Abstract
As the primary microtubule-organizing centers, centrosomes require γ-tubulin for microtubule nucleation and organization. Located in close vicinity to centrosomes, the Golgi complex is another microtubule-organizing organelle in interphase cells. CDK5RAP2 is a γ-tubulin complex-binding protein and functions in γ-tubulin attachment to centrosomes. In this study, we find that CDK5RAP2 localizes to the Golgi complex in an ATP- and centrosome-dependent manner and associates with Golgi membranes independently of microtubules. CDK5RAP2 contains a centrosome-targeting domain with its core region highly homologous to the Motif 2 (CM2) of centrosomin, a functionally related protein in Drosophila. This sequence, referred to as the CM2-like motif, is also conserved in related proteins in chicken and zebrafish. Therefore, CDK5RAP2 may undertake a conserved mechanism for centrosomal localization. Using a mutational approach, we demonstrate that the CM2-like motif plays a crucial role in the centrosomal and Golgi localization of CDK5RAP2. Furthermore, the CM2-like motif is essential for the association of the centrosome-targeting domain to pericentrin and AKAP450. The binding with pericentrin is required for the centrosomal and Golgi localization of CDK5RAP2, whereas the binding with AKAP450 is required for the Golgi localization. Although the CM2-like motif possesses the activity of Ca2+-independent calmodulin binding, binding of calmodulin to this sequence is dispensable for centrosomal and Golgi association. Altogether, CDK5RAP2 may represent a novel mechanism for centrosomal and Golgi localization.
Keywords: Calmodulin, Centrosome, Golgi, Molecular Dynamics, Trafficking, CDK5RAP2
Introduction
Centrosomes play a principal role in organizing the microtubule cytoskeleton of animal cells. They comprise a pair of centriolar cylinders surrounded by the pericentriolar material (PCM),2 wherein activities such as microtubule nucleation and organization, signal transduction, and cell cycle control take place. The PCM has been described as an amorphous mass of proteins, and little is known regarding its structural organization. Many of the proteins localizing to the PCM are large and contain multiple coiled-coils, a structural module known for protein-protein interactions (1). A centrosome-targeting sequence has been identified from the PCM proteins AKAP450 (also known as AKAP350, CG-NAP, and hyperion) and pericentrin (kendrin known as the human homolog) (2). This sequence, referred to as the pericentrin-AKAP450 centrosomal targeting (PACT) domain, contains a core region of ∼90 amino acids located near the protein C terminus. A centrosome-localizing signal is also delineated in the cell cycle regulatory protein cyclin E (3). This signal sequence, consisting of 20 residues with motifs conserved in cyclin E from different mammalian species and in different isoforms, is responsible for attaching cyclin E to the centrosomes during the G1 phase.
Within the centrosomes of animal cells and the spindle pole bodies of budding and fission yeasts, calmodulin (CaM) appears to be involved in various activities such as centrosome duplication (4). Several components of the PCM or spindle pole bodies display CaM-binding activities that serve as potential targets of CaM. For example, Spc110 of budding yeast and its related fission yeast protein Pcp1p contain a CaM-binding site at their C termini (5–8). Moreover, interaction with CaM is required for the stability and function of Spc110 (8, 9). In mammalian cells, the PACT domain of AKAP450 and pericentrin binds to CaM; however, this binding is unessential for the centrosomal localization of AKAP450 (2).
The Golgi complex plays a pivotal role in protein post-translational modification and cargo sorting. It also serves as a microtubule-nucleating center in interphase cells (10–13). The Golgi usually shows a crescent moon morphology surrounding the centrosome during interphase and becomes disassembled during mitosis. Closely associated with each other, the centrosome and the Golgi cooperate to function in various cellular processes such as polarity establishment (14, 15). A number of proteins, including AKPA450, FTCD (58K), and CAP350, localize to both centrosomes and the Golgi complex (16–18). It has been proposed that centrosomes play an important role in Golgi positioning and function (19, 20). In addition, Golgi activities such as GM130-dependent activation of Cdc42 regulate centrosomal organization and function (21).
CDK5RAP2, a protein whose mutations cause autosomal recessive primary microcephaly, is involved in the centrosomal attachment of γ-tubulin and centrosome cohesion (22–24). It contains a γ-tubulin complex-binding domain conserved in Drosophila centrosomin (Cnn) and fission yeast Mto1p and Pcp1p, the proteins known to bind to γ-tubulin complexes (23). This domain has also been referred to as Cnn Motif 1 (CM1) (25). In addition to centrosomal localization, CDK5RAP2 attaches to growing microtubule tips by associating with EB1 and regulates the plus-end dynamics of microtubules (26). In this report, we show the localization of CDK5RAP2 to the Golgi complex and characterize its Golgi association. The centrosome and Golgi-targeting domains are mapped in CDK5RAP2 to the C-terminal regions that contain a conserved motif, Cnn Motif 2 (CM2). The CM2-like sequence of CDK5RAP2 exhibits Ca2+-independent CaM-binding activity. We also investigate the mechanisms employed by CDK5RAP2 to target centrosomes and the Golgi complex and the role of the CM2-like sequence, as well as its CaM-binding activity, in such localizations.
EXPERIMENTAL PROCEDURES
Plasmids and Immunoreagents
Cloning of CDK5RAP2 fragments and deletion and point mutation variants were conducted by PCR methods using the human sequence (23) as template. The cDNA coding for human centrin 2 was amplified from a human ovary cDNA library (Clontech) and inserted into pEGFP-N3 for expression of centrin-GFP. The following siRNA duplexes were synthesized and used as described previously: si-CDK5RAP2, 5′-UGGAAGAUCUCCUAACUAA-3′ (23); si-pericentrin, 5′-GCAGCUGAGCUGAAGGAGA-3′ (27); and si-AKAP450, 5′-AACUUUGAAGUUAACUAUCAA-3′ (11).
A mouse monoclonal antibody was generated against 1–70 of CDK5RAP2. Clone 5F6 of the monoclonal antibody was selected to show high specificity of immunostaining. The anti-Odf2/hCenexin antibody (28) was a gift from Dr. Kyung S. Lee (National Cancer Institute, Bethesda, MD); the monoclonal antibody GT335 that labels centrioles (29) was from Dr. Carsten Janke (Insitut Curie, Orsay, France). The following antibodies were purchased: anti-γ-tubulin (GTU88; Sigma), anti-α-tubulin (Sigma), anti-centrin2 (N-17; Santa Cruz Biotechnology, Santa Cruz, CA), anti-mannosidase II (Millipore), anti-TGN46 (Serotec), anti-GM130 (BD Biosciences), monoclonal anti-AKAP450 (BD Biosciences), anti-pericentrin2 (Santa Cruz Biotechnology), anti-ϵ-tubulin (Sigma), anti-EGFP (FL, Santa Cruz Biotechnology), anti-His6 (H-15, Santa Cruz Biotechnology), and anti-FLAG (M2, Sigma).
Cell Culture and Fluorescence Microscopy
HeLa and MCF-7 cells in Dulbecco's modified Eagle's medium, hTERT-RPE-1 cells in Dulbecco's modified Eagle's medium/F12 (1:1), and MRC5 cells in minimal essential medium were cultured with 10% fetal bovine serum at 37 °C in 5% CO2. Transfection was performed with the Lipofectamine and Plus reagents (Invitrogen). Drug treatments were performed by maintaining cells at 37 °C in the complete media under one of the following conditions: 10 μg/ml brefeldin A (Sigma) for 1 h, 10 μm nocodazole (Sigma) for 1.5 h, or 50 mm 2-deoxy-d-glucose and 10 mm sodium azide for 1.5 h. Cells were fixed in methanol at −20 °C for 5 min and were then subjected to staining with antibodies as indicated. Secondary antibodies were Alexa Fluor 488 or Alexa Fluor 594-conjugated donkey antibodies (Invitrogen). Nuclei were labeled with Hoechst 33258 (Sigma).
Cell images were acquired using an epifluorescence microscope (Eclipse TE2000; Nikon). The quantification of fluorescence intensity was performed using the MetaMorph software (Molecular Devices) as reported previously (30, 31). The images selected for quantification were unsaturated and acquired under identical conditions. The average fluorescence intensity was measured within a ∼1-μm-diameter area over the centrosomes or placed at the cytoplasmic areas without any specific pattern. In each cell, three different areas in the cytoplasm were measured to obtain the average cytoplasmic intensity. Background intensity taken from untransfected cells was subtracted from the centrosomal and cytoplasmic measurements.
Centrosome Ablation
The laser ablation of centrosomes was performed as described by Khodjakov and co-workers (32) with modifications. Briefly, two-photon excited fluorescence images of centrosomes labeled with centrin-GFP in HeLa cells were obtained by excitation with a femotosecond laser with pulse energy of 0.06 nJ (5 milliwatts). Laser pulses of 800 nm, 120 fs, 76 MHz generated by a titanium:sapphire laser (Mira 900; Coherent) were focused onto the centrosome with a 60× and 1.20 NA objective lens. A 0.8-μm square area centered at the centrin dot was ablated with 8 nJ (620 milliwatts; 18 μs) femtosecond laser pulses by a 3 × 3 point scan. After ablation, cells were recovered at 37 °C in fresh medium for ∼2 h.
CaM Binding Assay
CDK5RAP2 fragments were expressed in fusion with a His6 tag in Escherichia coli BL21(DE3) or with GFP in HEK293T. Lysates were prepared with CaM-binding buffer (10 mm Tris-HCl, pH7.3, 150 mm NaCl, 1 mm MgCl2, 1 mm EDTA, 0.5% Triton X-100, and 1 mm phenylmethylsulfonyl fluoride) supplemented with 2 mm CaCl2 or 5 mm EGTA as indicated and clarified by centrifugation (25,000 × g; 30 min) at 4 °C. CaM binding was tested in assays as described previously (33). Briefly, CaM-conjugated Sepharose 4B (GE Healthcare), which was prewashed with respective buffers, was incubated in bacterial or cell extracts for 2 h at 4 °C with agitation. Blank Sepharose 4B (GE Healthcare) was used in controls instead of the CaM beads. The beads were then collected, washed, and boiled in the SDS-PAGE sample loading buffer.
RESULTS
Centrosome-dependent Localization to the Golgi Complex
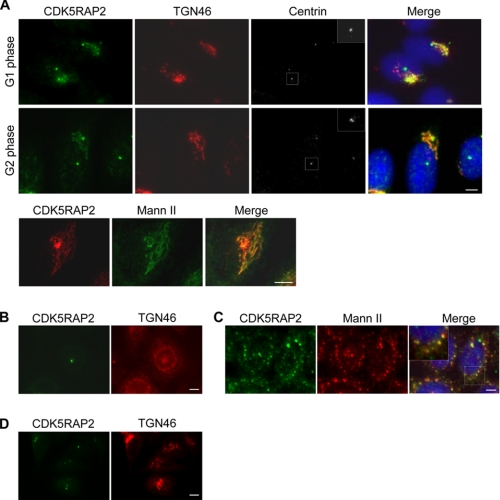
We produced and subsequently affinity-purified a monoclonal antibody recognizing an N-terminal region of CDK5RAP2. When HeLa cells were fixed in methanol at −20 °C and stained with the monoclonal antibody, CDK5RAP2 showed prominent localization to the centrosomes (Fig. 1A). This is similar to previously reported observations (22–24). In addition, the monoclonal antibody stained the Golgi complex, which was labeled with antibodies against TGN46 (trans-Golgi network) or mannosidase II (cis/medial-Golgi) (Fig. 1A). The Golgi localization lasted throughout the interphase until mitosis, at which point the Golgi was extensively fragmented. Golgi localization was also observed in several other examined human cell lines, including RPE-1 and MCF-7. Staining of both the Golgi and centrosomes was eliminated when the expression of CDK5RAP2 was suppressed using siRNAs (data not shown), confirming immunostaining specificity of the antibody.
FIGURE 1.
Golgi localization of CDK5RAP2. A, HeLa cells were stained with the monoclonal CDK5RAP2 antibody and other antibodies as labeled. Mann II, mannosidase II. Cell cycle stages were identified by the centrin patterns (insets). B–D, cells were subjected to treatment with brefeldin A (B), nocodazole (C), or 2-deoxy-d-glucose and sodium azide (D). Scale bars, 5 μm.
To confirm Golgi localization further, cells were treated with brefeldin A to induce Golgi fragmentation (34). The treatment abolished the Golgi-staining patterns of CDK5RAP2 without affecting its centrosomal staining (Fig. 1B). Golgi association was also characterized using the following procedures. Treatment with nocodazole, a microtubule-depolymerizing agent, caused Golgi fragmentation by inhibiting endoplasmic reticulum-to-Golgi anterograde transport (35). In the nocodazole-treated cells, CDK5RAP2 continued to co-localize with mannosidase II to the fragmented Golgi (Fig. 1C), indicating that CDK5RAP2 targets Golgi membranes in a microtubule-independent manner. When cells were depleted of the cellular ATP pool by treatment with 2-deoxy-d-glucose and sodium azide (36), CDK5RAP2 dissociated from the Golgi complex (Fig. 1D), revealing the energy dependence of CDK5RAP2 retention at the Golgi complex. The above treatments did not alter the centrosomal localization of CDK5RAP2.
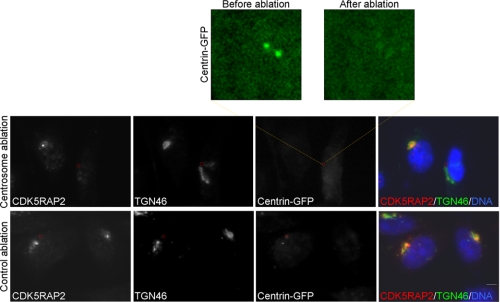
Given the localization of CDK5RAP2 to both the Golgi and centrosomes, we investigated whether Golgi localization is centrosome-dependent. To this end, centrosomes labeled with centrin-GFP were removed by short pulses of a laser beam. The ablation removed the centrosomal pattern of centrin-GFP fluorescence (Fig. 2, upper). After ablation, cells were maintained under culture conditions for a period of time (∼2 h) before they were subjected to immunostaining. The laser ablation eliminated the centrosomal staining of CDK5RAP2 (Fig. 2), confirming the physical removal of the centrosomes. The ablation did not disrupt the Golgi complex, as the Golgi morphology appeared normal in centrosome-ablated cells (Fig. 2). However, CDK5RAP2 completely dissociated from the Golgi complex in the ablated cells (Fig. 2). In the control cells, laser ablation performed at a cytoplasmic area under the same conditions did not affect the centrosomal and Golgi localization of CDK5RAP2 (Fig. 2). These assay results indicate that centrosomes play an indispensable role in the association of CDK5RAP2 with the Golgi complex.
FIGURE 2.
Centrosomes are required for the localization of CDK5RAP2 to the Golgi complex. HeLa cells expressing centrin-GFP were subjected to laser ablation of the centrosome or a cytoplasmic area (inside the boxed areas). Enlarged is the centrosomal area before and after ablation. After ablation, cells were cultured for 2 h and then processed for immunostaining. Shown are the representatives of five centrosome-ablated or control ablation cells from three separate experiments. Scale bar, 5 μm.
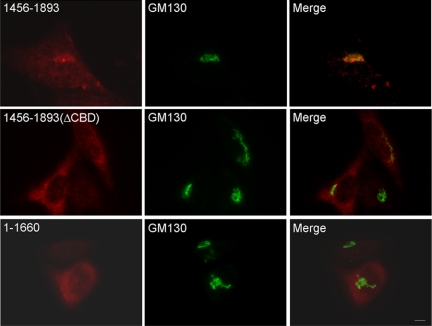
Various CDK5RAP2 fragments were produced to investigate their potential in Golgi targeting. The C-terminal fragment 1456–1893 displayed Golgi and centrosome patterns similar to those of the full-length protein, whereas the N-terminal construct 1–1660 did not show such properties (Fig. 3). A mutant of 1456–1893 was generated in which a short segment adjacent to the C terminus, 1861–1870, was deleted. The mutation was referred to as ΔCBD (see below). The ΔCBD mutant of 1456–1893 did not show any obvious Golgi pattern (Fig. 3), revealing the requirement of having 1861–1870 for localization. Truncation of 1456–1893 to 1726–1893 greatly diminished the Golgi-localizing activity (see Fig. 7A). Hence, Golgi localization requires a large C-terminal domain encompassing the region 1861–1870.
FIGURE 3.
Mapping the Golgi-targeting region. Cells transfected with CDK5RAP2 fragments or mutant (FLAG-tagged) were processed for anti-FLAG and anti-GM130 staining. 1456–1893(ΔCBD), 1456–1893 deleted from fragment(1861–1870). Representative images of three independent experiments are presented. Scale bar, 5 μm.
FIGURE 7.
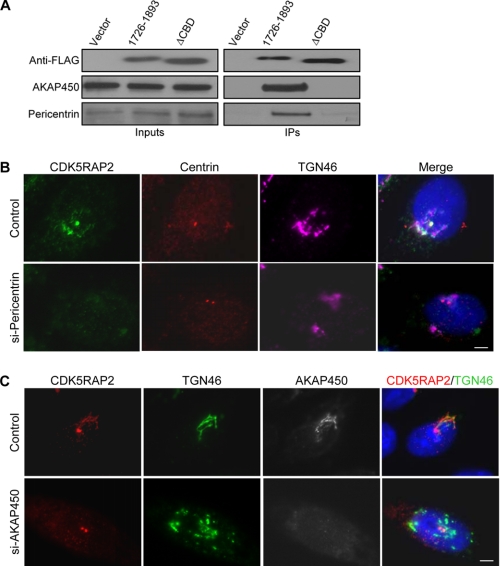
CM2-like motif is required for binding of several PCM/Golgi proteins to the C-terminal region of CDK5RAP5. A, HeLa cells transfected with FLAG-(1726–1893) or its ΔCBD mutant were subjected to anti-FLAG immunoprecipitation. The immunoprecipitates (IPs) and the extracts (Inputs) were analyzed on immunoblots. ΔCBD, fragment (1726–1893)(ΔCBD). B, cells transfected with control or pericentrin-targeting siRNA immunostained for CDK5RAP2, centrin2, and TGN46. C, HeLa cells transfected with control or AKAP450-targeting siRNA subjected to immunostaining. Data shown are representative from three independent experiments. Scale bars, 5 μm.
Conserved Motif at the C Terminus of CDK5RAP2
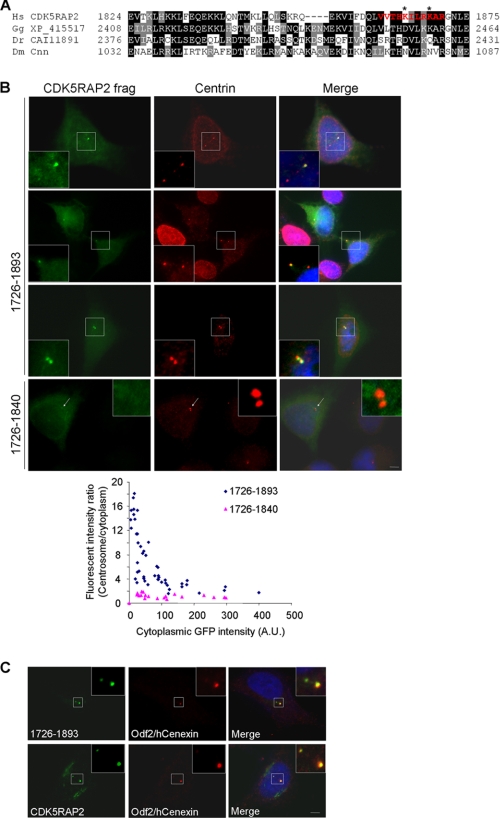
CDK5RAP2 and its related Drosophila protein Cnn share homology in two regions, CM1 and CM2, located at the N and C termini, respectively (23, 25, 37). We have shown previously that the CM1-like sequence of CDK5RAP2 is a γ-tubulin complex-binding domain (23). Sequences that are highly homologous to CM1 have been found in several proteins in lower organisms, including three putative animal proteins: one from chicken (XP_415517) and two from zebrafish (CAI11891 and AAH46878) (23, 37). Located within the Golgi-targeting region, the CM2-like motif comprises ∼50 amino acids adjacent to the C terminus of CDK5RAP2 (Fig. 4A). Chicken XP_415517 and zebrafish CAI11891 contain a homologous sequence, but zebrafish AAH46878 does not (Fig. 4A). Interestingly, sequence analysis via a Web-based method using the Calmodulin Target Data base (38) predicted a CaM-binding site spanning 1861–1870 of CDK5RAP2 (Fig. 4A). This short stretch is rich in hydrophobic and basic residues and is also predicted to exist in an α-helix configuration. Both are features of CaM-binding domains.
FIGURE 4.
CDK5RAP2 contains a conserved centrosome-targeting domain adjacent to the C terminus. A, sequence alignment of a C-terminal region from CDK5RAP2 and related proteins. Two putative proteins are from chicken (Gallus gallus; GenBank accession no. XP_415517) and zebrafish (Danio rerio; GenBank accession no. CAI11891). Sequence in red is the predicted CaM-binding motif. Asterisks mark Lys1865 and Lys1869 that are mutated for CaM-binding tests. B, C-terminal fragments of CDK5RAP2 transiently expressed in fusion with GFP at the N terminus. The cells were stained with a centrin antibody. Arrows denote centrosomes. Fluorescent intensities were determined at different expression levels of the proteins to derive the intensity ratios of the centrosomes to the cytoplasm. The cytoplasmic GFP signals are expressed in arbitrary units (A.U.). C, HeLa cells transfected with GFP-tagged 1726–1893 (upper) and stained for Odf2/hCenexin to identify mother centrioles. Untransfected cells (lower) were stained for endogenous CDK5RAP2 and Odf2/hCenexin. The same results were obtained in three independent experiments. Scale bars, 5 μm.
We generated several GFP fusion constructs derived from the C terminus of CDK5RAP2 and transfected them into HeLa cells to examine their subcellular localization. At low and medium expression levels, the fragment 1726–1893, a construct containing the entire CM2-like motif, showed strong localization to the centrosomes (Fig. 4B). These centrosomes were labeled for centrin, a centrosomal marker residing at the distal ends of the centrioles (39). The fragment 1726–1893 also showed Golgi-like patterns in the perinuclear region, with very low intensities (see Fig. 7A). To assess the centrosome-localizing activity further, we quantified the signals on the centrosomes, and in the cytoplasm of 1726–1893 from cells expressing the protein at various levels. The centrosome/cytoplasm ratios of the fragment showed the protein to be highly enriched on the centrosomes at low expression levels (Fig. 4B). When the expression was increased, the centrosome/cytoplasm ratios were quickly reduced accordingly until eventually, they were maintained at a low level (Fig. 4B). These results indicate that 1726–1893 attaches to the centrosomes with high affinity and specificity, and in a saturable manner.
The fragment 1726–1893 displayed stronger localization to one of the two centrioles in majority of the transfected cells (Fig. 4B). This phenotype was most evident in cytokinesis/early G1 cells (Fig. 4B). Double staining with an Odf2/hCenexin antibody, which specifically labels mother centrioles (28, 40), revealed the preferential localization of the transfected fragment to the mother centrioles (Fig. 4C). The fragment 1726–1893 became equally distributed to both centrioles in a small population of cells (Fig. 4B). The centrosome-localizing patterns of 1726–1893 were very similar to those of endogenous CDK5RAP2 (Fig. 4C). Note that the expression of this CDK5RAP2 fragment at high levels formed cytoplasmic aggregates and was identified as cytotoxic. The fragment 1726–1840, a construct with a truncated CM2-like motif, did not show any discernible centrosomal accumulation (Fig. 4B). Further truncation at the N terminus of 1726–1893 led to the formation of protein aggregates in various sizes in the cytoplasm, even at low expression levels. Hence, 1726–1893 specifically targets centrosomes with the CM2-like motif within the region essential for centrosomal targeting.
Binding of CaM with the CM2-like Motif
We tested putative CaM binding with the CM2-like motif of CDK5RAP2. Recombinant proteins derived from the C terminus of CDK5RAP2 and encompassing the CM2-like motif were expressed in bacteria with a His6 tag at the N terminus. The construct 1660–1893 was used for the binding test because of the poor solubility of 1726–1893 expressed in bacteria. The binding assays were performed in the presence of Ca2+ or EGTA. In the assays, bacterial extracts expressing 1660–1893 were incubated with CaM-conjugated or blank Sepharose. After binding, the immunoblotted proteins bound to the Sepharose beads revealed the association of 1660–1893 with the CaM of Ca2+ (Fig. 5A). In a control assay, a His6-tagged irrelevant protein did not bind to CaM (data not shown). This indicates that tag moiety does not conform to the CaM binding. Therefore, this CDK5RAP2 fragment displays Ca2+-independent and direct binding of CaM.
FIGURE 5.
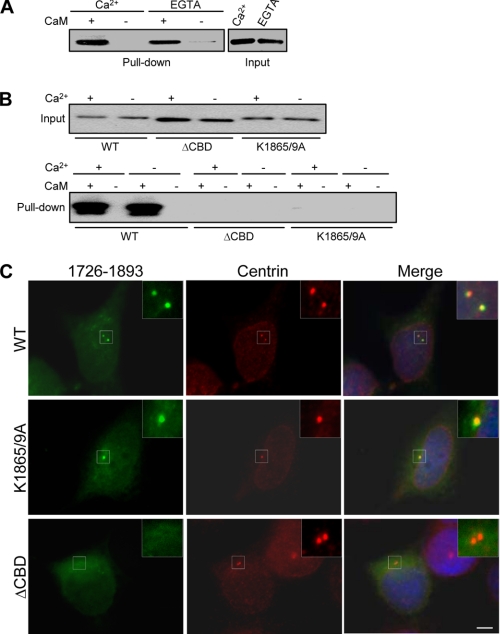
CaM binds to the CM2-like motif of CDK5RAP2. A, bacterial extracts expressing His6-tagged 1660–1893 were prepared in the presence of either 2 mm Ca2+ or 5 mm EGTA. After incubation of CaM-conjugated or blank Sepharose in the extracts, proteins bound to the beads were immunoblotted with an anti-His6 antibody. B, fragment(1660–1893) and its mutants were transiently expressed in HEK293T with a GFP tag. The cells were extracted either in the Ca2+-containing or in the EGTA-containing buffer. After binding of proteins in the extracts to CaM-conjugated or blank Sepharose, bound proteins and the inputs were analyzed on anti-GFP immunoblots. ΔCBD, 1660–1893(Δ1861–1870); K1865/9A, 1660–1893(K1865A/K1869A); WT, wild type. C, fragment(1726–1893) and its ΔCBD and K1865A/K1869A mutants were expressed in HeLa cells as GFP-tagged proteins. The cells were subjected to anti-centrin immunostaining. The same results were obtained in three independent experiments. Scale bar, 5 μm.
To validate the requirement of the predicted CaM-binding site for CaM binding, we generated a mutant of 1660–1893 with deleted 1861–1870 region and designated it as the CaM-binding domain-deleted mutant (ΔCBD). We also noted that two Lys residues within the predicted motif, Lys1865 and Lys1869, were not conserved in the related proteins of lower organisms (Fig. 4A). We tested the involvement of these two residues in CaM binding by substituting both lysines with Ala. The resulting mutant was designated as 1660–1893(K1865A/K1869A). The wild-type and mutant constructs were transiently expressed in HEK293T instead of in the bacteria for the CaM-binding test, as the mutant protein expressed in bacteria showed high background attachment to Sepharose. The HEK293T extracts were prepared under the condition either with or without Ca2+ but with EGTA. The wild-type protein specifically co-precipitated with CaM, both in the presence and in the absence of Ca2+ (Fig. 5B). This is in agreement with the binding results using the bacterially expressed protein. Under both conditions, neither 1660–1893(ΔCBD) nor 1660–1893(K1865A/K18699A) showed any detectable CaM-binding activity (Fig. 5B). These results reveal the crucial roles of 1861–1870, as well as the two lysines within this motif, in associating with CaM.
To investigate the potential role of CaM binding in the centrosomal targeting of the CM2-like motif, we examined the subcellular localizations of 1726–1893 and its K1865A/K1869A and ΔCBD mutants. The K1865A/K1869A mutant displayed intensive localization at the centrosomes, similar to the wild-type (Fig. 5C). However, the ΔCBD mutant was distributed throughout the cytoplasm without specific accumulation on the centrosomes (Fig. 5C). These results indicate that the sequence 1861–1870, but not the CaM-binding activity, is indispensable for centrosomal targeting. Considering the conservation of the CM2-like motif, but not of Lys1865 and Lys1869, in the related proteins of lower organisms, we reason that centrosomal targeting, but not CaM binding, is a conserved function of this domain.
Requirement of the CM2-like Motif for Centrosome and Golgi Targeting
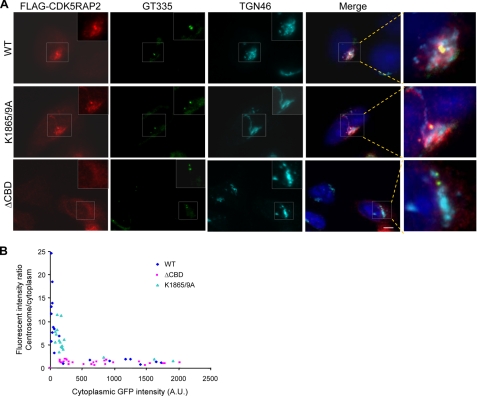
To assess the role of the CM2-like motif and its CaM-binding activity in CDK5RAP2 localization, we introduced the ΔCBD and K1865A/K1869A mutations into full-length CDK5RAP2. When expressed in HeLa cells, both the wild-type and K1865A/K1869A proteins exhibited centrosomal and Golgi localization (Fig. 6A). The ΔCBD mutant displayed mostly diffused patterns, without specific localization to the Golgi and centrosomes (Fig. 6A). The centrosome-localizing activities were further evaluated by determining the centrosome/cytoplasm ratios of fluorescent signals. The fluorescence intensities at the centrosomes and in the cytoplasm were measured in cells transfected at various levels to determine the ratios. The wild-type protein and the K1865A/K1869A mutant were incorporated into the centrosomes with similar affinities, whereas the ΔCBD mutant yielded low centrosome/cytoplasm intensity ratios across the concentration range (Fig. 6B). These results indicate the essential role of the CaM-binding domain, but not its CaM-binding function, within the CM2-like motif in targeting CDK5RAP2 to centrosomes and the Golgi complex.
FIGURE 6.
Centrosomal and Golgi localization of CDK5RAP2 and its mutants. A, CDK5RAP2 wild-type (WT) and the ΔCBD and K1865A/K1869A mutants expressed in HeLa cells. The cells were stained for FLAG-CDK5RAP2 (anti-FLAG), centrioles (GT335), TGN46, and DNA. Boxed areas are enlarged. ΔCBD, CDK5RAP2(ΔCBD); K1865/9A, CDK5RAP2(K1865A/K1869A). B, fluorescent intensity ratios of the centrosomes to the cytoplasm. Analyzed are cells expressing the wild type or the mutants at various levels. Representative results of three independent experiments for each panel are shown. Scale bar, 5 μm.
To explore the molecular basis underlying the centrosome targeting of CDK5RAP2, we searched for proteins that interact with its tail region. It has been shown that CDK5RAP2 and pericentrin display mutual dependence for centrosomal localization (41). In an immunoprecipitation experiment of 1726–1893, the co-immunoprecipitation of pericentrin was readily detected (Fig. 7A). In addition, AKAP450, a protein that localizes to both centrosomes and the Golgi complex (18, 42, 43), was also found to co-precipitate with 1726–1893 (Fig. 7A). In contrast, neither pericentrin nor AKAP450 was co-immunoprecipitated with the ΔCBD mutant (Fig. 7A). We also probed another centrosomal protein, ϵ-tubulin (44), but did not detect it in the immunoprecipitates (data not shown), revealing that the centrosomes were not pulled down by the CDK5RAP2 fragment.
We went further to suppress the expression of pericentrin and AKAP450 and examined their effects on the localization of CDK5RAP2. The expressions were inhibited by transfecting cells with siRNA duplexes against pericentrin or AKAP450 as described previously (11, 27). The inhibition of pericentrin expression blocked the localization of CDK5RAP2 to both the centrosomes and the Golgi complex (Fig. 7B). Interestingly, the depletion of AKAP450 eliminated the Golgi localization of CDK5RAP2, but did not affect its centrosomal localization (Fig. 7C). We also observed Golgi fragmentation upon AKAP450 depletion (Fig. 7C), in agreement with a previous report (45). These results indicate that CDK5RAP2 requires interaction with pericentrin for centrosomal and Golgi localization and requires binding with AKAP450 for Golgi localization.
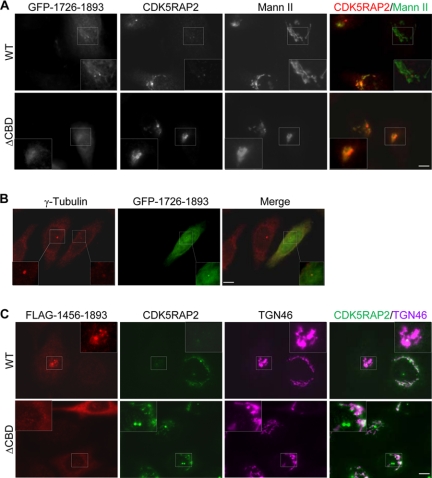
We overexpressed the centrosome-targeting fragment 1726–1893 to assess its effect on the localization of endogenous CDK5RAP2. The expression of 1726–1893, even at low levels, dramatically reduced the staining of endogenous CDK5RAP2 at the centrosomes and the Golgi networks (Fig. 7A). In contrast, the expression of its ΔCBD mutant did not show such effects (Fig. 8A). As previously mentioned, these expressions did not affect the centrosomal localization of centrin (Figs. 4B and 5C). However, the expression of the wild-type fragment inhibited the localization of γ-tubulin to the centrosomes (Fig. 8B), consistent with the function of CDK5RAP2 in recruiting γ-tubulin to centrosomes (23). We observed similar effects using the longer construct 1456–1893 (Fig. 8C and data not shown). These assays suggest that the expression of the centrosome-targeting domains displaces endogenous CDK5RAP2 from centrosomes and abolishes the Golgi localization of the endogenous protein.
FIGURE 8.
Overexpression of the C-terminal fragments delocalizes endogenous CDK5RAP2 from centrosomes and the Golgi complex. A, HeLa cells were transfected with the CDK5RAP2 fragment(1726–1893) or its ΔCBD mutant. The cells were stained for endogenous CDK5RAP2 and mannosidase II (Mann II). WT, wild type; ΔCBD, fragment(1861–1870) deletion mutant. B, cells expressing fragment(1726–1893) were stained for γ-tubulin. The centrosomal area is enlarged from an untransfected (solid line) and a transfected (dashed line) cell. C, cells were transfected with fragment(1456–1893) and then stained for endogenous CDK5RAP2 and TGN46. Representative data of three separate experiments for each panel are presented. Scale bars, 5 μm.
DISCUSSION
CDK5RAP2, a large protein with multiple coiled-coil domains, displays several functions in the control of microtubule organization. Such functions include assembly of γ-tubulin into centrosomes, centrosome cohesion, and microtubule plus-end regulation (23, 24, 26). In the present study, we have demonstrated the localization of CDK5RAP2 at the Golgi complex using a newly generated monoclonal antibody. Similarly, myomegalin (also named as phosphodiesterase 4D-interacting protein), a CDK5RAP2 homolog expressed in mammalian muscles, is a centrosomal and Golgi protein (46). A possible explanation as to why the Golgi localization of CDK5RAP2 has not been described previously is that the ability to detect Golgi association can be affected by antibody sensitivity and specificity, as well as by epitope location. Upon brefeldin A treatment, CDK5RAP2 completely disperses from the Golgi throughout the cytoplasm without detectable retention in the endoplasmic reticulum, implying that it is unlikely to be a luminal Golgi resident protein. CDK5RAP2 associates with Golgi membranes through a microtubule-independent mechanism, as it remains to associate with fragmented Golgi stacks in nocodazole-treated cells. We have also determined that ATP depletion quickly dissociates CDK5RAP2 from the Golgi, suggesting a dynamic association of CDK5RAP2 with the Golgi complex. Such ATP-dependent association is reminiscent of proteins residing in the cis-Golgi network that are actively involved in endoplasmic reticulum-to-Golgi trafficking (36).
At present, the function of CDK5RAP2 at the Golgi remains unclear. As a microtubule-organizing organelle, the Golgi complex contains γ-tubulin complexes to mediate microtubule nucleation (10). Given that CDK5RAP2 is a γ-tubulin complex-binding protein (23), CDK5RAP2 may act in the attachment of γ-tubulin complexes to, and microtubule nucleation at, the Golgi complex. It has been reported that GMAP-210 and AKAP450 are involved in such functions at cis-Golgi membranes (10, 11). CDK5RAP2 seems to be functionally related to these proteins at the Golgi complex.
The centrosome and the Golgi complex are two closely associated cellular organelles. The centrosome has been suggested as determining Golgi positions and playing an important role in its function (19, 20). We have found that CDK5RAP2 resides in both organelles. Moreover, using laser-based microsurgery, we have demonstrated that the Golgi association of CDK5RAP2 is dependent on centrosomes. This finding represents the first report of the indispensable role of centrosomes in protein assembly in the Golgi complex. It is plausible that CDK5RAP2 traffics from centrosomes to the Golgi complex; such trafficking would be one mode by which centrosomes affect Golgi organization.
In eukaryotes, γ-tubulin is essential in the microtubule-organizing function of centrosomes or their equivalents. CDK5RAP2 associates with the γ-tubulin ring complex through the conserved CM1-like motif at the protein N terminus, thus playing an important role in the centrosomal assembly of γ-tubulin (23). We have demonstrated here that the CM2-like motif and its flanking regions serve as a centrosome-targeting domain. Both CM1 and CM2 are conserved in the putative proteins of chicken (XP_415517) and zebrafish (CAI11891), implying that these proteins are functionally related to CDK5RAP2 and Cnn in the respective organisms. The CM2-like motif of CDK5RAP2 contains an unclassified CaM-binding motif and possesses a CaM-binding activity. However, such binding is not required for centrosomal and Golgi targeting of the protein. In fact, one or both lysines within the CaM-binding motif crucial for CaM binding are substituted in Drosophila Cnn and in the chicken and zebrafish sequences, suggesting that such binding property is not conserved in the corresponding sequence of these proteins.
The CDK5RAP2 fragment 1726–1893 containing the CM2-like motif associates with centrosomes with high affinity and in a specific manner. Within this fragment, the CM2-like motif plays a crucial role in centrosomal assembly. This conserved motif does not show significant homology to documented centrosome-localizing sequences, such as the PACT domain from pericentrin and AKAP450 and the centrosomal localization signal of cyclin E (2, 3). Therefore, CDK5RAP2 and related proteins may contain a distinct centrosome-targeting domain. The disruption of the CM2-like motif dramatically diminishes the centrosomal localization of CDK5RAP2, pointing to a principal role of this domain in the attachment of CDK5RAP2 to centrosomes. These results suggest that the centrosomal localization is through a conserved mechanism dictated by the C-terminal sequence.
Our data show that CDK5RAP2 appears to be assembled onto centrosomes through interacting with pericentrin, although the centrosome-targeting region of CDK5RAP2 interacts with both pericentrin and AKAP450. Pericentrin is a scaffold protein that serves to recruit a number of proteins to the PCM (47, 48). The interaction with pericentrin is consistent with our previous observation that the cytoplasmic aggregates formed by overexpressed CDK5RAP2 contains pericentrin (23). However, our results do not exclude that the possible association of CDK5RAP2 with other PCM proteins also contributes to its centrosomal assembly. In fact, we detected the binding of CDK5RAP2 to Cep170,3 a centrosomal protein with preferential localization to the mother centriole (49), in addition to pericentrin and AKAP450. Therefore, CDK5RAP2 may associate with multiple proteins on the centrosomes.
Compared with the centrosome-targeting sequence, a relatively large region (i.e. 1456–1893) containing the CM2-like motif is responsible for directing CDK5RAP2 to the Golgi complex. Within the Golgi-targeting region, the centrosome-targeting sequence (i.e. 1726–1893) has at least two roles in Golgi attachment. First, it plays an indirect role by determining the centrosomal assembly of CDK5RAP2, as the centrosomal targeting is requisite for the Golgi localization of CDK5RAP2. Second, it associates with the Golgi protein AKAP450. Such association is indispensable but inadequate for the Golgi localization. Thus, CDK5RAP2 employs a complex targeting mechanism in which the centrosome-targeting sequence plays a key role. Future analysis of how centrosomes mediate the Golgi targeting of CDK5RAP2 would provide more insights on the centrosome and Golgi coalition.
This work was supported by the Research Grants Council (General Research Fund and Collaborative Research Fund) and the University Grants Committee (Area of Excellence and Special Equipment Grant schemes) of Hong Kong.
Z. Wang, L. Shi, and R. Z. Qi, unpublished results.
- PCM
- pericentriolar material
- PACT
- pericentrin-AKAP450 centrosomal targeting
- CaM
- calmodulin
- Cnn
- centrosomin
- CM1
- centrosomin Motif 1
- CM2
- centrosomin Motif 2
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- ΔCBD
- CaM-binding domain-deleted mutant.
REFERENCES
- 1.Salisbury J. L. (2003) Curr. Biol. 13, R88–R90 [DOI] [PubMed] [Google Scholar]
- 2.Gillingham A. K., Munro S. (2000) EMBO Rep. 1, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto Y., Maller J. L. (2004) Science 306, 885–888 [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto Y., Maller J. L. (2002) Science 295, 499–502 [DOI] [PubMed] [Google Scholar]
- 5.Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N. (2002) Cell Growth Differ. 13, 47–58 [PubMed] [Google Scholar]
- 6.Spang A., Grein K., Schiebel E. (1996) J. Cell Sci. 109, 2229–2237 [DOI] [PubMed] [Google Scholar]
- 7.Geiser J. R., Sundberg H. A., Chang B. H., Muller E. G., Davis T. N. (1993) Mol. Cell. Biol. 13, 7913–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stirling D. A., Welch K. A., Stark M. J. (1994) EMBO J. 13, 4329–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundberg H. A., Goetsch L., Byers B., Davis T. N. (1996) J. Cell Biol. 133, 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ríos R. M., Sanchís A., Tassin A. M., Fedriani C., Bornens M. (2004) Cell 118, 323–335 [DOI] [PubMed] [Google Scholar]
- 11.Rivero S., Cardenas J., Bornens M., Rios R. M. (2009) EMBO J. 28, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. (2001) Mol. Biol. Cell 12, 2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P. M., Andreyeva N., Gleeson P., Galjart N., Maia A. R., McLeod I. X., Yates J. R., 3rd, Maiato H., Khodjakov A., Akhmanova A., Kaverina I. (2007) Dev. Cell 12, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magdalena J., Millard T. H., Machesky L. M. (2003) J. Cell Sci. 116, 743–756 [DOI] [PubMed] [Google Scholar]
- 15.Bisel B., Wang Y., Wei J. H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. (2008) J. Cell Biol. 182, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppeler-Lebel A., Celati C., Bellett G., Mogensen M. M., Klein-Hitpass L., Bornens M., Tassin A. M. (2007) J. Cell Sci. 120, 3299–3308 [DOI] [PubMed] [Google Scholar]
- 17.Hagiwara H., Tajika Y., Matsuzaki T., Suzuki T., Aoki T., Takata K. (2006) Histochem. Cell Biol. 126, 251–259 [DOI] [PubMed] [Google Scholar]
- 18.Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. (1999) J. Biol. Chem. 274, 17267–17274 [DOI] [PubMed] [Google Scholar]
- 19.Marie M., Dale H. A., Sannerud R., Saraste J. (2009) Mol. Biol. Cell 20, 4458–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller P. M., Folkmann A. W., Maia A. R., Efimova N., Efimov A., Kaverina I. (2009) Nat. Cell Biol. 11, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodani A., Kristensen I., Huang L., Sütterlin C. (2009) Mol. Biol. Cell 20, 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bond J., Roberts E., Springell K., Lizarraga S. B., Lizarraga S., Scott S., Higgins J., Hampshire D. J., Morrison E. E., Leal G. F., Silva E. O., Costa S. M., Baralle D., Raponi M., Karbani G., Rashid Y., Jafri H., Bennett C., Corry P., Walsh C. A., Woods C. G. (2005) Nat. Genet. 37, 353–355 [DOI] [PubMed] [Google Scholar]
- 23.Fong K. W., Choi Y. K., Rattner J. B., Qi R. Z. (2008) Mol. Biol. Cell 19, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graser S., Stierhof Y. D., Nigg E. A. (2007) J. Cell Sci. 120, 4321–4331 [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Megraw T. L. (2007) Mol. Biol. Cell 18, 4037–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong K. W., Hau S. Y., Kho Y. S., Jia Y., He L., Qi R. Z. (2009) Mol. Biol. Cell 20, 3660–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dammermann A., Merdes A. (2002) J. Cell Biol. 159, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soung N. K., Kang Y. H., Kim K., Kamijo K., Yoon H., Seong Y. S., Kuo Y. L., Miki T., Kim S. R., Kuriyama R., Giam C. Z., Ahn C. H., Lee K. S. (2006) Mol. Cell. Biol. 26, 8316–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobinnec Y., Khodjakov A., Mir L. M., Rieder C. L., Eddé B., Bornens M. (1998) J. Cell Biol. 143, 1575–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keryer G., Witczak O., Delouvée A., Kemmner W. A., Rouillard D., Tasken K., Bornens M. (2003) Mol. Biol. Cell 14, 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khodjakov A., Rieder C. L. (1999) J. Cell Biol. 146, 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khodjakov A., Cole R. W., Rieder C. L. (1997) Cell Motil. Cytoskeleton 38, 311–317 [DOI] [PubMed] [Google Scholar]
- 33.He L., Hou Z., Qi R. Z. (2008) J. Biol. Chem. 283, 13252–13260 [DOI] [PubMed] [Google Scholar]
- 34.Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole N. B., Sciaky N., Marotta A., Song J., Lippincott-Schwartz J. (1996) Mol. Biol. Cell 7, 631–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.del Valle M., Robledo Y., Sandoval I. V. (1999) J. Cell Sci. 112, 4017–4029 [DOI] [PubMed] [Google Scholar]
- 37.Sawin K. E., Lourenco P. C., Snaith H. A. (2004) Curr. Biol. 14, 763–775 [DOI] [PubMed] [Google Scholar]
- 38.Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 39.Paoletti A., Moudjou M., Paintrand M., Salisbury J. L., Bornens M. (1996) J. Cell Sci. 109, 3089–3102 [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa Y., Yamane Y., Okanoue T., Tsukita S., Tsukita S. (2001) Mol. Biol. Cell 12, 1687–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haren L., Stearns T., Lüders J. (2009) PLoS One 4, e5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witczak O., Skålhegg B. S., Keryer G., Bornens M., Taskén K., Jahnsen T., Orstavik S. (1999) EMBO J. 18, 1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt P. H., Dransfield D. T., Claudio J. O., Hawley R. G., Trotter K. W., Milgram S. L., Goldenring J. R. (1999) J. Biol. Chem. 274, 3055–3066 [DOI] [PubMed] [Google Scholar]
- 44.Chang P., Stearns T. (2000) Nat. Cell Biol. 2, 30–35 [DOI] [PubMed] [Google Scholar]
- 45.Larocca M. C., Shanks R. A., Tian L., Nelson D. L., Stewart D. M., Goldenring J. R. (2004) Mol. Biol. Cell 15, 2771–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verde I., Pahlke G., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S. L., Conti M. (2001) J. Biol. Chem. 276, 11189–11198 [DOI] [PubMed] [Google Scholar]
- 47.Delaval B., Doxsey S. J. (2010) J. Cell Biol. 188, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dictenberg J. B., Zimmerman W., Sparks C. A., Young A., Vidair C., Zheng Y., Carrington W., Fay F. S., Doxsey S. J. (1998) J. Cell Biol. 141, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guarguaglini G., Duncan P. I., Stierhof Y. D., Holmström T., Duensing S., Nigg E. A. (2005) Mol. Biol. Cell 16, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]