWSU5-V55 variant in the EIAVWSU5 virus stock, SGA and sequencing were performed. Of 25 single amplicons analyzed, 24 had the EIAVWSU5 SU sequence while 1 had the EIAVWSU5-V55 variant SU sequence (data not shown). Thus, the EIAVWSU5-V55 variant was present as a minor population in the EIAVWSU5 virus stock used to inoculate the SCID foals in this study. Since the EIAVWSU5 stock virus had been passed and amplified in cell culture several times over the years to prepare new stocks, it was of interest to determine if the EIAVWSU5-V55 variant was present in the original EIAVWSU5 seed stock prepared in 1988 (42). Interestingly, the EIAVWSU5-V55 variant was also identified in this stock by nested RT-PCR using the EIAVWSU5-V55-specific primers.
The EIAVWSU5-V55 SU variant was resistant to neutralization.
To determine the neutralization phenotype of the EIAVWSU5-V55 variant, virus was isolated from experimental SCID foal A2239 55-dpi plasma and propagated in cell culture. The isolate was confirmed to be EIAVWSU5-V55 by SGA analysis and sequencing. Serum neutralization assays against this EIAVWSU5-V55 isolate were then performed using A2239 55-dpi serum, A2240 48-dpi serum, and immune serum from A2150 (the source of immune plasma used for infusion). Compared to the ability of sera from these animals to neutralize EIAVWSU5, the EIAVWSU5-V55 isolate was neutralized very poorly (Fig. 6 a). Thus, EIAVWSU5-V55 was confirmed to be a neutralization-resistant SU variant.
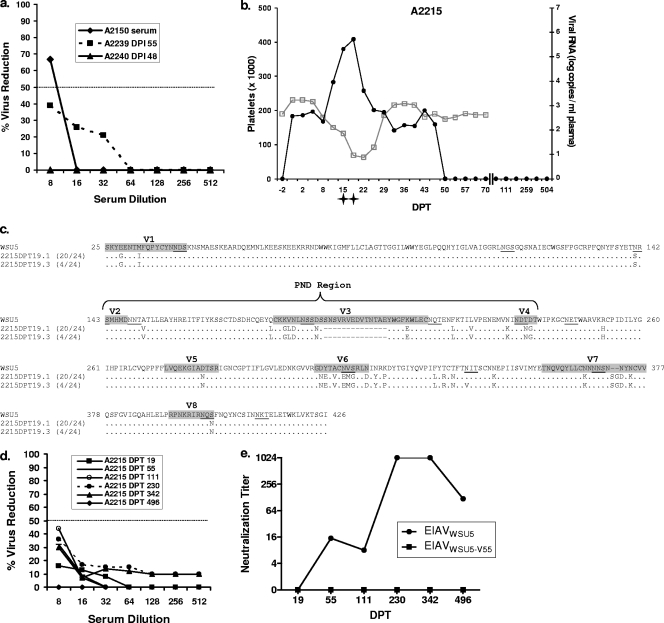
FIG. 6.
(a) Serum neutralization of the EIAVWSU5-V55 variant by A2150 immune serum, A2239 serum at 55 dpi, and A2240 serum at 48 dpi. The dashed line represents 50% virus reduction. (b) Peripheral blood platelet counts (open squares) and plasma viral loads (filled circles) for immunocompetent horse A2215 after inoculation with blood from SCID foal A2239. Stars indicate febrile days (rectal temperature of >101.5°F). Platelet counts were not obtained at missing data points. (c) Deduced amino acid sequences of single plasma viral RNA RT-PCR amplicons from A2215 at 19 dpt. The EIAVWSU5 consensus sequence for comparison is shown at the top. Other annotations are the same as described in the legend of Fig. 5. (d) Serum neutralization of the EIAVWSU5-V55 variant by A2215 serum at 19, 55, 111, 230, 342, and 496 dpt. The dashed line represents 50% virus reduction. (e) The 50% serum neutralization titers against EIAVWSU5 and EIAVWSU5-V55 by A2215 serum at 19, 55, 111, 230, 342, and 496 dpt.
The EIAVWSU5-V55 SU variant resulted in clinical disease in an immunocompetent horse but was efficiently controlled in the absence of type-specific NAb.
Blood inoculation into a susceptible horse with subsequent monitoring for clinical disease and seroconversion is one of the oldest and most sensitive methods for confirming EIAV infection (7). Whole blood from experimental SCID foal A2239 was obtained at 41 dpi (the first day of detectable plasma viral RNA), and 250 ml was inoculated i.v. to immunocompetent horse A2215. This horse developed a fever at 15 days post-blood transfer (dpt), which correlated with plasma viral RNA and thrombocytopenia (Fig. 6b). The fever subsided within 4 days, and no further viremic episodes occurred during 455 days of monitoring. Gag-specific antibodies were detected in serum collected at 30 dpt by AGID assay and ELISA. SGA analysis and sequencing of viral RNA obtained from A2215 on the day of peak viremia (dpt 19) identified the EIAVWSU5-V55 variant exclusively (24/24 single amplicons) (Fig. 6c).
Serum obtained from A2215 at 19, 55, 111, 230, 342, and 496 dpt was assayed for neutralizing activity against both EIAVWSU5-V55 (A2215 inoculum) and EIAVWSU5 (SCID foal inoculum). As was observed previously for the A2150, A2239, and A2240 sera (Fig. 6a), serum samples from A2215 poorly neutralized EIAVWSU5-V55 (Fig. 6d). Although 111-dpt serum had the highest level of EIAVWSU5-V55 neutralization, none of the A2215 sera achieved 50% virus reduction at dilutions as low as 1:8. Quite unexpectedly, however, A2215 serum neutralized EIAVWSU5 with a peak titer of 1,024 on dpt 230 (Fig. 6e). Overall, these results indicated that efficient and long-term control of the EIAVWSU5-V55 variant occurred in A2215 but that EIAVWSU5-V55 infection did not elicit a significant EIAVWSU5-V55-specific NAb response despite production of EIAVWSU5-specific NAbs.
Long-term control of EIAVWSU5-V55 was associated with EIAV Gag-specific CTL.
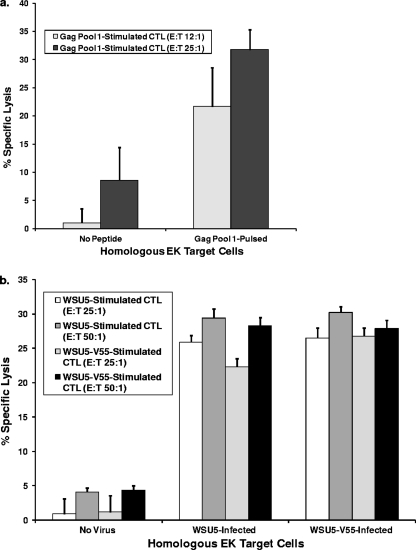
During the period of prolonged control of EIAVWSU5-V55 in A2215 (dpt 400 to 500), CTL activity against EIAV Gag and whole virus (EIAVWSU5 and EIAVWSU5-V55) was identified in both freshly isolated and cryopreserved PBMC. Memory CTL in PBMC stimulated with Gag peptide pool 1, which comprised 14 overlapping synthetic peptides containing immunodominant EIAV Gag p15 epitopes (34), specifically lysed Gag pool 1-pulsed homologous EK target cells (Fig. 7 a). In addition, memory CTL in PBMC stimulated with EIAVWSU5-V55 recognized both EIAVWSU5-V55- and EIAVWSU5-infected target cells with similar levels of specific lysis (Fig. 7b). Moreover, memory CTL stimulated with EIAVWSU5 also recognized both EIAVWSU5-V55- and EIAVWSU5-infected targets with similar levels of specific lysis (Fig. 7b). These results demonstrated that durable control of the EIAVWSU5-V55 variant was associated with Gag-specific CTL and that these CTL targeted conserved epitopes as both EIAVWSU5 and EIAVWSU5-V55 were recognized.
FIG. 7.
(a) Percent specific lysis of non-peptide-pulsed and Gag peptide pool 1-pulsed homologous EK target cells by CTL in Gag peptide pool 1-stimulated fresh PBMC isolated from horse A2215 on dpt 404. Results of two independent assays are shown, one performed with an effector/target (E/T) ratio of 12:1 and the other with an E/T ratio of 25:1. Error bars represent SE. (b) Percent specific lysis of uninfected, EIAVWSU5-infected, and EIAVWSU5-V55-infected homologous EK target cells by CTL in EIAVWSU5-stimulated and EIAVWSU5-V55-stimulated cryopreserved PBMC isolated from horse A2215 on dpt 504. Results of two independent assays are shown, one performed with an E/T ratio of 25:1 and the other with an E/T ratio of 50:1. Error bars represent SE.
DISCUSSION
In this study, we documented that immune plasma from a horse persistently infected with EIAVWSU5 contained antibodies capable of neutralizing homologous virus plus five other viruses containing heterologous SU PND regions derived from a pony persistently infected with wild-type EIAVWyo. When infused into four SCID foals prior to and after EIAVWSU5 challenge, this plasma resulted in clinical protection against EIAV-induced disease in all foals and prevented infection in one foal. Importantly, however, the breakthrough infections in three of the foals were caused by a neutralization-resistant SU variant that was present at low copy number in the inoculum. Durable control of this variant virus was achieved in the absence of type-specific NAbs in an immunocompetent horse and was associated with the presence of Gag-specific CTL. This work demonstrates for the first time in a large animal lentivirus model that broadly active NAbs can be protective in the complete absence of other adaptive immune responses, but the limitations of such an approach were also evident.
Although SU epitopes recognized by equine NAbs have not been precisely defined, three linear epitopes recognized by murine monoclonal NAbs have been mapped, with two in the PND V3 and one in the V5 region (1). Previous work has indicated that the V3 and V4 domains are the primary determinants of EIAV SU neutralization sensitivity and resistance (19). The immune plasma infused in this study neutralized several viruses containing SU regions V2 to V4 heterologous and contained NAbs that blocked homologous EIAVWSU5 infection in all treated SCID foals. In three foals, however, these NAbs did not prevent infection by EIAVWSU5-V55, a rare variant present in the EIAVWSU5 inoculum. The profound amino acid variation/deletion within V3 of EIAVWSU5-V55 suggested that NAbs in the infused plasma were targeted to this region. However, numerous amino acid changes, including those affecting PNLGS, also occurred within V4, V5, V6, and V7, with the most significant of these changes encompassing V6 and V7. Thus, the neutralization-resistant phenotype of EIAVWSU5-V55 may have resulted from changes that masked neutralizing epitopes within the PND and/or from variation in neutralizing epitopes located within V6/V7. Interestingly, we have observed SU changes similar to those above in immunocompetent horses following EIAVWSU5 infection. These included significant deletions and substitutions in V3 and substitutions in V2 and V4 (37, 38). More profound changes in V3 to V7 were observed in an EIAVWSU5-challenged SCID foal that had been reconstituted with EIAVWSU5-specific T and B lymphocytes (35). A significant number of these changes involved PNLGS. Although the SU changes observed in these previous studies were the result of immune selection, with some resulting in CTL escape (37, 38), it was not known whether the predominant selection force was NAbs, CTL, or both. In contrast, the current study provided a novel means to prove that NAbs alone constitute a significant selection force on the lentivirus envelope.
Based on the spacing of conserved cysteines, the EIAV SU V3/V4 region is structurally analogous to the HIV-1 gp120 V1/V2 stem-loop (17). The V1/V2 stem comprises part of the inner domain bridging sheet of HIV-1 gp120, which forms the CD4 binding pocket, and, along with the outer domain V3 loop, also binds the chemokine coreceptor (23). These binding sites are shielded by the V1/V2 and V3 loops (23). In addition to these structural similarities, some PNLGS are conserved between EIAV and HIV-1; two of these occur within and just downstream of EIAV V4 (17, 26). In the current study, the PNLGS within V4 (N233) was shifted two positions downstream in EIAVWSU5-V55 by a T235N substitution. This could have masked an NAb epitope or changed the conformation of SU such that an epitope was abolished. Unlike HIV-1, however, EIAV can utilize a single receptor, the equine lentivirus receptor 1 (ELR-1) (61). Although the EIAV SU binding site for ELR-1 remains to be defined, deletions in V4, V5, V6, V7, or V8 abolish binding while deletions in V1, V2, or V3 do not (58). Interestingly, though, a murine monoclonal antibody against an epitope in V3 did block ELR-1 binding (58). Thus, complicated and discontinuous sequences in EIAV SU mediate receptor binding, and the roles of V3, V4 and the other hypervariable regions in receptor binding and/or shielding are not completely understood. In the current study, the amino acid changes in V4 to V8 observed in EIAVWSU5-V55 could have contributed to its neutralization resistance just as much as, or more than, the changes in V3.
The appearance of the same neutralization-resistant variant in three separate SCID foals made it unlikely that the amino acid substitutions and deletions in EIAVWSU5-V55 arose de novo simultaneously in all three foals. Although our studies confirmed that EIAVWSU5-V55 was a rare variant in the EIAVWSU5 virus stock used for inoculation, the factors contributing to its origin and persistence in this stock are not clear. Regardless, infusion of convalescent immune plasma from a horse infected with EIAV for over 8 years exerted the selection force of a fully mature NAb response in a period of just a few weeks in foals devoid of adaptive immunity. This provided a rigorous test of the efficacy and limitations of NAbs alone in the protection against lentivirus infection. The fact that our challenge included two functionally heterologous viruses was important since humans can acquire more than one virus when infected with HIV-1 (51).
A previous study (25) detected an SU variant, designated EIAVΔPND, during the third febrile episode in a pony experimentally infected with the PV strain of EIAV (EIAVPV). EIAVΔPND has changes in the V3 and V4 regions similar to EIAVWSU5-V55, including a 14-amino-acid deletion in the PND. Consistent with our findings, autologous serum samples failed to neutralize the EIAVΔPND variant but displayed high levels of neutralization activity against the EIAVPV inoculum (25). Interestingly, two ponies inoculated with a chimeric virus containing the EIAVΔPND SU failed to develop high viral loads and clinical disease or NAbs; however, immunosuppression with dexamethasone resulted in higher viral loads and clinical disease, followed by the appearance of variant-specific NAbs (8). The type-specific NAb response mapped to the variant V3 and V4 regions, indicating the importance of these regions for neutralization by sera from these ponies (18). In contrast to those studies, the EIAVWSU5-V55 variant reported here replicated to a high enough level to cause clinical disease when inoculated into an immunocompetent horse, yet it was not capable of eliciting a significant type-specific NAb response. Unlike the previously reported EIAVΔPND, however, EIAVWSU5-V55 also contains significant changes in V6 and V7 which may have affected V3/V4 immunogenicity via conformational changes. Moreover, one of the murine monoclonal antibody neutralization epitopes in V3 (DNT) is intact in EIAVΔPND (18) but disrupted in EIAVWSU5-V55, and additional amino acid changes that are not present in EIAVΔPND occurred within and just upstream of V4 in EIAVWSU5-V55 (18). Regardless, critical epitopes were effectively abolished or masked by the SU changes in EIAVWSU5-V55 such that neutralizing B-cell responses against this variant could not be efficiently primed.
Although EIAVWSU5-V55 did not elicit a detectable type-specific NAb response in immunocompetent horse A2215, antibodies elicited against this virus were capable of neutralizing the parent EIAVWSU5 SCID foal inoculum. An intriguing explanation is that antibodies were elicited in A2215 against epitopes in the EIAVWSU5-V55 envelope that were shared with EIAVWSU5. However, because of conformational differences in the two envelopes, these antibodies did not efficiently interfere with the complex interactions between SU hypervariable regions and the ELR-1 receptor for EIAVWSU5-V55 but did so for EIAVWSU5. Despite similarity in epitopes, antibodies with slight differences in recognition of the HIV-1 gp120 CD4 binding site induce significant differences in gp120 conformation (4). Poorly neutralizing antibodies recognize conformational shifts that result in V1/V2 orientations that are poorly compatible with the functional viral spike. Thus, only the most accurately targeted CD4 binding site antibodies (such as b12) are able to neutralize resistant (tier 2) HIV-1 isolates (4). Whether the same holds true for EIAV receptor binding site antibodies remains to be determined. Nonetheless, identification of convalescent immune plasmas capable of neutralizing resistant heterologous lentiviruses such as EIAVWSU5-V55 poses a significant challenge, as will developing strategies to elicit efficient NAb responses against them.
It was not surprising that control of the EIAVWSU5-V55 variant in immunocompetent horse A2215 was associated with Gag-specific CTL, given our previous work indicating that the majority of EIAV-infected horses that effectively control infection mount Gag-specific CTL responses (30). Similarly, Gag-specific CTL play an important role in controlling HIV-1 replication in elite controllers (39). Although envelope SU-specific CTL are immunodominant in EIAV-infected horses, these CTL do not correlate with control of viremia due to SU variation and subsequent CTL escape (37, 38). Amino acid variation can occur in EIAV Gag (37, 62), but these changes do not lead to loss of recognition by CTL that correlate with control (37). Importantly, the p15 region of Gag targeted by A2215 CTL in the current study contains epitope clusters recognized by CTL from major histocompatibility complex class I (MHC-I)-disparate horses (5). These epitope clusters are targets for high-avidity CTL that correlate with long-term virus control (6). While the current study indicated that broadly reactive immune plasma was sufficient to control homologous virus in SCID foals, control of the EIAVWSU5-V55 variant was not consistently achieved. In contrast, this same variant was effectively controlled in an immunocompetent horse in the absence of EIAVWSU5-V55-specific NAbs when Gag p15-specific CTL were present. These observations highlight the importance of developing lentivirus vaccines that include conserved CTL immunogens in addition to eliciting broad NAb responses.
In summary, our results provide a strong basis for further dissection and fine mapping of NAb epitopes important in protection from EIAV infection and/or clinical disease. Since EIAV SU shares structural features with the SU of HIV-1, results of these studies could have important implications for the design of protective HIV-1 vaccines where experiments of this type are not possible.
Acknowledgments
We thank Robert Nelson, Emma Karel, Lori Fuller, Ashley Trtek, and Evan McQuirk for excellent technical assistance, as well as Rebecca Tallmadge, Wuwei Wu, and Liam Broughton-Neiswanger for contributing to useful discussions.
This work was supported in part by U.S. Public Health Service, National Institutes of Health grants AI073101 (R.H.M.) and CA128568 (S.C.).
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Ball, J. M., K. E. Rushlow, C. J. Issel, and R. C. Montelaro. 1992. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J. Virol. 66:732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, S., L. H. Evans, M. Sevoian, and B. Chesebro. 1987. Role of the host immune response in selection of equine infectious anemia virus variants. J. Virol. 61:3783-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpenter, S., L. H. Evans, M. Sevoian, and B. Chesebro. 1990. In vivo and in vitro selection of equine infectious anemia virus variants, p. 99-115. In E. Kurstak, R. G. Marusyk, F. A. Murphy, and M. H. V. Van Regenmortel (ed.), Applied virology research. Plenum Publishing Corp., New York, NY.
- 4.Chen, L., Y. D. Kwon, T. Zhou, X. Wu, S. O'Dell, L. Cavacini, A. J. Hessell, M. Pancera, M. Tang, L. Xu, Z. Y. Yang, M. Y. Zhang, J. Arthos, D. R. Burton, D. S. Dimitrov, G. J. Nabsel, M. R. Posner, J. Sodroski, R. Wyatt, J. R. Mascola, and P. D. Kwong. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, C., R. H. Mealey, and T. C. McGuire. 2004. CTL from EIAV carrier horses with diverse MHC class I alleles recognize epitope clusters in Gag matrix and capsid proteins. Virology 327:144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, C., R. H. Mealey, and T. C. McGuire. 2005. Evaluation of high functional avidity CTL to Gag epitope clusters in EIAV carrier horses. Virology 342:228-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coggins, L., N. L. Norcross, and S. R. Nusbaum. 1972. Diagnosis of equine infectious anemia by immunodiffusion test. Am. J. Vet. Res. 33:11-18. [PubMed] [Google Scholar]
- 8.Craigo, J. K., C. Leroux, L. Howe, J. D. Steckbeck, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2002. Transient immune suppression of inapparent carriers infected with a principal neutralizing domain-deficient equine infectious anaemia virus induces neutralizing antibodies and lowers steady-state virus replication. J. Gen. Virol. 83:1353-1359. [DOI] [PubMed] [Google Scholar]
- 9.Craigo, J. K., B. Zhang, S. Barnes, T. L. Tagmyer, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2007. Envelope variation as a primary determinant of lentiviral vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 104:15105-15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, T. B., K. J. Wardrop, S. J. Tornquist, E. Reilich, K. M. Meyers, and T. C. McGuire. 1996. A primary production deficit in the thrombocytopenia of equine infectious anemia. J. Virol. 70:7842-7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickover, R., E. Garratty, K. Yusim, C. Miller, B. Korber, and Y. Bryson. 2006. Role of maternal autologous neutralizing antibody in selective perinatal transmission of human immunodeficiency virus type 1 escape variants. J. Virol. 80:6525-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrantelli, F., K. A. Buckley, R. A. Rasmussen, A. Chalmers, T. Wang, P. L. Li, A. L. Williams, R. Hofmann-Lehmann, D. C. Montefiori, L. A. Cavacini, H. Katinger, G. Stiegler, D. C. Anderson, H. M. McClure, and R. M. Ruprecht. 2007. Time dependence of protective post-exposure prophylaxis with human monoclonal antibodies against pathogenic SHIV challenge in newborn macaques. Virology 358:69-78. [DOI] [PubMed] [Google Scholar]
- 13.Ferrantelli, F., R. Hofmann-Lehmann, R. A. Rasmussen, T. Wang, W. Xu, P. L. Li, D. C. Montefiori, L. A. Cavacini, H. Katinger, G. Stiegler, D. C. Anderson, H. M. McClure, and R. M. Ruprecht. 2003. Post-exposure prophylaxis with human monoclonal antibodies prevented SHIV89.6P infection or disease in neonatal macaques. AIDS 17:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Ferrantelli, F., R. A. Rasmussen, K. A. Buckley, P. L. Li, T. Wang, D. C. Montefiori, H. Katinger, G. Stiegler, D. C. Anderson, H. M. McClure, and R. M. Ruprecht. 2004. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian-human immunodeficiency virus by human anti-HIV monoclonal antibodies. J. Infect. Dis. 189:2167-2173. [DOI] [PubMed] [Google Scholar]
- 15.Floyd, R. A., J. E. Schneider, Jr., and D. P. Dittmer. 2004. Methylene blue photoinactivation of RNA viruses. Antiviral Res. 61:141-151. [DOI] [PubMed] [Google Scholar]
- 16.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotzel, I. 2003. Conservation of the human immunodeficiency virus type 1 gp120 V1/V2 stem/loop structure in the equine infectious anemia virus (EIAV) gp90. AIDS Res. Hum. Retroviruses 19:923-924. [DOI] [PubMed] [Google Scholar]
- 18.Howe, L., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2005. Specificity of serum neutralizing antibodies induced by transient immune suppression of inapparent carrier ponies infected with a neutralization-resistant equine infectious anemia virus envelope strain. J. Gen. Virol. 86:139-149. [DOI] [PubMed] [Google Scholar]
- 19.Howe, L., C. Leroux, C. J. Issel, and R. C. Montelaro. 2002. Equine infectious anemia virus envelope evolution in vivo during persistent infection progressively increases resistance to in vitro serum antibody neutralization as a dominant phenotype. J. Virol. 76:10588-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain, K. A., C. J. Issel, K. L. Schnorr, P. M. Rwambo, and R. C. Montelaro. 1987. Antigenic analysis of equine infectious anemia virus (EIAV) variants by using monoclonal antibodies: epitopes of glycoprotein gp90 of EIAV stimulate neutralizing antibodies. J. Virol. 61:2956-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 22.Kono, Y. 1988. Antigenic variation of equine infectious anemia virus as detected by virus neutralization. Brief report. Arch. Virol. 98:91-97. [DOI] [PubMed] [Google Scholar]
- 23.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leroux, C., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2001. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J. Virol. 75:4570-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leroux, C., C. J. Issel, and R. C. Montelaro. 1997. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J. Virol. 71:9627-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, H., X. Zhang, X. Fan, T. Shen, X. Tong, R. Shen, and Y. Shao. 2005. A conservative domain shared by HIV gp120 and EIAV gp90: implications for HIV vaccine design. AIDS Res. Hum. Retroviruses 21:1057-1059. [DOI] [PubMed] [Google Scholar]
- 27.Mahalanabis, M., P. Jayaraman, T. Miura, F. Pereyra, E. M. Chester, B. Richardson, B. Walker, and N. L. Haigwood. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascola, J. R. 2007. HIV/AIDS: allied responses. Nature 449:29-30. [DOI] [PubMed] [Google Scholar]
- 29.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuire, T. C., S. R. Leib, S. M. Lonning, W. Zhang, K. M. Byrne, and R. H. Mealey. 2000. Equine infectious anaemia virus proteins with epitopes most frequently recognized by cytotoxic T lymphocytes from infected horses. J. Gen. Virol. 81:2735-2739. [DOI] [PubMed] [Google Scholar]
- 31.McGuire, T. C., K. I. O'Rourke, and L. E. Perryman. 1990. Immunopathogenesis of equine infectious anemia lentivirus disease. Dev. Biol. Stand. 72:31-37. [PubMed] [Google Scholar]
- 32.McGuire, T. C., M. J. Poppie, and K. L. Banks. 1974. Combined (B- and T-lymphocyte) immunodeficiency: a fatal genetic disease in Arabian foals. J. Am. Vet. Med. Assoc. 164:70-76. [PubMed] [Google Scholar]
- 33.Mealey, R. H., D. G. Fraser, J. L. Oaks, G. H. Cantor, and T. C. McGuire. 2001. Immune reconstitution prevents continuous equine infectious anemia virus replication in an Arabian foal with severe combined immunodeficiency: lessons for control of lentiviruses. Clin. Immunol. 101:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mealey, R. H., S. R. Leib, M. H. Littke, B. Wagner, D. W. Horohov, and T. C. McGuire. 2009. Viral load and clinical disease enhancement associated with a lentivirus cytotoxic T lymphocyte vaccine regimen. Vaccine 27:2453-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mealey, R. H., S. R. Leib, S. L. Pownder, and T. C. McGuire. 2004. Adaptive immunity is the primary force driving selection of equine infectious anemia virus envelope SU variants during acute infection. J. Virol. 78:9295-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mealey, R. H., M. H. Littke, S. R. Leib, W. C. Davis, and T. C. McGuire. 2008. Failure of low-dose recombinant human IL-2 to support the survival of virus-specific CTL clones infused into severe combined immunodeficient foals: lack of correlation between in vitro activity and in vivo efficacy. Vet. Immunol. Immunopathol. 121:8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mealey, R. H., A. Sharif, S. A. Ellis, M. H. Littke, S. R. Leib, and T. C. McGuire. 2005. Early detection of dominant Env-specific and subdominant Gag-specific CD8+ lymphocytes in equine infectious anemia virus-infected horses using major histocompatibility complex class I/peptide tetrameric complexes. Virology 339:110-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mealey, R. H., B. Zhang, S. R. Leib, M. H. Littke, and T. C. McGuire. 2003. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology 313:537-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura, T., M. A. Brockman, A. Schneidewind, M. Lobritz, F. Pereyra, A. Rathod, B. L. Block, Z. L. Brumme, C. J. Brumme, B. Baker, A. C. Rothchild, B. Li, A. Trocha, E. Cutrell, N. Frahm, C. Brander, I. Toth, E. J. Arts, T. M. Allen, and B. D. Walker. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J. Virol. 83:2743-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr, H., J. Knuver-Hopf, U. Gravemann, A. Redecker-Klein, and T. H. Muller. 2004. West Nile virus in plasma is highly sensitive to methylene blue-light treatment. Transfusion 44:886-890. [DOI] [PubMed] [Google Scholar]
- 41.Motulsky, H. 1995. Intuitive biostatistics. Oxford University Press, New York, NY.
- 42.O'Rourke, K., L. E. Perryman, and T. C. McGuire. 1988. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J. Gen. Virol. 69:667-674. [DOI] [PubMed] [Google Scholar]
- 43.O'Rourke, K. I., L. E. Perryman, and T. C. McGuire. 1989. Cross-neutralizing and subclass characteristics of antibody from horses with equine infectious anemia virus. Vet. Immunol. Immunopathol. 23:41-49. [DOI] [PubMed] [Google Scholar]
- 44.Palmer, S., M. Kearney, F. Maldarelli, E. K. Halvas, C. J. Bixby, H. Bazmi, D. Rock, J. Falloon, R. T. Davey, Jr., R. L. Dewar, J. A. Metcalf, S. Hammer, J. W. Mellors, and J. M. Coffin. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perryman, L. E., T. C. McGuire, and T. B. Crawford. 1978. Maintenance of foals with combined immunodeficiency: causes and control of secondary infections. Am. J. Vet. Res. 39:1043-1047. [PubMed] [Google Scholar]
- 46.Perryman, L. E., K. I. O'Rourke, and T. C. McGuire. 1988. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J. Virol. 62:3073-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perryman, L. E., and R. L. Torbeck. 1980. Combined immunodeficiency of Arabian horses: confirmation of autosomal recessive mode of inheritance. J. Am. Vet. Med. Assoc. 176:1250-1251. [PubMed] [Google Scholar]
- 48.Quakkelaar, E. D., E. M. Bunnik, F. P. van Alphen, B. D. Boeser-Nunnink, A. C. van Nuenen, and H. Schuitemaker. 2007. Escape of human immunodeficiency virus type 1 from broadly neutralizing antibodies is not associated with a reduction of viral replicative capacity in vitro. Virology 363:447-453. [DOI] [PubMed] [Google Scholar]
- 49.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rybarczyk, B. J., D. Montefiori, P. R. Johnson, A. West, R. E. Johnston, and R. Swanstrom. 2004. Correlation between env V1/V2 region diversification and neutralizing antibodies during primary infection by simian immunodeficiency virus sm in rhesus macaques. J. Virol. 78:3561-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salazar-Gonzalez, J. F., E. Bailes, K. T. Pham, M. G. Salazar, M. B. Guffey, B. F. Keele, C. A. Derdeyn, P. Farmer, E. Hunter, S. Allen, O. Manigart, J. Mulenga, J. A. Anderson, R. Swanstrom, B. F. Haynes, G. S. Athreya, B. T. Korber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salinovich, O., S. L. Payne, R. C. Montelaro, K. A. Hussain, C. J. Issel, and K. L. Schnorr. 1986. Rapid emergence of novel antigenic and genetic variants of equine infectious anemia virus during persistent infection. J. Virol. 57:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheid, J. F., H. Mouquet, N. Feldhahn, M. S. Seaman, K. Velinzon, J. Pietzsch, R. G. Ott, R. M. Anthony, H. Zebroski, A. Hurley, A. Phogat, B. Chakrabarti, Y. Li, M. Connors, F. Pereyra, B. D. Walker, H. Wardemann, D. Ho, R. T. Wyatt, J. R. Mascola, J. V. Ravetch, and M. C. Nussenzweig. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 54.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204-210. [DOI] [PubMed] [Google Scholar]
- 55.Shin, E. K., L. E. Perryman, and K. Meek. 1997. A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J. Immunol. 158:3565-3569. [PubMed] [Google Scholar]
- 56.Siliciano, R. F., A. D. Keegan, R. Z. Dintzis, H. M. Dintzis, and H. S. Shin. 1985. The interaction of nominal antigen with T cell antigen receptors. I. Specific binding of multivalent nominal antigen to cytolytic T cell clones. J. Immunol. 135:906-914. [PubMed] [Google Scholar]
- 57.Sponseller, B. A., W. O. Sparks, Y. Wannemuehler, Y. Li, A. K. Antons, J. L. Oaks, and S. Carpenter. 2007. Immune selection of equine infectious anemia virus env variants during the long-term inapparent stage of disease. Virology 363:156-165. [DOI] [PubMed] [Google Scholar]
- 58.Sun, C., B. Zhang, J. Jin, and R. C. Montelaro. 2008. Binding of equine infectious anemia virus to the equine lentivirus receptor-1 is mediated by complex discontinuous sequences in the viral envelope gp90 protein. J. Gen. Virol. 89:2011-2019. [DOI] [PubMed] [Google Scholar]
- 59.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 60.Wiler, R., R. Leber, B. B. Moore, L. F. VanDyk, L. E. Perryman, and K. Meek. 1995. Equine severe combined immunodeficiency: a defect in V(D)J. recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. U. S. A. 92:11485-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, B., S. Jin, J. Jin, F. Li, and R. C. Montelaro. 2005. A tumor necrosis factor receptor family protein serves as a cellular receptor for the macrophage-tropic equine lentivirus. Proc. Natl. Acad. Sci. U. S. A. 102:9918-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, W., D. B. Auyong, J. L. Oaks, and T. C. McGuire. 1999. Natural variation of equine infectious anemia virus Gag protein cytotoxic T lymphocyte epitopes. Virology 261:242-252. [DOI] [PubMed] [Google Scholar]
- 63.Zolla-Pazner, S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]