Abstract
Various kinds of fatty acids are distributed in membrane phospholipids in mammalian cells and tissues. The degree of fatty acid unsaturation in membrane phospholipids affects many membrane-associated functions and can be influenced by diet and by altered activities of lipid-metabolizing enzymes such as fatty acid desaturases. However, little is known about how mammalian cells respond to changes in phospholipid fatty acid composition. In this study we showed that stearoyl-CoA desaturase 1 (SCD1) knockdown increased the amount of saturated fatty acids and decreased that of monounsaturated fatty acids in phospholipids without affecting the amount or the composition of free fatty acid and induced unfolded protein response (UPR), evidenced by increased expression of C/EBP homologous protein (CHOP) and glucose-regulated protein 78 (GRP78) mRNAs and splicing of Xbox-binding protein 1 (XBP1) mRNA. SCD1 knockdown-induced UPR was rescued by various unsaturated fatty acids and was enhanced by saturated fatty acid. Lysophosphatidylcholine acyltransferase 3 (LPCAT3), which incorporates preferentially polyunsaturated fatty acids into phosphatidylcholine, was up-regulated in SCD1 knockdown cells. Knockdown of LPCAT3 synergistically enhanced UPR with SCD1 knockdown. Finally we showed that palmitic acid-induced UPR was significantly enhanced by LPCAT3 knockdown as well as SCD1 knockdown. These results suggest that a decrease in membrane phospholipid unsaturation induces UPR.
Keywords: Fatty Acid, Fatty Acid Metabolism, Membrane, Membrane Lipids, Phospholipid, Phospholipid Metabolism, Lipotoxicity, Lysophospholipid Acyltransferase, Unfolded Protein Response
Introduction
Fatty acids in membrane phospholipids of mammalian cells and tissues exhibit considerable structural diversity, including varying chain lengths and degrees of unsaturation (1–3). The degree of fatty acid unsaturation in membrane phospholipids determines the biophysical properties of the membrane, which in turn influences many crucial membrane-associated functions, including the activities of membrane-bound proteins such as ion channels and endo- and exocytosis (4). The alteration of the degree of fatty acid unsaturation in cell membrane has also been implicated in a variety of disease states, including diabetes, obesity, hypertension, cancer, and neurological and heart diseases (5, 6).
The fatty acid composition of membrane phospholipids is likely to be affected by the exogenous fatty acids from the diet or by altered activities of lipid-metabolizing enzymes such as fatty acid desaturases. So far, several cellular responses to changes in phospholipid fatty acid composition have been found in microorganisms. In the cyanobacterium, the palladium-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene, which is responsible for the desaturation of fatty acids of membrane lipids (7). Gene-engineered rigidity of the membrane was also reported to enhance the inducible expression of cold-inducible genes (8). However, cellular responses to changes in phospholipid fatty acid composition have been poorly understood in mammalian cells.
Stearoyl-CoA desaturase 1 (SCD1)3 is a key enzyme in lipid and energy metabolism. It catalyzes the Δ9-desaturation of the saturated fatty acids (SFAs), palmitic acid (16:0) and stearic acid (18:0), to the monounsaturated fatty acids (MUFAs), palmitoleic acid (16:1n-7) and oleic acid (18:1n-9), respectively (9). Significant reductions in tissue triglycerides and cholesteryl esters have been observed in a mouse model with targeted disruption in the SCD1 gene (SCD1−/−) (10). Moreover, SCD1−/− mice are resistant to diet- and leptin deficiency-induced adiposity (11), which makes SCD1 a possible therapeutic target for the treatment of obesity. In addition to regulating lipid and energy metabolism, SCD1 plays a role in determining the SFA/MUFA balance in membrane phospholipids. Increases in the SFA/MUFA ratio in membrane phospholipids have been observed in SCD1−/− mice (12) and in SCD1 knockdown human adipocytes (13).
A growing body of evidence indicates that fatty acid composition in membrane phospholipids is also regulated by lysophospholipid acyltransferase (LPLAT). Glycerophospholipids, the major phospholipids in biological membrane, are synthesized de novo by the Kennedy and Weiss pathway (14). Subsequently, in the remodeling pathway (Lands cycle) (15, 16), cycles of deacylation (phospholipases) and reacylation (LPLATs) of glycerophospholipids modify the fatty acid composition to generate the mature membrane. Mammalian cells possess a number of LPLATs that exhibit distinct acyl-CoA donor and lysophospholipid acceptor specificities (17). For example, lysophosphatidylcholine acyltransferase 3 (LPCAT3)/membrane-bound O-acyltransferase 5 and lysophosphatidylinositol acyltransferase 1 (LPIAT1) 1/membrane-bound O-acyltransferase 7 incorporate polyunsaturated fatty acids (PUFAs) such as arachidonic acid (20:4) into phosphatidylcholine (PC) and phosphatidylinositol (PI), respectively (18–21), whereas LPCAT1 incorporates SFAs such as 16:0 into PC (22, 23). Knock-out or knockdown of these LPLATs changes the phospholipid fatty acid composition (18, 21).
In this study we show that SCD1 knockdown decreases membrane phospholipid unsaturation without affecting the amount or the composition of free fatty acids and causes caspase-dependent cell death and unfolded protein response (UPR). Moreover, among various LPLATs, LPCAT3 is up-regulated in SCD1 knockdown cells, and LPCAT3 knockdown synergistically enhances UPR with SCD1 knockdown. Overload of saturated long chain fatty acids within cells is known to cause lipotoxicity, which contributes to various pathological conditions such as insulin resistance, type 2 diabetes, and cardiovascular disease (24). We also show that LPCAT3 knockdown cells are very susceptible to palmitic acid-induced toxicity.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) unless otherwise stated. Palmitic acid (16:0), oleic acid (18:1n-9), linoleic acid (18:2), arachidonic acid (20:4), and eicosapentaenoic acid (20:5) were purchased from Cayman Chemicals (Ann Arbor, MI). Dulbecco's modified Eagle's medium was obtained from Invitrogen. Thapsigargin and anti-α-tubulin was obtained from Sigma. Anti-SCD1 was purchased from Advanced Targeting Systems (San Diego, CA). Anti-double-stranded RNA-activated kinase-like kinase (PERK) and anti-inositol-requiring 1α (IRE1α) were from Cell Signaling Technology (Beverly, MA). Benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (Z-VAD-FMK) and benzyloxycarbonyl-Phe-Ala-fluoromethyl ketone (Z-FA-FMK) were obtained from BioVision (Mountain View, CA). A control small interfering RNA (siRNA) and siRNAs directed against SCD1, LPCAT1, LPCAT2, LPCAT4, LPIAT1, IRE1α, and PERK were obtained from Nippon EGT (Toyama, Japan). An siRNA directed against LPCAT3 was purchased from Invitrogen.
Cell Culture and siRNA Transfection
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mm l-glutamine. siRNAs were transfected into HeLa cells with LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's protocol. The final concentration of siRNA was 10 nm. For fatty acid treatment the cells were incubated in the identical medium with fatty acid at indicated concentration or solvent (ethanol) for indicated time periods before analysis.
Lipid Analysis
Lipids were extracted by the method of Bligh and Dyer (25). Phospholipids were isolated by application of the lipid extract to a column of silica coupled with aminopropyl groups (Bond Elute® NH2, Varian, Lake Forrest, CA). Isolated phospholipids were methylated with 2.5% H2SO4 in methanol. The resulting fatty acid methyl esters were then extracted with hexane and subjected to gas chromatography-mass spectrometry (GC-MS) analysis. GC-MS analysis was performed by using an Agilent 7890A-5975C GC-MS network system (Agilent Technologies, Wilmington, DE) equipped with a DB-23 capillary column (60 m × 250 μm × 0.15 μm; Agilent Technologies). The oven temperature program was as follows; the initial temperature was 50 °C for 1 min, then raised to 175 °C at 25 °C/min, to 235 °C at 5 °C/min, and held for 5 min. The injector and detector temperatures were both set at 250 °C. The amounts of free fatty acids in samples were measured using an acyl-CoA oxidase-based colorimetric kit (WAKO FA-C; Wako Pure Chemicals, Osaka, Japan).
Total RNA Isolation and Quantitative Real-time PCR
Total RNA from cells was extracted using Isogen (Nippongene, Toyama, Japan) and reverse-transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR was performed using SYBR® Green PCR Master Mix (TaKaRa) and LightCycler 480 (Roche Diagnostics). The sequences of the oligonucleotides were as follows. SCD1 forward (CCGACGTGGCTTTTTCTTC) and reverse (CCTCCTCTGGAACATCACCA), SCD5 forward (CCAAGCCACTCACTCTGCTC) and reverse (CGACAGCCAGAAATATCCTCA), C/EBP homologous protein (CHOP) forward (AGCTGGAAGCCTGGTATGAG) and reverse (GTGCGTGTGACCTCTGTTGG), glucose-regulated protein 78 (GRP78) forward (ACCGCTGAGGCTTATTTGG) and GRP78 reverse (GCGTCTTTGGTTGCTTGG), GAPDH forward (GCCAAGGTCATCCATGACAACT) and reverse (GAGGGGCCATCCACAGTCTT), LPCAT1 forward (GATGAAGGGAGGAGAGAAGATAGG) and LPCAT1 reverse (CAGACAGGGCAACCACACAC), LPCAT2 forward (CTGTCTTGTGCAACCCTTCC) and reverse (ACATCAAGGTCAGGCACTCC), LPCAT3 forward (TGGGCCGCACCATCAC) and LPCAT3 reverse (AGTTGCCGGTGGCAGTGTA), LPCAT4 forward (GTTTGGCTTGCGAAATTCATG) and LPCAT4 reverse (CATGCGCCTTACAGCTAAATCC), and LPIAT1 forward (GCCCTCCCTGATGGAGAC) and LPIAT1 reverse (GTAGGTGCGGTAGCGGAAGA). Target gene expression was normalized on the basis of GAPDH content.
Western Blotting
HeLa cells were lysed in ice-cold lysis buffer (20 mm Tris-HCl, pH 7.4, 1% Triton X-100, 10% glycerol, 137 mm NaCl, 2 mm EDTA, 50 mm β-glycerophosphate) containing protease inhibitor (10 μg/ml leupeptin, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, and 1 mm phenylmethylsulfonyl fluoride) and phosphatase inhibitor (2 mm Na3VO4, 10 mm NaF) for 15 min on ice. After centrifugation at 12,000 × g for 20 min at 4 °C, the supernatants were used as total protein extracts. The protein concentrations of samples were determined by the bicinchoninic acid (BCA) assay (Pierce). Each total protein extract (20 μg/lane) was subjected to SDS-PAGE and immunoblotting. The following dilutions of primary antibodies were used for immunoblotting: anti-SCD1 (1:1000), anti-PERK (1:1000), anti-IRE1α (1:1000), and anti-α-tubulin (1:5000).
Determination of Cell Viability
Cell viability was determined using a cell counting kit-8 (Dojindo Laboratories, Kumamoto, Japan) in which 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt (WST-8) was used as a substrate. The relative number of surviving cells was determined in triplicate by estimating the value of control siRNA-transfected cells as 100%.
Measurement of Xbox-binding Protein 1 (XBP1) mRNA Splicing
Total RNA was isolated from HeLa cells as described above. 1 μg of total RNA was reverse-transcribed with PrimeScript® II 1st Strand cDNA synthesis Kit (TaKaRa Bio, Shiga, Japan). PCR fragments representing the unspliced and spliced forms of XBP1 were amplified with Ex Taq polymerase (TaKaRa Bio). Primer sequences used to amplify Human XBP1 were AAACAGAGTAGCAGCTCAGACTGC and TCCTTCTGGGTAGACCTCTGGGAG. PCR products were resolved on a 3% agarose gel and visualized using ethidium bromide.
Statistical Analysis
Data were converted to means ± S.E. values, and the Student's test was applied to determine significant differences between two samples (p < 0.01). Statistical differences between multiple treatment groups and a control group were determined using analysis of variance and Tukey-Kramer post hoc test.
RESULTS
SCD1 Knockdown Decreases Membrane Phospholipid Unsaturation and Induces Cell Death in HeLa Cells
Because SCD plays an important role in determining the SFA/MUFA balance in membrane phospholipids (12, 13), we examined how perturbation of SCD function affects phospholipid fatty acid composition and cellular functions in HeLa cells. Transfection of an siRNA against human SCD1 (siSCD1) in HeLa cells efficiently reduced protein expression of SCD1 (Fig. 1A). GC-MS analysis of phospholipid fatty acids showed that SCD1 knockdown led to increases of SFAs, 16:0 and 18:0, and decreases of MUFAs, 16:1n-7 and 18:1n-9, and cis-vaccenic acid (18:1n-7) in phospholipids compared with control (Table 1). These changes increased the SFA/MUFA ratio in phospholipids of SCD1 knockdown cells (supplemental Fig. S1B). Knockdown of SCD5, another isoform of human SCD (26), did not affect the SFA/MUFA ratio in phospholipids (supplemental Fig. S1, A and B). These results indicate that SCD1 is a major isoform of Δ9 desaturase in HeLa cells.
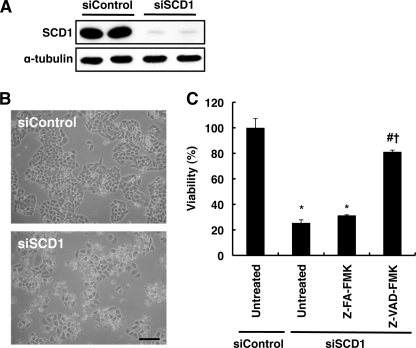
FIGURE 1.
SCD1 knockdown induces cell death in HeLa cells. A, a control siRNA (siControl) and SCD1 siRNA (siSCD1) were transfected into HeLa cells. After 48 h cell lysates were prepared and subjected to Western blot analysis using anti-SCD1 antibody. α-Tubulin expression was used as a loading control. B, cells were transfected with the indicated siRNA. At 72 h after transfection representative photographs were taken under phase-contrast microscopy. Bar, 200 μm. C, cells were transfected with the indicated siRNA. In the presence or absence of 20 μm Z-VAD-FMK, a pan-caspase inhibitor, cell viability was determined as described under “Experimental Procedures.” Z-FA-FMK, an inactive analog of Z-VAD-FMK, was used as a negative control. The data represent the mean ± S.E. of three experiments. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01), the number symbol indicates a significant difference compared with siSCD1-transfected cells that were untreated (p < 0.01), and the dagger indicates a significant difference compared with siSCD1-transfected cells that were treated with Z-FA-FMK (p < 0.01).
TABLE 1.
Fatty acid composition (mol %) of phospholipids and free fatty acids in normal and SCD1/LPCAT3 knockdown cells
Data represent the average ± S.E. of three independent experiments. ND, not detected (<0.5%).
| Fatty acid | Phospholipids |
Free fatty acids |
||||
|---|---|---|---|---|---|---|
| siControl | siSCD1 | siSCD1 siLPCAT3 | siControl | siSCD1 | siSCD1 siLPCAT3 | |
| 14:0 | 1.12 ± 0.01 | 0.88 ± 0.34 | 1.28 ± 0.16 | ND | ND | ND |
| 16:0 | 35.39 ± 0.08 | 40.09 ± 0.25a | 40.41 ± 0.38a | 44.00 ± 2.09 | 39.65 ± 3.56 | 40.70 ± 0.86 |
| 16:1 | 3.53 ± 0.22 | 1.20 ± 0.07a | 1.40 ± 0.14a | ND | ND | ND |
| 18:0 | 13.68 ± 0.94 | 20.64 ± 0.80a | 21.89 ± 0.63a | 55.99 ± 2.09 | 60.35 ± 3.56 | 59.30 ± 0.86 |
| 18:1n-9 | 30.62 ± 0.53 | 20.56 ± 0.45a | 20.29 ± 0.37a | ND | ND | ND |
| 18:1n-7 | 5.75 ± 0.17 | 3.54 ± 0.17a | 3.09 ± 0.21a | ND | ND | ND |
| 18:2 | 0.82 ± 0.01 | 1.20 ± 0.04a | 1.12 ± 0.01a,b | ND | ND | ND |
| 20:4 | 5.25 ± 0.13 | 6.98 ± 0.10a | 5.77 ± 0.09a,b | ND | ND | ND |
| 20:5 | 1.78 ± 0.04 | 2.09 ± 0.04a | 1.79 ± 0.06b | ND | ND | ND |
| 22:6 | 2.07 ± 0.06 | 2.82 ± 0.07a | 2.97 ± 0.01a | ND | ND | ND |
a p < 0.05 versus siControl.
b p < 0.05 versus siSCD1.
We next measured the amount and the composition of free fatty acids in SCD1 knockdown cells. HeLa cells contained very small amounts of free fatty acids (4.51 ± 0.29 nmol/mg of protein) compared with the amounts of phospholipids (150.36 ± 3.46 nmol/mg of protein). SCD1 knockdown did not significantly affect the amounts of free fatty acids (4.98 ± 0.12 nmol/mg of protein). SFAs such as 16:0 and 18:0 were predominantly detected in the free fatty acid fraction, and SCD1 knockdown did not cause a significant change in the fatty acid composition (Table 1). These results indicate that knockdown of SCD1 decreases membrane phospholipid unsaturation without affecting the amount and the composition of free fatty acids.
In addition to the changes in phospholipid fatty acids, we found that SCD1 knockdown markedly reduced cell viability when cells were incubated more than 72 h after siRNA transfection (Fig. 1, B and C), whereas SCD5 knockdown did not affect cell viability (data not shown). Moreover, Z-VAD-FMK, a pan-caspase inhibitor, significantly suppressed cell death induced by SCD1 knockdown, whereas Z-FA-FMK, an inactive analog of Z-VAD-FMK, did not (Fig. 1C). These results suggest that the observed cell death was mediated by apoptosis.
SCD1 Knockdown Induces UPR
Recent studies have revealed that chronic SFA exposure induces cell death and activates UPR in cultured cells (27–29). These studies led us to investigate whether UPR activation occurred in SCD1 knockdown cells in which SFAs in phospholipids were increased (Table 1).
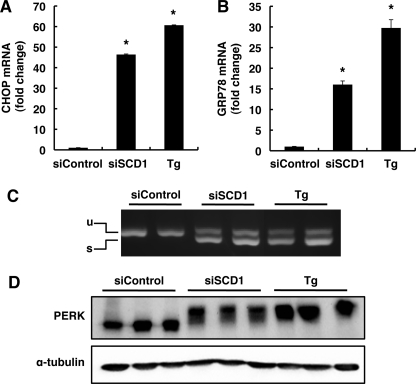
UPR is initiated by three endoplasmic reticulum (ER) transmembrane proteins, inositol-requiring 1, PERK, and activating transcription factor 6 (ATF6). UPR induces transcription of a set of genes whose protein products increase the capacity for protein folding and ER-associated degradation and induces apoptosis when the ER function is severely impaired (30). Of these genes, CHOP and glucose-regulated protein 78 (GRP78) were often used as UPR markers. Quantitative real-time PCR analysis showed that SCD1 knockdown strikingly induced the expression of CHOP and GRP78 mRNAs, which is comparable with that induced by 0.5 μm thapsigargin treatment for 12 h (Fig. 2, A and B). We also examined activations of the UPR sensor proteins IRE1 and PERK. Activation of IRE1 leads to alternative splicing of transcription factor XBP1, and PERK is activated by autophosphorylation, a modification that slows its electrophoretic mobility (31, 32). SCD1 knockdown induced XBP1 splicing to the same extent as thapsigargin treatment induced XBP1 splicing (Fig. 2C). Western blotting using anti-PERK revealed that SCD1 knockdown induced a shift in PERK protein mobility that was similar to the shift induced by thapsigargin treatment (Fig. 2D). These results indicate that SCD1 knockdown induces UPR activation.
FIGURE 2.
SCD1 knockdown induces UPR. HeLa cells were transfected with the indicated siRNA. At 72 h after transfection, total RNA and cell lysates were prepared. A and B, expression of CHOP (A) and GRP78 (B) mRNAs were detected by quantitative real-time PCR. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over siControl. Thapsigargin-treated cells (Tg) were used as a positive control. C, semiquantitative reverse transcription-PCR analysis of XBP1 spliced and unspliced mRNA is shown. The positions of the unspliced form (u) and spliced form (s) are indicated. Thapsigargin-treated cells were used as a positive control. D, shown is an immunoblot analysis of PERK. Thapsigargin-treated cells were used as a positive control, and α-tubulin expression was used as a loading control. The data represent the mean ± S.E. of three experiments. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01).
We also examined whether UPR activation contributes to the cell death induced by SCD1 knockdown. Transfection of siRNA against IRE1α or PERK efficiently reduced the protein expression of IRE1α or PERK, respectively (Fig. 3A and B). The cell death induced by SCD1 knockdown was significantly suppressed by knockdown of PERK but was not suppressed by knockdown of IRE1α (Fig. 3C). Consistent with these results, the induction of proapoptotic transcription factor CHOP in SCD1 knockdown cells was reduced by knockdown of PERK but enhanced by knockdown of IRE1α (Fig. 3D). These results suggest that the PERK-CHOP arm of the UPR is involved in the death of SCD1 knockdown cells.
FIGURE 3.
PERK contributes to the cell death induced by SCD1 knockdown. A and B, HeLa cells were transfected with the indicated siRNA. After 72 h, cell lysates were prepared and subjected to Western blot analysis using antibodies against IRE1α (A) and PERK (B). α-Tubulin expression was used as a loading control. siIRE1α and siPERK indicate siRNAs against IRE1α and PERK, respectively. C, cells were transfected with the indicated siRNA. At 66 h after transfection, cell viability was determined as described under “Experimental Procedures.” D, expression of CHOP mRNA was detected by quantitative real-time PCR. The expression level of each gene was normalized to expression of the GAPDH gene and is represented as -fold induction over siControl. The data represent the mean ± S.E. of three experiments. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01). The number symbols indicate significant differences compared with siSCD1-transfected cells (p < 0.05).
Polyunsaturated Fatty Acids Suppress UPR Induced by SCD1 Knockdown
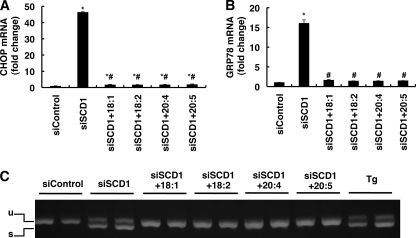
We next examined the effect of unsaturated fatty acid administration to the SCD1 knockdown cells on UPR activation. Administration of 18:1n-9, which is a major product of SCD1, completely suppressed the induction of CHOP and GRP78 expression and XBP1 splicing in SCD1 knockdown cells (Fig. 4, A–C). In contrast, the addition of a saturated fatty acid such as 16:0 caused severe cell death in SCD1 knockdown cells (data not shown). The suppression effects were also observed when PUFAs, linoleic acid (18:2), 20:4, or eicosapentaenoic acid (20:5) were administered (Fig. 4, A–C). Treatment with these unsaturated fatty acids also completely rescued SCD1 knockdown-induced cell death but had no effect on thapsigargin-induced alternate splicing of XBP1 (data not shown). Lipid analysis showed that the supplemented unsaturated fatty acids were efficiently incorporated into the phospholipid fraction (supplemental Fig. S2). These results indicate that various unsaturated fatty acids, in addition to 18:1n-9, are able to rescue the SCD1 knockdown phenotype.
FIGURE 4.
Polyunsaturated fatty acids suppress UPR induced by SCD1 knockdown. HeLa cells were transfected with the indicated siRNA. 48 h after transfection cells were further incubated for 24 h in media supplemented with the indicated fatty acid (50 μm) and then harvested. A and B, expression of CHOP (A) and GRP78 (B) mRNAs detected by real-time PCR is shown. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over siControl. C, semiquantitative reverse transcription-PCR analysis of XBP1 spliced and unspliced mRNA. The positions of the unspliced form (u) and spliced form (s) are indicated. Thapsigargin-treated cells (Tg) were used as a positive control. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01), and the number symbols indicate significant differences compared with siSCD1-transfected cells which were untreated (p < 0.01).
SCD1 Knockdown Increases Palmitic Acid-induced UPR
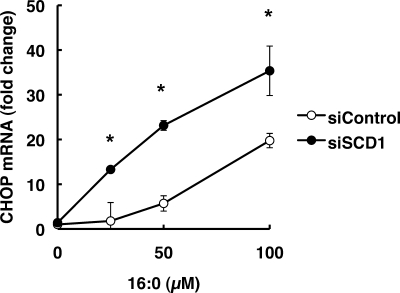
We also examined the effect of exposure of 16:0 in control and SCD1 knockdown cells. To examine CHOP expression in 16:0-treated cells, we harvested cells before excessive cell death occurred. In control cells, 50 μm 16:0 exposure for 12 h did not significantly increase CHOP expression. In SCD1 knockdown cells, however, significant increase in CHOP expression was observed after exposure to 50 μm 16:0 (Fig. 5).
FIGURE 5.
SCD1 knockdown increases palmitic acid-induced UPR. At 48 h after siRNA transfection, cells were further incubated for 12 h in media supplemented with 16:0. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over untreated siControl. The data represent the mean ± S.E. of three experiments. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01).
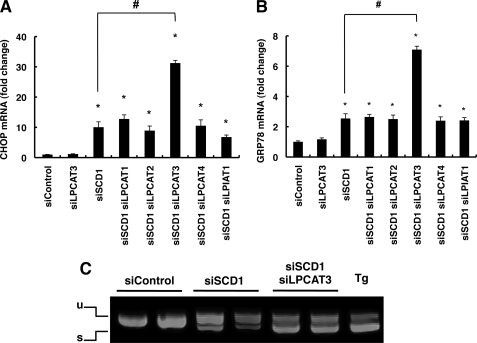
LPCAT3 Knockdown Enhances SCD1 Knockdown-mediated UPR
To check our finding that decreases of unsaturated fatty acids in phospholipids contribute to UPR activation in SCD1 knockdown cells, we performed double knockdown of SCD1 and various LPLATs, which are known to be involved in the incorporation of various fatty acids into membrane phospholipids via the remodeling pathway (17). For knockdown targets, we chose LPCAT1, -2, -3, and -4, which incorporate various fatty acids into PC, the most abundant phospholipid in the biological membrane (33). The siRNA against each LPLAT efficiently reduced the level of their target LPLAT mRNA (supplemental Fig. S3). As shown in Fig. 6, A and B, although LPCAT3 single-knockdown did not induce CHOP and GRP78 mRNA expressions, LPCAT3 knockdown significantly enhanced the SCD1 knockdown-induced CHOP and GRP78 mRNA expressions. Knockdown of other LPCATs did not significantly affect the expressions. Knockdown of LPIAT1, a major acyltransferase that incorporates 20:4 into PI, did not affect CHOP and GRP78 mRNA expressions in SCD1 knockdown cells. The production of the spliced form of XBP1 mRNA in SCD1 knockdown cells was also enhanced by LPCAT3 knockdown (Fig. 6C). On the other hand, LPCAT3 knockdown had no effect on the thapsigargin-induced alternate splicing of XBP1 (data not shown).
FIGURE 6.
LPCAT3 knockdown enhances SCD1 knockdown-mediated UPR. HeLa cells were transfected with the indicated siRNA. At 66 h after transfection total RNAs were prepared. A and B, expression of CHOP (A) and GRP78 (B) mRNAs were detected by quantitative real-time PCR. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over siControl. siLPCAT1, siLPCAT2, siLPCAT3, siLPCAT4, and siLPIAT1 indicate siRNAs against LPCAT1, LPCAT2, LPCAT3, LPCAT4, and LPIAT1, respectively. C, semiquantitative reverse transcription-PCR analysis of XBP1 spliced and unspliced mRNA. The positions of the unspliced form (u) and spliced form (s) are indicated. Thapsigargin-treated cells (Tg) were used as a positive control. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01), and the number symbols indicate significant differences compared with siSCD1-transfected cells (p < 0.01).
Lipid analysis revealed that SCD1/LPCAT3 double-knockdown did not alter the amount of SFAs but decreased that of PUFAs in phospholipids, as shown by significantly lower levels of 18:2, 20:4, and 20:5 than were observed in SCD1 single-knockdown cells (Table 1). In the free fatty acid fraction, the amount in SCD1/LPCAT3 double knockdown cells (5.18 ± 0.62 nmol/mg of protein) was comparable with that in SCD1 single-knockdown cells (4.98 ± 0.12 nmol/mg of protein), and the composition was not changed (Table 1).
Supplementation of a 0.5 μm PUFA mixture (25 nm 18:2, 380 nm 20:4, and 95 nm 20:5) slightly increased the phospholipid PUFA content in SCD1/LPCAT3 double-knockdown cells, in which the PUFA content is comparable with that of SCD1 single-knockdown cells (supplemental Fig. S4A). Under the same conditions, supplementation of the unsaturated fatty acid mixture canceled the effect of LPCAT3 knockdown on CHOP expression in SCD1 knockdown cells (supplemental Fig. S4B). Collectively, these results strongly suggest that a decrease in membrane phospholipids unsaturation induces UPR.
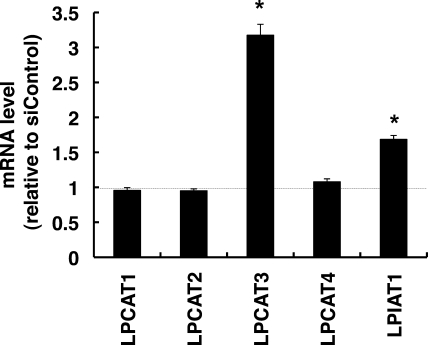
Interestingly, LPCAT3 was significantly up-regulated in SCD1 knockdown cells. Although the expressions of LPCAT1, -2, and -4 were not affected by SCD1 knockdown, LPIAT1 was also slightly up-regulated (Fig. 7).
FIGURE 7.
LPCAT3 is up-regulated in SCD1 knockdown cells. HeLa cells were transfected with the indicated siRNA. At 66 h after transfection total RNAs were prepared. The expression level of each gene was detected by quantitative real-time PCR. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over siControl. The data represents the mean ± S.E. of three experiments. The asterisks indicate significant differences compared with siControl-transfected cells (p < 0.01).
LPCAT3 Knockdown Cells Are Susceptible to Exogenous Palmitic Acid Toxicity
To test the possibility that decreases of unsaturated fatty acids in phospholipids increase the susceptibility to exogenous palmitic acid toxicity, we examined the effect of LPCAT3 knockdown on 16:0-induced UPR signaling. LPCAT3 knockdown alone did not induce UPR activation in control cells but greatly enhanced the CHOP and GRP78 mRNA expressions and splicing of XBP1 mRNA in 16:0-treated cells (Fig. 8, A–C). Lipid analysis showed that LPCAT3 knockdown reduced the amounts of unsaturated fatty acids, especially 18:1n-9, 18:1n-7, and 20:4 in phospholipids of 16:0-treated cells (Table 2). These results indicate that decreases of unsaturated fatty acids in phospholipids make cells more susceptible to palmitic acid toxicity.
FIGURE 8.
LPCAT3 knockdown cells are susceptible to palmitic acid exposure. HeLa cells were transfected with the indicated siRNA. At 60 h after transfection cells were further incubated for 12 h in media supplemented with 16:0 (100 μm) and then harvested. A and B, expressions of CHOP (A) and GRP78 (B) mRNAs were detected by quantitative real-time PCR. The expression level of each gene was normalized to the GAPDH gene and is represented as -fold induction over siControl treated with ethanol. C, semiquantitative reverse transcription-PCR analysis of XBP1 spliced and unspliced mRNA is shown. The positions of the unspliced form (u) and spliced form (s) are indicated. Thapsigargin-treated cells (Tg) were used as a positive control. The asterisks indicate significant differences compared with siControl-transfected cells, which were treated with ethanol (p < 0.01).
TABLE 2.
Fatty acid composition (mol %) of phospholipids in normal and LPCAT3 knockdown cells administered with 16:0
Data are the mol % and represent the average ± S.E. of three independent experiments.
| Fatty acid | siControl + 16:0 | siLPCAT3 + 16:0 |
|---|---|---|
| 14:0 | 0.51 ± 0.01 | 0.66 ± 0.01a |
| 16:0 | 43.18 ± 0.26 | 50.52 ± 0.30a |
| 16:1 | 12.50 ± 0.04 | 13.72 ± 0.10 |
| 18:0 | 7.80 ± 0.10 | 7.46 ± 0.36a |
| 18:1n-9 | 23.83 ± 0.04 | 19.27 ± 0.07a |
| 18:1n-7 | 5.99 ± 0.12 | 2.75 ± 0.05a |
| 18:2 | 0.58 ± 0.03 | 0.55 ± 0.01 |
| 20:4 | 3.29 ± 0.09 | 2.64 ± 0.02a |
| 20:5 | 1.10 ± 0.01 | 0.99 ± 0.03 |
| 22:6 | 1.21 ± 0.03 | 1.45 ± 0.01a |
a p < 0.05 versus siControl + 16:0.
Although LPCAT3 has been characterized as a principal enzyme that incorporates 20:4 into PC (18–20), the above results suggest that LPCAT3 also contributes to incorporating MUFAs into PC in cells. We, therefore, measured the in vitro acyltransferase activity using the membrane fractions of LPCAT3 knockdown cells. The membranes of LPCAT3 knockdown cells showed remarkably reduced LPCAT activity toward oleoyl-CoA, linoleoyl-CoA, and arachidonoyl-CoA, whereas the activity toward stearoyl-CoA acid was not changed (supplemental Fig. S5). These in vitro and in vivo data indicate that LPCAT3 contributes to incorporating MUFAs into PC.
DISCUSSION
In this study we demonstrated that knockdown of SCD1, a rate-limiting enzyme in the cellular synthesis of MUFAs from SFAs, successfully reduced membrane phospholipid unsaturation without affecting the amount or the composition of free fatty acid in HeLa cells. With this system, we also showed that the decrease in membrane phospholipid unsaturation induces UPR- and caspase-dependent cell death.
We examined in detail the fatty acid composition of cellular phospholipid and free fatty acid fractions in HeLa cells using GC-MS. We found that SCD1 knockdown led to increases of SFAs, 16:0 and 18:0, and decreases of MUFAs, 16:1n-7, 18:1n-9, and 18:1n-7, in phospholipids, which leads to a decrease in membrane phospholipid unsaturation. On the other hand, HeLa cells in culture possess very small amounts of free fatty acids, most of which are 16:0 and 18:0, and SCD1 knockdown did not affect the amounts or the composition in the free fatty acid fraction. We showed by using these SCD1 knockdown cells that the decrease in fatty acid unsaturation in the phospholipids induced the activation of UPR sensor proteins, IRE1 and PERK, and induced the mRNA expressions of CHOP and GRP78, which are downstream of the UPR pathway. Consistent with these results, Flowers et al. (34) showed that maintaining SCD1−/− mice on a very low fat diet induced ER stress in the liver, as indicated by enhanced splicing of XBP1 and elevated expression of several genes involved in the UPR pathways such as CHOP and GRP78.
In addition to UPR activation, SCD1 knockdown caused cell death in HeLa cells. Although cell death induced by SCD1 knockdown has been observed in human tumor cell lines (35), the mechanism has not been fully characterized. We showed that, using a pan-caspase inhibitor, apoptosis occurred in SCD1 knockdown cells. Moreover, we found the death of SCD1 knockdown cells is mediated at least in part by the PERK-CHOP arm of the UPR.
To further examine whether the UPR induction in SCD1 knockdown cells is due to the changes in specific fatty acid(s) or the degree of fatty acid unsaturation, we performed fatty acid supplementation experiments. Treatment of 18:1n-9, a major product of SCD1, completely suppressed the induction of UPR. The suppression effects were not specific to 18:1n-9, but similar effects were observed with PUFAs, confirming that UPR activation is induced by the degree of fatty acid unsaturation rather than by changes of the specific lipid products or substrate of SCD1.
The idea that the degree of unsaturation in the fatty acyl chains of membrane phospholipids is critical for stimulating UPR is further supported by the observation that knockdown of LPCAT3, which incorporates PUFAs into PC, synergistically enhanced UPR activation with SCD1 knockdown. Fatty acid composition of membrane phospholipids is determined by both the de novo pathway and the remodeling pathway (14–16). In the de novo pathway, glycerol 3-phosphate is sequentially acylated to form phosphatidic acid, which serves as a general precursor for all phospholipids. The newly synthesized phospholipids possess a mono- or di-unsaturated fatty acid at the sn-2 position. These phospholipids receive PUFAs to form mature membrane phospholipids by the remodeling pathway in which the sn-2 acyl chain is replaced with PUFAs by deacylation and reacylation. Several laboratories, including ours, have recently identified LPLATs that incorporate PUFAs into the sn-2 position of lysophospholipids (17). The acyltransferases involved in the remodeling pathway so far identified belong to either the 1-acylglycerol-3-phosphate O-acyltransferase family or membrane-bound O-acyltransferase family (36). LPCAT1 mainly incorporate SFA into PC to form disaturated PC, a component of lung surfactant (22, 23). LPCAT2 incorporates acetate or 20:4 into PC to form platelet-activating factor and its precursor (37). LPCAT3 exhibits broad substrate specificity in which PUFAs like 18:2 and 20:4 are incorporated into PC, phosphatidylethanolamine, and phosphatidylserine (18–20). LPCAT4 preferentially incorporates 18:1n-9 into PC and phosphatidylethanolamine (18). On the other hand, LPIAT1 is unique in that it incorporates 20:4 into only PI (21). Among these LPLATs, knockdown of only LPCAT3 synergistically enhanced UPR activation with SCD1 knockdown. LPCAT3 knockdown in SCD1 knockdown cells decreased 18:2, 20:4, and 20:5 in phospholipids, leading to a small but significant decrease in phospholipid unsaturation, which accounts for the increased UPR. The finding that LPCAT3 is a major acyltransferase that incorporates unsaturated fatty acids such as 18:1n-9, 18:2, and 20:4 into PC suggests that LPCAT3 has a role in determining membrane phospholipid unsaturation.
A more interesting finding was that the LPCAT3 mRNA level was significantly up-regulated by SCD1 knockdown. Consistent with this, the amounts of PUFAs such as 18:2 and 20:4, both of which are good substrates for LPCAT3, were increased in the phospholipid fraction of SCD1 knockdown cells, and LPCAT3 knockdown decreased the amount of 18:2 and 20:4. These results suggest that LPCAT3 compensates for the decreased synthesis of monounsaturated fatty acids from saturated fatty acids in SCD1 knockdown cells by incorporating PUFAs into PC. Although various LPLATs have recently been identified, little is known about how they are regulated. In a preliminary experiment we examined whether up-regulation of the LPCAT3 gene in SCD1 knockdown cells was dependent on UPR signaling, but single knockdown of either IRE1α, PERK, or activating transcription factor 6 did not cancel the LPCAT3 gene up-regulation. Activation of UPR is known to stimulate membrane phospholipid synthesis. Activation of XBP1 leads to an increase in choline cytidylyltransferase and choline phosphotransferase activities, both of which are involved in the cytidine-5′-diphosphate-choline pathway of PC synthesis (38, 39). Recent studies also revealed that ATF6α induces PC biosynthesis by increasing choline kinase and choline phosphotransferase activities (40). In fact, we also observed that the amount of phospholipid was increased about 1.3-fold in SCD1 knockdown cells (data not shown). Thus, activation of both de novo PC synthesis and LPCAT3-mediated fatty acid remodeling may compensate for the change in the fatty acid composition of phospholipids in SCD1 knockdown cells. Further studies are needed to clarify the signaling pathway that controls the LPCAT3 gene expression.
Accumulation of SFAs within cells causes lipotoxicity, which is considered as an underlying factor contributing to various pathological conditions such as insulin resistance, type 2 diabetes, and cardiovascular disease (24). A variety of tissue culture systems have been used to study mechanisms of lipotoxicity at the cellular level (28, 29, 41, 42). These studies generally implicate SFAs as inducers of cell death. Palmitic acid-induced cell death is characterized by markers of apoptosis, including cytochrome c release, caspase activation, and DNA fragmentation. These events may be initiated via several mechanisms, including decreased synthesis of cardiolipin, an abundant phospholipid in mitochondria (43), increased ceramide synthesis (44, 45), and increased generation of reactive oxygen species (29, 46). Palmitic acid may also act more directly at the level of ER to initiate a lipotoxic response. Borradaile et al. (47) reported that administered 16:0 is rapidly incorporated into lipid components of ER and impairs ER structure and integrity, suggesting that ER plays an important proximal role in palmitic acid-induced toxicity. In addition, SCD1, which is expressed mainly in the ER, is known to play a protective role in palmitic acid-induced toxicity. Increased expression of SCD1 suppresses palmitic acid-induced toxicity (48, 49), whereas inhibition of SCD1 enhances the toxicity (50). Consistent with these previous observations, SCD1 knockdown in HeLa cells enhanced palmitic acid-induced UPR (Fig. 5) and cell death (data not shown). Moreover, we demonstrated that LPCAT3 knockdown also synergistically enhanced palmitic acid-induced UPR. These results suggest that a decrease of the membrane phospholipid unsaturation contributes to palmitic acid-induced UPR. They also suggest that LPCAT3, in addition to SCD1, is a genetic factor that determines the extent of palmitic acid-induced toxicity.
Supplementary Material
Acknowledgments
We gratefully acknowledge technical assistance from Hideko Fukuda, Yuko Toyoda, and Michiko Kamio.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by Core Research for Evolutional Science and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S5.
- SCD
- stearoyl-CoA desaturase
- ATF6
- activating transcription factor 6
- CHOP
- C/EBP homologous protein
- ER
- endoplasmic reticulum
- GC-MS
- gas chromatography-mass spectrometry
- GRP78
- glucose-regulated protein 78
- IRE1
- inositol-requiring 1
- LPCAT
- lysophosphatidylcholine acyltransferase
- LPLAT
- lysophospholipid acyltransferase
- LPIAT1
- lysophosphatidylinositol acyltransferase 1
- MUFA
- monounsaturated fatty acid
- PC
- phosphatidylcholine
- PERK
- double-stranded RNA-activated kinase-like kinase
- PI
- phosphatidylinositol
- PUFA
- polyunsaturated fatty acid
- SFA
- saturated fatty acid
- siRNA
- small interfering RNA
- UPR
- unfolded protein response
- XBP1
- Xbox-binding protein 1
- X:Yn-Z
- fatty acid chain of X carbon atoms and Y methylene-interrupted cis bonds (Z indicates the position of the terminal double bond relative to the methyl end of the molecules)
- Z-VAD-FMK
- benzyloxycarbonyl-Val-Ala-Asp- fluoromethyl ketone
- Z-FA-FMK
- benzyloxycarbonyl-Phe-Ala-fluoromethyl ketone
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GRP78
- glucose-regulated protein 78.
REFERENCES
- 1.Lands W. E., Crawford C. G. (1976) Enzymes of Membrane Phospholipid Metabolism in Animals, 2nd Ed., pp. 3–85, Plenum Press, New York [Google Scholar]
- 2.Holub B. J., Kuksis A. (1978) Adv. Lipid Res 16, 1–125 [DOI] [PubMed] [Google Scholar]
- 3.MacDonald J. I., Sprecher H. (1991) Biochim. Biophys. Acta 1084, 105–121 [DOI] [PubMed] [Google Scholar]
- 4.Spector A. A., Yorek M. A. (1985) J. Lipid Res. 26, 1015–1035 [PubMed] [Google Scholar]
- 5.Clandinin M. T., Cheema S., Field C. J., Garg M. L., Venkatraman J., Clandinin T. R. (1991) FASEB J. 5, 2761–2769 [DOI] [PubMed] [Google Scholar]
- 6.Ntambi J. M. (1995) Prog. Lipid Res. 34, 139–150 [DOI] [PubMed] [Google Scholar]
- 7.Vigh L., Los D. A., Horváth I., Murata N. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9090–9094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba M., Suzuki I., Szalontai B., Kanesaki Y., Los D. A., Hayashi H., Murata N. (2003) J. Biol. Chem. 278, 12191–12198 [DOI] [PubMed] [Google Scholar]
- 9.Flowers M. T., Ntambi J. M. (2008) Curr. Opin. Lipidol. 19, 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki M., Kim Y. C., Gray-Keller M. P., Attie A. D., Ntambi J. M. (2000) J. Biol. Chem. 275, 30132–30138 [DOI] [PubMed] [Google Scholar]
- 11.Cohen P., Miyazaki M., Socci N. D., Hagge-Greenberg A., Liedtke W., Soukas A. A., Sharma R., Hudgins L. C., Ntambi J. M., Friedman J. M. (2002) Science 297, 240–243 [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki M., Man W. C., Ntambi J. M. (2001) J. Nutr. 131, 2260–2268 [DOI] [PubMed] [Google Scholar]
- 13.Collins J. M., Neville M. J., Hoppa M. B., Frayn K. N. (2010) J. Biol. Chem. 285, 6044–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy E. P., Weiss S. B. (1956) J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 15.Lands W. E. (1958) J. Biol. Chem. 231, 883–888 [PubMed] [Google Scholar]
- 16.Lands W. E. (2000) Biochim. Biophys. Acta 1483, 1–14 [DOI] [PubMed] [Google Scholar]
- 17.Shindou H., Hishikawa D., Harayama T., Yuki K., Shimizu T. (2009) J. Lipid Res. 50, S46–S51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2830–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y., Chen Y. Q., Bonacci T. M., Bredt D. S., Li S., Bensch W. R., Moller D. E., Kowala M., Konrad R. J., Cao G. (2008) J. Biol. Chem. 283, 8258–8265 [DOI] [PubMed] [Google Scholar]
- 20.Matsuda S., Inoue T., Lee H. C., Kono N., Tanaka F., Gengyo-Ando K., Mitani S., Arai H. (2008) Genes Cells 13, 879–888 [DOI] [PubMed] [Google Scholar]
- 21.Lee H. C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. (2008) Mol. Biol. Cell 19, 1174–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaffer J. E. (2003) Curr. Opin. Lipidol. 14, 281–287 [DOI] [PubMed] [Google Scholar]
- 25.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Yu L., Schmidt R. E., Su C., Huang X., Gould K., Cao G. (2005) Biochem. Biophys. Res. Commun. 332, 735–742 [DOI] [PubMed] [Google Scholar]
- 27.Kharroubi I., Ladrière L., Cardozo A. K., Dogusan Z., Cnop M., Eizirik D. L. (2004) Endocrinology 145, 5087–5096 [DOI] [PubMed] [Google Scholar]
- 28.Wei Y., Wang D., Topczewski F., Pagliassotti M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E275–E281 [DOI] [PubMed] [Google Scholar]
- 29.Borradaile N. M., Buhman K. K., Listenberger L. L., Magee C. J., Morimoto E. T., Ory D. S., Schaffer J. E. (2006) Mol. Biol. Cell 17, 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 32.Harding H. P., Zhang Y., Ron D. (1999) Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 33.van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flowers M. T., Keller M. P., Choi Y., Lan H., Kendziorski C., Ntambi J. M., Attie A. D. (2008) Physiol. Genomics 33, 361–372 [DOI] [PubMed] [Google Scholar]
- 35.Morgan-Lappe S. E., Tucker L. A., Huang X., Zhang Q., Sarthy A. V., Zakula D., Vernetti L., Schurdak M., Wang J., Fesik S. W. (2007) Cancer Res. 67, 4390–4398 [DOI] [PubMed] [Google Scholar]
- 36.Shindou H., Shimizu T. (2009) J. Biol. Chem. 284, 1–5 [DOI] [PubMed] [Google Scholar]
- 37.Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. (2007) J. Biol. Chem. 282, 6532–6539 [DOI] [PubMed] [Google Scholar]
- 38.Sriburi R., Jackowski S., Mori K., Brewer J. W. (2004) J. Cell Biol. 167, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sriburi R., Bommiasamy H., Buldak G. L., Robbins G. R., Frank M., Jackowski S., Brewer J. W. (2007) J. Biol. Chem. 282, 7024–7034 [DOI] [PubMed] [Google Scholar]
- 40.Bommiasamy H., Back S. H., Fagone P., Lee K., Meshinchi S., Vink E., Sriburi R., Frank M., Jackowski S., Kaufman R. J., Brewer J. W. (2009) J. Cell Sci. 122, 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo W., Wong S., Xie W., Lei T., Luo Z. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E576–E586 [DOI] [PubMed] [Google Scholar]
- 42.Almaguel F. G., Liu J. W., Pacheco F. J., Casiano C. A., De Leon M. (2009) J. Neurosci Res. 87, 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostrander D. B., Sparagna G. C., Amoscato A. A., McMillin J. B., Dowhan W. (2001) J. Biol. Chem. 276, 38061–38067 [DOI] [PubMed] [Google Scholar]
- 44.Sparagna G. C., Hickson-Bick D. L., Buja L. M., McMillin J. B. (2000) Am. J. Physiol. Heart Circ. Physiol. 279, H2124–H2132 [DOI] [PubMed] [Google Scholar]
- 45.Dyntar D., Eppenberger-Eberhardt M., Maedler K., Pruschy M., Eppenberger H. M., Spinas G. A., Donath M. Y. (2001) Diabetes 50, 2105–2113 [DOI] [PubMed] [Google Scholar]
- 46.Listenberger L. L., Ory D. S., Schaffer J. E. (2001) J. Biol. Chem. 276, 14890–14895 [DOI] [PubMed] [Google Scholar]
- 47.Borradaile N. M., Han X., Harp J. D., Gale S. E., Ory D. S., Schaffer J. E. (2006) J. Lipid Res. 47, 2726–2737 [DOI] [PubMed] [Google Scholar]
- 48.Peter A., Weigert C., Staiger H., Rittig K., Cegan A., Lutz P., Machicao F., Häring H. U., Schleicher E. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E339–E349 [DOI] [PubMed] [Google Scholar]
- 49.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Y., Wang D., Pagliassotti M. J. (2007) Mol. Cell Biochem. 303, 105–113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.