Abstract
To facilitate the release of infectious progeny virions, human immunodeficiency virus type 1 (HIV-1) exploits the Endosomal Sorting Complex Required for Transport (ESCRT) pathway by engaging Tsg101 and ALIX through late assembly (L) domains in the C-terminal p6 domain of Gag. However, the L domains in p6 are known to be dispensable for efficient particle production by certain HIV-1 Gag constructs that have the nucleocapsid (NC) domain replaced by a foreign dimerization domain to substitute for the assembly function of NC. We now show that one such L domain-independent HIV-1 Gag construct (termed ZWT) that has NC-p1-p6 replaced by a leucine zipper domain is resistant to dominant-negative inhibitors of the ESCRT pathway that block HIV-1 particle production. However, ZWT became dependent on the presence of an L domain when NC-p1-p6 was restored to its C terminus. Furthermore, when the NC domain was replaced by a leucine zipper, the p1-p6 region, but not p6 alone, conferred sensitivity to inhibition of the ESCRT pathway. In an authentic HIV-1 Gag context, the effect of an inhibitor of the ESCRT pathway on particle production could be alleviated by deleting a portion of the NC domain together with p1. Together, these results indicate that the ESCRT pathway dependence of HIV-1 budding is determined, at least in part, by the NC-p1 region of Gag.
Human immunodeficiency virus type 1 (HIV-1) and other retroviruses hijack the cellular Endosomal Sorting Complex Required for Transport (ESCRT) pathway to promote the detachment of virions from the cell surface and from each other (3, 21, 42, 44, 47). The ESCRT pathway was initially identified based on its requirement for the sorting of ubiquitinated cargo into multivesicular bodies (MVB) (50, 51). During MVB biogenesis, the ESCRT pathway drives the membrane deformation and fission events required for the inward vesiculation of the limiting membrane of this organelle (26, 29, 50, 51). More recently, it emerged that the ESCRT pathway is also essential for the normal abscission of daughter cells during the final stage of cell division (10, 43). Most of the components of the ESCRT pathway are involved in the formation of four heteromeric protein complexes termed ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III. Additional components include ALIX, which interacts both with ESCRT-I and ESCRT-III, and the AAA ATPase Vps4, which mediates the disassembly of ESCRT-III (29, 42).
The deformation and scission of endocytic membranes is thought to be mediated by ESCRT-III, which, together with Vps4, constitutes the most conserved element of the pathway (23, 26, 42). Indeed, it was recently shown that purified yeast ESCRT-III induces membrane deformation (52), and in another study three subunits of yeast ESCRT-III were sufficient to promote the formation of intralumenal vesicles in an in vitro assay (61). In mammals, ESCRT-III is formed by the charged MVB proteins (CHMPs), which are structurally related and tightly regulated through autoinhibition (2, 33, 46, 53, 62). The removal of an inhibitory C-terminal domain induces polymerization and association with endosomal membranes and converts CHMPs into potent inhibitors of retroviral budding (34, 46, 53, 60, 62). Alternatively, CHMPs can be converted into strong inhibitors of the ESCRT pathway and of HIV-1 budding through the addition of a bulky tag such as green fluorescent protein (GFP) or red fluorescent protein (RFP) (27, 36, 39, 54). Retroviral budding in general is also strongly inhibited by catalytically inactive Vps4 (22, 41, 55), or upon Vsp4B depletion (31), confirming the crucial role of ESCRT-III.
Retroviruses engage the ESCRT pathway through the activity of so-called late assembly (L) domains in Gag. In the case of HIV-1, the primary L domain maps to a conserved PTAP motif in the C-terminal p6 domain of Gag (24, 28) and interacts with the ESCRT-I component Tsg101 (15, 22, 40, 58). HIV-1 p6 also harbors an auxiliary L domain of the LYPxnL type, which interacts with the V domain of ALIX (20, 35, 39, 54, 59, 63). Interestingly, Tsg101 binding site mutants of HIV-1 can be fully rescued through the overexpression of ALIX, and this rescue depends on the ALIX binding site in p6 (20, 56). In contrast, the overexpression of a specific splice variant of the ubiquitin ligase Nedd4-2 has been shown to rescue the release and infectivity of HIV-1 mutants lacking all known L domains in p6 (12, 57). Nedd4 family ubiquitin ligases had previously been implicated in the function of PPxY-type L domains, which also depend on an intact ESCRT pathway for function (4, 32, 38). However, HIV-1 Gag lacks PPxY motifs, and the WW domains of Nedd4-2, which mediate its interaction with PPxY motifs, are dispensable for the rescue of HIV-1 L domain mutants (57).
ALIX also interacts with the nucleocapsid (NC) region of HIV-1 Gag (18, 49), which is located upstream of p6 and the p1 spacer peptide. ALIX binds HIV-1 NC via its Bro1 domain, and the capacity to interact with NC and to stimulate the release of a minimal HIV-1 Gag construct is shared among widely divergent Bro1 domain proteins (48). Based on these findings and the observation that certain mutations in NC cause a phenotype that resembles that of L domain mutants, it has been proposed that NC cooperates with p6 to recruit the machinery required for normal HIV-1 budding (18, 49).
NC also plays a role in Gag polyprotein multimerization, and this function of NC depends on its RNA-binding activity (5-8). It has been proposed that the role of the NC-nucleic acid interaction during assembly is to promote the formation of Gag dimers (37), and HIV-1 assembly in the absence of NC can indeed be efficiently rescued by leucine zipper dimerization domains (65). Surprisingly, in this setting the L domains in p6 also became dispensable, since particle production remained efficient even when the entire NC-p1-p6 region of HIV-1 Gag was replaced by a leucine zipper (1, 65). These findings raised the possibility that the reliance of wild-type (WT) HIV-1 Gag on a functional ESCRT pathway is, at least in part, specified by NC-p1-p6. However, it also remained possible that the chimeric Gag constructs engaged the ESCRT pathway in an alternative manner.
In the present report, we provide evidence supporting the first of those two possibilities. Particle production became independent of ESCRT when the entire NC-p1-p6 region was replaced by a leucine zipper, and reversion to ESCRT dependence was shown to occur as a result of restoration of p1-p6 but not of p6 alone. Furthermore, although the deletion of p1 alone had little effect in an authentic HIV-1 Gag context, the additional removal of a portion of NC improved particle production in the presence of an inhibitor of the ESCRT pathway. Together, these data imply that the NC-p1 region plays an important role in the ESCRT-dependence of HIV-1 particle production.
MATERIALS AND METHODS
Proviral constructs.
All HIV-1 Gag constructs used in this study were based on HXBH10, a vpu-positive version of the infectious HXB2 proviral clone of HIV-1. The protease (PR)-negative variant HXBH10-PR− (which was used to express Pr55gag), HXBH10ΔPTAPP, and the HXBH10-based chimeric Gag constructs ZWT and ZIL-p6 have been previously described (1). ZWT-NCp1 was obtained by fusing the HXBH10 NCp1 coding sequence to the 3′ end of the GCN4 sequence in the ZWT construct. The nucleotide sequence at the junction is 5′ AAG CTT GTG GGT GAG ctc ATG CAG AGA GGC AAT 3′ (with GCN4 sequences in underlined uppercase type and HIV-1 sequences in uppercase type without underlining). The NCp1 coding sequence is immediately followed by a stop codon, which in turn is followed by HIV-1 sequences, starting with nucleotide 5228 of HXBH10. ZWT-NCp1p6 and ZWT-NCp1p6ΔPTAPP were then obtained by replacing SphI-ApaI fragments in PR-negative variants of HXBH10 and HXBH10ΔPTAPP with the corresponding fragment from ZWT-NCp1. ZIL-p1p6 and its ΔPTAPP and Y36s versions were obtained by fusing HIV-1 sequences, starting with nucleotide 2076 of a PR-negative variant of HXBH10, HXBH10ΔPTAPP, or HXBH10-Y36s (49), to the 3′ end of the variant GCN4 sequence in the ZIL-p6 construct via an engineered BsrBI cloning site. The nucleotide sequence at the junction is 5′ AAA CTG ATC GGT GAG cgg CAG GCT AAT TTT TTA 3′ (with GCN4 sequences in underlined uppercase type and HIV-1 sequences in uppercase type without underlining). In addition to p1 and p6, ZIL-p1p6 encodes the last 4 amino acids of NC. ZIL-p6ΔPTAPP is a version of ZIL-p6 with an in-frame deletion of codons 7 through 11 of p6. The Δp1 version of HXBH10 lacks precisely the p1 coding sequence (NC-p1 codons 56 to 71). Versions of HXBH10-Y36s lacking NC-p1 codons 56 to 71 (here called Δp1), 52 to 71, 36 to 71, or 15 to 71 have been described previously (49).
Analysis of viral particle production.
293T cells (1.2 × 106) were seeded into T25 flasks and transfected 24 h later using a calcium phosphate precipitation technique. The cultures were transfected with 1.5 μg of proviral DNA together with expression vectors for CHMP3-RFP (54) or Vps4AE228Q (54) or the appropriate empty vectors (0.5 μg each). The total amount of transfected DNA was brought to 8 μg with carrier DNA (pBluescript). At 24 h posttransfection, the cells were lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and the culture supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm-pore-size filters. Virions or virus-like particles (VLP) released into the medium were then pelleted through 20% sucrose cushions by ultracentrifugation for 2 h at 27,000 rpm and 4°C in a Beckman SW41 rotor. Pelletable material and the cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, as described elsewhere (1), using rabbit anti-HIV CA serum (Advanced Biotechnologies) or 183-H12-5C anti-HIV CA antibody (11). Western blots were quantitated with ImageJ software.
Electron microscopy.
Transfected 293T cells were fixed in 2.5% glutaraldehyde, postfixed with 1% osmium tetroxide, dehydrated through a graded series of ethanol treatments, and embedded in resin. Ultrathin sections were stained with lead citrate and uranyl acetate and examined using a Philips CM 10 transmission electron microscope.
RESULTS
Replacing the NC-p1-p6 region of HIV-1 Gag overcomes inhibition by dominant-negative ESCRT proteins.
Although the NC domain of HIV-1 Gag is crucial for viral particle assembly, particle production could be rescued by replacing NC with a heterologous dimerization domain (1, 65). One such chimeric HIV-1 Gag construct, termed ZWT, efficiently produced VLP even though it has the entire NC-p1-p6 region replaced by the leucine zipper domain of yeast GCN4 and thus lacks evident binding sites for Tsg101 or ALIX (1). This observation raised the possibility that ZWT Gag does not depend on an L domain for its efficient release. Alternatively, it seemed possible that ZWT engages the ESCRT pathway through a cryptic L domain that is masked in the presence of NC-p1-6. For instance, ZWT may make more efficient use of Nedd4-2s, a member of the Nedd4 family of E3 ubiquitin ligases that is uniquely capable of rescuing HIV-1 release in the absence of all known L domains (12, 57).
To determine whether the release of ZWT Gag requires an intact ESCRT pathway, we made use of the fact that this pathway is effectively blocked by activated ESCRT-III components (53, 62). The CHMPs, which together form ESCRT-III, contain an autoinhibitory C-terminal domain. When this domain is removed or disrupted through the addition of a bulky tag such as GFP or red fluorescent protein (RFP), ESCRT-III components such as CHMP3 become inhibitors of the ESCRT pathway and of HIV-1 budding (39, 53, 54, 59, 62). We chose to use CHMP3-RFP because we had previously observed that this fusion protein potently blocks HIV-1 budding (54).
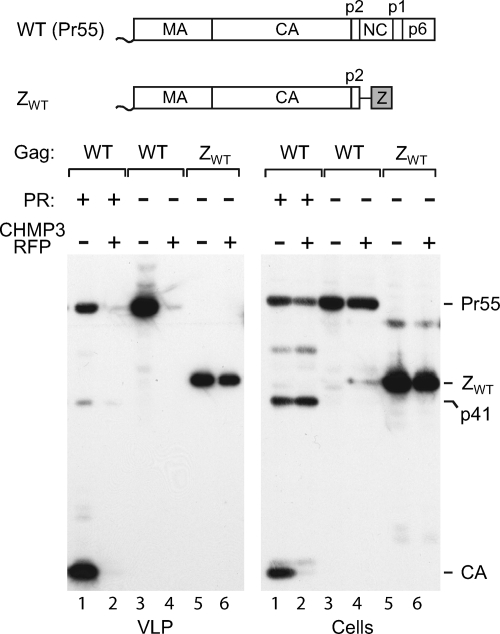
When an expression vector for CHMP3-RFP was cotransfected into 293T cells together with WT HIV-1 proviral DNA, viral particle production was, as expected, almost completely abolished (Fig. 1, lanes 1 and 2). Because the ZWT construct lacks the coding sequence for PR and thus produces only unprocessed Gag, we also examined whether the effect of CHMP3-RFP on the release of WT HIV-1 Gag depended on the presence of an active PR. We found that the release of viral particles by full-length HIV-1 was blocked by CHMP3-RFP even when the proteolyic processing of Gag was prevented by a point mutation that inactivated PR (Fig. 1, lanes 3 and 4).
FIG. 1.
Replacement of NC-p1-p6 by the GCN4 leucine zipper confers resistance to dominant-negative CHMP3. 293T cells were transfected with 1.5 μg of WT HIV-1 proviral DNA, of a PR-defective HIV-1 provirus, or of the ZWT proviral construct, along with 0.5 μg of a vector expressing RFP alone or CHMP3-RFP. VLP pellets and the cell lysates were analyzed by Western blotting with anti-CA serum. The GCN4 zipper (Z) domain is indicated by a gray box.
In the experiment represented by Fig. 1, the ZWT construct released about 60% of the amount of particulate Gag obtained with HXBH10-PR−. Importantly, CHMP3-RFP only slightly reduced particle production by the ZWT construct (Fig. 1, lanes 5 and 6). Quantification indicated that CHMP3-RFP reduced the production of ZWT particles by about 10%, taking the cellular levels of ZWT Gag into account. Similarly, the production of ZWT particles was little affected by catalytically inactive Vps4AE228Q (data not shown), a dominant-negative mutant that blocks the release of viruses that use PTAP, LYPxnL, or PPxY-type L domains (22, 41). Taken together, these results indicated that the release of HIV-1 Gag no longer depended on functional ESCRT machinery when the NC-p1-p6 region was replaced by a heterologous dimerization domain.
Role of NC-p1 in the requirement for L domains.
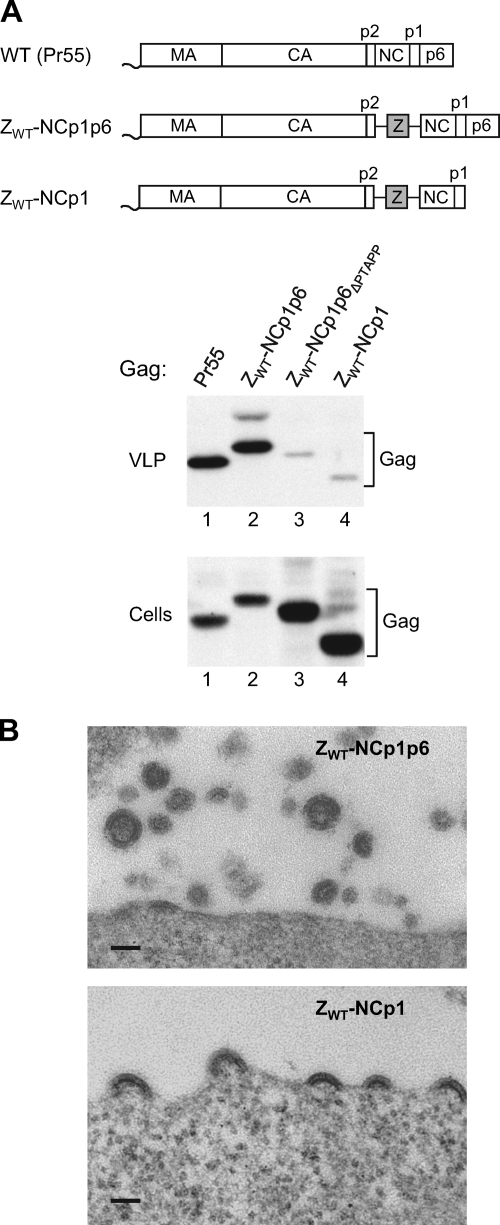
The results described above suggested that the NC-p1-p6 region, apart from mediating the interaction between HIV-1 Gag and ESCRT pathway components, also confers a need to engage the ESCRT machinery for efficient virus release. However, it also remained possible that the presence of the GCN4 leucine zipper domain downstream of CA-p2, rather than the absence of the NC-p1-p6 region, was responsible for the insensitivity of ZWT Gag to inhibitors of the ESCRT machinery. In an effort to distinguish between these possibilities, we asked whether an L domain is required for efficient particle release when NC-p1-p6 is added back to ZWT Gag. To this end, we made a version of ZWT that has the NC-p1-p6 region appended to the C terminus of the GCN4 zipper sequence (Fig. 2A). Although the resulting ZWT-NCp1p6 construct contained an intact pol gene, proteolytic processing of Gag was prevented by a mutation in PR. As shown in Fig. 2A, particle production by the ZWT-NCp1p6 proviral construct was comparable to that observed with authentic HIV-1 encoding an inactive PR (lanes 1 and 2). In contrast, particle production was inefficient when the Tsg101 binding site in the p6 region of ZWT-NCp1p6 was deleted (ΔPTAPP); instead, the chimeric Gag molecule accumulated in the transfected cells (Fig. 2A, lane 3). This phenotype was similar to that observed for a version of ZWT that had NC-p1 rather than NC-p1-p6 fused to its C terminus (Fig. 2A, lane 4). Thin-section electron microscopy confirmed that 293T cells transfected with the ZWT-NCp1p6 construct released particles, although these often appeared to be incomplete and smaller than particles formed by authentic HIV-1 Gag (Fig. 2B). On the other hand, an examination of 293T cells transfected with the ZWT-NCp1 construct by electron microscopy revealed an accumulation of budding structures on the cell surface (Fig. 2B). Since the parental ZWT construct efficiently releases spherical particles (1, 14), these results suggested that NC-p1 caused a budding arrest in the ZWT context and that this arrest could be mitigated through the engagement of the ESCRT machinery via p6.
FIG. 2.
The chimeric ZWT Gag construct exhibits L domain dependence when NC-p1 is fused to its C terminus. (A) 293T cells were transfected with a PR-defective HIV-1 provirus expressing WT Pr55gag or with the indicated Gag constructs. VLP pellets and the cell lysates were analyzed by Western blotting with anti-CA serum. (B) Thin sections of 293T cells transiently expressing the ZWT-NCp1p6 or ZWT-NCp1 proviral construct were examined by transmission electron microscopy. Scale bars, 100 nm.
The p1 spacer peptide can block particle production when the ESCRT machinery is inhibited.
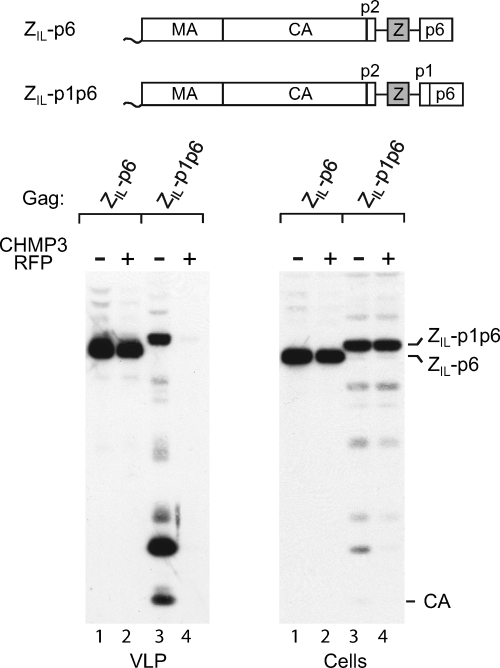
We previously reported that fusing p6 to the C terminus of ZWT Gag had little effect on particle production, consistent with the notion that ZWT Gag does not require an L domain (1). However, particle production by a variant of ZWT with a mutant zipper thought to induce trimerization (termed ZIL) appeared to benefit from the presence of a p6 domain (1). We therefore asked whether the ZIL-p6 Gag construct, illustrated in Fig. 3, is sensitive to inhibition by a dominant-negative ESCRT-III component. As shown in Fig. 3 (lanes 1 and 2), CHMP3-RFP had little if any effect on the release of ZIL-p6 Gag under conditions in which the release of WT HIV-1 Gag was nearly completely prevented (see Fig. 1). Indeed, quantification indicated that, when normalized for a modest inhibitory effect of CHMP3-RFP on the expression levels of ZIL-p6 Gag, the efficiency of particle production was identical in the presence and absence of CHMP3-RFP. Since the ZIL-p6 Gag construct produces particles with close to WT efficiency (1), its insensitivity to CHMP3-RFP strongly argues against the possibility that ESCRT independence is a byproduct of inefficient release.
FIG. 3.
The addition of p1-p6 but not of p6 alone to a chimeric Gag construct confers ESCRT pathway dependence. 293T cells were transfected with 1.5 μg of the indicated Gag constructs together with 0.5 μg of a vector expressing RFP alone or CHMP3-RFP. VLP pellets and the cell lysates were analyzed by Western blotting with anti-CA serum.
To determine whether p1 plays a role in conferring sensitivity to CHMP3-RFP, we also examined its effect on ZIL-p1p6, which differs from ZIL-p6 only by the inclusion of the p1 spacer peptide between the leucine zipper sequence and p6 (Fig. 3). Of note, the presence of p1 was predicted to restore frameshifting into the pol frame and thus proteolytic processing of the chimeric Gag precursor. Remarkably, CHMP3-RFP completely abolished particle production by ZIL-p1p6 (Fig. 3, lanes 3 and 4). Since ZIL-p6 lacks the frameshift signal within the p1 coding sequence and thus produces only unprocessed Gag, whereas ZIL-p1p6 Gag was partially processed as expected, we also examined the effect of CHMP3-RFP on a PR-negative version of ZIL-p1p6. When proteolytic processing was prevented, CHMP3-RFP inhibited the release of ZIL-p1p6 Gag about 5-fold in the experiment represented by Fig. 4 A (lanes 1 and 2) and more than 10-fold in a repeat experiment (data not shown). We infer that the presence of p6 did not by itself confer a requirement for an intact ESCRT pathway for efficient particle release. However, the ESCRT machinery became crucial when p1 was also present.
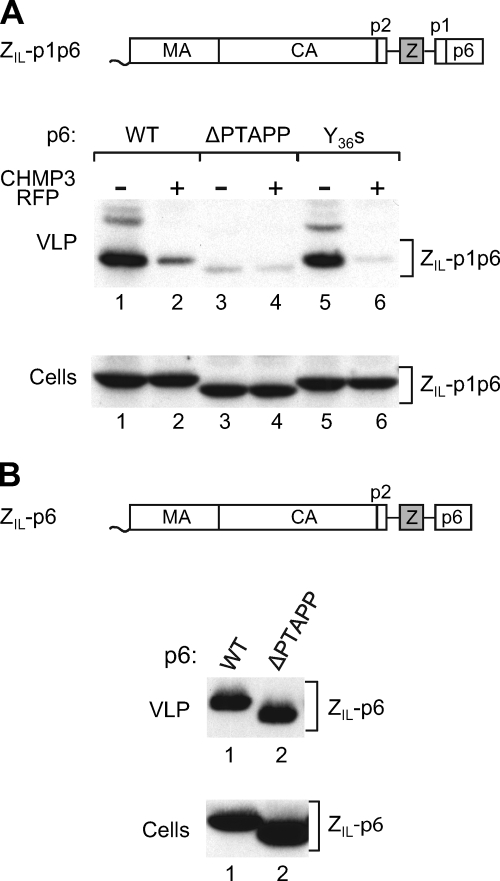
FIG. 4.
The addition of p1-p6 but not of p6 alone to a chimeric Gag construct confers L domain dependence. (A) Effect of deleting the Tsg101 or the ALIX binding site from a chimeric Gag construct containing p1. 293T cells were transfected with 1.5 μg of WT ZIL-p1p6 or the indicated mutants, along with 0.5 μg of a vector expressing RFP alone or CHMP3-RFP. VLP pellets and the cell lysates were analyzed by Western blotting with anti-CA serum. (B) Effect of deleting the Tsg101 binding site from a chimeric Gag construct lacking p1. 293T cells were transfected with 1.5 μg of WT ZIL-p6 or a ΔPTAPP version.
The p1 spacer peptide can confer a requirement for Tsg101 binding.
The sensitivity of ZIL-p1p6 to CHMP3-RFP indicated that efficient particle production by this construct required the presence of an L domain. On the other hand, the lack of an effect of CHMP3-RFP on the release of ZIL-p6 Gag suggested that particle production by ZIL-p6 did not depend on the Tsg101 binding site in p6. To test these predictions, we deleted the Tsg101 binding site from the p6 domains of PR-negative ZIL-p1p6 and of ZIL-p6.
As shown in Fig. 4A, ZIL-p1p6ΔPTAPP released about 15-fold less particulate Gag than the parental ZIL-p1p6 construct (compare lanes 1 and 3). Moreover, the small amount of Gag that was released by ZIL-p1p6ΔPTAPP was only marginally further reduced by CHMP3-RFP (Fig. 4A; compare lanes 3 and 4), indicating that the Tsg101 binding site played a predominant role in engaging the ESCRT machinery, as is the case for WT HIV-1 Gag (20). Consistent with this notion, the Y36s mutation, which removes the entire ALIX binding site from p6, only moderately reduced particle production by ZIL-p1p6 (Fig. 4A, lane 5). Furthermore, the ALIX binding site mutant remained highly sensitive to inhibition by CHMP3-RFP (Fig. 4A; compare lanes 5 and 6). As expected, the Tsg101 binding site in p6 was largely dispensable for particle production by ZIL-p6, because the parental ZIL-p6 Gag construct and the Tsg101 binding site mutant ZIL-p6ΔPTAPP produced comparable amounts of particles (Fig. 4B). Thus, at least in this chimeric Gag context, the presence or absence of p1 determined whether a Tsg101 binding site was required for efficient particle release.
Removal of NC-p1 coding sequences from authentic HIV-1 alleviates sensitivity to dominant-negative CHMP3.
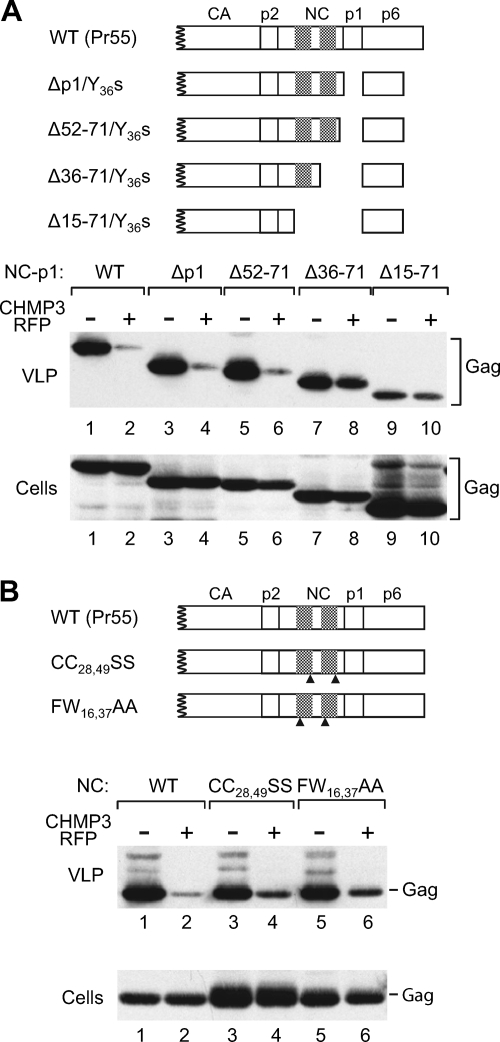
The results described above were obtained with chimeric Gag constructs that contained a heterologous dimerization domain. To determine whether the NC-p1 region affects the sensitivity of HIV-1 Gag to CHMP3-RFP in the absence of foreign sequences, we made use of a panel of previously described proviral NC-p1 deletion mutants (49). Because these mutants were originally used to map the ALIX binding site in NC-p1, they also harbored the Y36s mutation near the C terminus of p6 to remove the LYPxnL-type binding site for ALIX (49). Since the NC-p1 deletions removed the frameshift signal required for the expression of PR, a PR-negative version of HXBH10 was used for comparison.
As shown in Fig. 5A, the Δp1/Y36s and Δ52-71/Y36s mutants produced amounts of particles similar to those produced by the parental HIV-1 provirus (HXBH10-PR−). CHMP3-RFP reduced particle production by HXBH10-PR− about 20-fold in this experiment and had comparable effects on particle production by the Δp1/Y36s and Δ52-71/Y36s mutants (lanes 1 to 6). Thus, in the presence of all or of most of NC, deleting p1 did not reduce the requirement for a functional ESCRT pathway. We also note that the ALIX binding site in p6 was dispensable for the inhibitory effect of CHMP3-RFP.
FIG. 5.
The removal of NC-p1 sequences from authentic HIV-1 Gag alleviates the effect of dominant-negative CHMP3 on particle production. (A) 293T cells were transfected with 1.5 μg of a PR-defective HIV-1 provirus expressing WT Pr55gag or with the indicated NC-p1 deletion mutants, along with a vector expressing RFP alone or CHMP3-RFP. VLP pellets and the cell lysates were analyzed by Western blotting with anti-CA serum. (B) Effect of CHMP3-RFP on the indicated NC zinc finger mutants. Cross-hatched boxes indicate the positions of the zinc fingers in NC, and arrowheads indicate the positions of residues that were substituted. Western blotting was performed with 183-H12-5C anti-HIV CA antibody.
Quantitation of the amount of VLP-associated Gag relative to the amount of cell-associated Gag indicated that the Δ36-71/Y36s mutant released Gag with an efficiency that was approximately 80% of the WT level. Interestingly, CHMP3-RFP reduced the production of Δ36-71/Y36s mutant particles only about 1.5-fold (Fig. 5A, lanes 7 and 8). As a consequence, the Δ36-71/Y36s mutant, which lacks the second zinc finger of NC together with p1, produced more particles in the presence of CHMP3-RFP than the parental provirus or the Δp1/Y36s and Δ52-71/Y36s mutants. The inhibitory effect of CHMP3-RFP on particle production was also severely diminished by the more extensive Δ15-71 deletion, which removed both zinc fingers of NC together with p1 (Fig. 5A, lanes 9 and 10). However, the Δ15-71 deletion also caused a significant assembly defect.
Because of the role of NC in viral RNA incorporation, we also determined the effects of CHMP3-RFP on variants of HXBH10-PR− with point mutations in NC that are known to impair genomic RNA encapsidation. The CC28,49SS mutation disrupts zinc-coordinating residues in both zinc fingers of NC and reduces the packaging of genomic viral RNA into HIV-1 virions more than 20-fold (17). The FW16,37AA mutation targets aromatic residues at position n + 1 of each zinc finger that are each absolutely essential for virus replication, and the F16A mutation alone reduces genomic HIV-1 RNA encapsidation to 15% of wild-type levels (17). Although the CC28,49SS mutation and, to a lesser extent, the FW16,37AA mutation caused an accumulation of Gag in the cell lysates, both mutations had only small effects on viral particle production in the HXBH10-PR− context (Fig. 5B). Particle production by the CC28,49SS and FW16,37AA mutants was only moderately less sensitive to CHMP3-RFP than particle production by the parental provirus (Fig. 5B). Nevertheless, results similar to those represented in Fig. 5B were obtained in three independent experiments, indicating that the mutated NC residues contribute to the sensitivity of HIV-1 Gag to inhibition of the ESCRT pathway. Taken together, these results support the notion that the requirement for intact ESCRT machinery during HIV-1 budding is at least in part imposed by NC-p1.
DISCUSSION
Depending on the producer cell type examined, HIV-1 p6 mutants with defective PTAP L domains exhibit severe defects in either virion-cell or virion-virion detachment (16). In adherent cell lines and in primary human macrophages, the defect at the level of virion-cell detachment predominates, resulting in inefficient virus release (16, 22, 24). It was therefore surprising that certain chimeric HIV-1 Gag constructs did not require p6 for the efficient production of VLP in adherent cells (1, 65). In these Gag constructs, HIV-1 NC was replaced by foreign protein-protein interaction domains to substitute for its assembly function. Remarkably, this led to near wild-type levels of particle production even in cases where the entire NC-p1-p6 region was replaced and all consensus L domain motifs in HIV-1 Gag were therefore eliminated (1, 64).
One possible explanation for these observations was that the NC-p1-p6 region of HIV-1 Gag has an inhibitory effect on virus release that is counteracted by the L domain function of p6, and the results of the present study support this model. First, we found that replacing NC-p1-p6 with a heterologous dimerization domain in a Gag construct termed ZWT caused resistance to dominant-negative CHMP3 or Vps4 under conditions where particle production by WT HIV-1 Gag was nearly abolished. We note that in a previous study, dominant-negative Vps4 had no effect on p6-deficient HIV-1 lacking active PR (19). However, in our hands p6-deficient HIV-1 exhibited significant defects in particle formation even after PR was inactivated, whereas the ZWT Gag construct produced nearly wild-type levels of particles in our study (1). Since dominant-negative versions of the CHMPs or of Vps4 block the function of all known L domains, our observations argue against the use of an alternative L domain by ZWT Gag that would explain its efficient release despite the absence of p6.
In principle, the apparent L domain independence of the chimeric ZWT Gag construct could have been caused by the presence of a foreign leucine zipper domain rather than by the absence of C-terminal HIV-1 Gag sequences. However, L domain dependence was restored in a Gag construct that harbored both the leucine zipper domain and NC-p1-p6, since efficient particle production by this construct depended on the Tsg101 binding site in p6. Moreover, particle production was substantially inhibited when NC-p1 rather than NC-p1-p6 was added back to ZWT. Together, these results suggested that the presence or absence of NC-p1 determined whether an L domain was needed to promote particle release.
Our data suggest that the p6 domain itself does not confer a requirement for an L domain, since the Tsg101 binding site within p6 was not required for efficient particle production by HIV-1 Gag when NC-p1 was replaced by a leucine zipper. Furthermore, particle production by this p6-containing construct was largely resistant to CHMP3-RFP, indicating that ESCRT-mediated membrane scission was not required. Additionally, the finding that p6 by itself did not confer sensitivity to CHMP3-RFP suggested that dominant-negative ESCRT pathway components do not simply arrest budding by tethering Gag to aberrant ESCRT complexes via the Tsg101 and ALIX binding sites in p6.
Remarkably, replacing only the NC domain by a leucine zipper had effects very different from replacing NC-p1. In the former case, the Tsg101 binding site in p6 became crucial for efficient particle production, and CHMP3-RFP had a profound inhibitory effect on particle release. Thus, the presence of p1-p6 but not of p6 alone at the C terminus of the leucine zipper sequence conferred both L domain dependence and ESCRT pathway dependence that resembled that exhibited by authentic HIV-1 Gag. Although the mechanism by which p1 contributed to this phenotype remains unknown, it does not appear to involve the role of the p1 coding sequence in ribosomal frameshifting, because the slippery sequence at which frameshifting occurs (30) was not required to confer sensitivity to CHMP3-RFP (data not shown). We cannot exclude the possibility that the presence of p1 positioned the p6 domain in a manner that allowed the entrapment of Gag by aberrant ESCRT polymers induced by CHMP3-RFP. However, we consider it more likely that p1 conferred a bona fide requirement for a functional ESCRT pathway in the chimeric Gag context, since in the presence of p1 the Tsg101 binding site in p6 became critical for particle production.
A recent three-dimensional analysis of immature HIV-1 particles and of HIV-1 budding sites indicated that ESCRT-III assists Gag in the membrane bending required to complete the budding process (9). Consistent with this model, upon overexpression certain ESCRT-III components have been shown to assemble into circular arrays that bend the plasma membrane into buds that emerge from the cell surface (25). It is possible that certain Gag-leucine zipper chimeras do not need the assistance of ESCRT-III because the zipper domain improves the ability of Gag to bend membranes on its own. According to this scenario, the fusion of NC-p1 or of p1-p6 to the zipper domain might simply compromise this ability and thus restore ESCRT dependence. However, we also observed that the sensitivity of authentic HIV-1 Gag to inhibition of the ESCRT pathway could be reduced by certain deletions in the NC-p1 region. In the absence of ESCRT pathway inhibition, these deletions reduced the efficiency of Gag release, which argues against the possibility that ESCRT pathway independence was merely a consequence of an improved ability to assemble into a lattice capable of bending membranes.
Particle production by authentic HIV-1 Gag became only moderately less sensitive to inhibition of the ESCRT pathway when specific NC residues crucial for the encapsidation of the viral genomic RNA were mutated. However, it was previously demonstrated that cellular mRNA can substitute for viral genomic RNA during retroviral assembly (45). In addition to its ability to specifically recognize genomic viral RNA, NC exhibits a nonspecific RNA binding activity that is critical for its function in particle assembly (5, 13). This nonspecific RNA binding activity depends on clusters of basic residues but not on the integrity of the zinc fingers in NC. Notably, it has been shown that the CC28,49SS and FW16,37AA mutations have only a minor effect on the ability of NC to bind RNA in vitro, despite their drastic effects on the packaging of viral genomic RNA (13). It is thus likely that the CC28,49SS and FW16,37AA mutant particles packaged cellular RNA rather than viral RNA. Therefore, our results do not exclude the possibility that the role of NC in the ESCRT pathway dependence of budding is related to its function in RNA encapsidation. We note that particles produced by the ZWT Gag construct, which are released in a fully ESCRT-independent manner (this study), contain no detectable cellular RNA and are therefore thought to assemble without incorporating any form of nucleic acid (14).
In the authentic Gag context, deleting p1 alone had little effect on the inhibition of particle production by dominant-negative CHMP3. Rather, it was necessary to additionally delete a portion of NC to mitigate the effect of CHMP3-RFP. On the other hand, p1 conferred sensitivity to dominant-negative CHMP3 in a chimeric Gag context lacking NC. Together, these observations suggest that NC and p1 can independently contribute to a requirement for ESCRT pathway engagement for optimal particle production. An understanding of how NC and p1 confer such a requirement may provide further insights into why retroviruses use L domains, given that efficient particle release can in principle be achieved in an ESCRT pathway-independent manner.
Acknowledgments
We thank Stewart Craig for technical assistance and Gregory Hendricks for performing the electron microscopy. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Bruce Chesebro and Kathy Wehrly: HIV-1 p24 monoclonal antibody (183-H12-5C).
This work was supported by grant R37AI029873 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 28 April 2010.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajorek, M., H. L. Schubert, J. McCullough, C. Langelier, D. M. Eckert, W. M. Stubblefield, N. T. Uter, D. G. Myszka, C. P. Hill, and W. I. Sundquist. 2009. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, L. A., J. A. Briggs, B. Glass, J. D. Riches, M. N. Simon, M. C. Johnson, B. Muller, K. Grunewald, and H. G. Krausslich. 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908-1912. [DOI] [PubMed] [Google Scholar]
- 11.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, H. Y., E. Morita, U. von Schwedler, B. Muller, H. G. Krausslich, and W. I. Sundquist. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82:4884-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crist, R. M., S. A. Datta, A. G. Stephen, F. Soheilian, J. Mirro, R. J. Fisher, K. Nagashima, and A. Rein. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dussupt, V., M. P. Javid, G. Abou-Jaoude, J. A. Jadwin, J. de La Cruz, K. Nagashima, and F. Bouamr. 2009. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 5:e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, Y., N. Wu, X. Gan, W. Yan, J. C. Morrell, and S. J. Gould. 2007. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841-852. [DOI] [PubMed] [Google Scholar]
- 21.Fujii, K., J. H. Hurley, and E. O. Freed. 2007. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat. Rev. Microbiol. 5:912-916. [DOI] [PubMed] [Google Scholar]
- 22.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 23.Ghazi-Tabatabai, S., T. Obita, A. V. Pobbati, O. Perisic, R. Y. Samson, S. D. Bell, and R. L. Williams. 2009. Evolution and assembly of ESCRTs. Biochem. Soc. Trans. 37:151-155. [DOI] [PubMed] [Google Scholar]
- 24.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson, P. I., R. Roth, Y. Lin, and J. E. Heuser. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson, P. I., S. Shim, and S. A. Merrill. 2009. Cell biology of the ESCRT machinery. Curr. Opin. Cell Biol. 21:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard, T. L., D. R. Stauffer, C. R. Degnin, and S. M. Hollenberg. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114:2395-2404. [DOI] [PubMed] [Google Scholar]
- 28.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurley, J. H. 2008. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20:4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 31.Kieffer, C., J. J. Skalicky, E. Morita, I. De Domenico, D. M. Ward, J. Kaplan, and W. I. Sundquist. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15:62-73. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 32.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lata, S., M. Roessle, J. Solomons, M. Jamin, H. G. Gottlinger, D. I. Svergun, and W. Weissenhorn. 2008. Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378:816-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lata, S., G. Schoehn, A. Jain, R. Pires, J. Piehler, H. G. Gottlinger, and W. Weissenhorn. 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science 321:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, S., A. Joshi, K. Nagashima, E. O. Freed, and J. H. Hurley. 2007. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14:194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, Y., L. A. Kimpler, T. V. Naismith, J. M. Lauer, and P. I. Hanson. 2005. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280:12799-12809. [DOI] [PubMed] [Google Scholar]
- 37.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald, B., and J. Martin-Serrano. 2009. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 122:2167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 45.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muzioł, T., E. Pineda-Molina, R. B. Ravelli, A. Zamborlini, Y. Usami, H. Gottlinger, and W. Weissenhorn. 2006. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10:821-830. [DOI] [PubMed] [Google Scholar]
- 47.Pincetic, A., and J. Leis. 2009. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009:6239691-6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2009. Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J. Virol. 83:7185-7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 82:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raiborg, C., and H. Stenmark. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445-452. [DOI] [PubMed] [Google Scholar]
- 51.Saksena, S., J. Sun, T. Chu, and S. D. Emr. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32:561-573. [DOI] [PubMed] [Google Scholar]
- 52.Saksena, S., J. Wahlman, D. Teis, A. E. Johnson, and S. D. Emr. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shim, S., L. A. Kimpler, and P. I. Hanson. 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8:1068-1079. [DOI] [PubMed] [Google Scholar]
- 54.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 55.Tanzi, G. O., A. J. Piefer, and P. Bates. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 77:8440-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Usami, Y., S. Popov, E. Popova, and H. G. Gottlinger. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 60.Whitley, P., B. J. Reaves, M. Hashimoto, A. M. Riley, B. V. Potter, and G. D. Holman. 2003. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5 bisphosphate dependent endosome compartmentalization. J. Biol. Chem. 278:38786-38795. [DOI] [PubMed] [Google Scholar]
- 61.Wollert, T., C. Wunder, J. Lippincott-Schwartz, and J. H. Hurley. 2009. Membrane scission by the ESCRT-III complex. Nature 458:172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zamborlini, A., Y. Usami, S. R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. U. S. A. 103:19140-19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai, Q., R. D. Fisher, H. Y. Chung, D. G. Myszka, W. I. Sundquist, and C. P. Hill. 2008. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43-49. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]