Abstract
Mixed lineage kinases (MLKs) have been implicated in cytokine signaling as well as in cell death pathways. Our studies show that MLK3 is activated in leukocyte-infiltrated islets of non-obese diabetic mice and that MLK3 activation compromises mitochondrial integrity and induces apoptosis of beta cells. Using an ex vivo model of islet-splenocyte co-culture, we show that MLK3 mediates its effects via the pseudokinase TRB3, a mammalian homolog of Drosophila Tribbles. TRB3 expression strongly coincided with conformational change and mitochondrial translocation of BAX. Mechanistically, MLK3 directly interacted with and stabilized TRB3, resulting in inhibition of Akt, a strong suppressor of BAX translocation and mitochondrial membrane permeabilization. Accordingly, attenuation of MLK3 or TRB3 expression each prevented cytokine-induced BAX conformational change and attenuated the progression to apoptosis. We conclude that MLKs compromise mitochondrial integrity and suppress cellular survival mechanisms via TRB3-dependent inhibition of Akt.
Keywords: Cell Death, Cytokine Action, Diabetes, Mitochondria, Pancreatic Islet, Akt, BAX, MLK3, TRB3, TRIB3
Introduction
In type 1 diabetes, the autoimmune destruction of pancreatic beta cells is driven by leukocyte infiltration and the damaging effects of locally secreted cytokines. Cytokines activate MAPKs3 JNK and p38, via signaling modules that involve the sequential activation of a MAP3K, MAP2K, and MAPK, all scaffolded by a single protein (1). The existence of several families of MAP3Ks raises the possibility that each MAP3K may be activated by specific classes of stimuli. The serine-threonine MAP3K mixed lineage kinase-3 (MLK3) is activated by cytokines (2, 3) and assembles a signaling module consisting of MKK7, JNK, and the scaffold protein JIP1 (4, 5). Fibroblasts with a targeted deletion of either MKK7 or MLK3 are attenuated in their response to cytokines (6, 7). Elevation of MLK3 has been linked to induction of apoptosis in neurons (8–10), and inhibition of MLKs can delay progression of neurodegenerative diseases (reviewed in Ref. 11 and studies quoted therein). The striking parallels between the beta cell and neuronal phenotypes, coupled with the ability of cytokines to activate MLK3, prompted us to examine whether MLKs participate in cytokine-induced beta cell death.
Here we show that MLK3 is markedly elevated in leukocyte-infiltrated islets of the non-obese type 1 diabetic (NOD) mouse. To investigate the potential role of MLK3 in beta cell death, we devised an ex vivo system for co-culture of primary islets with immune-activated splenocytes. Compared with static culture with purified cytokines, this system is likely to be more representative of the milieu encountered by islets in autoimmune diabetes. We observed rapid activation of MLK3, and MLK3 was required for cytokine-mediated apoptosis via BAX, a proapoptotic member of the BCL-2 protein family. MLK3 mediated its effects via the pseudokinase TRB3 (TRIBBLES homolog 3), originally identified as an inducible factor in neuronal cell death (12) and subsequently shown to be a potent negative regulator of the prosurvival kinase Akt (13). We found that MLK3-mediated stabilization of TRB3 led to a conformational change of BAX and the ensuing mitochondrial outer membrane permeabilization (14–16). For the vast majority of cells, mitochondrial outer membrane permeabilization has been identified as the commitment point for cellular apoptosis (17, 18). In our studies, the strong correlation between the induction of MLK3 and TRB3, with BAX conformational change, suggests that MLK3 activation may play a central role in committing the beta cell to an apoptotic fate.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies used include mouse anti-porin (MitoSciences, Eugene, OR), β-tubulin, and HA (Covance, Berkeley, CA); rabbit anti-MLK3, pMLK3, BAX, cytochrome c, JNK, phospho-JNK, GST, AKT, phospho-AKT, Myc tag, FLAG tag, (Cell Signaling, Beverly, MA), MLK1, and MLK2 (Abgent, San Diego, CA) and PDX-1 (19) for Western blotting; and sheep anti-insulin (The Binding Site, San Diego, CA), mouse anti-BAX clone 6A7 (BD Biosciences), caspase-6 (20), TRB3 (Marc Montminy, Salk Institute), MLK3 (A. Rana, Loyola University Chicago), and pMLK3 (Cell Signaling, Beverly, CA) used for immunostaining after standardizing in cell lines (supplemental Fig. 7). Other antibodies used to screen the splenocyte-islet co-culture (SICC) arrays were caspase-3, -7, -8, and -9 (20); Smarc and Traf3 (21); p53 and Fas (22); and Bcl-XS, BAK, tBid, Bid and BAD (20). Mouse anti-HA-agarose beads (Sigma) and Myc-agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and glutathione-Sepharose beads (Amersham Biosciences) were used for immunoprecipitation and pull-down experiments. Fluorescence and Western blotting employed fluorescent or horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA), the latter detected using Supersignal chemiluminescence reagents (Pierce). Other reagents include the In Situ TUNEL kit (Roche Applied Science), MitoTraker and JC-1 dye (Invitrogen), CEP11004 (Cephalon Inc., Frazer, PA), Jnk-II inhibitor SP600125, cycloheximide (EMD Biosciences, San Diego, CA), and human IL-1β, human tumor necrosis factor-α, mouse interferon-γ, and mouse IL-2 (Peprotech, Rocky Hill, NJ).
Plasmids, Constructs, and siRNA
PEBG-MLK3 plasmids were a gift from Dr. Ajay Rana, and TRB3 deletion constructs have been described elsewhere (23). The MLK3 and GL3-luciferase shRNA sequences GGGCAGCGATGTCTGGAGC (24) and CTTACGCTGAGTACTTCGA were used to generate a double-stranded oligonucleotide that was subcloned downstream of the U6 promoter in Bluescript vector pBS-U6. pcDNA3-RFP-BAX was generated by amplifying BAX cDNA from CMVSport6 BAX (Open Biosystems, Huntsville, AL) and subcloned in frame with RFP into pcDNA3-dsRED (Invitrogen). Myc-Akt was generated by PCR-subcloning Akt2 (Addgene) into the EcoRI site of pcDNA3Myc (Stratagene) and sequence-verified. CMVSport6 expression vectors for ASK1 and TAK1 were from Open Biosystems, pcDNA3-MEKK1 was a gift from Dr. R. Davis (University of Massachusetts), and pcDNA3-TPL2, was a gift from P. Tsichlis (Tufts University, Boston, MA). For bacterial expression vectors, BamHI digests of full-length wild type and kinase-dead MLK3 from PEBG expression vectors were subcloned in frame into PGEX-4T1 vector.
Generation of Recombinant Akt Adenovirus
Myristolated AKT (T308D/S473D) and AKT-KD (K179M) were subcloned into pAdTrack-CMV, and adenoviruses were generated using the AdEasy system as described previously (25).
SICC and SICC Cell Array
Diabetic NOD spleens were crushed and passed through a 70-μm mesh. Red blood cells were lysed in 0.15 m NH4Cl, and 1.5 × 106 splenocytes/well were plated in 24-well dishes in RPMI supplemented with 60 units/ml mouse IL-2 and 10% heat-inactivated FBS. Splenocytes were stimulated with plate-bound anti-CD3 and exogenous anti-CD28 antibodies (10 and 1 μg/ml, respectively; BD Biosciences) for 3 days as described previously (26). Islets were isolated 2 days later, rested overnight, and cultured with transwell filters in the presence of unstimulated or stimulated splenocytes, with or without 500 nm CEP11004 pretreatment and collected over a time course of 24 h. At each time point, islets were fixed, spun into pellets, and embedded in agarose plugs. Individual plugs from an entire time course were laid out on a grid and processed for paraffin embedding in a single block as described (27).
Tissue Preparation, Immunohistochemistry, and Immunofluorescence
Isolated islets and pancreata were fixed in Bouins fixative for 30 min and 4 h, respectively, washed in phosphate-buffered saline, postfixed in Z-Fix (Anatech, Battle Hill, MI), processed for paraffin embedding, and immununostained as described previously (20, 27, 28). For staining, slides were pretreated with targeted retrieval solution, high pH (DAKO, Carpinteria, CA), as per the manufacturer's instructions or in 110 mm citrate buffer, pH 6.0, and primary antibodies were revealed using a diaminobenzidine-based detection method employing either an avidin-biotin complex reagent (Vector Laboratories, Burlingame, CA) or the Envision plus horseradish peroxidase system (DAKO). For immunofluorescence, Min6 cells were fixed in 4% paraformaldehye for 5 min and permeabilized with 0.1% Triton X-100, and primary antibodies were visualized with species-specific secondary antibodies conjugated to fluorescent probes.
Human Adult Islets
Human adult islets were provided by the Islet Cell Resource Center Basic Science Human Islet Distribution Program and the Islet Transplant Program (University of Illinois, Chicago, IL). Islets were hand-picked and cultured overnight in CRML supplemented with 10% FBS. Prior to stimulation with cytokines, islets were cultured for at least 8 h in RPMI 1640 supplemented with 10% FBS.
Mouse Islet Isolation
Islets were isolated using a Ficoll gradient as described (29).
TUNEL Assay
Dewaxed sections of the SICC array were pretreated with 10 μg/ml Proteinase K for 30 min at 37 °C, whereas Min6 cells were fixed in 2% paraformaldehyde prior to TUNEL labeling as per the manufacturer's instructions (Roche Applied Science).
Mitochondrial Potential
For confocal microscopy, cells were incubated with JC-1 (2.5 μg/ml) for 15 min at 37 °C following the indicated treatments and examined immediately. For quantification, cells were processed using the Guava MitoPotential kit. Cell populations were gated based on JC-1 fluorescence and quantified using the Guava EasyCyte flow cytometer (Guava Technologies, Hayward, CA).
Mitochondrial Fractionation
Using a modified method (30), cells were harvested and allowed to swell for 30 min in hypotonic buffer (10 mm Hepes, 5 mm MgCl2, 40 mm KCl, 10 μg/ml aprotinin, and leupeptin). After 30 passes through a 25-gauge needle, sucrose was added to a final concentration of 250 mm, along with sodium orthovanadate (0.5 mm) and phenylmethylsulfonyl fluoride (1 mm), and nuclei and cell debris were removed by centrifugation at 800 × g for 10 min at 4 °C. The supernatant was centrifuged at 10,000 × g for 20 min at 4 °C to separate mitochondrial and cytosolic fractions, and 15 μg of protein was used for SDS-PAGE.
Cell Culture, Transfection, Infection, and FACS Sorting
Min6 cells (passages 15–18 only) were grown in Dulbecco's modified Eagle's medium containing 25 mm glucose supplemented with 4% heat-inactivated FBS and 50 μm β-mercaptoethanol. HEPG2 cells were grown in a 1:1 mix of Dulbecco's modified Eagle's medium and F12K and 5% heat-inactivated FBS. COS-7 cells were grown in Dulbecco's modified Eagle's medium and 10% heat inactivated FBS. Transfections for GST pull-down assays, immunoprecipitation, and BAX-RFP translocation in Min6, HEPG2, and COS-7 cells were performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions and were treated as described 48 h post-transfection. Min6 cells were infected with Ad Akt-myristoylated (Myr) or Ad Akt-kinase-dead (KD) (multiplicity of infection = 25) for 6 h in growth medium and 40 h postinfection were treated as described. For endogenous MLK3 knockdown, due to the low transfection efficiency of beta cell lines, beta TC-3 cells were nucleofected with U6 shRNA constructs and pcDNA3-RFP reporter construct, used as a co-transfection marker. Cell sorting (FACS-DiVa; BD Biosciences) gated for RFP expression was followed by Western blotting analysis of MLK3 protein.
Nucleofection of shRNA Constructs and Transfection of siRNA
For shRNA rescue experiments, 1 × 106 Min6 cells were electroporated using Nucleofector kit V (Amaxa GmbH, Cologne, Germany) with 2 μg of plasmid DNA encoding shRNA or MLK3-KD, and 0.4 μg of pcDNA3-RFP. The indicated treatments were performed 40–48 h postnucleofection.
BAX-RFP Translocation
COS-7 cells transfected with pcDNA3-RFP, pcDNA3-BAX-RFP, pcDNA3, and pcDNA3-HA-TRB3, as indicated, were incubated with Mitotracker dye (200 nm) for 30 min at 37 °C, fixed, and analyzed immediately.
Cycloheximide Chase, Western Blotting, and Insulin Stimulation
These experiments were performed as described (29).
GST Pull-down Assay and Immunoprecipitation
Mammalian expression vectors encoding GST or HA- or Myc-tagged fusion proteins were expressed in Min6 or HepG2 cells followed by GST pull-downs with glutathione-Sepharose (Amersham Biosciences) or immunoprecipitations as described (29).
In Vitro Protein Interactions
Bacterial MLK3 proteins were expressed using BL21-RIL cells (Stratagene, La Jolla CA) and purified with glutathione-Sepharose beads (Amersham Biosciences) according to the manufacturer's instructions. TRB3 protein was generated by the TNT-coupled reticulocyte lysate system (Promega Corp., Madison, WI) using T7 polymerase and pcDNA3-HA-TRB3 as template with a full complement of amino acids. Glutathione-Sepharose-bound GST-MLK3 wild type (WT) or KD proteins were incubated with equal amounts of in vitro synthesized TRB3 protein in NETN buffer and a full complement of protease and phosphatase inhibitors. The GST pull-downs were washed using NETN and subjected to SDS-PAGE, and the top and bottom halves of the transferred blots were probed using pMLK3 (125 kDa) and TRB3 (48 kDa) antibodies, respectively. MLK3 input was detected using Gel-Code Blue stain reagent (Thermo-Pierce).
Microscopy and Image Acquisition
Fluorescent and bright field images were acquired on either an inverted Olympus IX81 microscope attached to the Radiance 2100MP laser-scanning system (Bio-Rad) or a ScanScope CS digital slide scanner (Aperio Technologies, Vista, CA), respectively. All images were assembled in Photoshop 10 (Adobe Systems Inc., San Jose, CA).
Statistics
Quantification was performed on 10 randomly selected fields of view using the ×40 or ×60 objective from three separate experiments. Total cell counts were obtained using propidium iodide (0.05 μg/ml), 4′,6-diamidino-2-phenylindole (1 μg/ml), RFP, GFP, or insulin for cell specificity in SICC arrays. Quantification of caspase-6 was performed on images deconvolved using ImageScope software (Aperio Technologies, Vista, CA), and the caspase-6-positive pixels were expressed as a percentage of total pixels. Differences between means were examined using analysis of variance (ANOVA) followed by a Bonferroni post hoc comparison. In all cases, p values less than or equal to 0.05 were considered significant. Analysis was performed using StatView 5.0 statistical software.
RESULTS
Cytokines Induce Expression of MLK3 in the Pancreatic Beta Cell
NOD mice spontaneously develop autoimmune diabetes around 15–18 weeks of age. Pancreas sections from 6- or 18-week-old NOD female mice with respective ambient glucose levels of 110 and 280 mg/dl were stained for MLK3 and its active form (pMLK3). Both total and phosphorylated MLK3 were strongly induced in leukocyte-infiltrated islets (Fig. 1A) but not detected in uninfiltrated islets from the same mouse or in islets of prediabetic NOD mice. These data suggested a correlation between local secretion of cytokines and induction and activation of MLK3. A widely used mixture of cytokines for study of beta cell death includes the three components IL-1β, tumor necrosis factor-α, and interferon-γ (31). Using each cytokine to stimulate Min6 cells, we found that in the beta cell, IL-1β rapidly induced total MLK3 protein levels within 20–30 min (Fig. 1B). Similarly, in human islets, IL-1β acutely induced the MLK3 isoform of the mixed lineage kinases by more than 4-fold. A smaller effect on MLK1 and -2 (supplemental Fig. 1) was observed in human islets. Of note, the rapid up-regulation of MLK3 protein was not due to increased transcription because no increase in MLK3 mRNA was observed (supplemental Fig. 2). Our data mirrored a key finding in a previous report where apoptogenic stimuli were shown to rapidly up-regulate MLK3 protein (32) by increasing protein stability.
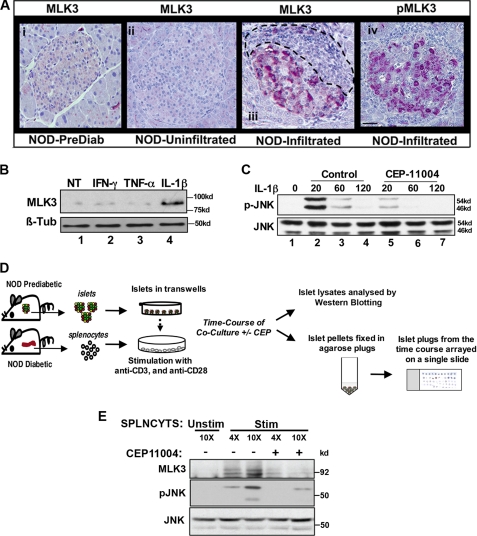
FIGURE 1.
MLK3 is activated in leukocyte-infiltrated islets of NOD mice, by IL-1β in Min6 cells, and in SICC. A, pancreatic sections from prediabetic (panel i) and diabetic NOD mice (panels ii–iv) were stained for MLK3 (panels i–iii) and phospho-MLK3 (panel iv) as described. Leukocyte infiltration is outlined. Bar, 20 μm. B, Western blotting of extracts (60 μg of protein) from Min6 cells treated for 20 min with 10 nm tumor necrosis factor-α, 40 ng/ml interferon-γ, or 20 ng/ml IL-1β using anti-MLK3 antibodies and β-tubulin used as a control. C, Western blots for time-dependent JNK activation from Min6 cells treated with IL-1β (20 ng/ml) in the absence (lanes 2–4) or presence of MLK3 inhibitor CEP11004 (lanes 5–7). D, schematic of SICC. E, Western blots for total MLK3 and phospho-JNK (pJNK) from of SICC islets co-cultured with increasing numbers of splenocytes (4- and 10-fold for 30 min) in the absence (lanes 1–3) or presence of CEP11004 (lanes 4 and 5). Total JNK levels serve as control. NT, not treated.
Because MLK3 is a MAP3K, which preferentially activates JNK (33), we examined whether IL-1β induced JNK activity in an MLK3-dependent manner. Min6 insulinoma cells were treated with IL-1β, in the presence and absence of 500 nm CEP11004, a potent inhibitor of the mixed lineage kinase family (34), whose specificity was first ascertained as shown (supplemental Fig. 3). IL-1β rapidly induced JNK activation in the beta cell, and this effect was strongly attenuated by pretreatment with CEP11004 (Fig. 1C).
To demonstrate cytokine-mediated induction of MLK3 in a physiologically relevant system, we devised an ex vivo SICC system, which would mimic the islet response to leukocyte invasion in insulitis. Unlike the events in vivo, this system (depicted by a schematic in Fig. 1D) permits the study of the earliest molecular events in cytokine signaling in a relatively controlled environment while maintaining the complex cytokine environment of insulitis. Splenocytes from diabetic NOD mice were isolated and stimulated with anti-CD3 and anti-CD28 antibodies to trigger production of the full repertoire of cytokines (supplemental Table 1). During co-culture, islets were suspended in transwells such that they were immersed in stimulated medium but had no direct contact with splenocytes. To study the effect of MLK3, islets were co-cultured with or without pretreatment with CEP11004. Again MLK3 was rapidly (30 min) activated by cytokines, and Western blotting of islet extracts from SICC (Fig. 1E) showed that increasing numbers of splenocytes induced MLK3 and activated JNK in a dose-dependent manner. MLK3 and JNK were both inhibited using CEP11004, strongly suggesting that, similar to IL-β, conditions that mimic insulitis also activate MLK3 and its downstream kinase JNK.
MLKs Mediate Cytokine-dependent Beta Cell Death
To examine whether MLKs were required for inducing autoimmune destruction of beta cells, we employed the SICC system described above. A time course of SICC islets from all experimental groups was arrayed on a single slide to enable a clear comparison across various samples. Similar to data from Western blotting, (Fig. 1E), MLK3 induction was also detected by immunostaining, and pretreatment with CEP11004 markedly inhibited this induction (Fig. 2A, column 1, rows ii and iii). Within 24 h of co-culture, 15% of the insulin-expressing cells in the islet were positive for TUNEL (column 2, row ii), which detects DNA strand breaks in end stage cell death. Pretreatment with CEP11004 effectively reduced TUNEL staining to unstimulated levels (column 2, row iii). CEP11004 treatment similarly inhibited the cleavage and activation of caspase-6 (supplemental Fig. 4). Quantification of TUNEL is shown in a graph (Fig. 2B).
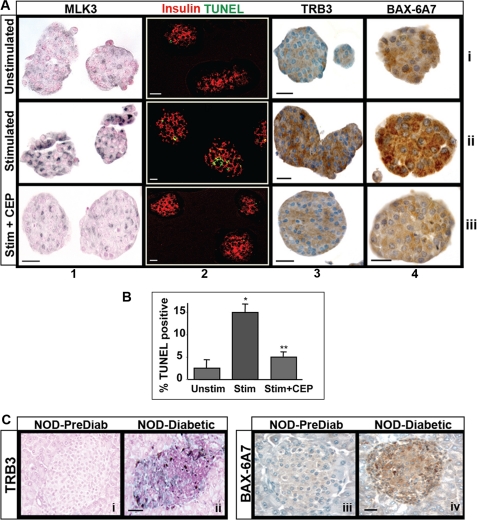
FIGURE 2.
MLK-dependent induction of proapoptotic markers in SICC. A, SICC for 24 h with unstimulated (row i) and stimulated splenocytes pretreated with DMSO (row ii) or with CEP11004 (row iii) were used to detect MLK3, TUNEL, TRB3, and conformationally altered BAX using monoclonal antibody clone 6A7 as described (columns 1–4, respectively). B, quantification of TUNEL (*, p < 0.001 versus unstimulated media; **, p < 0.001 versus stimulated medium as tested by ANOVA followed by Bonferroni post hoc test) represents means ± S.E. (error bars) of three independent experiments. C, pancreatic sections from prediabetic and diabetic NOD mice were immunostained for TRB3 (panels i and ii) or BAX-6A7 (panels iii and iv). Bar, 20 μm.
TRB3 and BAX Are Downstream Effectors of MLK3
The arrayed time course of SICC islets was screened for shifts in expression of several apoptosis markers (see “Experimental Procedures”). Distinct MLK-dependent induction was observed for only two markers. The first was the pseudokinase TRB3 (Fig. 2A, column 3), which has been implicated in stress-induced apoptosis (12, 35, 36). The second marker was BAX (Fig. 2A, column 4), a proapoptotic member of the BCL-2 family of proteins, known to be critically required for pancreatic beta cell apoptosis (37). Apoptotic events have been shown to trigger a conformational change in BAX, which marks a critical transition toward its insertion in the mitochondrial membrane to initiate apoptosis. This structural change exposes a stretch of seven amino acids, normally buried in a hydrophobic pocket under quiescent conditions (38). Using a monoclonal antibody (6A7) that specifically detects this exposed region, immunostaining of SICC islets (Fig. 2B, column 4) revealed intense punctate staining of BAX (row ii), and both TRB3 and BAX-6A7 induction were attenuated by pretreatment with the MLK inhibitor CEP11004 (row iii). Similar induction of TRB3 (Fig. 2C, panels i and ii) and BAX conformational change (panels iii and iv) were also detected in leukocyte-infiltrated islets of NOD mice.
TRB3 Induction Correlates with BAX Translocation and a Compromise in Mitochondrial Function
To understand the mechanistic connection between MLKs, TRB3, and BAX, we used activated splenocyte-derived conditioned or “stimulated” medium to treat Min6 mouse insulinoma cells, followed by immunofluorescent staining at 8 h (Fig. 3A). For clarity, an outline of the stained cells is presented below each image (bottom). As seen in Fig. 3A (top), TRB3 was induced in 20% of Min6 cells treated with stimulated medium (Fig. 3A, top, column 2). Remarkably, co-staining with 6A7 antibody detected conformationally altered BAX in most TRB3-positive cells (column 3), with little signal in TRB3-negative cells (see bottom panel), suggesting a close relationship between TRB3 induction and BAX conformational change. Indeed, overexpression of TRB3 was sufficient to induce mitochondrial translocation of coexpressed BAX-RFP fusion protein in cultured COS-7 cells. As seen in Fig. 3B, unlike the diffused localization of BAX-RFP in the presence of CMV control vector, the presence of TRB3 resulted in a strong overlap of BAX-RFP signal with the green Mitotracker dye. We next examined whether TRB3 protein induction was MLK-dependent. Using Western blotting, we found that stimulated medium (Fig. 3C) and IL-1β (Fig. 3D) both strongly induced TRB3 protein, and pretreatment with MLK inhibitor CEP11004 completely abrogated this effect. TRB3 protein induction was not accompanied by a change in TRB3 mRNA (supplemental Fig. 2), suggesting a post-transcriptional mechanism for TRB3 regulation.
FIGURE 3.
Cytokines up-regulate TRB3, compromise mitochondrial function, and induce apoptosis in an MLK-dependent manner. A, colocalization of endogenous TRB3 with conformationally altered BAX (row i) in Min6 cells treated for 8 h with unstimulated (column 1) or stimulated (columns 2–4) medium. The arrowheads show colocalization of TRB3 (green) and BAX-6A7 (red). Bar, 10 μm. Schematic representation of TRB3 and BAX-6A7 colocalization is shown in row ii. B, confocal microscopy of COS-7 cells transfected with RFP or BAX-RFP fusion constructs in the presence of CMV control or CMV HA-TRB3. Mitochondria were tracked with deep red Mitotracker dye (pseudocolored green). Bar, 1 μm. C and D, Western blotting of 60 μg of protein from Min6 cells treated with stimulated medium (4 h) from SICC (C) or 20 ng/ml IL-1β (D) in the absence (B, lane 2; C, lanes 2–4) or presence of CEP11004 (B, lane 3; C, lanes 5–7). E, Western blotting (15 μg of protein) of cytoplasmic (Cyto) and mitochondrial (Mito) fractions for BAX or cytochrome c, from Min6 cells treated for 8 h with unstimulated (−) or stimulated (+) medium, with or without CEP11004. Tubulin and porin served as controls for cytoplasmic and mitochondrial fractions, respectively. F, J-aggregates in Min6 cells incubated for 16 h with unstimulated (panel i) or stimulated medium, with DMSO (panel ii) or CEP11004 (panel iii), followed by JC-1 dye treatment for 15 min. Bar, 2 μm. Change in mitochondrial potential was assessed by FACS analysis of J-aggregates (panel iv). Shown is TUNEL detection (panels v and vi; bar, 40 μm) and quantification (panel viii) of apoptotic Min6 cells treated as described for panels i–iii (*, p < 0.05 versus unstimulated media; **, p < 0.05 versus stimulated medium, as tested by ANOVA followed by Bonferroni post hoc test). Error bars, means +/− S.D. NT, not treated.
To test whether the observed conformational change in BAX led to membrane insertion and mitochondrial outer membrane permeabilization, we examined levels of BAX in subcellular fractions from Min6 cells treated with stimulated medium for 8 h. Western blots showed an increase in mitochondrial BAX and a corresponding reciprocal drop in cytoplasmic BAX (Fig. 3D) as early as 4 h and up to 12 h (not shown). In parallel, an inverse pattern of cytochrome c localization was observed, which indicated a breach in mitochondrial membrane integrity (39). The presence of CEP11004 inhibited both BAX translocation and cytochrome c leaching, linking both of these events to activation of MLKs. In the beta cell, mitochondria are central for glucose sensing and coupled insulin secretion. Because actively respiring cells have over 1000 mitochondria/cell, we examined whether the detected mitochondrial breach was sufficient to impact cellular respiration. In intact, actively respiring mitochondria, JC-1 dye forms j-aggregates that fluoresce red. Both confocal analysis and flow cytometry showed that an 8–16-h incubation of Min6 cells with cytokine-rich stimulated medium resulted in a ∼30% drop in red fluorescence, (Fig. 3E, panels ii and iv), which was almost completely reversed in the presence of CEP11004 (panels iii and iv).
In Min6 cells, CEP11004 treatment also efficiently inhibited stimulated medium-mediated activation of caspase-6 (supplemental Fig. 4) and end stage apoptosis measured by TUNEL (Fig. 3E, panels v–viii). Similar to data for primary islets, apoptosis induced by conditioned medium in Min6 cells was also inhibited by CEP11004 pretreatment. Overall, a contemporaneous induction and colocalization of endogenous TRB3 protein with BAX translocation suggested a tight functional correlation between the two proteins, thus linking MLK3 and TRB3 to the mitochondrial death machinery via BAX.
MLK3 and TRB3 Knockdown Inhibit BAX Conformational Change and Beta Cell Death
Next, we used shRNA directed against MLK3 (shRNA validation shown in supplemental Fig. 5) and TRB3 (23) as an alternate means of inhibiting stimulated medium-induced apoptosis. Due to inefficient DNA uptake by Min6 cells, shRNA plasmids were cotransfected/nucleofected (Amaxa Biosystems) with plasmid expressing CMV-red florescence protein (pseudocolored blue) to accurately track cells that took up shRNA (Fig. 4A, column 1). Cells treated for 4–8 h with unstimulated (row i) and stimulated medium (rows ii–iv) were immunostained using TRB3 (column 2; detected with Alexa-fluor 488, green) and conformation-specific anti-Bax-6A7 (column 3; detected with Alexa-fluor 647, pseudocolored red) antibodies. For clarity, a schematic diagram of shRNA-expressing cells in blue and the TRB3 and BAX-6A7 double positive cells in yellow is shown in column 5. Induction of endogenous TRB3 protein again strongly coincided with BAX conformational change (denoted by arrowheads and in yellow in column 5). Expression of control shRNA (U6-GL3; rows i and ii) did not inhibit induction of 6A7-BAX and TRB3 (marked by an arrow; row ii, column 4). Accordingly, as represented in column 5, TRB3 and BAX6A7 double positive cells (yellow) showed a clear overlap with control plasmid expressing cells in blue. In contrast, both MLK3 (U6-MLK3, row iii) and TRB3 shRNA (U6-TRB3, row iv) attenuated induction of TRB3 and BAX conformational change, as evidenced by the exclusion of BAX-6A7 and TRB3 from the shRNA-transfected, pseudocolored blue cells (columns 4 and 5, rows iii and iv). The percentage of transfected (RFP+, pseudocolored blue) cells positive for co-expression of BAX-6A7 and TRB3 is shown in graph format in Fig. 4C.
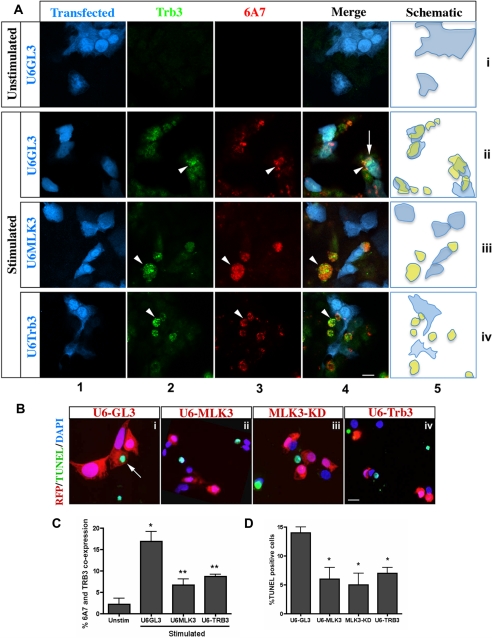
FIGURE 4.
Cytokine-induced BAX conformational change, and apoptosis requires MLK3 and TRB3. A, Min6 cells were cotransfected with shRNA constructs U6-GL3 (row ii), U6-MLK3 (row iii), or U6-TRB3 (row iv), with RFP (pseudocolored blue, column 1) to track transfected cells. Induction of endogenous TRB3 (green, column 2) and its colocalization with BAX-6A7 (column 3, pseudocolored red) was detected following an 8-h treatment with unstimulated (row i) or stimulated (rows ii–iv) medium. Column 4 shows a merge of RFP (pseudocolored blue) marker to track shRNA uptake, BAX-6A7, and TRB3 signals. Arrowheads, colocalization of TRB3 and BAX-6A7 in the absence of shRNA; arrows, TRB3 and BAX-6A7 in the presence of shRNA control U6-GL3. Bar, 10 μm. A schematic representation of the overlapping expression of shRNA (blue) and TRB3/BAX-6A7 double positive cells (yellow) is shown in column 5. B, TUNEL analysis of Min6 cells co-transfected with RFP reporter and either U6-GL3 (panel i), U6-MLK3 (panel ii), MLK3-KD (panel iii), or U6-TRB3 (panel iv) were treated with stimulated medium for 24 h. Arrows, TUNEL, shRNA/RFP double positive. Bar, 1 μm. C, quantification (means ± S.D. (error bars) of three independent experiments) of shRNA-positive cells co-expressing BAX-6A7 and TRB3 (*, p < 0.001 versus unstimulated media; **, p < 0.001 versus U6-GL3, as tested by ANOVA followed by Bonferroni post hoc test). D, TUNEL quantification of RFP positive cells (mean ± S.D. of three experiments; *, p < 0.001 versus U6-GL3, as tested by ANOVA followed by Bonferroni post hoc test).
Using the same approach as in Fig. 4A, we examined whether knockdown of MLK3 and TRB3 also attenuated end stage apoptosis. Min6 cells were nucleofected, using RFP to track the cells expressing shRNA constructs. After a 24-h incubation with stimulated medium, transfected RFP and TUNEL (green) double positive cells were quantified and expressed as a percentage of the total transfected cells. The percentage of untransfected TUNEL-positive cells was comparable across treatments. Compared with U6-GL3 control (i), KD MLK3 (iii) or knockdown of either MLK3 (ii) or TRB3 (iv) all decreased apoptosis by ∼60% (Fig. 4, B and D).
MLK3 Interacts with and Stabilizes TRB3
In the absence of a discernible effect of cytokine-dependent induction of TRB3 mRNA at time points when TRB3 protein induction was observed (supplemental Fig. 2), we examined whether MLK3 associated with TRB3. Using bacterially expressed WT and KD proteins, we show that GST-MLK3-WT interacted more avidly with in vitro transcribed and translated TRB3 compared with GST-MLK3-KD (Fig. 5A, bottom, compare lanes 3 and 4). Having observed that MLK3 could directly bind TRB3 in vitro, we co-transfected HepG2 cells with HA-TRB3 and a mammalian vector for GST-MLK3. Western blots of GST pull-downs (Fig. 5B) showed that, similar to the in vitro data, TRB3 interacted with GST-MLK3-WT and less avidly with GST-MLK3-KD (panel iii, compare lanes 2 and 5). Importantly, MLK3 protein levels appeared to correlate with its own phosphorylation status, and a 60-min exposure to CEP11004 abrogated phosphorylation and destabilized MLK3-WT (lanes 2 and 3, panels i and ii). Notably, total TRB3 protein levels (panel iv, compare lane 2 with lanes 3 and 5) tracked with MLK3 protein levels (panel ii), and as shown by others (32), catalytically active MLK3 (lanes 1 and 2) was more stable compared with catalytic inactivation of MLK3 using CEP11004 (lane 3) or using kinase-dead MLK3-KD (lanes 4 and 5). Co-transfected β-tubulin control remained constant (panel iv).
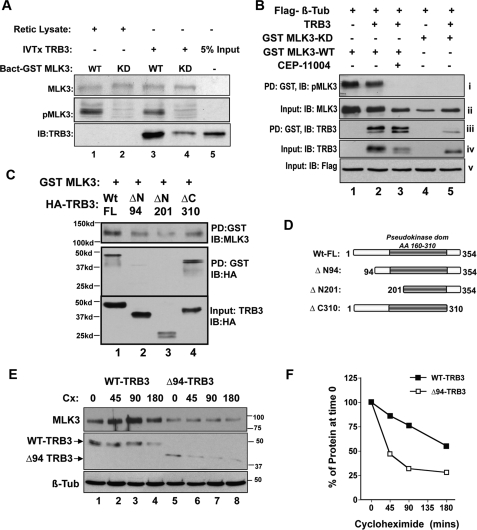
FIGURE 5.
MLK3 binds and stabilizes TRB3 protein. A, in vitro protein-protein interaction using bacterially expressed GST-MLK3-WT (lanes 1 and 3) and GST-MLK3-KD (lanes 2 and 4) to pull down in vitro transcribed and translated (IVT) HA-TRB3 (lanes 3–5) or reticulocyte lysate programmed with empty vector (lanes 1 and 2), followed by Western immunoblotting (IB) for TRB3 (top). Total input of MLK3 as Coomassie-stained proteins (middle) and their phosphorylation status (bottom) are shown. B, TRB3 and MLK3 interactions were assessed from lysates of cells co-transfected with HA-TRB3-WT (lanes 2, 3, and 5) and either GST-MLK3-WT (lanes 1–3) or GST-MLK3-KD (lanes 4 and 5) by GST pull-down (PD) of cellular extracts (PD), followed by Western blotting. Blots of GST pull-downs (panels i and iii) or 7.5% input (panels ii, iv, and v) using antibodies against pMLK3 (panel i), TRB3 (panels iii and iv), total MLK3 (panel ii), and co-transfected control FLAG-β-tubulin (panel v) are shown. Lane 3, effect of 60′ treatment with MLK inhibitor CEP11004. C, MLK3 interacts with the N terminus of TRB3. Shown are GST pull-downs from cells co-transfected with GST-MLK3-WT (lanes 1–4) and HA-TRB3-WT (lane 1) or TRB3 mutants ΔN94, ΔN201, ΔC310 (lanes 2–4, respectively), followed by Western blotting using anti-MLK3 (top) and anti-HA (middle) antibodies. Expression levels of TRB3 constructs were assessed using 7.5% input of total cellular lysate using anti-HA antibodies (bottom). D, schematic of TRB3 deletion mutants. E, time-dependent cycloheximide chase of HA-TRB3-WT (lanes 1–4) and HA-TRB3-Δ94 (lanes 5–8) immunoprecipitated using anti-HA beads from cells co-expressing GST-MLK3-WT followed by Western blotting using anti-MLK3 and anti-HA antibodies. Co-transfected FLAG-β-tubulin was used as a stable control. F, densitometric quantification of time-dependent TRB3 loss to determine t½ of TRB3-WT (closed circles; t½ = 3 h) or TRB3-Δ94 (open circles; T½ = 45 min) in the presence of catalytically active MLK3.
To further qualify the interaction between MLK3 and TRB3, we sought to map the MLK3 interaction domain in TRB3. We cotransfected HepG2 cells with GST-MLK3-WT, and full-length or truncation mutants of TRB3 and analyzed the interaction using GST pull-down assays (Fig. 5C). MLK3 was unable to bind N-terminal deletion mutants of TRB3 (lanes 2 and 3) with interaction localized to the N-terminal 94 amino acids of TRB3. Divergence of the TRB3 N terminus from other members of the TRIBBLES family of proteins attests to the likely specificity of the MLK3-TRB3 interaction.
In addition to demonstrating protein-protein interaction, data from Fig. 5B suggested other attributes of MLK3-TRB3 interaction. Based on its ability to dimerize, catalytically active MLK3 has been shown to be more stable (32), as evidenced by higher levels of WT-MLK3 in Fig. 5B. Interestingly, total TRB3 protein levels appeared to parallel those of MLK3 protein itself. Therefore, we hypothesized that binding MLK3 may result in increased TRB3 protein stability. We used cycloheximide chase experiments to determine the stability of HA-TRB3-WT, and MLK3 interaction-defective HA-TRB3-Δ94. As seen in Fig. 5, E and F, in the presence of GST-MLK3-WT, the half-life (t½) of TRB3-WT was more than 3 h, whereas t½ of HA-TRB3-Δ94 was sharply reduced to 45 min. These data showed that interaction with MLK3 increased the stability of TRB3.
Knockdown of TRB3 Rescues Cytokine-mediated Inhibition of Akt
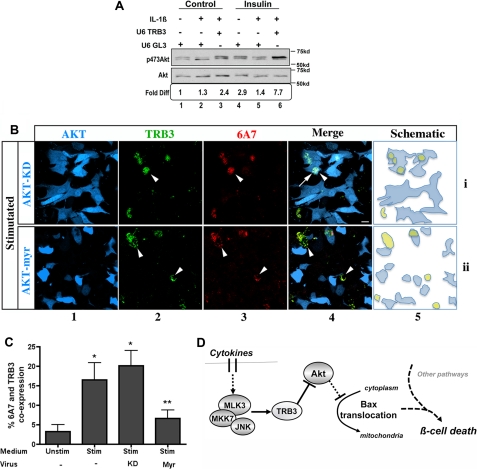
Akt has been identified as a survival factor based on its ability to preserve mitochondrial integrity (40, 41). Other studies have shown that Akt activity can be inhibited by proinflammatory cytokines (42, 43). We therefore examined whether TRB3 knockdown attenuated the ability of cytokines to inhibit Akt Ser473 phosphorylation (Fig. 6A) in Min6 cells. In the presence of control U6-GL3, insulin treatment activated Akt 3-fold (lanes 1 and 4), which was suppressed by a 4-h pretreatment with MLK3-agonist IL-1β (compare lanes 2 and 5 with lanes 1 and 4). Co-transfection of TRB3 shRNA not only prevented the inhibitory effect of IL-β on Akt phosphorylation but also boosted basal Akt activation, for a 7-fold increase in Akt phosphorylation in the presence of insulin (lane 6).
FIGURE 6.
Cytokine-induced expression of TRB3 inhibits Akt activity and results in BAX conformational change. A, TRB3 knockdown rescues cytokine-dependent inhibition of Akt. Myc immunoprecipitates from cells transfected with Myc-Akt and shRNA constructs U6-GL3 (lanes 1 and 2 and lanes 4 and 5) or U6-TRB3 (lanes 3 and 6) were analyzed for phospho-Ser473-Akt and total Myc-Akt using Western blotting. Prior to immunoprecipitation, cells were treated with vehicle or 20 ng/ml IL-1β for 4 h (lanes 2 and 3 and lanes 5 and 6) followed by a 20-min stimulus using 20 nm insulin (lanes 4–6). Densitometric quantification of -fold difference in Akt phosphorylation is included below each lane. B, constitutive Akt activity rescues cytokine-induced TRB3 expression and BAX conformational change. Min6 cells transduced with adenovirus expressing GFP-Akt-KD (row i) or GFP-Akt-Myr (myristoylated) (row ii) were treated with stimulated medium for 4 h and immunostained for induction of endogenous TRB3 (column 2; pseudocolored green for clarity) and conformationally altered BAX (6A7) (column 3). GFP expression (column 1; pseudocolored blue for clarity) marked the transduced cells. Column 4 shows TRB3/BAX-6A7/GFP merge. Arrowheads, colocalization of TRB3 with BAX-6A7; arrows, TRB3 and BAX-6A7 colocalization with Akt-KD. Bar, 10 μm. A schematic representation of the overlapping expression of transduced cells (blue) and TRB3/BAX-6A7-double positive cells (yellow) is shown in column 5. C, quantification of GFP+ cells co-expressing BAX-6A7 and TRB3 (means ± S.D. (error bars) of three independent experiments (*, p < 0.001 versus unstimulated medium; **, p < 0.001 versus Akt-KD, as tested by ANOVA followed by Bonferroni post hoc test). D, schematic of MLK and TRB3 in cytokine-activated beta cell apoptosis. The solid and dotted lines indicate direct and indirect effects, respectively.
Constitutively Active Akt Suppresses TRB3 Expression and BAX Conformational Change
Finally, we examined whether overexpression of Akt could compensate for inhibitory effects of TRB3 and rescue cytokine-dependent, TRB3-mediated, BAX conformational change (Fig. 6B). Adenoviruses encoding KD or constitutively active Myr Akt (rows i and ii) were expressed in Min6 cells and tracked by coexpressed GFP (pseudocolored blue, column 1). For visual clarity of TRB3 and BAX colocalization in yellow, TRB3 florescent signal was pseudocolored green (column 2), and BAX-6A7 was visualized using Alexa-fluor 568 (column 3). Stimulated medium again resulted in induction of TRB3 and co-staining with BAX-6A7 (arrowheads, column 4). As depicted in a schematic representation in column 5, more of the TRB3, BAX-6A7 double positive cells (in yellow) were excluded from the Akt-Myr-expressing cells (row ii) compared with Akt-KD (row i) expressors. TRB3 and BAX-6A7 double positive cells were quantified and expressed as a percentage of total virus-expressing (GFP+) population (Fig. 6C). Taken together and as represented in Fig. 6D, our data suggest that cytokine-dependent induction of MLK3 and TRB3 work together to inhibit the protective effects of Akt on mitochondria in the beta cell and weaken cellular resistance to other cytokine-activated effectors.
DISCUSSION
In this study, we have identified MLK3 as a pivotal kinase in cytokine-activated beta cell death, using both primary islets and beta cell lines. MLK3 was induced by the complex mix of cytokines secreted by activated splenocytes as well as by purified IL-1β. Because static incubation of islets with a mixture of cytokines results in little (44) or late induction (5–10 days) of beta cell death (45), we devised a system that would better represent the milieu of insulitis encountered by islets in vivo. Cell death pathways were examined using an ex vivo system where islets were co-cultured with activated splenocytes. Using this system, we found that MLK action was focused on compromising mitochondrial integrity in the early stages (4–16 h) of beta cell death, by inducing conformational change and mitochondrial translocation of BAX. More importantly, we identified TRB3, a negative regulator of Akt, as a novel and key downstream mediator of MLK3 effects.
Akt can neutralize proapoptotic events by multiple mechanisms, including inhibiting conformational change and mitochondrial translocation of BAX (15, 16). Accordingly, in Min6 cells, in response to cytokines, more than 95% of TRB3-expressing cells were also positive for BAX conformational change, and both were inhibited by the knockdown of either MLK3 or TRB3 (schematic for model represented in Fig. 6D). These data highlight a close correlation between TRB3 induction and BAX conformational change and show that in the beta cell, both events are downstream of MLK3. Thus, our findings demonstrate that in the pancreatic beta cell, activation of MLK3 not only regulates JNK activation (Fig. 1, C and E) and its potential apoptotic effects (reviewed in Refs. 46 and 47), but as reported here, MLK3 also lowers Akt-dependent resistance to apoptosis via TRB3. Mechanistically, we found that MLK3 binds and stabilizes and raises cellular levels of TRB3 to inhibit the survival kinase Akt.
In the beta cell, MLK3 and TRB3 were required for cytokine-mediated mitochondrial translocation of BAX. Permeabilization of the outer mitochondrial membrane following insertion of conformationally altered BAX oligomer often represents a commitment to cell death (17, 18). Akt increases cell survival by deploying multiple mechanisms that converge on preserving mitochondrial integrity, and some of these directly engage BAX (48, 49). However, other pathways have been shown to contribute toward beta cell apoptosis, including events driven by NF-κB and FoxO transcription factors (31, 50). A key target for these pathways is iNOS, which increases oxidative stress by inducing reactive oxygen species and nitrosylation of proteins (51). Interestingly, in our model, iNOS induction at 8–12 h was unresponsive to MLK inhibition (supplemental Fig. 6) and occurred later than the effects of MLKs on the mitochondria (at 4–8 h; Fig. 3, A–D). The focused effect of MLKs on compromising mitochondrial function, with little effect on some of the above events, suggests that additional MAP3Ks may be involved in regulating parallel aspects of the apoptosis program.
Proinflammatory cytokines have been shown to induce pancreatic beta cell death via apoptosis as well as necrosis, and compromise in mitochondrial function has been proposed to be a prominent event in beta cell death (52). Permeabilization of the mitochondrial membrane results in activation of potent proteases and caspases and secondary activation of endonucleases, leading to apoptosis (53). On the other hand, mitochondrial permeabilization has been associated with a decrease in bioenergetic capacity, dysfunction of the electron transport, and production of superoxide anions (54). Our finding that MLK3 impacts mitochondria early in the death cascade is likely to increase the effectiveness of later events, irrespective of the mode of cell death (53, 54). We interpret these data to suggest that MLK- and TRB3-driven mitochondrial instability may be important to confer “death competence” to the beta cell.
Robust activation of MLK3 in islets of NOD diabetic mice suggests a role for MLK3 in type 1 diabetes. In the beta cell, cytokine-mediated activation of JNK was MLK-dependent (supplemental Fig. 5B). However, the ability of MLK3 to induce cellular apoptosis is context-dependent, as evidenced by its ability to regulate cell proliferation via ERK kinases (55, 56) and tissue morphogenesis, including regulation of neural tube closure during development (57). In the mature beta cell, we find that in addition to JNK, the pseudokinase TRB3 is a novel and critical non-kinase effector of MLK action. Thus, in our model, actions of JNK and TRB3 converge on BAX, and both are regulated by MLKs in the beta cell, raising the profile of MLKs as potential targets in treatment of type 1 diabetes.
Supplementary Material
Acknowledgments
We thank Cephalon Inc. for MLK inhibitor CEP11004 and Drs. Natasha Hill (The Scripps Research Institute, La Jolla, CA), Marianne Martinic, Damien Bresson, and Matthias Von Herrath (La Jolla Insititute for Immunology and Allergy, La Jolla, CA) for advice on developing the splenocyte co-culture model and help with cytokine enzyme-linked immunosorbent assays.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK080147. This work was also supported by a University of California San Diego-UCLA Diabetes Endocrinology Research Center Pilot and feasibility grant, a Juvenile Diabetes Research Foundation grant, and a Hillblom Foundation network grant (to U. S. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–7.
- MAPK
- mitogen-activated protein kinase
- SICC
- splenocyte-islet co-culture
- MLK
- mixed lineage kinase
- NOD
- non-obese diabetic
- GST
- glutathione S-transferase
- GFP
- green fluorescence protein
- RFP
- red fluorescence protein
- Myr
- myristolylated
- ANOVA
- analysis of variance
- JNK
- c-Jun N-terminal kinase
- HA
- hemagglutinin
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- IL
- interleukin
- FBS
- fetal bovine serum
- shRNA
- short hairpin RNA
- ANOVA
- analysis of variance
- pMLK3
- phospho-MLK3.
REFERENCES
- 1.Widmann C., Gibson S., Jarpe M. B., Johnson G. L. (1999) Physiol. Rev. 79, 143–180 [DOI] [PubMed] [Google Scholar]
- 2.Sathyanarayana P., Barthwal M. K., Kundu C. N., Lane M. E., Bergmann A., Tzivion G., Rana A. (2002) Mol. Cell 10, 1527–1533 [DOI] [PubMed] [Google Scholar]
- 3.Korchnak A. C., Zhan Y., Aguilar M. T., Chadee D. N. (2009) Cell. Signal. 21, 1620–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitmarsh A. J., Cavanagh J., Tournier C., Yasuda J., Davis R. J. (1998) Science 281, 1671–1674 [DOI] [PubMed] [Google Scholar]
- 5.Yasuda J., Whitmarsh A. J., Cavanagh J., Sharma M., Davis R. J. (1999) Mol. Cell. Biol. 19, 7245–7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tournier C., Dong C., Turner T. K., Jones S. N., Flavell R. A., Davis R. J. (2001) Genes Dev. 15, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancho D., Ventura J. J., Jaeschke A., Doran B., Flavell R. A., Davis R. J. (2005) Mol. Cell. Biol. 25, 3670–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z., Maroney A. C., Dobrzanski P., Kukekov N. V., Greene L. A. (2001) Mol. Cell. Biol. 21, 4713–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J., Zhang Q. G., Zhang G. Y. (2005) Neuroscience 131, 147–159 [DOI] [PubMed] [Google Scholar]
- 10.Bodner A., Maroney A. C., Finn J. P., Ghadge G., Roos R., Miller R. J. (2002) J. Neurochem. 82, 1424–1434 [DOI] [PubMed] [Google Scholar]
- 11.Wang L. H., Besirli C. G., Johnson E. M., Jr. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 451–474 [DOI] [PubMed] [Google Scholar]
- 12.Mayumi-Matsuda K., Kojima S., Suzuki H., Sakata T. (1999) Biochem. Biophys. Res. Commun. 258, 260–264 [DOI] [PubMed] [Google Scholar]
- 13.Du K., Herzig S., Kulkarni R. N., Montminy M. (2003) Science 300, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 14.Kennedy S. G., Kandel E. S., Cross T. K., Hay N. (1999) Mol. Cell. Biol. 19, 5800–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi H., Wang H. G. (2001) Oncogene 20, 7779–7786 [DOI] [PubMed] [Google Scholar]
- 16.Rathmell J. C., Fox C. J., Plas D. R., Hammerman P. S., Cinalli R. M., Thompson C. B. (2003) Mol. Cell. Biol. 23, 7315–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheridan C., Martin S. J. (2008) Trends Cell Biol. 18, 353–357 [DOI] [PubMed] [Google Scholar]
- 18.Chang L. K., Putcha G. V., Deshmukh M., Johnson E. M., Jr. (2002) Biochimie 84, 223–231 [DOI] [PubMed] [Google Scholar]
- 19.Sharma S., Jhala U. S., Johnson T., Ferreri K., Leonard J., Montminy M. (1997) Mol. Cell. Biol. 17, 2598–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krajewska M., Rosenthal R. E., Mikolajczyk J., Stennicke H. R., Wiesenthal T., Mai J., Naito M., Salvesen G. S., Reed J. C., Fiskum G., Krajewski S. (2004) Exp. Neurol. 189, 261–279 [DOI] [PubMed] [Google Scholar]
- 21.Krajewski S., Zapata J. M., Krajewska M., VanArsdale T., Shabaik A., Gascoyne R. D., Reed J. C. (1997) J. Immunol. 159, 5841–5852 [PubMed] [Google Scholar]
- 22.Krajewski S., Thor A. D., Edgerton S. M., Moore D. H., 2nd, Krajewska M., Reed J. C. (1997) Clin. Cancer Res. 3, 199–208 [PubMed] [Google Scholar]
- 23.Kato S., Du K. (2007) Biochem. Biophys. Res. Commun. 353, 933–938 [DOI] [PubMed] [Google Scholar]
- 24.Roy S. K., Shuman J. D., Platanias L. C., Shapiro P. S., Reddy S. P., Johnson P. F., Kalvakolanu D. V. (2005) J. Biol. Chem. 280, 24462–24471 [DOI] [PubMed] [Google Scholar]
- 25.He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bresson D., Togher L., Rodrigo E., Chen Y., Bluestone J. A., Herold K. C., von Herrath M. (2006) J. Clin. Invest. 116, 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krajewska M., Zapata J. M., Meinhold-Heerlein I., Hedayat H., Monks A., Bettendorf H., Shabaik A., Bubendorf L., Kallioniemi O. P., Kim H., Reifenberger G., Reed J. C., Krajewski S. (2002) Neoplasia 4, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krajewska M., Mai J. K., Zapata J. M., Ashwell K. W., Schendel S. L., Reed J. C., Krajewski S. (2002) Cell Death Differ. 9, 145–157 [DOI] [PubMed] [Google Scholar]
- 29.Humphrey R. K., Yu S. M., Flores L. E., Jhala U. S. (2010) J. Biol. Chem. 285, 3406–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danial N. N., Gramm C. F., Scorrano L., Zhang C. Y., Krauss S., Ranger A. M., Datta S. R., Greenberg M. E., Licklider L. J., Lowell B. B., Gygi S. P., Korsmeyer S. J. (2003) Nature 424, 952–956 [DOI] [PubMed] [Google Scholar]
- 31.Donath M. Y., Størling J., Maedler K., Mandrup-Poulsen T. (2003) J. Mol. Med. 81, 455–470 [DOI] [PubMed] [Google Scholar]
- 32.Xu Z., Kukekov N. V., Greene L. A. (2005) Mol. Cell. Biol. 25, 9949–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison D. K., Davis R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 34.Murakata C., Kaneko M., Gessner G., Angeles T. S., Ator M. A., O'Kane T. M., McKenna B. A., Thomas B. A., Mathiasen J. R., Saporito M. S., Bozyczko-Coyne D., Hudkins R. L. (2002) Bioorg. Med. Chem. Lett. 12, 147–150 [DOI] [PubMed] [Google Scholar]
- 35.Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian B., Wang H., Men X., Zhang W., Cai H., Xu S., Xu Y., Ye L., Wollheim C. B., Lou J. (2008) J. Endocrinol. 199, 407–416 [DOI] [PubMed] [Google Scholar]
- 37.Dansen T. B., Whitfield J., Rostker F., Brown-Swigart L., Evan G. I. (2006) J. Biol. Chem. 281, 10890–10895 [DOI] [PubMed] [Google Scholar]
- 38.Hsu Y. T., Youle R. J. (1998) J. Biol. Chem. 273, 10777–10783 [DOI] [PubMed] [Google Scholar]
- 39.Chao D. T., Korsmeyer S. J. (1998) Annu. Rev. Immunol. 16, 395–419 [DOI] [PubMed] [Google Scholar]
- 40.Dijkers P. F., Birkenkamp K. U., Lam E. W., Thomas N. S., Lammers J. W., Koenderman L., Coffer P. J. (2002) J. Cell Biol. 156, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plas D. R., Talapatra S., Edinger A. L., Rathmell J. C., Thompson C. B. (2001) J. Biol. Chem. 276, 12041–12048 [DOI] [PubMed] [Google Scholar]
- 42.Bouzakri K., Ribaux P., Halban P. A. (2009) J. Biol. Chem. 284, 27892–27898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venieratos P. D., Drossopoulou G. I., Kapodistria K. D., Tsilibary E. C., Kitsiou P. V. (2010) Cell. Signal. 22, 791–800 [DOI] [PubMed] [Google Scholar]
- 44.Collier J. J., Fueger P. T., Hohmeier H. E., Newgard C. B. (2006) Diabetes 55, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 45.Kim S., Millet I., Kim H. S., Kim J. Y., Han M. S., Lee M. K., Kim K. W., Sherwin R. S., Karin M., Lee M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1913–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho S. G., Choi E. J. (2002) J. Biochem. Mol. Biol. 35, 24–27 [DOI] [PubMed] [Google Scholar]
- 47.Dhanasekaran D. N., Reddy E. P. (2008) Oncogene 27, 6245–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parcellier A., Tintignac L. A., Zhuravleva E., Hemmings B. A. (2008) Cell. Signal. 20, 21–30 [DOI] [PubMed] [Google Scholar]
- 49.Miyamoto S., Murphy A. N., Brown J. H. (2009) J. Bioenerg. Biomembr. 41, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cnop M., Welsh N., Jonas J. C., Jörns A., Lenzen S., Eizirik D. L. (2005) Diabetes 54, Suppl. 2, S97–S107 [DOI] [PubMed] [Google Scholar]
- 51.Pacher P., Beckman J. S., Liaudet L. (2007) Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szabadkai G., Duchen M. R. (2009) Apoptosis 14, 1405–1423 [DOI] [PubMed] [Google Scholar]
- 53.Kroemer G., Galluzzi L., Brenner C. (2007) Physiol. Rev. 87, 99–163 [DOI] [PubMed] [Google Scholar]
- 54.Lemasters J. J., Qian T., He L., Kim J. S., Elmore S. P., Cascio W. E., Brenner D. A. (2002) Antioxid. Redox Signal. 4, 769–781 [DOI] [PubMed] [Google Scholar]
- 55.Chadee D. N., Kyriakis J. M. (2004) Cell Cycle 3, 1227–1229 [DOI] [PubMed] [Google Scholar]
- 56.Chadee D. N., Kyriakis J. M. (2004) Nat. Cell Biol. 6, 770–776 [DOI] [PubMed] [Google Scholar]
- 57.Stronach B., Perrimon N. (2002) Genes Dev. 16, 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.