Abstract
Sindbis virus (SINV) is the prototype member of the Alphavirus genus, whose members cause severe human diseases for which there is no specific treatment. To ascertain host factors important in the replication of the SINV RNA genome, we generated a SINV expressing nsP4, the viral RNA-dependent RNA polymerase, with an in-frame 3×Flag epitope tag. Proteomic analysis of nsP4-containing complexes isolated from cells infected with the tagged virus revealed 29 associated host proteins. Of these, 10 proteins were associated only at a later time of infection (12 h), 14 were associated both early and late, and five were isolated only at the earlier time (6 h postinfection). These results demonstrate the dynamic nature of the virus-host interaction that occurs over the course of infection and suggest that different host proteins may be required for the multiple functions carried out by nsP4. Two related proteins found in association with nsP4 at both times of infection, GTPase-activating protein (SH3 domain) binding protein 1 (G3BP1) and G3BP2 were also previously identified as associated with SINV nsP2 and nsP3. We demonstrate a likely overlapping role for these host factors in limiting SINV replication events. The present study also identifies 10 host factors associated with nsP4 6 h after infection that were not found to be associated with nsP2 or nsP3. These factors are candidates for playing important roles in the RNA replication process. Identifying host factors essential for replication should lead to new strategies to interrupt alphavirus replication.
Alphaviruses are important human and animal pathogens, causing fever, rash, arthritis, encephalitis, and death (reviewed in reference 18). These enveloped positive-strand RNA viruses cycle in nature through small vertebrate reservoir hosts and invertebrate vectors, most commonly mosquitoes. Infection of larger mammals, including humans, through the bite of an infected mosquito can lead to severe disease, for which there is no specific therapy. Viruses in the Alphavirus genus share common genomic organization and replication strategies, and much has been learned through study of Sindbis virus (SINV), the prototype virus in this genus. SINV, like the other alphaviruses, enters cells by receptor-mediated endocytosis and after fusion in the endosome releases the positive-strand RNA genome, which is translated in the cytoplasm to generate a nonstructural polyprotein, P123 (reviewed in reference 36). Translational readthrough of an opal termination codon also results in the production of the polyprotein P1234. The activity of a cysteine protease residing in nsP2 mediates polyprotein processing, which results in the generation of P123 and nonstructural protein 4 (nsP4), the viral RNA-dependent RNA polymerase. Together, P123 and nsP4 produce a full-length negative-strand copy of the genomic RNA. Further regulated sequential processing of the polyprotein results in replication complexes active in both positive and negative strand RNA synthesis. After complete proteolytic processing, replication complexes containing the individual nsP1, nsP2, nsP3, and nsP4 proteins are capable of only positive-strand RNA production, including both genomic RNA and a subgenomic RNA colinear with the 3′ end of the genome and encoding the viral structural proteins.
The roles of the individual viral proteins in viral RNA replication have been investigated by examination of temperature-sensitive mutants, as well as targeted mutagenesis (reviewed in reference 20). nsP1 interacts with cellular membranes and plays a role in capping the newly generated genomic and subgenomic transcripts. In addition to its proteolytic activity, nsP2 possesses nucleotriphosphatase and helicase activities and is involved in regulation of RNA synthesis. The nsP2 protein of SINV, as well as that of Semliki Forest virus, also has known activity in the alteration of host cellular processes, such as the shutoff of host translation and transcription (10, 12, 14, 16, 29). Mutational analysis has demonstrated a role for nsP3 in RNA synthesis, although the precise function is not understood. Several recent nsP3 interactome studies (6, 11, 17) have identified nsP3-associated cellular factors that suggest possible roles for this viral protein in modulation of host cellular pathways. The alphavirus nsP4 is the RNA-dependent RNA polymerase with recent biochemical studies confirming and characterizing its activity (32, 38, 39).
Although roles in RNA replication for the virus-encoded nonstructural proteins (nsPs) have been elucidated (reviewed in references 20 and 36), little is known regarding how host factors may participate in the replication process. To address this, we and others have taken proteomic approaches to identify candidate host factors that may be involved in SINV replication. Using SINV expressing nsPs tagged in frame with the green fluorescent protein (GFP), host proteins that interact with nsP2 (1) and nsP3 (6, 11, 17) have been identified. The results from those studies allow for the generation of hypotheses as to how SINV utilizes host factors for viral replication processes. Many alphaviruses, including SINV, express P123 at levels higher than nsP4, the RNA-dependent RNA polymerase, due to an opal termination codon between nsP3 and nsP4 (35) and due to rapid degradation of nsP4 (8). We reasoned, therefore, that nsP1, nsP2, and nsP3 likely have multiple functions within the infected cell and that host proteins previously identified as interacting with nsP2 and nsP3 may play roles in important processes that might not be directly related to replication of the viral RNA. In fact, evidence has recently been found for multiple types of nsP3-containing complexes in SINV-infected cells (17), suggesting that the functions of these viral proteins and their interacting host proteins may be multifaceted. We therefore undertook studies to identify host factors that interact with nsP4, the viral RNA-dependent RNA polymerase, to identify candidate host factors that might participate more directly in viral RNA replication.
In the present study we isolated a Flag-tagged version of the viral RNA-dependent RNA polymerase nsP4 from infected vertebrate cells at two different times of infection, a late time (12 h) and an earlier time (6 h), and identified the interacting host proteins by mass spectrometry. Some proteins, e.g., protein arginine methyltransferase 5 (PRMT5) and unc-84 homology B (UNC84), were detected in association with nsP4 only at the earlier time point. Others, e.g., adenine nucleotide translocator 1 (ANT1) and F1-ATPase, were observed only at the late time point. Some host factors, e.g., methylosome protein 50 (MEP50), heat shock protein 90 (HSP90), and the Ras-GTPase-activating protein SH3-domain-binding protein 1 (G3BP1) and G3BP2, were associated with nsP4 at both the early and late time points. The identified proteins may play direct roles in viral RNA replication, or may be participating in host responses aimed at limiting viral replication. Two proteins, G3BP1 and G3BP2, identified in nsP4-containing complexes were also identified in nsP2 (1)- and nsP3 (6, 11, 17)-containing complexes in prior studies. We demonstrate here a role for these proteins in limiting SINV replication events.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses.
Rat2 (rat fibroblast) and BHK-21 (hamster kidney) cells were maintained as previously described (2). 293T (human embryonic kidney) cells were maintained in Dulbecco modified eagle medium supplemented with 10% fetal bovine serum at 37°C in a humidified chamber containing 5% CO2. All virus stocks were generated by electroporation of in vitro-transcribed RNA into BHK-21 cells and were titered as described previously (2). For infectious center assays, dilutions of electroporated BHK-21 cells were added to wells containing naive BHK-21 cells and, after allowing the cells to settle, a semisolid agarose overlay was applied. Plaques were enumerated and measured 2 days later after fixation, removal of the agarose overlay, and crystal violet staining. Unless otherwise specified, the multiplicity of infection (MOI) was calculated based on titers obtained on BHK-21 cells.
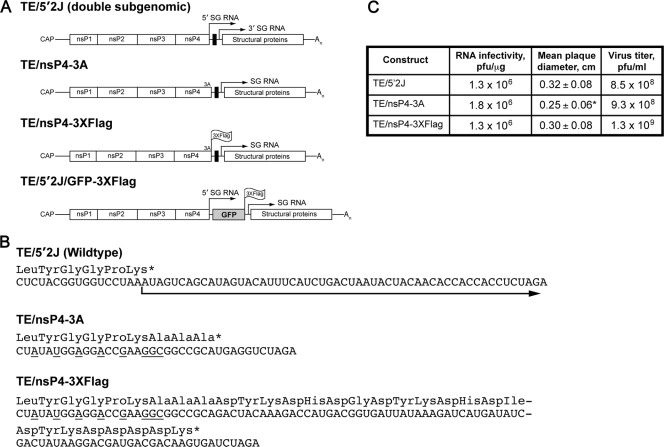
Plasmid TE/5′2J, a double-subgenomic (SG) SINV expression vector (described in reference 19) is designed for the expression of a gene of interest from SG RNA derived from the 5′ SG promoter, while the SINV structural genes are encoded by RNA derived from the 3′ SG promoter. The nucleotides encoding the carboxyl-terminal portion of nsP4, as well as nucleotides in the 5′ SG promoter important for promoter function (42), were mutated by replacing the BamHI-XbaI fragment of pTE/5′2J with sequences derived by PCR amplification with Oligo 1 (upstream of the BamHI site) and Oligo 2 (containing the desired mutations and XbaI site, see Table 1 ). Virus generated from the resultant plasmid, pTE/nsP4-3A, expresses nsP4 with three additional alanine residues at the carboxyl terminus. Plasmid pTE/nsP4-3A was used as a template for PCR amplification using Oligo 1 and Oligo 3, which contains nucleotides encoding a 3×Flag tag (44) and XbaI site. The resultant BamHI-XbaI fragment was used to replace the BamHI-XbaI fragment of pTE/5′2J to generate pTE/nsP4-3×Flag. Virus derived from this plasmid contains the 5′ SG promoter mutations and expresses nsP4 with a carboxyl-terminal 3×Flag epitope tag. GFP encoding sequences from plasmid pEGFP-C1 (Clontech, Mountain View, CA) were amplified by PCR using Oligo 4 (containing an XbaI site, Kozak consensus, and EGFP N terminus) and Oligo 5 (containing sequences encoding the C terminus of EGFP fused to a 3×Flag epitope tag and Bsp120I site). The XbaI-Bsp120I fragment from the resultant DNA was inserted into the XbaI-Bsp120I site of pTE/5′2J to generate pTE/5′2J/GFP-3×Flag. Virus derived from this plasmid was used as a control for the immunoisolations, since it expresses free GFP with a 3×Flag epitope tag. All DNA derived by PCR amplification was sequenced. Plasmid sequences are available upon request. SINV Toto1101 (wild-type SINV) has been previously described (31). SINV Toto1101/Luc, expressing firefly luciferase as an in-frame fusion within nsP3 and Toto1101/Luc:ts110 containing a temperature sensitive mutation (glycine 324 to glutamic acid) in nsP4 have been described previously (2).
TABLE 1.
Oligonucleotide primers used for PCR amplification for plasmid construction
| Plasmid | Primer | Sequence (5′ to 3′) |
|---|---|---|
| pTE/nsP4-3A | Oligo 1 | GGATATCGCATCATTCGACAA |
| Oligo 2 | CAGTCTAGACCTCATGCGGCCGCCTTCGGTCCTCCATATAGATGCTTTATTTCC | |
| pTE/nsP4-3×Flag | Oligo 3 | CAGTCTAGATCACTTGTCGTCATCGTCCTTATAGTCGATATCATGATCTTTATAA |
| TCACCGTCATGGTCTTTGTAGTCTGCGGCCGCCTTCGGTCCTCC | ||
| pTE/5′2J/GFP-3×Flag | Oligo 4 | GGTCTAGAGCCACCATGGTGAGCAAGGGCGAG |
| Oligo 5 | CAGGGCCCTCACTTGTCGTCATCGTCCTTATAGTCGATATCATGATCTTTATAAT | |
| CACCGTCATGGTCTTTGTAGTCCTTGTACAGCTCGTCCATG |
siRNA silencing and virus infections.
G3BP1 and/or G3BP2 were silenced using ON-TARGETplus SMARTpool siRNAs (L-012099-00 and L-015329-01; Dharmacon). ON-TARGETplus siCONTROL nontargeting pool (D-001810-10; Dharmacon) was used as an irrelevant control. Silencing was performed using the manufacturer's instructions (Invitrogen) for reverse transfection of 293T cells using Lipofectamine RNAiMAX and a final small interfering RNA (siRNA) concentration of 10 nM, unless indicated otherwise. For silencing in the 24-, 12-, and 6-well formats, 50,000, 125,000, and 250,000 cells were utilized per well, respectively. Silencing was allowed to proceed for 2 days prior to harvest and/or infection. To assess polyprotein expression, silenced cells were infected with Firefly luciferase expressing SINV (Toto1101/Luc), and the cells were harvested for luciferase activity (see below). To assess virion production, growth curves were performed on silenced 293T cells infected with SINV Toto1101 (MOI of 0.01). After the 1-h infection, the inoculum was removed, and the cells were washed twice with warm medium. At the time of harvest, the medium was removed and stored at −80°C until assay. The samples were titered by plaque assay on permissive BHK-21 cells as previously described (2). To assess the effect of G3BP silencing on SINV RNA levels, silenced cells were infected with Toto1101/Luc (MOI of 5). At various times after infection, the cells were washed and RNA was harvested for quantitative reverse transcription-PCR (RT-PCR) analysis (see below).
Luciferase and cell viability assays.
Luciferase assays were performed on cells silenced and infected in a 24-well format. After being washed with phosphate-buffered saline (PBS), the cells were lysed directly in 100 μl of 1× passive lysis buffer (Promega), and the luciferase activity in 10 μl of lysate was determined by using the firefly luciferase assay system (Promega) according to the manufacturer's recommendation. Readings were performed for 10 s in a Berthold LB960 or LB9507 luminometer. Cell viability after silencing was determined by using the Cell Titer Glo (Promega) assay. Culture medium was removed from the silenced cells in 24-well plates, and 150 μl of fresh medium was added to each well. An equal volume of Cell Titer Glo reagent was added, the plate was placed on an orbital shaker for 2 min, and the lysates were transferred to an opaque luminometer plate. Light production was measured in a Berthold luminometer for 0.1 s.
Analysis of SINV RNA production by metabolic labeling.
293T cells seeded in 35-mm dishes were mock infected or infected with Toto1101/Luc:ts110 (MOI of 10) in the presence of 1 μg of actinomycin D/ml at a permissive (28°C) or nonpermissive (40°C) temperature. After the 1-h infection, fresh medium containing 10 μCi of [5-3H]uridine (Amersham TRK178, 28 Ci/mmol)/ml and 1 μg of actinomycin D/ml was added. Total RNA was isolated 23 h later using TRIzol reagent (Gibco) and was size separated by denaturing agarose gel electrophoresis. Newly synthesized viral RNA was detected by fluorography.
nsP4 immunoisolation.
Rat 2 cells were infected with SINV TE/nsP4-3×Flag virus or the TE/5′2J/GFP-3×Flag control virus at an MOI of ∼30. The MOI was calculated based on the titer of the stock on Rat2 cells, or on an estimated titer based on the ratio (1/3.25) of the number of plaques obtained with the same stock on Rat2 compared to BHK-21 cells, respectively. Immunoaffinity purifications from cells infected for 6 or 12 h were carried out as previously described (6, 7). Briefly, magnetic beads (M-270 epoxy beads; Invitrogen) were conjugated with 10 μg of anti-FLAG antibody (M2; Sigma, St. Louis, MO) per mg of beads. Infected cells (grown in 150-mm plates) were washed in PBS containing 0.5 mM EDTA and harvested by scraping into PBS using 10 ml/plate. After pooling and collection by centrifugation, the cells were weighed; for each sample, including control and nsP4 samples at 6 and 12 h postinfection (hpi), enough plates were utilized to yield ∼1 g of cells (15 to 25 plates). The cells were resuspended in 20 mM HEPES containing 1.2% polyvinylpyrrolidone and 1/100 (vol/vol) protease inhibitor mixture (Sigma), using 100 to 200 μl per g of cells. After freezing the cell suspension by dropping it into liquid nitrogen, the frozen cells were cryogenically disrupted in a Retch MM301 ball mill (Retch) using 8 to 10 cycles of 3 min at 30 Hz, with intermittent cooling in liquid nitrogen. The resulting frozen cell powder samples were suspended in 6 to 8 ml of lysis buffer consisting of 20 mM K-HEPES (pH 7.4), 110 mM potassium acetate, 2 mM MgCl2, 0.1% Tween 20, 1% Triton X-100, 0.5% deoxycholate, 500 mM NaCl, 1/100 (vol/vol) protease inhibitor mixture (Sigma), and 25 U of DNase and RNase/ml. Each resulting cell lysate was incubated with 20 mg of conjugated beads for 1 h at 4°C. Purified complexes were eluted with 700 μl of 0.5 N NH4OH-0.5 mM EDTA by shaking for 20 min at room temperature, dried by vacuum centrifugation, and suspended in protein electrophoresis sample buffer.
SDS-PAGE and mass spectrometric analyses.
Proteins were resolved by SDS-PAGE (NuPAGE 4 to 12% Bis-Tris; Invitrogen) and stained with Coomassie blue (GelCode Blue; Pierce). Each entire gel lane was cut into ∼66 sections 1 mm in length. Adjacent sections were then combined, based on the pattern of Coomassie staining, into approximately 30 samples, and the proteins were digested with 12.5 ng of sequencing-grade modified trypsin (Promega)/μl. The resulting peptides were extracted on reversed-phase resin (Poros 20 R2; PerSeptive Biosystems) and eluted with 50% (vol/vol) methanol, 20% (vol/vol) acetonitrile, and 0.1% (vol/vol) trifluoroacetic acid, containing 20 mg of 2,5-dihydroxybenzoic acid matrix/ml. Mass spectrometric analyses on the samples derived from cells infected with SINV for 12 h were performed by matrix-assisted laser desorption ionization (MALDI) MS (prOTOF; Perkin-Elmer) and MALDI MS/MS (vMALDI LTQ XL; Thermo), as previously described (3, 6). The mass spectrometric analyses for the control samples, cells infected for 6 h, and selected MS/MS analyses performed on cells infected for 12 h were performed on a MALDI LTQ Orbitrap XL (Thermo Electron, Bremen, Germany) (27, 37). Mass spectra were visualized and processed in Qual Browser (version 2.0.7; Thermo Fisher Scientific). Protein candidates were identified by database searching against the National Center for Biotechnology Information nonredundant protein database, version 06/10/16, using the XProteo computer algorithm. Search parameters for MS data were as follows: species Rattus norvegicus (38,954 sequences) or viruses (346,953 sequences), accordingly; protein mass of 0 to 500 kDa; protein pI ranging from 1 to 14; mixture search, auto; display top, 50; enzyme, trypsin; miscleavage, 1; mass type, monoisotopic; charge state, MH+; mass tolerance, 0.02 Da; fixed modification, carbamidomethylation of Cys; and variable modifications, oxidation of Met and phosphorylations on Ser, Thr, or Tyr. Candidate peptides were confirmed by manual or automatic CID MS/MS analyses. Additional parameters used for searching MS/MS data were as follows: data type, MS/MS; precursor tolerance, 0.02 Da; fragment tolerance, 0.5 Da; and instrument, MALDI_I_TRAP. All XProteo assignments made from MS/MS data acquired in automated mode were checked manually. The XProteo probability scores, based on an improved version of the ProFound Bayesian algorithm (45), indicate d′ (discriminability) values for each candidate protein as the normalized distance between the score distribution (of the candidate protein) and the distribution of randomly matched proteins (in units of standard deviation). A d′ score of 4 corresponds to a true-positive rate of 0.99 and a false-positive rate of 0.05.
Confocal microscopy.
Rat2 cells were seeded onto poly-l-lysine (Sigma, 100 μg/ml)-coated coverslips and the next day were mock infected or infected with SINV (TE/nsP4-3×Flag) at an MOI of ∼30 (based on titers obtained on Rat2 cells). After 6 or 12 h, the cells were fixed in PBS containing 1% paraformaldehyde and processed for confocal microscopy as previously described (6). Each sample was incubated with both antibody to G3BP1 (1:1,000, mouse monoclonal antibody 61126; BD Transduction Laboratories) and antiserum raised against SINV nsP4 (1:6,000; nsP4-1 rabbit polyclonal antibody, described in reference 25). Alexa Fluor 488 donkey anti-mouse IgG (1:1,000, Molecular Probes, catalog no. A21202) and Rhodamine Red-X conjugated donkey anti-rabbit IgG (1:200; Jackson Immunoresearch Laboratories, catalog no. 711 295 152) were utilized as secondary antibodies. Nuclei were stained with 5 μM TO-PRO-3 in PBS. Cells were mounted in PBS with 90% glycerol and visualized in the Rockefeller University Bio-Imaging Resource Center utilizing a Zeiss LSM 510 upright Axioplan confocal microscope equipped with a HeNe laser (633 line) and a krypton/argon laser (488 and 568 lines) and a 40×/1.2 NA C-Apochromat water immersion objective lens. For the red channel, as well as the green channel, consistent laser intensity and gain settings were used to acquire images across all samples using the Zeiss LSM 510 version 3.2 software. Brightness and contrast settings on the far-red channel were adjusted after acquisition using the LSM software to allow better visualization of nuclei; no adjustments were made on the green and red channels.
Quantitative RT-PCR.
Total RNA was harvested from cells using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. One microgram of total RNA was transcribed into cDNA by using SuperScript III first strand synthesis (Invitrogen) with random hexamers as primers according to the manufacturer's instructions. cDNA was diluted 1:5 or 1:10 and amplified with the Qiagen QuantiTect SYBR green PCR kit or with Roche LightCycler 480 SYBR green Master Mix; products were detected with a LightCycler 480 (Roche). Enzyme activation occurred at 95°C for 15 min (Qiagen SYBR green) or 5 min (Roche SYBR green), followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Primers for amplification of human G3BP1 and human G3BP2 were obtained from Qiagen (QuantiTect primer assays QT00001897 and QT00049581, respectively). Samples were normalized based on the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA present, as determined by real-time PCR using a QuantiTect primer assay QT00309099. Relative levels were determined from samples, each assayed in triplicate PCRs, using the comparative threshold method. The efficiency of G3BP1, G3BP2, and GAPDH amplification was tested and found to be approximately equal in the cDNA dilution range utilized in this assay.
For quantification of SINV RNA, cDNA was synthesized as described above from 1 μg of total RNA and diluted 1:10 to 1:100 for amplification as described above using the primers 5′-CCCAGGAACCCGCAAGTATG-3′ and 5′-CGTGAGGAAGATTGCGGTTC-3′ and the Roche LightCycler 480 SYBR green Master Mix. The 173-nucleotide amplicon corresponds to nucleotides 3365 to 3537 of the Toto1101 SINV genome (within nsP2). cDNA derived from in vitro-transcribed RNA using XhoI-linearized plasmid pToto1101/Luc/Pol− (2) as a template was utilized to generate a standard curve. For each cell sample, cDNA synthesis was performed in duplicate and utilized in duplicate PCRs.
Western blotting and antibodies.
Cells were lysed in 2× Laemmli sample buffer and proteins were separated by SDS-8% PAGE and transferred to Hybond ECL nitrocellulose membranes (Amersham). Blots were incubated with primary and secondary antibodies as described previously (28). Anti-G3BP1 mouse monoclonal antibody (catalog no. 611126; BD Transduction Laboratories) was utilized at a 1:750 dilution, anti-β-actin mouse monoclonal antibody AC-15 (A5441, Sigma) was utilized at 1:5,000 dilution, and horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce) was utilized at 1:20,000 dilution. Chemiluminescent detection was performed with SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's instructions.
RESULTS
SINV expressing a 3×Flag epitope-tagged RNA-dependent RNA polymerase is viable.
To facilitate immunoisolation of nsP4 and interacting factors, we generated SINV expressing nsP4 with a carboxy-terminal epitope tag. Because the RNA sequences at the C terminus of nsP4 also function as the promoter for subgenomic (SG) RNA synthesis (reviewed in reference 36), mutations in the C terminus of nsP4 have the potential to alter SG promoter function. Therefore, we utilized a SINV (TE/5′2J, Fig. 1 A) with a duplicated SG promoter and expressing the viral structural genes from the 3′ SG promoter. This allowed alteration of nsP4 (and the 5′ SG promoter) without affecting expression of the viral structural proteins. Our strategy included mutation of specific residues known to be important for SG promoter function (42) to block production of SG RNA from the upstream promoter while maintaining the nsP4 coding sequence (Fig. 1B). We first generated a virus with NotI and XbaI restriction sites for cloning epitope tag-encoding sequences as fusions to the carboxyl terminus of nsP4. The resulting virus, designated SINV TE/nsP4-3A, expresses nsP4 with three additional alanine residues at the C terminus. This virus is viable, and replicates similarly to the parental virus, SINV TE/5′2J, which expresses wild-type nsP4 (Fig. 1C). Having successfully utilized GFP as a tag for visualization and immunoisolation of nsP3 (6), we first attempted to generate a virus that expressed nsP4 with a C-terminal GFP fusion. However, the resultant virus was nonviable (data not shown). We reasoned that a smaller tag might be better tolerated and so generated a virus expressing a 3×Flag epitope tag (44). This virus (SINV TE/nsP4-3×Flag) expresses nsP4 with a C-terminal 3×Flag tag, is viable, and replicates similarly to TE/5′2J and TE/nsP4-3A. RNA infectivity of the three virus constructs, as determined by infectious center assay, was similar (Fig. 1C). Although the mean size of the TE/nsP4-3A infectious center plaques was slightly smaller (difference from TE/5′2J = 0.07 ± 0.02 cm), the titers of all three resulting virus stocks were similar.
FIG. 1.
Characteristics of viruses used in the present study. (A) Schematic diagrams of the viruses utilized in this study are shown. TE/5′2J expresses two subgenomic (SG) RNAs, indicated by arrows above the diagram; the 5′ SG RNA express foreign genes inserted using restriction sites (black box) between the two SG transcriptional start sites, while the 3′ SG RNA encodes the viral structural proteins. An indicates the poly(A) tail. Three alanine residues expressed by SINV TE/nsP3-3A are indicated at the carboxyl terminus of nsP4, the viral RNA-dependent RNA polymerase. The location of the 3×Flag tag at the carboxyl termini of nsP4 and GFP in SINV TE/nsP4-3×Flag and TE/5′2J/GFP-3×Flag, respectively, is indicated. (B) The RNA sequences of the parental (TE/5′2J) and derived viruses are shown, with the amino acid sequence shown above. An asterisk indicates a stop codon. The arrow indicates the SG RNA derived from the 5′ SG promoter. Nucleotides mutated to disrupt SG promoter function are underlined. (C) Comparison of the infectivity of the viral constructs. BHK-21 cells were electroporated with 1 μg of each RNA in duplicate (or triplicate, TE/nsP4-3×Flag), and the number of infectious centers was determined. Mean plaque diameter, as assessed by measurement of 30 to 40 plaques for each virus, is shown, with the standard deviation indicated. The asterisk indicates a statistically different mean diameter from the TE/5′2J parental virus (P = 0.0015, unpaired t test). The remaining electroporated cells were placed in culture and 1 day later the medium overlying the cells was removed and pooled to generate a virus stock. The titer of infectious virus, determined by standard plaque assay, is indicated.
Affinity isolation of the RNA polymerase nsP4 reveals common and unique host interacting partners at early and late times of infection.
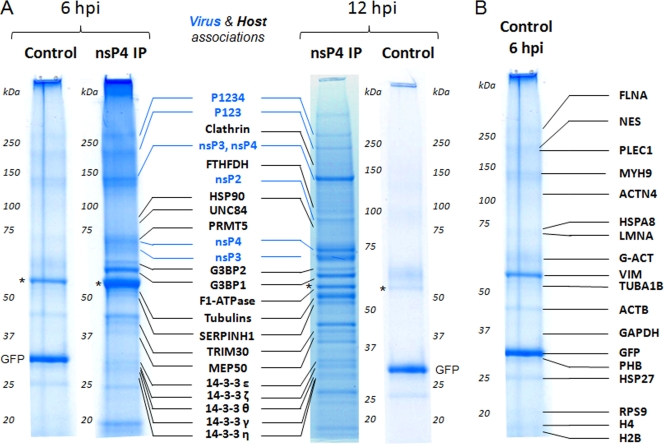
Having a SINV expressing an epitope-tagged nsP4 allowed us to isolate nsP4 and identify candidate host factors that might play a role in SINV RNA replication. We chose to utilize Rat2 fibroblasts for the present study, in order to allow direct comparison to our previous study, which identified the host factors interacting with nsP3 in infected Rat2 cells (6). Rat2 cells were infected with TE/nsP4-3×Flag or the control virus TE/5′2J/GFP-3×Flag, which expresses GFP containing a 3×Flag epitope tag and serves as a control for proteins nonspecifically binding to the beads or to the Flag antibody. We chose to analyze samples at a late time after infection (12 h), as well as an earlier time (6 h) chosen as a compromise to allow detection of host factors involved in early processes, while still allowing adequate material for analysis. At 6 and 12 h after infection, the cells were harvested, and nsP4-containing complexes were affinity isolated by using magnetic beads conjugated to anti-Flag antibodies. Figure 2 shows the isolated proteins, as assessed by SDS-PAGE, and the identity of the highest-scoring proteins, as determined by mass spectrometry. A list of the proteins identified with significant scores by database searching using the MS data and confirmed by MS/MS analyses with a minimum of two peptides is shown in Table 2. Table S1 in the supplemental material presents the complete list of identified proteins, together with their protein sequence coverage, database scores, and the sequences of the peptides confirmed by MS/MS. Proteins found in the control samples at either time point were considered likely contaminants; the identity of the proteins isolated in the 6-hpi control sample is shown in Fig. 2B. Although these putative contaminants were detected in both 6- and 12-h control isolations, the proteins were more prominent in the 6-h isolation and the data (scores, sequence coverage, and peptides confirmed) from this sample are therefore included in Table S1 in the supplemental material as the control sample. As expected, nsP4-containing complexes contained other SINV proteins involved in replication (nsP2, nsP3, and P123), as well as the complete nonstructural polyprotein, P1234. Of the identified proteins, 29 host proteins were absent in the control samples but present in nsP4-containing complexes isolated from SINV-infected cells. Of these, 14 were found in association with nsP4 at both 6 and 12 h: G3BP1, G3BP2, MEP50, HSP90, tripartite motif protein 30 (TRIM30), thymidylate synthase (TYMS), tubulin beta 5 (TUBB5), ribosomal protein S3 (RPS3), serine (or cysteine) proteinase inhibitor (clade H) member 1 (SERPINH1), and five isoforms of tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (14-3-3). Five host proteins were isolated with nsP4-containing complexes at 6 h only: unc-84 homolog B (UNC84), protein arginine methyltransferase 5 (PRMT5), guanine nucleotide binding protein, beta polypeptide 2-like 1 (GNB2L1), splicing factor, arginine/serine-rich 10 (SFRS10), and annexin A1 (ANXA1). The remaining 10 nsP4-associated host proteins, which included F1-ATPase, adenine nucleotide translocator 1 (ANT1), clathrin, insulin receptor (INSR), regulatory particle triple-A ATPase 4 (RPT4), fibrillarin (FBL), cell division cycle 40 (CDC40), 10-formyltetrahydrofolate dehydrogenase (FTHFDH), and ribosomal proteins L6 and L18, were present only in the sample harvested 12 h after infection.
FIG. 2.
Identification of nsP4-associated host factors using isolations on magnetic beads and mass spectrometry. Rat2 cells were infected with SINV TE/5′2J/GFP-3×Flag (control) virus expressing Flag-tagged GFP or SINV TE/nsP4-3×Flag (nsP4) virus expressing Flag-tagged nsP4 and at the indicated times after infection the Flag-tagged proteins and associated factors were isolated by affinity purification using magnetic beads conjugated to anti-Flag antibodies as described in Materials and Methods. The isolated proteins were resolved by SDS-PAGE and stained with Coomassie blue, and the proteins present in each entire lane were identified by mass spectrometry. (A) The identity of viral (blue) and prominent host (black) proteins in the nsP4 samples is indicated. Vimentin (indicated with an asterisk) was predominant in both the control and nsP4 samples. GFP, isolated due to the 3×Flag tag present on the GFP-3×Flag, is indicated in the control samples. (B) The identity of the proteins present in the control sample harvested at 6 hpi is indicated, forming a list of likely contaminants for our nsP4 isolations. Table 2 and Table S1 in the supplemental material provide the complete list of identified proteins.
TABLE 2.
Isolated proteins considered in this studya
| Virus, host, or likely contaminant | Isolated protein | Description | gi no. | Presence or absence |
||
|---|---|---|---|---|---|---|
| At 6 hpi | At 12 hpi | In control | ||||
| Virus (SINV) | nsP2 | nsP2 nonstructural protein | 25121507 | + | + | - |
| nsP3 | nsP3 nonstructural protein | 25121508 | + | + | - | |
| nsP4 | nsP4 nonstructural protein | 25121505 | + | + | - | |
| P123 | p230 nonstructural polyprotein | 9790318 | + | - | - | |
| P1234 | p270 nonstructural polyprotein | 9790317 | + | + | - | |
| Host (Rattus norvegicus) | G3BP1 | GTPase activating protein (SH3 domain) binding protein 1 | 109488113 | + | + | - |
| G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 | 50927029 | + | + | - | |
| MEP50 | Ac2-269; methylosome protein 50 | 32527757 | + | + | - | |
| HSP90 | Heat shock protein 90-kDa alpha (cytosolic), class B member 1 | 51243733 | + | + | - | |
| TRIM30 | Similar to tripartite motif protein 30-like | 109462593 | + | + | - | |
| 14-3-3ζ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 1051270 | + | + | - | |
| 14-3-3ɛ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon | 1469948 | + | + | - | |
| 14-3-3η | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta | 6981710 | + | + | - | |
| 14-3-3θ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta | 6981712 | + | + | - | |
| 14-3-3γ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma | 9507245 | + | + | - | |
| TYMS | Thymidylate synthase | 9507217 | + | + | - | |
| TUBB5 | Tubulin, beta 5 | 27465535 | + | + | - | |
| RPS3 | Ribosomal protein S3 | 57164151 | + | + | - | |
| SERPINH1 | Serine (or cysteine) proteinase inhibitor, clade H, member 1 | 55824765 | + | + | - | |
| UNC84 | unc-84 homolog B | 109481014 | + | - | - | |
| PRMT5 | Protein arginine methyltransferase 5 | 184160976 | + | - | - | |
| Gnb2l1 | Guanine nucleotide binding protein, beta polypeptide 2-like 1 (protein kinase C receptor) | 18543331 | + | − | − | |
| SFRS10 | Splicing factor, arginine/serine-rich 10 | 1255683 | + | - | - | |
| ANXA1 | Annexin A1 | 6978501 | + | - | - | |
| F1-ATPase | Chain A, rat liver F1-ATPase | 6729934 | - | + | - | |
| ANT1 | Solute carrier family 25, member 4 (adenine nucleotide translocator) | 461475 | - | + | - | |
| Clathrin | Clathrin, heavy chain (Hc) | 9506497 | - | + | - | |
| INSR | Insulin receptor | 8393621 | - | + | - | |
| RPT4 | Proteasome 26S ATPase subunit 4 (regulatory particle triple A ATPase 4) | 25742677 | - | + | - | |
| FBL | Fibrillarin | 71043704 | - | + | - | |
| CDC40 | Cell division cycle 40 homolog | 157817551 | - | + | - | |
| FTHFDH | Aldehyde dehydrogenase 1 family, member L1 (10-formyltetrahydrofolate dehydrogenase) | 1346044 | - | + | - | |
| RPL6 | Ribosomal protein L6 | 38511552 | - | + | - | |
| RPL18 | Ribosomal protein L18 | 89573867 | - | + | - | |
| Likely contaminants (Rattus norvegicus) | TUBA1B | Tubulin, alpha-1b | 34740335 | + | + | + |
| GAPDH | Similar to glyceraldehyde-3-phosphate dehydrogenase | 109484558 | + | + | + | |
| RPS9 | Ribosomal protein S9 | 110347598 | - | + | - | |
| VIM | Vimentin | 14389299 | + | + | + | |
| MYH9 | Myosin, heavy chain 9, nonmuscle | 6981236 | + | + | + | |
| NES | Nestin | 6981262 | + | + | + | |
| PHB | Prohibitin | 6679299 | + | + | + | |
| HSP27 | Heat shock protein β1 (heat shock 27-kDa protein) | 1170367 | + | + | + | |
| G-ACT | Similar to actin, cytoplasmic 2 (gamma-actin) | 109492380 | + | + | + | |
| ACTB | Beta actin | 4501885 | + | + | + | |
| LMNA | Lamin A | 453180 | + | - | + | |
| PLEC1 | Plectin 1 | 46800552 | + | - | + | |
| ACTN4 | Alpha actinin 4 | 77539778 | + | - | + | |
| HIS1H4B | Histone cluster 1, H4b | 12083635 | + | - | + | |
| H2B | Histone H2B | 223096 | + | - | + | |
| HSPA8 | Similar to heat shock protein 8 (heat shock cognate 71-kDa protein) | 109492762 | + | - | + | |
| FLNA | Filamin, alpha (actin binding protein 280) | 197386807 | - | + | + | |
| ACTN1 | Actinin, alpha 1 | 13591902 | - | - | + | |
The tagged protein was nsP4.
G3BP1 and 2 interact with both nsP3 and nsP4 and together exert negative effects on SINV polyprotein expression.
Our previous studies (6), as well as those of others (11, 17), identified G3BP1 and G3BP2 as interacting with SINV nsP3-containing complexes. In addition, G3BP was identified as a factor present in SINV nsP2-containing complexes (1). Although we (6) and recently others (17) demonstrated colocalization of G3BP1 with nsP3, the consequences of the interaction remained unclear. Our first attempts to investigate the role of G3BP in SINV replication by reducing levels of G3BP1 using siRNA-mediated silencing failed to significantly alter virus production in infected cells (data not shown). This initially suggested to us, and as has been proposed by others (17), that G3BP1 might not play a direct role in viral RNA replication. However, our present studies identified G3BP1 and G3BP2 as factors present in nsP4-containing complexes, suggesting a possible direct role in viral RNA replication.
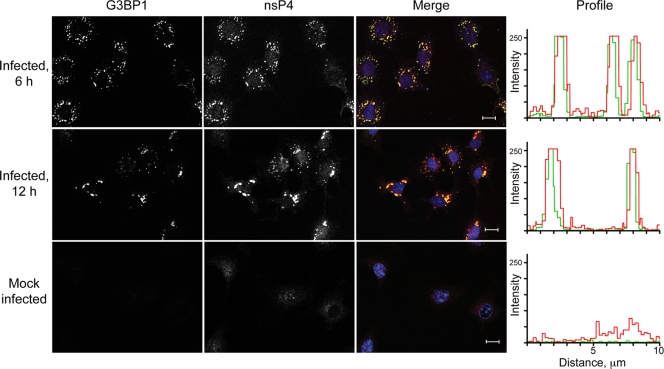
To verify the presence of G3BP1 in nsP4-containing complexes, we analyzed infected Rat2 cells by confocal microscopy. As illustrated in Fig. 3, G3BP1 colocalized with nsP4 at both early (6 h) and late (12 h) times after infection. As we have previously demonstrated in studies examining the colocalization of G3BP1 and SINV nsP3 (6), G3BP1 was undetectable in uninfected Rat2 cells. Upon infection, G3BP1 became detectable in cytoplasmic foci that colocalized with SINV nsP4. Mock-infected cells demonstrated a faint nonspecific signal (both nuclear and cytoplasmic) with the nsP4 antiserum. Overall, however, the nsP4 signal was of increased intensity in the infected cells and overlapped with the G3BP1 signal, as seen in the intensity profiles of the red and green channels (Fig. 3). These findings demonstrate that a substantial amount of SINV nsP4 is colocalized with G3BP1 in infected cells.
FIG. 3.
G3BP1 colocalizes with nsP4 in SINV-infected cells. Rat2 cells were infected with SINV for the indicated times or were mock infected. After fixation the cells were analyzed by indirect immunofluorescence confocal microscopy as described in Materials and Methods. The individual G3BP1 and nsP4 signals are shown (left 2 panels, white), as well as the merged G3BP1 (green) and nsP4 (red) images. Nuclei are shown in blue in the merged images. Bars, 10 μm. A representative fluorescence intensity profile is shown on the right for each sample.
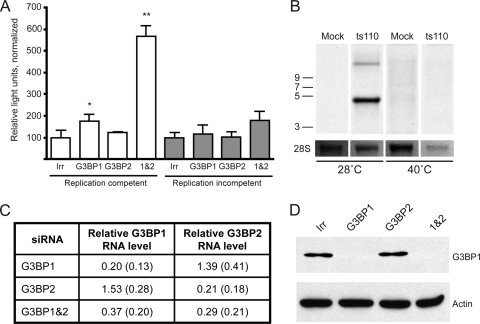
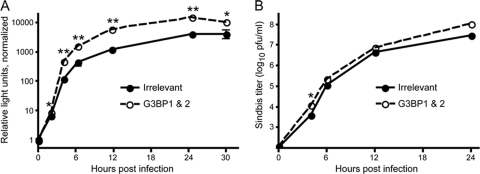
Given the homologous nature of G3BP1 and G3BP2 (reviewed in reference 23), we wondered whether they might carry out redundant functions. To assess this, we used siRNA-mediated silencing to reduce levels of G3BP1, G3BP2, or both and measured the effect on virus replication. For these studies, we utilized 293T cells (a human fibroblast line), which are readily transfectable, and infection with SINV Toto1101/Luc (2), which is a replication-competent virus expressing firefly luciferase as an in-frame fusion within nsP3. Detection of luciferase activity allows for sensitive and facile quantification of early replication events, with luciferase activity reflecting nsP3 levels derived from incoming genome translation, as well as newly replicated viral genomes that are subsequently translated. The results from one representative experiment are shown in Fig. 4 A. Reducing levels of G3BP1 resulted in a statistically significant enhancement of early replication (P = 0.04, two-tailed t test) as measured 5 h after infection (Fig. 4A, indicated as “replication competent”). In four of six experiments (including Fig. 4A), silencing of G3BP1 resulted in a slight (∼2-fold) statistically significant increase in SINV replication (data not shown). In the experimental results depicted in Fig. 4A, silencing of G3BP2 did not result in increased SINV replication, although in two of the six experiments it resulted in a slight statistically significant increase. On the other hand, silencing of both G3BP1 and G3BP2 together resulted in a larger (>5-fold) statistically significant increase in luciferase activity (P = 0.0002, two-tailed t test); the enhancement (ca. 3- to 8-fold) was reproducible across all six experiments (data not shown). In the experiment shown in Fig. 4A, treatment of the 293T cells with siRNAs against both G3BP1 and G3BP2 resulted in decreases in both G3BP1 and G3BP2 RNA. The relative level of G3BP1 RNA, compared to the irrelevant siRNA treated sample, was 0.60, while the relative level of G3BP2 RNA was 0.52, as measured by quantitative real-time RT-PCR. Interestingly, while silencing of G3BP1 or G3BP2 individually was also effective, in each case the RNA levels of the other G3BP increased; after silencing G3BP1 the relative G3BP2 RNA level was 1.70 and after silencing G3BP2 the relative G3BP1 RNA level was 1.41. This suggests that G3BP1 and G3BP2 likely have compensatory functions and may be coordinately controlled. The mean relative G3BP RNA levels (relative to the irrelevant siRNA-treated sample) from three experiments are shown in Fig. 4C. It is likely that the variability in silencing and compensatory changes in G3BP levels account for the variable effects on SINV replication that occur when the G3BPs are individually silenced. That silencing of G3BP1 resulted in reduction of G3BP1 protein was also verified by Western blot analysis and a representative blot with robust protein reduction is shown in Fig. 4D; in some experiments the G3BP1 protein, although reduced, was still detectable in the G3BP1 and G3BP1&2 silenced samples.
FIG. 4.
Silencing of G3BP1 and G3BP2 enhances SINV polyprotein expression. (A) 293T cells were transfected with an irrelevant siRNA (Irr) or siRNA smartpools targeting G3BP1, G3BP2, or both G3BP1 and G3BP2 as indicated. Two days later, the cells were infected for 1 h (MOI of ∼5) with SINV Toto1101/Luc at 37°C (replication competent, open bars) or Toto1101/Luc:ts110 at 40°C (replication incompetent, shaded bars). Fresh medium was added, and the cells were incubated for an additional 5 h at the respective temperatures prior to lysis for determination of luciferase activity. Bars indicate the mean luciferase activity, normalized for cell viability; error bars indicate the standard deviation of triplicate samples. Asterisks indicate values significantly different than the respective control (Irr) for each virus, which was set at 100% (unpaired t test; *, P < 0.05; **, P < 0.01). The results shown are representative of several similar experiments. (B) The temperature sensitive nature of Toto1101/Luc:ts110 in 293T cells was confirmed by metabolic labeling. RNA was harvested from mock-infected or Toto1101/luc:ts110-infected cells (MOI of ∼10) that had been incubated 24 h at permissive (28°C) or nonpermissive (40°C) temperatures in the presence of actinomycin D and [5-3H]uridine. Total RNA (3 μg) was size separated by denaturing agarose electrophoresis, and the newly synthesized RNA was visualized by fluorography. A 23 h (28°C) and 12 days (40°C), exposure of the dried gel was obtained. Below, a reverse image of the ethidium-stained gel obtained prior to fluorography shows rRNA as a loading control. (C) G3BP1 and G3BP2 RNA silencing efficacy was determined in samples treated as in panel A, but harvested prior to infection. The table indicates the G3BP RNA levels relative to levels present in the irrelevant siRNA-treated sample (set at 1.0). GAPDH RNA levels were used for normalization. The data are the mean relative levels and standard deviations from three independent experiments. (D) Western blot analysis confirms reductions in G3BP1 protein levels. Equal volumes of lysates from cells treated as in C with the indicated siRNAs were loaded onto 8% polyacrylamide gels and subjected to Western blot analysis using anti-G3BP1 (top) or anti-β-actin (bottom) antibodies.
To investigate whether the enhanced luciferase activity seen upon silencing of the G3BPs was due to increased translation or increased RNA replication, we tested the effect of silencing on translation of incoming genome, using Toto1101/Luc:ts110 (2). This virus carries a mutation in nsP4, rendering it temperature sensitive for RNA production (34), and thus luciferase levels reflect translation of the initial incoming viral genome. Metabolic labeling of cells infected with Toto1101/Luc:ts110 (Fig. 4B) confirmed the temperature-sensitive nature of this virus; no viral RNA was detected when cells were incubated at the nonpermissive temperature (40°C). Infection with this virus is indicated as “replication incompetent” in Fig. 4A, which represents one of four similar experiments. Although not reaching statistical significance in this experiment, silencing of G3BP1 or G3BP1 and G3BP2 together resulted in a trend toward increased luciferase activity in three of the four experiments, each reaching statistical significance once. Silencing of G3BP2 alone failed to result in any statistically significant luciferase difference in any of the four experiments. The trend in G3BP silenced cells toward increased luciferase expression in the absence of RNA replication, together with the statistically significant increase seen with replication competent virus (which would amplify the number of RNA templates for translation), suggested that the G3BPs might play a role in limiting viral polyprotein expression.
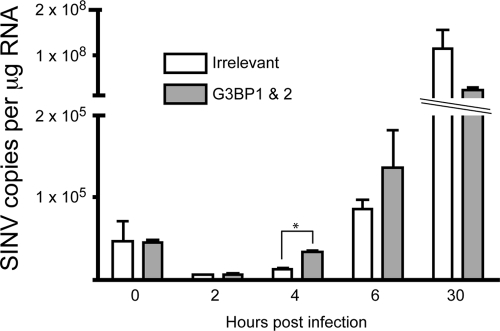
We wondered whether, in addition to effects on polyprotein expression, the G3BPs could be influencing RNA levels, either directly or through enhanced polyprotein expression. To test this, SINV RNA in siRNA-treated cells was quantified by real-time RT-PCR at various times after infection with Toto1101/Luc virus (Fig. 5). In the same experiment, both luciferase activity and RNA levels were quantified. Luciferase activity in cells treated with siRNAs targeting G3BP1 and G3BP2 was increased 3.4-, 5.8-, and 3.9-fold at 4, 6, and 30 h, respectively. Although there was a trend toward increased SINV RNA levels at 4 and 6 h postinfection, the effect was less than that seen on polyprotein (luciferase) expression with only a 2.5- and 1.5-fold increase at 4 and 6 h, respectively. Moreover, the increased luciferase activity detected after silencing of both G3BP1 and G3BP2 could not be fully explained by the altered SINV RNA levels. In particular, after 30 h of infection, the cells treated with the G3BP targeting siRNAs contained lower levels of SINV RNA, despite having an almost 4-fold increase in luciferase activity. Based on these findings, taken all together, we conclude that in combination G3BP1 and G3BP2 exert a controlling influence on early SINV replication events, affecting polyprotein expression, which, in these cells has minimal effects on SINV RNA levels.
FIG. 5.
Silencing of the G3BPs has minimal effects on SINV RNA levels. 293T cells were treated with irrelevant siRNA (□) or siRNAs targeting both G3BP1 and G3BP2 (░⃞) using a final siRNA concentration of 80 nM. After 2 days, the cells were infected with Toto1101/Luc (MOI of ∼5). At the indicated times after infection, the cells were washed, and RNA was harvested and utilized for quantification of SINV RNA levels by quantitative real-time RT-PCR. Known copy numbers of in vitro-transcribed SINV RNA were used to generate a standard curve. For each condition, RNA from a single well was used to prepare duplicate cDNA samples, which were assayed in duplicate; bars represent the mean values obtained from the cDNAs ± the standard errors of the means. The asterisk indicates values significantly different than the irrelevant silenced sample at a given time point (unpaired t test, P < 0.05). Similar results were obtained in another independent experiment, although there was no statistically significant difference at the 4-h time point.
To determine whether the enhanced polyprotein expression seen upon silencing of the G3BPs would be maintained during the course of infection, 293T cells were treated with an irrelevant siRNA or siRNAs targeting G3BP1 and G3BP2 and 2 days later were infected with SINV (Toto1101/Luc) and luciferase activity was measured at various times after infection. As can be seen in Fig. 6 A, a statistically significant enhanced replication, first detected at 2 h after infection, was maintained for the remainder of the infection (P < 0.05, two-tailed t test at each time point 2 to 30 h). The enhancement seen from 4 to 24 h ranged from 3.5- to 5-fold. In a similar experiment, we determined whether increased SINV replication as a result of silencing the G3BPs would result in a change in virus production. 293T cells (irrelevant siRNA- or G3BP1 and G3BP2 siRNA-treated) were infected with SINV (Toto1101), and virion production was determined. As shown in Fig. 6B, silencing of the G3BPs resulted in a trend toward enhancement of virus production. However, aside from the 2-h time point, the differences in titers were not statistically significant. Moreover, the magnitude of the effect on virion production was less dramatic than the effect on polyprotein expression, with only an ∼2-fold increase seen at times between 4 and 24 hpi. This suggests that in these cells, virion production is not completely limited by polyprotein expression levels, and other steps in the virus life cycle may contribute to the overall number of infectious particles generated.
FIG. 6.
Silencing of the G3BPs has a sustained effect on SINV polyprotein expression and a small enhancing effect on SINV virion production. 293T cells were treated with an irrelevant siRNA (•) or siRNAs targeting G3BP1 and G3BP2 (○) and were infected 2 days later with SINV. (A) Cells seeded in 24-well plates were infected with Toto1101/Luc (MOI of ∼5) and were harvested at the indicated times. The data are the mean luciferase activities relative to the activity of the 0-h irrelevant silenced sample; error bars indicate the standard deviation of triplicate samples. Similar results were obtained in one other independent experiment. (B) Cells seeded in 12-well plates were infected with Toto1101 (MOI of ∼0.01), and the amount of virus released into the medium of duplicate samples was determined by plaque assay titration on BHK-21 cells. Separate wells were utilized for each time point. The data are mean log titers ± the standard errors of the means; error bars are obscured by the symbols. Similar results were obtained in another independent experiment. For both panels A and B, asterisks indicate values significantly different than the irrelevant silenced sample at a given time point (unpaired t test; *, P < 0.05; **, P < 0.01).
DISCUSSION
In this study we have isolated and identified 29 host factors that associate with nsP4, the viral RNA-dependent RNA polymerase, at early (6 h) and late (12 h) times of infection with SINV. The use of a SINV expressing 3×Flag epitope-tagged nsP4 enabled the identification of host factors most likely to directly influence RNA replication and transcription. Some of the 29 proteins we found associated with nsP4 may be important for the SINV life cycle and in the present study we identified G3BP1 and G3BP2 as playing a role in the SINV replication process. Control isolations using cells infected with a virus expressing GFP containing a 3×Flag epitope tag (TE/5′2J/GFP-3×Flag) were used to distinguish nonspecific association to the Flag tag, anti-Flag antibodies, or magnetic beads. However, during incubation of the cell lysate with magnetic beads, isolated nsP4 complexes can bind to additional nonspecific proteins. Although we utilized a relatively rapid (1-h) affinity isolation incubation time to minimize nonspecific interactions (7), it is unlikely that all of the identified nsP4-interacting factors are specifically associated with nsP4. In addition to proteins isolated in control samples (marked as likely contaminants in Table 2 and Table S1 in the supplemental material), other potential nonspecifically interacting proteins can be identified based on previous experience and the literature. Highly abundant cytoskeletal (tubulin) or ribosomal proteins have been noted as possible contaminants in Flag affinity isolations (4), and we previously identified other proteins (glyceraldeyde-3-phosphate dehydrogenase, prohibitin, and fibrillarin [data not shown]) in control affinity isolations from unrelated experiments using the Flag epitope tag. Thus, most of the proteins identified in the current study should be considered as potential factors that could be important for SINV replication and its regulation. In the present study we show that G3BP1 and G3BP2 play roles in SINV replication; additional studies are required to confirm the other interactions and elucidate which factors are important for SINV replication.
Five host factors—UNC84, PRMT5, GNB2L1, SFRS10, and ANXA1—were found to be associated with nsP4 only at 6 hpi, not being observed in complexes isolated at 12 h. Aside from GNB2L1, these proteins were not previously detected in affinity isolations of nsP2 (1) or nsP3 (6, 11, 17). Since both nsP2 and nsP3 are present in early replication complexes, the failure to identify UNC84, PRMT5, SFRS10, and ANXA1 in previous studies is rather surprising. However, given the excess of P123 synthesis compared to nsP4 in SINV-infected cells, it is possible that, in previous studies, the host factors interacting with the replicase-associated nsP2 and nsP3 were obscured by host factors associated with the relatively larger amounts of nsP2 and nsP3 residing outside of replication complexes. This in fact was a major consideration that led us to utilize a tagged nsP4 for the isolation of host factors possibly involved in viral RNA replication. The finding of five factors in association with nsP4 only at early times after infection suggests their involvement in early, but not late RNA replication processes. For example, these factors might aid the formation of viral replication complexes, facilitate minus-strand synthesis or mediate the switch from minus to plus strand synthesis. Interestingly, annexins are involved in membrane reorganization (reviewed in reference 15) and UNC84 (also known as SUN2) is a nuclear membrane protein proposed to be involved in positioning of the nucleus of muscle cells through protein-protein interactions (24). One can speculate that these proteins may be involved in mediating the proper membrane topology and localization to facilitate the formation of the SINV replication complexes, which are found in association with host membranes (13, 17). PRMT5 (also found only at 6 h) is known to associate with MEP50 (found at both 6 and 12 h) as components of the methylosome, which modifies arginine residues in Sm splicosome proteins to dimethylarginine, facilitating their recruitment to the survival of motor neurons complex and the formation of small nuclear ribonucleoprotein particles (9). Although one can conjecture a role for arginine methylation of host or viral proteins in the formation of SINV replication complexes, this protein modification has not been studied in the context of SINV infection. Another protein identified only at 6 h, SFRS10, also known as Tra2, is involved in splicing regulation (33) and might participate in replicase formation or the regulation of RNA replication through its RNA binding activity. However, in our experience splicing factors are common contaminants in Flag affinity isolations (data not shown). In addition, MEP50 and PRMT5 have been noted as common contaminants in Flag affinity isolations (4). Therefore, in future studies it will be important to confirm the specificity of these interactions.
Ten proteins were found associated with nsP4 only at late times (12 h) after infection. Although the number of proteins identified precludes discussing their individual potential roles in SINV replication, we speculate that some of these proteins may play roles in later events of the replication process, possibly including regulation of assembly processes, such as diverting genomic RNA toward assembly rather than translation. Since SINV infection of most vertebrate cells results in severe alterations in morphology, cytopathic effect, and death by apoptosis (26), a role for these factors in regulating the morphological changes and cell death caused by SINV could also be postulated. Of the factors isolated only at 12 h, the two ribosomal proteins were also identified as associating within nsP3-containing complexes, while the other eight were not previously identified in association with nsP2 (1) or nsP3 (6, 11, 17). Determination of the functions of these proteins in SINV replication will require additional study.
The factors found at both early and late times are candidates for host factors important for minus and plus strand RNA synthesis, as well as subgenomic RNA transcription. Interestingly, 14 host factors were found in association with nsP4 at both early and late times after infection. These included G3BP1 and G3BP2, which were previously identified in nsP3-containing complexes (6, 11, 17), as well as in association with nsP2 (1). We also identified multiple 14-3-3 proteins in association with nsP4 at both early and late times. 14-3-3 proteins have also previously been found in association with nsP3 (6, 11, 17). In our previous study (6) several 14-3-3 isoforms were found associated with nsP3 after 6 h of infection and longer but not at times earlier than 6 h (6). Six proteins—MEP50, HSP90, TRIM30, 14-3-3θ, TYMS, and SERPINH1—found associated with nsP4 at both 6 and 12 h after infection were not previously identified in complexes isolated by immunoprecipitation of nsP2 or nsP3.
In the present study we chose to follow up on a possible functional role for G3BP1 and G3BP2 in SINV replication, since these proteins were previously detected in association with other SINV nsPs and demonstrated a robust association with nsP4 (see Coomassie blue-stained bands, Fig. 2). Our data suggest that G3BP1 and G3BP2 function to limit SINV polyprotein expression. Limiting SINV polyprotein expression could be beneficial to virus replication or could represent a host cell response to limit virus replication. Although silencing of the G3BPs had minimal effects on virion production in cell culture (Fig. 6B), it is possible that the recruitment of G3BP1 and G3BP2 is critical for regulating SINV replication in vivo, for example, in specific cell types, and may play a role in the pathogenesis of this virus. G3BP1 assembles stress granules (40), sites of mRNA triage for continued translation or degradation (21, 22). As has been suggested by others (17), SINV sequestration of G3BP1 may serve to interfere with the host cell stress response. Interestingly, poliovirus, another positive-strand RNA virus, cleaves G3BP1 with a resultant interference in stress granule formation (41). G3BP1 has also been found in association with replication complexes of the RNA virus hepatitis C; however, in this case G3BP1 knock down was associated with decreased RNA replication (43). G3BP2 has been implicated in control of NF-κB signaling by retaining IκB/NF-κB complexes in the cytoplasm (30). Thus, the recruitment of G3BP2 into nsP4-containing complexes might serve to limit G3BP2's effects on NF-κB signaling.
We demonstrate here, using a luciferase-expressing SINV, that silencing of G3BP1 and G3BP2 results in increased polyprotein expression from the viral genomic RNA (Fig. 4A). This effect could be direct or, alternatively, could be due to effects on viral RNA replication. Although we detected a trend toward increased polyprotein expression using a temperature-sensitive virus defective in RNA replication (Fig. 4A), we were unable to reproducibly detect significantly increased luciferase activity. We found no evidence of leaky replication of the temperature-sensitive mutant (Fig. 4B) to account for the occasional statistically significant increase in luciferase activity. Although the lack of a reproducible significant effect on replication incompetent SINV RNA translation suggested that the G3BPs might directly affect RNA replication, we also did not detect significantly increased RNA levels upon G3BP silencing. It is possible that reduction of G3BP levels results in a subtle effect on polyprotein expression, which, upon replication of the viral RNA gets amplified to statistically significant levels as the new RNA templates enter the translational pool. Interestingly, upon silencing of the G3BPs, a 3- to 5-fold enhancement of polyprotein expression from replication competent virus was maintained over time after infection (Fig. 6A) but did not result in substantial increases in viral RNA (Fig. 5) or virion production (Fig. 6B). We interpret the small increases in viral RNA levels and virion production seen upon G3BP silencing to be a result of the enhanced genome polyprotein expression. However, a small direct effect on RNA replication or virion production cannot be excluded, since experiments to measure RNA replication and virion production independently from polyprotein expression are not possible.
Given the RNA binding activity of the G3BPs and their role in the formation of stress granules, sites where mRNA translation and degradation are regulated (21), our results suggest that G3BP1 and G3BP2 might normally function to limit SINV genome translation by recruitment of the viral RNA into the stress granule pathway. It is somewhat surprising that reducing the level of these proteins did not have a more dramatic effect. One possible explanation for the modest effect we detected (3- to 8-fold) is that the recruitment of G3BPs by nsPs that are not directly involved in RNA replication results in a functional depletion of G3BPs to which our siRNA-mediated silencing adds little. The fact that we can demonstrate an effect highlights a possible role for the stress granule pathway in the host cell response to SINV infection. Alternatively, it is possible that a subtle reduction in polyprotein expression is beneficial for the virus, in which case SINV may have usurped this pathway to its advantage. An alternative possibility is that the G3BPs recruit SINV RNA out of the translating pool of RNA and into nsP4-containing replication complexes. Interestingly, since G3BP1 was identified as a helicase capable of unwinding both DNA and RNA substrates (5), a role in clearing the viral RNA of proteins in preparation for the switch from translation to replication is possible. Although this would predict a delay or reduction in the formation of replication complexes in G3BP silenced cells, any newly replicated RNA that is produced would be predicted to produce more nsPs due to enhanced time in the translating pool. Understanding whether the G3BPs play a role in replication template recruitment and/or more direct inhibitory roles on translation or polyprotein stability requires further investigation.
In the present study we successfully generated a viable SINV expressing an epitope-tagged version of the viral RNA-dependent RNA polymerase (nsP4). Affinity isolation of the tagged nsP4 led to the identification of 29 proteins that were associated within the nsP4-containing complexes. While testing for which of these host proteins play active roles in SINV RNA replication and understanding the details of the host factor-nsP4 interaction require further investigation, knowledge in this area could lead to new approaches to disrupt these critical interactions and limit the devastating diseases caused by alphaviruses.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health grants AI063233 (M.R.M.), DP1DA026192 (I.M.C.), RR00862 (B.T.C.), and RR02220 (B.T.C.); Princeton University startup funding (I.M.C.); the Irma T. Hirschl/Monique Weill-Caulier Trust; the Greenberg Medical Research Institute; and the Starr Foundation.
We thank John-William N. Carroll and Cristian Cruz for technical assistance in various stages of this work and Zhigang Yi for help with the quantitative assay for SINV RNA. We also thank the reviewers for their insightful comments.
Footnotes
Published ahead of print on 14 April 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Atasheva, S., R. Gorchakov, R. English, I. Frolov, and E. Frolova. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 81:5046-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bick, M. J., J.-W. N. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. MacDonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blethrow, J. D., C. Tang, C. Deng, and A. N. Krutchinsky. 2007. Modular mass spectrometric tool for analysis of composition and phosphorylation of protein complexes. PLoS One 2:e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G. I., and A. C. Gingras. 2007. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Costa, M., A. Ochem, A. Staub, and A. Falaschi. 1999. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 27:817-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristea, I. M., J.-W. N. Carroll, M. P. Rout, C. M. Rice, B. T. Chait, and M. R. MacDonald. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269-30278. [DOI] [PubMed] [Google Scholar]
- 7.Cristea, I. M., R. Williams, B. T. Chait, and M. P. Rout. 2005. Fluorescent proteins as proteomic probes. Mol. Cell Proteomics 4:1933-1941. [DOI] [PubMed] [Google Scholar]
- 8.de Groot, R. J., T. Rumenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. U. S. A. 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277:8243-8247. [DOI] [PubMed] [Google Scholar]
- 10.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova, E., R. Gorchakov, N. Garmashova, S. Atasheva, L. A. Vergara, and I. Frolov. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 80:4122-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerke, V., C. E. Creutz, and S. E. Moss. 2005. annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell. Biol. 6:449-461. [DOI] [PubMed] [Google Scholar]
- 16.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorchakov, R., N. Garmashova, E. Frolova, and I. Frolov. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 82:10088-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin, D. E. 2007. Alphaviruses, p. 1023-1067. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 19.Hahn, C. S., Y. S. Hahn, T. J. Braciale, and C. M. Rice. 1992. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc. Natl. Acad. Sci. U. S. A. 89:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kääriäinen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acids Res. Mol. Biol. 71:187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fritzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy, D., J. French, E. Guitard, K. Ru, B. Tocque, and J. Mattick. 2001. Characterization of G3BPs: tissue specific expression, chromosomal localization and rasGAP(120) binding studies. J. Cell Biochem. 84:173-187. [DOI] [PubMed] [Google Scholar]
- 24.Lei, K., X. Zhang, X. Ding, X. Guo, M. Chen, B. Zhu, T. Xu, Y. Zhuang, R. Xu, and M. Han. 2009. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed]
- 25.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 27.Luo, Y., T. Li, F. Yu, T. Kramer, and I. M. Cristea. 2010. Resolving the composition of protein complexes using a MALDI LTQ orbitrap. J. Am. Soc. Mass Spectrom. 21:34-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald, M. R., M. W. Burney, S. B. Resnick, and H. I. Virgin. 1999. Spliced mRNA encoding the murine cytomegalovirus chemokine homolog predicts a beta chemokine of novel structure. J. Virol. 73:3682-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prigent, M., I. Barlat, H. Langen, and C. Dargemont. 2000. IκBα and IκBα/NF-κB complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 275:36441-36449. [DOI] [PubMed] [Google Scholar]
- 31.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubach, J. K., B. R. Wasik, J. C. Rupp, R. J. Kuhn, R. W. Hardy, and J. L. Smith. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla, S., and S. A. Fisher. 2008. Tra2beta as a novel mediator of vascular smooth muscle diversification. Circ Res. 103:485-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 1976. Mutants of Sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology 74:154-168. [DOI] [PubMed] [Google Scholar]
- 35.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. U. S. A. 80:5271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strupat, K., V. Kovtoun, H. Bui, R. Viner, G. Stafford, and S. Horning. 2009. MALDI produced ions inspected with a linear ion trap-Orbitrap hybrid mass analyzer. J. Am. Soc. Mass Spectrom. 20:1451-1463. [Google Scholar]
- 38.Thal, M. A., B. R. Wasik, J. Posto, and R. W. Hardy. 2007. Template requirements for recognition and copying by Sindbis virus RNA-dependent RNA polymerase. Virology 358:221-232. [DOI] [PubMed] [Google Scholar]
- 39.Tomar, S., R. W. Hardy, J. L. Smith, and R. J. Kuhn. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 80:9962-9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tourriere, H., K. Chebli, L. Zekri, B. Courselaud, J. M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.White, J. P., A. M. Cardenas, W. E. Marissen, and R. E. Lloyd. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295-305. [DOI] [PubMed] [Google Scholar]
- 42.Wielgosz, M. M., R. Raju, and H. V. Huang. 2001. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J. Virol. 75:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi, Z., C. Fang, T. Pan, J. Wang, P. Yang, and Z. Yuan. 2006. Subproteomic study of hepatitis C virus replicon reveals Ras-GTPase-activating protein binding protein 1 as potential HCV RC component. Biochem. Biophys. Res. Commun. 350:174-178. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, L., R. Hernan, and B. Brizzard. 2001. Multiple tandem epitope tagging for enhanced detection of protein expressed in mammalian cells. Mol. Biotechnol. 19:313-321. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.