Abstract
DDX6 (Rck/p54) is an evolutionarily conserved member of the SF2 DEAD-box RNA helicase family that contributes to the regulation of translation and storage and the degradation of cellular mRNAs. It interacts with multiple proteins and is a component of the micro-RNA (miRNA)-induced silencing complex (miRISC). Since miRNA-122 (miR-122) is essential for efficient hepatitis C virus (HCV) replication, we investigated the requirement for DDX6 in HCV replication in cultured hepatoma cells. Small interfering RNA (siRNA)-mediated knockdown of DDX6 and rescue with an siRNA-resistant mutant demonstrated that DDX6 expression is indeed required for optimal HCV replication. However, DDX6 knockdown did not impair miR-122 biogenesis or alter HCV responsiveness to miR-122 supplementation. Overexpression of DDX6 fused to EYFP (EYFP-DDX6) enhanced replication, whereas a helicase-deficient mutant with a substitution in the conserved DEAD-box motif II (DQAD) had a dominant-negative effect, reducing HCV yields. Coimmunoprecipitation experiments revealed an intracellular complex containing DDX6, HCV core protein, and both viral and cellular RNAs, the formation of which was dependent upon the C-terminal domain of DDX6 but not DDX6 helicase activity. However, since DDX6 abundance influenced the replication of subgenomic HCV RNAs lacking core sequence, the relevance of this complex is uncertain. Importantly, DDX6 knockdown caused minimal reductions in cellular proliferation, generally stimulated cellular translation ([35S]Met incorporation), and did not impair translation directed by the HCV internal ribosome entry site. Thus, DDX6 helicase activity is essential for efficient HCV replication, reflecting essential roles for DDX6 in HCV genome amplification and/or maintenance of cellular homeostasis.
Persistent hepacivirus infection is associated with progressive liver fibrosis and the development of hepatocellular carcinoma (23). Worldwide, over 130 million people are infected with hepatitis C virus (HCV), which is estimated to cause over 350,000 cirrhosis and cancer deaths annually (33). Chronic hepatitis C is thus a major threat to human health. Current IFN-based treatments are effective in <50% of patients infected with the most prevalent viral genotypes, and there is a need for more effective therapies. Although good progress is being made in the development of new antiviral compounds that target major enzymatic activities expressed by HCV (a serine protease and an RNA-dependent RNA polymerase), resistance emerges rapidly to small-molecule inhibitors of these viral enzymes due to the highly replicative nature of the infection coupled with error-prone viral RNA synthesis (40). Unfortunately, efforts to develop more effective therapeutic measures are handicapped by the fact that many aspects of the biology and molecular virology of this pathogen remain poorly defined. A better understanding of its interaction with the host cell and, in particular, its dependence on host cell proteins and micro-RNAs (miRNAs) for replication, may point the way to alternative cellular targets for antiviral therapies. Although far from certain, therapeutics targeting such host factors may be less likely to engender the development of resistance.
Several host cell proteins have been implicated in HCV genome replication, including in particular the SNARE-like vesicle-associated membrane protein-associated host protein, VAP-A (also known as hVAP-33), as well as VAP-B (11, 13); a geranylgeranylated F box protein, FBL2 (43); and cyclophilin B (CypB), the latter of which interacts with the NS5B polymerase and is targeted by several candidate antiviral compounds now in clinical development (45). In addition, recent studies show that a highly abundant, liver-specific miRNA, miR-122, facilitates replication of HCV, both positively regulating the abundance of autonomously replicating HCV RNAs in Huh-7 hepatoma cells (21) and enhancing the replication of infectious virus (36). The requirement for miR-122 in HCV replication is strongly supported by a recent study demonstrating that pharmacologic sequestration of miR-122 has a dramatic antiviral effect in HCV-infected chimpanzees (22). How miR-122 promotes replication is incompletely understood, although some data suggest it does so by increasing the efficiency of translation of viral RNA, which is driven by a cap-independent process involving internal entry of 40S ribosomes directed by an internal ribosome entry site (IRES) in the 5′ untranslated RNA (UTR) segment of the genome (15).
DDX6 (Rck/p54), is a cellular RNA helicase with ATP-dependent RNA-unwinding activity (46). It is a member of an evolutionarily conserved family of DExD/H helicases that are typically found as components of large messenger ribonucleoprotein (mRNP) complexes. DDX6 and its homologs are thought to function in remodeling mRNPs so as to facilitate multiple aspects of mRNA metabolism, including transcription, splicing, translation, degradation, and storage (8, 26). Human DDX6 was first identified as a 54-kDa protein encoded by a gene located at a chromosomal breakpoint in a cell line, RC-K8, derived from a B-cell lymphoma (hence its alternative name, Rck/p54) (28). It has been suggested to function as a proto-oncogene and is overexpressed in some colorectal cancers (14). Both it and its closely related yeast homologue, Dhh1p, function as general translational repressors (7). DDX6 interacts with the initiation factor eIF-4E and other cellular protein partners to repress the translational activity of mRNPs. It is a component of stress granules but localizes primarily to processing bodies (P bodies) in which its mRNP remodeling activities may contribute to mRNA storage, degradation, and recycling for translation (46). DDX6 directly interacts with Argonaute-1 (Ago1) and Ago2 in miRNA-induced silencing complexes (miRISC), and it plays a role in miRNA-mediated translational repression (6).
Since miR-122 is required for HCV replication and DDX6 is a miRISC component, we considered the possibility that DDX6 could functionally regulate the replication of HCV. Viral interactions with two other members of the DExD-box RNA helicase family (e.g., DDX3 and DDX5/p68) have been implicated in the replication of HCV in cell culture. DDX3 interacts with the HCV core protein (32) and is required for efficient HCV replication (3, 4), while the viral polymerase, NS5B, interacts with DDX5, leading to its redistribution from the nucleus to the cytoplasm (12). DDX6 may also be relevant to hepatitis C since it is upregulated within the liver during chronic infection (30). Our interest in studying its role in HCV replication was stimulated by the fact that its yeast homolog, Dhh1p, is required for efficient translation and replication of the plant brome mosaic virus (BMV) in yeast, a well-established model for replication of positive-strand RNA viruses (2). Consistent with the strong evolutionary conservation of DDX6 function, DDX6 is capable of substituting for Dhh1p in supporting BMV replication in yeast cells.
Against this background, we set out to determine whether DDX6 is involved in HCV replication. While our studies were in progress, Scheller et al. (37) reported that RNA interference-mediated silencing of Rck/p54 and several related proteins substantially impairs the replication of HCV in cell culture, ostensibly due to an impairment in HCV translation in the absence of DDX6 expression. Our results confirm that DDX6 expression is required for optimal replication of HCV RNA but argue against this being due to a requirement for DDX6 during IRES-directed translation. We show that miR-122 facilitates HCV RNA replication independently of DDX6 and that mutations that ablate the ATPase and unwindase activities of DDX6 cause a relocalization of DDX6 within the cell and also negate the ability of DDX6 to support viral replication.
MATERIALS AND METHODS
Cell lines.
Huh-7, FT3-7 (a clonal derivative of Huh-7 cells), and Huh-7.5 cells (kindly provided by Charles Rice, Rockefeller University) were cultured as described previously (50). Several G418-resistant Huh-7-derived replicon cell lines were used in these studies, including Htat2ANeo/NS3-5B/QR/VI/KR/KR5A/SI cells, which contain a subgenomic (NS3-NS5B), genotype 1a (H77) replicon (48); Htat2ANeo/C-5B/QR/KR/FV/SI, a related cell line in which the replicon contains the complete HCV polyprotein-coding sequence (48); Ntat2ANeo/C-5B/2-3, which contains a similar full-length genotype 1b (HCV-N) replicon (17); and Sg-2a cells, which contain a subgenomic genotype 2a (JFH-1) replicon. Replicon cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, 4 mM l-glutamine, and 0.25 mg of G418/ml. Huh-7-191/20 cells conditionally express HCV core protein under tight regulation by the Tet-Off promoter and were cultured as described previously (24). FRhK-4 cells, used for experiments with hepatitis A virus (HAV), were cultured as described previously (47).
HCV.
Experiments with cell culture-infectious HCV were carried out with HJ3-5 virus (vH-NS2/NS3-J/Y361H/Q1251L) (49), an intergenotypic chimeric virus produced in cell culture that is also infectious in chimpanzees (M. Yi and S. M. Lemon, unpublished data). The HJ3-5 genome is comprised of sequence encoding core-NS2 of the genotype 1a H77c virus placed within the background of the genotype 2a JFH-1 virus genome (49). It contains two adaptive mutations, Y361H and Q1251L, in E1 and NS3, respectively, that facilitate its replication (49). Virus infections and infectious virus titrations were carried out as described previously (50).
HAV.
HAV infections were carried out using HM175/18f virus, a cytopathic, cell-culture-adapted virus variant, as described previously (47). Infectious virus titers were determined by using a modified infrared fluorescence focus assay (9) as follows. First, 1.5 × 105 FRhK-4 cells were plated in a six-well plate. The following day, the cells were inoculated with serial dilutions of virus (600 μl) prepared in DMEM with 2% FBS, and the virus was allowed to adsorb at 36°C for 1.5 h. Cells were then overlaid with DMEM containing 0.5% low-melting-point agarose and incubated for 7 days at 36°C in 5% CO2. The overlays were gently removed, and the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 25 min, washed twice with PBS for 5 min each time, and permeabilized with 0.2% Triton X-100 in PBS at room temperature for 12 min. After further washing, the cells were blocked with 10% normal goat serum for 1 h at room temperature and incubated overnight with a murine monoclonal anti-HAV (6A5) antibody at a 1:600 dilution (600 μl per well) in 3% milk at 4°C overnight. The cells were washed four times with PBS containing 0.1% Tween 20 (PBS-T) and incubated for 1 h with a 1:800 dilution (600 μl per well) of goat anti-mouse Alexa Fluor 680 antibody (Invitrogen) in 3% milk at room temperature. The plates were extensively washed, dried, and scanned with an Odyssey infrared imaging system (LI-COR, Lincoln, NB). Foci of viral replication were counted, and the results were calculated as focus-forming units (FFU)/ml.
Plasmids.
pHJ3-5 is a T7 expression vector that contains the sequence of HJ3-5 virus, while pHJ3QL-ΔE1-p7 is a related vector containing an in-frame deletion of the E1-p7 coding sequence (49). pHJ3-5-ΔC61-148 expresses HJ3-5 RNA with an in-frame deletion of amino acids 61 to 148 of core. pHJ-3-5-ΔC21-p7 contains an in-frame deletion from core to p7 but retains the first 20 amino acids of core. pHJ3-5-ΔC61-148 and pHJ-3-5-ΔC21-p7 were constructed from pHJ3-5 by standard PCR-based mutagenesis techniques. Similar methods were used to construct pHJ3-5/RLuc2A, which contains the Renilla luciferase sequence, fused at its 3′ end to the foot-and-mouth disease virus 2A autoprotease sequence, inserted between the p7 and NS2 sequences of pHJ3-5 (18). To eliminate its ability to replicate, pHJ3-5/RLuc2A was further modified by creation of an Asp-to-Asn substitution within the GDD motif of the polymerase active site, resulting in pHJ3-5/RLuc2A-GND.
pEYFP-C1-Rck (referred to hereafter as pEYFP-DDX6) was kindly provided by T. M. Rana, University of Massachusetts Medical School, Worcester, MA, and encodes N-terminal enhanced yellow fluorescent protein (EYFP)-tagged, full-length DDX6 (6). pEYFP-DDX6-m6 contains six silent point mutations that ablate the binding of DDX6-1 small interfering RNA (siRNA; see below) but maintain the wild-type amino acid sequence of DDX6. It was generated by using the QuikChange site-directed mutagenesis kit (Stratagene). pEYFP-DDX6-ΔC, generated by standard molecular biology techniques, contains a deletion of sequence encoding the C-terminal 183 amino acids of the DDX6 protein (C-terminal domain 2) (1). pEYFP-DDX6-EQ is an additional mutant that encodes Gln in lieu of Glu within the DEAD-box motif II of DDX6 and thus lacks helicase activity (46). pEYFP encodes only the EYFP and was used as a negative control in transfection experiments.
pRLHL expresses a dicistronic RNA containing Renilla luciferase sequence in the first cistron, and firefly luciferase sequence in its second cistron, separated by the HCV internal ribosome entry site (IRES) sequence (16). A PCR product carrying a T7 promoter at its 5′ end and a poly(A) tail of 30 adenosines at its 3′ end was used as a template for in vitro transcription of a capped and polyadenylated RNA encoding firefly luciferase.
Antibodies.
The antibodies used in these studies included mouse monoclonal antibody (MAb) to HCV core protein (C7-50; Affinity BioReagents); polyclonal rabbit anti-NS5A (a gift from Craig E. Cameron, Pennsylvania State University, University Park); mouse monoclonal 9E10 anti-NS5A antibody (a generous gift from Brett D. Lindenbach, Yale University School of Medicine, New Haven, CT); rabbit anti-HAV 2A protein (a gift from Verena Gauss-Muller, University of Lubeck, Lubeck, Germany); 6A5 anti-HAV MAb (a gift from J. Hughes, Merck Sharp and Dohme Research Laboratories, West Point, PA); anti-dsRNA MAb (J2, Scicons, Hungary); rabbit polyclonal anti-DDX6 (Bethyl Labs); mouse anti-green fluorescent protein (anti-GFP) MAb (Clontech); rabbit polyclonal anti-GFP (Living Colors Full-length A.V. anti-GFP; Clontech); and rabbit polyclonal anti-calnexin (Sigma).
siRNA and miRNA oligonucleotides.
RNA oligonucleotides and siRNAs were custom synthesized or purchased from Dharmacon. The sequences were as follows: miR-122wt, 5′-UGGAGUGUGACAAUGGUGUUUGU-3′; miR-122*, 5′-AAACGCCAUUAUCACACUAAAUA-3′ (21); miR-124, 5′-UUAAGGCACGCGGUGAAUGCCA-3′; and miR-124*, 5′-CCGUGUUCACAGCGGACCUUGA-3′. Mature miR-122 and miR-124 duplexes were generated by annealing equal molar amounts of miR-122wt and miR-122* and of miR-124 and miR-124*, respectively.
Two DDX6-specific siRNAs, DDX6-1 and DDX6-3, targeting different regions of DDX6 mRNA, were used to ensure specificity of RNA interference experiments. Control siRNAs, used to monitor off-target effects of siRNA knockdown, included DDX6-1m and DDX6-3m, sequence-related siRNAs each containing two mutations in the DDX6-1 and DDX6-3 siRNA sequences, respectively. The sequences of these siRNAs were as follows: DDX6-1, 5′-GCAGAAACCCUAUGAGAUUUU-3′ and 5′-AAUCUCAUAGGGUUUCUGCUU-3′ (6); DDX6-1m, 5′-GCAGAAACCGAAUGAGAUUUU-3′ and 5′-AAUCUCAUUCGGUUUCUGCUU-3′; DDX6-3, 5′-CCAAAGGAUCUAAGAAUCAdTdT-3′ and 5′-UGAUUCUUAGAUCCUUUGGdTdT-3′ (39); and DDX6-3m, 5′-CCAAAGGAUGAAAGAAUCAdTdT-3′ and 5′-UGAUUCUUUCAUCCUUUGGdTdT-3′ (mutated sequences are underlined in the control siRNAs). Additional siRNA controls included the ON-TARGETplus nontargeting pool of 4 siRNAs (Dharmacon catalog no. D001810-10-20; hereafter referred to as Ctrl), which has no known targets within mammalian genomes.
Transfections.
siRNAs (50 nM) and miRNAs (50 nM) were transfected into FT3-7 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommended procedures. Lipofectamine 2000 was also used for transfection of the plasmid DNA. HCV RNA and FLuc RNA transfections were carried out by using a TransIT mRNA transfection kit (Mirus Bio) for 5 h according to the manufacturer's protocol.
siRNA-mediated knockdown of DDX6 and HCV replication.
FT3-7 cells grown in 24-well plates were transfected with siRNA (50 nM) as described above. At 48 h, the cells were infected with HJ3-5 virus at a multiplicity of infection (MOI) of 1.0. One day later, the cells were washed twice with PBS and refed with fresh media. Supernatant culture fluids were collected subsequently at 24-h intervals (days 2 and 3 postinfection), and the infectious virus titers were determined by FFU assay as described previously (50). On day 3 postinfection, cell lysates were prepared for immunoblot analysis. For experiments involving rescue of DDX6 knockdown, the cells were transfected with DDX6-1 or DDX6-1m siRNAs as described above and supertransfected, 24 h later, with pEYFP-DDX6-m6 (20, 100, and 500 ng/well) or pEYFP (500 ng/well) DNA using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were infected with HJ3-5 virus (MOI = 0.5), and the infection was monitored as described above. Cell lysates, prepared 3 days after infection, were analyzed in immunoblots with DDX6, calnexin, HCV core, and NS5A-specific antibodies.
DDX6 knockdown and HAV replication.
FT3-7 cells were transfected with siRNAs as described above and infected 6 h later with HM175/18f virus at an MOI of 1.0. Virus was allowed to adsorb to cells for 1.5 h in antibiotic-free DMEM with 2% FBS; the cells were then refed with fresh media, followed by incubation at 36°C in a 5% CO2 environment. On day 5 postinfection, the cell lysates were prepared for immunoblot analysis, while virus was harvested from supernatant fluids and by repetitive freeze-thawing of a parallel set of cultures. The cell lysate was sonicated, extracted with an equal volume of chloroform, and stored at −80°C for virus titration.
WST-1 assays.
FT3-7 cells (104 cells/well of a 96-well plate) were transfected with DDX6 siRNAs as described above. At 0, 24, 48, 72, and 96 h posttransfection, 10 μl of WST-1 reagent (2-[2-methoxy-4-nitrophenyl]-3-[4-nitrophenyl]-5-[2,4-disulfo-phenyl]-2H-tetrazolium monosodium salt; Chemicon International, Inc.) was added to triplicate sets of wells, and the cells were incubated at 37°C for 1 h prior to measurement of the absorbance at 450 nm.
Coimmunoprecipitation experiments.
FT3-7 cells were infected with HJ3-5 HCV at an MOI of 0.2. When >90% positive for HCV core expression, as determined by immunostaining, the cells were transfected with EYFP-DDX6 or EYFP expression vectors as described above. After 48 h, the cells were treated with trypsin, washed twice with ice-cold PBS, and resuspended in immunoprecipitation lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM EDTA, 1% NP-40, 10% glycerol, freshly added 2 mM dithiothreitol, and protease inhibitor cocktail [Roche]) on ice for 30 min. The lysate was centrifuged at 14,000 rpm at 4°C for 10 min to remove debris and preclarified with protein G-Sepharose 4 Fast Flow beads (GE Healthcare) at 4°C overnight. The lysates were then subjected to immunoprecipitation (500 μg of protein in 500 μl of IP lysis buffer) with rabbit anti-GFP antibody (1:200 dilution) and protein G-Sepharose 4 Fast Flow beads. After extensive washing in immunoprecipitation lysis buffer (10 min each time, four times, at 4°C), the proteins were eluted by boiling in 1× sodium dodecyl sulfate loading buffer and subjected to immunoblotting. For reverse coimmunoprecipitation, HCV core protein was precipitated from HJ3-5-infected FT3-7 cell lysates using a murine anti-core MAb (1:100 dilution, C7-50; Affinity BioReagents) and immunoblotted with rabbit polyclonal anti-DDX6 (Bethyl Labs). An anti-GFP MAb (Clontech) was used as an isotype control.
Confocal fluorescence microscopy.
HCV-infected FT3-7 cells grown in eight-well glass chamber slides were washed once with PBS and fixed with 4% paraformaldehyde (in PBS) for 25 min at room temperature. After washing with PBS-glycine (100 mM, for 15 min at room temperature), the cells were permeabilized with digitonin (50 μg/ml) for 5 min at room temperature. After three washes of 5 min each with PBS, the cells were incubated with the primary antibodies anti-core (1:600 dilution in 3% bovine serum albumin), anti-dsRNA (1:500), or rabbit anti-DDX6 (1:200) overnight at 4°C. The cells were then washed three times with PBS-T and incubated with the secondary antibodies Alexa Fluor 488-conjugated goat anti-mouse IgG and Alexa Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen, 1:200 each) for 1 h at room temperature. After three washes of 20 min each with PBS-T, the slides were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; 1:1,000 dilution) for 5 min, washed with PBS-T, mounted in VectaShield mounting fluid (Vector Laboratories, Burlingame, CA), and examined with a Zeiss LSM510 Meta laser scanning confocal microscope.
For localization of various mutant forms of DDX6, FT3-7 cells were transfected with EYFP-DDX6 or EYFP expression vectors. At 48 h, the cells were washed once with PBS, fixed with 4% paraformaldehyde, and incubated with PBS-glycine (100 mM, for 15 min at room temperature) as described above. After being rinsed with PBS, slides were mounted in VectaShield mounting fluid, and examined as described above.
Northern blots for HCV RNA.
Total RNA was isolated from cells by using the RNeasy minikit (Qiagen) and analyzed using reagents supplied with the NorthernMax kit (Applied Biosystems). Briefly, 5 μg of each RNA sample was resolved on a 0.9% denaturing formaldehyde agarose gel, transferred to a BrightStar-Plus nylon membrane (Applied Biosystems) by downward capillary transfer and hybridized overnight at 68°C with 32P-labeled antisense riboprobes complementary to the genotype 2a HCV (JFH1) 5′-untranslated region (5′UTR; 340 nucleotides) or β-actin (as a loading control). After extensive washing, the membranes were scanned on a Personal Molecular Imager (Bio-Rad), and the band densities quantified with Quantity One software (Bio-Rad).
Detection of miR-122.
Total cellular RNA (10 μg), extracted using Trizol reagent (Invitrogen), was resolved on a 15% polyacrylamide-8 M urea gel and transferred to a positively charged nylon membrane (Applied Biosystems) by downward capillary transfer using NorthernMax transfer buffer (Applied Biosystems) for 3 h. After UV cross-linking, the membrane was prehybridized in ULTRAhyb-Oligo hybridization buffer (Applied Biosystems) for an hour at 42°C and hybridized with 32P-labeled riboprobes specific for miR-122 and 5S-rRNA at 42°C overnight. The membranes were washed twice with wash buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% sodium dodecyl sulfate) at 42°C for 30 min each time and then scanned on a Storm 860 PhosphorImager (Molecular Dynamics).
HCV translation assays.
Lysates of pRLHL-transfected cells were prepared 24 and 48 h after transfection and assayed for firefly and Renilla luciferase reporter activities by using a dual luciferase assay kit (Promega). Firefly luciferase activity (HCV IRES-mediated translation) was normalized to the Renilla luciferase activity (cap-dependent translation). To study translation from authentic monocistronic HCV genomes, cells were cotransfected with HJ3-5/RLuc2A-GND HCV RNA (1.25 μg per well, six-well plate) and polyadenylated firefly luciferase (FLuc) mRNA (0.25 μg per well) by using a TransIT mRNA transfection kit (Mirus Bio) for 5 h according to the manufacturer's protocol. Cell lysates were prepared 8 h later and assayed for Renilla (HCV IRES-mediated translation) and firefly luciferase (cap-dependent translation) activities as described above.
[35S]methionine incorporation.
Cells were incubated in methionine- and cysteine-free DMEM containing 2% FBS for 1 h before the addition of 100 μCi of Tran35S-label (MP Biomedicals)/ml for 0, 15, 30, and 60 min. The cells were harvested in PBS containing 1% NP-40, 0.1% sodium dodecyl sulfate, 0.03% sodium deoxycholate, and protease inhibitor cocktail (Roche Applied Sciences). 35S incorporation in trichloroacetic acid-precipitated cell lysates (25 to 50 μl) prepared from equivalent numbers of cells was quantified by scintillation counting (6), and values are presented as the means ± the standard deviations (SD) from three independent experiments.
RESULTS
DDX6 knockdown impairs HCV replication.
We investigated the role of DDX6, an miRISC component that is upregulated in the liver during chronic hepatitis C, in the replication of HJ3-5 virus, an intergenotypic, chimeric HCV that replicates efficiently in cultured human hepatoma cells (49). The structure of the genome of this chimeric virus is shown in Fig. 1A. To silence the expression of DDX6, we transfected cells with two different siRNAs—DDX6-1 (6) and DDX6-3 (39)—that target distinct sequences in human DDX6 mRNA. To control for the specificity of gene silencing, we designed two additional siRNAs, representing DDX6-1 and DDX6-3, with point mutations at nucleotides 9 and 10, respectively (DDX6-1m and DDX6-3m). As an additional control, the cells were transfected with a pool of four other siRNAs (designated “Ctrl”) that have no known targets in mammalian genomes (Dharmacon). FT3-7 cells, which are derived from Huh-7 human hepatoma cells and efficiently support HCV replication, were transfected with these siRNAs as described in Materials and Methods, and 2 days later the cells were infected with HJ3-5 virus at an MOI of 1.0.
FIG. 1.
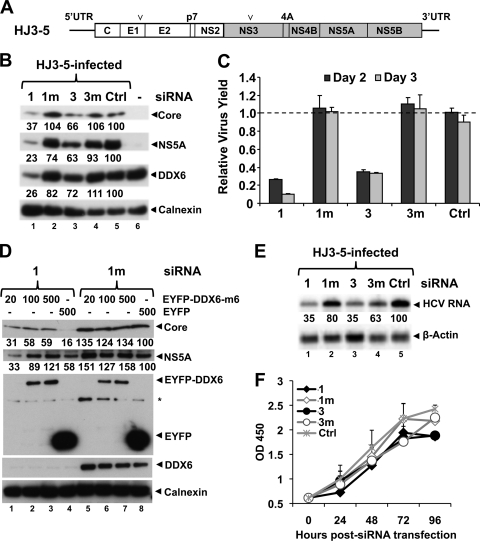
DDX6 knockdown impairs HCV replication. (A) Schematic diagram of genomic organization of chimeric HCV genotype 1a/2a (HJ3-5) RNA that contains core, E1, E2, p7, and NS2 sequence derived from H77s (genotype 1a, white box) and NS3-5B and both UTRs from JFH1 (genotype 2a, gray box). Arrowheads indicate the positions of two adaptive mutations, one each in E1 (Y361H) and NS3 (Q1251L) genes that enhance its replication. (B) FT3-7 cells were transfected with the indicated DDX6-specific or control siRNAs: 1, DDX6-1; 1m, DDX6-1m; 3, DDX6-3; 3m, DDX6-3m; and Ctrl. At 48 h, cells were infected with HJ3-5 HCV at an MOI of 1, and cell lysates prepared 3 days after infection were subjected to immunoblotting. Various band intensities were measured by using AlphaEaseFC Software version 4.0.0 (Alpha Innotech Corp.), normalized to calnexin levels, and represented as a percentage of Ctrl siRNA (Ctrl)-treated cells. (C) Virus supernatants collected 2 and 3 days postinfection were titrated on naive Huh-7.5 cells in triplicate. The results from two independent experiments are presented here as means ± the SD. (D) FT3-7 cells were transfected with 1 (DDX6-1) or 1m (DDX6-1m) siRNA. At 24 h, cells were transfected with DNA encoding siRNA-resistant form of EYFP-DDX6 or pEYFP and were infected with HJ3-5 HCV (MOI = 0.5) on the next day. Cell lysates, prepared 3 days after infection, were immunoblotted. Band densities were quantitated as described above and are represented as percentages of that of DDX6-1m/pEYFP-transfected cells. (E) FT3-7 cells were transfected with the indicated siRNAs and infected with HJ3-5 HCV at an MOI of 1.0 at 48 h posttransfection. Total RNA was isolated 3 days later and subjected to Northern blotting. PhosphorImager quantitations are represented as a percentage of that of Ctrl siRNA-treated cells. (F) FT3-7 cells were transfected with the indicated siRNAs, followed by spectrophotometric measurements of WST-1 activity at 450 nm at the indicated time points (mean optical density at 450 nm ± the SD, n = 2).
Transfection of either the DDX6-1 or DDX6-3 siRNAs decreased the abundance of DDX6 protein by 5 days posttransfection (Fig. 1B, lanes 1 and 3). DDX6-1 siRNA was more efficient and reduced DDX6 levels to 26% of Ctrl-transfected cells, while DDX6-3 reduced DDX6 expression only to 72% based on the quantitation of immunoblots. As expected, the mutant siRNAs, DDX6-1m and DDX6-3m, and the Ctrl siRNA pool had little effect on DDX6 abundance (Fig. 1B, lanes 2, 4, and 5, 82 to 111% of normal), supporting the specificity of DDX6 knockdown, particularly by DDX6-1. Importantly, transfection of DDX6-1 also reduced the abundance of HCV core protein (37% that of Ctrl siRNA-treated cells), as well as NS5A (23%) (Fig. 1B, lanes 1 and 3). The reduction in core and NS5A abundance correlated well with the efficiency of DDX6 knockdown. To assess the effect of DDX6 knockdown on production of infectious virus, we collected cell culture supernatant fluid at 24-h intervals (replacing it completely with fresh media) and determined infectious virus yields by using a standard infectious focus assay (50). This revealed that DDX6-1 transfection also reduced infectious virus yields by 8-fold on day 3 postinfection (Fig. 1C), which is concordant with the degree of DDX6 knockdown in these cells. DDX6-3, on the other hand, reduced virus yields by only 3-fold, a finding consistent with its less efficient silencing of DDX6. Neither of the control siRNAs had any effect on virus yield, suggesting that the observed reductions in virus replication were related specifically to reduced DDX6 expression. Northern blotting for HCV RNA indicated that DDX6 silencing also specifically reduced viral RNA abundance (Fig. 1E, lanes 1 and 3). Taken together, these results suggest that efficient HCV replication is closely linked to DDX6 expression in these hepatoma cells.
To rigorously test this conclusion and show that impaired viral replication results specifically from DDX6 silencing and not an off-target effect of the siRNA, we constructed a mutated DDX6 expression vector (pEYFP-DDX6-m6) that contains six base mismatches within the site targeted by the DDX6-1 siRNA without changing the amino acid sequence of the protein. We silenced the expression of endogenous DDX6 by transfection of the DDX6-1 siRNA as in Fig. 1B and, 24 h later, retransfected the cells with pEYFP-DDX6-m6 or a related empty vector (pEYFP, see Materials and Methods). The cells were then infected with HJ3-5 virus at an MOI of 0.5, and cell lysates were prepared 3 days later for immunoblotting. As in the prior experiments, transfection of the DDX6-1 siRNA resulted in decreased expression of endogenous DDX6, as well as reduced abundance of core and NS5A proteins expressed by HCV, compared to cells transfected with the DDX6-1m siRNA (Fig. 1D, compare lanes 4 and 8). Ectopic expression of the siRNA-resistant EYFP-DDX6-m6 partially restored the levels of core, as well as NS5A, in the cells in which endogenous DDX6 had been silenced (compare lanes 1 to 3 with lane 4). In addition, EYFP-DDX6-m6 overexpression slightly increased the abundance of NS5A and core proteins in cells transfected with the inactive, control DDX6-1m siRNA (compare lanes 5 to 7 with lane 8). These results confirm that the reduction in viral protein expression is caused specifically by DDX6 silencing and not by any spurious off-target effects of siRNA transfection.
DDX6 knockdown and global changes in cellular metabolism and proliferation.
DEAD-box RNA helicases, and in particular DDX6 and its various homologs in different vertebrate and invertebrate species, are involved in multiple aspects of RNA metabolism, including transcription, splicing, RNA processing, translation, storage, and degradation (8, 26, 46). Therefore, DDX6 silencing could have pleiotropic effects on cellular metabolism and growth. Since HCV replication is linked to the proliferation status of cells (31, 38), we assessed the impact of DDX6 silencing on the growth and metabolism of FT3-7 cells in a WST-1 reduction assay. Transfection of the DDX6-specific siRNAs resulted in minor reductions in WST-1 reduction (20 to 25%) after 72 to 96 h, compared to cells transfected with the siRNA controls (Fig. 1F). Similarly, DDX6 knockdown reduced cellular growth by 25% after 96 h, as determined by directly the counting cells (see below). Thus, silencing DDX6 results in only a slight reduction in the growth of cells.
As an additional measure of the general health of FT3-7 cells and their ability to support viral replication after DDX6 silencing, we evaluated the replication of an unrelated hepatotropic virus, HAV. Cells were transfected with DDX6-1 or DDX6-1m siRNA as described above and then infected with the HM175/18f strain of HAV at an MOI of 1.0. In contrast to the results we obtained with HCV, we observed no effect of DDX6 silencing on the cellular abundance of the HAV VP1-2A protein 5 days later (data not shown). Moreover, the yield of infectious HAV was not significantly reduced by DDX6 silencing (2.06 × 106 FFU/ml versus 1.81 × 106 FFU/ml). Combined with the data shown in Fig. 1F, these results suggest that DDX6 is specifically required for efficient replication of HCV and that the reductions in HCV replication observed after DDX6 silencing are unlikely to result from a general defect in cellular metabolism.
Overexpression of DDX6 stimulates HCV replication.
To determine whether overexpression of DDX6 enhances HCV replication, we monitored replication of virus in cells in which DDX6 was overexpressed as an N-terminal EYFP-fusion protein. There was little discernible difference in core, NS5A (data not shown), or NS3 expression levels in immunoblots (relative to a β-actin loading control, Fig. 2 A), but overexpression of EYFP-DDX6 resulted in a 2-fold increase in virus yields compared to cells transfected with a control EYFP vector (Fig. 2B). These results were reproducible in repeat experiments and support the findings in Fig. 1. While they confirm that DDX6 expression levels correlate with the efficiency of HCV replication, the small increases in virus yield observed with DDX6 overexpression suggest that the abundance of endogenous DDX6 is generally not limiting for viral replication in these cells.
FIG. 2.
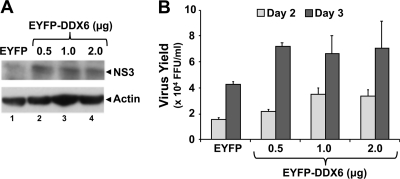
DDX6 overexpression stimulates HCV replication. FT3-7 cells were transfected with pEYFP-DDX6 or pEYFP plasmid DNA. Cells were then infected with HJ3-5 HCV at an MOI of 0.2 at 24 h posttransfection and were fed with fresh media every 24 h. (A) Cell lysates, prepared 3 days postinfection, were immunoblotted. (B) Virus supernatants collected 2 and 3 days postinfection were titrated on naive Huh-7.5 cells, in triplicate. The results from two independent experiments are presented here as means ± the SD.
DDX6 is not required for miR-122 facilitation of HCV replication.
A direct interaction of miR-122, a liver-specific miRNA, with HCV RNA is required for efficient genome amplification (19, 21). It is not known whether this also requires miR-122 recruitment of miRISC complexes, but silencing miRISC components such as the argonaute proteins (Ago1 to Ago4) inhibits HCV replication (36). Since DDX6 interacts with Ago1 and Ago2 (6), we considered the possibility that DDX6 might be required for miR-122 to promote replication. First, to determine whether DDX6 abundance influences miR-122 biogenesis, we isolated total cellular RNA 4 days after transfection of the siRNAs and subjected it to Northern blotting with 32P-labeled miR-122- and 5S rRNA-specific probes. As shown in Fig. 3A, we observed no changes in miR-122 abundance after transfection of the DDX6-specific siRNA (top panel), despite substantial reduction of DDX6 expression in these experiments (bottom panel). We next sought to determine whether the ability of miR-122 to promote HCV RNA accumulation is sustained in cells in which DDX6 expression had been silenced. We transfected FT3-7 cells with the DDX6 siRNAs described above and then supplemented the cells with miR-122, or with miR-124 as a negative control (21), by transfecting synthetic miRNA duplexes 48 h later (see Materials and Methods). At 54 h, the cells were infected with virus at an MOI of 0.2 and supplemented with miR-122 or miR-124 again at 90 h. Cell lysates were prepared at 120 h and subjected to immunoblotting. As anticipated, miR-122 supplementation substantially enhanced the abundance of the core and NS5A proteins in cells transfected with the control siRNA compared to similarly transfected cells supplemented with miR-124 (Fig. 3B, compare lanes 3 and 4). Core protein expression was reduced, as expected, in cells transfected with the DDX6-specific siRNA (Fig. 3B, compare lanes 2 and 4) but was nonetheless strongly upregulated by miR-122 supplementation (compare lanes 1 and 2) despite a high degree of DDX6 silencing. NS5A expression was also increased. These results indicate that the enhancement observed in viral replication after supplementation with miR-122 is not dependent upon DDX6, and miR-122 and DDX6 facilitate replication via independent mechanisms. Importantly, miR-122 supplementation fully compensated for the loss of DDX6 in the knockdown cells, bringing both core and NS5A abundance to levels exceeding that in the control cells (Fig. 3B, compare lanes 1 versus 4).
FIG. 3.
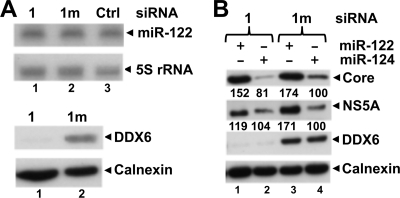
DDX6 is not required for miR-122 facilitation of HCV replication. (A) FT3-7 cells were transfected with 1 (DDX6-1) or 1m (DDX6-1m) siRNA, and cell lysates were prepared 4 days later. Total RNA was subjected to Northern blotting to detect miR-122 and 5S rRNA (used as a loading control) using 32P-labeled riboprobes (top panel). Protein extracts were immunoblotted for DDX6 and calnexin (bottom panel) (B) FT3-7 cells were transfected with 1 (DDX6-1) or 1m (DDX6-1m) siRNA. After 48 h, the cells were transfected with miR-122 or miR-124 (control miRNA) and infected with HJ3-5 HCV (MOI = 0.2) at 54 h. On day 4, the cells were transfected with another dose of miR-122 or miR-124. Cell lysates were prepared on the next day and subjected to Western blotting. Band intensities were measured as described in Fig. 1B and are represented as a percentage of DDX6-1m/miR-124 transfected cells.
DDX6 forms a complex containing HCV RNA and core protein.
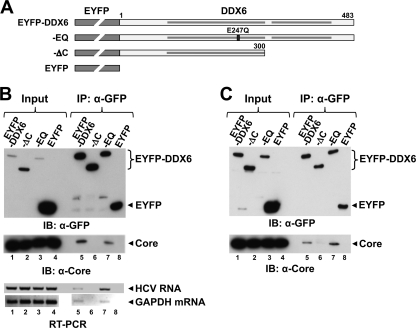
To better understand the mechanism by which DDX6 facilitates the replication of HCV RNAs, we carried out coimmunoprecipitation experiments to assess whether DDX6 might interact with one or more viral proteins. FT3-7 cells were infected with HJ3-5 virus and cultured until more than 90% of the cells were positive for core protein expression as determined by immunostaining. The cells were then transfected with the pEYFP-DDX6-wt or pEYFP expression vectors, and cell lysates were prepared 2 days later for coimmunoprecipitation experiments. EYFP-DDX6 was efficiently immunoprecipitated with polyclonal rabbit anti-GFP antibody (Fig. 4A, left panel, lane 3). Immunoblotting of these precipitates with HCV-specific antibodies revealed the presence of core protein (Fig. 4A, bottom right panel, lane 3), but not NS3 (Fig. 4A, top right panel, lanes 3 and 4), NS5A, or NS5B (data not shown). Under the stringent conditions used for this coimmunoprecipitation experiment, only a small fraction of the total intracellular core protein coimmunoprecipitated with DDX6. However, the coimmunoprecipitation of core was specific to DDX6, since it was not observed in EYFP precipitates (Fig. 4A, right panel, compare lanes 3 and 4). We next confirmed the existence of a complex containing endogenous DDX6 and core protein by immunoprecipitating the core protein from lysates of virus-infected cells using a murine monoclonal antibody and blotting these precipitates with antibody to DDX6. As expected, endogenous DDX6 coimmunoprecipitated with the core protein (Fig. 4B, bottom panel, lane 2), while an irrelevant isotype control antibody precipitated neither the core protein nor DDX6 (lane 3).
FIG. 4.
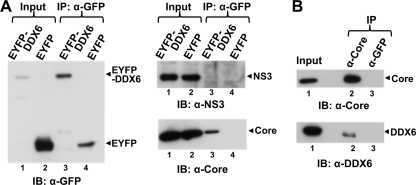
DDX6 forms a complex with the HCV core protein. (A) HJ3-5 HCV-infected FT3-7 cells were transfected with DNA vector expressing wild-type EYFP-DDX6 or EYFP alone. Cell lysates were prepared 2 days later and subjected to coimmunoprecipitation with anti-GFP (rabbit polyclonal; Clontech) antibody. Coimmunoprecipitation samples were immunoblotted with GFP (mouse monoclonal; Clontech). Anti-GFP precipitates were probed with antibody to NS3 (top panel) or HCV core (bottom right panel). Input represents 1/20 of the IP for the anti-GFP blot and 1/80 of the IP for the NS3 and core blots. (B) Cell lysates prepared from HJ3-5 HCV-infected FT3-7 cells were subjected to coimmunoprecipitation with HCV core (C7-50; Affinity BioReagents) or GFP-specific (mouse monoclonal [Clontech]; used as an isotype control) antibody and immunoblotted for HCV core (upper panel) or endogenous DDX6 (lower panel). Input represents 1/20 and 1/80 of the IP for the anti-HCV core and anti-DDX6 blots, respectively.
To further characterize this complex, we generated an expression vector encoding a C-terminal DDX6 deletion mutant (pEYFP-DDX6-ΔC or “ΔC”) lacking the C-terminal 183 amino acid residues (1). This deletion removes the second of two RecA-like P-loop NTPase superfamily domains in DDX6 and, based on studies with related DEAD-box family members, would be expected to ablate ATPase and helicase activity (46). We also constructed a second mutant (pEYFP-DDX6-EQ or “EQ”) that lacks helicase activity due to a point mutation that replaces the Glu in the DExD-box motif (motif II) with Gln (46) (Fig. 5A). We transfected the wild-type (wt, pEYFP-DDX6-wt), ΔC, and EQ mutant expression vectors into virus-infected cells as described above and prepared lysates 2 days later for coimmunoprecipitation studies. Similar to the results shown in Fig. 4A, wt DDX6 coimmunoprecipitated with core, whereas the ΔC mutant did not (Fig. 5B, lane 5 versus lane 6), indicating that the C-terminal 183 amino acids are required for complex formation with the core protein. In contrast, the helicase active-site mutant (EQ) was coimmunoprecipitated in a fashion similar to wt DDX6 (Fig. 5B, lane 7). To investigate whether other viral proteins are required for DDX6 to form a complex with core, we carried out a similar experiment using Huh-7/191-20 cells that conditionally express only the core protein (amino acids 1 to 191 of the HCV polyprotein) under the control of the Tet-Off promoter (24). After inducing the cells to express core protein, they were transfected with the DDX6 expression vectors as described above. Both the wt and the EQ mutant were coimmunoprecipitated with the core protein, whereas the ΔC mutant was not (Fig. 5C, lanes 5 and 7 versus lane 6). Thus, there is no requirement for other viral proteins to form complexes containing DDX6 and core, and (as indicated above) nonstructural viral proteins known to be important for viral replication (NS3, NS5A, and NS5B) do not appear to participate in this complex.
FIG. 5.
The interaction of DDX6 with HCV core and viral RNA is dependent on the C-terminal domain of DDX6. (A) Schematic representation of EYFP-DDX6 and related mutants, ΔC (C-terminal deletion mutant lacking 183 amino acids), EQ (mutation in the DEAD-box helicase motif II involving the substitution of Glu-247 by Gln), and empty vector expressing EYFP alone. The ΔC mutation removes the second of two conserved RecA domains, identified by the shaded bars within the DDX6 sequence. (B) HJ3-5 HCV-infected FT3-7 cells were transfected with DNA vector expressing various forms of EYFP-DDX6 or just the EYFP. Cell lysates were prepared 2 days later and subjected to coimmunoprecipitation with anti-GFP (rabbit polyclonal; Clontech) antibody. Coimmunoprecipitation samples were immunoblotted with GFP (top panel) and core-specific (bottom panel) antibodies. Input represents 1/20 of the immunoprecipitation (IP) for anti-GFP blot and 1/80 of the IP for HCV core blot. Total RNA isolated from immunoprecipitates using an RNeasy minikit (Qiagen) was subjected to RT-PCR for detection of genomic HCV RNA (primers targeting the NS3 region) and GAPDH mRNA. (C) Huh-7-191/20 cells were induced to express HCV core protein by removing tetracycline from the medium for 3 days. Cells were then transfected with various DNAs and subjected to coimmunoprecipitation exactly as described for panel B.
To determine whether the DDX6-core complex also contains viral RNA, we extracted RNA from immunoprecipitates prepared from virus-infected and pEYFP-DDX6-transfected cells using anti-GFP and subjected it to reverse transcription-PCR (RT-PCR) using HCV-specific primers targeting the NS3 region of the genome (49). Although the amount varied between individual experiments, HCV RNA was consistently detected in precipitates of the wt and EQ mutant EYFP-DDX6 proteins but not in precipitates of the ΔC mutant or EYFP (Fig. 5B, bottom panel). However, additional RT-PCR assays demonstrated that cellular (GAPDH) mRNA was also present in the wt and EQ mutant EYFP-DDX6 precipitates (Fig. 5B, bottom panel), indicating that the association of DDX6 with RNA is not specific to HCV RNA.
Taken together, the data shown in Fig. 4 and 5 indicate that DDX6 forms complexes containing core protein as well as viral and cellular RNAs in HCV-infected cells and that the formation of these complexes is dependent on the C-terminal domain of DDX6 but independent of helicase activity. Although this may reflect a direct interaction between core and DDX6, it is also possible that the coimmunoprecipitation of core and DDX6 may reflect binding to a common set of viral or cellular RNAs since the core protein has independent RNA-binding activity (41). To assess this possibility, we digested cell extracts with RNase prior to antibody precipitation. Unfortunately, the results of these experiments were inconclusive, since RNase digestion substantially enhanced nonspecific interactions of core and resulted in its precipitation with irrelevant isotype control antibodies (data not shown). We also cannot exclude the possibility that core and DDX6 are bridged by a third, host-encoded protein partner. This is not unlikely, since DDX6 and its closely related homologs interact with numerous cellular proteins (46).
DDX6 facilitation of viral replication is independent of core protein expression.
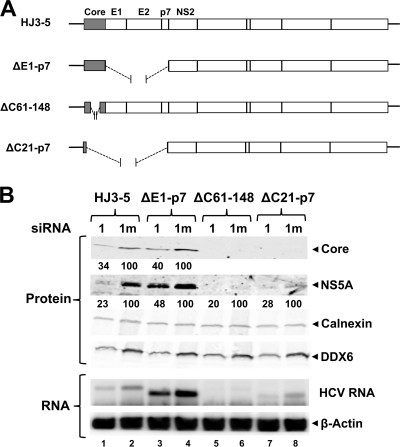
To ascertain whether the influence of DDX6 expression on HCV replication might be functionally related to the DDX6-core protein complex identified above, we studied the effect of DDX6 silencing on replication of several subgenomic HCV RNAs that contain in-frame deletions removing all or part of the core-coding sequence. These included in-frame deletions of amino acid residues 61 to 148 within the core protein (ΔC61-148) (35) or sequence extending from amino acid residue 21 of core to the end of p7 (ΔC21-p7) (Fig. 6A). As a control, we also studied a mutant with an in-frame deletion spanning the E1 to p7 sequence but leaving the core sequence intact (ΔE1-p7) (49). Importantly, the core protein is not required for HCV genome amplification (27). Thus, we reasoned that we would observe no effect of DDX6 silencing on replication of viral RNAs that do not encode core if DDX6 facilitation of viral replication was mediated through the core-DDX6 complex described above.
FIG. 6.
DDX6 promotes HCV replication by enhancing genome amplification. (A) Schematic representation of wild-type HJ3-5, HJ3-5-ΔE1-p7 (lacks E1, E2, and p7), HJ3-5-ΔC61-148 (lacks amino acids 61 to 148 of core protein), and ΔC21-p7 (containing the first 20 amino acids of core fused in-frame with NS2 to 5B genes). (B) FT3-7 cells were transfected with 1 (DDX6-1) or 1m (DDX6-1m) siRNA. After 48 h, cells were transfected with the indicated HJ3-5 HCV RNA using TransIT mRNA transfection reagent (Mirus Bio). Immunoblotting of cell lysates prepared 3 days post-HCV RNA transfection is shown. HCV core and NS5A expression levels were quantitated on an Odyssey infrared imaging system (LI-COR) and are represented as the ratio of that in 1 (DDX6-1) siRNA-treated cells and in control, 1m (DDX6-1m) siRNA-treated cells (set as 100) after normalization to calnexin levels.
We transfected FT3-7 cells with the DDX6-1 and DDX6-1m siRNAs and then supertransfected the cells 2 days later with the full-length or mutant subgenomic RNAs. As before, DDX6 knockdown reduced the abundance of core (34% of control siRNA-treated cells) and NS5A (23%) expressed by wt HJ3-5 RNA (Fig. 6B, compare lane 1 with lane 2). Surprisingly, the ΔC61-148 RNA, which contains a large internal deletion in the core gene, replicated very poorly even in the absence of DDX6 knockdown, as evidenced by very low level expression of NS5A protein and little accumulation of viral RNA (Fig. 6B, compare lane 6 to lane 2). However, the levels of NS5A were reduced even further after DDX6 knockdown (20% of that of the DDX6-1m siRNA-treated cells) (Fig. 6B, compare lanes 5 and 6), suggesting that DDX6 silencing further impairs the replication efficiency of this RNA despite the large internal deletion within core. The ΔC21-p7 RNA, which expresses only the first 20 amino acids of core, which were retained to facilitate IRES activity (44), replicated more efficiently than ΔC61-148 (Fig. 6B, compare lanes 6 and 8) but was also impaired by DDX6 silencing, which reduced both viral protein and RNA levels (Fig. 6B, compare lane 7 to lane 8). These results suggest that the facilitation of HCV replication by DDX6 occurs independently of core expression and is thus independent of the ability of DDX6 to form a complex with core.
The ΔE1-p7 RNA expresses the full-length core protein (Fig. 6A) but lacks the envelope protein sequence and cannot produce infectious virus (49). Its replication, judged by expression of core and NS5A, was also reduced by DDX6 silencing (Fig. 6B, compare lane 3 to lane 4). Since neither this RNA nor the other core deletion variants described above can produce infectious virus, these results also show that DDX6 functions primarily to enhance genome amplification by promoting the translation, transcription, or stability of the viral RNA and not by facilitating other steps in the viral life cycle (such as viral assembly, release, entry, etc.).
DDX6 helicase activity is essential to promote HCV replication.
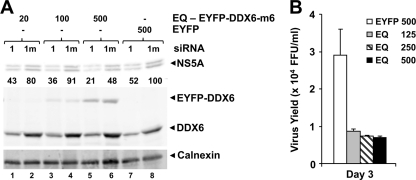
Although the EYFP-DDX6-EQ mutant, which lacks helicase activity (46), was capable of forming a complex with the core protein (Fig. 5), overexpression of this protein failed to enhance HCV replication (Fig. 7), as we had observed previously with wt EYFP-DDX6 (Fig. 2). In contrast, transient overexpression of EYFP-DDX6-EQ led to a reproducible decrease in HCV replication, as demonstrated by reduced levels of HCV core and NS5A protein expression (data not shown) and decreased virus yields (Fig. 7B). Reduced production of infectious virus was also observed in EYFP-DDX6-m6-EQ-expressing cells in which endogenous DDX6 had been silenced by transfection of DDX6-1 siRNA (Fig. 7A, compare lanes 1, 3, and 5 to lane 7). In aggregate, these data indicate that the helicase activity of DDX6 is essential to its ability to facilitate viral replication and furthermore suggest that the DDX6-EQ mutant exerts a dominant-negative effect on viral replication. In contrast, overexpression of the EYFP-DDX6 ΔC mutant had no significant effect, positive or negative, on viral replication (data not shown).
FIG. 7.
DDX6-EQ has a dominant-negative effect on HCV replication. (A) FT3-7 cells were transfected with DDX6-1 (lanes 1) or DDX6-1m (lanes 1m) siRNA and 48 h later retransfected with increasing quantities of expression vectors encoding siRNA-resistant EYFP-DDX6-m6-EQ or EYFP. One day later, the cells were infected with HJ3-5 HCV (MOI = 0.5). Cell lysates prepared 3 days after infection were immunoblotted for HCV NS5A, calnexin, and DDX6 proteins. NS5A band densities were quantified as described in Fig. 6A and are presented as the percent expression relative to DDX6-1m/pEYFP-transfected cells. (B) FT3-7 cells were transfected with pEYFP-DDX6-EQ or pEYFP plasmid DNA. Cells were infected with HJ3-5 HCV at an MOI of 0.2 at 24 h posttransfection and fed with fresh medium every 24 h. Virus supernatants collected 2 and 3 days postinfection were titrated on naive Huh-7.5 cells, in triplicate. Representative results from one of multiple experiments are presented here as means ± the SD.
The DDX6-EQ and ΔC mutants have altered cytoplasmic distribution.
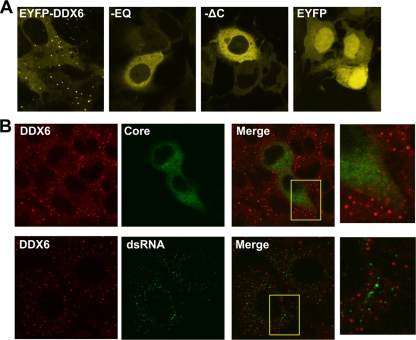
To better understand the inability of the EYFP-DDX6 mutants to facilitate virus replication, we utilized confocal laser scanning microscopy to determine their subcellular localization in transfected FT3-7 cells. DDX6 is a component of P-bodies, cytoplasmic structures that are intimately involved in mRNA metabolism, degradation, and storage (10). Consistent with this, we found that wt EYFP-DDX6 localized primarily to perinuclear, punctate P-body-like structures with only low-level, diffuse background staining in the cytoplasm. We confirmed that the punctuate structures were P-bodies by demonstrating colocalization of EYFP-DDX6 with N-terminal ECFP-tagged Ago2 (ECFP-Ago2) in cotransfected FT3-7 cells (data not shown). In contrast, both the EQ and the ΔC EYFP-DDX6 mutants were distributed diffusely throughout the cytoplasm, with little if any localization to distinct P-body-like structures (see EQ and ΔC panels, Fig. 8A). These results differ from a recent report (29), which suggested that the mutation of motif II (EQ) in the Xenopus homolog of DDX6, Xp54, causes only a reduction and not the complete loss of DDX6 localization to P-bodies that we observed. This discrepancy may reflect differences in the cell types studied and the dynamics of P-body turnover. Xp54-EQ did not support the assembly of new P-bodies and yet remained associated with previously assembled P bodies (29). Importantly, the lack of localization of DDX6-EQ to P-bodies in our studies indicates that the DDX6-core complex (which EYFP-DDX6-EQ retains the ability to form, as shown in Fig. 5) is not likely to be associated with P-bodies.
FIG. 8.
DDX6-EQ and DDX6-ΔC have altered subcellular distributions. (A) FT3-7 cells were transfected with pEYFP-DDX6, pEYFP-DDX6-EQ, pEYFP-DDX6-ΔC, or pEYFP vector DNA. The cells were fixed with 4% paraformaldehyde 2 days later and imaged by confocal laser scanning microscopy for EYFP fluorescence. Wild-type DDX6 was localized to punctate structures identified as P-bodies, while both mutants were distributed diffusely within the cytoplasm and EYFP alone entered nuclei. (B) FT3-7 cells were infected with HJ3-5 virus at an MOI of 0.2. Two days later, the cells were fixed with 4% paraformaldehyde, permeabilized with digitonin, and stained with antibodies specific for DDX6 and HCV core (top panels) or DDX6 and dsRNA (bottom panels). In the top row of panels, HCV-infected cells are identified by cytoplasmic staining for core antigen (green) and demonstrated a moderate reduction in the number of P-bodies (identified by staining for endogenous DDX6). In the bottom row of panels, punctate staining for dsRNA (green) shows the location of replicating RNA in infected cells (no such staining was observed in uninfected cells). Importantly, dsRNA did not localize to P-bodies. Expanded views of areas from the merged images (yellow boxes) are shown to the right.
We next determined the intracellular localization of endogenous DDX6 in relation to viral proteins and replicating HCV double-stranded RNA (dsRNA). Virus-infected cells were fixed and permeabilized, immunolabeled with antibodies to core and DDX6, and studied by confocal laser scanning microscopy. As with ectopically expressed wt EYFP-DDX6 (Fig. 8A), these studies revealed endogenous DDX6 to be localized to intensely stained, punctuate perinuclear cytoplasmic structures consistent with P-bodies (Fig. 8B). However, low-level, diffuse DDX6-specific labeling was also evident in the cytosol, again reflecting what was observed with EYFP-DDX6 (compare Fig. 8A and B). Consistent with previous studies, core was expressed with a granular, cytoplasmic distribution (Fig. 8B). Despite the evidence for formation of core-DDX6 complexes in coimmunoprecipitation experiments (Fig. 4 and 5), there was no significant colocalization of DDX6 with core in these images. This suggests that only a small fraction of the total core protein resides in complexes with DDX6 (which is consistent with the immunoprecipitation results in Fig. 4) and, as described in the preceding paragraph, the fact that the DDX6-core complex is unlikely to be associated with P bodies. As might be expected from the coimmunoprecipitation experiments, we also observed no colocalization of DDX6 with the viral NS3 or NS5A proteins (data not shown).
dsRNA is produced during replication of HCV. It can be detected by labeling infected cells with dsRNA-specific antibodies and colocalizes with membrane-bound viral replicase complexes at the site of active viral RNA synthesis (42). HCV-specific dsRNA has also been demonstrated by microscopy within hepatocytes in frozen sections of liver biopsies from patients with chronic hepatitis C (25). We probed infected cells for the presence of dsRNA using a monoclonal antibody that recognizes dsRNA segments greater than 40 bp in length in a sequence-independent fashion (5) and that does not label any dsRNA in uninfected cells (data not shown). Dual staining of infected FT3-7 cells for DDX6 and dsRNA (Fig. 8B) revealed most dsRNA to be localized to punctate cytoplasmic foci which were distinct from the P bodies in which the DDX6 abundance was concentrated. These results suggest that HCV replication does not occur in close association with P-bodies, despite the localization of DDX6 to P-bodies and the influence of DDX6 on HCV replication. However, these studies do not exclude the presence of small amounts of DDX6 at sites of new HCV RNA synthesis.
DDX6 does not promote HCV translation.
The results described above indicate that the helicase activity of DDX6 facilitates HCV replication but leave unresolved the mechanism by which this occurs. One possible explanation would be the specific promotion of viral translation by DDX6 as suggested recently by Scheller et al. (37). HCV translation is directed by an IRES located within the 5′UTR that requires few canonical host translation initiation factors (34). To assess its efficiency, we used a reporter plasmid (pRLHL) (16) that produces dicistronic transcripts under the control of the cytomegalovirus promoter in which the upstream cistron encodes Renilla luciferase (RLuc) and the downstream cistron encodes firefly luciferase (FLuc), with the HCV IRES placed in the intercistronic space (Fig. 9A). The 3′ end of the pRLHL transcript is polyadenylated. RLuc is thus translated by cap-dependent translation, whereas the translation of FLuc is directed by the HCV IRES. We transfected FT3-7 cells with the DDX6-1 or mutant DDX6-1m siRNAs, supertransfected the cells with the reporter plasmid, and prepared cell lysates for testing in a dual luciferase assay when the silencing of DDX6 was maximal. We found that DDX6 knockdown resulted in a decrease in both RLuc and FLuc expression, with IRES-directed translation affected to a greater extent (Fig. 9B). In these experiments, the ratio of FLuc to RLuc reflects the activity of the HCV IRES relative to cap-dependent translation of the same transcript and thus provides a measure of IRES activity that is independent of any differences in transfection efficiency. Concerns over possible changes in RNA stability are also eliminated since both FLuc and RLuc are expressed from the same RNA molecule. This analysis suggested that IRES activity was reduced by no more than 25 to 30% relative to cap-dependent translation after DDX6 silencing (Fig. 9B). We obtained similar results in Huh-7/191-20 cells that express core conditionally, and found no consistent effect of core protein expression on either HCV translation or the response to DDX6 knockdown (data not shown).
FIG. 9.
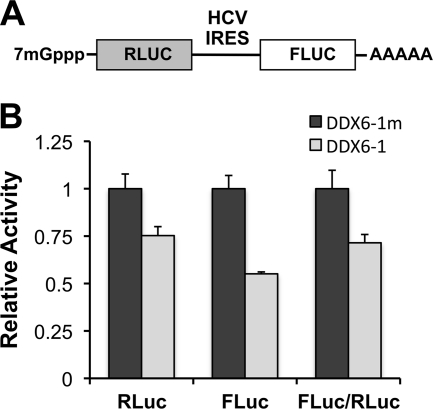
Influence of DDX6 knockdown on translation of a dicistronic RNA containing the HCV IRES. (A) Schematic representation of the pRLHL plasmid that expresses a dicistronic RNA containing Renilla luciferase sequence in the first cistron and firefly luciferase sequence in its second cistron, separated by the IRES of HCV. (B) pRLHL DNA was transfected into FT3-7 cells 64 h after transfection of the indicated siRNAs. Cells were harvested 48 h later for dual luciferase assays. The results shown represent RLuc (cap-dependent translation) and FLuc (IRES-directed translation) activities and the FLuc/RLuc ratio (relative IRES activity), normalized to that in DDX6-1m-transfected cells (n = 3, means ± the SD).
To further explore the effect of DDX6 silencing on HCV translation, we inserted the RLuc coding sequence (fused at its C terminus to the foot-and-mouth disease virus 2A autoprotease) between the p7 and NS2 coding sequences of HJ3-5 to generate a replication-competent, full-length HCV genome expressing the reporter protein as part of its polyprotein (HJ3-5/RLuc2A). We then constructed a replication-defective variant by creating an Asn substitution within the GDD motif of the NS5B polymerase (HJ3-5/RLuc2A-GND) (Fig. 10A). After silencing DDX6 expression by transfection of DDX6-specific siRNA (Fig. 10B), we transfected FT3-7 cells with this genome-length, replication-defective reporter RNA in combination with a capped, polyadenylated mRNA encoding FLuc as an internal control for transfection and cap-dependent translation. Under these conditions, DDX6 silencing reduced FLuc activity expressed from the capped mRNA by ca. 50% (Fig. 10C). In contrast, RLuc expression from the HCV RNA was maintained at close to 100% that of cells transfected with the control DDX6-1m siRNA. When the RLuc activities were normalized to the FLuc values, to account for potential differences in transfection efficiency, the results suggested a 2-fold increase in the relative translational efficiency of the HCV IRES after DDX6 knockdown (Fig. 10C). The explanation for the difference in the results shown in Fig. 9B and 10C (a small decrease versus a 2-fold increase in IRES-directed translation relative to cap-dependent translation) is uncertain, but it may reflect differences in the 3′UTR sequences of the RNA transcripts used in these experiments. Similar experiments were done in HeLa cells, in which DDX6 knockdown was very efficient (Fig. 10B). We observed no significant changes in expression of either FLuc or RLuc in HeLa cells, and no apparent change in the relative efficiency of translation directed by the HCV IRES.
FIG. 10.
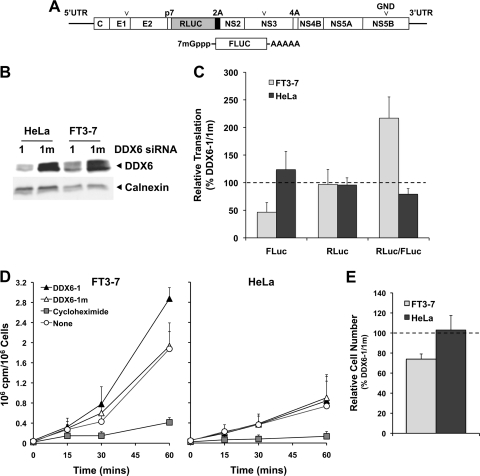
DDX6 regulation of HCV translation in FT3-7 and HeLa cells. (A) Schematic showing the genome organization of the HJ3-5/RLuc2A HCV which contains an in-frame insertion of Renilla luciferase (RLuc) sequence, fused at its C terminus to the foot-and-mouth disease virus 2A autoprotease, between the p7 and NS2 coding sequences of HJ3-5 virus. The RNA also contains a replication-lethal mutation (GND) in the NS5B polymerase sequence. The 5′ and 3′ ends of this RNA are the authentic viral UTRs. Below is shown the capped FLuc mRNA that contains a 3′ poly(A) tail of 30 adenosine residues that was used as an internal control for transfection and translation efficiencies. (B) Immunoblot showing efficient DDX6 knockdown after transfection of either FT3-7 or HeLa cells with the DDX6-1 siRNA. (C) Impact of DDX6 knockdown on HCV IRES-directed translation. FT3-7 or HeLa cells were transfected with DDX6 siRNAs and 4 days later were retransfected with HJ3-5/RLuc2A/GND and FLuc mRNAs. Cells were harvested 8 h later and assayed for FLuc and RLuc activities. The results shown represent the mean percentages ± the range of FLuc and RLuc activities and the RLuc/FLuc ratio, obtained in duplicate DDX6-1-transfected cultures, normalized to those obtained in DDX6-1m-transfected cells, and are representative of results from multiple experiments. (D) Metabolic labeling of FT3-7 (left panel) and HeLa (right panel) cells after DDX6 knockdown. Cells, transfected 4 days previously with siRNAs as described in panel C, were cultured for 1 h in methionine- and cysteine-free medium and then incubated with 100 μCi of Tran35S-label (MP Biomedicals)/ml. Cells were harvested at the times indicated, and [35S]Met present in trichloroacetic acid precipitate measured in a scintillation counter. The results from three independent experiments are presented as cpm incorporated/106 cells (means ± the SD). (E) Impact of DDX6 knockdown on the proliferation of FT3-7 and HeLa cells. Cells were enumerated 96 h after transfection with DDX6 siRNAs. The results are shown as the ratio of the number of cells in the DDX6-1-transfected cultures relative to those transfected with the mutant DDX6-1m siRNA at the end of this growth period (means ± the SD, n = 3).
Because the decrease observed in translation of capped mRNAs in FT3-7 cells in both Fig. 9 and Fig. 10 is contrary to the notion that DDX6 is a general translational repressor (6, 7), we examined the overall effect of DDX6 knockdown on cellular translation during metabolic labeling with [35S]Met (6). Despite the reduction observed in translation of the transfected capped reporter RNAs, DDX6 knockdown resulted in a 50% increase in [35S]Met incorporation compared to FT3-7 cells transfected with the control siRNA, DDX6-1m (Fig. 10D). These results confirm that DDX6 exerts a general repressive effect on cellular translation in FT7-3 cells. The decreased FLuc activity observed with the transfected reporter RNAs is likely to reflect enhanced competition for ribosomes or translation factors by endogenous mRNAs after DDX6 knockdown. DDX6 knockdown had little effect on [35S]Met incorporation in HeLa cells (Fig. 10D), a finding consistent with the absence of an effect on translation of the capped RNAs in these cells (Fig. 10C). In aggregate, however, the data shown in Fig. 10C indicate that translation directed by the HCV IRES, when placed naturally within the context of genomic RNA with authentic 5′UTR and 3′UTR sequences, is less sensitive to changes in DDX6 abundance than cap-dependent translation of a polyadenylated transcript. Thus, the inhibition of virus replication and infectious virus yield that is observed in cells subjected to siRNA-mediated DDX6 knockdown (Fig. 1C) is not associated with a specific impairment of HCV RNA translation.
In addition to measuring [35S]Met incorporation, we assessed the impact of DDX6 knockdown on the proliferation of FT3-7 and HeLa cells in these experiments. Interestingly, despite the increase observed in [35S]Met incorporation, FT3-7 cells were either slowed in their growth or had an increased rate of death, as we observed a 25% reduction in the number of cells present after a 96-h incubation period compared to cells transfected with the control siRNA (Fig. 10E). This result is highly concordant with the WST-1 reduction assay results described above (Fig. 1F). Interesting, DDX6 knockdown had no effect on the proliferation or survival of HeLa cells.
DISCUSSION
The studies we describe here provide evidence that DDX6, a member of the DEAD-box RNA helicase family, is required for efficient replication of HCV in cultured hepatoma cells. DDX6 is evolutionarily highly conserved from yeast to vertebrates and involved in multiple aspects of RNA metabolism. We used RNA interference to knockdown DDX6 expression, and found that this substantially reduced HCV replication in Huh-7 cells (Fig. 1B-C and 1E). We confirmed a specific requirement for DDX6 by rescuing viral replication by ectopically expressing an siRNA-resistant DDX6 mutant in cells in which the expression of endogenous DDX6 had been silenced (Fig. 1D). We also found the requirement of DDX6 to be HCV-specific, as viral protein expression and infectious virus yields of another hepatotropic, positive-strand RNA virus, HAV (a picornavirus), were not reduced by DDX6 knockdown. DDX6 promotes genome amplification and not other aspects of the viral life cycle, since knockdown of DDX6 degrades the replication efficiency of viral RNAs with deletion mutations in structural proteins that preclude assembly of virus particles (Fig. 6).
DDX6 plays an important but incompletely understood role in cellular RNA metabolism, and an important concern in evaluating these results was that its knockdown by siRNA might have pleiotropic effects. This concern is magnified by the fact that HCV RNA synthesis is typically dependent on cellular proliferation (31, 38). Metabolic assays demonstrated that DDX6 knockdown has a relatively modest effect on the growth of Huh-7 cells, reducing both WST-1 reduction and cell proliferation over a 96 h period by only ∼25% (Fig. 1F and 10D). Whether this degree of cytotoxicity is sufficient to account for the 8-fold reduction in infectious virus yield observed in these cells (Fig. 1C) is difficult to determine, yet central to the interpretation of these results. To shed light on this question, we carried out further experiments aimed at determining whether DDX6 interacts specifically with any viral proteins, or is colocalized in cells with components of the viral replicase.
Coimmunoprecipitation experiments indicated that DDX6 forms an intracellular complex that involves the HCV core protein (Fig. 4), and that the C-terminal domain 2 (but not the helicase activity) of DDX6 is essential for this (Fig. 5). These complexes form in virus-infected cells, and also in cells expressing only core protein. They do not contain nonstructural proteins of the virus (NS3, NS5A, or NS5B), but are associated with both viral and cellular RNAs (Fig. 5). The existence of these complexes is of doubtful relevance to the requirement for DDX6 in HCV genome amplification, as subgenomic HCV RNAs that do not express core protein also demonstrate dependence on DDX6 expression in transient replication assays (Fig. 6) as well as in stable cell lines expressing HCV replicons (data not shown). This provides an interesting parallel to DDX3, another DEAD-box RNA helicase that also interacts with core (32). Although DDX3 is essential for efficient HCV replication, recent studies indicate that this is not dependent on its interaction with core (3, 4). Importantly, core expression leads to the redistribution of DDX3 within the cytoplasm and colocalization with core. We did not see any significant change in the cellular distribution of DDX6 in cells expressing core, nor could we demonstrate colocalization of these proteins by confocal microscopy (Fig. 8). In both infected and uninfected cells, DDX6 was primarily localized to discrete, punctuate, perinuclear structures identified as P-bodies. The number and size of the P-bodies appeared to be decreased in cells infected with HCV (see Fig. 8B), but there was no association of core protein or viral dsRNA with these structures. On the other hand, both the EYFP-DDX6-EQ as well as EYFP-DDX6-ΔC mutants demonstrated a diffuse cytoplasmic distribution, and were only poorly localized to P-bodies (Fig. 8). This is consistent with a recent study showing that multiple conserved domains within the Xenopus homolog of DDX6, Xp54, are involved in recruitment of the protein to P-bodies (29).
Like the wild-type EYFP-DDX6, both viral and cellular RNAs coimmunoprecipitated from infected cell lysates with EYFP-DDX6-EQ, which lacks helicase activity, while this was not the case with EYFP-DDX6-ΔC (Fig. 5B). The RNA-binding activity of DEAD-box helicases is generally nonspecific, functionally localized to the C-terminal RecA homology domain, and not affected by mutations such as the EQ mutation in motif II of the protein (8). Since core is also a promiscuous RNA-binding protein (41), it seems likely that the complex we observed reflects binding to common RNAs rather than true protein-protein interactions. Our efforts to demonstrate an RNA-independent interaction between DDX6 and core were inconclusive, as the core protein precipitated in a nonspecific fashion with several different antibodies following RNase A or T1 digestion of lysates (data not shown). Both wild-type DDX6 and the EQ mutant formed a complex with core (Fig. 5B and C), but only wild-type DDX6 upregulated viral replication (Fig. 2). In contrast, EYFP-DDX6-EQ appeared to have a dominant-negative effect on viral replication, as its overexpression resulted in reduced viral protein abundance and infectious virus yields (Fig. 7). This indicates that the helicase activity of DDX6 somehow facilitates replication of HCV RNA, but leaves unanswered the question whether this is a direct effect on the replicase or one that is exerted through one or more changes in cellular homeostasis resulting from loss of DDX6 helicase activity. Importantly, the capacity of DDX6 to facilitate HCV replication may be linked to its ability to localize to P bodies, since both the EQ and ΔC mutants failed to localize to P bodies.
How could DDX6 facilitate HCV replication? DDX6 is a part of the miRISC complex and miR-122, a liver-specific miRNA, is required for efficient replication of HCV (20, 21). However, DDX6 knockdown did not affect the abundance of miR-122 (Fig. 3A), and miR-122 supplementation reversed the defect in HCV replication observed after DDX6 knockdown (Fig. 3B). We conclude from these results that the ability of miR-122 to stimulate the accumulation of HCV RNA is not dependent upon DDX6, and that these two host factors promote HCV replication by independent and possibly redundant mechanisms.
While this work was on-going, Scheller et al. (37) reported that the knockdown of several proteins involved in regulating the fate of cellular mRNA, including PatL1, Lsm1-7, and Rck/p54 (DDX6), impaired the replication of HCV RNAs. In transient-transfection experiments, DDX6 knockdown caused a 5-fold reduction in replication of subgenomic RNAs and an ∼30-fold decrease in infectious virus yield that these authors attributed at least in part to impaired translation directed by the HCV IRES (37). Our results do not support this conclusion. While we found that DDX6 knockdown decreased translation of a reporter protein under the control of the HCV IRES placed in a dicistronic RNA, translation of the upstream cistron of this capped RNA was also decreased, resulting in only a modest net decrease in relative IRES activity (Fig. 9). More importantly, translation directed by the HCV IRES in its natural context in a genome-length RNA with authentic 5′ and 3′UTR sequences, monitored by expression of an reporter protein embedded in the polyprotein, was not reduced by DDX6 knockdown in either Huh7 or HeLa cells (Fig. 10). To control for variation in transfection efficiency in this experiment, we cotransfected a capped and polyadenylated reporter mRNA with the viral RNA. In Huh-7 cells we consistently observed decreases in FLuc expression from this control RNA following DDX6 knockdown, but no changes in IRES-directed translation of RLuc embedded in the viral polyprotein, suggesting that IRES-directed translation may actually be favored in these cells after knockdown of DDX6. DDX6 knockdown caused little change in translation of either reporter in HeLa cells (Fig. 10). In aggregate, the results shown in Fig. 10 argue strongly against a specific requirement for DDX6 for HCV IRES-mediated translation. These results are consistent with the observation that DDX6 knockdown has no effect on translation directed by the classical swine fever virus IRES (29), which is closely related both structurally and functionally to the HCV IRES. We also found that DDX6 knockdown had no effect on the replication of HAV, a picornavirus in which translation is directed by an IRES that is heavily dependent upon cellular translation factors for its activity.
To determine the overall impact of DDX6 knockdown on cellular homeostasis, we monitored [35S] incorporation as a measure of cellular translation and also determined the effect of gene knockdown on WST-1 reduction and cellular proliferation. Interestingly, in Huh7 cells, DDX6 knockdown upregulated translation generally but somewhat paradoxically slowed the rate of cellular proliferation (Fig. 1F, 10D, and 10E). This is likely to reflect a general perturbation in cellular control of translation and perhaps P-body-related functions of mRNA storage and degradation. It is easy to envision how virus replication could become less efficient in this setting, making it possible that the impaired HCV replication observed both by us and by Scheller et al. (37) may stem from such general effects of DDX6 knockdown.
An alternative possibility is suggested by the fact that Dhh1p, the yeast homologue of DDX6, is required both for translation of BMV proteins as well as replication of the BMV genome in yeast cells (2). Human DDX6 can complement the loss of Dhh1p in yeast cells, but is more efficient in complementing the defect in replication than the defect in BMV translation in Dhhlp-deficient cells (2). This is consistent with our results, which suggest that DDX6 knockdown degrades RNA replication rather than IRES-directed HCV translation. Since DDX6 coimmunoprecipitates with HCV RNA (Fig. 5B), it remains possible that the helicase activity of DDX6 may promote viral replication by becoming engaged in remodeling of the viral RNA at some step in the HCV life cycle. This would explain not only the requirement that we have observed for DDX6 helicase activity in the promotion of HCV replication, but also the apparent dominant-negative activity of the helicase-defective DDX6-EQ mutant (Fig. 7). This latter observation is particularly difficult to reconcile with the hypothesis that DDX6 facilitates replication via indirect effects. However, the extensive experimental data that we present in this communication do not allow a clear distinction to be drawn between these two possibilities, which are not mutually exclusive. Rather, they point to the challenges of determining whether host proteins act directly or indirectly in the HCV life cycle when modulation of their abundance is shown to affect viral replication.
Acknowledgments
We are grateful to T. M. Rana, University of Massachusetts Medical School, for providing the pEYFP-Rck/p54, pECFP-Ago1, and pECFP-Ago2 plasmids and to Charles M. Rice, Rockefeller University, for the Huh-7.5 cells. We also thank David R. McGivern, Zongdi Feng, and Aditya Hindupur for helpful discussions.
R.K.J. was a recipient of a James W. McLaughlin Predoctoral Fellowship. This study was supported in part by grants from the National Institutes of Health (U19-AI40035 and P20-CA150343) to S.M.L.
Footnotes
Published ahead of print on 14 April 2010.
REFERENCES
- 1.Akao, Y., H. Yoshida, K. Matsumoto, T. Matsui, K. Hogetu, N. Tanaka, and J. Usukura. 2003. A tumour-associated DEAD-box protein, rck/p54 exhibits RNA unwinding activity toward c-myc RNAs in vitro. Genes Cells 8:671-676. [DOI] [PubMed] [Google Scholar]
- 2.Alves-Rodrigues, I., A. Mas, and J. Diez. 2007. Xenopus Xp54 and human RCK/p54 helicases functionally replace yeast Dhh1p in brome mosaic virus RNA replication. J. Virol. 81:4378-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angus, A. G., D. Dalrymple, S. Boulant, D. R. McGivern, R. F. Clayton, M. J. Scott, R. Adair, S. Graham, A. M. Owsianka, P. Targett-Adams, K. Li, T. Wakita, J. McLauchlan, S. M. Lemon, and A. H. Patel. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 91:122-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariumi, Y., M. Kuroki, K. Abe, H. Dansako, M. Ikeda, T. Wakita, and N. Kato. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81:13922-13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonin, M., J. Oberstrass, N. Lukacs, K. Ewert, E. Oesterschulze, R. Kassing, and W. Nellen. 2000. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA 6:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, C. Y., and T. M. Rana. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4:e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17-37. [DOI] [PubMed] [Google Scholar]
- 9.Counihan, N. A., L. M. Daniel, J. Chojnacki, and D. A. Anderson. 2006. Infrared fluorescent immunofocus assay (IR-FIFA) for the quantitation of non-cytopathic and minimally cytopathic viruses. J. Virol. Methods 133:62-69. [DOI] [PubMed] [Google Scholar]
- 10.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of posttranscriptional pathways. Nat. Rev. Mol. Cell. Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 11.Gao, L., H. Aizaki, J. W. He, and M. M. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goh, P. Y., Y. J. Tan, S. P. Lim, Y. H. Tan, S. G. Lim, F. Fuller-Pace, and W. Hong. 2004. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J. Virol. 78:5288-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, Y. Matsuura, K. L. Norman, and P. Sarnow. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto, K., Y. Nakagawa, H. Morikawa, M. Niki, Y. Egashira, I. Hirata, K. Katsu, and Y. Akao. 2001. Co-overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis 22:1965-1970. [DOI] [PubMed] [Google Scholar]
- 15.Henke, J. I., D. Goergen, J. Zheng, Y. Song, C. G. Schuttler, C. Fehr, C. Junemann, and M. Niepmann. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jopling, C. L. 2008. Regulation of hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 36:1220-1223. [DOI] [PubMed] [Google Scholar]
- 20.Jopling, C. L., S. Schţz, and P. Sarnow. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 22.Lanford, R. E., E. S. Hildebrandt-Eriksen, A. Petri, R. Persson, M. Lindow, M. E. Munk, S. Kauppinen, and H. Orum. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemon, S. M., C. Walker, M. J. Alter, M. Yi, D. Knipe, P. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus. 2007. Hepatitis C viruses, p. 1253-1304. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 24.Li, K., T. Prow, S. M. Lemon, and M. R. Beard. 2002. Cellular response to conditional expression of hepatitis C virus core protein in Huh7 cultured human hepatoma cells. Hepatology 35:1237-1246. [DOI] [PubMed] [Google Scholar]
- 25.Liang, Y., T. Shilagard, S.-Y. Xiao, N. Snyder, D. Lau, L. Cicalese, H. Weiss, G. Vargas, and S. M. Lemon. 2009. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology 137:1448-1458. [DOI] [PubMed] [Google Scholar]
- 26.Linder, P. 2006. Dead-box proteins: a family affair-active and passive players in RNP-remodeling. Nucleic Acids Res. 34:4168-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 28.Lu, D., and J. J. Yunis. 1992. Cloning, expression, and localization of an RNA helicase gene from a human lymphoid cell line with chromosomal breakpoint 11q23.3. Nucleic Acids Res. 20:1967-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minshall, N., M. Kress, D. Weil, and N. Standart. 2009. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization, and assembly. Mol. Biol. Cell 20:2464-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyaji, K., Y. Nakagawa, K. Matsumoto, H. Yoshida, H. Morikawa, Y. Hongou, Y. Arisaka, H. Kojima, T. Inoue, I. Hirata, K. Katsu, and Y. Akao. 2003. Overexpression of a DEAD box/RNA helicase protein, rck/p54, in human hepatocytes from patients with hepatitis C virus-related chronic hepatitis and its implication in hepatocellular carcinogenesis. J. Viral Hepat. 10:241-248. [DOI] [PubMed] [Google Scholar]
- 31.Nelson, H. B., and H. Tang. 2006. Effect of cell growth on hepatitis C virus (HCV) replication and a mechanism of cell confluence-based inhibition of HCV RNA and protein expression. J. Virol. 80:1181-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owsianka, A. M., and A. H. Patel. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330-340. [DOI] [PubMed] [Google Scholar]
- 33.Perz, J. F., G. L. Armstrong, L. A. Farrington, Y. J. Hutin, and B. P. Bell. 2006. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J. Hepatol. 45:529-538. [DOI] [PubMed] [Google Scholar]
- 34.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan, T., R. K. Beran, C. Peters, I. C. Lorenz, and B. D. Lindenbach. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall, G., M. Panis, J. D. Cooper, T. L. Tellinghuisen, K. E. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. D. Lindenbach, M. Chien, D. B. Weir, J. J. Russo, J. Ju, M. J. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. M. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheller, N., L. B. Mina, R. P. Galao, A. Chari, M. Gimenez-Barcons, A. Noueiry, U. Fischer, A. Meyerhans, and J. Diez. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517-13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholle, F., K. Li, F. Bodola, M. Ikeda, B. A. Luxon, S. M. Lemon, A. Serman, R. F. Le, C. Aigueperse, M. Kress, F. Dautry, D. Weil, H. Lerat, H. L. Kammoun, I. Hainault, E. Merour, M. R. Higgs, C. Callens, S. M. Lemon, F. Foufelle, and J. M. Pawlotsky. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serman, A., R. F. Le, C. Aigueperse, M. Kress, F. Dautry, and D. Weil. 2007. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 35:4715-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimakami, T., R. E. Lanford, and S. M. Lemon. 2009. Hepatitis C: recent successes and continuing challenges in the development of improved treatment modalities. Curr. Opin. Pharmacol. 9:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 44.Wang, T.-H., R. C. A. Rijnbrand, and S. M. Lemon. 2000. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J. Virol. 74:11347-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 46.Weston, A., and J. Sommerville. 2006. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation, and RNA degradation. Nucleic Acids Res. 34:3082-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, Y., Y. Liang, L. Qu, Z. Chen, M. Yi, K. Li, and S. M. Lemon. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U. S. A. 104:7253-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi, M., and S. M. Lemon. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]