Abstract
The thioesterase FlK from the fluoroacetate-producing Streptomyces cattleya catalyzes the hydrolysis of fluoroacetyl-coenzyme A. This provides an effective self-defense mechanism, preventing any fluoroacetyl-coenzyme A formed from being further metabolized to 4-hydroxy-trans-aconitate, a lethal inhibitor of the tricarboxylic acid cycle. Remarkably, FlK does not accept acetyl-coenzyme A as a substrate. Crystal structure analysis shows that FlK forms a dimer, in which each subunit adopts a hot dog fold as observed for type II thioesterases. Unlike other type II thioesterases, which invariably utilize either an aspartate or a glutamate as catalytic base, we show by site-directed mutagenesis and crystallography that FlK employs a catalytic triad composed of Thr42, His76, and a water molecule, analogous to the Ser/Cys-His-acid triad of type I thioesterases. Structural comparison of FlK complexed with various substrate analogues suggests that the interaction between the fluorine of the substrate and the side chain of Arg120 located opposite to the catalytic triad is essential for correct coordination of the substrate at the active site and therefore accounts for the substrate specificity.
Keywords: Acetyl-Coenzyme A, Bacterial Metabolism, Coenzyme A, Crystal Structure, Enzyme Mechanisms, Protein Structure, Site-directed Mutagenesis, Fluoroacetyl-Coenzyme A, Thioesterase
Introduction
In 1986, Sanada et al. (1) discovered that Streptomyces cattleya is able to produce fluoroacetate (FAc)8 from fluoride. Attempts to elucidate the mechanism of this unusual transformation have led to the discovery of a fluorinase enzyme catalyzing the formation of 5′-fluoro-5′-deoxyadenosine from S-adenosyl-l-methionine and fluoride (2–6). We have cloned the fluorinase gene flA (5) and subsequently identified a gene cluster (fl) surrounding the fluorinase gene from S. cattleya (7). Characterization of the 5′-fluoro-5′-deoxyadenosine phosphorylase (3), the enzyme responsible for the second step of the FAc biosynthesis encoded by the flB gene located next to the flA, has confirmed the involvement of the fl gene cluster in the biosynthesis of FAc (7). In our efforts to further elucidate the functions of the fl gene cluster, the protein encoded by the flK gene located close to the 3′-end of the cluster was characterized as a thioesterase that cleaves fluoroacetyl-coenzyme A (FAcCoA) to produce FAc and CoA. It is noteworthy that acetyl-coenzyme A (AcCoA) is not hydrolyzed by FlK (7). Previous studies have shown that FAc can be used by AcCoA synthase to synthesize FAcCoA, and citrate synthase can catalyze formation of 2-fluorocitrate from FAcCoA and oxaloacetate (8). Lauble et al. (9) have shown that the (−)-erythro diastereomer of 2-fluorocitrate is further converted to 4-hydroxy-trans-aconitate, a lethal competitive inhibitor of aconitase, thereby blocking the tricarboxylic acid cycle. The presence of a FAcCoA-hydrolyzing enzyme that has no effect on the metabolically important intermediate AcCoA may therefore serve as a self-resistance mechanism for the FAc-producing S. cattleya.
FlK is homologous to many predicted thioesterases and hypothetical proteins according to protein-protein BLAST searches against a non-redundant protein sequence data base (7). However, none of these putative homologues has a defined function. Two groups of thioesterases have been identified, thioesterases I and thioesterases II, based on differences in their amino acid sequence, protein folds, and catalytic mechanisms. Type I thioesterases have evolved within the α/β-hydrolase fold superfamily and are usually present as a product-releasing domain of multidomain proteins, such as type I fatty acid synthases, polyketide synthases, and nonribosomal peptide synthases. Type I thioesterases use serine or cysteine as the catalytic residue (10, 11). Type II thioesterases are discrete proteins associated with activities of hydrolyzing short- or medium-chain acyl-CoAs and exist either as a homodimer of a single hot dog fold (12) or as a monomer comprising a duplicated hot dog fold (13). It has been shown that the active sites of type II thioesterase enzymes contain either a glutamate or an aspartate residue acting as a general base or nucleophile responsible for the catalytic function (14, 15), although the precise catalytic mechanism has remained controversial. FlK, a discrete protein catalyzing hydrolysis of a short-chain acyl-CoA (fluoroacetyl CoA), apparently belongs to the type II thioesterase family and is therefore expected to have a carboxylic side chain at the active site. Protein sequence alignment analysis has revealed that nine amino acid residues in FlK (Gly8, Thr42, Glu50, Leu61, Gly69, His76, Ala78, Gly83, and Gly116) are all conserved in homologues that have a similar protein size to FlK (ranging from 127 to 180 amino acids). The presence of Thr42 and His76 together with Glu50 among the conserved residues led us to speculate that FlK may employ a variation of the classical catalytic triad-type mechanism with Thr42 (instead of a Ser or a Cys), His76, and Glu50 (instead of an Asp). His76 would then serve as a base to deprotonate Thr42. The deprotonated Thr42-Oγ1 is proposed to act as a nucleophile attacking the fluoroacetyl carbonyl carbon of FAcCoA to initiate the hydrolysis reaction and the carboxylic side chain of Glu50 to neutralize the charge developed on the His76 side chain during the transition state of the reaction by hydrogen bonding with the imidazole nitrogen of His76 (7). Although a hydroxyl derived from threonine rather than from a serine residue has been found to act as the catalytic nucleophile in some proteinases, including the individual activities of the proteosome (16), to the best of our knowledge, no functionally characterized thioesterase has been reported to use threonine in this way.
We have carried out site-directed mutagenesis targeting the putative catalytic residues and defined structures by x-ray crystallography of the wild type and mutant FlK proteins both with and without bound analogues of FAcCoA. Our studies provide insights into the catalytic mechanism of FlK and the high substrate specificity of FlK for FAcCoA over the closely related AcCoA.
EXPERIMENTAL PROCEDURES
Cells, Reagents, and Equipment
All oligonucleotide primers were synthesized by MWG-Biotech. Restriction enzymes were purchased from New England Biolabs, T4 DNA ligase was from Fermentas Life Sciences, and cloned Pfu DNA polymerases were from Stratagene. The following reagents were purchased from Sigma: AcCoA, 5,5′-dithio-bis(2-nitrobenzoic acid) (Ellman's reagent), CHES, ethylene glycol, polyethylene glycol 4000, polyethylene glycol monomethyl ether 2000, potassium chloride (KCl), l-selenomethionine (l-SeMet), trisodium citrate, Tris, sodium acetate, and sodium fluoroacetate. Amino acids used for cell cultures were from DUCHEFA Biochemie. FAcCoA was synthesized as described previously (7). An ÅKTA FPLC system and a HiLoad Superdex S200 column (16 × 60 cm) (GE Healthcare) were used for gel filtration purification of proteins. The protein concentration was determined using the Bradford method (Sigma). DNA sequencing was performed in the Department of Biochemistry DNA Sequencing Facility (University of Cambridge) on an Applied Biosystems 3730xl DNA analyzer. Escherichia coli NovaBlue or DH10B cells were used for cloning, and E. coli RosettaTM (DE3)pLysS cells were used for protein expression.
Cloning of FlK and Mutants
The cloning of the recombinant His-tagged wild-type FlK (WtFlK) in pET28a(+) (Novagen) was described in our previous study (7). Mutants of FlK each containing a single amino acid mutation were created by PCR using the WtFlK-expressing plasmid as a template. For each mutant, two PCR amplifications were carried out using two primer pairs: T7-27/3′-5′ primers for PCR-1 and T7T-27/5′-3′ primers for PCR-2. The T7–27 primer (5′-TAATACGACTCACTATAGGGGAATTGT-3′) and T7T-27 primer (5′-GCTAGTTATTGCTCAG CGGTGGCAGCA-3′) hybridize, respectively, to the upstream or downstream regions of the multicloning site in pET28a(+). Mutations were created in either the 3′-5′ primer or the 5′-3′ primer or both, depending on the specific design for each individual mutant.
PCR was carried out using cloned Pfu DNA polymerase (Stratagene) with 25 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min plus a final extension at 72 °C for 10 min. The resulting PCR products were digested with appropriate restriction enzymes, purified by gel extraction (Qiagen), and inserted into pET28a(+) vector between the NdeI and BamHI sites. The inserts of the recombinant plasmids were verified by DNA sequencing. Constructs with the correct inserts were introduced into E. coli RosettaTM (DE3)pLysS competent cells (Novagen) for protein expression.
Expression and Purification of FlK and FlK Mutants
An overnight culture (10 ml) from a single colony harboring the desired plasmid was used for inoculation of 1 liter of fresh LB medium containing kanamycin (50 μg/ml). The culture was incubated at 37 °C, 250 rpm until A600 = 0.5–0.8 was reached. Overexpression of the proteins was induced by 0.2–0.3 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 16 °C with shaking at 220 rpm before being harvested by centrifugation. For incorporation of l-SeMet into FlK, 1 liter of fresh medium containing 6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 1 g of NH4Cl, 0.5 g of NaCl, 1 mm MgSO4, 4 g of glucose, 0.5 mg of thiamine vitamin B1, and 50 mg of kanamycin was inoculated with cells from a 10-ml overnight culture and incubated at 37 °C with shaking at 250 rpm. When the cell density reached A600 = 0.3–0.5, a mixture of l-lysine (100 mg), l-phenylalanine (100 mg), l-threonine (100 mg), l-leucine (50 mg), l-isoleucine (50 mg), l-valine (50 mg), and l-SeMet (50 mg) was added into the culture, and incubation was continued for 30 min at 37 °C. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 1 mm to induce protein overexpression overnight at 16 °C, 200 rpm. The cells were broken by sonication for 4 min (1 s on, 10 s off) to release overexpressed recombinant protein. The supernatant of the cell lysate was applied to a His-Bind® column (Novagen) charged with Co2+ ions. Nonspecific proteins were removed by washing the column with washing buffer (0.5 m NaCl, 40 mm imidazole, 20 mm Tris-HCl, pH 7.9), and the recombinant protein was eluted with elution buffer (0.5 m NaCl, 200 mm imidazole, 20 mm Tris-HCl, pH 7.9). The buffer containing the purified proteins was exchanged with a buffer containing 100 mm KCl and 20 mm Tris-HCl, pH 7.5, and concentrated at 4 °C to a total volume of 1–2 ml. The His tag of the proteins was removed by thrombin (restriction grade; Novagen) digestion at room temperature overnight. The tag-free proteins were further purified by gel filtration on an ÄKTA Explorer FPLC system using a HiLoadTM 16/60 SuperdexTM 200 Prep Grade column and a mobile phase containing 100 mm KCl and 20 mm Tris-HCl, pH 7.5. The purity of the proteins was checked by SDS-PAGE. The protein masses were confirmed by liquid chromatography-electrospray ionization-mass spectrometry on a 2.0 × 250-mm Jupiter 5-μm C4 column (Phenomenex) using a Finnigan LCQ (Thermo Finnigan) coupled with an HP1100 high pressure liquid chromatography system (Agilent) with a flow rate of 0.2 ml/min and the following mobile phase gradient: 0–5 min, 35–45% B; 5–25 min, 45–75% B; 25–30 min, 75–95% B (buffer A: 0.1% trifluoroacetic acid in H2O; buffer B: 0.1% trifluoroacetic acid in acetonitrile).
Enzymatic Assays
The thioesterase activity of FlK and its mutants on FAcCoA was measured by monitoring spectrophotometrically the increase in absorbance at 412 nm due to the reaction of released CoASH with 5,5′-dithio-bis(2-nitrobenzoic acid with a CARY 100 Bio UV-visible spectrophotometer (Varian) as described previously (7). All assays were performed at 25 °C in a total volume of 0.5 ml in a spectrophotometer cuvette with a 1-cm light path.
Preparation of Fluoroacetyl Carba(dethia)-pantetheine and Fluoroacetyl Oxa(dethia)-pantetheine
To prepare fluoroacetyl carba(dethia)-pantetheine (FAcCPan), monofluorination of (O,O′-diacetyl)-malonyl carba(dethia)-pantetheine methylester with SelecfluorTM was carried out, followed by base-catalyzed loss of the protecting groups and concomitant deprotection and decarboxylation of the malonyl moiety. Fluoroacetyl oxa(dethia)-pantetheine (FAcOPan) was obtained from 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide coupling of O,O′-isopropylidene-oxa(dethia)-pantetheine with sodium fluoroacetate, followed by acetal deprotection with Dowex 50-X8-400.
Crystallization of SeMet FlK in the Apo-form
The crystallization trials with the SeMet FlK protein at a concentration of 20 mg/ml in 50 mm Tris-HCl and 100 mm KCl, pH 7.5, were carried out using a Cartesian HONEYBEETM 81 crystallization robot (Genomic Solutions), and protein crystallization was screened using the crystallization conditions from SM1 (Nextal) and the Classics (Qiagen) crystallization solution kits. To each reservoir of 96-well CrystalQuick crystallization plates with square sitting drop position (Molecular Dimensions) were added 100 μl of the crystallization solution and 100 nl of protein solution. The best crystals appeared after approximately 1 week in 0.1 m Tris, pH 7.5, and 20% polyethylene glycol monomethyl ether 2000.
Crystallization of Wild-type FlK (WtFlK) and Its Mutants (T42SFlK and T42AFlK) in Complex with Acetate (Ac) and FAc
Initially, crystallization trials were carried out using hanging drop vapor diffusion at 293 K. The protein at 10 mg/ml in 50 mm Tris-HCl buffer, pH 7.5, containing 100 mm KCl was screened against mother liquor solutions from the Classics suite (Qiagen) and kept at 20 °C. The initial crystals were obtained in 30% polyethylene glycol 4000, 0.1 m sodium acetate, and 0.1 m Tris-HCl, pH 7.0. After optimization, the best crystals were obtained with protein at 5 mg/ml in the same crystallization condition. The drops comprised 1 μl of protein solution and 1 μl of crystallization solution, and the crystals appeared after 3 or 4 days. To crystallize the FlK in complex with FAc, the sodium acetate present in the crystallization solution was replaced by 0.2 m sodium fluoroacetate. The crystals for both complexes were flash-frozen in liquid nitrogen to perform the data collection after soaking in cryoprotectant solution composed of 25–30% ethylene glycol and 75–70% well solution. Both mutants (T42SFlK and T42AFlK) and WtFlK were crystallized under the same conditions.
Crystallization of T42SFlK in Complex with AcCoA
After ∼6 months, crystals of T42SFlK·AcCoA complex grew in conditions similar to those for the WtFlK·Ac complex except where the acetate was replaced with 20 mm AcCoA.
Crystallization of WtFlK in Complex with Analogues of Fluoroacetyl Pantetheine
The protein was incubated with 5 mm either FAcCPan or FAcOPan. Initial trials were carried out using a Cartesian HONEYBEETM 81 crystallization robot (Genomic Solutions), and the protein was screened using the crystallization conditions from SM1 (Nextal), Classics, pHclear, and pHclear II (Qiagen) crystallization solution kits. In each reservoir of the 96-well CrystalQuick crystallization plate with square sitting drop position (Molecular Dimensions), 100 μl of the crystallization solution was added. The drops contained 100 nl of the reservoir solution and 100 nl of protein solution at 10 mg/ml in 50 mm Tris-HCl and 100 mm KCl, pH 7.5. The best hits without acetate appeared in a condition that comprised 100 mm CHES, pH 9.5, and 1 m trisodium citrate. After optimization, the best crystals were obtained in a condition containing 100 mm CHES, pH 9.5, and 0.8 m trisodium citrate with a protein concentration of 10 mg/ml in the presence of 10 mm FAcCPan or FAcOPan.
Data Collection and Structure Determination
Data for apo-form WtFlK and SeMet FlK were collected at beamline ID14.4, European Synchrotron Radiation Facility (Grenoble, France). The data sets were processed using DENZO and SCALEPACK (version 1.97) (17). The data were truncated and converted to structure factors using TRUNCATE (18) from the CCP4 suite (19). X-ray data for complexes of WtFlK·FAc, T42AFlK·FAc, T42SFlK·AcCoA, WtFlK·FAcCPan, and WtFlK·FAcOPan were collected at the Swiss Light Source, and the WtFlK·Ac, T42SFlK·Ac, and T42SFlK·FAc were collected at the European Synchrotron Radiation Facility. The data sets were processed using the program Mosflm (20) and scaled by SCALA (21). The structure of SeMet FlK in its apo-form was solved using single-wavelength anomalous diffraction and the Phenix program (22), and the other structures were solved by molecular replacement using the apo-structure as probe search in the program AMoRe (23) in the CCP4 suite (19). The refinement was carried out using the program REFMAC 5.2 (24). Visual inspection and water addition were performed using XtalView/xfit (25) and Coot (26). The quality of the model was assessed using PROCHECK (27). The figures were made using PyMOL (28).
RESULTS
Site-directed Mutagenesis
To investigate the reaction mechanism of FlK, mutants of FlK were created at residues Thr42, Glu50, and His76, candidates for the catalytic triad in the FlK protein. To assess the involvement of these residues in the catalytic mechanism, the activities of the mutants were assayed and compared with that of WtFlK. Four mutants of the FlK protein carrying single amino acid mutations (T42S, T42A, E50A, and H76D, respectively) were cloned in pET28a(+) and overexpressed in E. coli RosettaTM (DE3)pLysS cells as His-tagged proteins. All proteins were purified by Co2+-charged His-Bind resin (Novagen) followed by gel filtration. Both WtFlK and T42SFlK were expressed as soluble proteins with a yield of over 10 mg from 1 liter of culture. T42AFlK and E50AFlK mutants yielded only 2 mg of soluble enzyme/liter of culture, although their expression levels were comparable with those of WtFlK and T42SFlK. During storage at 4 °C T42SFlK and T42AFlK also showed a much higher tendency to aggregate than WtFlK. The recombinant H76DFlK protein expressed in E. coli was completely insoluble, so it was not used for further studies. All of the mutations seemed have disturbed protein folding, with the H76D mutant being the most affected. The molecular weights of the purified proteins were determined by liquid chromatography-electrospray ionization-mass spectrometry. Retention times on gel filtration through a Superdex S200 column indicate that purified WtFlK and the T42AFlK, T42SFlK, and E50AFlK mutants are all dimers.
The enzymatic characterization of WtFlK using FAcCoA as substrate has been reported previously (7). The present study demonstrated that replacing the Thr42 with Ala42 in FlK abolishes the enzyme activity, supporting our hypothesis that the Thr42-Oγ1 may act as the catalytic nucleophile that attacks the carbonyl thioester carbon of FAcCoA to initiate the hydrolysis reaction. Not surprisingly, T42SFlK is still able to hydrolyze FAcCoA with a kcat/Km value of 27.3 mm−1 s−1, which is about 19 times higher than that for WtFlK (1.47 mm−1 s−1), and a relatively smaller Km value (15 μm, 50% lower than that for WtFlK (30 μm)). It is noteworthy that T42SFlK activity was inhibited at a substrate concentration above 12 μm. These results suggest that the Ser42-Oγ in T42SFlK is able to carry out the nucleophilic attack and that the absence of the Thr42-Cγ in the T42S mutant may have led to decreased ability to ensure correct substrate binding, resulting in the substrate inhibition. The kcat/Km value of E50AFlK (0.54 mm−1 s−1) is only 37% of that for WtFlK, whereas the Km (206 μm) is much higher than that for WtFlK. E50AFlK also exhibited substrate inhibition when the FAcCoA concentration was above 19 μm, suggesting that Glu50 plays an important role in substrate recognition (Table 1).
TABLE 1.
Comparison of wild-type and mutant FlK
Enzyme reactions were carried out using FAcCoA as the substrate. Errors quoted represent the S.E. of curve fitting (Lineweaver-Burk plot). Each data point is the average of four (WtFlK), three (T42SFlK), or two E50AFlK measurements.
| Protein | Soluble protein | kcata | Km | kcat/Kmb | FAcCoA concentration above which substrate inhibition was observed |
|---|---|---|---|---|---|
| mg/liter | s−1 | μm | mm−1s−1 | μm | |
| WtFlK | ∼10–12 | 0.044 ± 0.001 | 30 ± 1.3 | 1.47 ± 0.10 | |
| T42SFlK | ∼10–12 | 0.409 ± 0.159 | 15 ± 1.2 | 27.3 ± 12.8 | 12 |
| T42AFlK | ∼2 | ||||
| E50AFlK | ∼2 | 0.111 ± 0.041 | 206 ± 67 | 0.54 ± 0.37 | 19 |
a The units are number of CoA released/s/enzyme molecule.
b The error values given for kcat/Km are calculated from the sum of the relative errors on values in the numerator and denominator.
FlK Crystal Structure Determination
In order to understand the catalytic mechanism of FlK, in particular how FlK distinguishes between FAcCoA and the structurally very similar AcCoA, 10 crystal structures of WtFlK, T42SFlK, and T42AFlK with or without various ligands were determined. The WtFlK structures were solved in the apo-form and in complex with either the substrate analogue AcCoA or the substrate fragment analogues FAcCPan and FAcOPan, the product FAc, or the product analogue Ac. Although attempts to crystallize T42S and T42A mutants in their apo-forms failed, crystals of T42SFlK in complex with Ac, FAc, or AcCoA and T42AFlK with bound FAc were obtained, and their structures were solved.
Overall Fold and Oligomeric Association of FlK
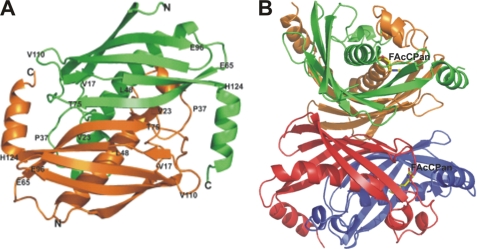
The crystals of FlK diffracted at resolutions between 2.35 and 1.5 Å and belong to space group C2 or P21 with two, four, or eight FlK molecules in the asymmetric unit, depending on the nature of the complex. Those complexes that crystallized in C2 (WtFlK·Ac, WtFlK·FAc, T42SFlK·Ac, T42SFlK·FAc, and T42AFlK·FAc) have a dimer in the asymmetric unit with a local 2-fold symmetry axis. The crystals in P21 have either dimers (two in T42SFlK·AcCoA and apo-SeMet WtFlK) in the asymmetric unit, also with local 2-fold symmetry, or two tetramers (WtFlK·FAcCPan and WtFlK·FAcOPan) with orthogonal 2-fold axes giving 222 pseudosymmetry (Fig. 1, A and B). Operation of the crystallographic 2-fold axis on the dimers in the asymmetric unit in space group C2 generates a tetramer, identical to that observed in the WtFlK·FAcPan or WtFlK·FAcOPan crystal structures. The observation that these crystal structures contain discrete tetramers suggests that the protein may have dimer-tetramer equilibrium in solution in which tetramers predominate at high concentrations.
FIGURE 1.
Overall structure of FlK depicted in a schematic representation. A, structure of WtFlK dimer. The two protomers of the dimer are shown in green and orange, respectively. B, tetrameric structure (dimer of dimers) of WtFlK in complex with the substrate analogue FAcCPan. The FAcCPan molecule is shown as sticks. The two protomers of the dimer on the top are indicated in green and orange, respectively. A red/blue color scheme is used for the dimer at the bottom.
The protomer of FlK has a hot dog fold that comprises a five-strand antiparallel β-sheet (the bun) wrapping around a central α-helix (the sausage) (Fig. 1A). The hot dog fold was first observed for the structure of β-hydroxydecanoyl thiol ester dehydratase (12). A diverse range of enzymes with distinct catalytic activities has been found to have this fold. Thioesterases are important members of the hot dog superfamily and include 4-hydroxybenzoyl-CoA thioesterases from Pseudomonas sp. CBS-3 (29) and from Arthrobacter sp. SU (30), Escherichia coli thioesterases II (13), and acyl-CoA thioesterases YciA from Hemophilus influenzae (HI0827) (31). Despite the conserved hot dog fold, these thioesterases share a very low degree of identity at amino acid sequence level. The FlK asymmetric unit contains a dimer with pseudosymmetry about a 2-fold axis as observed previously in the structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. (29). and (R)-specific enoyl-CoA hydratase from Aeromonas caviae (32). Most other thioesterases with the hot dog fold form tetramers, although hexameric structures have also been observed (31, 33). The main contacts between the two protomers in the FlK dimer are through strand 2 (residues 70–76) of the two protomers. The β-strands of the two protomers form together a 10-strand β-sheet with the putative active site at the interface between the protomers. A disulfide bond formed by their respective Cys73 residues covalently connects the two subunits. Wrapping around the disulfide bond are the Ile72 and the Val74 from both protomers with their side chains within hydrophobic interaction distance of each other. The Cγ2 of the putative catalytic Thr42 also makes a hydrophobic contact with Ile72-Cγ2 of the partner protomer. All of these interactions contribute to bringing the two protomers together. Furthermore, there are two salt bridges formed between the two protomers; the Lys21-Nζ interacts with the Glu29-Oϵ2, and Lys135-Nζ interacts with the carboxyl Oδ2 groups of Asp111 and Asp108 in the partner protomer. Hydrogen bonds formed by Ser30-Oγ and Glu32-Oϵ1 with Trp51-Nϵ1, Gly43-N with Glu50-Oϵ2 through a water molecule, and Tyr27-OH with Phe44-O also serve to stabilize the FlK dimer. The contact area between two protomers is ∼1750 Å2, corresponding to 30% of the accessible surface area of each protomer, suggesting strong interactions between them. These observations have led us to assume that the minimal active unit of FlK is a dimer.
The tetrameric structure observed for WtFlK·FAcCPan is formed by a dimer of dimers with pseudo 222 symmetry (Fig. 1B). The main interaction regions are the 10-strand β-sheets present in the dimers, which are positioned back to back. Non-polar interactions predominate, but additionally there are some water-mediated hydrogen bonds. Similar tetramers have been observed in other thioesterases (13, 30, 34, 35). The contact area between two dimers is ∼3300 Å2, corresponding to about 30% of the total solvent-accessible area of each dimer, further suggesting the possibility of an equilibrium between dimers and tetramer in solution.
The Active Site
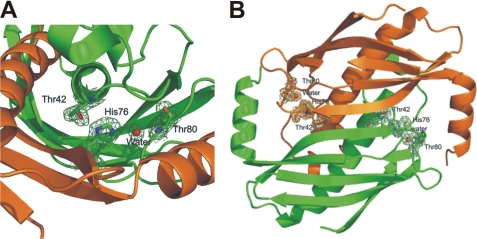
Consistent with the results of the site-directed mutagenesis of FlK described above, the crystal structure of apo-form WtFlK shows that side chains of the candidate catalytic residues Thr42 and His76 in one protomer and that of Glu50 from the adjacent protomer cluster between the dimer interface into the region between the “bun” and the “sausage” (Fig. 2, A and B). As expected, the Oγ1 of Thr42 and the Nδ1 of His76 are within hydrogen bonding distance (2.74 Å). To our surprise, we observed that the carboxylic group of Glu50 is located on the opposite side of Thr42, where it cannot have any direct contact with His76. However, a water molecule (Wat1) is in a perfect position to form a hydrogen bond with the Nδ2 of His76 (2.79 Å). This water molecule is conserved in the crystal structures of wild-type and mutant FlK with or without bound ligands. Although this observation was unexpected, it is in good agreement with our site-directed mutagenesis results. A water molecule replacing the acidic residue in this type of catalytic triad was also found in the HAV-3C gene product, an α-chymotrypsin-like protease produced by the hepatitis A picornavirus (36). The Thr42-His76-Wat1 network is further stabilized by a hydrogen bond between Wat1 and Thr80 side chain hydroxyl and another conserved water molecule (Wat2) that also forms a hydrogen bond with the Phe40-N. Around the His76-Wat1-Thr80/Wat2 network, the side chains from Val39, Phe40, Ala78, Ala79, and Ile113 form a hydrophobic shield, protecting the network from attack by solvent molecules. Based on our structural evidence, we now propose that the catalytic triad in FlK comprises Thr42-His76-water (Fig. 2A), located at the interface between two protomers with two active sites in each FlK dimer (Fig. 2B).
FIGURE 2.
Catalytic center of FlK. A, the catalytic triad of FlK is composed of Thr42, His76 (shown as sticks), and a conserved water molecule (Wat) (shown as a sphere). Green and orange colors indicate the two protomers of the dimer, respectively. B, the catalytic triad is located at the dimer interface. The electron density map used was 2Fo − 2Fc.
Substrate Binding Pocket
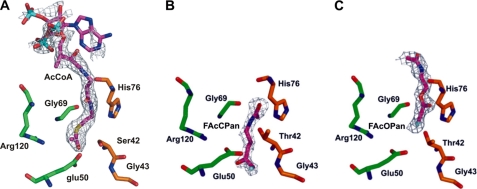
Although attempts to co-crystallize FlK with FAcCoA, CoA, or pantothenic acid failed, we successfully obtained crystals of T42SFlK with bound AcCoA and WtFlK in complex with two analogues of fluoroacetyl-pantetheine, FAcCPan, and FAcOPan. The sulfur atom of the thioester is replaced by a methylene in FAcCPan and by an oxygen atom in FAcOPan (Fig. 3). Comparison of the electron density maps of the crystals obtained from co-crystallization of WtFlK with FAc and CoA indicates that only FAc was bound. Comparisons of the structure of WtFlK without bound ligand with those of WtFlK·FAcCPan, WtFlK·FAcOPan, and T42SFlK·AcCoA have provided interesting insights into the structural basis of substrate recognition by FlK.
FIGURE 3.
Active sites of ligand-bound FlK with 2Fo − 2Fc electron density map for T42SFlK·AcCoA (A), WtFlK·FAcCPan (B), and WtFlK·FAcOPan (C). The molecules of AcCoA, FAcCPan, and FAcOPan are all shown in a deep purple color. Green and orange colors indicate residues from the two protomers of the dimer.
In the ligand-free WtFlK crystal structure, three well conserved water molecules (designated Wat3, Wat4, and Wat5) mediate a hydrogen bonding network linking Thr42 and Gly43 in one protomer to Glu50, Arg120, and Gly69 in the neighboring protomer; Wat3 links the Gly43-N with the Glu50-Oϵ2, and Wat4 and Wat5 are within hydrogen bonding distance of each other and of the Thr42-N and Gly69-N, respectively. The Arg120-NH1 joins the network by salt bridging with Glu50-Oϵ1 and hydrogen bonding with Wat5. Converting Arg120 to Ala120 yielded completely insoluble protein, as did converting Glu50 to Ala50, suggesting the importance of these interactions in maintaining the correct protein folding and/or the correct architecture of the active site cavity.
In the T42SFlK·AcCoA complex, the AcCoA is sandwiched between the long β-sheet and residues 17–42 that form two small α-helices (Fig. 4B). However, only the acetyl and β-mercaptoethylamine moieties from acetyl-CoA are buried in the protein, whereas the pantothenic acid and the 3′-phosphoryl-ADP are exposed to solvent, allowing considerable flexibility (Fig. 4, A and B). Three of the four protomers present in the asymmetric unit have a bound AcCoA, and in one of them, it was possible to fit the complete molecule, despite the poor electron density for the 3′-phosphoryl-ADP moiety, possibly due to the high flexibility in the solvent. The overall structure of FlK does not change significantly as a result of the T42S point mutation and the binding of AcCoA, but the AcCoA has forced Wat4 and Wat5 out of the active site cavity. The acetyl methyl group of the AcCoA occupies the position of Wat5, and the thioester carbonyl oxygen interacts via a hydrogen bond to Ser42-N. The orientations of the three bound AcCoA molecules at the active site are significantly different, reflecting a dynamic AcCoA-FlK interaction process and the flexibility of the substrate binding site. The acetyl carbonyl carbons of the three bound AcCoA molecules are all in the wrong orientation with respect to Oγ of Ser42 to allow nucleophilic attack. This explains the inability of FlK to hydrolyze AcCoA and implies that the presence of the fluorine in FAcCoA is crucial for substrate recognition.
FIGURE 4.
Overall structure of T42SFlK in complex with AcCoA. The two protomers of the T42SFlK dimer are represented in green and orange colors, respectively. A, 2Fo − 2Fc electron density map shows that AcCoA is bound between two protomers of the active FlK dimer. B, AcCoA is sandwiched between the long β-sheet and two small α-helices formed by residues 17–42. The long β-sheet and two small α-helices of protomer A and B are shown in light blue and light purple, respectively. C, representation of the molecular surface shows that only the acetyl and β-mercaptoethylamine moieties from AcCoA are buried in the active site of the protein.
The positions of FAcCPan and FAcOPan in WtFlK differ significantly from that of AcCoA in the T42SFlK·AcCoA structure. Compared with the bound AcCoA, the bound FAcCPan is buried deeper in the active site, and the FAcOPan is located closer to the entrance of the tunnel (Fig. 3B). In a way that resembles the binding of AcCoA in T42SFlK, the carbonyl carbons of both FAcCPan and FAcOPan in WtFlK are in the wrong position and are even further away (3.88, 3.94, and 5.26 Å, respectively) from the Oγof Thr42. Furthermore, the binding coordinates of the same molecule also vary in different FlK dimers, suggesting that the presence of the nucleotide part of the CoA moiety is required for the correct overall engagement of the substrate at the substrate-binding site.
In all of the FAc or Ac-bound FlK complexes, more than one molecule of FAc or Ac is bound in the large cavity of each active site. This may be a consequence of the high ligand concentrations (0.2 m) used in the crystallization solution and of the absence of the substrate. Interestingly but not surprisingly, at each active site, there is always one ligand situated at a position to where either the acetyl group of the AcCoA or the equivalent moiety of FAcCPan or FAcOPan is bound. The negatively charged carboxylic oxygen moieties of the ligands and the fluorine in the case of FAc are usually in contact with Thr42-N and/or Gly69-N in a seemingly random manner in terms of hydrogen bond donor-acceptor matching, suggesting that the backbone amide groups of Thr42 and Gly69 may be crucial binding interactions available at the active site to bind the substrate.
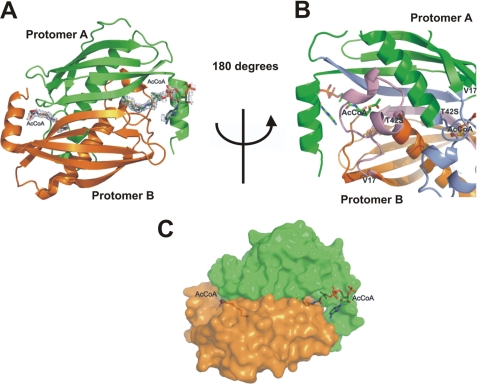
Computational Docking of FAcCoA to FlK
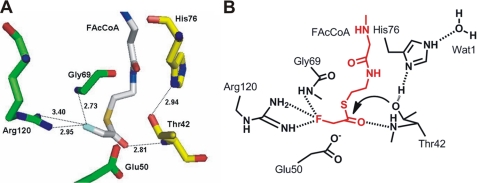
We have modeled a FAcCoA molecule into WtFlK (Fig. 5), based on the crystal structures of FlK in complex with various ligands, especially with AcCoA. The minimum energy was calculated using the SYBYL 8.1.1.09097 program. In a similar way to the AcCoA bound in T42SFlK, the 3′-phosphoryl-ADP region of the docked FAcCoA is at the enzyme surface in front of the entrance of the tunnel leading to the active site. The rest of the molecule extends into the tunnel, where the pantetheinyl moiety interacts through its N12, O9, N14, and C19, respectively, with the main chain oxygen and the side chain of His76, Ala79-N, and Phe128-Cϵ1. The hydrogen bond between the side chains of Arg120 and Glu50 is broken due to the flip of the carboxyl group of the Glu50. The fluorine is placed at a hydrogen bonding distance from Gly69-N and the guanidinium group of Arg120. Mutation of the Arg120 to Ala120 yielded a completely insoluble protein (data not shown), consistent with the involvement of Arg120 in maintaining the conformation of FlK. Thr42-Oγ is now located closer (3.79 Å) to the thioester carbonyl carbon and at a better, although not optimal, position for a possible nucleophilic attack. The Thr42-N that is close (2.81 Å) to the thioester carbonyl oxygen could act as an oxyanion hole for stabilization of the tetrahedral acyl-enzyme intermediate (Fig. 5).
FIGURE 5.
Proposed FAcCoA coordination at the active site of FlK. A, a stick model with FAcCoA shown in silver and residues from the two protomers represented in yellow and green colors, respectively. B, a scheme with FAcCoA (red) and active site residues (black). Hydrogen bonding interactions are denoted by dotted lines with distance in Å.
Increased Flexibility at the Active Site Resulting from Mutations of Thr42
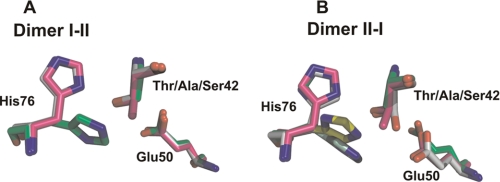
The hydrophobic interactions between Thr42-Cγ2 and Ile72-Cγ2 in the adjacent protomer provide a “dry” environment around the catalytic Oγ1 of Thr42, protecting it from attacks by solvent molecules, a feature observed for other enzymes that have similar catalytic triad mechanisms (37). The Thr42-Cγ2 may also restrain the freedom of the substrate in the active site. Loss of this restraint imposed by the Cγ2 in the T42S mutant may allow random misbinding of the substrate, which in turn may result in the substrate inhibition observed in the enzyme assays. In support of this notion, the imidazole group of the His76 in the complex T42AFlK·FAc has a rotational disorder of ∼90° or exists in two alternative conformers not observed in WtFlK structures, and the Ser42 in one of the protomers of the T42SFlK·Ac structure shows a double conformation, demonstrating the increased flexibility of these residues in the absence of the Thr42-Cγ2 and/or -Oγ2 (Fig. 6).
FIGURE 6.
Structural differences observed in the catalytic site of FlK due to different mutations of Thr42. The active site residues Thr42, His76, and Glu50 of WtFIK are superimposed with their counterpart residues in T42AFIK and T42SFIK. A, Thr/Ala/Ser42 and His76 from protomer I and the Glu50 from protomer II; B, Thr/Ala/Ser42 and His76 from protomer II and the Glu50 from protomer I. Pink, WtFlK; green, T42AFlK; white, T42SFlK. The alternative conformations of His76 in T42A (pale green) and in T42S (yellow) are indicated.
Conformational Change for Product Release
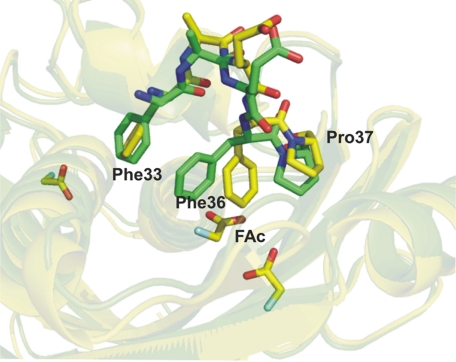
The positions of the main chains are almost completely conserved in all FlK structures with or without mutations and/or bound ligands. Local conformational changes, especially in the hydrophobic loop 33FAEFP37, are observed. The side chain conformations of Phe33, Phe36, and Pro37 in the structures with bound Ac or FAc differ from those of unbound WtFlK and the structures in complex with AcCoA, FAcCPan, or FAcOPan (Fig. 7). In the unbound WtFlK, the planes of the phenyl rings of Phe33 and Phe36 are nearly perpendicular to each other resembling a closed two-door gate blocking the “exit” surrounded by the major helix (the “sausage”), the β-sheet (the “bun”), and the 33FAEFP37 loop. Binding of the substrate analogues, AcCoA, FAcCPan, or FAcOPan, does not lead to any significant change in the positions of the “doors.” When the active site is occupied by the product FAc or its analogue Ac, the “gate” in one of the protomers is wide open mainly due to a dramatic swing and flip of the side chain of Phe36, and the gate in the other protomer of the same dimer is also open although to a lesser extent. It is tempting to speculate that this opened gate is the exit for the product FAc. The negatively charged carboxyl group of Glu50, which has been “freed” from the hydrogen bond with the Arg120 side chain due to substrate binding, is located near the gate and may facilitate expulsion of the negatively charged FAc from the active site.
FIGURE 7.
Representation of the hydrophobic loop 33FAEFP37 present in the active site of FlK. Shown are the differences in the side chain for residues Phe33, Phe36, and Pro37 for WtFlK·FAc (yellow) and apo-WtFlK (green).
DISCUSSION
FlK, a thioesterase from fluoroacetate producing S. cattleya, hydrolyzes the thioester bond of FAcCoA but not that of AcCoA. The selectivity of FlK to distinguish FAcCoA from AcCoA is remarkable because it ensures that AcCoA, a key intermediate of the primary metabolism is not cleaved by mistake. The only difference between the two compounds is the presence of a fluorine atom in FAcCoA but not in AcCoA. A combination of x-ray crystallography with site-directed mutagenesis experiments allowed us to gain insights into both catalytic mechanism and the unique substrate specificity of FlK.
Our investigations suggest that FlK utilizes a catalytic triad composed of a threonine (Thr42), a histidine (His76), and a water molecule. The imidazole side chain of His76 acts as a base to deprotonate the hydroxyl of Thr42. The nucleophile Thr42-Oγ attacks the fluoroacetyl carbonyl carbon, forming a tetrahedral transition intermediate. The positive charge generated on the His76 residue is stabilized by the hydrogen bonding between the imidazole ring and the conserved water molecule. Reformation of the carbonyl double bond breaks the carbon-sulfur bond. A second tetrahedral intermediate is then formed between the -OH part of a water molecule and the carbonyl group of the fluoroacetyl-FlK intermediate. The remaining proton of the water is bonded to His76. When the carbonyl bond reforms, the bond to the Thr42-Oγ is broken, releasing the fluoroacetate. The His76-bonded hydrogen is transferred to Thr42-Oγ, reestablishing the hydrogen bonding between Thr42 and His76. The Glu50, which is conserved in most type II thioesterases as the catalytic residue, is not a part of the catalytic mechanism of FlK but plays an important role in maintaining the configuration of the active site.
It is not unexpected that T42SFlK is active because the Ser42-Oγ can still form a hydrogen bond with the imidazole side chain of His76 and therefore has the potential to act as a nucleophile as the Thr42-Oγ in WtFlK. When FAcCoA concentration was higher than 12 μm, however, excess-substrate inhibition (38) was observed for the mutant. In other words, high concentrations of FAcCoA inhibited the activity of T42SFlK. We speculate that the absence of the Thr42-Cγ in T42SFlK may have created more space, allowing an easier access of substrate to the active site, especially at low substrate concentrations, giving rise to the lower Km value and the higher turnover of T42SFlK reaction as compared with WtFlK activity. However, the same mutation also increased protein flexibility, as shown by the double confirmations of the active site residues in some of the protomers of the mutant (Fig. 6). The enzyme-substrate complex formed between these abnormal active sites and substrate could be non-productive. At high substrate concentrations, the chance of non-productive enzyme-substrate complex formation increases when more and more normal active sites are occupied by FAcCoA. The formation of non-productive enzyme-substrate complex at the active site of one protomer may also interfere with the catalytic activity of the partner protomer of the same dimer. These may explain the excess-substrate inhibition phenomenon observed for T42SFlK and E50AFlK, but the exact mechanism remains to be elucidated.
FlK is a homodimer with a hot dog fold similar to other type II thioesterases, although it does not show extensive sequence similarity to any type II thioesterases with known function. A common feature shared by nearly all functionally characterized hot dog fold thioesterases is the presence of an acidic residue, aspartate or glutamate, providing general base catalysis. One exception is FcoT, a long-chain fatty acyl-CoA thioesterase from Mycobacterium tuberculosis, which has a hot dog fold but uses an active site completely different from that of either type I or type II thioesterases. FcoT has been proposed to represent a new class of thioesterases, type III thioesterases (39). With a type II thioesterase fold, the catalytic machinery of FlK (Thr-His-water) resembles those of type I thioesterases (Ser/Cys-base-acid), exhibiting an unconventional combination of protein fold and catalytic mechanism. Superposition of FlK with 18 structures of type II thioesterases from the Protein Data Bank reveals that the Glu50 in FlK is well conserved in most of these thioesterases, although in some cases, it is an aspartate instead of a glutamate (data not shown).
Among all of the hot dog fold proteins in the Protein Data Bank, only two hypothetical proteins, TTHA0967 (Protein Data Bank entry 2CWZ) from Thermus thermophilus and TM0581 (Protein Data Bank entry 2Q78) from Thermotoga maritime, have shown head-to-tail sequence similarity to FlK (33 and 22% identity, respectively). Superposition of FlK with these two structures has revealed a very similar putative active site in 2CWZ and 2Q78. Most of the residues lining the active site of FlK, Thr42, Glu50, Gly69, and His76, are well conserved in 2CWZ (Thr36, Glu44, Gly63, and His76) and in 2Q78 (Thr38, His72, and Arg116) with the following differences. First, the counterpart of the Arg120 of FlK in 2CWZ is a glutamine (Gln115). The Arg120 in the unliganded FlK interacts through its guanidinium group with the carboxyl side chain of Glu50. The amide side chain of Gln115 in CWZ2, however, is pointing in the opposite direction with no possibility of forming any direct contact with Glu44. A large empty space can be observed in the putative active site of 2CWZ due to this change of Arg to Gln. If 2CWZ is also an acyl-CoA thioesterase, the substrate should have a larger acyl moiety than a fluoroacetyl group. Second, the equivalents of FlK Glu50 and Gly69 in 2Q78 are His46 and Val65, respectively. The imidazole side chain of the His46 and the isopropyl group of the Val65 at the active site provide a binding motif quite different from that in FlK.
The significant differences in the overall positions of FAcCPan and FAcOPan in the active site of FlK as compared with that of the acetyl-pantheteinyl segment of the T42SFlK-bound AcCoA (Fig. 3) suggest that binding of the nucleotide portion of the substrate on the enzyme surface dictates the depth to which the fluoroacetyl pantetheinyl arm extends into the active site cleft, ensuring the correct interaction between the arm with the amino acid residues in the vicinity of the active site. Although the sulfur atom of the thioester is considered a poor hydrogen bond acceptor, the transient dipole interaction between the sulfur and the main chain amides along the substrate binding tunnel may also contribute to positioning of the substrate. In the absence of the bulky nucleotide tail and the sulfur atom, FAcCPan moved further into the active site. FAcOPan is retained near the tunnel entrance because the oxygen atom replacing the sulfur atom has a stronger tendency to interact with the main chain amides of the substrate binding tunnel.
Although the 3′-phosphoryl ADP tail of FAcCoA is required for the overall positioning of FAcCoA in the FlK active site, the accurate orientation of the fluoroacetyl thioester moiety must be fine tuned by the hydrogen bonding of the fluorine with the side chain of Arg120 and the main chain amide of Gly69 in order to assume the required catalytic coordination. The guanidinium group of Arg120 and the main chain amide of Gly69 appear to be the key determinants for the unusual high substrate specificity of FlK, which serve to recognize the only structural difference between FAcCoA and AcCoA through their interactions with the fluorine of FAcCoA. Moreover, the thioester carbonyl carbon in FAcCoA is more positively charged than that in AcCoA due to the strong electronegativity of fluorine. The interactions between the fluorine and Arg120/Gly69 also help to stabilize this positive charge, making the carbonyl carbon a better target for nucleophilic attack by Thr42-Oγ (Fig. 5).
Acknowledgment
We thank Dr. David O'Hagan (University of St. Andrews) for helpful discussions.
- FAc
- fluoroacetate
- Ac
- acetate
- AcCoA
- acetyl-coenzyme A
- FAcCoA
- fluoroacetyl-coenzyme A
- FAcCPan
- fluoroacetyl carba(dethia)-pantetheine
- FAcOPan
- fluoroacetyl oxa(dethia)-pantetheine
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- WtFlK
- wild type FlK
- SeMet
- selenomethionine.
REFERENCES
- 1.Sanada M., Miyano T., Iwadare S., Williamson J. M., Arison B. H., Smith J. L., Douglas A. W., Liesch J. M., Inamine E. (1986) J. Antibiot. 39, 259–265 [DOI] [PubMed] [Google Scholar]
- 2.O'Hagan D., Schaffrath C., Cobb S. L., Hamilton J. T., Murphy C. D. (2002) Nature 416, 279. [DOI] [PubMed] [Google Scholar]
- 3.Schaffrath C., Deng H., O'Hagan D. (2003) FEBS Lett. 547, 111–114 [DOI] [PubMed] [Google Scholar]
- 4.Deng H., O'Hagan D., Schaffrath C. (2004) Nat. Prod. Rep. 21, 773–784 [DOI] [PubMed] [Google Scholar]
- 5.Dong C., Huang F., Deng H., Schaffrath C., Spencer J. B., O'Hagan D., Naismith J. H. (2004) Nature 427, 561–565 [DOI] [PubMed] [Google Scholar]
- 6.Zhu X., Robinson D. A., McEwan A. R., O'Hagan D., Naismith J. H. (2007) J. Am. Chem. Soc. 129, 13597–14604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang F., Haydock S. F., Spiteller D., Mironenko T., Li T. L., O'Hagan D., Leadlay P. F., Spencer J. B. (2006) Chem. Biol. 13, 475–484 [DOI] [PubMed] [Google Scholar]
- 8.Peters R., Wakelin R. W. (1953) Proc. R. Soc. Lond. B Biol. Sci. 140, 497–506 [DOI] [PubMed] [Google Scholar]
- 9.Lauble H., Kennedy M. C., Emptage M. H., Beinert H., Stout C. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13699–13703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J., Sussman J. L., Verschueren K. H. G., Goldman A. (1992) Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 11.Pleiss J., Fischer M., Schmid R. D. (1998) Chem. Phys. Lipids 93, 67–80 [DOI] [PubMed] [Google Scholar]
- 12.Leesong M., Henderson B. S., Gillig J. R., Schwab J. M., Smith J. L. (1996) Structure 4, 253–264 [DOI] [PubMed] [Google Scholar]
- 13.Li J., Derewenda U., Dauter Z., Smith S., Derewenda Z. S. (2000) Nat. Struct. Biol. 7, 555–559 [DOI] [PubMed] [Google Scholar]
- 14.Dillon S. C., Bateman A. (2004) BMC Bioinformatics 5, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pidugu L. S., Maity K., Ramaswamy K., Surolia N., Suguna K. (2009) BMC Struct. Biol. 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. (1995) Science 268, 579–582 [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18.French G. S., Wilson K. S. (1978) Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 19.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 20.Leslie A. G. (2006) Acta Crystallogr. D 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 21.Evans P. (2006) Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 22.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. (2001) Acta Crystallogr. D 57, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 28.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 29.Benning M. M., Wesenberg G., Liu R., Taylor K. L., Dunaway-Mariano D., Holden H. M. (1998) J. Biol. Chem. 273, 33572–33579 [DOI] [PubMed] [Google Scholar]
- 30.Thoden J. B., Zhuang Z., Dunaway-Mariano D., Holden H. M. (2003) J. Biol. Chem. 278, 43709–43716 [DOI] [PubMed] [Google Scholar]
- 31.Willis M. A., Zhuang Z., Song F., Howard A., Dunaway-Mariano D., Herzberg O. (2008) Biochemistry 47, 2797–2805 [DOI] [PubMed] [Google Scholar]
- 32.Hisano T., Tsuge T., Fukui T., Iwata T., Miki K., Doi Y. (2003) J. Biol. Chem. 278, 617–624 [DOI] [PubMed] [Google Scholar]
- 33.Yokoyama T., Choi K. J., Bosch A. M., Yeo H. J. (2009) Biochim. Biophys. Acta 1794, 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kunishima N., Asada Y., Sugahara M., Ishijima J., Nodake Y., Sugahara M., Miyano M., Kuramitsu S., Yokoyama S., Sugahara M. (2005) J. Mol. Biol. 352, 212–228 [DOI] [PubMed] [Google Scholar]
- 35.Song F., Zhuang Z., Finci L., Dunaway-Mariano D., Kniewel R., Buglino J. A., Solorzano V., Wu J., Lima C. D. (2006) J. Biol. Chem. 281, 11028–11038 [DOI] [PubMed] [Google Scholar]
- 36.Allaire M., Chernaia M. M., Malcolm B. A., James M. N. (1994) Nature 369, 72–76 [DOI] [PubMed] [Google Scholar]
- 37.Dodson G., Wlodawer A. (1998) Trends Biochem. Sci. 23, 347–352 [DOI] [PubMed] [Google Scholar]
- 38.Kühl P. W. (1994) Biochem. J. 298, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Langley R., Gulten G., Wang L., Sacchettini J. C. (2007) Chem. Biol. 14, 543–551 [DOI] [PubMed] [Google Scholar]