Abstract
The microtubule-associated protein tau plays a central role in the pathogenesis of Alzheimer disease (AD) and abnormally accumulates as neurofibrillary tangles; therefore, the pathways by which tau is degraded have been examined extensively. In AD brain tau is abnormally truncated at Asp421 (tauΔC), which increases its fibrillogenic properties and results in compromised neuronal function. Given the fact that the accumulation of tauΔC is a pathogenic process in AD, in this study we examined whether full-length tau and tauΔC are degraded through similar or different mechanisms. To this end a tetracycline-inducible model was used to show that tauΔC was degraded significantly faster than full-length tau (FL-tau). Pharmacological inhibition of the proteasome or autophagy pathways demonstrated that although FL-tau is degraded by the proteasome, tauΔC is cleared predominantly by macroautophagy. We also found that tauΔC binds C terminus of Hsp70-interacting protein more efficiently than tau. This interaction leads to an increased ubiquitylation of tauΔC in a reconstituted in vitro assay, but surprisingly, tau (full-length or truncated) was not ubiquitylated in situ. The finding that tauΔC and FL-tau are differentially processed by these degradation systems provides important insights for the development of therapeutic strategies, which are focused on modulating degradation systems to preferentially clear pathological forms of the proteins.
Keywords: Alzheimer Disease, Autophagy, Proteasome, Protein Degradation, Tau
Introduction
Alzheimer disease (AD)2 is characterized by the gradual onset of dementia manifested as amnesia, cognitive decline, aphasia, and eventually an inability of an afflicted individual to carry out everyday activities (1). The primary pathological hallmarks of AD are plaques composed of β-amyloid and neurofibrillary tangles composed of the microtubule-associated protein tau (2). Tau has been shown to self-assemble in vitro (3) and in situ (4) into microscopically visible aggregates. This propensity for self-assembly is enhanced when tau is either pathologically phosphorylated (5), pseudophosphorylated (6), or cleaved by caspases in the C terminus at Asp421 (7). In addition to visible aggregates, electron microscopy studies have revealed the existence of pre-microscopically visible forms of tau self-assembly, which are in a more soluble state than the microscopic aggregates (8). The existence of these oligomeric soluble forms combined with the observations that 1) in a tauopathy mouse model, the toxic agent is in a soluble form (as opposed to an insoluble tangle) (9) and 2) in a related neurodegenerative aggregation disease, visible aggregates are actually cytoprotective (10) suggests that these soluble oligomeric forms of tau may be the toxic agents of tauopathies. More strikingly, other work has shown that the expression of tau is critically important for the sensitization of neurons to β-amyloid-induced cell death in both a neuron model (11) as well as β-amyloid -induced memory loss in the mouse (12). In addition, recent work (13) has placed the cleavage of tau at Asp421 early in the pathological cascade leading to neurofibrillary tangle formation.
Tau truncated at Asp421 due to caspase cleavage (tauΔC) is present in AD brain (7), and previous studies have demonstrated the aggregative and toxic nature of tauΔC (7, 14). Our recent studies have also determined that the truncation of tau at Asp421 modulates its ability to bind microtubules and influence their stability (15). It also sensitizes cells to thapsigargin-induced cell death (16) and mitochondrial dysfunction (17). This tauΔC-induced sensitization to stressors occurs in the absence of microscopically detectable aggregates (17) and, thus, may indicate that it is an early, contributing event in AD pathogenesis. Given the toxic nature of tauΔC, it is of critical importance to understand the pathways that regulate its turnover and clearance from the cell.
The co-localization of ubiquitin with neurofibrillary tangles in AD brain was first reported more than 20 years ago (18). These early findings demonstrating the presence of ubiquitin immunoreactivity at the level of the neurofibrillary tangles was in part the impetus for subsequent studies investigating the potential role of the proteasome system in mediating tau degradation both in physiological and pathological conditions. However, the findings have not been conclusive as they are sometimes seemingly contradictory. Tau has been described as a proteasomal substrate, degraded both through ubiquitin-dependent (19, 20) and -independent (21, 22) means. Tau interaction with the co-chaperone C terminus of Hsp70 interacting protein (CHIP) has been well documented (19, 20, 23), and either overexpression (24) or ablation (25) of CHIP has been shown to lead to changes in tau stability. In general CHIP appears to be involved in enhancing cell survival and attenuating cell death in response to cell stress including ischemic insult (26) and heat shock (27). CHIP has been suggested to be an E3 ligase of tau (19, 20), although it is important to note that the particular E2 activating enzyme involved in the E2/CHIP partnership may differentially drive CHIP enzymatic activity (28).
In AD neurons there is an accumulation of autophagic vacuoles (AVs) (29). This finding of AV accumulation is coupled with indications of disruptions in the macroautophagic pathway, including lowered Beclin-1 levels and structural perturbations to lysosomal morphology (30). Tau has previously been described as a substrate of lysosomal degradation (31) and more recently as a substrate of autophagy (32, 33) (and more specifically, chaperone-mediated autophagy (34)). Despite these findings, the initiating events and pathways that regulate the proteasomal or lysosomal degradation of tau have not been clearly described.
In this study we found that a C-terminal-truncated form of tau that mimics tau cleaved at Asp421 (tauΔC) is removed from the cell much more rapidly than full-length tau (FL-tau) and that the removal of tauΔC from the cell is driven by macroautophagic and lysosomal mechanisms, whereas FL-tau degradation is more dependent on the proteasome. We also found that tauΔC more strongly interacts with CHIP compared with full-length tau and that CHIP preferentially directs the ubiquitylation of tauΔC in vitro. However, we found that neither full-length (FL-tau) nor tauΔC is ubiquitylated in situ. Given these data and the known differences in folding state and aggregative potential (35) between FL-tau and tauΔC, we propose that the loss of the last 20 amino acids that occurs during AD progression may cause a “switch” between the proteasomal and autophagic degradation pathways and could account for the evidence suggesting the involvement of both pathways in the degradation of tau.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Epoxomicin, 3-methyladenine (3-MA), and bafilomycin A1 were purchased from Calbiochem. Chloroquine and resazurin were purchased from VWR. Recombinant E1, UbcH5b, HA-ubiquitin, and Hsp70 were purchased from Boston Biochem. The Tau5 antibody was kindly provided by Dr. L Binder (36), and the polyclonal tau antibody was purchased from Dako. The HA antibody HA-7 (Sigma) was used for immunoprecipitation, and the polyclonal HA antibody used for immunoblotting was purchased from Abcam. Other antibodies used were anti-CHIP (Calbiochem), anti-β-actin (Chemicon), anti-myc clone 9B11 (Cell Signaling), anti-LC3 (Calbiochem), anti-p62 (Biomol), and anti-Hsc/p70 (Assay Designs).
Plasmids and Recombinant Protein
The plasmid for myc-CHIP was a gift from Dr. Douglas Cyr. The plasmid for HA-ubiquitin was a gift from Dr. Scott Wilson. To make the plasmid for GFP-LC3, RNA was extracted from naïve CN1.4 cells (37) using Trizol (Invitrogen) according to the manufacturer's protocol. cDNA was created by reverse transcription using Superscript III (Invitrogen), and PCR was performed. The product was digested and inserted into the EcoRI site of pEGFP-C1. To make GST-CHIP, a cassette containing CHIP was subcloned from myc-CHIP.pCMV, digested, and inserted into the BamHI and EcoRI sites of pGEX-6P-2. The identities of all constructs were confirmed by DNA sequencing. Protein purification was performed exactly as described (38).
Cell Culture and Transfections
Immortalized CN1.4 mouse cortical neurons (CN) that stably express inducible forms of FL-tau (containing four microtubule repeats and no N-terminal inserts) and tauΔC (truncated at Asp421) have been described previously (16, 39). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum and 2 mm l-glutamine. HEK 293-TN cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% Fetal Clone II and 2 mm l-glutamine. Cells were maintained at 33 °C in a humidified atmosphere containing 5% CO2.
To induce the expression of tau, the stably transfected cells were seeded on 60-mm dishes for 36 h before being treated with doxycycline. The amount of doxycycline used to induce tau expression was experimentally optimized to result in equivalent levels of tau expression between cell lines and ranged from 0.5 to 2 μg/ml. After 12 h, the cells were moved to 37 °C, the culture media were replaced with Dulbecco's modified Eagle's medium supplemented with 1% fetal bovine serum, 2 mm l-glutamine, and doxycycline, and the cells were cultured for an additional 36 h. For degradation experiments, at time = 0 the doxycycline-containing media was removed, and the cells were carefully washed twice with prewarmed PBS and replenished with doxycycline-deficient media. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) following manufacturer protocols. Cells were cultured for 48 h after transfection before use.
Cell Lysis, Immunoprecipitation, and Immunoblotting
For direct blotting of lysates, the cells were collected in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40 (Nonidet P-40), 5 mm EDTA, 0.5% sodium deoxycholate, 0.1% SDS) containing 1 mm phenylmethylsulfonyl fluoride and 10 μg/ml each of aprotinin, leupeptin, and pepstatin, and the cells were briefly sonicated and centrifuged at 20,000 × g for 10 min at 4 °C. Protein concentration was determined by the bicinchoninic acid assay, and samples were prepared for immunoblotting by dilution in 2× sample buffer (0.25 m Tris-HCl, pH 7.5, 2% SDS, 25 mm dithiothreitol, 5 mm EDTA, 5 mm EGTA, 10% glycerol, and 0.01% bromphenol blue).
For immunoprecipitation (IP), the cells were collected in IP lysis buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm EGTA, 1 mm EDTA, 0.2 mm sodium vanadate, 0.5% Nonidet P-40) containing 1 mm phenylmethylsulfonyl fluoride and 10 μg/ml leupeptin, aprotinin, and pepstatin. After sonication, centrifugation, and determination of protein content, the samples were diluted to 1 mg/ml with IP lysis buffer. The cells were incubated with mouse monoclonal Tau5 or HA antibodies preconjugated with sheep anti-mouse magnetic beads (Dynal) for 3 h on a rotational shaker at 4 °C. After incubation, the beads were washed either 4 times with low stringency wash buffer (PBS supplemented with 300 mm NaCl and 0.5% Nonidet P-40) or 7 times with high stringency wash buffer (50 mm Tris-HCl, pH 7.4, 1.1 m NaCl, 1% Nonidet P-40, 0.1% SDS, and 1 mm EDTA) before elution with 2× sample buffer.
For immunoblotting, samples were resolved on 7.5 or 10% polyacrylamide gels and transferred to nitrocellulose membranes. After a 1-h incubation in 5% nonfat dry milk in TBST (20 mm Tris-HCl, pH 7.6, 137 mm NaCl, and 0.05% Tween 20), blots were incubated overnight with primary antibody. Blots were washed with TBST and incubated with a secondary antibody for 1 h. After thorough washing with TBST, blots were developed with chemiluminescence (40).
In Vitro Ubiquitylation
FL-tau or tauΔC were immunoprecipitated from cells incubated with doxycycline, and the immunoprecipitates were washed with high stringency wash buffer before a final wash in assay buffer (50 mm Tris-HCl, pH 7.5, 2.5 mm MgCl2, 2 mm dithiothreitol). The ubiquitylation was then carried out essentially as described (41). The magnetic beads were then incubated with combinations of recombinant Hsp70 (1 μg), HA-ubiquitin (10 μg), E1 (200 ng), UbcH5b (400 ng), CHIP (1 μg), and ATP (2 mm) in a final volume of 50 μl. The samples were incubated for 15 or 60 min. To stop the reactions, 2× sample buffer was added, and the samples were boiled.
Viability Determination
For the determination of cell viability, cells were seeded in 12-well dishes and cultured as described. Cells were incubated in 400 μm resazurin for 1 h, and fluorescence was measured using a plate reader (BioTek Synergy HT Multi-Detection Microplate Reader) at excitation/emission wavelengths of 530/590 nm.
Proteasome Activity Assay
Cells were collected and lysed in IP lysis buffer as described. Protein content was determined using the bicinchoninic acid assay, and equal amounts of protein were incubated with 25 μm proteasome substrate benzoyl-VGR-amidomethylcoumarin (Biomol), an indicator of trypsin-like activity. Fluorescence output was measured kinetically at excitation/emission wavelengths of 360/460 nm. To determine the specificity of the substrate for proteasome activity, parallel samples were spiked with 1 μm epoxomicin.
Imaging and Autophagic Measurements
Cells were seeded on 35-mm glass-bottom dishes, transfected with GFP-LC3, and cultured as described above. Images were captured with an AxioCamMR3 camera (Zeiss Corp., Thornwood, NY) connected to a Zeiss Axio Observer D1 microscope with an EC Plan-Neofluor 40×/1.3 oil M27 objective and exported in TIFF format using AxioVision software. Determination of autophagic phenotype was determined as described (42, 43). Briefly, cells were treated in either complete media or Hanks' balanced salt solution supplemented with 20 nm bafilomycin A1 to induce maximal numbers of GFP-LC3 puncta. The number of puncta per cell was counted for each condition to determine a threshold autophagy level. Cells with >30 puncta/cell were scored as “autophagic.” and cells with <30 puncta/cell were scored as “non-autophagic.”
Statistics
Degradation rates (Figs. 1–3) were analyzed using one-way analysis of variance and post-hoc t tests. Comparisons in Fig. 6 were performed using Student's t test. A threshold of p < 0.05 was used to establish significance.
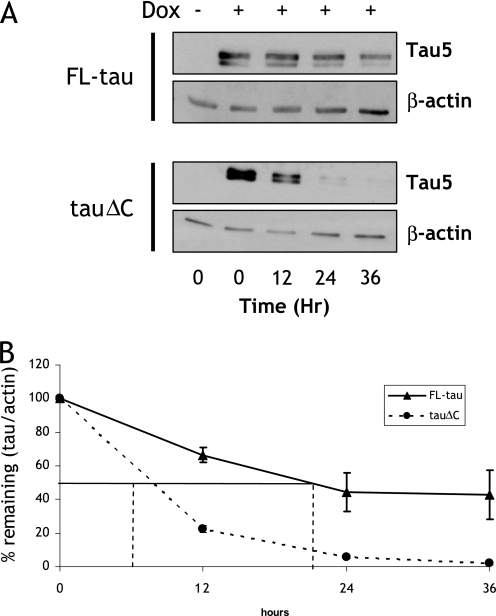
FIGURE 1.
TauΔC is degraded faster than FL-tau. A, CN cells stably expressing either inducible FL-tau or tauΔC were treated with doxycycline (Dox, or vehicle) for 48 h. At time = 0, cells were washed twice with PBS, and the media were replaced with doxycycline-deficient media. Lysates were collected every 12 h thereafter. Collected lysates were immunoblotted for total tau using Tau5 antibody. The membranes were also reprobed with a β-actin antibody as a loading control. B, shown is quantification of relative levels of tau remaining after cessation of doxycycline treatment described in A. The amount of tau was normalized to β-actin levels and then expressed as the percentage compared with tau/actin at t = 0. t½ is the time at which 50% of FL-tau or tauΔC is remaining and is indicated as vertical droplines. For FL-tau, t½ = 20.5 h; for tauΔC, t½ = 7.7 h. Data points are the means ± S.E. collected from seven independent experiments. p < 0.05 between FL-tau and tauΔC at all data points.
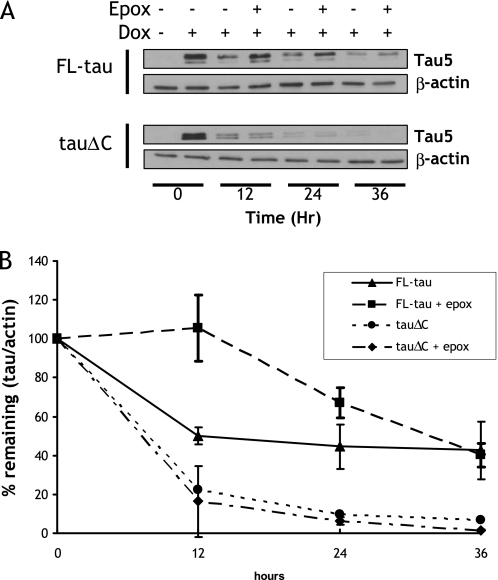
FIGURE 2.
Treatment with a proteasome inhibitor attenuated the degradation of FL-tau but not tauΔC. A, CN cells stably expressing either inducible FL-tau or tauΔC were treated with doxycycline (Dox, or vehicle) for 48 h. At time = 0, cells were washed twice with PBS, and the media were replaced with doxycycline-deficient media with either vehicle or 12.5 nm epoxomicin (epox). Lysates were collected every 12 h and immunoblotted for total tau using Tau5 antibody. The membranes were also reprobed with a β-actin antibody as a loading control. B, shown is quantification of the data described in A. Results are shown as means ±S.E. collected from four independent experiments. There was a significant difference in FL-tau levels (p < 0.05) between vehicle and epoxomicin-treated FL-tau sample groups at 12 and 24 h.
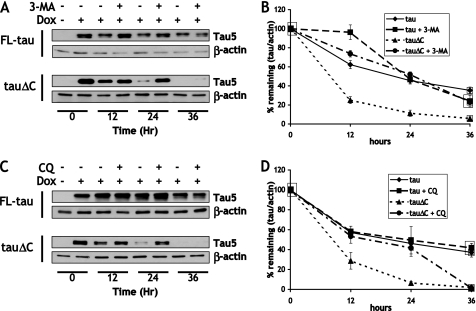
FIGURE 3.
Treatment with macroautophagy inhibitors preferentially attenuates the degradation of tauΔC. A, CN cells stably expressing either inducible FL-tau or tauΔC were treated with doxycycline (Dox, or vehicle) for 48 h. At time = 0, cells were washed twice with PBS, and the media was replaced with doxycycline-deficient media with either vehicle or 10 mm 3-MA. Lysates were collected every 12 h and immunoblotted with Tau5 antibody and anti-β-actin. B, shown is quantification of the data described in A. Results are the means ±S.E. from four independent experiments. There was a significant difference in FL-tau levels (p < 0.05) between vehicle and treated groups at 12 h and vehicle and treated tauΔC groups at 12 and 24 h. C, data were collected as in A, except cells were treated with 10 μm chloroquine (CQ) or vehicle upon doxycycline washout. D, shown is quantification of data described in C. Results are the means ±S.E. from four independent experiments. Treatment groups are significantly different (p < 0.05) for tauΔC at 12 and 24 h.
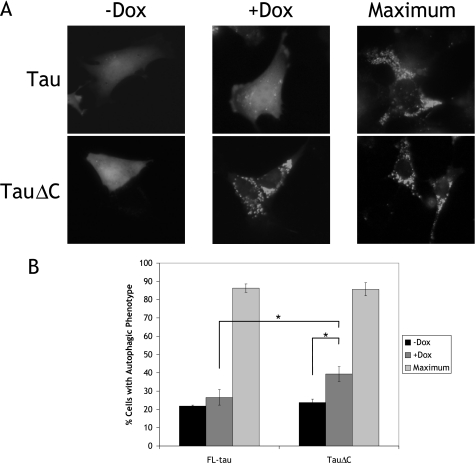
FIGURE 6.
Expression of tauΔC induces an autophagic phenotype. A, CN cells were transiently transfected with GFP-LC3, and doxycycline (Dox) was added to induce expression of FL-tau or tauΔC. As a control cells were treated with vehicle only (−Dox). Maximal LC3 puncta were induced 1 h before imaging by treatment with 20 nm bafilomycin A1 diluted in Hanks balanced salt solution. Forty-eight hours after incubation in the absence or presence of doxycycline, cells were imaged, and the phenotype was scored as autophagic or non-autophagic as described under “Experimental Procedures.” B, shown is the quantification of data described in A. For each experiment, 60–75 cells were scored. Results are the means ±S.E. from three independent experiments. *, p < 0.05.
RESULTS
TauΔC Is Cleared More Rapidly from the Cell Than FL-tau
A Tet-on turnover assay was used to determine the relative rates of clearance of FL-tau and tauΔC. Cells were incubated with doxycycline for 48 h followed by a thorough wash with PBS and continued incubation in doxycycline-free media. Lysates were collected at the indicated time points over the next 36 h and immunoblotted for tau (Fig. 1A). In addition, mRNA levels for FL-tau and tauΔC were also analyzed by quantitative real-time-PCR after doxycycline withdrawal, and no differences in the decay rate of mRNA were detected (data not shown). Furthermore, tau mRNA was completely absent from the cell 6 h after doxycycline withdrawal, well before the first assay point. The amount of FL-tau or tauΔC remaining in the lysate was normalized to the amount present at the point doxycycline was removed (t = 0). TauΔC was cleared from the cell much more quickly than FL-tau (t½ = 7.7h versus 20.5 h for FL-tau). This half-life for FL-tau compares favorably with previous pulse-chase assays that were carried out in PC12 cells (44), validating the model for use in experiments to further define the differences in the turnover of FL-tau and tauΔC.
Epoxomicin Attenuates the Degradation of FL-tau but Not tauΔC
Previous studies have suggested that tau can be degraded by the proteasome in an ubiquitin-dependent (19, 20) and -independent manner (22). To determine the contribution of proteasome to the degradation of FL-tau and tauΔC, the turnover assay was carried out in the absence or presence of 12.5 nm epoxomicin. Epoxomicin was selected because it is an irreversible, potent, and selective inhibitor of the proteasome (45). Before carrying out the turnover studies, the effect of epoxomicin on cell viability was determined. The viability of cells was 95.2 ± 3.2% of untreated control after 36 h of treatment with epoxomicin, indicating that toxicity would not be a confounding factor. Treatment of cells with epoxomicin decreased the degradation of FL-tau at 12 and 24 h but did not affect the degradation of tauΔC (Fig. 2). To ensure that there were similar amounts of proteasome activity between cell lines, lysates were incubated with the substrate of proteasomal trypsin-like activity, benzoyl-VGR-amidomethylcoumarin. Surprisingly, the relative proteasome activity of tauΔC cells were slightly elevated (FL-tau, 100 ± 4.2%; tauΔC, 125.7 ± 5.3%), The proteasome activities of both cell lines were equally suppressed by the addition of epoxomicin. This further underscores the specificity of the proteasomal degradation of FL-tau.
Similar results were obtained with 1 μm MG132 (i.e. the degradation of FL-tau but not tauΔC was inhibited); however, we did not include these data due to the fact that this treatment was toxic to the cells (viability was 29.1 ± 3.5% after 36 h of treatment) (supplemental Fig. S1).
Inhibition of Macroautophagy Attenuates the Degradation of tauΔC but Not FL-tau
It has been reported that tau can be degraded through the lysosome (33, 46), and more recently a report has suggested that an aggregate-prone fragment of tau is degraded through chaperone-mediated autophagy (34). To determine the role of the lysosomal degradation pathway in the clearance of FL-tau and tauΔC, an inhibitor of macroautophagic induction, 3-MA, was used in the turnover assay (Fig. 3A). Treatment of FL-tau or tauΔC expressing cells with 10 mm 3-MA for 36 h resulted in viability of 67.1 ± 3.8% compared with untreated cells. Treatment with 3-MA resulted in the inhibition of turnover of tauΔC at all time points but failed to significantly decrease the rate of FL-tau degradation (Fig. 3B). There was an anomalous, but significant, increase of FL-tau levels 12 h after 3-MA application, but this was not sustained for the remaining assay period. To further confirm the selective role of autophagy in the clearance of tauΔC, but not FL-tau, chloroquine was also used in the turnover assay (Fig. 3C). Chloroquine is a downstream inhibitor of the fusion of autophagic vacuoles with lysosomes and, therefore, can be used to assess the accumulation of substrates normally removed by lysosomal degradation (47). Cell viability was not significantly affected by treatment with 10 μm chloroquine for 36 h (91.5 ± 4.2% viability compared with untreated controls). Like 3-MA, chloroquine treatment attenuated the clearance of tauΔC but had no effect of the clearance of FL-tau (Fig. 3D). The ability of both cell lines to appropriately respond to both 3-MA and chloroquine was verified by immunoblotting LC3-II and p62. The amounts of both autophagic markers increased with the addition of chloroquine, and this increase was blocked by concomitant treatment with 3-MA (supplemental Fig. S2, upper panel).
TauΔC Is Preferentially Ubiquitylated by CHIP in Vitro
Previous reports have proposed that tau is ubiquitylated by a complex that includes CHIP (20). To determine if tauΔC is ubiquitylated, in vitro ubiquitylation assays were carried out using CHIP as an E3 ligase. FL-tau and tauΔC immunoprecipitated from cell lysates were used as the substrates to capture the physiological post-translational modifications, which would be absent if non-eukaryotic production/purification systems were used. When FL-tau and tauΔC were incubated with a reconstituted ubiquitylation complex, tauΔC was clearly ubiquitylated much more readily than FL-tau (Fig. 4), with immunoreactive bands of equivalent molecular weights on membranes blotted for both HA-ubiquitin and tau. Control reactions in which tau, ATP, and/or CHIP were not added confirmed the specificity of the ubiquitylation reaction. In addition, reactions terminated after 15 min revealed differentially ubiquitylated forms of tauΔC, whereas after 60 min the majority of the tauΔC was converted into an insoluble aggregate that was detected as a single tau- and HA-immunoreactive band present in the stacking gel (Fig. 4).
FIGURE 4.
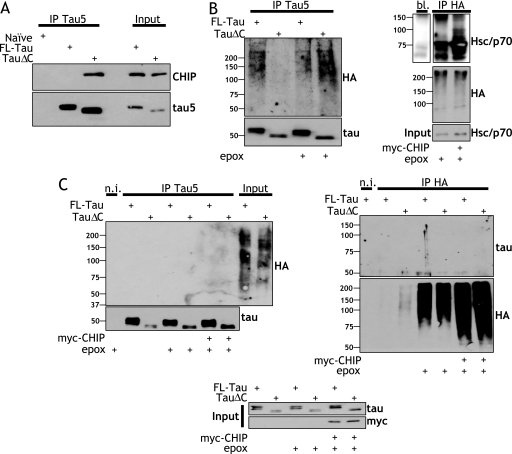
In vitro tauΔC is preferentially ubiquitylated by CHIP. Cell lysates from naïve CN cells or CN cells stably expressing FL-tau or tauΔC were immunoprecipitated with Tau5 antibody and washed. The immunoprecipitates were then incubated with reaction mixtures containing the indicated combinations of ATP and recombinant CHIP as well as E1, E2, Hsp70, and HA-ubiquitin. The mixtures were incubated at 37 °C for the indicated lengths of time. Samples were resolved by electrophoresis and immunoblotted using Tau5 and HA antibodies. The positions of molecular weight standards (kDa) are indicated at the left. st., stacking gel.
TauΔC Preferentially Interacts with CHIP
Previous studies have determined that tau interacts with CHIP (19, 20); therefore, co-immunoprecipitation experiments were carried out using this cell model to both corroborate these previous findings and also determine whether tauΔC interacts with CHIP. To our surprise, tauΔC interacted much more strongly with endogenous CHIP than did FL-tau (Fig. 5A). Lanes loaded with equal amounts of cell lysate are shown on the right-hand side of blots to demonstrate that the cells express the same amount of endogenous CHIP.
FIGURE 5.
CHIP preferentially interacts with tauΔC, and FL-tau and tauΔC are not ubiquitylated in situ. A, TauΔC preferentially interacts with CHIP. Cell lysates from naïve CN cells or CN cells expressing FL-tau or tauΔC were immunoprecipitated with Tau5 antibody before immunoblotting for CHIP and, subsequently, tau. B, less stringent washing of tau immunoprecipitates results in coimmunoprecipitation of ubiquitylated proteins. Left, cells were transiently transfected with plasmids expressing FL-tau or tauΔC and expressing HA-ubiquitin and treated for 12 h with 50 nm epoxomicin (epox) or vehicle as indicated. Lysates were collected, and immunoprecipitation was performed using Tau5 antibody. The immunoprecipitates were washed with PBS, resolved, and immunoblotted with polyclonal antibodies toward HA and tau. Right, as a positive control for CHIP activity, 293TN cells were transiently transfected without or with myc-CHIP and HA-ubiquitin and treated for 12 h with 50 nm epoxomicin. Lysates or buffer alone (bl.) were immunoprecipitated with an anti-HA antibody, and the immunoprecipitates were blotted with a polyclonal antibody toward Hsp/c70 or HA. C, stringent washing of immunoprecipitates reveals the lack of tau ubiquitylation. CN cells were transiently transfected with plasmids expressing FL-tau or tauΔC and expressing HA-ubiquitin with or without myc-CHIP. Cells were treated with 50 nm epoxomicin or vehicle 12 h before collection. Lysates were immunoprecipitated with either a non-immune (n.i.) mouse antibody, Tau5, or anti-HA as indicated. The immunoprecipitates were washed seven times with a high stringency buffer containing 50 mm Tris, 1.1 m NaCl, 1% Nonidet P-40, 0.1% SDS, and 1 mm EDTA. The immunoprecipitates were immunoblotted with either a polyclonal tau antibody or a polyclonal HA antibody. Inputs are also shown. The positions at which molecular weight standards (kDa) migrated are indicated to the left.
Neither FL-tau nor tauΔC Is Ubiquitylated in Situ
Previous studies have shown that tau co-immunoprecipitates with ubiquitin (20) in situ, and this co-immunoprecipitation may be enhanced due to either tau phosphorylation (19) or the inclusion of a fourth microtubule binding repeat (48). In those previous studies, the co-immunoprecipitation of ubiquitin immunoreactivity with tau was interpreted as an indication that tau was ubiquitylated despite the absence of high molecular weight tau immunoreactive bands. Given that tauΔC is more efficiently ubiquitylated in vitro when compared with FL-tau, an in situ ubiquitylation assay was next performed to assess the physiological relevance of our previous in vitro results. First, immunoprecipitation with the tau5 antibody was carried out in FL-tau- and tauΔC-expressing cells that were transfected with HA-ubiquitin and incubated in the absence or presence of epoxomicin. After washing immunoprecipitates with low stringency wash buffer, we detected readily observable high molecular weight HA-ubiquitylated species, although the HA immunoreactive bands did not overlap the tau immunoreactive bands (Fig. 5B, left panels). To verify that the E3-ligase activity of exogenous myc-CHIP was present, we also carried out in situ ubiquitylation assays targeted toward Hsc/p70, which has been previously identified as a CHIP substrate (49). HEK cells transiently transfected with myc-CHIP and incubated in the absence and presence of epoxomicin and immunoprecipitations were carried out with an HA antibody (Fig. 5B, right panels). The immunoprecipitates were blotted for Hsc/p70, and discrete bands were visible just above the unmodified protein from cells transfected with myc-CHIP, indicating ubiquitylation. These bands, which are at a lower molecular weight and discrete, were distinct from the high molecular weight ubiquitin apparent in the tau immunoprecipitates.
To test if similar discrete HA- and tau-immunopositive bands could be detected when tau-expressing cells were treated with epoxomicin in the presence of CHIP, naïve cortical cells were transiently transfected with plasmids for FL-tau or tauΔC as well as HA-ubiquitin in the absence or presence of myc-CHIP. After immunoprecipitation of tau, precipitates were thoroughly washed with a high stringency wash buffer to completely remove co-precipitating proteins which could contribute to the presence of ubiquitin immunoreactivity that was not due to the ubiquitylated state of tau. The reverse immunoprecipitation was also performed using an HA antibody. Both approaches demonstrated that neither FL-tau nor tauΔC was ubiquitylated in the cells (Fig. 5C), as was evident in the absence of co-precipitating immunoreactivity as well as the absence of high molecular weight species in both tau and HA blots.
TauΔC Expression Induces Autophagy
These findings indicate that tauΔC is being degraded through macroautophagy. Given that other aggregative proteins can induce autophagy merely through their expression (50), the autophagic levels of cortical cells expressing FL-tau or tauΔC were determined. Cells were transfected with GFP-LC3, and FL-tau or tauΔC expression was induced. After 48 h of expression, some cells were further treated with 20 nm bafilomycin A1 in nutrient starvation conditions (Hanks balanced salt solution) to both induce autophagy and inhibit autophagic vacuole fusion in order to determine a maximal level of autophagy (Fig. 6A). Cells were scored for autophagic phenotype as described under “Experimental Procedures,” and the fraction of transfected cells with an autophagic phenotype was determined (Fig. 6B). Both cell lines exhibited relatively low and equal amounts of autophagic cells in basal, uninduced conditions (FL-tau, 21.9 ± 0.48%; tauΔC, 23.7 ± 2%). Although induction of FL-tau did not significantly increase the amount of autophagic cells, induction of tauΔC did (uninduced, 23.7 ± 2%; induced, 39.3 ± 4.0%). Moreover, there was a significant difference in the number of autophagic cells present in induced FL-tau and induced tauΔC cells (FL-tau, 26.6 ± 4.2%; tauΔC, 39.3 ± 4.0%). Both cell types displayed equally high amounts of autophagic cells upon Hanks balanced salt solution /bafilomycin A1 treatment.
DISCUSSION
Abnormal posttranslational processing of tau is a key feature of AD. Tau truncated at Asp421 (tauΔC) is readily detectable in AD, but not control, brains (7), and there are significant data to suggest that this form of tau is toxic to cells (13, 16, 17). Therefore, understanding the processes that regulate the turnover of this pathologically modified form of tau is of importance. In this study we provide clear evidence that FL-tau and tauΔC undergo turnover in the cell through different pathways. Removal of the last 20 amino acids of tau is sufficient to shift the clearance pathway from one that is primarily dependent on the proteasome to one that is more dependent on autophagy; this 20-amino acid truncation also substantially decreases the half-life of tau. Autophagy pathways have been reported to be compromised in AD brain (52), which could, thus, contribute to the pathological accumulation of tauΔC. Our studies are in agreement with previous findings suggesting that although tau is degraded by the proteasome, it is in a ubiquitylation-independent manner (21, 22).
The shifting degradation pathways described are accompanied by the increased association of CHIP and tauΔC and the absence of in situ ubiquitylation of both FL-tau and tauΔC. At this point it is still unclear why increased co-immunoprecipitation of tauΔC and CHIP is associated with a decrease in the dependence of tauΔC clearance on proteasome-mediated degradation. A previous study showed that tauΔC accumulates in the absence of CHIP in a knock-out mouse model, although it should be noted that the accumulation of tauΔC in this study was accompanied by a concomitant increase in the level of activated caspase-3 (25). Therefore, it cannot be ruled out that in that study the increase in tauΔC attributed to the deletion of CHIP is not due to changes in degradation but, rather, to an increase of tauΔC production.
Although lysosomal and autophagic degradation pathways have long been considered to be the cell's preferred means of clearing long-lived proteins (53), studies have indicated that proteins with short half-lives are also turned over through autophagy pathways (54). Conversely, the proteasomal pathway that is often involved in the rapid and tightly regulated degradation of proteins involved in transcription and the cell cycle (55) can also contribute to clearance of more stable long-lived proteins (56). Accordingly, we describe a model in which the rapid clearance of tauΔC (Fig. 1) can be clearly attributed to macroautophagy (Fig. 3), whereas the slower turnover of FL-tau is attributed to proteasomal degradation (Fig. 2). More specifically, the slower proteasomal degradation of FL-tau is due to ubiquitin-independent degradation by the 20 S proteasome, a finding that parallels the previous description of the slow 20 S proteasome-dependent clearance of oxidized proteins (57).
It can be suggested that the increased aggregative potential and/or modified microtubule binding capacity displayed by tauΔC (15) could contribute to the fact that it is primarily degraded through the autophagy rather than the proteasome pathway. Evidence for the increased aggregative capacity of tauΔC has been demonstrated through its increased MC1 immunoreactivity (7), which surprisingly occurs concurrently with decreased PHF-1 immunoreactivity (15). Previous studies have suggested that a decrease in the microtubule binding capacity of tau isoforms may lead to an increase in tau turnover (44). Therefore, it can be suggested that that if tauΔC binds microtubules less efficiently than FL-tau, then this may increase its potential to oligomerize, and this misfolding and oligomerization may make tauΔC a favorable macroautophagy substrate, as occurs with huntingtin (58).
Previous studies have clearly demonstrated that tau and CHIP interact (19, 20), and in this study we provide evidence that CHIP interacts with tauΔC to an even greater extent than FL-tau. Those previous studies focused primarily on the role of CHIP as an E3 ligase with tau as the substrate. Although this is certainly the case in vitro, it needs to be considered that this may not be the primary function of CHIP in situ. For example, it is well documented that CHIP can act as a stabilizing factor (59). Therefore, CHIP may be interacting with tau to maintain it in a proper conformation, especially given its natively unfolded nature and propensity for aggregation (3, 60), which is even more of a factor with tauΔC (35). In support of the supposition that CHIP has other roles in the cell besides ubiquitylating substrates to target them to proteasomal degradation is the finding that substrates previously identified as clients of CHIP are degraded just as readily in its absence (61). The same study also described the in vitro ubiquitylation of neuronal nitric-oxide synthase, a previously identified CHIP substrate, as occurring readily in the presence of a CHIP-null extract. In another study, the CHIP protective role in response to heat shock was shown to be independent of its ubiquitin ligase activity, and that interaction with chaperones such as Hsp70 and Hsp90 was far more important to its protective function (62). In fact, it has been documented that tau readily interacts in complex with Hsp70 and Hsp90 (63). These studies combined with the finding that tau and tauΔC are ubiquitylated in vitro but not in situ by CHIP suggest the possibility that the non-E3 ligase functions (e.g. its ability to recruit co-chaperones (64)) of CHIP may be of more significance when evaluating the tau-CHIP interactions. If this is the case, it would not be surprising that CHIP preferentially bound the less stable and more aggregative tauΔC compared with FL-tau.
The specific ubiquitin activating enzyme, or E2, that is associated with CHIP may also affect its substrate selection, and the identity of the elements of this E2/E3 protein complex may also vary the specific class of isopeptide linkages formed in the ubiquitin chains. In particular, it has been found that CHIP complexed with UbcH5, a particular class of E2 enzymes, results in forked ubiquitin chains that are unable to be digested by the proteasome (28). Furthermore, this same study demonstrated that CHIP readily catalyzes the formation of Lys63-linked ubiquitin chains in the presence of a different E2, UbcH13/Uev1a. The E2 UbcH5b was identified as the E2 that associates with tau and CHIP in vivo (19), indicating that if CHIP is ubiquitylating tau in the AD brain, this may in fact decrease its ability to be degraded by the proteasome. It is important to note that most studies of CHIP-mediated tau ubiquitylation (including our own) use the UbcH5 family of E2 ligases to catalyze ubiquitin chain formation.
Although pharmacological inhibition of the proteasome prevented tau degradation, it had no effect on the degradation of tauΔC. However, autophagic inhibitors significantly inhibited the turnover of tauΔC. 3-MA, one of the few commercially available inhibitors that functions as an upstream inhibitor of macroautophagic induction, was used in addition to the downstream inhibitor of the fusion of AVs and lysosomes, chloroquine. Given that these two different autophagy inhibitors gave the same result strongly implicates macroautophagy as the preferred pathway of tauΔC clearance. A recent study has suggested a role for chaperone-mediated autophagy (34) in tau clearance, although it is important to note that in this study a tau construct of just the microtubule binding domains was the primary substrate. Another study provided data indicating that the lysosome played a role in the clearance of tauΔC through the use of lysosomal protease inhibitors after treatment with the inflammatory product prostaglandin J2 (33). Our results are in concordance with both of these studies in that there appears to be a clear role for the lysosome in the degradation of tauΔC, which is more aggregative and induces stress more than FL-tau.
Our findings also demonstrate that expression of tauΔC is sufficient to increase the autophagic phenotype as defined by the appearance of cells with increased GFP-LC3 puncta (Fig. 6). These findings suggest that the generation of tauΔC in AD may contribute to the increase in the presence of AVs in AD brain. Because the expression of FL-tau in the same cell model does not drive a similar increase of autophagic activity, this is likely not a general response due simply to protein overexpression or other confounding factors. Furthermore, it argues for the specificity of the autophagic response to tauΔC. The possibility that the cell lines utilized had differing maximal capacities for autophagic activity was invalidated by inducing the maximal appearance of GFP-LC3 puncta with a combination of nutrient deprivation and pharmacological blockage of AV fusion (Fig. 6), which results in identical numbers of cells with increased puncta.
Our study demonstrates that a physiological form of tau is preferentially degraded by the proteasome, whereas a pathological form that is more aggregative and toxic (tauΔC) is apparently not processed by the proteasome but is instead degraded by the lysosome through macroautophagy. This pathway shift takes place in the complete absence of the direct ubiquitylation of tau. Though here the clearance of tauΔC readily occurs, it should be noted that the autophagic response in the cell model is robust and responsive to cell stress. As noted, close ultrastructural examination of AD brain has shown an accumulation of autophagic vacuoles, which may indicate a flaw in the vesicle fusion step of the autophagic process (29). In addition, lower levels of Beclin-1, a key macroautophagy regulator, have been described in AD brain (30). Finally, microtubule dynamics have reported to be compromised in AD, which would impair the ability of autophagic vacuoles to migrate to lysosomes for degradation (51). Taken together, these key findings indicate an inability of macroautophagy to proceed to completion in AD brain. If autophagy cannot proceed to completion, then the processes we describe here for the removal of tauΔC would be subverted and lead to the accumulation of tauΔC in compromised neurons. This accumulation could contribute further to AD neurodegeneration.
Supplementary Material
Acknowledgments
We thank Drs. Douglas Cyr (University of North Carolina, Chapel Hill), Scott Wilson (University of Alabama, Birmingham), and Lester Binder (Feinberg School of Medicine, Northwestern University) for providing the myc-CHIP construct, HA-ubiquitin construct, and Tau5 antibody, respectively. We also thank Dr. Anthony Filiano for helpful discussions.
Note Added in Proof
A recent study by Grune et al. (65) also reported the lack of tau ubiquitylation in cell models, confirming our results.
This work was supported, in whole or in part, by National Institutes of Health Grant NS051279. This work was also supported by a grant from the Alzheimer's Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- AD
- Alzheimer disease
- CHIP
- C terminus of Hsp70-interacting protein
- AV
- autophagic vacuole
- 3-MA
- 3-methyladenine
- IP
- immunoprecipitation
- CN
- cortical neuron
- FL
- full-length
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Alzheimer A., Stelzmann R. A., Schnitzlein H. N., Murtagh F. R. (1995) Clin. Anat. 8, 429–431 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J. (1986) Neurobiol. Aging 7, 425–432 [DOI] [PubMed] [Google Scholar]
- 3.von Bergen M., Friedhoff P., Biernat J., Heberle J., Mandelkow E. M., Mandelkow E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeTure M., Ko L. W., Easson C., Yen S. H. (2002) Am. J. Pathol. 161, 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso A., Zaidi T., Novak M., Grundke-Iqbal I., Iqbal K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rankin C. A., Sun Q., Gamblin T. C. (2005) Brain Res. Mol. Brain Res. 138, 84–93 [DOI] [PubMed] [Google Scholar]
- 7.Rissman R. A., Poon W. W., Blurton-Jones M., Oddo S., Torp R., Vitek M. P., LaFerla F. M., Rohn T. T., Cotman C. W. (2004) J. Clin. Invest. 114, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeganathan S., von Bergen M., Mandelkow E. M., Mandelkow E. (2008) Biochemistry 47, 10526–10539 [DOI] [PubMed] [Google Scholar]
- 9.Santacruz K., Lewis J., Spires T., Paulson J., Kotilinek L., Ingelsson M., Guimaraes A., DeTure M., Ramsden M., McGowan E., Forster C., Yue M., Orne J., Janus C., Mariash A., Kuskowski M., Hyman B., Hutton M., Ashe K. H. (2005) Science 309, 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 11.Rapoport M., Dawson H. N., Binder L. I., Vitek M. P., Ferreira A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6364–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberson E. D., Scearce-Levie K., Palop J. J., Yan F., Cheng I. H., Wu T., Gerstein H., Yu G. Q., Mucke L. (2007) Science 316, 750–754 [DOI] [PubMed] [Google Scholar]
- 13.de Calignon A., Fox L. M., Pitstick R., Carlson G. A., Bacskai B. J., Spires-Jones T. L., Hyman B. T. (2010) Nature 464, 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung C. W., Song Y. H., Kim I. K., Yoon W. J., Ryu B. R., Jo D. G., Woo H. N., Kwon Y. K., Kim H. H., Gwag B. J., Mook-Jung I. H., Jung Y. K. (2001) Neurobiol. Dis. 8, 162–172 [DOI] [PubMed] [Google Scholar]
- 15.Ding H., Matthews T. A., Johnson G. V. (2006) J. Biol. Chem. 281, 19107–19114 [DOI] [PubMed] [Google Scholar]
- 16.Matthews-Roberson T. A., Quintanilla R. A., Ding H., Johnson G. V. (2008) Brain Res. 1234, 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintanilla R. A., Matthews-Roberson T. A., Dolan P. J., Johnson G. V. (2009) J. Biol. Chem. 284, 18754–18766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki H., Murayama S., Tomonaga M., Izumiyama N., Shimada H. (1987) Acta Neuropathol. 75, 156–159 [DOI] [PubMed] [Google Scholar]
- 19.Shimura H., Schwartz D., Gygi S. P., Kosik K. S. (2004) J. Biol. Chem. 279, 4869–4876 [DOI] [PubMed] [Google Scholar]
- 20.Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., Kim J., Dillmann W. H., Browne S. E., Hall A., Voellmy R., Tsuboi Y., Dawson T. M., Wolozin B., Hardy J., Hutton M. (2004) Hum. Mol. Genet. 13, 703–714 [DOI] [PubMed] [Google Scholar]
- 21.Carrettiero D. C., Hernandez I., Neveu P., Papagiannakopoulos T., Kosik K. S. (2009) J. Neurosci. 29, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David D. C., Layfield R., Serpell L., Narain Y., Goedert M., Spillantini M. G. (2002) J. Neurochem. 83, 176–185 [DOI] [PubMed] [Google Scholar]
- 23.Grelle G., Kostka S., Otto A., Kersten B., Genser K. F., Müller E. C., Wälter S., Böddrich A., Stelzl U., Hänig C., Volkmer-Engert R., Landgraf C., Alberti S., Höhfeld J., Strödicke M., Wanker E. E. (2006) Mol. Cell. Proteomics 5, 234–244 [DOI] [PubMed] [Google Scholar]
- 24.Sahara N., Murayama M., Mizoroki T., Urushitani M., Imai Y., Takahashi R., Murata S., Tanaka K., Takashima A. (2005) J. Neurochem. 94, 1254–1263 [DOI] [PubMed] [Google Scholar]
- 25.Dickey C. A., Yue M., Lin W. L., Dickson D. W., Dunmore J. H., Lee W. C., Zehr C., West G., Cao S., Clark A. M., Caldwell G. A., Caldwell K. A., Eckman C., Patterson C., Hutton M., Petrucelli L. (2006) J. Neurosci. 26, 6985–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C., Xu Z., He X. R., Michael L. H., Patterson C. (2005) Am. J. Physiol. Heart Circ. Physiol. 288, H2836–H2842 [DOI] [PubMed] [Google Scholar]
- 27.Qian S. B., McDonough H., Boellmann F., Cyr D. M., Patterson C. (2006) Nature 440, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 29.Nixon R. A., Wegiel J., Kumar A., Yu W. H., Peterhoff C., Cataldo A., Cuervo A. M. (2005) J. Neuropathol. Exp. Neurol. 64, 113–122 [DOI] [PubMed] [Google Scholar]
- 30.Pickford F., Masliah E., Britschgi M., Lucin K., Narasimhan R., Jaeger P. A., Small S., Spencer B., Rockenstein E., Levine B., Wyss-Coray T. (2008) J. Clin. Invest. 118, 2190–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bednarski E., Lynch G. (1996) J. Neurochem. 67, 1846–1855 [DOI] [PubMed] [Google Scholar]
- 32.Hamano T., Gendron T. F., Causevic E., Yen S. H., Lin W. L., Isidoro C., Deture M., Ko L. W. (2008) Eur. J. Neurosci. 27, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 33.Arnaud L. T., Myeku N., Figueiredo-Pereira M. E. (2009) J. Neurochem. 110, 328–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Martinez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E. M., Cuervo A. M., Mandelkow E. (2009) Hum. Mol. Genet. 18, 4153–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry R. W., Abraha A., Lagalwar S., LaPointe N., Gamblin T. C., Cryns V. L., Binder L. I. (2003) Biochemistry 42, 8325–8331 [DOI] [PubMed] [Google Scholar]
- 36.Carmel G., Mager E. M., Binder L. I., Kuret J. (1996) J. Biol. Chem. 271, 32789–32795 [DOI] [PubMed] [Google Scholar]
- 37.Bongarzone E. R., Foster L., Byravan S., Casaccia-Bonnefil P., Schonmann V., Campagnoni A. T. (1998) J. Neurosci. Res. 54, 309–319 [DOI] [PubMed] [Google Scholar]
- 38.Mi K., Dolan P. J., Johnson G. V. (2006) J. Biol. Chem. 281, 4787–4794 [DOI] [PubMed] [Google Scholar]
- 39.Shelton S. B., Krishnamurthy P., Johnson G. V. (2004) J. Neurosci. Res. 76, 110–120 [DOI] [PubMed] [Google Scholar]
- 40.Thorpe G. H., Kricka L. J. (1986) Methods Enzymol. 133, 331–353 [DOI] [PubMed] [Google Scholar]
- 41.Moore D. J., West A. B., Dikeman D. A., Dawson V. L., Dawson T. M. (2008) J. Neurochem. 105, 1806–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brady N. R., Hamacher-Brady A., Yuan H., Gottlieb R. A. (2007) FEBS J. 274, 3184–3197 [DOI] [PubMed] [Google Scholar]
- 44.Drubin D., Kobayashi S., Kellogg D., Kirschner M. (1988) J. Cell Biol. 106, 1583–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng L., Mohan R., Kwok B. H., Elofsson M., Sin N., Crews C. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10403–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi X., Haque T. S., Zhou J., Skillman A. G., Lin B., Lee C. E., Kuntz I. D., Ellman J. A., Lynch G. (2000) J. Neurochem. 74, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 47.Ahlberg J., Marzella L., Glaumann H. (1982) Lab. Invest. 47, 523–532 [PubMed] [Google Scholar]
- 48.Hatakeyama S., Matsumoto M., Kamura T., Murayama M., Chui D. H., Planel E., Takahashi R., Nakayama K. I., Takashima A. (2004) J. Neurochem. 91, 299–307 [DOI] [PubMed] [Google Scholar]
- 49.Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., Patterson C. (2001) J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 50.Wong E. S., Tan J. M., Soong W. E., Hussein K., Nukina N., Dawson V. L., Dawson T. M., Cuervo A. M., Lim K. L. (2008) Hum. Mol. Genet. 17, 2570–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B., Chohan M. O., Grundke-Iqbal I., Iqbal K. (2007) Acta Neuropathol. 113, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nixon R. A. (2006) Trends Neurosci. 29, 528–535 [DOI] [PubMed] [Google Scholar]
- 53.Schworer C. M., Shiffer K. A., Mortimore G. E. (1981) J. Biol. Chem. 256, 7652–7658 [PubMed] [Google Scholar]
- 54.Henell F., Berkenstam A., Ahlberg J., Glaumann H. (1987) Exp. Mol. Pathol. 46, 1–14 [DOI] [PubMed] [Google Scholar]
- 55.Schrader E. K., Harstad K. G., Matouschek A. (2009) Nat. Chem. Biol. 5, 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 57.Shringarpure R., Grune T., Mehlhase J., Davies K. J. (2003) J. Biol. Chem. 278, 311–318 [DOI] [PubMed] [Google Scholar]
- 58.Kaganovich D., Kopito R., Frydman J. (2008) Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., Patterson C. (1999) Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., von Bergen M., Mandelkow E. M., Mandelkow E. (2002) J. Biol. Chem. 277, 41390–41400 [DOI] [PubMed] [Google Scholar]
- 61.Morishima Y., Wang A. M., Yu Z., Pratt W. B., Osawa Y., Lieberman A. P. (2008) Hum. Mol. Genet. 17, 3942–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C. B., Patterson C., Dickson D. W., Nahman N. S., Jr., Hutton M., Burrows F., Petrucelli L. (2007) J. Clin. Invest. 117, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murata S., Chiba T., Tanaka K. (2003) Int. J. Biochem. Cell Biol. 35, 572–578 [DOI] [PubMed] [Google Scholar]
- 65.Grune T., Botzen D., Engels M., Voss P., Kaiser B., Jung T., Grimm S., Ermak G., Davies K. J. (2010) Arch. Biochem. Biophys., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.