Abstract
The induction of expression of many cellular immediate early genes (IEG) involves the transcription factor serum response factor (SRF). Two families of SRF coactivators have also been implicated in IEG induction, the ternary complex factors (TCFs), ELK1, Sap1, and Net, and the myocardin-related factors, MKL1 and MKL2. We found that serum induction of some SRF target genes is preferentially regulated by MKL1/2, whereas others are redundantly activated by both TCFs and MKL1/2. Yet ELK1 can also repress transcription. Binding of ELK1 and MKL1 to SRF has been found to be mutually exclusive in vitro, suggesting that ELK1 could repress expression of IEGs by blocking MKL1 binding. We characterized the in vivo binding of MKL1 and ELK1 to target genes and found an inverse relationship of serum-induced MKL1 binding and serum-decreased ELK1 binding. However, experiments with short hairpin RNA-mediated MKL1/2 depletion and expression of a nuclear MKL1 (N100) variant in stably transfected cells failed to alter ELK1 binding, suggesting that ELK1 binding to target genes is regulated independently of MKL1/2. Nevertheless, we found that short interfering RNA-mediated depletion of TCFs increased target gene expression in cells containing the N100 MKL1 activator, most notably in cells under continuous growth conditions. These results indicate that the TCFs can function both as activators and repressors of target gene expression depending upon the cellular growth conditions.
Keywords: Coregulator Transcription, Ets Family Transcription Factor, Fos, Signal Transduction, Transcription Factors, ELK1, MKL1, Serum Response Factor, Immediate Early Genes, Myocardin
Introduction
Extracellular signals stimulate cell growth through the activation of signaling pathways that orchestrate the expression of cellular immediate early genes (IEGs).2 IEGs are genes whose expression does not require new protein synthesis suggesting that signaling pathways relatively directly activate their expression without the synthesis of new mRNA or protein (1–4). Examples of IEGs are c-fos, Nur77, Egr1, and Egr2, which are all transcription factors (5). Cytoskeletal genes are also activated, in particular actin genes and vinculin (6). Experimentally IEGs are induced in tissue culture cells by brief serum treatment of serum-starved cells. Many IEGs are known to contain serum response elements (SRE) in their promoters, which are bound by serum response factors (SRF) (7, 8). At least two signaling pathways are involved in activating SRF target genes. One is the MAPK signaling pathway (9, 10) and the other is the RhoA-actin pathway (11). Both pathways affect the activity of SRF transcriptional coregulators. Although serum does not affect SRF binding to DNA, it can elicit different responses by modulating the activities of coregulators of SRF under various biological situations (12, 13).
In the MAPK pathway, signals phosphorylate SRF-transcriptional coregulators known as ternary complex factors (TCF) or p62TCF (9, 10). The TCF family is composed of three Ets-related genes, ELK1, SAP1 (ELK4), and NET (ELK3, SAP2, and ERP), and contains four closely related domains as follows: an Ets-DNA binding domain at their N terminus; a B box region involved in SRF interaction; a phosphorylation-activated transcriptional activation domain located in the C-terminal region; and a MAPK-docking site in a D domain (14–19). Phosphorylation of ELK1 by ERK1/2 in the transcriptional activation domain activates this function (20). ELK1 binds constitutively to SRE target sites and is associated with the transcriptional coactivators CREB-binding protein and/or p300 (21–23). Phosphorylation of ELK1 alters the complex with p300 to facilitate transcriptional activation (22). Besides its role as a transcriptional activator, TCFs have also been shown to repress transcription. Net appears to be the strongest repressor among the three family members and contains two repression domains known as NID (24) and CID (25). In the case of ELK1, a repression domain has been mapped that functions in serum-starved conditions and requires SUMO modification (26). Shortly after being activated by mitogenic stimuli, ELK1 returns back to the repressor state by recruiting the mSin3A-HDAC1 corepressor complex to its Ets-like DNA binding domain (27, 28).
The second pathway for activation of SRF/SRE is through activation of G protein-coupled receptors that signal to the RhoA GTPase to induce actin polymerization. Actin can directly regulate another SRF coregulator, the megakaryoblastic leukemia (MKL) family proteins (29). There are three homologous MKL proteins, myocardin, MKL1 (MRTF-A, MAL, and BSAC), and MKL2 (MRTF-B) (29–34). MKL1 was found in a gene fusion with the RBM15 gene in megakaryoblastic leukemia cells, hence its name (35, 36). The RBM15-MKL1 fusion protein is activated for SRE activation compared with the MKL1 gene, suggesting that its activation of target genes is involved in causing megakaryoblastic leukemia (30, 37). Myocardin is expressed in cardiac and smooth muscle cells, although MKL1 and MKL2 are relatively ubiquitously expressed (30–32, 34, 38). Activated RhoA causes MKL1/2 translocation from the cytoplasm to the nucleus. This is accomplished by RhoA stimulation of F-actin filaments and the consequent reduction of G-actin monomers (29, 39). G-actin monomers bind MKL1 through N-terminal RPEL motifs and increase its nuclear export resulting in predominantly cytoplasmic MKL1 (40, 41). When RhoA causes a reduction in G-actin, less is bound to MKL1, nuclear export decreases, and MKL1 becomes mostly nuclear (41). Once in the nucleus, MKL1/2 forms a complex with SRF and activates SRF target gene expression, including the vinculin gene and the SRF gene itself (29, 30). Dominant-negative forms of MKL1 and shRNA-mediated reduction in MKL1 and MKL2 levels suppressed activation of target gene expression by the RhoA pathway (29, 30).
The relative contribution of these pathways to subsets of target gene expression has been a focus of interest. TCF binding to SREs is dictated by a short purine-rich sequence near the CArG box SRF-binding site (42–44). However, the position of this sequence can be variable such that it is difficult to predict which SREs will bind TCFs (44). MKL factors bind to SRF, but any further DNA binding specificity is unclear. DNase I footprinting suggests that MKL1 may bind to the flanking sequence of the SRE along with binding SRF (45). In addition, mapping of myocardin activation of the telokin promoter suggested that there may be some SRE sequences that allow myocardin activation of the gene beyond binding of SRF (46). TCFs require interactions with SRF to bind to the SRE, whereas MKLs can bind only to SRF (13, 42, 45, 47). Both ELK1 and MKLs bind to the same hydrophobic pocket in the SRF DNA binding domain such that their binding to SRF is mutually exclusive (29, 45, 48). The differential effects of the SRF cofactors have been most notably shown in smooth muscle cell differentiation where platelet-derived growth factor activation of ELK1 caused proliferation and blocked differentiation. In contrast, in the absence of ELK1 activation, myocardin was able to activate smooth muscle gene expression and cause differentiation (48). In this system, myocardin activated target genes much more strongly than even phosphorylated ELK1, such that ELK1 inhibition of myocardin binding appeared to block gene activation rather than any repressor function. Yoshida et al. (49) also found that platelet-derived growth factor could increase ELK1 and reduce MKL1 binding to the target smooth muscle α-actin promoter in smooth muscle cells, suggesting the control of MKL1 binding by ELK1. These studies and the mutually exclusive nature of TCF and MKL binding to SRF in vitro suggest that these cofactors may play antagonistic roles and that the levels, activation, and specificity of these cofactors may determine target gene activation and subsequent cell fate decisions.
In this study, we aimed to examine the roles of TCFs and MKLs in the regulation of expression of cellular IEGs. We have investigated how these factors are required for activation or repression of IEGs and whether their mutually exclusive binding to SRF is significant in vivo. We found that some IEGs require only MKLs for serum induction, although others can utilize either the TCFs or MKLs. Although binding of MKLs and TCFs to target promoters is inversely related, experiments manipulating MKL levels do not support the hypothesis of direct regulation by MKL of TCF binding. Finally, we have found evidence for a repressor role for TCFs on specific target gene expression by showing that depletion of TCFs leads to elevated target gene expression in the presence of a constitutively active form of MKL1.
EXPERIMENTAL PROCEDURES
Plasmids
The pRevTRE retroviral expression vector (Clontech) and the N-terminal deletion mutant lacking amino acids 1–100 of MKL1 in pRevTRE, N100, were described previously (50). Human ELK1 cDNA was cloned into the cytomegalovirus expression vector pCGN with an N-terminal hemagglutinin epitope tag to generate (pCGN-hELK1). Point mutations were introduced into pCGN-hELK1 with the QuikChange multisite-directed mutagenesis kit (Stratagene). The amino acid mutations in ELK1 were as follows: Ets domain R62E, R65E, and Y66D; B box L158P; R box K249A, V250A, and E251A; transcriptional activation domain S383A. Human SAP1 (clone ID 4364006) and mouse Net (Clone ID 3155850) cDNAs in pCMV-SPORT6 expression vectors were purchased from Open Biosystems.
The pFosWT-GL3 luciferase reporter gene contains an SRE sequence in the mouse c-fos enhancer upstream of the minimal promoter region of human c-fos as described previously (51). The lentiviral pLKO.1 vector (52, 53) was used for shRNA expression. For knockdown of both MKL1 and MKL2, the sequence CATGGAGCTGGTGGAGAAGAA that is common in both the mouse MKL1 and MKL2 genes was cloned into pLKO.1 using the forward and reverse sequences 5′-CCGGCATGGAGCTGGTGGAGAAGAACTCGAGTTCTTCTCCACCAGCTCCATGTTTTTG-3′ and 5′-TTAAGTTTTTGTACCTCGACCACCTCTTCTTGAGCTCAAGAAGAGGTGGTCGAGGTAC-3′.
Production of MKL1/2 shRNA Cell Line
Plasmids expressing shRNA targeting MKL1 and MKL2 (as above) or control shRNA (non-target shRNA control vector, SHC002, from Sigma) were transfected into 293 cells for lentiviral production along with packaging plasmids (pD8.9 and pVSVG). Tet-Off NIH3T3 (TO3T3) cells (Clontech) were then infected with the viral supernatant and selected using 5 μg/ml puromycin (InvivoGen). Individual puromycin-resistant colonies were picked and analyzed for MKL1 and MKL2 levels.
siRNA-mediated Depletion
Mouse-specific ELK1, SAP1, and Net siRNAs as well as nonspecific control siRNA were purchased from Santa Cruz Biotechnology. Two more siRNA sequences (siRNA1 and siRNA2) were synthesized by Integrated DNA Technologies to further reduce TCF gene expression. The TCF siRNA sequences were as follows: siRNA1, forward 5′-UCGUAAUUCAUGUUGGUCUUGUUCUUG-3′ and reverse 5′-AGAACAAGACCAACAUGAAUUACGA-3′; siRNA2, forward 5′-GACACAAACUUGUAGACAAACUUCUGG-3′ and reverse 5′-AGAAGUUUGUCUACAAGUUUGUGTC-3′. The siRNA1 sequence matches 27/27 nucleotides of ELK1 and 26/27 of Net and 21/24 of SAP1. The siRNA2 sequence matches 27/27 nucleotides of ELK1, 22/23 of SAP1 and 19/20 of Net.
siRNA duplexes for mSin3A-1 and mSin3A-2, mHDAC1, mHDAC2, mHDAC3, and mUBC9 were synthesized by Integrated DNA Technologies. Two separate duplexes were used for depletion of mSin3A. siRNA sequences were as follows: siRNA for mSin3A-1, forward 5′-ACAGAAUCAACCUUCUCAGCAGUUGUU-3′ and reverse 5′-CAACUGCUGAGAAGGUUGAUUCUGT-3′; siRNA for mSin3A-2, forward 5′-CCAGGAAUCACCAGUGUAUGCAGCC-3′ and reverse 5′-GGCUGCAUACACUGGUGAUUCCUGGUC-3′; siRNA for mHDAC1, forward 5′-UGGGUAGUUCACAGCAUAGUACUUGCC-3′ and reverse 5′-CAAGUACUAUGCUGUGAACUACCCA-3′; siRNA for mHDAC2, forward 5′-CCAUAAUUUAGCAGCAAGUUAUGAGUC-3′ and reverse 5′-CUCAUAACUUGCUGCUAAAUUAUGG-3′; siRNA for mHDAC3, forward 5′-CAAUGACAUAGUAAUUGGUAUCCTG-3′ and reverse 5′-CAGGAUACCAAUUACUAUGUCAUUGAC-3′; siRNA for mUBC9, forward 5′-GUAGCUGUCCCAACAAAGAACCCTG-3′ and reverse 5′-CAGGGUUCUUUGUUGGGACAGCUACAA-3′. Each siRNA (30 pmol) was diluted in 100 μl of PowerFect Transfection buffer (SignaGen). The diluted siRNA was complexed with 4 μl of PowerFect siRNA transfection reagent (SignaGen) for 6-well plates. The cells were collected for RNA extraction 2 days after transfection. The Sap1 and Net siRNAs (10 pmol each) from Santa Cruz Biotechnology and 10 pmol of combined ELK1 siRNA1 and -2 were used for TCF family depletion. All experiments were performed in triplicate.
Cell Culture and Luciferase Assays
The MEF/3T3 Tet-Off (TO3T3) cells from Clontech stably expressing either an empty vector (pRevTRE, TRE cells) or N100 MKL1 were described previously (50). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 50 μg/ml hygromycin (InvivoGen), 100 μg/ml G418 (InvivoGen), and 1 μg/ml tetracycline (Sigma) at 37 °C in a 5% CO2 incubator.
For luciferase assays, cells were plated onto 24-well plates and the following day transfected with 100 ng of pRL-SV40P (with the SV40 promoter driving the Renilla luciferase gene), 100 ng of the pFosWT-GL3 luciferase reporter gene, and with or without 10 ng of pRevN100 (expressing MKL1 N100) and 100 ng of pCGN-hELK1 or mutants using Lipofectamine 2000 according to the manufacturer's instructions. The next day, the cells were serum-starved in DMEM containing 0.2% FBS and then harvested at 48 h post-transfection. Luciferase activities were measured with the Dual-Luciferase reporter system (Promega). The Renilla luciferase activities were used as an internal standard of transfection efficiency.
RNA Extraction, cDNA Synthesis, and Quantitative Real Time PCR Analysis
RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's directions. To make cDNAs, RNA (1 μg) was primed with random hexamers and reverse-transcribed with ImprompII reverse transcriptase (Promega) according to the manufacturer's directions. Expression levels of specific cDNAs were measured using a SYBR Green master mix (Fermentas) and an ABI PRISM 7900HT sequence detection system (Applied Biosystems). The relative mRNA level was calculated using 18 S rRNA as a control and the difference in Ct values between serum-starved and stimulated cells. Results are expressed as the fold change in stimulated compared with starved cells. At least three biological replicates were performed for each sample.
Immunoblotting
Cells grown on 10-cm tissue culture plates were rinsed with ice-cold phosphate-buffered saline (PBS) and then lysed in 160 μl of RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 0.1% SDS) containing a protease inhibitor mixture (catalog no. 539134, Calbiochem). The lysate was kept on ice for 30 min and then centrifuged at 11,000 × g for 10 min at 4 °C. The supernatant was boiled with Laemmli sample buffer (63 mm Tris-HCl, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.0025% bromphenol blue, pH 6.8), and one-fourth of the total cell lysate was loaded on to an SDS-polyacrylamide gel. The antibodies used to detect MKLs were rabbit anti-MKL1 (H-140) and goat anti-MKL2 (C-19) from Santa Cruz Biotechnology and rabbit anti-ELK1 (1277-1) from Epitomics. Each was used at a 1:500 dilution. Mouse monoclonal anti-Hsp90 antibody (Santa Cruz Biotechnology) or polyclonal rabbit anti-actin antibody (Sigma) was used for a loading control. Fluorescently labeled secondary antibodies (Alexa Fluor 680 goat anti-rabbit IgG from Invitrogen and IRDye 800 donkey anti-goat IgG from LI-COR Biosciences) were used to visualize the proteins and detected with a Li-Cor Odyssey infrared imaging system. The MEK1/2 inhibitor PD0325901 (ChemieTek) (54) was dissolved in dimethyl sulfoxide and added to cells at a final concentration of 5 μm for 30 min prior to stimulation with 20% serum for 5 min. Phorbol 12-myristate 13-acetate (5 μm; Sigma) was added to serum-starved cells for 30 min.
Immunostaining
Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. The fixed cells were then permeabilized with 0.2% Triton X-100 in PBS for 10 min and blocked with 1% bovine serum albumin in PBS for 30 min at 37 °C. The cells were incubated with goat anti-MKL1 antibody (1:100) or mouse anti-FLAG M2 antibody (Sigma) (1:500) in PBS for 1 h at room temperature. The cells were then washed twice with PBS and incubated with secondary antibody (Alexa Fluor 488 from Molecular Probes, Inc.) at a dilution of 1:500 in PBS for 30 min at room temperature. Next, the cells were washed three times with PBS and mounted with Fluoromount-G from Southern Biotech. The cells were analyzed with a Nikon Diaphot 300 epifluorescent microscope.
Chromatin Immunoprecipitation Assay
Cells on 140-mm plates (∼1 × 107 cells) were fixed with ice-cold 1% formaldehyde in PBS for 15 min at 22 °C. Glycine (2.5 m) was added directly to a final concentration of 0.125 m for 5 min to stop fixation, and the cells were then rinsed twice in ice-cold PBS. The cells were scraped into 1 ml of ChIP-RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 5 mm EDTA) and then sonicated with the tubes in an ice bath with an Ultrasonics sonicator at setting 1 for 1.5 min. The samples were diluted to a protein concentration of 1 mg/ml with ChIP-RIPA, and 100 μl of the lysates were saved for input. The samples (1 ml) were then cleared by centrifugation for 15 min at 4 °C at 13,000 rpm. The chromatin solution was incubated with 1 μg of affinity-purified antibody and rotated at 4 °C overnight. Antibodies used were goat anti-MKL1 (SC-21558, Santa Cruz Biotechnology) and rabbit anti-ELK1 (1277-1, Epitomics). Immune complexes were immunoprecipitated with 30 μl of 50% protein G-Sepharose 4B (Zymed Laboratories Inc.) for MKL1 and 30 μl of 50% protein A-agarose (Invitrogen) for ELK1 at 4 °C for 1 h. The immunoprecipitated complexes were washed twice with ChIP-RIPA, three times with IP buffer (100 mm Tris-HCl, pH 8.0, 0.5 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate), twice again with RIPA, and twice with TE (10 mm Tris-HCl, pH 8.0, 1 mm EDTA) and eluted in elution buffer (46 mm Tris-HCl, pH 8.0, 1% SDS, 0.6 mm EDTA) by incubation for 10 min at 65 °C. The cross-linking of the samples was then reversed by the addition of NaCl to a final concentration of 200 mm and incubation at 65 °C overnight. A QIAquick PCR purification kit (Qiagen) was used to purify DNA from the samples and eluted into 45 μl of autoclaved water. For each qPCR, 6 μl of DNA samples were used for each gene. To normalize the DNA binding values, the amount of each gene in immunoprecipitated DNA was divided by the amount in input DNA.
RESULTS
MKL1 and MKL2 Are Required for Serum-induced Expression of Some SRF Target Genes
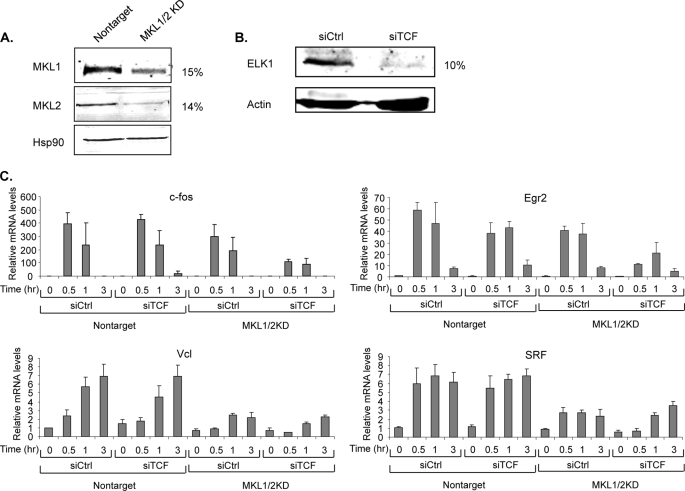
We previously identified MKL-dependent immediate early genes by examining with microarrays the serum-induced expression in NIH3T3 cells with and without a stably expressed dominant-negative mutant of MKL1 (6). To confirm the microarray results for the MKL target genes, we generated NIH3T3 cells with an shRNA lentiviral vector targeting a common sequence in mouse MKL1 and MKL2. These “double knockdown” cells of MKL1 and MKL2 are referred to here as MKL1/2KD. We found no effect on immediate early gene expression with shRNAs that targeted only MKL1 or MKL2, such that double knockdown was required to observe the effects (data not shown). NIH3T3 cells containing a lentiviral vector with an shRNA sequence that does not target known mouse genes were used as control cells. Expression of both MKL1 and MKL2 was down-regulated by 80–85% at both RNA and protein levels (Fig. 1A and data not shown). We tested by qPCR the transcript levels of 18 immediate early genes that are candidates for MKL regulation. The MKL dependence of these IEGs varied from little or none to greater than 75% reduced serum induction (Table 1). Among them, SRF and vinculin genes were significantly down-regulated in these cells, whereas there were much smaller effects on c-fos and early growth response 2 (Egr2) expression (Fig. 1C, compare expression in siCtrl conditions for MKL1/2KD cells compared with nontarget cells). Along with our previous study (6), these results show that serum induction of certain IEGs is MKL-dependent.
FIGURE 1.
Utilization of MKL and TCF cofactors for serum induction of immediate early genes. A, MKL1 and MKL2 levels in TO3T3 cells stably expressing either control shRNA or shRNA targeting MKL1 and MKL2 were measured by immunoblotting with anti-MKL1 and -MKL2 antibodies. Hsp90 levels detected with anti-Hsp90 antibodies served as a loading control. B, ELK1 levels in TRE cells transfected with TCF siRNAs or control siRNAs were measured by immunoblotting with anti-ELK1. Actin levels were detected with anti-actin antibodies as a loading control. The levels of MKL1/2 and ELK1 proteins in the si/shRNA lanes compared with control lanes are indicated to the right of the immunoblots. C, nontarget and MKL1/2 knockdown (KD) cells were transfected with either control siRNA or siRNA targeting the TCF family, ELK1, Sap1, and Net. The next day, the cells were starved in media supplemented with 0.2% FBS overnight and stimulated for the indicated times with media containing 20% FBS. Total RNA was isolated from cells, and the levels of the indicated genes were analyzed by quantitative real time RT-PCR. 18 S rRNA levels were measured to normalize the RNA amount. Means of three experiments are shown with standard deviations.
TABLE 1.
Effect of depletion of both MKL1 and MKL2 on serum-induced expression of immediate early genes
Tet-Off NIH3T3 cells expressing a control shRNA (nontarget cells) or MKL1/2 shRNA (MKL1/2KD cells) were analyzed for serum induction of immediate early gene expression. The cells were serum-starved with 0.2% newborn calf serum for 24 h and then stimulated with 20% serum for 0.5, 1, and 2 h. The fold induction for the time with the highest levels of expression is shown. Values in MKL1/2KD cells were compared with those with nontarget cells. MKL dependency is indicated as follows: −, <25% reduction; +, 25–50% reduction; ++, 50–75% reduction; +++, 75–95% reduction. Each experiment was performed three times. p values indicate the statistical significance of the difference of induced levels in MKL1/2KD cells compared with nontarget cells.
| Gene name | Alternative gene name | Fold induction |
MKL-dependent | p value | |
|---|---|---|---|---|---|
| Nontarget | MKL1/2KD | ||||
| Serpine1 | PAI-1 | 107 | 115 | − | 0.100* |
| Hbegf | Heparin-binding EGF-like growth factor | 56 | 46 | − | 0.169* |
| Fos | c-fos | 112 | 86 | − | 0.604* |
| Cyr61 | Cysteine-rich protein 61 | 10 | 7 | + | 0.068* |
| Btg2 | B cell translocation gene 2, antiproliferative | 50 | 28 | + | 0.028 |
| Ereg | Epiregulin | 62 | 33 | + | 0.045 |
| Errfi1 | Mig6 | 33 | 17 | + | 0.037 |
| ITGA5 | Integrin α5 | 16 | 8 | + | 0.016 |
| Phlda1 | Pleckstrin homology-like domain, family A, member1 | 29 | 14 | + | 0.032 |
| Nr4a1 | Nur77 | 380 | 182 | ++ | 0.038 |
| Ctgf | Connective tissue growth factor | 21 | 9 | ++ | 0.039 |
| Egr2 | Krox20 | 91 | 40 | ++ | 0.021 |
| Cdc42ep3 | Cdc42ep3 | 3 | 1 | ++ | 0.066* |
| Thbs1 | Thrombospondin1 | 3 | 1 | ++ | 0.001 |
| Srf | Serum response factor | 5 | 2 | ++ | 0.018 |
| Vcl | Vinculin | 4 | 1 | ++ | 0.033 |
| Ptgs2 | Cox-2 | 215 | 50 | +++ | 0.014 |
| IL6 | Interleukin 6 | 37 | 6 | +++ | 0.099* |
a p values >0.05 indicate lack of statistical significance.
Redundancy of MKL and TCF Cofactors for Serum Induction of Specific Immediate Early Genes
Because the Ets-related SRF cofactors ELK1, Sap1, and Net have been implicated in serum induction of SRF target genes, we tested whether they are required for serum induction of IEGs that do not require MKL1. These Ets-related factors form a ternary complex with SRF and its SRE DNA-binding site, such that they are termed ternary complex factors. We reduced expression of all three TCF members in MKL1/2KD and control cells by transiently expressing siRNAs targeting each member. (As with the MKL factors, reduction of a single factor did not affect IEG expression (data not shown).) We found that the siRNAs resulted in about a 90% reduction in Elk-1 protein levels (Fig. 1B). qPCR analysis showed that mRNA levels of Elk-1, Sap1, and Net were reduced by 53–84% (data not shown). Despite trying several commercial antibodies, we were not able to measure Sap1 and Net protein levels. We measured the effect of the TCF siRNAs on IEG expression by qPCR. We found little effect alone on serum induction of c-fos, Egr2, vinculin, and SRF genes (Fig. 1C, compare siTCF to siCtrl in nontarget cells). There was also little further reduction of SRF and vinculin expression in MKL1/2KD cells where expression was already reduced by the MKL1/2 shRNAs. However, serum stimulated induction of c-fos and Egr2 was significantly impaired in the MKL1/2KD cells with siRNAs targeting TCFs (Fig. 1C, compare siTCF in MKL1/2KD cells to that with nontarget cells). These results indicate that serum induction of the IEGs c-fos and Egr2 can be mediated by either of the SRF cofactor families, MKL1/2 or the TCF family. Serum induction of other IEGs, however, such as vinculin and SRF, is mediated preferentially by MKL1/2.
Deletion of Actin Binding Domain of MKL1 Causes Its Nuclear Localization but Not Activation of Target Genes
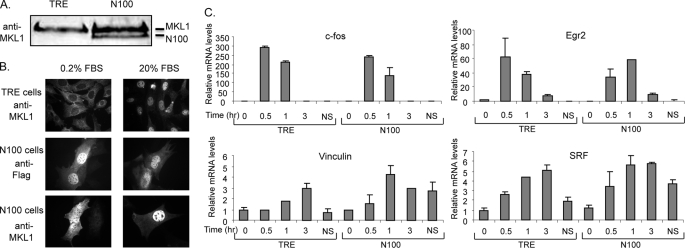
To test the importance of actin binding to MKL1, we generated a mutant, N100, lacking amino acids 1–100 spanning the actin-binding RPEL motifs (50). Cells (N100 cells) were generated that stably express N100 in Tet-Off NIH3T3 cells (TO3T3 cells) or that contain an empty vector (TRE cells). Although N100 was expressed on a tetracycline-regulated promoter, we did not observe tetracycline regulation (data not shown). Expression of N100 was about half that of endogenous MKL1 (Fig. 2A). Although MKL1 is normally cytoplasmic in serum-starved cells and moves to the nucleus upon serum induction (Fig. 2B, top), we found, using anti-FLAG antibodies to an epitope tag on N100, that N100 is constitutively nuclear (Fig. 2B, middle). MKL1 is both cytoplasmic and nuclear in serum-starved N100 cells due to the presence of both endogenous MKL1 and N100 (Fig. 2B, bottom). In transient reporter studies, overexpression of N100 strongly activated SRE-containing reporter genes (Fig. 3A). However, we previously found that N100 was not sufficient to activate endogenous expression of vinculin in serum-starved cells despite its nuclear localization (50). Here, we further examined the expression of MKL1 target genes in N100 cell lines in serum-starved and -stimulated conditions as well as in continuously growing conditions. Transcript levels of c-fos, Egr2, vinculin, and SRF were tested by qPCR. There was little effect of N100 on serum-starved or -stimulated expression of these IEGs (Fig. 2C). There was, however, an effect in nonstarved cells, continuously grown in media containing 10% serum. There was modest elevation of expression of c-fos (2.1-fold, p = 0.021) and Egr2 (2.9-fold, p = 0.008) in N100 cells compared with TRE cells, although these levels are small compared with serum-stimulated levels. There was a more notable elevation of the MKL-dependent IEGs vinculin (3.5-fold, p = 0.013) and SRF (2.0-fold, p value 0.008). The elevation of expression of these genes was close to the levels of expression in serum-induced cells (Fig. 2B).
FIGURE 2.
Nuclear MKL1 does not affect IEG expression. A, endogenous MKL1 levels in TRE (TO3T3 cell containing an empty pRevTRE vector) and N100 cells (TO3T3 cells containing pRevN100) were assessed by immunoblotting using anti-MKL1 antibodies. B, cellular localization of MKL1 in TRE and N100 cells was detected by immunofluorescence with anti-MKL1 or anti-FLAG antibodies. Cells were either serum-starved or stimulated for 5 min and analyzed as described under “Experimental Procedures.” C, TRE and N100 cells were serum-starved in media containing 0.2% FBS overnight and then stimulated with 20% FBS for the indicated times. TRE and N100 cells continuously grown in media containing 10% FBS are indicated as nonstarved (NS). Endogenous expression levels of c-fos, Egr2, Vcl, and SRF were assessed by quantitative real time RT-PCR. Means of three experiments are shown with standard deviations.
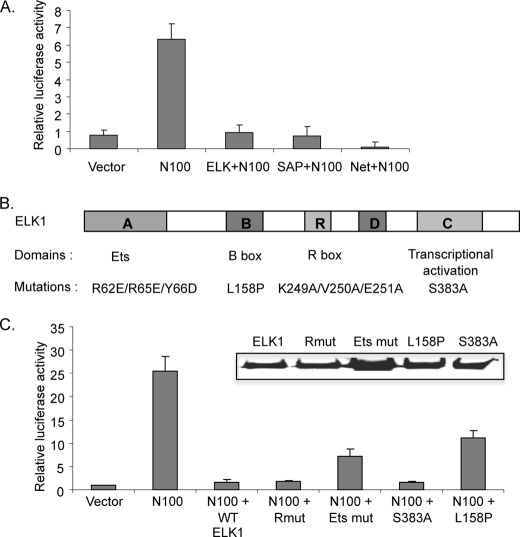
FIGURE 3.
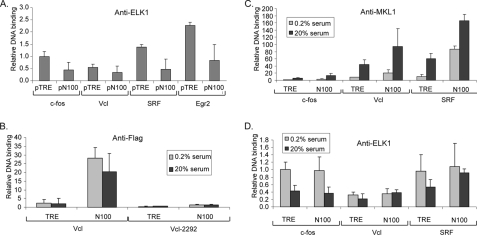
Repression of MKL1 activation by ELK1. A, TRE cells were transfected with pFosWTGL3 reporter containing the SRE region of the mouse c-fos promoter (79), pRevN100 expressing the MKL1 N100 deletion mutant, and each TCF family expression plasmid. As an internal control pRLSV40P, with the SV40 promoter driving Renilla luciferase gene, was also transfected. Cells were harvested 48 h after transfection, and Firefly and Renilla luciferase activities were measured. Firefly luciferase levels were normalized to Renilla levels to control for transfection efficiencies. Results were then normalized as fold activation relative to the level measured in TRE cells transfected with empty vectors. Data are the mean value from three experiments with error bars showing the standard deviations. B, schematic diagram of the ELK1 protein domains and mutations. The mutations in ELK1 are indicated below each domain. C, cells were transfected as in A with the indicated ELK1 mutants. Inset, the expression of wild type and each ELK1 mutant was detected by immunoblotting with anti-hemagglutinin antibodies.
These results demonstrate that nuclear N100 is not sufficient to activate expression of endogenous target genes in serum-starved cells. One explanation would be that the target genes are under the control of a repressor under these conditions. However, in nonstarved conditions, some activation by N100 was observed, suggesting that the repressor is less active under these conditions. The differential effect of N100 on the MKL-dependent vinculin and SRF genes, compared with the MKL- and TCF-dependent c-fos and Egr2 genes, suggests that the latter genes may be under further control mechanisms to ensure their proper regulation.
ELK1 Inhibits Transcriptional Activation by MKL1
We considered the possibility that the TCF cofactors of SRF might be involved in repression of MKL target genes and whether it could account for the lack of activation of target gene expression by N100. Prior studies have shown that TCFs have repressor activity (24, 25, 27). In addition, binding of MKL1 and the related protein myocardin to SRF was found to be mutually exclusive to binding by TCFs to SRF (29, 45, 48). Repression by ELK1 has been shown to function in serum-starved cells by recruiting HDAC2 and requires a specific repression domain in ELK1 (27, 28). We tested whether the TCF family members can inhibit transcriptional activation by N100 in transient transfection experiments with an SRE reporter gene. N100 increased expression of the reporter gene, and this increase was strongly blocked by overexpression of each TCF member (ELK1, Sap1, and Net) (Fig. 3A). To further evaluate which domain in ELK1 is required to suppress MKL1 activity, ELK1 mutant constructs were generated. Mutations were used that had been shown previously to affect the Ets DNA binding domain, the B box SRF interaction domain, the R box repression domain, or the C box transcriptional activation domain (Fig. 3B) (9, 27, 55–58). The expression of each of these mutants was similar to wild type ELK1 expression (Fig. 3C, inset). We found no effect of the R box and transcriptional activation domain mutants on repression by ELK1, suggesting that these domains are not required for ELK1 repression of transcriptional activation by N100. The Ets DNA binding domain and B box mutants relieved repression by ∼30 and 45%, respectively. As each of these mutants would be predicted to reduce binding of ELK1 to the SRE with SRF, these results suggest that the inhibitory role of ELK1 is mediated by blocking MKL1 binding to SRF, consistent with previous results showing mutually exclusive binding in vitro.
Binding of MKL1 and ELK1 to Target Genes
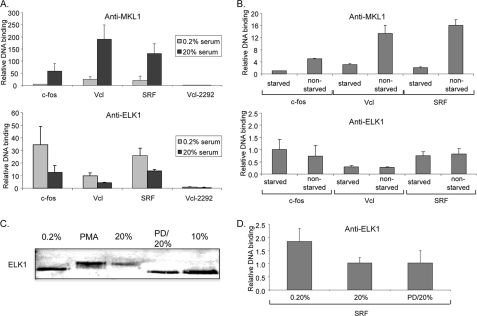
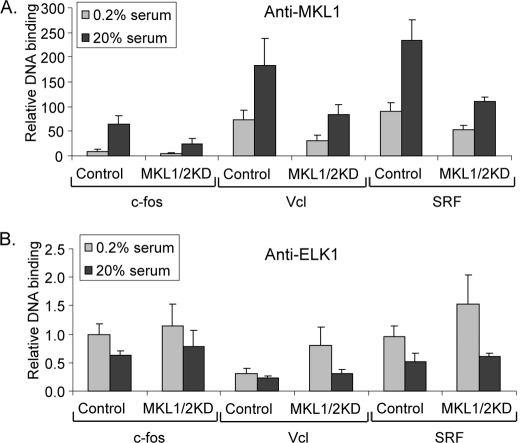
Because ELK1 can repress activation by MKL1 and this may occur because of mutually exclusive binding to SRF, we tested whether we could detect differences in binding to target genes in vivo by chromatin immunoprecipitation (ChIP). We would expect inducible binding of MKL1 to target promoters because of its translocation from the cytoplasm to the nucleus. Previous studies have not shown an inducible change in ELK1 binding to target SREs, but this was not done with inducers that would activate MKL1 (59). Moreover, mutually exclusive binding of MKL1 and ELK1 to SRF would dictate that ELK1 binding should decrease upon MKL1 activation and translocation to the nucleus. In fact, we found that MKL1 binding to the three target genes tested, c-fos, vinculin, and SRF, was serum-inducible (6.4–11.8-fold) consistent with nuclear translocation of MKL1 (Fig. 4A, top). In addition, ELK1 binding to the target promoters decreased 41–70% after serum stimulation of serum-starved cells (Fig. 4A, bottom). As a control, we found equal immunoprecipitation of ELK1 in serum-starved and -stimulated cells, indicating that the differences in ELK1 binding to the target genes are not due to changes in binding to the antibody used (data not shown). The inverse binding of MKL1 and ELK1 to the target promoters is consistent with mutually exclusive binding of the factors to SRF. The extent of MKL1 and ELK1 binding to the target promoters, which all contain SREs, varied but in all cases was above background defined by binding to an upstream site in the vinculin promoter (Vcl-2292; Fig. 4A). Although the ELK1-binding site in the vinculin SRE is not clear, significant binding was still observed with this promoter. It is notable that ELK1 bound to the vinculin and SRF promoters even though TCFs were neither required nor sufficient for induction of these promoters (Fig. 1C).
FIGURE 4.
Binding of MKL1 and ELK1 to target promoters. A, TRE cells were serum-starved for 24 h in DMEM containing 0.2% FBS and then stimulated with 20% FBS for 5 min. Chromatin immunoprecipitation was performed with anti-MKL1 or anti-ELK1 antibodies and the indicated gene probes by qPCR. Vcl-2292, probe for upstream region of the Vcl gene used as a control. To obtain the relative DNA binding values, the amount of signal for each gene in immunoprecipitated DNA was divided by the amount in input DNA. B, levels of MKL1 and ELK1 binding to starved and nonstarved TRE cells were determined as in A. Nonstarved TRE cells were continuously grown in DMEM containing 10% FBS. C, immunoblot of TRE cells with anti-ELK1 antibodies. Cells were serum-starved (0.2%) or induced with phorbol 12-myristate 13-acetate (PMA; 100 ng/ml), serum (20%) as above, pretreated with the MEK1/2 inhibitor PD0325901 (PD, 5 μm) for 30 min, and serum-stimulated for 5 min (PD/20%), or grown continually in media containing serum (10%). D, chromatin immunoprecipitation for binding of ELK1 to the SRF promoter in TRE cells treated as in C was performed with anti-ELK1 antibodies as in A.
Given the different levels of expression of SRF and vinculin genes in N100 cells under nonstarved conditions, we tested the binding of MKL1 and ELK1 to the target promoters under these conditions. MKL1 binding to the target promoters was higher in nonstarved cells than in serum-starved cells suggesting that MKL1 is at least partially nuclear under these conditions (Fig. 4B, top). ELK1 binding, however, was unchanged in serum-starved and nonstarved cells (Fig. 4B, bottom). This suggests that MKL1 binding is not sufficient to preclude ELK1 binding under the nonstarved conditions and that both factors may exert effects on target gene expression in nonstarved cells.
We tested whether serum-induced phosphorylation of ELK1 was required for the reduction of ELK1 binding to the target promoters seen by chromatin immunoprecipitation (Fig. 4A, bottom). As expected, we observed that serum and phorbol 12-myristate 13-acetate, an activator of protein kinase C and subsequently ERK1/2, induced a shift in mobility of ELK1 in immunoblots because of its phosphorylation (Fig. 4C). The serum-induced shift was blocked by the MEK1/2 inhibitor PD0325901. However, PD0325901 had no effect on the serum-induced reduction in ELK1 binding to the SRF promoter (Fig. 4D), indicating that ELK1 phosphorylation is not required for the reduction of binding. Although there was a small serum-induced decrease in ELK1 levels in Fig. 4C, no consistent decrease was observed in other experiments (data not shown). In addition, no decrease was observed with serum induction and PD0325901 in Fig. 4C even though a reduction of binding to the SRF promoter was seen in Fig. 4D. It is also interesting that there was no phosphorylation of ELK1 in nonstarved cells grown in 10% serum suggesting that ELK1 can repress transcription under these conditions (Fig. 4C).
Effect of MKL1/2 on ELK1 Binding to Target Genes
Because of the inverse relation of MKL1 and ELK1 binding to SREs upon serum induction, we hypothesized that if serum-induced nuclear translocation of MKL1 causes displacement of ELK1 binding to SRF, depletion of MKL proteins in MKL1/2KD cells should reduce the effect of serum on ELK1 binding to target genes. To test this idea, we evaluated MKL1 and ELK1 binding in control and MKL1/2KD cells by chromatin immunoprecipitation. MKL1 binding was at least 50% reduced in MKL1/2KD cells for the c-fos, SRF, and vinculin genes in both serum-starved and -stimulated conditions (Fig. 5A). We still observed a serum-induced increase in MKL1 binding in MKL1/2KD cells, presumably because of the nuclear translocation of residual MKL1. In general, there was little effect of MKL1/2 reduction on ELK1 binding to the target promoters (Fig. 5B). With all three genes, there was still a serum-induced reduction in ELK1 binding in the MKL1/2KD cells. Because the levels of ELK1 binding were not increased in MKL1/2KD cells under serum-stimulated conditions, these results do not support the hypothesis that MKL1/2 binding to SRF blocks ELK1 binding. Because MKL1 binding was reduced by the shRNAs (Fig. 5A), a corresponding increase of ELK1 binding would have been expected. It is still possible, however, that the reduction of MKL1/2 by shRNAs was not sufficient to observe a change in ELK1 binding. There was a significant increase in ELK1 binding to the vinculin and SRF promoters in the MKL1/2KD serum-starved cells compared with control serum-starved cells. Although MKL1 is predominantly cytoplasmic in serum-starved cells, the anti-MKL1 ChIP shows that there is some binding to the vinculin and SRF target genes under these conditions, such that the reduction in MKL1 binding could result in increased ELK1 binding under these conditions.
FIGURE 5.
Effect of MKL1/2 depletion on MKL1 and ELK1 binding to target promoters. Control (nontarget) and MKL1/2KD cells were analyzed for MKL1 and ELK1 binding to the indicated target gene promoters by ChIP. The cells were serum-starved overnight and then stimulated for 5 min. ChIP analysis of MKL1 and ELK1 was as described for Fig. 4.
Effect of Nuclear MKL1 on ELK1 Binding to Target Promoters
Conversely to the reduction of MKL1/2 by shRNAs, we tested the effect of N100 MKL1 on ELK1 binding to target promoters. Because N100 is constitutively nuclear (Fig. 2A), if MKL1 blocks ELK1 binding to promoters by binding competitively to SRF, we would expect a reduction in ELK1 binding. We first tried transient transfection of N100 (pN100) or control vector (pTRE) and measured endogenous ELK1 binding to the target promoters (Fig. 6A). As predicted, transfection of pN100 reduced binding of ELK1 to each of the target promoters. We next tested the effect of N100 in the stable cell line. Using anti-FLAG antibodies to the epitope tag on N100, we found by chromatin immunoprecipitation that N100 bound constitutively to the vinculin promoter (Fig. 6B). Little binding was observed in TRE cells that lack N100 or at an upstream site in the vinculin promoter (Vcl-2292) showing the specificity of N100 binding (Fig. 6B).
FIGURE 6.
Effect of nuclear MKL1 on ELK1 binding to target promoters. A, vector expressing MKL1 N100 (pN100) or control vector (pTRE) was transfected into TRE cells, and 48 h after transfection, ELK1 binding to target promoters was determined by chromatin immunoprecipitation as in Fig. 4. B, DNA binding of N100 to target promoters in stably transfected N100 and TRE cells was analyzed by ChIP using antibodies to the FLAG epitope tag on N100. Primers for the Vcl promoter and for a control region 2292 bp upstream of the Vcl gene transcriptional initiation site were used. C and D, relative in vivo binding of MKL1 (C) and ELK1 (D) in TRE and N100 cell lines was detected by ChIP as in Fig. 4. Cells were serum-starved or stimulated for 5 min as indicated.
Using anti-MKL1 antibodies, which can immunoprecipitate both N100 and endogenous MKL1, we found that MKL1 binding was elevated in N100 serum-starved cells compared with control TRE cells at each of the target promoters (Fig. 6C). Serum induction caused an increase in MKL1 binding, presumably because of endogenous MKL1. The increase in binding of MKL1 in N100 serum-starved cells compared with control TRE cells was particularly notable for the SRF promoter.
We next tested the binding of ELK1 to the target promoters in N100 cells. Despite the increase in MKL1 binding in serum-starved and -stimulated cells, there was little if any reduction in ELK1 binding in N100 compared with control TRE cells (Fig. 6D). As with the MKL1/2KD cells (Fig. 5), these results do not support the hypothesis that competitive MKL1 binding affects ELK1 binding and accounts for the serum-induced reduction in ELK1 binding. Although overexpression of N100 can reduce ELK1 binding (Fig. 6A), lower levels (similar to half of endogenous levels of MKL1) were not sufficient to affect ELK1 binding. In particular, note that binding of MKL1 to the SRF promoter in serum-starved N100 cells was high, but ELK1 binding to the same promoter was not reduced (Fig. 6, C and D).
siRNA-mediated Suppression of TCFs Increases Expression in N100 Cells
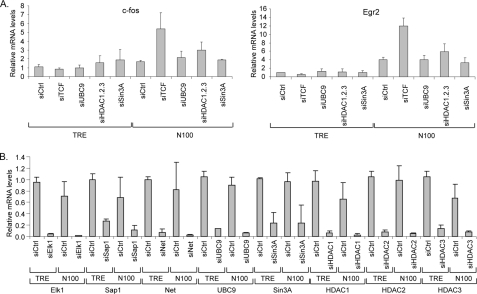
Given the evidence for TCF repressor activity (Fig. 3) (24–28) and the reduction of ELK1 binding by serum stimulation (Fig. 4), we sought to determine whether TCFs act as repressors of endogenous IEG expression. We reduced TCF family member (ELK1, Sap1, and Net) expression by transient expression of siRNAs as in Fig. 1. Because loss of a repressor might not activate expression without an activator, we also tested for expression in N100 cells where the MKL1 activator is constitutively nuclear. We found small but significant changes because of the TCF siRNAs on the four IEGs tested (Fig. 7).
FIGURE 7.
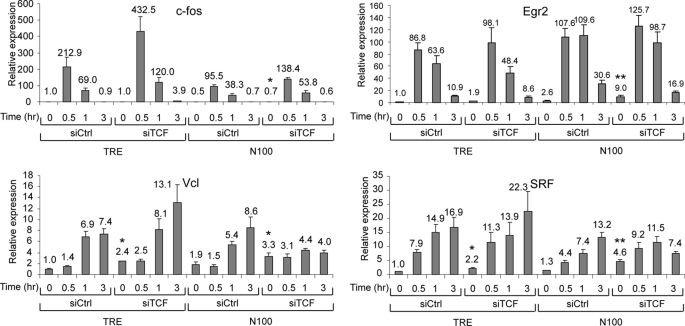
Suppression of TCFs increases IEG expression in serum-starved N100 cells. TRE and N100 cells were transiently transfected with siRNAs targeting the TCF family members or control siRNAs as in Fig. 1B. One day after transfection, the cells were serum-starved overnight followed by serum stimulation for 0.5, 1, or 3 h. Levels of the IEG mRNAs were determined by qPCR as in Fig. 1. The means were calculated from three independent experiments. *, p = 0.05 to 0.01; **, p < 0.01 for values compared with control TRE siCtrl levels.
Serum induction of c-fos expression was lower in N100 cells than in control cells, but there was no significant effect of TCF siRNAs in N100 cells. For Egr2, however, there was a significant increase of 9.0-fold in serum-starved N100 cells with TCF siRNAs compared with TRE cells with control siRNAs (p = 0.007) (Fig. 7). This level of expression, however, is small compared with the serum-induced levels of about 100-fold. For vinculin and SRF, IEGs that are dependent upon MKLs for their activation (Fig. 1) and that are less strongly induced, there was a significant increase of expression in serum-starved N100 cells with TCF siRNAs compared with control siRNAs and TRE cells. For instance, SRF levels were induced 4.6-fold in N100, siTCF serum-starved cells compared with 1.3-fold with control siRNAs and 1.0-fold in TRE cells with control siRNAs (p = 0.009). Less activation was seen by TCF siRNAs in TRE cells. These results suggest that TCFs act as repressors of the SRF gene but that increased expression is only clearly observed when the N100 activator is present. It is also notable that whereas basal vinculin expression was elevated in N100 cells, subsequent serum induction was nearly abolished. This level of expression is intermediate between that of serum-starved cells and peak serum induction such that TCF siRNAs cause both elevated expression and block further serum induction. It is possible that there is a feedback response that limits further vinculin expression once its expression is elevated, which would be consistent with TCFs acting as repressors under these conditions.
We observed stronger effects of TCF siRNAs under nonstarved, continuously growing cellular conditions (Fig. 8). N100 and control TRE cells were grown in media with 10% serum, without starving, and RNA was isolated and analyzed by qPCR. Under these conditions, expression of vinculin and SRF was elevated in N100 compared with TRE cells, but there was no effect of TCF siRNAs. However, for the c-fos and Egr2 genes, there was only a modest increase of expression in N100 cells with control siRNAs compared with TRE cells. The combination of N100 cells and TCF siRNAs resulted in a strong increase of expression for c-fos and Egr2 (Fig. 8). Thus, under these nonstarved continuous growth conditions, TCFs appear to act as repressors of expression of the c-fos and Egr2 gene with expression dependent on the presence of the N100 MKL1 activator.
FIGURE 8.
siRNA-mediated depletion of TCFs increases expression of c-fos and Egr2 in N100 cells under continuous growth conditions. TRE and N100 cells grown in media containing 10% FBS were transfected with control siRNA and TCF siRNAs for 48 h. Levels of c-fos, Egr2, Vcl, and Srf mRNAs were analyzed by qPCR as in Fig. 1.
Test of TCF Corepressors for Repression of Target Genes
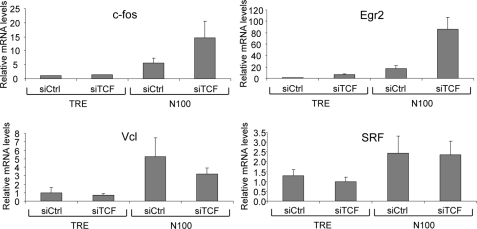
Because our results do not support repression by the TCFs acting by blocking MKL1 binding (Figs. 5 and 6), we tested the model that TCF represses because of its interaction with the corepressor mSin3a and HDAC1 or by sumoylation and binding to HDAC2 as reported previously (28, 60). The repression by sumoylation was found to occur in serum-starved cells, and mSin3a repression occurred temporally after serum activation. We used siRNAs to deplete expression of these repressive factors. Depletion of Ubc9, an E2 SUMO ligase, was used to block sumoylation (61). We tested the effects of the siRNAs on c-fos and Egr2 expression in TRE cells and cells expressing the N100 activated form of MKL1. No effect was observed in TRE cells (Fig. 9A). In N100 cells, we found that depletion of the TCFs increased c-fos and Egr2 expression as found in Fig. 8. However, there was no similar increase of expression by depletion of mSin3a or Ubc9. We further tested the role of histone deacetylases by depleting three of the class I histone deacetylases, HDAC1–3. There was a modest 1.76-fold increase in c-fos expression; however, this effect was not statistically significant (p = 0.064). We confirmed the depletion by the siRNAs of their targets by qPCR and found strong depletion in both TRE and N100 cells (Fig. 9B). These results argue against a role for mSin3a or sumoylation and their associated histone deacetylases in repression by TCFs of the c-fos and Egr2 target genes.
FIGURE 9.
siRNA-mediated depletion of possible TCF corepressive factors. TRE and N100 cells grown in media containing 10% FBS were transfected with control siRNA and the indicated siRNAs for 48 h. A, levels of c-fos and Egr2 mRNAs were analyzed by qPCR as in Fig. 1. B, relative mRNA levels of the indicated siRNA target genes were determined by qPCR.
DISCUSSION
Redundancy of SRF Cofactors MKL and TCF
SRF plays an important role in many biological processes such as proliferation and smooth muscle differentiation by working with transcriptional coregulators (30, 31, 62–64). We have confirmed previously identified MKL-dependent genes by expression of shRNAs targeting both MKL1 and MKL2. Serum stimulation of SRF and vinculin expression was strongly blocked by MKL1/2 depletion. However, serum induction of other IEGs, such as c-fos and Egr2, did not require MKL1/2. These IEGs also did not require the TCF SRF cofactors (ELK1, Sap1, and Net) for serum induction as seen by their siRNA depletion. However, depletion of both these groups of factors strongly reduced c-fos and Egr2 induction, indicating that serum induction of these genes can be stimulated by either MKL1/2 or TCFs. Serum induction of c-fos was not impaired in cells from ELK1-deficient mice (65). This could be explained by the redundancy of other TCF family members (Sap1 and Net), but our results suggest that the further redundancy with MKL family members allows for serum induction in the absence of ELK1.
Repression of IEGs
We tested whether activation of MKL1, i.e. its nuclear localization, was sufficient for activation of target genes. We used a variant of MKL1 (N100) lacking its N-terminal actin-binding motifs. Surprisingly, target genes were not expressed more highly in serum-starved cells expressing N100. Chromatin immunoprecipitation showed that N100 is bound equally to the target genes in serum-starved and -induced cells, showing that the lack of expression is not due to lack of promoter occupancy. This suggests that either there is negative regulation of the target genes in serum-starved cells, which blocks transcriptional activation by N100, or that another positively acting step is required for increased transcription. We found increased expression of the vinculin and SRF genes in N100 cells when they were grown under nonstarved conditions (i.e. continually in media with 10% serum). This suggests that the second function, whether the inhibition of a repressor or activation of another step, is activated under these conditions. The other IEGs tested, c-fos and Egr2, were relatively unaffected by N100 under any of the growth conditions. This may be due to other regulatory mechanisms for these genes but also suggests the action of a repressor.
The TCF family members have been found to have repressor activity such that we considered whether they might be responsible for the lack of activation of target gene expression by N100. Net was the first TCF family member to be found with repressor activity that could be reversed by treatment of cells with oncogenes such as H-ras (66). ELK1 was subsequently found to repress gene expression through an mSin3a-HDAC1 complex or by SUMO modification and association with HDAC2 (27, 28, 60). In addition, Kasza et al. (59) previously found that overexpression of ELK1 could decrease reporter gene activation by MKLΔN, which is similar to N100. The antagonism of ELK1 and MKL1 may be accounted for by their competition for binding to SRF. In vitro binding studies have shown that binding of MKL1 and myocardin to SRF is mutually exclusive to that of ELK1 due to binding to a common surface on SRF (29, 45, 48). We were able to repeat the inhibition by ELK1 of MKL1 activation of SRE reporter gene expression. Mutational analysis of ELK1 suggests that the inhibition is due to competitive binding to SRF rather than the activity of the ELK1 repression domain. However, these studies were performed with overexpression of ELK1, and we sought to determine whether endogenous levels of the TCFs function to repress target gene expression in wild type or N100 cells.
Requirement of TCFs for Repression
We reduced endogenous TCF levels with siRNAs targeting ELK1, Sap1, and Net. We observed little if any effect on expression of IEGs in 3T3 cells. We reasoned that even if repression by TCFs was removed, expression of target genes might not be observed without a transcriptional activator present. Therefore, we tested for the effect of TCF siRNAs in N100 cells where the N100 form of MKL1 is bound to target genes in serum-starved and -stimulated cells. In this situation, we observed a modest increase in expression of the IEGs consistent with the TCFs functioning as repressors (Fig. 7). This effect was most notable for expression of SRF in serum-starved cells (4.6-fold increase in expression compared with control cells and 3.5-fold increase compared with serum-starved N100 cells with control siRNAs). Vinculin expression was also increased by the TCF siRNAs in N100 cells, and serum induction in these cells was also notably curtailed. As the level of vinculin expression was intermediate between control serum-starved and -induced levels, it is difficult to distinguish whether TCFs are required for serum induction in N100 cells or whether there is a feedback mechanism blocking further vinculin induction. Because the TCFs were not required for vinculin expression in control cells, we favor the latter explanation.
There was still strong serum induction of c-fos and Egr2 expression in N100 cells with TCF siRNAs, suggesting that TCFs are not sufficient to repress expression of these genes. These results imply that there must be other mechanisms to block MKL1 activation of c-fos and Egr2 expression. There could be another repressor or need for another activating step as discussed below. However, when cells were grown under nonstarved conditions, there was a strong increase of expression of c-fos and Egr2 that required N100 and TCF siRNAs (Fig. 8). Under these conditions, the repressive function of the TCFs is particularly clear. These results point to the importance of TCFs in maintaining the low levels of expression of target genes under continual growth conditions, in addition to its activating role in the acute induction of target genes following the addition of inducers such as serum.
It is interesting that we found differing requirements for TCF repression in serum-starved cells and “nonstarved” cells continually grown in 10% serum. The greater effect of TCF siRNAs in nonstarved cells suggests that additional mechanisms are acting in serum-starved cells to limit gene activation. These differing conditions are likely to occur physiologically where cells are more quiescent, similar to the serum-starved conditions, and poised for growth factor activation. Other cells may have more continual exposure to growth factors and be more similar to the nonstarved condition. The differential requirements for TCFs for repression suggest that there are additional mechanisms regulating expression, and it will be important to elucidate additional repressive or activating factors.
There was no effect of TCF siRNAs on vinculin and SRF expression in nonstarved N100 cells. As N100 activated expression of the vinculin and SRF genes under these conditions, this suggests that TCFs are not able to repress activation of these genes by N100 under continual growth conditions. The specificity of TCFs as repressors of the c-fos and Egr2 genes, but not vinculin and SRF under these conditions, is consistent with the activity of TCFs as serum-induced activators of c-fos and Egr2 but not vinculin and SRF (Fig. 1). This is surprising because chromatin immunoprecipitations suggested similar binding of ELK1 to c-fos and SRF promoters (Figs. 4–6). The basis for the specificity of these factors is not clear. TCFs bind to a purine-rich sequence near the CArG SRF-binding site. This sequence minimally only requires the sequence GGA and can be on either strand and up to 27 bases away from the CArG box (44). This makes it difficult to predict true TCF recognition sites because many false sites can be identified by these criteria. Although ELK1 was found to bind to an Ets site upstream of SRF sites in the SRF promoter, the requirement of ELK1 for serum induction of SRF was not previously tested (67). The specificity of TCF activity for the c-fos gene compared with the SRF gene may still require specific TCF-binding sites for proper function, but the rules for distinguishing these sites remain to be elucidated.
Modulation of MKL and TCF Binding to Target Genes
We examined by chromatin immunoprecipitations whether MKL1 and ELK1 binding to target promoters in cells varies with serum stimulation. As expected, MKL1 binding was serum-inducible, consistent with its movement from the cytoplasm to the nucleus. ELK1 binding was reciprocally reduced by serum induction. This was not previously seen using other inducers of IEG expression such as 12-O-tetradecanoylphorbol-13-acetate and epidermal growth factor (59, 68). In contrast, in rat aortic cells ELK1 binding to the c-fos SRE was increased after serum induction from undetectable levels (69). Oxidized phospholipids also increased ELK1 binding to the smooth muscle α-actin promoter in rat aortic smooth muscle cells, suggesting that ELK1 binding to the target genes is regulated differently in smooth muscle cells than in fibroblasts (70). A recent genomic ELK1 ChIP analysis found that ELK1 was bound to over 1000 promoter regions in serum-starved HeLa cells, showing that ELK1 is generally bound to target genes before induction (71).
The binding of ELK1 to the promoters studied here did not correspond to their requirement for serum induction. For instance, ELK1 bound to the SRF promoter (Fig. 4A) but was not required for serum induction of SRF (Fig. 1C). It is possible that the ChIP is detecting ELK1 binding on the SRF promoter at a site separate from SRF or in a nonfunctional position. The specific binding of ELK1 to a site in the SRF promoter might position it in a conformation that is not conducive for transcriptional activation. This could be similar to the results with the glucocorticoid receptor whose transcriptional activation was dependent upon its conformation on specific binding sites rather than its overall binding level (72).
The serum-induced reduction of ELK1 binding we found would be expected due to increased MKL1 binding and the mutual exclusiveness of their binding to SRF in vitro (29, 45, 48). We tested whether the reduction in ELK1 binding was due to MKL1 binding. Surprisingly, neither reducing MKL1/2 levels with shRNAs nor expressing a constitutively nuclear form of MKL1 affected levels of ELK1 binding to target promoters. Although we were able to show that transient overexpression of nuclear MKL1 could reduce ELK1 binding to target promoters, likely by competitive binding to SRF, endogenous levels of MKL1 do not appear to regulate ELK1 binding.
Overexpression of ELK1 reduced activation of SRE reporter genes by MKL1, and mutants of ELK1 suggest that this is by a competitive mechanism. We were not able to measure MKL1 DNA binding levels in cells with the TCF siRNAs, because this requires a large amount of cells and transient transfection of the siRNAs. However, the changes in MKL1 cellular localization suggest that serum-induced increases of MKL1 binding to target promoters are more likely due to its translocation to the nucleus than any reduction in ELK1 binding. Nevertheless, reduced ELK1 binding may still contribute to increased MKL1 binding. Because ELK1 contains a transcriptional repression domain, reduction of ELK1 binding may be important in reducing repression by this domain rather than by affecting MKL1 binding. As discussed above, reducing TCF levels increased target gene expression, depending upon the cellular growth conditions and the specific target. These results suggest that the repression activity of the TCFs is indeed significant to endogenous target gene expression.
Because binding of ELK1 and MKL1 to SRF is mutually exclusive in vitro, it is surprising that reciprocal binding is not always seen by ChIP in vivo. It is possible that binding by these SRF cofactors is relatively low in cells such that target promoters in a population of cells can have either TCFs or MKL1/2 bound, albeit to different promoters. Alternatively, the binding may be transient enough to have these factors cycling on and off the same promoter. In this scenario, the ChIP results may reflect the shift in the equilibrium to more binding of either factor, but both factors can bind. Finally, it is possible that SRF cofactors interact with other factors in a larger complex such that direct binding to SRF is only one determinant of binding and that TCFs and MKL1/2 can both be present in the complex at the same time. It will be interesting to determine whether there are any interactions among the TCFs and MKL1/2 or any other cofactors identified in the future.
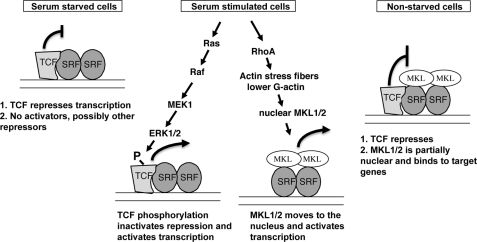
Model for Regulation of IEGs by MKL1/2 and TCFs
Our results support a model whereby immediate early genes are regulated by the TCF and MKL SRF coregulators (Fig. 10). In serum-starved and nonstarved cells, the TCFs repress transcription. Our results do not support this repression occurring by blocking MKL binding or recruiting HDAC1–3. TCF phosphorylation by the ERK1/2 pathway switches it from a repressor to an activator in serum-stimulated cells. The RhoA pathway causes an increase in F-actin (39, 73). The resulting decrease in G-actin results in its dissociation from MKL1 and localization of MKL1 to the nucleus (29, 74). We found that some IEGs (c-fos and Egr2) could be activated by either the TCF or MKL pathway, although others (SRF and vinculin) could only be activated by the MKL pathway. Similarly, only the c-fos and Egr2 genes were sensitive to TCF repression in nonstarved cells. In serum-stimulated cells, TCF repression is inactivated, although both TCF and MKL are activated to increase transcription. In nonstarved cells, even though we find that MKL1 is bound to the target promoters, little expression is seen, partially due to TCF repression because TCF is unphosphorylated in this state. SRF and vinculin were not sensitive to TCF repression in nonstarved cells, possibly because TCF binds less productively to their promoters. It is unclear how repression by the TCFs is mediated. Despite previous results showing mutually exclusive binding of ELK1 and MKL1 to SRF in vitro, we did not find that changes in MKL1 levels supported this mode of inhibition in vivo. Repression by TCFs through either sumoylation and recruitment of HDAC2 or recruitment of mSin3a and HDAC1 has also been reported (24–28); however, our depletion of these factors by siRNAs suggests that they are not major factors for TCF inhibition in nonstarved NIH3T3 cells. Surprising, depletion of the TCFs in serum-starved N100 cells (that have activated MKL1) had little effect on repression, suggesting that there are other limiting factors under these conditions.
FIGURE 10.
Model for TCF and MKL regulation of immediate early gene expression. A model for regulation of c-fos and Egr2 expression is shown in serum-starved, serum-stimulated cells or nonstarved cells. The curved lines with perpendicular lines at the ends indicate transcriptional repression, and those with arrowheads indicate transcriptional activation. See text for details.
In reporter gene assays, we found that TCFs repressed activation by MKL1, and this was dependent upon the Ets and B box domains of ELK1. Both of these mutations affect ELK1 binding to SRF and the SRE; however, they may also affect interactions with a corepressor. The R domain of ELK1 is sumoylated and required for repression in reporter gene assays (27, 60). Mutation of this domain of ELK1 did not affect activation by MKL1 coexpression, consistent with our lack of effect of Ubc9 and histone deacetylase siRNAs on c-fos and Egr2 expression. The domain(s) of ELK1 required for repression of endogenous target genes such as c-fos is unknown and will require careful reconstitution experiments.
The importance of control of MKL1 activation by TCFs or other factors is apparent from the activation of MKL1 in megakaryoblastic leukemia (35, 36). In this disease, MKL1 is fused to the RBM15 protein and is activated due to its constitutive nuclear localization (30, 37).
Despite our increasing understanding of serum regulation of SRF target genes, our results show that additional mechanisms must be required to explain the observed expression of some IEGs. In particular, we find here that serum stimulation of c-fos and Egr2 is relatively normal even in the absence of TCFs and the presence of the nuclear form of MKL1 (N100) (Fig. 7). This suggests that there is still another step(s) required to allow activation of target gene expression by N100 in serum-starved cells. This may be a novel repressor whose activity would be relieved by serum induction, activation of a coactivator, or an activating modification of MKL1. We previously found that phosphorylation of MKL1 serves to inhibit its activity, such that this modification does not appear to be required for MKL1 activation (50). SUMO modification of MKL1 and myocardin has been reported, however, with opposite effects on transcriptional activation by MKL1 and myocardin (75, 76). We have observed SUMO modification of MKL1 with overexpression of SUMO and the SUMO-conjugating enzyme Ubc9. However, we found that a moderate increase of MKL1 activation of SRE reporter genes caused by SUMO and Ubc9 was nonspecific, as several other reporters were activated as well.3
Besides the TCFs, there have also been other repressors of SRF activity identified. The osteogenic transcription factor Runx2 was found to repress myocardin activation of gene expression in C3H10T1/2 cells by binding to SRF and blocking myocardin binding (77). However, we found that overexpression of Runx2 had no effect on MKL1 activation of SRE reporter genes in 3T3 cells.4 Recently, SCAI was also identified as a repressor of MKL1 activity (78). SCAI binds directly to MKL1, and overexpression of SCAI reduced not only MKL1 activation of SRE reporter genes but also serum and RhoA activation. It is not known yet whether SCAI is required for proper regulation of endogenous target genes or whether its activity is regulated by signaling pathways. It will be interesting to determine whether and how these or other factors are involved in serum regulation of SRF target genes.
This work was supported, in whole or in part, by National Institutes of Health Grant CA50329 (to R. P.).
T. C. Lewis and R. Prywes, unpublished results.
S.-M. Lee, M. Vasishtha, and R. Prywes, unpublished results.
- IEG
- immediate early gene
- SRF
- serum-response factor
- SRE
- serum-response element
- TCF
- ternary complex factor
- shRNA
- short hairpin RNA
- siRNA
- short interfering RNA
- ChIP
- chromatin immunoprecipitation
- qRT-PCR
- quantitative reverse transcription RT-PCR
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- MKL
- megakaryoblastic leukemia
- MAPK
- mitogen-activated protein kinase
- KD
- knockdown
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1.Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. (1988) Mol. Cell. Biol. 8, 2140–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cochran B. H., Zullo J., Verma I. M., Stiles C. D. (1984) Science 226, 1080–1082 [DOI] [PubMed] [Google Scholar]
- 3.Müller R., Bravo R., Burckhardt J., Curran T. (1984) Nature 312, 716–720 [DOI] [PubMed] [Google Scholar]
- 4.Greenberg M. E., Ziff E. B. (1984) Nature 311, 433–438 [DOI] [PubMed] [Google Scholar]
- 5.Bravo R. (1990) Cell Growth Differ. 1, 305–309 [PubMed] [Google Scholar]
- 6.Selvaraj A., Prywes R. (2004) BMC Mol. Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treisman R. (1987) EMBO J. 6, 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman C., Runswick M., Pollock R., Treisman R. (1988) Cell 55, 989–1003 [DOI] [PubMed] [Google Scholar]
- 9.Janknecht R., Ernst W. H., Pingoud V., Nordheim A. (1993) EMBO J. 12, 5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchwalter G., Gross C., Wasylyk B. (2004) Gene 324, 1–14 [DOI] [PubMed] [Google Scholar]
- 11.Hill C. S., Wynne J., Treisman R. (1995) Cell 81, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 12.Messenguy F., Dubois E. (2003) Gene 316, 1–21 [DOI] [PubMed] [Google Scholar]
- 13.Cen B., Selvaraj A., Prywes R. (2004) J. Cell. Biochem. 93, 74–82 [DOI] [PubMed] [Google Scholar]
- 14.Hassler M., Richmond T. J. (2001) EMBO J. 20, 3018–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shore P., Sharrocks A. D. (1994) Mol. Cell. Biol. 14, 3283–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducret C., Maira S. M., Lutz Y., Wasylyk B. (2000) Oncogene 19, 5063–5072 [DOI] [PubMed] [Google Scholar]
- 17.Yang S. H., Yates P. R., Whitmarsh A. J., Davis R. J., Sharrocks A. D. (1998) Mol. Cell. Biol. 18, 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S. H., Whitmarsh A. J., Davis R. J., Sharrocks A. D. (1998) EMBO J. 17, 1740–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janknecht R., Zinck R., Ernst W. H., Nordheim A. (1994) Oncogene 9, 1273–1278 [PubMed] [Google Scholar]
- 20.Yang S. H., Shore P., Willingham N., Lakey J. H., Sharrocks A. D. (1999) EMBO J. 18, 5666–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janknecht R., Nordheim A. (1996) Biochem. Biophys. Res. Commun. 228, 831–837 [DOI] [PubMed] [Google Scholar]
- 22.Li Q. J., Yang S. H., Maeda Y., Sladek F. M., Sharrocks A. D., Martins-Green M. (2003) EMBO J. 22, 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissen L. J., Gelly J. C., Hipskind R. A. (2001) J. Biol. Chem. 276, 5213–5221 [DOI] [PubMed] [Google Scholar]
- 24.Maira S. M., Wurtz J. M., Wasylyk B. (1996) EMBO J. 15, 5849–5865 [PMC free article] [PubMed] [Google Scholar]
- 25.Criqui-Filipe P., Ducret C., Maira S. M., Wasylyk B. (1999) EMBO J. 18, 3392–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D. (2003) Mol. Cell 12, 63–74 [DOI] [PubMed] [Google Scholar]
- 27.Yang S. H., Bumpass D. C., Perkins N. D., Sharrocks A. D. (2002) Mol. Cell. Biol. 22, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S. H., Vickers E., Brehm A., Kouzarides T., Sharrocks A. D. (2001) Mol. Cell. Biol. 21, 2802–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miralles F., Posern G., Zaromytidou A. I., Treisman R. (2003) Cell 113, 329–342 [DOI] [PubMed] [Google Scholar]
- 30.Cen B., Selvaraj A., Burgess R. C., Hitzler J. K., Ma Z., Morris S. W., Prywes R. (2003) Mol. Cell. Biol. 23, 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvaraj A., Prywes R. (2003) J. Biol. Chem. 278, 41977–41987 [DOI] [PubMed] [Google Scholar]
- 32.Wang D. Z., Li S., Hockemeyer D., Sutherland L., Wang Z., Schratt G., Richardson J. A., Nordheim A., Olson E. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasazuki T., Sawada T., Sakon S., Kitamura T., Kishi T., Okazaki T., Katano M., Tanaka M., Watanabe M., Yagita H., Okumura K., Nakano H. (2002) J. Biol. Chem. 277, 28853–28860 [DOI] [PubMed] [Google Scholar]
- 34.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 35.Ma Z., Morris S. W., Valentine V., Li M., Herbrick J. A., Cui X., Bouman D., Li Y., Mehta P. K., Nizetic D., Kaneko Y., Chan G. C., Chan L. C., Squire J., Scherer S. W., Hitzler J. K. (2001) Nat. Genet. 28, 220–221 [DOI] [PubMed] [Google Scholar]
- 36.Mercher T., Coniat M. B., Monni R., Mauchauffe M., Nguyen Khac F., Gressin L., Mugneret F., Leblanc T., Dastugue N., Berger R., Bernard O. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5776–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Descot A., Rex-Haffner M., Courtois G., Bluteau D., Menssen A., Mercher T., Bernard O. A., Treisman R., Posern G. (2008) Mol. Cell. Biol. 28, 6171–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Mol. Cell. Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sotiropoulos A., Gineitis D., Copeland J., Treisman R. (1999) Cell 98, 159–169 [DOI] [PubMed] [Google Scholar]
- 40.Posern G., Miralles F., Guettler S., Treisman R. (2004) EMBO J. 23, 3973–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vartiainen M. K., Guettler S., Larijani B., Treisman R. (2007) Science 316, 1749–1752 [DOI] [PubMed] [Google Scholar]
- 42.Sharrocks A. D., Brown A. L., Ling Y., Yates P. R. (1997) Int. J. Biochem. Cell Biol. 29, 1371–1387 [DOI] [PubMed] [Google Scholar]
- 43.Shore P., Whitmarsh A. J., Bhaskaran R., Davis R. J., Waltho J. P., Sharrocks A. D. (1996) Mol. Cell. Biol. 16, 3338–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treisman R., Marais R., Wynne J. (1992) EMBO J. 11, 4631–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaromytidou A. I., Miralles F., Treisman R. (2006) Mol. Cell. Biol. 26, 4134–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J., Herring B. P. (2005) J. Biol. Chem. 280, 10861–10869 [DOI] [PubMed] [Google Scholar]
- 47.Shaw P. E., Schröter H., Nordheim A. (1989) Cell 56, 563–572 [DOI] [PubMed] [Google Scholar]
- 48.Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. (2004) Nature 428, 185–189 [DOI] [PubMed] [Google Scholar]
- 49.Yoshida T., Gan Q., Shang Y., Owens G. K. (2007) Am. J. Physiol. Cell Physiol. 292, C886–C895 [DOI] [PubMed] [Google Scholar]
- 50.Muehlich S., Wang R., Lee S. M., Lewis T. C., Dai C., Prywes R. (2008) Mol. Cell. Biol. 28, 6302–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Prywes R. (2000) Oncogene 19, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 52.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D. (2003) RNA 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 54.Solit D. B., Garraway L. A., Pratilas C. A., Sawai A., Getz G., Basso A., Ye Q., Lobo J. M., She Y., Osman I., Golub T. R., Sebolt-Leopold J., Sellers W. R., Rosen N. (2006) Nature 439, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mo Y., Vaessen B., Johnston K., Marmorstein R. (2000) Nat. Struct. Biol. 7, 292–297 [DOI] [PubMed] [Google Scholar]
- 56.Ling Y., Lakey J. H., Roberts C. E., Sharrocks A. D. (1997) EMBO J. 16, 2431–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price M. A., Rogers A. E., Treisman R. (1995) EMBO J. 14, 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gille H., Kortenjann M., Thomae O., Moomaw C., Slaughter C., Cobb M. H., Shaw P. E. (1995) EMBO J. 14, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasza A., O'Donnell A., Gascoigne K., Zeef L. A., Hayes A., Sharrocks A. D. (2005) J. Biol. Chem. 280, 1149–1155 [DOI] [PubMed] [Google Scholar]
- 60.Yang S. H., Sharrocks A. D. (2004) Mol. Cell 13, 611–617 [DOI] [PubMed] [Google Scholar]
- 61.Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 62.Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 63.Treisman R. (1994) Curr. Opin. Genet. Dev. 4, 96–101 [DOI] [PubMed] [Google Scholar]
- 64.Shore P., Sharrocks A. D. (1995) Eur. J. Biochem. 229, 1–13 [DOI] [PubMed] [Google Scholar]
- 65.Cesari F., Brecht S., Vintersten K., Vuong L. G., Hofmann M., Klingel K., Schnorr J. J., Arsenian S., Schild H., Herdegen T., Wiebel F. F., Nordheim A. (2004) Mol. Cell. Biol. 24, 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giovane A., Pintzas A., Maira S. M., Sobieszczuk P., Wasylyk B. (1994) Genes Dev. 8, 1502–1513 [DOI] [PubMed] [Google Scholar]
- 67.Spencer J. A., Major M. L., Misra R. P. (1999) Mol. Cell. Biol. 19, 3977–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrera R. E., Shaw P. E., Nordheim A. (1989) Nature 340, 68–70 [DOI] [PubMed] [Google Scholar]
- 69.Zhou J., Hu G., Herring B. P. (2005) Mol. Cell. Biol. 25, 9874–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshida T., Gan Q., Owens G. K. (2008) Am. J. Physiol. Cell Physiol. 295, C1175–C1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boros J., Donaldson I. J., O'Donnell A., Odrowaz Z. A., Zeef L., Lupien M., Meyer C. A., Liu X. S., Brown M., Sharrocks A. D. (2009) Genome Res. 19, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ridley A. J., Hall A. (1992) Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 74.Guettler S., Vartiainen M. K., Miralles F., Larijani B., Treisman R. (2008) Mol. Cell. Biol. 28, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakagawa K., Kuzumaki N. (2005) Genes Cells 10, 835–850 [DOI] [PubMed] [Google Scholar]
- 76.Wang J., Li A., Wang Z., Feng X., Olson E. N., Schwartz R. J. (2007) Mol. Cell. Biol. 27, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanaka T., Sato H., Doi H., Yoshida C. A., Shimizu T., Matsui H., Yamazaki M., Akiyama H., Kawai-Kowase K., Iso T., Komori T., Arai M., Kurabayashi M. (2008) Mol. Cell. Biol. 28, 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brandt D. T., Baarlink C., Kitzing T. M., Kremmer E., Ivaska J., Nollau P., Grosse R. (2009) Nat. Cell Biol. 11, 557–568 [DOI] [PubMed] [Google Scholar]
- 79.Wang Y., Falasca M., Schlessinger J., Malstrom S., Tsichlis P., Settleman J., Hu W., Lim B., Prywes R. (1998) Cell Growth Differ. 9, 513–522 [PubMed] [Google Scholar]