Abstract
Fe/S clusters are part of the active site of many enzymes and are essential for cell viability. In eukaryotes the cysteine desulfurase Nfs (IscS) donates the sulfur during Fe/S cluster assembly and was thought sufficient for this reaction. Moreover, Nfs is indispensable for tRNA thiolation, a modification generally required for tRNA function and protein synthesis. Recently, Isd11 was discovered as an integral part of the Nfs activity at an early step of Fe/S cluster assembly. Here we show, using a combination of genetic, molecular, and biochemical approaches, that Isd11, in line with its strong association with Nfs, is localized in the mitochondrion of T. brucei. In addition to its involvement in Fe/S assembly, Isd11 also partakes in both cytoplasmic and mitochondrial tRNA thiolation, whereas Mtu1, another protein proposed to collaborate with Nfs in tRNA thiolation, is required for this process solely within the mitochondrion. Taken together these data place Isd11 at the center of these sulfur transactions and raises the possibility of a connection between Fe/S metabolism and protein synthesis, helping integrate two seemingly unrelated pathways.
Keywords: Parasitology, Protein/Iron-Sulfur, RNA/Interference/RNAi, RNA/Modification, RNA/Transfer RNA, Subcellular Organelles/Mitochondria, Isd11, Trypanosoma
Introduction
Every extant archaeal, bacterial, and eukaryotic cell contains proteins whose functions depend on iron-sulfur (Fe/S) clusters. In a typical eukaryotic cell these ancient co-factors are part of more than 100 different proteins, the assembly of Fe/S clusters is invariably essential for viability. In particular, several mitochondrial proteins involved in electron transport, such as subunits of respiratory complexes I, II, and III, and ferredoxin contain numerous Fe/S clusters. Moreover, nuclear and cytosolic proteins, including the ribosomal protein Rli1, primase Pri2, and xanthine oxidoreductase, to name just a few, depend functionally on these co-factors (for recent reviews, see Refs. 1 and 2).
A key component of the Fe/S cluster assembly (ISC) machinery is the cysteine desulfurase (Nfs), which removes sulfur from cysteine, converting it into alanine (3). Nfs binds to a highly conserved scaffold protein termed IscU, forming a complex, upon which the Fe/S cluster is assembled with the sulfur provided by Nfs and iron coming from an additional protein, likely frataxin (4, 5). However, the exact mechanism of cluster formation has yet to be fully understood. Somewhat unexpectedly, a third component of this Fe/S cluster assembly complex, a small protein named Isd11, was identified as a stable and essential binding partner of the eukaryotic Nfs protein (6–9). Based on in vitro experiments, Nfs was originally thought to catalyze sulfur transfer by itself (10), but it was recently shown to require the assistance of Isd11 for the formation of Fe/S clusters on the IscU scaffold (6, 7). Co-expression in Escherichia coli of Nfs and Isd11 from the microsporidian Trachipleistophora hominis confirmed that it is indeed a complex of both tightly bound proteins that represents the functional cysteine desulfurase (11).
The Isd11 gene has also been found in the ancestral protists that contain hydrogenosomes and mitosomes (12, 13). Isd11 belongs to the LYR family (9) and in yeast stabilizes mitochondrial Nfs1, which is in its absence prone to aggregation. Although in yeast Isd11 seems to be confined to the mitochondrion (6, 7), it is equally abundant in the mitochondrion and nucleus of human cells, where its depletion resulted in inactivation of Fe/S cluster-containing cytosolic and mitochondrial aconitases. Furthermore, its down-regulation in human cells disrupts iron homeostasis (9).
Moreover, an interconnection between Fe/S cluster biogenesis and 2-thiouridine (s2U) modification of tRNAs has been found in yeast and human cells. In these organisms, tRNA thiolation occurs in two places: the cytoplasm and mitochondria. In both compartments, modification of mitochondrial and cytosolic tRNAs requires the participation of components of the mitochondrial (ISC) and cytosolic Fe/S cluster assembly machineries (CIA), respectively (1). This is likely due to the direct involvement of some of these proteins in tRNA modifications and by the fact that Tyw1 and Elp3 proteins themselves need Fe/S clusters for their function (14). However, thiolation of tRNAs in both cellular compartments of the yeast cell was shown to directly depend on mitochondrial Nfs1 (15–17) but does not require the involvement of Fe/S-containing enzymes. This suggests a role for this desulfurase in sulfur-relay, independent of its function in Fe/S cluster assembly.
The factors that relay the sulfur from Nfs1 to tRNAs differ in the two compartments. In mitochondria, Mtu1, a homolog of bacterial MnmA, acts as the tRNA-specific 2-thiouridylase (17). Disruption of this gene in yeast not only eliminated the thiolation of mitochondrial tRNAs but also led to reduced respiratory activity and impaired mitochondrial protein synthesis. Furthermore, the down-regulation of the human homologue of MTU1 in HeLa cells by RNA interference (RNAi)2 resulted in a phenotype observed in patients afflicted with myoclonus epilepsy associated with ragged-red fibers (17).
In the cytoplasm, Urm1p, the earliest-diverging ubiquitin-like protein, was shown to act as a sulfur carrier in tRNA thiolation, providing a possible evolutionary link between ubiquitin-like proteins and sulfur transfer. Specifically, five genes (URM1, UBA4, NCS2, NCS6, and YOR251c) have been identified in a genome-wide screen of Saccharomyces cerevisiae to be responsible for 2-thiouridine synthesis (18–20). In vitro assays indicate that Ncs6p binds to tRNA, whereas Uba4p first adenylates and then directly transfers sulfur onto Urm1p (18). However, tRNA thiolation still cannot be efficiently reconstituted in vitro in any of these systems, suggesting the involvement of additional components.
Trypanosoma brucei and related flagellates are responsible for human African sleeping sickness and numerous other serious diseases that afflict millions in tropical regions. Within the last decade this early branching eukaryote also became approachable by numerous methods of forward and reverse genetics. In these cells, both the mitochondrion-targeted Nfs and a nucleus-localized Nfs-like protein have cysteine desulfurase and selenocysteine lyase activities. However, of these two proteins only Nfs is essential for ISC (21, 22). Due to the complete loss of tRNA genes from its organellar DNA, the single mitochondrion of T. brucei imports all tRNAs from the cytoplasm (23). We have recently shown that in the insect stage of the parasite Nfs is indispensable for thiolation of both cytosolic and mitochondrial tRNAs. Furthermore, thiolation has important implications for cytoplasmic tRNA stability of normally thiolated tRNAs and mitochondrial C to U editing of tRNATrp (24).
In this work we demonstrate that Isd11 is essential for Fe/S cluster assembly of both mitochondrial and cytosolic proteins of T. brucei, like in other eukaryotes (6, 7, 9, 11, 13). We also show for the first time that Isd11 is required for thiolation of both cytosolic and mitochondrial tRNAs and that Isd11 is not solely an Nfs chaperone for Fe/S cluster assembly, whereas Mtu1 is essential for mitochondrial tRNA thiolation only. Taken together, these findings provide further evidence for the association of the two pathways and suggest a possible link between Fe/S cluster assembly and translation mediated by tRNA thiolation. The involvement of Isd11 in tRNA thiolation also provides yet another missing key component in the complex tRNA thiolation pathway of eukaryotic cells.
EXPERIMENTAL PROCEDURES
Plasmid Constructs, Transfections, Cloning, RNAi Induction, and Cultivation
A 300-nucleotide long part of the Isd11 gene (Tb10.389.1785) was PCR amplified using primers Isd-F (5′-CTCGAGATGACAAACACCGTCAAACACT) and Isd-R (5′-ACTAGTTTACTCTTCTTCTTCTTGCGTCAC) (added XhoI and SpeI restriction sites are underlined) from total genomic DNA of T. brucei strain 29-13. The amplicon was cloned into the p2T7-177 vector, which was upon NotI-mediated linearization introduced into procyclic T. brucei 29-13 cells and selected as described elsewhere (25). The same strategy was used to prepare cells in which the Mtu1 mRNA was targeted by RNAi. A 501-nucleotide long fragment of the Mtu1 gene (Tb927.8.1830) was amplified via primers Mtu-F (5′-CTCGAGGCAACACGCTCAACGTTG) and Mtu-R (5′-GGATCCCCGCGTGAACACCTTCTC) (added XhoI and BamHI restriction sites are underlined), cloned, and stably integrated into the T. brucei 29-13 PF cells as described above. RNAi was triggered by the addition of 1 μg/ml of tetracycline to the SDM-79 medium. Cell density was measured every 24 h using the Beckman Z2 Coulter counter over a period of 8 (Isd11)/14 (Mtu1) days after the induction of double-stranded RNA synthesis. The inducibly expressed C-terminal tandem affinity purification (TAP)-tagged Isd11 protein was created by PCR amplification of the full-size Isd11 gene and inserted into the pLew-79-MHTAP plasmid (26). Similarly, to obtain cells expressing TAP-tagged IscU and frataxin proteins, the full-size genes encoding the scaffold protein IscU (Tb09.21.2830) and frataxin (Tb927.3.1000) were PCR-amplified as described above. The NotI-linearized constructs were electroporated into the 29-13 cells and phleomycine-resistant clones were selected following a protocol described previously (25). The bloodstream T. brucei strain 920 was cultured at 37 °C as described elsewhere (27).
Northern Blot Analyses
Total RNA was isolated using TRIzol (Sigma) from procyclic cells cultivated in SDM-79 medium. Approximately 10 μg/lane of total RNA was loaded on a 1% agarose-formaldehyde gel, blotted, and cross-linked. Hybridization was performed in NaPi solution using probes labeled by random priming with [α-32P]dATP (MP Biomedicals) overnight at 55 °C. Membranes were washed and the radioactive signal was detected as described previously (28).
Expression and Purification of Recombinant Isd11
The full-size Isd11 gene was amplified by PCR with primers IsdE-F (5′-CACCATGACAAACACCGTCAAA) and IsdE-R (5′-ACTAGTTTACTCTTCTTCTTCTTGCGTCAC) (containing the stop codon (underlined)). The amplicon was gel-purified and cloned into the pET100 expression vector (Invitrogen). The resulting expression plasmid encoding His6-tagged Isd11 was transformed into the E. coli strain BL21(DE3) (Novagen). Insoluble protein was obtained from induced bacterial cells (incubation at 30 °C for 2 h) under denaturing conditions using affinity chromatography.
Digitonin Fractionation and Immunolocalization
Digitonin fractionation was performed following a protocol described elsewhere (21). Subcellular localization of the expressed tagged proteins within the cell was determined by IFA using polyclonal anti-Myc antibodies (Invitrogen) as described previously (29). Co-localization analysis was performed using monoclonal antibody mAB61 against the MRP1/2 complex (30) coupled with Texas Red X-conjugated secondary antibody (Invitrogen).
Preparation of Antibodies and Western Blot Analyses
Polyclonal antibody was prepared by immunizing rabbits at 2-week intervals with four subcutaneous injections of 0.5 mg of purified recombinant Isd11 protein emulsified with complete (1st injection) and incomplete (following injections) Freund's adjuvant. Another polyclonal antibody was raised by immunizing a rat at 2-week intervals by Cocalico Biologicals Inc. (Reamstown, PA). Cell lysates corresponding to 5 × 106 cells/lane were separeted on 15% SDS-polyacrylamide gel, blotted, and probed. The rabbit serum (1:1,000) against Isd11 was used for further experiments because it contained more specific antibodies. The polyclonal rabbit antibodies against the T. brucei frataxin (31), Nfs and IscU (21), Gap1 (27), and enolase (kindly provided by P. A. M. Michels) were used at 1:500, 1:1,000, 1:1,000, 1:1,000, and 1:100,000 dilutions, respectively.
TAP Purification and Mass Spectrometry Analysis
The TAP protocol was adapted from the published method (32). The tagged complexes were purified from 1 × 109 cells lysed with 0.25% Nonidet P-40, cleared by low speed centrifugation, and the supernatant was further lysed with 1.25% Nonidet P-40 and cleared by high speed centrifugation (40,000 × g at 4οC in a Sorvall SW-55 rotor for 40 min). The tagged complexes were isolated by sequential binding to IgG and calmodulin affinity columns. This method was adapted from the published protocol (33). Purified complexes were analyzed by mass spectrometry basically as described previously (32).
Measurement of Mitochondrial Membrane Potential
After centrifugation, exponentially growing procyclic cells were resuspended in 1 ml of fresh SDM-79 medium (at a concentration 5 × 106 cells/ml) and mitochondrial inner membrane potential was measured by the uptake of 0.4% tetramethylrhodamine ethyl ester (Molecular Probes) for 20 min at 27 °C. After staining, the cells were resuspended in a dye-free medium and instantly measured by flow cytometry using an Epics XL flow cytometer (Coulter).
Measurement of Enzymatic Activities
The activities of fumarase and aconitase were measured in total cell lysates, as well as in subcellular fractions obtained by digitonin treatment, the purity of which was controlled by compartment-specific antibodies. Fumarase activity was determined in procyclic cells resuspended in Hanks' balanced salt solution (Invitrogen) containing 0.1% Triton X-100 and incubated for 5 min on ice. After centrifugation, the activity was measured spectrophotometrically at 240 nm as the rate of production of fumarate. The activities of aconitase and threonine dehydrogenase were established as described elsewhere (34).
APM Gel Electrophoresis and Northern Blot Analysis
Thiolation of different tRNAs was analyzed using the [(N-acryloylamino)-phenyl]mercuric chloride (APM) gel, and blotted as described elsewhere (35). In the control reactions the thiol group was removed by treatment in 0.4% hydrogen peroxide, 1 mm EDTA, 100 mm phosphate buffer (pH 8.0) for 1 h at room temperature. The percentage of thiolation was established using the volume of band(s) corresponding to the thiolated and non-thiolated species. To determine ratios of total tRNA in each sample the volume of the whole lane was used and compared between RNA isolated from the non-induced and RNAi-induced cells. Ratios were then standardized using tRNAs that do not get thiolated.
RESULTS
Isd11 Is a Mitochondrial Protein
We have used the Isd11 protein sequence of S. cerevisiae as a query to search the T. brucei genome. We have identified three putative Isd11 homologues, Isd11-1, Isd11-2, and Isd11-3 (calculated molecular masses 11.77, 23.69, and 18.79 kDa, respectively) (Fig. S1). Although all three genes contain the LYR/K domain, only Isd11-1 (hence Isd11) shares high sequence similarity with Isd11 proteins from plants, human, and other eukaryotes (Fig. S2). Disruption of Isd11-2 by RNAi was not lethal and preliminary results indicate its association with respiratory complexes rather than with Fe/S cluster assembly.3 Isd11–3 was not studied further.
On the basis of its N-terminal leader sequences, MitoProt predicted mitochondrial localization of Isd11 with a 0.879 (∼88%) probability. The predicted cleavage site of the T. brucei mitochondrial processing peptidase was at position 13 (Figs. S3 and S4). To further assess its predicted localization, Isd11 was expressed in E. coli and rabbit polyclonal antibodies were raised against the purified recombinant protein. Fractionation by treatment with the detergent digitonin allowed us to obtain cytosolic and mitochondrial fractions of the T. brucei procyclics, the purity of which was confirmed by antibodies against compartment-specific markers enolase (cytoplasmic) and the Gap1 protein (mitochondrial) (Fig. 1). In agreement with the in silico predictions, the Isd11 protein was localized to the mitochondrion (Fig. 1A, left panel). Next, we were interested on whether Isd11 is also present in the bloodstream (=mammalian) stage of T. brucei, whose single mitochondrion is largely down-regulated in terms of size and activity. Indeed, also in the bloodstream stage, all detectable Isd11 protein appears to be confined to the mitochondrion (Fig. 1A, right panel). Immunofluorescence assays using anti-Myc polyclonal antibodies showed that tagged IscU and Isd11 localize to the reticulated mitochondrion and that both proteins are equally distributed throughout the organelle (Fig. 1B). These results also showed that the TAP tags did not interfere with mitochondrial import and thus provide validity for purifying the Isd11-Nfs-IscU complex.
FIGURE 1.
A, Isd11 is localized in the mitochondrion of the procyclic (PF) and bloodstream stages (BF). Cytosolic and mitochondrial fractions were obtained from both developmental stages by fractionation using digitonin. Upon resolution by SDS-PAGE the lysates were blotted and immunoprobed with α-Isd11 antibodies. Antibodies against enolase and GAP1 proteins were used as cytosolic and mitochondrial markers, respectively. Total cell lysates (T), cytosolic (C), and mitochondrial (M) fractions were screened in both stages. Positions of the protein markers are indicated on the right. B, tagged Isd11 and IscU were visualized by fluorescence microscopy using polyclonal anti-c-Myc antiserum coupled with fluorescein isothiocyanate-conjugated secondary antibody. Co-localization immunofluorescence was performed with monoclonal antibody mAb61 against the mitochondrial MRP1/2 complex (30). The first column (phase) shows phase-contrast light microscopy of the T. brucei procyclic cells; the second column (Dapi) shows staining of nuclear and mitochondrial (=kinetoplast) DNA with 4,6-diamidino-2-phenylindole (DAPI); the third column (c-Myc) shows localization of the TAP-tagged proteins using the polyclonal α-Myc antibody; the fourth column (mAb61) shows co-localization of the mitochondrial MRP1/2 complex using monoclonal α-mAb61 antibody; the last column (merge) documents merged fluorescence images. C, proteins were identified by mass spectrometry analysis of TAP tags from cells expressing TAP-tagged Isd11, IscU, or frataxin. Only proteins identified in both IscU and Isd11 TAP tag purifications and at least by two unique peptides are shown. The number indicates the total number of unique peptides identified.
Isd11 Forms a Stable Complex with Nfs and IscU
In yeast, microsporidian, and human cells, Isd11 was shown to be associated with the Nfs protein (6, 7, 9, 11, 13). To explore whether similar interactions of Isd11 occur in trypanosomes, a TAP-tagged version of this protein was generated and expressed under control of a tetracycline-inducible promoter. Clarified cell lysate obtained from these cells was subjected to tandem affinity purification. SYPRO Ruby staining of an aliquot of a TAP eluate containing Isd11, separated by SDS-PAGE, revealed the presence of only two visible protein bands (data not shown). Mass spectrometry analysis of the tagged Isd11 complex identified, besides Isd11, Nfs, and IscU, both with at least two unique doubly tryptic peptides (Fig. 1C). To exclude the possibility of nonspecific protein association, IscU was selected for reciprocal TAP tagging. Reassuringly, the TAP-tagged IscU pulled down Isd11 and Nfs, confirming that these three proteins form a stable complex. Finally, we have also TAP-tagged frataxin to assay its putative interaction with the above proteins. As shown in Fig. 1C, under the conditions used, mass spectrometry analysis of the purified TAP-tagged frataxin did not reveal the presence of any other protein, implying that in T. brucei frataxin does not form a stable complex with the above proteins.
Isd11 Is Required for Fe/S Cluster Assembly
To establish the role of Isd11 in T. brucei procyclics, we performed RNAi-mediated knockdown of the transcript using a tetracycline-inducible RNAi construct. Upon the addition of the antibiotics, proliferation of the clonal cell line was markedly inhibited starting with day 3 (Fig. 2A), which is in good agreement with the decrease in the levels of the target protein (see below). The growth inhibition persisted for 8 days post-induction when the growth curve was terminated (Fig. 2A). Northern blotting after 72 h of RNAi induction showed depletion of the target Isd11 mRNA (Fig. 2B). This result was also confirmed by Western blot analysis that showed a dramatic decrease in Isd11 protein levels by day 3 of RNAi induction. The protein was virtually eliminated by day 5 and became almost undetectable in lysates isolated from cells 1 week after RNAi induction (Fig. 3). From the Western blot it is also apparent that no leaky transcription occurred, as the level of Isd11 remained unchanged and at comparable levels in the 29-13 wild type as well as the non-induced cells (Fig. 3).
FIGURE 2.
A, effects of Isd11 RNAi on the growth of the parasite. Growth curves of non-induced (TET−; triangles) and induced (TET+; squares) knockdown for Isd11. The y axis is labeled by a log scale and represents the products of the measured cell densities and total dilutions. Cell densities were measured using the Beckman Z2 cell counter. The arrow indicates the sampling time point for the latter experiments. B, Isd11 mRNA levels were analyzed by blotting total RNA extracted from non-induced cells (−) and cells harvested 3 days of RNAi induction against Isd11 (+). The position of the targeted mRNA and double-stranded RNA synthesis following induction are indicated with open and closed arrows, respectively. As a loading control, the gel was stained with ethidium bromide to visualize rRNA bands. WT, wild type.
FIGURE 3.

Silencing of Isd11 affects stability of other Fe/S cluster assembly proteins in procyclic T. brucei. Expression level of Isd11, cysteine desulfurase (Nfs), scaffold protein (IscU), and frataxin as determined by immunoblot analysis of whole cell lysates of the parental 29-13 cell line (wt), non-induced cells (−), and cells 3, 5, and 7 days of RNAi induction. Enolase was used as a cytosolic loading control. Positions of the protein markers are indicated on the right.
The observed association between Isd11, Nfs, and IscU revealed by TAP tag analysis led us to assess the levels of each of these proteins in cells depleted of Isd11 by RNAi. Indeed, as its general binding partner, Nfs followed the expression pattern of Isd11, becoming substantially down-regulated 3 days of RNAi induction, although it does not seem to be totally eliminated in cells collected in the studied time points (Fig. 3). A very similar result was obtained using the anti-IscU antibody, whereas the levels of frataxin, which does not interact with the other three proteins (Fig. 1C), remained unaltered in the Isd11 knockdowns (Fig. 3).
To assess the putative role of Isd11 in the Fe/S cluster assembly, we have measured the activities of marker Fe/S cluster-containing aconitase and fumarase. Both enzymes have a dual localization in the T. brucei procyclics (31, 34), which allows separate evaluation of the impact of RNAi against Isd11 on cytosolic and mitochondrial proteins. The purity of cytosolic and mitochondrial fractions obtained from the non-induced and RNAi-induced procyclic T. brucei by digitonin treatment was first assessed by Western blot analysis using antibodies against Gap1 and enolase, as described above (Fig. 4). When compared with the non-induced cells, activities of both metabolic enzymes dropped by 45 to 60% in cells depleted for Isd11 (Fig. 4). The activity of mitochondrial threonine dehydrogenase, a protein lacking Fe/S clusters, remained unaltered in both the non-induced and RNAi-induced cells (Fig. 4).
FIGURE 4.
Activity of fumarase and aconitase (Fe/S cluster-containing proteins) is significantly reduced after down-regulation of Isd11. The percentage of specific aconitase and fumarase activity in total cell lysate (total), cytosol (cyto), and mitochondrion (mito) in the non-induced and RNAi-induced cells is shown. Values for the non-induced cells were considered as 100% activity. The mean ± S.D. values of six independent induction experiments are shown. The purity of cellular fractions obtained by digitonin fractionation from the non-induced (−) and RNAi-induced cells (+) was controlled with the α-enolase (cytosolic marker) and α-Gap1 antibodies (mitochondrial marker).
Isd11 Is Essential for Cytosolic and Mitochondrial tRNA Thiolation
Besides playing a role in Fe/S assembly, the Nfs protein is also the key desulfurase for tRNA thiolation in E. coli, as well as in S. cerevisiae and T. brucei (15, 24, 36). These observations raised the question of whether Isd11 is also essential for tRNA thiolation. Because Isd11 is a binding partner for Nfs, we decided to explore its possible role in tRNA metabolism further. Total RNA was isolated from the non-induced cells and from cells depleted for Isd11 (collected 72 h after RNAi induction). These RNA samples were separated on mercury-containing APM gels (see “Experimental Procedures”). In these gels, the thiolated tRNA species migrate slower than the non-thiolated ones, and the thiolation status of tRNAs is thus reflected in their differential mobility. We performed Northern blots of APM gel-separated RNAs obtained from the non-induced and RNAi-induced cells, using radioactive probes specific for two cytosolic tRNAs (tRNAUUGGln and tRNAUUCGlu) that get thiolated at the wobble position. In cells with down-regulated Isd11 (+ lanes), thiolation of tRNAUUGGln and tRNAUUCGlu was reduced by 60 and 50%, respectively (Fig. 5A).
FIGURE 5.
Isd11 is essential for cytosolic tRNA thiolation. RNA isolated from non-induced (−) and RNAi-induced Isd11 cells (+) was separated on an APM gel followed by Northern blot analysis (A). Bands corresponding to thiolated and non-thiolated tRNAs are indicated. H2O2 is used as a control to show complete oxidation of thiolation, eliminating the mobility shift. The ratio of total tRNA isolated from RNAi-induced (+) to non-induced (−) cells after standardizing with non-thiolated tRNAs (B) is shown below each panel, where a ratio of 1 would indicate no change, less than 1 indicates lower steady-state levels of a particular tRNA.
Importantly, the levels of two non-thiolated cytoplasmic tRNA species used as controls, tRNAUCCGly and tRNAUAUIle, were unaltered regardless of the amount of Isd11 in the procyclic cells (Fig. 5B). These data indicate that Isd11 is indispensable for the thiolation of cytoplasmic tRNAs.
S. cerevisiae contains two separate pathways involving multiple protein factors that work on the sulfur-relay mechanism essential for tRNA thiolation. In the cytoplasm this pathway requires Nfs1 (the yeast homolog of Nfs) and several additional ubiquitin-like proteins, such as Uba24, Urm1, and Ncs6/Ncs2. The mitochondrial sulfur-relay system requires at least Nfs1 and Mtu1, the latter factor being implicated in the sulfur transfer step onto the tRNA. In light of the mitochondrial localization of Isd11 in T. brucei and other eukaryotes, we decided to explore its possible involvement in mitochondrial thiolation using a radioactively labeled probe specific for tRNATrp, the only known tRNA to undergo mitochondrion-specific thiolation in these cells. We probed Northern blots of total RNA isolated from the mitochondria of the wild type and RNAi-induced cells separated by the APM gels. We found that similar to the cytosolic case for tRNAGln and tRNAGlu, tRNATrp thiolation is also down-regulated in cells depleted for Isd11 (Fig. 7A).
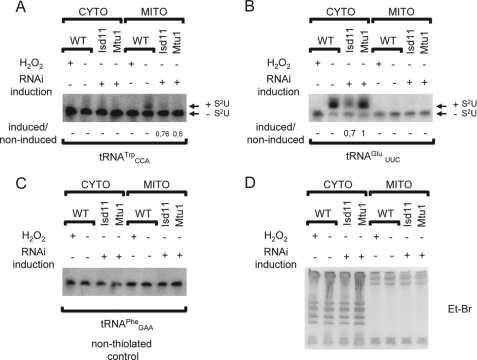
FIGURE 7.
Isd11 and Mtu1 are essential for mitochondrial thiolation of tRNACCATrp. Cytosolic and mitochondrial RNA from wild type cells (WT) and RNAi-induced cells was separated by APM acrylamide gel electrophoresis. A and B, Northern blot analysis probing for tRNATrp and tRNAGlu. Bands corresponding to thiolated and non-thiolated tRNAs are indicated. H2O2 is used as a control to show complete oxidation of thiolation, eliminating the mobility shift. The level of tRNA thiolation is shown below the panel and calculated as described previously. The tRNAPhe (C) is used as a loading control and ethidium bromide shows the purity of mitochondrial fractions (D).
Mtu1 Is Essential for Mitochondrial tRNA Thiolation
To assess possible involvement of Mtu1 in the thiolation reaction in trypanosomes, we performed a Blast search, using the S. cerevisiae Mtu1 protein as a query, which identified a single homologue for the Mtu1/MnmA gene in the T. brucei genome (Tb927.8.1830). The Mtu1 protein has a predicted molecular mass of 61.28 kDa, and its inclusion into the alignment of several eukaryotic Mtu1 genes (17) revealed the presence of virtually all conserved residues and 29 and 45% sequence identity and similarity, respectively, with its S. cerevisiae homologue (Fig. S3).
Given its essentiality in other systems, we anticipated a similar situation in the T. brucei procyclics. The addition of tetracycline induced a massive synthesis of double-stranded RNA, resulting in a significant down-regulation of the targeted Mtu1 mRNA (Fig. 6B). However, cells ablated for Mtu1 grow very well in the SDM-79 medium, their growth curve being virtually indistinguishable from those of the wild type 29-13 cells (data not shown) and the non-induced cells even after 2 weeks of cultivation (Fig. 6A). Nonetheless, we tested whether Mtu1 is also involved in mitochondrial thiolation. We found that Mtu1 RNAi led to a decrease in tRNATrp thiolation (Fig. 7A) comparable with that caused by the depletion of Isd11. Significantly, the function of Mtu1 is relegated to mitochondrial thiolation, because no decrease in cytoplasmic thiolation was observed with RNA isolated from the Mtu1 RNAi cells (Figs. 6C and 7B). In addition, tRNAGln and tRNAGlu, which are thiolated in the cytoplasm, show no thiolation following import into the mitochondria (Fig. 7B). Although this observation may seem puzzling, it has been shown very recently that these two tRNAs undergo de-thiolation following import by an as yet unknown mechanism (37).
FIGURE 6.
Silencing of Mtu1 affects neither growth phenotype nor cytosolic tRNA thiolation. A, growth curves of non-induced (TET−; triangles) and RNAi-induced (TET+; squares) knockdown for Mtu1 are presented as in Fig. 2A. Dotted and solid arrows indicate time points for RNA isolation and mitochondrial membrane potential measurements, respectively. B, Mtu1 mRNA levels were analyzed by blotting total RNA extracted from the non-induced cells (−) and cells harvested 3 days of RNAi induction against Mtu1 (+). Northern blot and the stained gel are as described in the legend to Fig. 2B. Mtu1 probe 1 was prepared from a fragment of the gene other than the one used for RNAi. Mtu1 probe 2 corresponds precisely to the gene fragment cloned into the RNAi vector. C, RNA isolated from the non-induced (−) and RNAi-induced Mtu1 cells (+) was separated on an APM gel followed by Northern blot analysis. Bands corresponding to thiolated and non-thiolated tRNAs are indicated. H2O2 was used as a control to show complete oxidation of thiolation, eliminating the mobility shift. The level of tRNA thiolation is shown below the panel and calculated as described previously. D, the tRNAGly and tRNAIle were used as loading controls.
Taken together, these data demonstrate that unlike Mtu1, which is mitochondrion-specific, Isd11 is essential for both cytosolic and mitochondrial thiolation. This behavior is similar to that observed with Nfs and leads to the conclusion that both proteins are essential for tRNA thiolation in both the cytoplasm and mitochondria. Significantly, Mtu1 itself does not contain an Fe/S cluster, implying that Isd11 plays a direct role in thiolation beyond its essential role in Fe/S assembly. Therefore the Nfs-Isd11 complex is an integral part of a more general sulfur-relay system not just limited to the Fe/S cluster assembly.
Effect of Isd11 and Mtu1 RNAi on Mitochondrial Membrane Potential
In the procyclic stage, mitochondrial inner membrane potential is maintained by the activity of the cytochrome-mediated respiratory chain (38). Quantification of the uptake of tetramethylrhodamine ethyl ester by flow cytometry enabled us to measure the potential in non-induced and RNAi-induced cells. As shown in Fig. 8A, a dramatic decrease of membrane potential was observed in the Isd11-depleted cells at day 3 of RNAi induction, likely a consequence of the core subunits of respiratory complexes being depleted for the Fe/S clusters. However, numerous measurements of the membrane potential at 3, 6, 9, and 12 days upon the induction of RNAi against Mtu1 did not reveal differences between the non-induced cells and those depleted for Mtu1 (Fig. 8B).
FIGURE 8.
Effect of RNAi against either Isd11 or Mtu1 on mitochondrial membrane potential. Mitochondrial membrane potential was measured in non-induced cells (thick black line), as well as in cells with down-regulated Isd11 (A) or Mtu1 (B) (thin gray line) that were, after incubation with 0.4% tetramethylrhodamine ethyl ester (TMRE), analyzed using flow cytometry. Ablated cell lines were analyzed 3 days of RNAi induction in the case of Isd11 and 3, 6, 9, and 12 days in case of Mtu1, respectively. The distribution of fluorescence was plotted as frequency histogram.
DISCUSSION
Isd11, a eukaryotic invention in the Fe/S cluster assembly pathway, is present in all sequenced eukaryotic genomes as a single-copy gene (6, 7, 12). Interestingly, the model flagellate T. brucei represents an exception, because a Blast search identified three putative homologs of Isd11 in its genome. Based on the presence of the LYR/K sequence motif found in subunits of respiratory complex I, Isd11 might originally have served as a subunit of this complex, but was at an early stage of eukaryotic evolution abducted for novel functions. Indeed, whereas the Isd11 protein functionally analyzed in this study is clearly associated with the Fe/S cluster assembly, another homologue (Isd11-2) appears to have a function unrelated with this process.3 In yeasts, microsporidia, and humans, Isd11 is a binding partner of Nfs, essential for its desulfurase activity in vivo and thus indispensable for cell survival (6, 7, 11, 13).
Using mass spectrometry and Western analysis, we have shown herein that in the unicellular parasite T. brucei, Isd11 also interacts with Nfs and the Fe/S cluster assembly chaperon IscU. Furthermore, we have demonstrated that the studied protein assists in the ISC pathway, because the Fe/S cluster-dependent activities of aconitase and fumarase were significantly decreased in the Isd11-deficient background.
There is one important difference between yeast and human knockdowns for Isd11. Although in human cells, down-regulation of this protein causes destabilization not only of its direct binding partner Nfs1, but also IscU (13), it is only Nfs1 that is eliminated from the Isd11 mutant yeast cells (7). The results obtained in trypanosomes are reminiscent of those obtained with human cells, as both proteins are clearly eliminated in the absence of Isd11. Human and trypanosome cells also share another aspect of the Isd11 function not seen with yeast. Although Isd11 depletion leads to defects of both mitochondrial and cytosolic Fe/S cluster proteins (Ref. 9 and this work), similar experiments in yeast only affect the organellar ISC pathway (6, 7).
Given the mitochondrial localization and strong association of Isd11 with Nfs shown in this work, intuitively, one would expect that Isd11 is also essential for thiolation, as previously suggested (1). We therefore tested this hypothesis further and undertook a detailed analysis of tRNAs in cells depleted for Isd11. Indeed, this protein is indispensable for T. brucei tRNA thiolation, a modification that is conserved, widespread, and essential in most organisms. Isd11, however, is needed not only for mitochondrial but also for cytosolic tRNA thiolation, despite the fact that in trypanosomes Isd11, analogous to Nfs, is detectable by Western blotting and immunolocalization only within the organelle (Ref. 24 and this work). The possible presence of Isd11 in the cytosol in amounts that escape selection by Western blot but that are sufficient to generate thiolated tRNAs in the cytoplasm via the cytoplasmic sulfur relay system (18, 39, 40) is highly unlikely for the following reasons. Another component of the Fe/S assembly, Atm1, is in all examined organisms strictly confined to the mitochondrion (1, 2, 41). However, we have shown that the same as Isd11, down-regulation of Atm1 affects thiolation of cytosolic tRNAs in T. brucei.4 This strongly implies that it is the extrusion, via Atm1, of an unknown sulfur component into the cytoplasm (1, 2) that is decreased in Isd11 RNAi knockdowns, which is responsible for the drop in cytosolic tRNA thiolation. Notably, Elp3 is an iron-sulfur containing protein, which was proposed to be one of the components of the sulfur-relay system for thiolation in yeast (18). This creates a scenario in which both proteins may be present in amounts in the cytosol that escape Western detection but are sufficient to generate thiolated tRNAs in the cytoplasm via the cytoplasmic sulfur-relay system (18, 39, 40).
In mitochondria, besides Nfs1 at least one more protein is required for thiolation, namely Mtu1, the eukaryotic homolog of MnmA (20, 37). As expected, we found that down-regulation of Mtu1 in T. brucei leads to the loss of mitochondrial but not cytoplasmic thiolation. Mtu1, however, is dispensable for cell growth and the unaltered mitochondrial membrane potential in cells lacking Mtu1 shows that organellar translation in T. brucei remains functional even with decreased thiolation, allowing the assembly of respiratory complexes. This is certainly unexpected, because defects in this gene are thought to be responsible for the serious human disorder myoclonus epilepsy associated with ragged-red fibers (17). Surprisingly, the conserved nucleotide binding motif (SGGVDS), which in other organisms is located in the N-terminal part of the Mtu1 protein, is in the T. brucei homologue found within the predicted mitochondrial signal peptide. We can only speculate that in this case the in silico prediction is incorrect and that the nucleotide binding motif remains present in the Mtu1 protein.
What then is the function of mitochondrial thiolation? In the T. brucei mitochondrion, tRNATrp undergoes thiolation and contains s2U at an unusual U33 position of the anticodon loop. This modification is commonly found at U34 (the first position of the anticodon) in tRNAGlu, tRNAGln, and tRNALys in bacteria and eukarya (42). tRNATrp also undergoes C to U editing at the first position of the anticodon. However, less than 100% molecules are edited and both the UCA and CCA anticodon-containing isoacceptors co-exist in the mitochondrion (24). These two tRNAs are then presumably dedicated to decoding the UGA and UGG codons. We hypothesized that thiolation serves to maintain the levels of these two isoacceptors in the organelle. In a previous study (24), we found that down-regulation of Nfs leads to reduced thiolation of tRNAs in both cellular compartments. Surprisingly, low thiolation produces two different phenotypic outcomes: (i) it triggers destabilization of only those specific cytosolic tRNAs, which are normally thiolated; (ii) in the mitochondrion, the lack of thiolation leads to excessive editing of tRNATrp (24).
Interestingly, tRNATrp is made in the nucleus and transits through the cytoplasm, where a portion of it is maintained for cytoplasmic translation and another portion is imported into the mitochondrion. In the cytoplasm, tRNATrp is not thiolated but gets the unusual U33 thiolation following mitochondrial import. This suggests that the cytoplasmic system is unable to thiolate U33 and is specific for U34, as in the case of tRNAGln, tRNAGlu, and tRNALys in most organisms (15, 17, 24, 43). This then implies that the mitochondrial thiolation system has gained the capacity to thiolate U33 and this adaptation may either be the result of the need to regulate tRNA editing, differential compartmentalization of enzyme and substrate, or both. To what extent these processes are exploited by these parasites for fine-tuning gene expression and metabolic rates remains an open question, but intracellular compartmentalization played a central role in the evolution of these pathways. It clearly impacts the way the maturation of various substrates is specified, be it for the purpose of assembly of an ancient Fe/S co-factor or an s2U tRNA modification. We suggest that perhaps the connection between thiolation and Fe/S cluster assembly is not just a moonlighting function for the enzymes involved. Our observations raise the possibility of a cross-talk between these very important pathways in an attempt to coordinate translation with the Fe/S-dependent metabolism.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AI065935 (to K. D. S.) and GM084065 and National Science Foundation Grant MCB-0620707 (to J. D. A.), Czech Science Foundation Grant 204/09/1667, Ministry of Education of the Czech Republic Grants LC07032, 2B06129, and 6007665801, and a Praemium Academiae award (to J. D. L.).

The on-line version of this article (available at http://jbc.org) contains supplemented Figs. S1–S4.
Z. Vávrová and J. Lukeš, unpublished results.
P. Flegontov, M. Obornik, P. Changmai, Z. Paris, and J. Lukeš, unpublished results.
- RNAi
- RNA interference
- TAP
- tandem affinity purification
- APM
- [(N-acryloylamino)-phenyl]mercuric chloride.
REFERENCES
- 1.Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 2.Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 3.Zheng L., White R. H., Cash V. L., Dean D. R. (1994) Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 4.Gerber J., Mühlenhoff U., Lill R. (2003) EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramazzotti A., Vanmansart V., Foury F. (2004) FEBS Lett. 557, 215–220 [DOI] [PubMed] [Google Scholar]
- 6.Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006) EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Mühlenhoff U., Lill R., Pfanner N. (2006) EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. (2008) J. Biol. Chem. 283, 25178–25185 [DOI] [PubMed] [Google Scholar]
- 9.Shi Y., Ghosh M. C., Tong W. H., Rouault T. A. (2009) Hum. Mol. Genet. 18, 3014–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., Lill R. (2004) J. Biol. Chem. 279, 36906–36915 [DOI] [PubMed] [Google Scholar]
- 11.Goldberg A. V., Molik S., Tsaousis A. D., Neumann K., Kuhnke G., Delbac F., Vivares C. P., Hirt R. P., Lill R., Embley T. M. (2008) Nature 452, 624–628 [DOI] [PubMed] [Google Scholar]
- 12.Richards T. A., van der Giezen M. (2006) Mol. Biol. Evol. 23, 1341–1344 [DOI] [PubMed] [Google Scholar]
- 13.Shan Y., Napoli E., Cortopassi G. (2007) Hum. Mol. Genet. 16, 929–941 [DOI] [PubMed] [Google Scholar]
- 14.Esberg A., Huang B., Johansson M. J., Byström A. S. (2006) Mol. Cell 24, 139–148 [DOI] [PubMed] [Google Scholar]
- 15.Nakai Y., Umeda N., Suzuki T., Nakai M., Hayashi H., Watanabe K., Kagamiyama H. (2004) J. Biol. Chem. 279, 12363–12368 [DOI] [PubMed] [Google Scholar]
- 16.Nakai Y., Nakai M., Lill R., Suzuki T., Hayashi H. (2007) Mol. Cell. Biol. 27, 2841–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. (2005) J. Biol. Chem. 280, 1613–1624 [DOI] [PubMed] [Google Scholar]
- 18.Leidel S., Pedrioli P. G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. (2009) Nature 458, 228–232 [DOI] [PubMed] [Google Scholar]
- 19.Huang B., Lu J., Byström A. S. (2008) RNA 14, 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noma A., Sakaguchi Y., Suzuki T. (2009) Nucleic Acids Res. 37, 1335–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smíd O., Horáková E., Vilímová V., Hrdy I., Cammack R., Horváth A., Lukeš J., Tachezy J. (2006) J. Biol. Chem. 281, 28679–28686 [DOI] [PubMed] [Google Scholar]
- 22.Poliak P., Van Hoewyk D., Oborník M., Zíková A., Stuart K. D., Tachezy J., Pilon M., Lukeš J. (2010) FEBS J. 277, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simpson A. M., Suyama Y., Dewes H., Campbell D. A., Simpson L. (1989) Nucleic Acids Res. 17, 5427–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wohlgamuth-Benedum J. M., Rubio M. A., Paris Z., Long S., Poliak P., Lukeš J., Alfonzo J. D. (2009) J. Biol. Chem. 284, 23947–23953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vondrusková E., van den Burg J., Zíková A., Ernst N. L., Stuart K. D., Benne R., Lukeš J. (2005) J. Biol. Chem. 280, 2429–2438 [DOI] [PubMed] [Google Scholar]
- 26.Jensen B. C., Kifer C. T., Brekken D. L., Randall A. C., Wang Q., Drees B. L., Parsons M. (2007) Mol. Biochem. Parasitol. 151, 28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimi H., Čičová Z., Novotná L., Wen Y. Z., Lukeš J. (2009) RNA 15, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimi H., Zíková A., Panigrahi A. K., Stuart K. D., Lukeš J. (2008) RNA 14, 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zíková A., Panigrahi A. K., Dalley R. A., Acestor N., Anupama A., Ogata Y., Myler P. J., Stuart K. (2008) Mol. Cell. Proteomics 7, 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panigrahi A. K., Zíková A., Dalley R. A., Acestor N., Ogata Y., Anupama A., Myler P. J., Stuart K. D. (2008) Mol. Cell. Proteomics 7, 534–545 [DOI] [PubMed] [Google Scholar]
- 31.Long S., Jirku̇ M., Ayala F. J., Lukeš J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13468–13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panigrahi A. K., Schnaufer A., Carmean N., Igo R. P., Jr., Gygi S. P., Ernst N. L., Palazzo S. S., Weston D. S., Aebersold R., Salavati R., Stuart K. D. (2001) Mol. Cell. Biol. 21, 6833–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 34.Saas J., Ziegelbauer K., von Haeseler A., Fast B., Boshart M. (2000) J. Biol. Chem. 275, 2745–2755 [DOI] [PubMed] [Google Scholar]
- 35.Crain P. F., Alfonzo J. D., Rozenski J., Kapushoc S. T., McCloskey J. A., Simpson L. (2002) RNA 8, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihara H., Kato S., Lacourciere G. M., Stadtman T. C., Kennedy R. A., Kurihara T., Tokumoto U., Takahashi Y., Esaki N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6679–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruske E. I., Sendfeld F., Schneider A. (2009) J. Biol. Chem. 284, 36491–36499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Besteiro S., Barrett M. P., Rivière L., Bringaud F. (2005) Trends Parasitol. 21, 185–191 [DOI] [PubMed] [Google Scholar]
- 39.Johansson M. J., Esberg A., Huang B., Björk G. R., Byström A. S. (2008) Mol. Cell. Biol. 10, 3301–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlieker C. D., Van der Veen A. G., Damon J. R., Spooner E., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 47, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zutz A., Gompf S., Schägger H., Tampé R. (2009) Biochim. Biophys. Acta 1787, 681–690 [DOI] [PubMed] [Google Scholar]
- 42.Agris P. F., Söll D., Seno T. (1973) Biochemistry 12, 4331–4337 [DOI] [PubMed] [Google Scholar]
- 43.Kamenski P., Kolesnikova O., Jubenot V., Entelis N., Krasheninnikov I. A., Martin R. P., Tarassov I. (2007) Mol. Cell 26, 625–637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.