Abstract
Hepatic glycogen content is important for glucose homeostasis and exhibits robust circadian rhythms that peak at the end of the active phase in mammals. The activities of the rate-limiting enzymes for glycogenesis and glycogenolysis also show circadian rhythms, and the balance between them forms the circadian rhythm of the hepatic glycogen content. However, no direct evidence has yet implicated the circadian clock in the regulation of glycogen metabolism at the molecular level. We show here that a Clock gene mutation damps the circadian rhythm of the hepatic glycogen content, as well as the circadian mRNA and protein expression of Gys2 (glycogen synthase 2), which is the rate-limiting enzyme of glycogenesis in the liver. Transient reporter assays revealed that CLOCK drives the transcriptional activation of Gys2 via two tandemly located E-boxes. Chromatin immunoprecipitation assays of liver tissues revealed that CLOCK binds to these E-box elements in vivo, and real time reporter assays showed that these elements are sufficient for circadian Gys2 expression in vitro. Thus, CLOCK regulates the circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2.
Keywords: Cell Metabolism, Glucose Metabolism, Glycogen, Glycogen Synthase, Transcription, Transcription Regulation, CLOCK, Circadian Rhythms, Clock Genes
Introduction
Many organisms display physiological and behavioral rhythms that are entrained to the 24-h cycle of light and darkness on Earth. The mammalian master clock that resides in the suprachiasmatic nucleus controls most physical and physiological circadian rhythms (1, 2). The generation of circadian rhythms is dependent on the concerted co-expression of specific clock genes. Clock was the first clock gene in vertebrates to be identified by forward mutagenesis using N-ethyl-N-nitrosourea in a behavioral screening (3). The Clock gene encodes a basic helix-loop-helix-Per-Arnt-Sim transcription factor (3, 4). CLOCK binds DNA and activates transcription after dimerization with BMAL1 (1, 2). The CLOCK-BMAL1 heterodimer drives the rhythmic transcription of other clock genes (such as period and cryptochrome) and circadian clock-controlled genes through an E-box (CACGTG). Because the Clock allele of Clock mutant mice is truncated and causes a deletion of 51 amino acids, the mutation presumably would not have a significant effect on the N-terminal basic helix-loop-helix and Per-Arnt-Sim domains, leaving CLOCK dimerization and DNA binding intact. The mutant CLOCK protein can still form heterodimers with BMAL1 that bind to DNA, but these heterodimers are deficient in transactivation (2). Genes relevant to the clock genes are also expressed in peripheral tissues such as the liver, heart, and lung (5). The mammalian circadian clock is an intracellular, transcriptional-translational mechanism that shares the same molecular components in the suprachiasmatic nucleus and in peripheral cells.
Hundreds of tissue-specific circadian genes that regulate an impressive diversity of biological processes have been identified using DNA microarray technology (6–8), and we identified putative CLOCK target genes in the mouse liver using microarray analyses (9). The screened genes encoded various key physical molecules associated with glucose and lipid metabolism. Recent studies have shown that mutations or deletions of the Clock and Bmal1 genes result in not only circadian disturbances but also metabolic abnormalities of glucose and lipid metabolism (10–13). These findings suggested that Clock is involved in glucose and lipid metabolism in peripheral tissues in addition to being a core component of the circadian clock.
The liver is mainly responsible for maintaining blood glucose levels within a narrow range by its ability to store glucose as glycogen and to produce glucose from glycogen breakdown or gluconeogenic precursors. Many genes associated with glucose metabolism in the liver exhibit robust circadian regulation (7, 8, 14), suggesting that the circadian clock plays a significant role in hepatic glucose metabolism. However, little direct evidence implicates the circadian clock in the regulation of glucose metabolism at the molecular level. Hepatic glycogen contents are important for glucose homeostasis, and they exhibit a circadian rhythm that peaks during the dark-light transition in nocturnal rodents (15). The activities of glycogen synthase and glycogen phosphorylase (the rate-limiting enzymes for glycogenesis and glycogenolysis) also show circadian variation, and the balance between them forms the basis for circadian variation in the hepatic glycogen content (15). Glycogen synthase 2 (Gys2) is the rate-limiting enzyme for glycogenesis, and it is predominantly expressed in the liver, whereas another isoform, Gys1, is widely expressed in fat, heart, and muscle but not at all in the liver (16). The activity of GYS2 displays circadian rhythms that peak late at night in nocturnal rodents. The activity is controlled by the reversible phosphorylation of several serine residues that results in inactivation (17). The rhythmic modification of GYS2 through hormonal signals such as those of insulin, glucagon, and glucocorticoids might regulate the circadian variation of hepatic glycogen synthesis (18). However, circadian variation in hepatic glycogen synthesis activity continues in starved intact and adrenalectomized rats (15, 19). Thus, the circadian regulation of glycogen synthesis is not fully understood at the molecular level. Here, we show that CLOCK regulates the circadian variation of hepatic glycogen synthesis through the direct transcriptional activation of Gys2.
EXPERIMENTAL PROCEDURES
Animals
Male Jcl:ICR (Clea Japan Inc., Tokyo, Japan) and homozygous Clock mutant mice on a Jcl:ICR background (20) at 7–10 weeks of age were maintained under a 12 h of light/12 h of dark cycle (lights on at 0:00 and lights off at 12:00) for at least 2 weeks before the day of the experiments. The mice were fasted overnight and sequentially sacrificed for some experiments. All of the animal experiments, care, and handling proceeded with the approval of our institutional Animal Care and Use Committee (Permission Number 2009-020).
Hepatic Glycogen Content
Frozen liver tissues (∼200 mg) were homogenized in 10% trichloroacetic acid, and then 1 ml of homogenate was separated by centrifugation at 17,800 × g for 10 min at 4 °C. The supernatant (300 μl) was mixed with 600 μl of ethanol, placed on ice for 5 min, and then separated again by centrifugation at 19,000 × g for 10 min at 4 °C. The precipitate was dissolved in 100 μl of distilled water and then mixed with 100 μl of 4 m H2SO4. Each sample was incubated for 30 min at 95 °C, then 200 μl of 2 m NaOH was added, and the mixture was neutralized with 1 m Tris-HCl (pH 8.0). The hydrolyzed free glucose concentration was evaluated using a glucose C-test Wako (Wako Pure Chemical Industries, Osaka, Japan) and corrected by liver weight.
Blood Metabolic Parameters
Plasma glucose was measured using a glucose C-test Wako (Wako Pure Chemical Industries) according to the manufacturer's protocol.
Histology
Liver tissues from wild-type and Clock mutant mice at ZT 2 were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Japan Co., Ltd., Tokyo) and frozen. Glycogen accumulation was analyzed in 10-μm cryosections of frozen liver using the periodic acid-Schiff's reaction kit (Muto Pure Chemicals Co. Ltd., Tokyo, Japan).
Isolation of RNA and Real Time Quantitative Reverse Transcription-PCR
Total RNA was isolated from liver tissues using RNAiso (TAKARA Bio Inc., Shiga, Japan) and then reverse-transcribed using the PrimeScript RT reagent kit (TAKARA Bio) according to the manufacturer's protocol. The cDNA levels of genes of interest were measured by real time quantitative PCR using a LightCycler (Roche Applied Science) with SYBR Premix Ex Taq (TAKARA Bio). Table 1 shows the sequences of the primer pairs. The amount of mRNA was corrected relative to that of β-actin. The maximal value for wild-type mice is expressed as 100%. The values are the means ± S.E. (n = 3).
TABLE 1.
Primer sequences for real time reverse transcription-PCR
| Gene | Forward and reverse sequences |
|---|---|
| Gys2 | 5′-CCAGCTTGACAAGTTCGACA-3′ |
| 5′-ATCAGGCTTCCTCTTCAGCA-3′ | |
| Glycogen phosphorylase (Gp) | 5′-CACTTACCAGCTGGGCTTGGACAT-3′ |
| 5′-AAAGCAAGCTGCCAGGCGTC-3′ | |
| Glucokinase (Gk) | 5′-GATCCGGGAAGAGAAGCAAG-3′ |
| 5′-GACAGGGATGAGGGACAGAG-3′ | |
| Phosphoenolpyruvate carboxykinase (Pepck) | 5′-GTGGGCGATGACATTGCC-3′ |
| 5′-ACTGAGGTGCCAGGAGCAAC-3′ | |
| Glucose-6-phosphatase (G6pase) | 5′-TGGTAGCCCTGTCTTTCTTTG-3′ |
| 5′-TTCCAGCATTCACACTTTCCT-3′ | |
| Glucose transporter 2 (Glut2) | 5′-GCAACTGGGTCTGCAATTTT-3′ |
| 5′-CCAGCGAAGAGGAAGAACAC-3′ | |
| mPer1 | 5′-CCAGATTGGTGGAGGTTACTGAGT-3′ |
| 5′-GCGAGAGTCTTCTTGGAGCAGTAG-3′ | |
| mPer2 | 5′-TTCCACTATGTGACAGCGGAGG-3′ |
| 5′-CGTATCCATTCATGTCGGGCTC-3′ | |
| D site albumin promoter binding protein (Dbp) | 5′-GGAACTGAAGCCTCAACCAAT-3′ |
| 5′-CTCCGGCTCCAGTACTTCTCA-3′ | |
| β-actin | 5′-CACACCTTCTACAATGAGCTGC-3′ |
| 5′-CATGATCTGGGTCATCTTTTCA-3′ |
Western Blotting
Frozen livers were homogenized in ice-cold lysis buffer (5 mm Tris-HCl, pH 7.5, 15 mm NaCl, 0.1% Nonidet-40, and proteinase inhibitor mixture (Roche Applied Science)). The lysates were boiled in 2× SDS sample buffer at 95 °C for 5 min. Total protein (100 μg) was resolved by SDS-PAGE on 7.5% polyacrylamide gels and then transferred onto nitrocellulose membranes (Bio-Rad). Nonspecific protein binding was blocked using 3% dried skim milk in phosphate-buffered saline (17). The proteins on the membranes were immunoblotted against anti-GYS2 antiserum (gift from Banyu Pharmaceutical Co., Ltd.) and anti-ACTIN antibody (Nihon Millipore K.K, Tokyo, Japan). Immunoreactive bands were visualized using horseradish peroxidase-conjugated secondary antibodies and the ECL detection system (GE Healthcare). The amount of protein was corrected relative to that of ACTIN. The maximal value for wild-type mice is expressed as 100%. The values are described as the means ± S.E. (n = 3).
Cell Culture
We cultured NIH3T3 cells in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and a mixture of penicillin and streptomycin at 37 °C under a humidified 5% CO2 atmosphere.
Transient Luciferase Assays
The first intron of Gys2 containing two E-boxes (+1170 to +2046) and their mutations were cloned into the pGL3-Promoter vector (Promega). Mouse CLOCK, BMAL1, and CRY1 expression plasmids were provided by Dr. T. Todo (21). Constructs (500 ng) were co-transfected with 1 ng of pRL-CMV (Promega) as the internal control into NIH3T3 cells (24-well plates) using HilyMax (DOJINDO Laboratories, Kumamoto, Japan) according to the manufacturer's protocols. Luciferase activities were measured using a dual luciferase reporter assay system (Promega) and a Luminometer model TD-20/20 (Turner Designs, Sunnyvale, CA). The transcriptional activities were normalized relative to Renilla luciferase activities.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation was adapted from the reported procedure (22). Fresh liver tissues from mice at ZT14 were cross-linked and homogenized in 1% formaldehyde for 15 min at room temperature. Cross-linking was stopped by adding 125 mm glycine. The homogenates were lysed with 25 mm Tris-HCl (pH 8.0), 140 mm NaCl, 1% Triton X-100, 0.1% SDS, 3 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride (lysis buffer) on ice for 30 min. The lysates were sonicated on ice and then incubated with anti-CLOCK antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) overnight at 4 °C, followed by protein A/G-agarose (Santa Cruz Biotechnology). Chromatin immunocomplexes were washed once each at 4 °C for 5 min with wash buffers 1, 2, and 3 (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% SDS, 1% Triton X-100 and 2 mm EDTA; 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 0.1% SDS, 1% Triton X-100 and 2 mm EDTA; and 10 mm Tris-HCl, pH 8.0, 0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, and 1 mm EDTA, respectively). The samples were finally washed twice with TE buffer. The immunocomplexes were removed with 1% SDS and 0.1 m NaHCO3 and then heated overnight at 65 °C to reverse the cross-links. The cross-links of DNA input samples were similarly reversed. Sample DNA was isolated from the immunoprecipitates and then amplified by PCR using the following primer sets: upstream region (from −2864 to −2620), 5′-TCA CTG TTT GCT CTA CTT TAT ATC C-3′ and 5′-AAG AGT CTT AAC GAA TAC TCA GCC-3′; E1E2 site on the first intron (from +1541 to +1804), 5′-TGT CTC ACA AAG CAA AGT CAA CAG G-3′ and 5′-CTG AGG GCG TCT AAA CAG AGG AGC-3′; and E-box like that of Per2 promoter (23), 5′-GGT TCC GCC CCG CCA GTA TGC-3′ and 5′-CCG TCA CTT GGT GCG CTC GGC-3′.
Real Time Luciferase Assays
The first intron of Gys2 containing two E-boxes and their mutations were cloned into SV40-dLuc that contains the SV-40 promoter and a rapid degradation domain modified from mouse ornithine decarboxylase at the C-terminal end of firefly luciferase (8). The Per2 promoter regions were cloned into pGL3-dLuc (24), and then 2 μg of reporter plasmids were transfected into NIH3T3 cells (35-mm collagen type Ι-coated dish) using HilyMax (DOJINDO Laboratories, Kumamoto, Japan). The cells were stimulated with 100 nm dexamethasone (Sigma-Aldrich) for 2 h in serum-free Dulbecco's modified Eagle's medium, and then the medium was replaced with fresh Dulbecco's modified Eagle's medium containing 100 μm luciferin (Wako Pure Chemical Industries), 25 mm HEPES (pH 7.2), and 10% fetal bovine serum. Bioluminescence was measured and integrated for 1 min at intervals of 10 min using Kronos AB-2500 (ATTO, Tokyo, Japan). The cells were cultured in a luminometer for >3 days to evaluate bioluminescence. To compare the phase and amplitude of Gys2-WT-dLuc and each mutant, the data were detrended by subtracting an average of 12 h from the data.
Statistics
Group variations were statistically analyzed using the one-way or two-way analysis of variance followed by the post-hoc Student's t test. All of the data are expressed as the means ± S.E., with a statistically significant difference defined as a value of p < 0.05.
RESULTS
Clock Mutation Affects Temporal Expression of Genes Associated with Glucose Metabolism
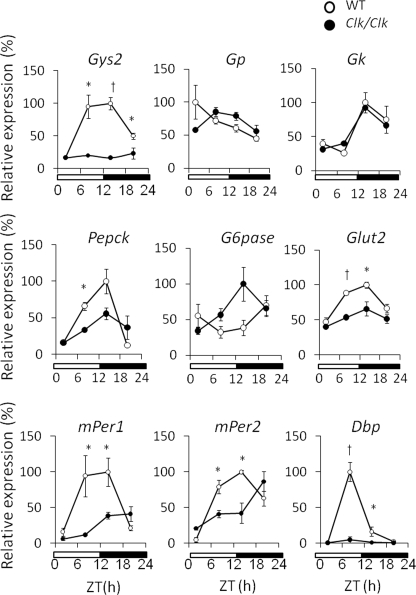
We examined the temporal expression profile of genes associated with glucose metabolism in the liver of wild-type and Clock mutant mice to determine the relationship between the circadian clock and glucose metabolism (Fig. 1). The circadian expression of mPeriod genes was attenuated and/or phase-shifted in the Clock mutant mouse liver as described (25), whereas that of Dbp was completely damped. Most genes associated with glucose metabolism were expressed in a circadian manner in the livers of wild-type mice. The circadian expression of Gys2 was remarkably damped, whereas the expression of Pepck and Glut2 was slightly damped in the livers of Clock mutant mice. Rhythmic expression of metabolic genes was not always damped in Clock mutant mice. For example, the temporal expression profile of G6pase was almost antiphasic compared with that in wild-type mice.
FIGURE 1.
Clock mutation affects the temporal expression of genes associated with glucose metabolism. The mice were maintained under a 12 h of light/12 h of dark cycle (lights on at 0:00 and lights off at 12:00). Total RNA was extracted from the livers of wild-type (WT) and Clock mutant mice, and the mRNA levels were quantified by real time reverse transcription-PCR. Open and closed symbols, wild-type and Clock mutant mice, respectively. Amount of mRNA was corrected relative to that of β-actin. The maximal value for wild-type mice is expressed as 100%. The values are the means ± S.E. (n = 3). The significant differences compared with values from wild-type mice at each time point are indicated. *, p < 0.05; †, p < 0.01. The p values were calculated using Student's t test.
Clock Mutation Damps Circadian Variation in GYS2 Protein under Both Fed and Fasted States
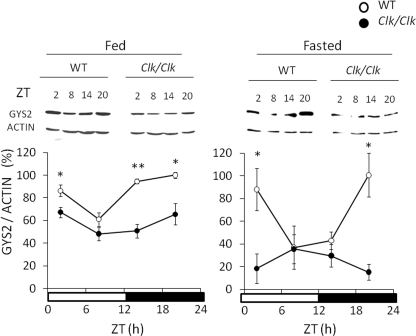
We isolated proteins from the livers of wild-type and Clock mutant mice to determine circadian variation in GYS2 protein using Western blotting analysis (Fig. 2). Levels of GYS2 protein fluctuated in a robust circadian manner that increased during the night in the wild-type mouse liver. Circadian fluctuation of GYS2 was significantly attenuated at low levels in Clock mutant mice. We examined the effects of fasting on the circadian variation of GYS2 protein, which was also maintained in a CLOCK-dependent manner under the fasting state (Fig. 2), and starvation increased the levels during the dark-light transition compared with the fed state (supplemental Fig. S1).
FIGURE 2.
Clock mutation damps circadian variation of GYS2 protein in both fed and fasted mice. Wild-type (WT) and Clock mutant mice were maintained under a 12 h of light/12 h of dark cycle (lights on at 0:00 and lights off at 12:00) and fed (left panel) or fasted (right panel) overnight. Protein was extracted from the livers of these mice, and then the GYS2 protein level was determined by Western blotting using anti-GYS2 antiserum. Open and closed symbols, wild-type and Clock mutant mice, respectively. The maximal value for wild-type mice is expressed as 100%. The values are the means ± S.E. (n = 3). The significant differences compared with the values from wild-type mice at each time point are indicated. *, p < 0.05; †, p < 0.01. The p values were calculated using Student's t test.
Clock Mutation Damps Circadian Variation of Hepatic Glycogen Contents
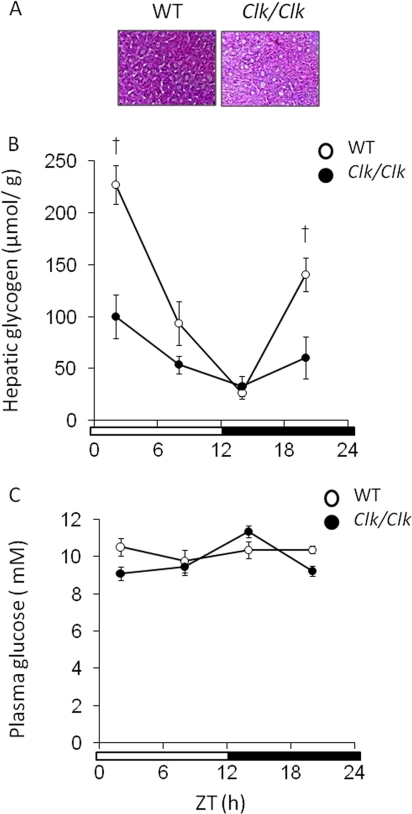
We examined the hepatic glycogen contents and plasma glucose levels in wild-type and Clock mutant mice (Fig. 3). Clock mutation damped the robust circadian rhythm of hepatic glycogen contents that peaked at the time of the dark-light transition in wild-type mice (Fig. 3, A and B). Plasma glucose levels remained constant in wild-type mice (Fig. 3B; one-way analysis of variance, p = 0.941; 38) but significantly fluctuated in Clock mutant mice (Fig. 3B; one-way analysis of variance, p = 0.034).
FIGURE 3.
Clock mutation damps circadian variation of hepatic glycogen contents. The mice were maintained under a 12 h of light/12 h of dark cycle (lights on at 0:00 and lights off at 12:00). Open and closed symbols, wild-type (WT) and Clock mutant mice, respectively. A, periodic acid-Schiff's staining of liver sampled at ZT 2 of wild-type mice and Clock mutant mice (n = 3). B, hepatic glycogen contents in wild-type mice and Clock mutant mice were measured at indicated times. The values are the means ± S.E. (n = 8). C, plasma glucose concentrations. The values are the means ± S.E. (n = 8). The significant differences compared with values from wild-type mice at each time point are indicated. †, p < 0.01. The p values were calculated using Student's t test.
Circadian Expression of Gys2 Continues during Fasting
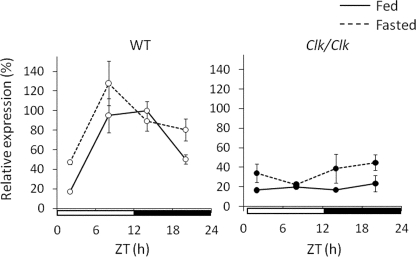
We examined the effects of fasting on the circadian expression of Gys2 (Fig. 4). Expression levels of Gys2 mRNA were not statistically different between fed and fasted wild-type mice throughout the day (two-way analysis of variance, p = 0.141). The circadian expression of Gys2 continued in the livers of wild-type mice under both fed and fasted conditions, whereas Clock mutation damped such expression under both conditions.
FIGURE 4.
Circadian expression of Gys2 continues during fasting. Wild-type (WT) and Clock mutant mice were maintained under a 12 h of light/12 h of dark cycle (lights on at 0:00 and lights off at 12:00) and fed (left panel) and fasted (right panel) overnight. Total RNA extracted from the livers of these mice was simultaneously quantified by real time reverse transcription-PCR. The data from fed mice of both genotypes are identical to those shown in Fig. 1. Open and closed symbols, wild-type and Clock mutant mice, respectively. The solid and dotted lines indicate fed and fasted mice, respectively. The amount of mRNA was corrected relative to that of β-actin. The maximal value for wild-type mice is expressed as 100%. The values are the means ± S.E. (n = 3).
CLOCK Drives the Transcriptional Activation of Gys2 via Two E-boxes in the First Intron
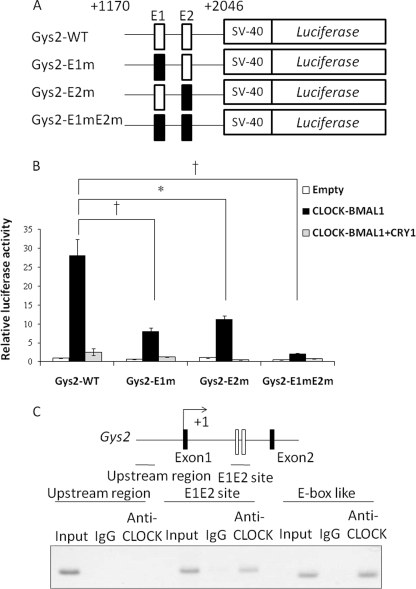
Database analysis could not identify a perfect E-box (CACGTG) within 3 kb of the 5′-flanking region of Gys2 gene. However, we found two perfect tandemly located E-box motifs (sites E1 and E2) in the first intron of the Gys2 gene (Fig. 5A). We therefore analyzed the functions of these E-box elements using a sequence of this region fused to the luciferase reporter plasmid. The transcriptional activity of an 877-bp fragment containing the E-box elements in the first intron was increased >28-fold following CLOCK and BMAL1 overexpression (Fig. 5B). This increase was severely suppressed by co-expression with CRY1, which is the negative component of CLOCK-BMAL1-dependent transcriptional activation (1). Mutation in either of the E1 and E2 sites decreased, and both mutations significantly suppressed CLOCK-BMAL1-dependent transcriptional activation. We analyzed the binding affinity of CLOCK for the putative DNA sequence in vivo using chromatin immunoprecipitation assays of the mouse liver at ZT14. The positive control for chromatin immunoprecipitation using an anti-CLOCK antibody was an E-box like that of Per2 (23). The results showed that CLOCK bound to the first intron region of Gys2 containing the two E-boxes (Fig. 5C).
FIGURE 5.
CLOCK drives transcriptional activation of Gys2 via two E-boxes in the first intron. A, schematic representation of mutant constructs of Gys2 first intronic region. Each or both E-boxes (sites E1 and E2) were mutated (E1 site, 5′-CACGTG-3′ to 5′-CACCAC-3′; E2 site, 5′-CACGTG-3′ to 5′-CACCAC-3′). B, analysis of E1 and E2 sites in the first Gys2 intronic region. Transcriptional assays included the indicated mutant constructs. Gys2-WT, wild type of first intron on Gys2 gene; Gys2-E1m, mutant E1 site; Gys2-E2m, mutant E2 site; Gys2-E1mE2m, mutant E1 and E2 sites. The normalized expression level was calculated relative to luciferase activity in empty vector. The values are described as the means ± S.E. (n = 3). Significant differences are indicated. *, p < 0.05; †, p < 0.01. p values were calculated using Student's t tests. The results are representative of three independent experiments. C, CLOCK binds to the first intronic region containing E1 and E2 sites in vivo. The chromatin immunoprecipitation assays were performed using livers from mice at ZT14. The region containing two E-boxes is shown as an open box. Horizontal bars, amplified regions (upstream region, from −2864 to −2620; E1E2 site on first intron, from +1541 to +1804). The E-box (CACGTT) on Per2 promoter known as CLOCK binding region served as a positive control. IgG, with mouse normal IgG; Anti-CLOCK, with anti-CLOCK antibody.
E1 and E2 Sites Are Sufficient for the Circadian Expression of Gys2
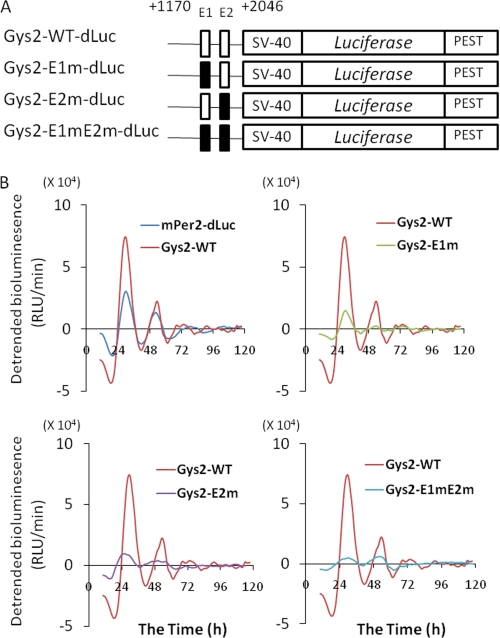
We performed real time reporter assays using NIH3T3 cells containing functional circadian clock components to clarify how the E1 and E2 sites are involved in the circadian regulation of Gys2 expression (26, 27). The transcriptional activity of this construct containing two E-boxes (Gys2-WT-dLuc) exhibited circadian oscillation as well as Per2-dLuc (Fig. 6B). On the other hand, the amplitude was robustly reduced by mutations of the E-boxes (Fig. 6B). These findings showed that both E-box elements are sufficient for the circadian expression of Gys2 as well as Per2 in NIH3T3 cells.
FIGURE 6.
Sites E1 and E2 are sufficient for circadian expression of Gys2. A, schematic representation of mutant constructs of Gys2 first intronic region. Each or both E-boxes (sites E1 and E2) were mutated (E1, 5′-CACGTG-3′ to 5′-CACCAC-3′; E2, 5′-CACGTG-3′ to 5′-CACCAC-3′). B, real time reporter assay. NIH3T3 cells were transfected with indicated mutant constructs and incubated with 100 nm of dexamethasone, and then bioluminescence was measured. The results are representative of three independent experiments that generated similar outcomes. Gys2-WT-dLuc, wild type of first intron on Gys2 gene; Gys2-E1m-dLuc, mutant E1 site; Gys2-E2m-dLuc, mutant E2 site; Gys2-E1mE2m-dLuc, mutant E1 and E2 sites (see “Experimental Procedures” for details of methods for analyzing circadian rhythms).
DISCUSSION
Recent studies have shown that mutations of the Clock and Bmal1 genes result not only in circadian behavioral abnormalities but also in metabolic disruption of glucose homeostasis (11, 28). However, a role for the circadian clock in the regulation of glucose metabolism at the molecular level has not been thoroughly substantiated. Here we found that the circadian expression of Gys2 (both mRNA and protein levels) was damped in the livers of Clock mutant, compared with wild-type mice. Circadian variations in the hepatic glycogen contents of wild-type mice were damped in Clock mutant mice. We also showed that two E-boxes located in the first intron of Gys2 are involved in the circadian expression of Gys2. These results suggest that the circadian clock in the liver regulates hepatic glycogen synthesis through the circadian transcriptional activation of Gys2.
GLUT2 is an insulin-independent glucose transporter. The circadian expression of Glut2 was damped in the livers of Clock mutant, compared with wild-type mice (Fig. 1), a situation resembling that of liver-specific Bmal1 knock-out mice (12). Tef (thyrotroph embryonic factor) is involved in Glut2 expression (29). We showed that the circadian rhythm of Tef expression is damped in the livers of Clock mutant, compared with wild-type mice (9). These findings suggest that CLOCK and BMAL1 regulate the circadian expression of Glut2 via the circadian regulation of Tef expression. Pepck encodes the key rate-limiting enzyme of gluconeogenesis (30, 31), and the activity is blunted in the Clock mutant mouse liver (11). We found that the Clock mutation slightly damped the circadian Pepck expression in the wild-type mouse liver (Fig. 1). Gluconeogenesis might be impaired in Clock mutant animals. The temporal expression profiles of several genes associated with glucose metabolism such as Gk and G6pase remained rhythmic in Clock mutant mice, suggesting that CLOCK is not always essential for the rhythmic expression of these genes, at least in vivo.
The expression of mPer2 remains circadian in the Clock mutant mouse liver (Ref. 25 and the present study). One possible explanation for this is that NPAS2, an analog of CLOCK protein, compensates for mutated CLOCK in these mice as described (32, 33). Another possibility is that the molecular components of the peripheral clock are not necessarily essential for the circadian expression of some clock genes such as mPer1 and mPer2 in vivo (12).
Gys2 encodes the key rate-limiting enzyme of glycogenesis (18, 34). The Clock mutation obviously damped circadian expression of Gys2 mRNA to the lowest levels and also damped GYS2 protein in the livers of wild-type mice (Fig. 2). The circadian rhythm of GYS2 activity continues during starvation in the rat liver (15), suggesting that the circadian regulation of GYS2 activity is governed by the endogenous circadian clock. Consistent with this notion, circadian Gys2 expression (both mRNA and protein levels) was maintained under starvation in a CLOCK-dependent manner (Figs. 2 and 4). These results suggest that the transcriptional activation of Gys2 by the circadian clock is involved in the circadian variation of GYS2 activity.
Here, we simultaneously evaluated the mRNA and protein levels of GYS2 accompanied by hepatic glycogen levels (supplemental Table S1). The acrophase of mRNA, protein, and glycogen levels obviously differed (12.1, 19.2 and 1.1 h, respectively). Feeding-dependent hormonal regulation of GYS2 activity seems to also be important for the circadian profile of hepatic glycogen contents. The acrophase of circadian Gys2 expression in wild-type mice seems to be similar to that of Dbp and Per2. The circadian expression of Dbp and Per2 is directly regulated by CLOCK via E-box elements (21, 35). These facts imply that CLOCK is responsible for the circadian expression of Gys2 via E-box elements. A database analysis did not identify a perfect E-box within 3 kb of the 5′-flanking region of the Gys2 gene. However, we found two E-boxes within the first intron of Gys2 (Fig. 5A). Recent studies suggest that intronic regions play important roles in the circadian expression of CLOCK-BMAL1 target genes such as Dbp (32) and Pparα (peroxisome proliferator-activated receptor α) (36). Therefore, we analyzed the function of these E-boxes and found that they are additively involved in CLOCK-BMAL1-dependent transcriptional activation (Fig. 5B). We also found that CLOCK bound to this region in the mouse liver (Fig. 5C), suggesting that CLOCK regulates Gys2 expression via two E-boxes in vivo (Fig. 5, B and C).
PPARα is a ligand-activated transcriptional factor belonging to the superfamily of nuclear receptors, and it is involved in the regulation of glucose and lipid metabolism (37). Fasting activates PPARα (37). One study has suggested that PPARα up-regulates Gys2 expression via the PPAR response element on the first intron of Gys2 (38). Expression levels of Gys2 are obviously reduced in the livers of Pparα knock-out mice only when fasting (39). We found that a Pparα deficiency did not affect the day/night fluctuation of Gys2 in the liver in fed mice (supplemental Fig. S2), suggesting that PPARα is not involved in circadian Gys2 expression, at least in the livers of fed mice.
Comparative genomics approaches to analyzing E-boxes have recently highlighted the importance of both the core consensus and flanking sequences in circadian expression control (40), yet genomic E-box elements are not always functional for circadian transcription (7). Therefore, we performed real time reporter assays to determine whether the two E-boxes are functional for the circadian expression of Gys2. We found that the bioluminescence of Gys2-WT exhibits circadian rhythm at phases similar to those of Per2-dLuc in a manner dependent on both intronic E-boxes (Fig. 6), suggesting that they are sufficient for the circadian expression of Gys2.
The liver is mostly responsible for maintaining blood glucose levels within a narrow range by its ability to store glucose as glycogen and to produce glucose from glycogen breakdown or gluconeogenic precursors (41, 42). Therefore, hepatic glycogen content is important for glucose homeostasis. We showed here that a Clock mutation affects hepatic glycogen metabolism. Circadian changes in hepatic glycogen contents were damped in the livers of Clock mutant mice (Fig. 3). Plasma glucose levels in Clock mutant mice significantly fluctuated, whereas they remained constant in wild-type mice (Fig. 3C). These results imply that a Clock mutation disrupts glucose homeostasis. Lamia et al. (12) notably found that resting blood glucose is significantly lower and that hepatic glycogen contents are possibly reduced in liver-specific Bmal1 knock-out mice. The circadian rhythms of glycogen contents might be damped in these Bmal1 knock-out mice, because the glycogen contents were examined only at the time of the trough (ZT 7–9). The circadian clock in the liver might partly contribute glucose homeostasis through the regulation of hepatic glycogen contents.
Although the circadian expression of Glut2 is damped in the livers of liver-specific Bmal1 knock-out mice as well as in Clock mutant mice (Fig. 1), glycogen levels are normal in the livers of Glut2 knock-out mice (43). These observations suggest that a reduction in Glut2 expression does not affect glycogen levels in the livers of liver-specific Bmal1 knock-out and Clock mutant mice.
Notably, plasma glucagon levels were significantly increased late at night in Clock mutant mice compared with those in wild-type mice (supplemental Fig. S3). Blood glucose levels in Clock mutant mice might be affected by inducing glucagon secretion. Insulin plays an important role in hepatic glycogen synthesis by inactivating glycogen synthase kinase-3 by phosphorylation, which results in less GYS2 phosphorylation, thus contributing to enhanced glycogen synthesis (18). Our results revealed similar insulin levels in Clock mutant and wild-type mice (supplemental Fig. S3). Hepatic glycogen levels slightly fluctuated in Clock mutant mice, although the mRNA expression levels of Gys2 were completely damped. Feeding-dependent rhythmic insulin secretion might also be involved in the fluctuation of hepatic glycogen levels in the Clock mutant mouse liver, whereas the circadian clock might regulate hepatic glycogen synthesis through the circadian regulation of glycogen synthase kinase-3β activity. We showed that glycogen synthase kinase-3β phosphorylation exhibits circadian rhythm not only in vivo but also in vitro (44), suggesting that the endogenous clock is involved in the circadian regulation of glycogen synthase kinase-3β activity. The circadian clock seems to regulate circadian glycogen synthesis by post-translationally regulating GYS2 as well as transcriptionally activating its mRNA expression through clock molecules.
Thus, our results suggest that the circadian clock in liver regulates hepatic glycogen synthesis through the circadian transcriptional activation of Gys2. CLOCK-BMAL1-dependent transcriptional activation drives the circadian variation of GYS2 protein that peaks during the night in accord with GYS2 activation via the insulin signal, thereby producing effective hepatic glycogen synthesis.
Recent studies have revealed an association between the circadian clock and metabolic diseases such as metabolic syndrome and diabetes (10, 45). The rates of glycogen synthesis and the glycogen contents in the livers of patients with diabetes are significantly decreased (46). Our findings might provide new insight into understanding the relationships between circadian clock and metabolic diseases.
Supplementary Material
Acknowledgments
We are grateful to Banyu Pharmaceutical Co., Ltd. for providing anti-GYS2 antiserum and to Dr. Takahashi Todo (Kyoto University, Kyoto, Japan) for providing the expression plasmids. We thank Drs. Yoshiaki Onishi and Koyomi Miyazaki (Biological Clock Research Group, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan) for helpful discussions.
Footnotes
This work was supported in part by Grant-in-Aid for Young Scientists (B) 18770057 (to K. O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
REFERENCES
- 1.Pando M. P., Sassone-Corsi P. (2001) Nature 410, 311–313 [DOI] [PubMed] [Google Scholar]
- 2.Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 [DOI] [PubMed] [Google Scholar]
- 3.Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., Takahashi J. S. (1994) Science 264, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King D. P., Zhao Y., Sangoram A. M., Wilsbacher L. D., Tanaka M., Antoch M. P., Steeves T. D., Vitaterna M. H., Kornhauser J. M., Lowrey P. L., Turek F. W., Takahashi J. S. (1997) Cell 89, 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto K., Nagase T., Fukui H., Horikawa K., Okada T., Tanaka H., Sato K., Miyake Y., Ohara O., Kako K., Ishida N. (1998) J. Biol. Chem. 273, 27039–27042 [DOI] [PubMed] [Google Scholar]
- 6.Akhtar R. A., Reddy A. B., Maywood E. S., Clayton J. D., King V. M., Smith A. G., Gant T. W., Hastings M. H., Kyriacou C. P. (2002) Curr. Biol. 12, 540–550 [DOI] [PubMed] [Google Scholar]
- 7.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 8.Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., Hashimoto S. (2002) Nature 418, 534–539 [DOI] [PubMed] [Google Scholar]
- 9.Oishi K., Miyazaki K., Kadota K., Kikuno R., Nagase T., Atsumi G., Ohkura N., Azama T., Mesaki M., Yukimasa S., Kobayashi H., Iitaka C., Umehara T., Horikoshi M., Kudo T., Shimizu Y., Yano M., Monden M., Machida K., Matsuda J., Horie S., Todo T., Ishida N. (2003) J. Biol. Chem. 278, 41519–41527 [DOI] [PubMed] [Google Scholar]
- 10.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., Fitzgerald G. A. (2004) PLoS Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamia K. A., Storch K. F., Weitz C. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oishi K., Atsumi G., Sugiyama S., Kodomari I., Kasamatsu M., Machida K., Ishida N. (2006) FEBS Lett. 580, 127–130 [DOI] [PubMed] [Google Scholar]
- 14.Miller B. H., McDearmon E. L., Panda S., Hayes K. R., Zhang J., Andrews J. L., Antoch M. P., Walker J. R., Esser K. A., Hogenesch J. B., Takahashi J. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa K., Shimazu T. (1976) Life Sci. 19, 1873–1878 [DOI] [PubMed] [Google Scholar]
- 16.Ferrer J. C., Favre C., Gomis R. R., Fernández-Novell J. M., García-Rocha M., de la Iglesia N., Cid E., Guinovart J. J. (2003) FEBS Lett. 546, 127–132 [DOI] [PubMed] [Google Scholar]
- 17.Kadotani A., Fujimura M., Nakamura T., Ohyama S., Harada N., Maruki H., Tamai Y., Kanatani A., Eiki J., Nagata Y. (2007) Arch. Biochem. Biophys. 466, 283–289 [DOI] [PubMed] [Google Scholar]
- 18.Bollen M., Keppens S., Stalmans W. (1998) Biochem. J. 336, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouillon D. J., Berdanier C. D. (1981) J. Nutr. 111, 1462–1474 [DOI] [PubMed] [Google Scholar]
- 20.Oishi K., Miyazaki K., Ishida N. (2002) Biochem. Biophys. Res. Commun. 298, 198–202 [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y., Ishikawa T., Hirayama J., Daiyasu H., Kanai S., Toh H., Fukuda I., Tsujimura T., Terada N., Kamei Y., Yuba S., Iwai S., Todo T. (2000) Genes Cells 5, 725–738 [DOI] [PubMed] [Google Scholar]
- 22.Boyd K. E., Wells J., Gutman J., Bartley S. M., Farnham P. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13887–13892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno T., Onishi Y., Ishida N. (2007) Nucleic Acids Res. 35, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo T., Tamagawa T., Shibata S. (2009) J. Circ. Rhythms 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchiya Y., Akashi M., Nishida E. (2003) Genes Cells 8, 713–720 [DOI] [PubMed] [Google Scholar]
- 27.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 28.Kennaway D. J., Owens J. A., Voultsios A., Boden M. J., Varcoe T. J. (2007) Am. J. Physiol. 293, R1528–R1537 [DOI] [PubMed] [Google Scholar]
- 29.Allaman-Pillet N., Roduit R., Oberson A., Abdelli S., Ruiz J., Beckmann J. S., Schorderet D. F., Bonny C. (2004) Mol. Cell. Endocrinol. 226, 59–66 [DOI] [PubMed] [Google Scholar]
- 30.O'Brien R. M., Granner D. K. (1990) Diabetes Care 13, 327–339 [DOI] [PubMed] [Google Scholar]
- 31.Miyake K., Ogawa W., Matsumoto M., Nakamura T., Sakaue H., Kasuga M. (2002) J. Clin. Invest. 110, 1483–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertolucci C., Cavallari N., Colognesi I., Aguzzi J., Chen Z., Caruso P., Foá A., Tosini G., Bernardi F., Pinotti M. (2008) Mol. Cell. Biol. 28, 3070–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeBruyne J. P., Weaver D. R., Reppert S. M. (2007) Nat. Neurosci. 10, 543–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaslow H. R., Lesikar D. D., Antwi D., Tan A. W. (1985) J. Biol. Chem. 260, 9953–9956 [PubMed] [Google Scholar]
- 35.Ripperger J. A., Schibler U. (2006) Nat. Genet. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 36.Oishi K., Shirai H., Ishida N. (2005) Biochem. J. 386, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre P., Chinetti G., Fruchart J. C., Staels B. (2006) J. Clin. Invest. 116, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandard S., Stienstra R., Escher P., Tan N. S., Kim I., Gonzalez F. J., Wahli W., Desvergne B., Müller M., Kersten S. (2007) Cell. Mol. Life Sci. 64, 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandsma R. H., Van Dijk T. H., Harmsel At A., Kok T., Reijngoud D. J., Staels B., Kuipers F. (2004) J. Biol. Chem. 279, 8930–8937 [DOI] [PubMed] [Google Scholar]
- 40.Kumaki Y., Ukai-Tadenuma M., Uno K. D., Nishio J., Masumoto K. H., Nagano M., Komori T., Shigeyoshi Y., Hogenesch J. B., Ueda H. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radziuk J., Pye S. (2001) Diabetes 17, 250–272 [DOI] [PubMed] [Google Scholar]
- 42.Bryson J. M., Cooney G. J., Wensley V. R., Blair S. C., Caterson I. D. (1993) Biochem. J. 295, 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burcelin R., del Carmen Muñoz M., Guillam M. T., Thorens B. (2000) J. Biol. Chem. 275, 10930–10936 [DOI] [PubMed] [Google Scholar]
- 44.Iitaka C., Miyazaki K., Akaike T., Ishida N. (2005) J. Biol. Chem. 280, 29397–29402 [DOI] [PubMed] [Google Scholar]
- 45.Ando H., Ushijima K., Yanagihara H., Hayashi Y., Takamura T., Kaneko S., Fujimura A. (2009) Clin. Exp. Hypertens. 31, 201–207 [DOI] [PubMed] [Google Scholar]
- 46.Krssak M., Brehm A., Bernroider E., Anderwald C., Nowotny P., Dalla Man C., Cobelli C., Cline G. W., Shulman G. I., Waldhäusl W., Roden M. (2004) Diabetes 53, 3048–3056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.