Abstract
Translation re-initiation provides the molecular basis for translational control of mammalian ATF4 and yeast GCN4 mediated by short upstream open reading (uORFs) in response to eIF2 phosphorylation. eIF4G is the major adaptor subunit of eIF4F that binds the cap-binding subunit eIF4E and the mRNA helicase eIF4A and is also required for re-initiation in mammals. Here we show that the yeast eIF4G2 mutations altering eIF4E- and eIF4A-binding sites increase re-initiation at GCN4 and impair recognition of the start codons of uORF1 or uORF4 located after uORF1. The increase in re-initiation at GCN4 was partially suppressed by increasing the distance between uORF1 and GCN4, suggesting that the mutations decrease the migration rate of the scanning ribosome in the GCN4 leader. Interestingly, eIF4E overexpression suppressed both the phenotypes caused by the mutation altering eIF4E-binding site. Thus, eIF4F is required for accurate AUG selection and re-initiation also in yeast, and the eIF4G interaction with the mRNA-cap appears to promote eIF4F re-acquisition by the re-initiating 40 S subunit. However, eIF4A overexpression suppressed the impaired AUG recognition but not the increase in re-initiation caused by the mutations altering eIF4A-binding site. These results not only provide evidence that mRNA unwinding by eIF4A stimulates start codon recognition, but also suggest that the eIF4A-binding site on eIF4G made of the HEAT domain stimulates the ribosomal scanning independent of eIF4A. Based on the RNA-binding activities identified within the unstructured segments flanking the eIF4G2 HEAT domain, we discuss the role of the HEAT domain in scanning beyond loading eIF4A onto the pre-initiation complex.
Keywords: Amino Acid, Eukaryote, Translation Control, Translation Initiation Factors, Yeast Genetics, HEAT Domain, eIF4A Helicase, eIF4F, Re-initiation
Introduction
Translation initiation in eukaryotes entails stepwise processes culminating in the assembly of 80 S ribosomal initiation complex (IC),2 with Met-tRNAiMet base-paired to the mRNA start codon composed of rAUG base triplet (for review, see Refs. 1, 2). Eukaryotic initiation factors (eIF) 1, 1A, 2, 3, and 5 together form a 43 S pre-IC (PIC) together with the 40 S ribosomal subunit. To recruit m7G-capped mRNA to the PIC, the mRNA is “primed” by the cytoplasmic cap-binding complex eIF4F, composed of the cap-binding subunit eIF4E, the mRNA helicase eIF4A, and the large adaptor subunit eIF4G. This process involves the binding of eIF4E to the cap structure and the ATP-dependent unwinding of the cap-proximal RNA by eIF4A. The mRNA interacts with the 40 S subunit over the specific path on the 40 S subunit that includes the decoding sites located between its head and body/platform domains (3). eIF4F and the attendant eIF4A-dependent unwinding remove the secondary structure for a sufficient stretch of the mRNA 5′ region to allow stable formation of the 48 S PIC at the mRNA 5′-end (4). The 48 S PIC migrates toward the mRNA 3′-end, scanning for the start codon.
The subsequent process of the 48 S PIC formation on the start codon is tightly regulated by nearly all the components making up the PIC (reviewed in Ref. 5). eIF2 is a heterotrimeric factor that directly binds Met-tRNAiMet in a GTP-dependent fashion. eIF5 promotes GTP hydrolysis for eIF2 (6). Recent studies propose that this eIF5-stimulated GTP hydrolysis occurs at an earlier stage of PIC formation, with the resulting GDP and Pi remaining on eIF2, until AUG is encountered by the anticodon of Met-tRNAiMet located at the 40 S P-site (7). eIF1 binding to the PIC near the E-site appears to suppress the transition from the “open,” scanning-competent conformation to the “closed,” scanning-incompetent conformation (8, 9). In response to the anticodon pairing to the AUG, the release of eIF1 is proposed to trigger the transition to the closed conformation, thereby allowing Pi to dissociate from eIF2 (10). These events lead to the dissociation of eIF2-GDP, eIF5, and perhaps other eIFs, which otherwise prevent binding of eIF5B and the eIF5B-mediated 60 S subunit joining that results in formation of the 80 S IC (11).
The aforementioned pathway of canonical eukaryotic translation initiation is designed to start translation from the 5′-proximal AUG of the capped mRNA. However, ∼45–50% of mammalian mRNAs (or ∼13% of yeast mRNAs) have at least one short upstream ORF (uORF) that either is bypassed by leaky scanning, prevents translation of the downstream ORF, or, alternatively, allows its re-initiation after translation of the short uORF (for review, see Ref. 12). The re-initiation is possible, if the ribosome that has finished translation of the short uORF is prevented from dissociating the mRNA and resumes scanning for a downstream start codon. Re-initiation after a short uORF in rabbit reticulocyte lysates requires eIF4F that has been loaded onto the mRNA by translation of the uORF, demonstrating that eIF4F is directly involved in scanning (13).
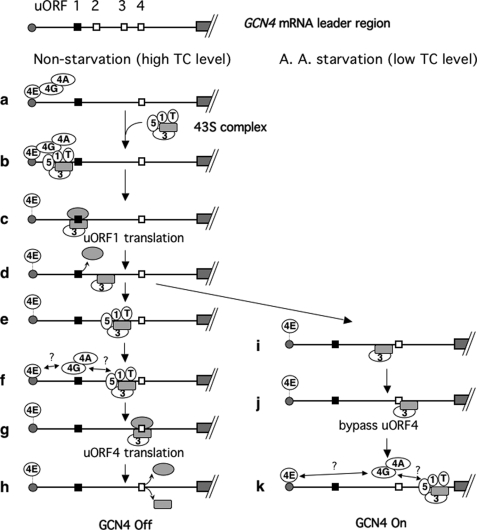
In the present study, we investigated the requirement for different activities of eIF4F in translation re-initiation of yeast GCN4 mRNA. Gcn4p is a transcription factor governing the general amino acid control response. Translation of the GCN4 ORF is activated in response to amino acid starvation and the attendant Gcn2p-catalyzed phosphorylation of eIF2 (14). This unique translational regulation is mediated by uORFs present in the leader region of GCN4 mRNA (Fig. 1, top panel). When amino acids are plentiful, interaction of the eIF3 a-subunit with a cis element found upstream of uORF1 prevents the 40 S ribosome from dissociating the mRNA after uORF1 translation (Fig. 1d) (15). Consequently, the ribosomes resume scanning, bind appropriate eIFs, and re-initiate translation at one of the inhibitory uORFs 2–4, thereby inhibiting GCN4 translation (Fig. 1, e–g). When yeast is starved for amino acids, however, the Gcn2p-catalyzed eIF2 phosphorylation inhibits guanine nucleotide exchange for eIF2, decreasing the level of eIF2/GTP/Met-tRNAiMet ternary complex (TC). The limitation of TC results in slower PIC formation with the 40 S subunits migrating after uORF1 translation, allowing the 40 S to bypass uORFs 2–4 and re-initiate at GCN4 instead (Fig. 1, i–k).
FIGURE 1.
Model for GCN4 translational control. Lines indicate the GCN4 mRNA leader with the gray boxes to the right (followed by diagonal hashed lines) representing the GCN4 coding region and the gray circle to the left representing the 5′ cap. Of the four uORFs, shown on top, two (uORFs 1 and 4) have been shown to be necessary and sufficient for regulation of GCN4 expression, and are depicted as filled and open squares, respectively, on the second line (panel a) and below (panels b–k). The figure illustrates the ribosome movement on the leader region with the focus on its association with eIF4G (4G), eIF4E (4E), eIF4A (4A), eIF1 (1), eIF3 (3), eIF5 (5), and eIF2 TC (T). eIF4E is associated with the cap loosely but strongly enough to protect it from decapping (dotted line between the cap and eIF4E). The cap interaction with eIF4E is enhanced by eIF4E binding to eIF4G (direct contact of the cap with eIF4E in panels a, b, e, f, and k). The 40 S and 60 S subunits are drawn as a gray round rectangle and gray oval, respectively. Under non-starvation conditions (left column), the preinitiation complex scans for and translates uORF1. After uORF1 translation, eIF3 bound to post-uORF1 termination 40 S prevents mRNA dissociation from the 40 S. As a consequence, a population of 40 S subunits remains associated with the mRNA and resumes scanning after reacquiring TC and other eIFs (panels e and f). Subsequent translation of uORF4 dissociates the ribosome, shutting off GCN4 translation (panels g and h). Under amino acid starvation conditions (right column), Gcn2p kinase is activated and phosphorylates eIF2. This phosphorylation renders eIF2 into a competitive inhibitor of GDP/GTP exchange activity (catalyzed by eIF2B; not shown), thereby reducing the level of eIF2/GTP and hence TC levels. uORF4 is bypassed due to low TC levels (panels i and j). TC and eIF4G can be re-acquired during scanning of the uORF4-GCN4 interval, resulting in translation of GCN4 (panel k). “?” in the figure indicates the uncertainty of the interaction of eIF4G with the cap-bound eIF4E, and the 48 S PIC migrating after uORF1 translation, which are addressed in this study.
eIF4G is a modular protein with binding sites for poly(A)-binding protein, eIF4E, and eIF4A that are conserved between yeast and humans (12) (Fig. 2A). The eIF4E-binding site comprises the conserved LLXXXLΦ motif, and the affinity of the cap to eIF4E is increased greatly by binding of this and the surrounding segment of eIF4G to eIF4E in yeast (16) (Fig. 1 also). The eIF4A-binding site forms the α-helical structure called the HEAT domain(s) (17). Mammalian eIF4G1 and eIF4G2 possess three HEAT domains (HEAT1–3) of which HEAT1 is the primary eIF4A-binding site, whereas HEAT2 is involved in regulation of eIF4A activity (4). Yeast eIF4G1 and eIF4G2 possess only HEAT1 (1). We previously showed that tif4632-1 altering the eIF4G2 HEAT1 weakly but significantly increases the frequency of translation initiation from a non-canonical UUG start codon, in a manner suppressed by eIF1 overexpression (18). These results plus the demonstration of physical interaction of eIF4G1 with eIF1 and eIF5 suggested that eIF4G is an important component of the scanning PIC, linking eIF1 to the 40 S subunit presumably in the scanning-competent conformation (18, 19). In the present study, we used variously modified GCN4-lacZ reporter constructs (20) to provide evidence that the interactions of eIF4G with both eIF4E and eIF4A (making up eIF4F), and probably their cap-binding and mRNA helicase activities promote precise selection of the canonical start codon of uORF1 as well as a start codon utilized by re-initiating 40 S subunits on the GCN4 mRNA. These and other results provide genetic evidence that eIF4F promotes re-initiation in yeast and provide insights into the mechanism by which different eIF4F activities promote mRNA recruitment and scanning.
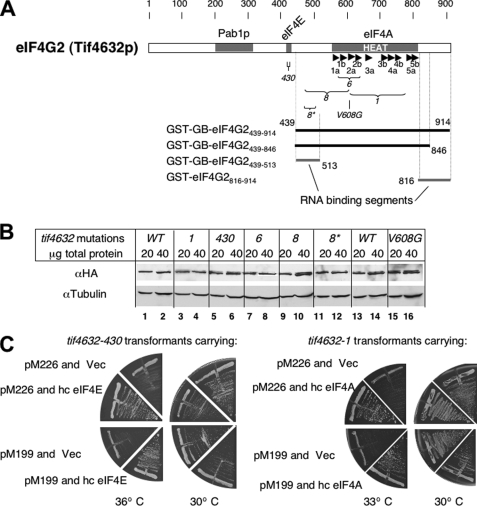
FIGURE 2.
eIF4G mutations used in this study. A, primary structure of eIF4G2 (Tif4632p) with defined binding sites for Pab1p, eIF4E, and eIF4A (gray boxes). Arrowheads denote α-helices numbered according to the structure of mammalian eIF4GII HEAT1 (17). Brackets indicate the regions of tif4632 mutations used in this study (see Table 3 for specific amino acid changes). Thick lines to the bottom represent the portion of eIF4G2 polypeptide carried by the four GST fusion constructs used here and indicated to the left (Table 1). Numbers indicate the eIF4G2 residues at the boundary of each construct. Thick gray lines indicate the RNA binding segments identified in this study (Fig. 7). B, immunoblot indicating equal abundance of HA-eIF4G2 mutants expressed from strains used in this study. Antibodies used are indicated to the left of each gel. Anti-HA detects HA-eIF4G2 and its mutants. Anti-tubulin is used as a loading control. Total protein amounts of cell extracts used are listed under mutations. Strain used are YAS1955 (WT), KAY108 (tif4632-1), KAY109 (tif4632-430), KAY110 (tif4632-6), KAY111 (tif4632-8), KAY872 (tif4632-8*), KAY220 (his4 WT), and KAY910 (his4 tif4632-V608G). C, plate assays demonstrating Ts− phenotypes of tif4632-430 (KAY109, left panels) and tif4632-1 (KAY108, right panels) transformants carrying GCN4-lacZ reporter plasmids (pM226 and pM199 used in Figs. 3 and 4, respectively) and their suppression by YEpA-TIF45 (hc eIF4E) and YEpA-TIF2 (hc eIF4A), respectively (Vec, an empty vector). Transformants carrying indicated plasmids were streaked out on plates made of synthetic defined medium and incubated at temperature listed at the bottom for 3 days.
MATERIALS AND METHODS
Plasmid Construction
Plasmids and oligodeoxyribonucleotides used in this study are listed in Tables 1 and 2, respectively. For integration of the his4-303AUU/UUG allele into yeast genome, the DNA fragment corresponding to the 5′-half of his4-303 (his4-303Δ) was amplified by PCR using oligonucleotides HIS4-Kpn/Xho and HIS4Bm (Table 2), and p440 (Table 1) as template, treated with KpnI and BamHI, and cloned into pRS306, generating pKA625. YEpA-TIF2 and -TIF45 were constructed by subcloning the 2.5-kb PstI TIF2 fragment of pAS3434 (21) and the BamHI-SalI 1.4-kb fragment of YCp-TIF45 encoding yeast eIF4E under the natural promoter3 into pRS422 (2μ ADE2), respectively. To prepare YDpU-SUI3 and -SUI3-2, we first cloned the XbaI-BamHI 0.8-kb fragment of p2122 (CEN LEU2 SUI3) (6) into pRS416 (CEN URA3) to make YDpU-SUI3(XB). Then the BamHI-HindIII 1.1-kb fragment of p2122 or p2192 (CEN LEU2 SUI3-2) (6) was cloned into YDpU-SUI3(XB) to make YDpU-SUI3 and YDpU-SUI3-2, respectively. To create plasmid to express tif4632-8* mutant, the 1.6-kb NdeI-NruI fragment of pAS3042 (HA-tif4632-8 TRP1) was cloned into the same site of pAS2068 (TIF4632 TRP1) (22), yielding YCpW-4G2–8*.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| p440 | Low copy his4–303AUU/UUG::lacZ URA3 plasmid | (44) |
| pKA625 | Integration his4–303AUU/UUGΔURA3 plasmid | This study |
| pM226 | GCN4::lacZ URA3 plasmid used in Fig. 3 | (20) |
| pM199 | GCN4::lacZ URA3 plasmid used in Fig. 4 | (20) |
| pA77 | GCN4::lacZ URA3 plasmid used in Fig. 5 | (45) |
| p235 | GCN4::lacZ URA3 plasmid used in Fig. 5 | (26) |
| pG4 | GCN4::lacZ URA3 plasmid used in Fig. 5 | (20) |
| YEpA-TIF2 | High copy TIF2 ADE2 plasmid | This study |
| YEpA-TIF45 | High copy TIF45 ADE2 plasmid | This study |
| YDpU-SUI3 | Low copy SUI3 URA3 plasmid | This study |
| YDpU-SUI3-2 | Low copy SUI3-2 URA3 plasmid | This study |
| pAS2077 | Single-copy HA-TIF4632 TRP1 plasmid | (22) |
| pAS3194 | pAS2077 carrying tif4632-V608G | A. B. Sachs |
| YCpW-4G2–8* | pAS2077 carrying tif4632-8* | This study |
| pGEX-GB-4G2ΔS-His | GST-GB-eIF4G2439–914 fusion plasmid | This study |
| pGEX-GB-4G2ΔSX | GST-GB-eIF4G2439–846 fusion plasmid | This study |
| pGEX-GB-4G2ΔSNr | GST-GB-eIF4G2439–513 fusion plasmid | This study |
| pGEX-GB-4G2ΔSNr-8* | pGEX-GB-4G2ΔSNr carrying tif4632-8* | This study |
| pGEX-4G2ΔAf | GST-eIF4G2816–914 fusion plasmid | This study |
TABLE 2.
Oligodeoxyribonucleotides used in this study
| Name | Sequence (5′ to 3′), restriction sites underlined | Description |
|---|---|---|
| 4G2-Bm-F | GGC CGG ATC CCC TGA TCC TGC GTG GGT TG | TIF4632 ORF nt 1315 to 1333 |
| 4G2-Bm-R | GGC CGG ATC CAT CAC TGT CCC CAT CGT TAT TC | TIF4632 ORF nt 2742 to 2721 |
| 4G2–816F | GGA TCC AAG AAA GAC GCC GGA CCA AAG | TIF4632 ORF nt 2446 to 2466 |
| 4G2–3′Xho | CTC GAG TTA ATC ACT GTC CCC ATC GTT A | TIF4632 ORF nt 2745 to 2724 |
| HIS4-Kpn/Xho | CGC GGT ACC TGA CCA ACA AGT GAA ACG TAT TCC TTC TTA CTA TTC CTC GAG GCC AGA TCA TCA ATT AAC GGT AGA ATC G | HIS4 ORF nt 85 to 11 |
| HIS4-Bm | GGC GGA TCC GAT CAA AGA AAA CGA AGA GAG AAA TC | 734 to 709 nt upstream of HIS4 start codon |
| HIS4–5′-Sac | CAA CAA TTG GAG CTC GAA CGC AG | 835 to 813 nt upstream of HIS4 start codon |
| HIS4–3′-RV | CTC TTT GGA GAA CTG GAG AAT CTC | HIS4 ORF nt 138 to 115 |
| HIS4-FW | ATG ACT ATG AAC AGT AGT ATA CTG | 149 to 126 nt upstream of HIS4 start codon |
| LacZ-F | AAC GTC GTG ACT GGG AAA AC | pMC1871a nt 3657 to 3676 |
| LacZ-R | GGC GGA TTG ACC GTA ATG | PMC1871a nt 3940 to 3957 |
a The GenBankTM accession number is L08936.
Three of the GST-eIF4G2 fusion constructs used in this study carry the B1 domain of streptococcal protein G (GB) as a solubility enhancement tag (23), aiming to better express the recombinant eIF4G2 segment in bacteria. To construct pGEX-GB-4G2ΔS-His, we first generated pGB-4G2ΔS by amplifying a 1.5-kb segment with PCR using oligonucleotides 4G2-Bm-F and 4G2-Bm-R and a TIF4632 DNA as template, digesting it with BamHI, followed by cloning into pGBfusion1 (23). The 1.6-kb NdeI-SalI fragment of pGB-4G2ΔS encompassing the GB-eIF4G2439–914 fusion-coding region was cloned into pGEX-4T-1 to create pGEX-GB-4G2ΔS-His. We confirmed that the insertion of GB-moiety between GST and eIF4G2 segments greatly increased the stability and expression of full-length GST-GB-eIF4G2439–914 fusion protein (also see Fig. 7A, lane 4, top Coomassie-stained gel), compared with the original GST-eIF4G2439–914 construct that was expressed from pGEX-4G2ΔS (18).
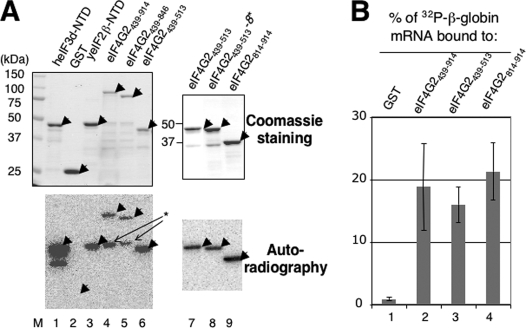
FIGURE 7.
Determination of RNA binding activity carried by the interdomain segments of yeast eIF4G2. A, Northwestern blot experiment. Recombinant eIF4G2 (lanes 4–8) or control (lanes 1–3) proteins indicated across the top are immobilized on a nitrocellulose membrane after SDS-PAGE and incubated with 32P-β-globin mRNA, as described (27). After washing, the autoradiography was taken with a PhosphorImager (bottom panels). As a control, the same amount of proteins used in this experiment was subjected for SDS-PAGE and stained with Coomassie Blue (top gels). Arrowheads indicate the expected location of the full-length products. The asterisk indicates the RNA-binding activity of a proteolytically cleaved product, derived from GST-GB-eIF4G2439–914 or GST-GB-eIF4G2439–846. M, size standards. eIF4G2 proteins used here are described in Fig. 2A and were derived from pGEX-4G2 plasmids in Table 1. heIF3d-NTD and yeIF2β-NTD, which are fused to GST, were expressed from pGEX-p66ΔE (28) and pKA883,4 respectively. B, GST-eIF4G2 proteins or GST alone indicated at the top were allowed to bind 32P-β-globin mRNA (∼20,000 cpm) and the percentage of the RNA co-precipitated by a glutathione resin (average from three independent experiments) was determined by scintillation counting and presented with bars indicating S.D.
pGEX-GB-4G2ΔSX was constructed by replacing the 0.8-kb HindIII-SalI fragment of pGEX-GB-4G2ΔS-His with the 0.6-kb HindIII-SalI fragment of pT7–4G2ΔSX (18) carrying the C-terminal deletion. To make pGEX-GB-4G2ΔSNr, pGEX-GB-4G2ΔS was digested with NruI (which cuts at the 3′ boundary of eIF4G2439–513-coding region) and SalI (which cuts at the vector cloning site 3′ of eIF4G2439–914-coding region), and the larger NruI-SalI fragment of the reaction was self-ligated after filling in with the Klenow enzyme. pGEX-GB-4G2ΔSNr -8* was made similarly from pGEX-GB-4G2ΔS-8.4 The 314-bp BamHI-XhoI fragment generated from PCR using oligonucleotides 4G2–816F and 4G2–3′Xho and a TIF4632 DNA as template was cloned into the same sites of pGEX-4T-1, creating pGEX-4G2ΔAf.
Yeast Strains
In this study, two series of yeast strains carrying eIF4G2 mutations were created and employed, as listed under columns 5 and 6 in Table 3. The first series includes YAS1955 as the wild-type parent (22), whereas the second one includes a new strain, KAY220 carrying the his4-303AUU/UUG allele. To introduce his4-303, an XhoI-linearized pKA625 DNA was used for transformation of YAS1951 to obtain Ura+ His− transformants carrying the his4Δ::URA3::his4-303 allele, which were then converted to Ura− by selecting for cells resistant to 5-fluoroorotic acid. The resulting clones with URA3 DNA looped out were either His+ or His− depending on the site of homologous recombination (due to generation of HIS4 or his4-303 on the chromosome, respectively). Thus, Ura− His− progenies were selected and analyzed further. The his4-303 allele of a Ura− His− progeny was confirmed by amplifying the DNA segment from the 5′ half of HIS4, using oligonucleotides HIS4–5′ Sac and HIS4–3′ RV, which locate outside of the area corresponding to the pKA625-borne his4-303Δ allele, and by sequencing it with oligonucleotide HIS4-FW. SUI-dependent expression of His4p from his4-303 was confirmed by His+ phenotypes, which were observed with its transformants with plasmids carrying dominant SUI alleles. The confirmed clone was designated KAY169. To perform plasmid shuffling, we introduced a TIF4632 URA3 plasmid to KAY169 and Ura+ Trp− clones were selected from the resulting transformant grown for generations in a media lacking uracyl but containing tryptophan. One such Ura+ Trp− clone was designated KAY173. The tif4632 mutant derivatives of YAS1955 and KAY169 (Table 3) were generated by introducing wild-type or mutant HA-TIF4632 TRP1 plasmid to YAS1948 (22) and KAY173, respectively, and evicting the resident TIF4632 URA3 plasmid by the growth of the resulting transformants in the presence of 5-fluoroorotic acid (plasmid shuffling).
TABLE 3.
S. cerevisiae tif4632 strains used in this study
| Tif4632 allele | Amino acid change | Mutated domain | Interaction impaired in vivo | GCN2+ strainsa | his4AUU/UUG strainsb |
|---|---|---|---|---|---|
| 1 WT | Wild-type | YAS1955 | KAY220 | ||
| 2 tif4632-1 | V608G, K644E, E756G, L783S | HEATc | eIF4A, eIF1 | KAY108 | KAY221 |
| 3 tif4632-6 | L574S, L637P | HEAT | eIF4A | KAY110 | KAY222 |
| 4 tif4632-8 | R473I, K491R, R501S, V608D | HEAT, linker | eIF4A | KAY111 | KAY223 |
| 5 tif4632-430 | L428A, L429A | LLXXXLΦ | eIF4E | KAY109 | KAY224 |
| 6 tif4632-8* | R473I, K491R, R501S | Linker | None found | KAY872 | KAY863 |
| 7 tif4632-V608G | V608G | HEAT | None found | N.C.d | KAY910 |
a Derivatives of YAS1955 (MATa ade2-1 his3–11,15 leu2–3, ura3-1 trp1-1 pep4::HIS3 tif4631::LEU2 tif4632::ura3 p[HA-TIF4632 TRP1]) carrying indicated mutations (18, 22).
b Derivatives of KAY220 (MATa ade2-1 his3–11,15 his4–303AUU/UUG leu2–3, ura3-1 trp1-1 pep4::HIS3 tif4631::LEU2 tif4632::ura3 p[HA-TIF4632 TRP1]) carrying indicated mutations.
c HEAT domain made of five α-helical HEAT repeats (aa 557–812 of 914 aa-long eIF4G2).
d N.C., not constructed for this study.
Genetic and Biochemical Methods
Standard yeast molecular biology methods, including β-galactosidase assay, were used throughout as described previously (24). Northern blotting was performed as essentially described (25), using 32P-labeled lacZ DNA probe generated by PCR using oligonucleotides LacZ-F and LacZ-R (Table 2) and p180 (26) as template. Northwestern blot determination of RNA-binding activity was performed with 32P-labeled β-globin mRNA, as described previously (27, 28).
During the analysis of GCN4-lacZ expression, we used a two-tailed t test to evaluate observed differences, if necessary. When comparing two β-galactosidase activity values (in Figs. 3C, 4B, and 5C), we grew a pair of transformants to be compared (e.g. eIF4A overexpression versus vector control) together for the same time, and collected and disrupted cells. The resulting cell extract from each transformant was assayed twice, and the average value was used for a single set of comparison (which counts as n = 1). Five to eight sets of such experiments, using >3 different transformants, were performed to calculate p values. When comparing the -fold increase of expression from pM199 or pA77 due to tif4632 mutations, a single set of experiments contained three to four independent cultures, each, of transformants of wild-type or tif4632 mutant carrying a given reporter plasmid (pM199 or pA77). The -fold increase was calculated by dividing the average GCN4-lacZ expression from the plasmid in the mutant background by that in the wild-type background. Four to five sets of such experiments were performed to obtain p values.
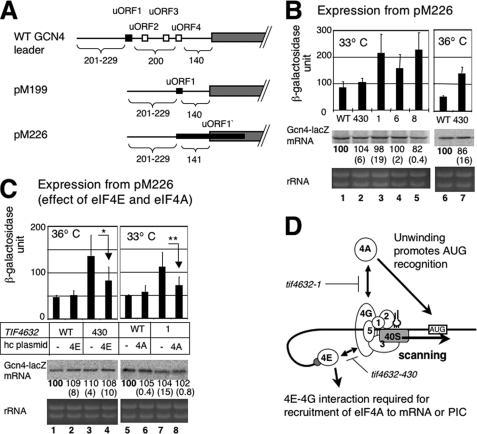
FIGURE 3.
Effect of tif4632 mutations on GCN4-lacZ expression from pM226. A, the primary structure of GCN4 leader region, either wild-type (top) or one found in pM199 (middle) or pM226 (bottom). Filled box, uORF1, the positive regulatory element. Open boxes, uORF2–4, the negative regulatory elements. Gray box, GCN4 ORF. Numbers under the schematic indicate the distance in nucleotides between 5′-end of the transcript and uORF1 (numbers to the left) and that between major elements of the leader region. B, transformants of YAS1955 (WT) and its tif4632-1 (1), -6 (6), -8 (8), and -430 (430) derivatives (Table 3) carrying pM226 were grown at indicate temperatures in synthetic complete medium lacking uracil (SC-ura) and assayed for β-galactosidase activity (24). Graphs to the top indicate average values of the β-galactosidase activity units with bars denoting S.D. (see “Materials and Methods” for statistics). Middle and bottom gels: 40 μg of total RNA isolated from the same transformants grown under the same conditions was resolved by agarose gel electrophoresis and stained by ethidium bromide (bottom gel), followed by Northern blotting with 32P-lacZ probe (middle gel). Values under the middle gel indicate the relative levels of GCN4-lacZ mRNA determined by the band 32P intensity, averaged from two independent experiments. Values in parentheses are S.D. from these experiments. C, double transformants of yeast strains as defined in B carrying pM226 and a hc ADE2 plasmid carrying eIF4E (4E), eIF4A (4A) (Table 1) or an empty vector (−) grown in SC-ura-ade were assayed for β-galactosidase activity and mRNA levels, and the data are presented as described in B. p values for differences indicated by arrows are 0.016 (n = 5) for * and 0.015 (n = 8) for **. D, model of the eIF interactions in the pre-initiation complex during the process of scanning for the uORF1 start codon (box labeled AUG). Thick line indicates mRNA, with gray circle representing 5′ m7G cap. Empty circles labeled with numbers denote relevant eIFs. The plug touching eIF2 is Met-tRNAiMet with anticodon. Stopped bars indicate interactions (arrows) impaired by tif4632 mutations.
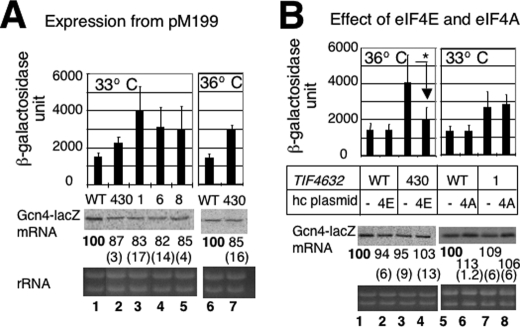
FIGURE 4.
Effect of tif4632 mutations on GCN4-lacZ expression from pM199. Transformants of YAS1955 (WT) or its tif4632 derivatives (Table 3) carrying pM199 in the presence (B) or absence (A) of additional plasmid overexpressing eIF4A (4A) and eIF4E (4E) were assayed for β-galactosidase and Northern blotting, and the data were presented as described in Fig. 3B. In B, p values for difference indicated by * is 0.007 (n = 8).
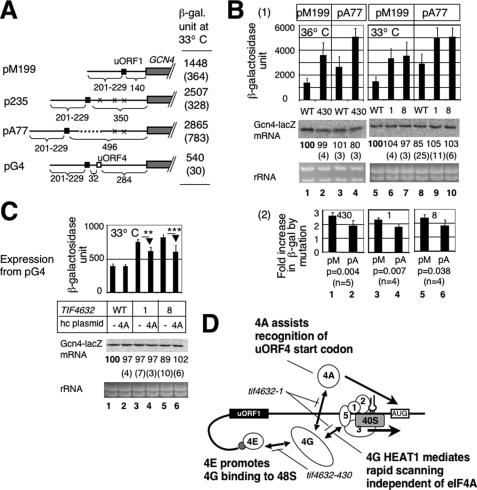
FIGURE 5.
Effect of tif4632 mutations on GCN4-lacZ expression from pA77 and pG4. A, modified GCN4 leader structures for the re-initiation reporter constructs used in this study, drawn with symbols defined in Fig. 3A. X indicates the presence of a start codon mutation in the corresponding uORF. Dotted line denotes a foreign sequence lacking start or stop codons. The table to the right summarizes β-galactosidase activity from YAS1955 transformants carrying the plasmids grown in SC-ura at 33 °C with S.D. in parenthesis. B and C, transformants of YAS1955 (WT) or its tif4632 derivatives (Table 3) carrying the indicated GCN4-lacZ plasmid in the presence (C) or absence (B and C) of an additional plasmid overexpressing eIF4A (4A) were assayed for β-galactosidase activity and mRNA levels; the data are presented as described in Fig. 3B. In B, panel 2 shows the ratio of β-galactosidase activity from pM199 (pM) or pA77 (pA) in the indicated mutant to that in wild type, based on the data in panel 1. p values for differences between the ratio with pM199 and one with pA77 are indicated below the graph. In C, p values for differences indicated by arrows are <0.001 (n = 6) for ** and 0.002 (n = 6) for ***. D, model of the eIF interactions in the pre-initiation complex during the process of scanning for a start codon (box labeled AUG) after uORF1 (filled box) translation. Symbols are as defined in Fig. 3D.
RESULTS
To dissect the requirement for cap-binding and mRNA helicase activities of eIF4F, we utilized temperature-sensitive (Ts−) mutations mapping in the eIF4E and eIF4A-binding sites of eIF4G2 (Tif4632p). We used yeast Saccharomyces cerevisiae strains deleted for chromosomal eIF4G1 (Tif4631p) and eIF4G2, the two isologues of eIF4G, but carrying a plasmid expressing the wild-type or mutant form of eIF4G2 as the sole source of eIF4G (Table 3). As shown in Fig. 2A, tif4632-430 alters the eIF4E-binding site, whereas tif4632-1, -6, and -8 altering the eIF4A-binding site (21, 22). In agreement with the identification of HEAT1 as the major eIF4A-binding site (17, 29), tif4632-1 and -6 alter four and two different amino acids, respectively, of the HEAT domain (Table 3). However, tif4632-8 not only alters one amino acid (Val-608) of HEAT1, but also changes three amino acids in the linker region between the eIF4E- and eIF4A-binding sites (Table 3). All four substitutions of tif4632-8 are required to show its Ts− phenotype (21). We created an allele, tif4632-8*, that contains only the three tif4632-8 mutations mapping in the linker region (Table 1) and confirmed that this allele confers wild-type growth without altering the expression of eIF4G2 (Fig. 2B). We also obtained a plasmid carrying tif4632-V608G (provided by Dr. Alan Sachs, see Table 1). These mutations are used to dissect the requirement for different residues altered by tif4632-1 or -8 in the specific steps of translation initiation. We confirmed that all the tif4632 mutations used in this study did not alter the abundance of HA-eIF4G2 (Fig. 2B).
For the GCN4-lacZ reporter assays, we transformed these strains with a derivative of p180 (GCN4-lacZ URA3) and if necessary, high copy (hc) plasmids encoding eIF4E (Tif45p also known as Cdc33p) or eIF4A (Tif2p) (see Table 2 for plasmids used). Ts− phenotypes of tif4632-430 and tif4632-1, -6, and -8 were suppressed by overexpression of eIF4E and eIF4A, respectively, independent of the presence of the GCN4-lacZ plasmids (pM199 and pM226, etc.; Fig. 2C and data not shown).
The eIF4G Interaction with eIF4E and eIF4A Promotes Accurate Selection of uORF1 Start Codon in Vivo
To address whether the eIF4G interaction with eIF4A and eIF4E is required for accurate AUG selection, we first measured GCN4-lacZ expression from pM226 in the tif4632 mutants. pM226 is derived from pM199, which carries the original uORF1 preceding GCN4 (20). pM199 was created by deleting the region immediately downstream of uORF1 stop codon until the stop codon of uORF4 from a parental GCN4-lacZ plasmid (Fig. 3A). Frameshift and other mutations introduced to pM199 elongated the uORF1-coding region (uORF1′) to overlap with GCN4, creating pM226: As the consequence, the expression from GCN4 was reduced by ∼20-fold in pM226 compared with expression from pM199 (20). As shown in Fig. 3B, tif4632 mutations disrupting either the eIF4A-(tif4632-1, -6, and -8) or eIF4E-(tif4632-430)-binding sites significantly increased expression of the GCN4-lacZ reporter at the semi-restrictive temperature (33 °C for the former, columns 1–5; 36 °C for the latter, columns 6–7) (p < 0.008), indicating increased frequencies of bypassing (leaky scanning) the uORF1 start codon. Importantly, overexpression of eIF4A and eIF4E from hc plasmids suppressed the increased expression by tif4632-1 and -430, respectively (Fig. 3C), as well as the Ts− growth of the transformants carrying pM226 (Fig. 2C). Moreover, these changes were observed without altering the level of GCN4-lacZ mRNA, as examined by Northern blot with a 32P-lacZ probe (Fig. 3, B and C, middle panel showing autoradiography). Thus, the interactions of eIF4G with both eIF4A and eIF4E increase the accuracy of recognition of the first AUG in the mRNA. Because eIF4A unwinds mRNA in a manner dependent on eIF4G, the requirement for eIF4G-eIF4A interaction is likely to unwind mRNA 5′ untranslated region to stimulate PIC recognition of the start codon. The requirement for eIF4G/eIF4E interaction is likely to recruit eIF4G, and hence eIF4A, to the mRNA 5′-untranslated region via its m7G-cap bound by eIF4E (Fig. 3D, also see “Discussion”). Note that these experiments do not address whether it is crucial for eIF4G to be physically linked to the scanning PIC, or whether the cap- and eIF4G/eIF4A-dependent unwinding occurs independent of the PIC function. These questions are addressed below, using other GCN4-lacZ reporters.
An Intact eIF4G HEAT1 and eIF4E/eIF4G Interaction Is Required for Normal Re-initiation of GCN4 mRNA
To address the role of eIF4G interaction with eIF4E and eIF4A in re-initiation, we used pM199, encoding GCN4 preceded by a single uORF1 in the leader region (Fig. 3A). We found that all the tif4632 Ts− mutations tested increased GCN4-lacZ expression from pM199 at their semi-restrictive temperatures (p < 0.04) without changing GCN4-lacZ mRNA level (Fig. 4B). Although these mutations increased GCN4-lacZ expression from pM226 by ∼80–120 β-galactosidase units due to leaky scanning of uORF1 (Fig. 3B), much larger increase of >1000 units observed with pM199 in these mutants suggests that the increase is not simply due to the leaky scanning mechanism. Rather, the tif4632 mutations appear to increase the frequency of translational re-initiation following uORF1. (This comparison requires that Gcn4-lacZ mRNA is expressed at the same level from pM226 and pM199. Indeed, Northern blot indicates that the amount of Gcn4-lacZ mRNA from pM226 is 92 + 7% (n = 3) of that from pM199 in wild-type cells under our growth conditions.) As shown in Fig. 4C, columns 3 and 4, hc eIF4E suppressed the increase in expression from pM199 caused by tif4632-430, suggesting that the observed change is due to defective interaction with eIF4E. In contrast, hc eIF4A did not suppress the increase caused by tif4632-1 (Fig. 4C, columns 7 and 8). This result supports the idea that the leaky scanning of uORF1 may not account for the increase in expression from pM199, because GCN4-lacZ expression from pM226 was suppressed by hc eIF4A (Fig. 3C). The inability of hc eIF4A to suppress the increase in expression from pM199 in turn suggests that tif4632-1 impaired another function of the eIF4G HEAT1 beyond promoting the interaction with eIF4A (see below).
In contrast to the eIF3a mutation that decreased expression from pM199 (15), the increased expression or re-initiation efficiency in the tif4632 mutants observed here was puzzling. Previous studies indicated that only half or fewer of the 40 S subunits migrating from uORF1 to GCN4 could re-acquire eIFs and re-initiate at GCN4 when the intergenic distance from uORF1 was 140 bases (as in pM199); the efficiency of translation re-initiation progressively increased as the intergenic distance was increased, suggesting that the migrating 40 S would have longer time to re-acquire eIFs (20). Indeed, we confirmed this trend with additional constructs, p235 and pA77, with the intergenic distance of 350 and 496 bases, respectively, under our experimental conditions (Fig. 5A). Thus, the increase in re-initiation frequency observed in Fig. 4 can be interpreted as a decrease in the migration rate of the scanning PIC. To test if this is the case, we examined whether the increase in the intergenic distance can mask the effect of the mutations on the re-initiation frequency. As shown in Fig. 5B, panel 1, we found that the magnitude of increase in GCN4-lacZ expression from pA77, with a wider intergenic distance, by each of the tif4632 mutations tested was significantly lower than that of increase in expression from pM199 (see panel 2 for quantification). These changes were observed without altering the level of GCN4-lacZ mRNA (Fig. 2C, panel 1, bottom two gels). Thus, the increase in the intergenic distance at least partially masks the effect of tif4632 mutations on expression from pM199.
To exclude the possibility that the observed effect of increasing the intergenic distance is an artifact due to insertion of a foreign sequence into pA77, we examined the effect of eIF4G mutations on GCN4-lacZ expression from p235 without the foreign sequence (see Fig. 5A for their GCN4-lacZ leader structure). As shown in supplemental Fig. S1, tif4632-6, -8, and -430 increased the expression only <1.5-fold at their semi-restrictive temperatures (columns 2, 5, and 6), in contrast to significant ∼2- to 2.5-fold increase by the same mutations, which were observed with pM199 (Figs. 4B and 5B). tif4632-1 increased Gcn4-lacZ activity from p235 by 2.5-fold, to the same extent as from pM199 (supplemental Fig. S1, column 4). Although further statistical examination needs to establish the significance of this finding, it could be attributed to the shorter intergenic distance of this construct than that of pA77. These results together support the model that the eIF4G mutations decrease the migration rate of the 40 S between uORF1 and GCN4.
That hc eIF4E suppressed the increased expression from pM199 caused by tif4632-430 (Fig. 4C, columns 3 and 4) suggests that the eIF4G interaction with eIF4E promotes the presumed rapid scanning. This would provide the first genetic or in vivo evidence that the 48 S complex during the course of scanning (re-initiation) must be linked to m7G-cap via eIF4E-eIF4G interaction. In contrast, hc eIF4A did not suppress the increase caused by tif4632-1 (Fig. 4C, columns 7 and 8). This result can be interpreted as suggesting that the eIF4G HEAT1, altered by this mutation, plays an eIF4A-independent role in promoting mRNA scanning (see below Fig. 6). In agreement with an eIF4A-independent role, the increase in expression from pM199 in tif4632-1, -6, and -8 mutations was observed also at their permissive temperature of 30 °C, and in addition, tif4632-8* carrying the three tif4632-8 mutations mapping in the linker region did not alter expression from pM199 at 30° or 36 °C.5
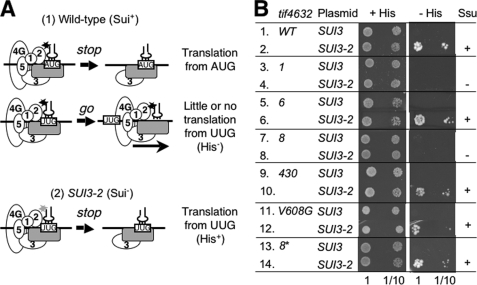
FIGURE 6.
tif4632 mutations suppress relaxed start codon selection caused by an eIF2β mutation (SUI3-2). A, model of Sui− phenotypes. A, panel 1, wild-type PIC fully loaded with different eIFs (circles with a number) can efficiently distinguish between AUG and UUG codons (boxed) on the mRNA (horizontal line), in part due to tight interaction (asterisk) between eIF2 and tRNAiMet (plug). If it encounters AUG, the PIC responds (stop) and releases eIFs to bind the 60 S, but if it encounters UUG, the PIC does not respond and resumes scanning (go). A, panel 2, when SUI3-2 impairs eIF2 interaction with tRNAiMet (light gray asterisk), the PIC encountering UUG erroneously responds (stop) and starts translation (Sui−). B, 5 μl of 0.15 A600 unit cultures of transformants of KAY220 (his4-303) and its tif4632 mutant derivatives carrying YDpU-SUI3 (SUI3) or YDpU-SUI3-2 (SUI3-2) and their 10-fold dilution were spotted onto SC-ura (+His) and SC-ura-his (−His) plates and incubated at 30 °C for 3 and 7 days, respectively.
eIF4G-eIF4A Interaction Also Stimulates Re-initiation at uORF4
In the absence of the evidence for the requirement for eIF4A/eIF4G interaction in re-initiation at GCN4 after uORF1 translation, we examined whether this interaction stimulates re-initiation at uORF4 instead of GCN4. The placement of uORF4 at 32 nucleotides downstream of uORF1 effectively represses re-initiation at GCN4 in the plasmid pG4, compared with re-initiation efficiency measured with p235, which lacks uORF4 but carries an equivalent distance between uORF1 and GCN4 (20) (also see Fig. 5A). Thus, ∼80% (Fig. 7B of Ref. 20 or (2507–540)/2507 from the values in Fig. 5A) of the 40 S, which was linked to the mRNA after uORF1 translation, re-initiated at uORF4 in this construct, despite the short distance from uORF1 (see “Discussion”). The eIF4A-binding site mutations tif4632-1 and -8 increased expression from pG4 (p < 0.0001) by ∼400–500 β-galactosidase units without altering mRNA abundance (Fig. 5C, columns 3 and 5). Because the leaky scanning mechanism may not explain this large increase (Fig. 3), this increase is most likely due to inhibition of re-initiation at uORF4, as expected. Furthermore, this increase was partially suppressed by hc eIF4A (Fig. 5C, columns 4 and 6). Thus, re-initiation at uORF4 at least partially depends on eIF4A/eIF4G interaction (Fig. 5D).
Together, the results shown in Figs. 4 and 5 suggest that, during re-initiation by the post-uORF1-termination 40 S, (i) eIF4E/eIF4G interaction and an intact HEAT domain are required for its rapid scanning for GCN4 start codon and (ii) the eIF4A/eIF4G interaction and hence, likely eIF4A-dependent mRNA unwinding, assist start codon recognition at uORF4. The presumed slow scanning phenotype caused by the HEAT1 mutation, tif4632-1, appeared to be independent of eIF4A (Fig. 4C), even though the same mutation seemed to impair re-initiation at uORF4 in a manner dependent on eIF4A.
eIF4A-independent Role of eIF4G HEAT1 in Regulation of Start Codon Selection
In an effort to understand the molecular basis of the eIF4A-independent role of the eIF4G HEAT domain, we studied the genetic relationship of tif4632 mutations with a mutation known to relax stringency of AUG selection. The SUI3-2 mutation altering eIF2β reduces eIF2 interaction with tRNAiMet, thereby increasing the spontaneous release of eIF2 from the scanning ribosome (6), and hence, the frequency of initiating translation from non-canonical UUG start codon (Suppressor of initiation codon mutation or Sui− phenotype, see Fig. 6A, panels a and b). Thus, a SUI3-2 plasmid dominantly suppresses the His− phenotype of the parental strain carrying the his4-303AUU/UUG allele. In this allele the HIS4 AUG start codon is changed to AUU (Fig. 6B, rows 1 and 2) and allows translation to initiate from the third UUG codon of HIS4 (6). We introduced the his4-303AUU/UUG allele to YAS1955 creating KAY220 and generated its derivatives carrying the tif4632 mutations (see “Materials and Methods” and Table 3). As shown in Fig. 6B, rows 4 and 8, tif4632-1 and tif4632-8 suppressed the His+ (Sui−) phenotype caused by SUI3-2 at the permissive temperature of 30 °C (Ssu−; suppressor of Sui phenotype), genetically relating the function of eIF4G with that of eIF2. However, the Ssu− phenotypes were not suppressed by hc eIF4A.5 These results support an eIF4A-independent role of the HEAT1 in regulation of AUG selection.
Because both the Ssu− mutations, tif4632-1 and -8, alter Val-608 within the HEAT domain (Fig. 2 and Table 3), we introduced tif4632-V608G to the his4-303 strain and found that this mutation did not alter the Sui− phenotype caused by SUI3-2 (Fig. 6B, rows 11 and 12). Moreover, tif4632-8* carrying the three amino acid substitutions of tif4632-8 located within the linker region was Ssu+ (Fig. 6B, rows 13 and 14). Therefore, the Ssu phenotype depends on the magnitude of disruption of the HEAT1 and its surrounding structure, rather than a specific residue of HEAT1.
Polypeptides Flanking Yeast eIF4G2 HEAT1 Bind RNA
Among many possibilities, a model to interpret the eIF4A-independent role of the HEAT1 during mRNA scanning is that it antagonizes re-folding of the mRNA by binding mRNA in a single-stranded state. The defect in such activity would destabilize the scanning PIC, impairing its propensity to scan rapidly or initiate at a suboptimal start codon. In support of this idea, RNA-binding activities were observed with the segment N-terminal to mammalian eIF4GI and the segments flanking yeast eIF4G1 (Tif4631p) HEAT1 (30, 31). It is possible that the intact HEAT1 is required for the coordination of these RNA-binding activities.
To test if the segments flanking the yeast eIF4G2 HEAT1 also bind RNA, we constructed, purified, and immobilized various truncated forms of eIF4G2 to a membrane after SDS-PAGE and allowed them to bind 32P-labeled β-globin mRNA (Northwestern blotting). As shown in Fig. 7A, bottom autoradiograph, the RNA was bound to a GST fusion form of the entire C-terminal half (aa 439–914) of yeast eIF4G2 containing the region N-terminal to the eIF4E-binding site (lane 4), as well as the shorter GST-fused constructs eIF4G2 (aa 439–846) (lane 5), eIF4G2 (aa 439–513) (lanes 6 and 7), and eIF4G2 (aa 816–914) (lane 9), of which the last two are segments N- or C-terminal to the HEAT1, respectively (see Fig. 2A). As controls, the NTDs of human eIF3d (28) and yeast eIF2β (32) subunits, which are known to bind the RNA, indeed bound to the 32P-mRNA (lanes 1 and 3). However, the GST alone (lane 2) or GST-eIF2β lacking the RNA-binding site (32)6 did not bind the 32P-mRNA.
To rule out the possibility that the RNA binding is the artifact due to denaturing of the proteins during SDS-PAGE, we performed GST pulldown assays of the 32P-mRNA. We confirmed that the eIF4G2(439–914), eIF4G2(439–513), and eIF4G2(816–914) polypeptides specifically co-precipitated the 32P-mRNA, when attached to a glutathione resin (Fig. 7B). Thus, we localized the RNA-binding activity to at least two distinct locations within yeast eIF4G2 outside of HEAT1 (see Fig. 2A for summary).
Because the RNA binding segment N-terminal to HEAT1 is characterized as being Arg-rich, and all three tif4632-8 mutations in this area alter Arg or Lys (Table 3), we tested their effect on the ability of eIF4G2 (aa 439–513) to bind RNA. However, the eIF4G segment with these mutations bound RNA as efficiently as wild-type (Fig. 7A, lane 8). Thus, the defect caused by tif4632-8 may not include the defective binding of RNA by this segment of eIF4G2.
In conclusion, our results demonstrating RNA-binding activities within the linker or C-terminal segments flanking eIF4G2 HEAT1 support the model that the eIF4A-independent role of eIF4G HEAT1 during the scanning process is to increase the integrity of the PIC-mRNA complex by binding unwound mRNA (see “Discussion”).
DISCUSSION
In the present study, we showed using GCN4-lacZ reporter assays that the interactions of eIF4G with both eIF4E and eIF4A stimulate the recognition of the start codon of uORF1 (Fig. 3) and a start codon following uORF1, either one for uORF4 or GCN4, during re-initiation (Figs. 4 and 5). The involvement of the interactions with eIF4A and eIF4E in selecting the first start codon of the mRNA (Fig. 3) provides in vivo evidence that cap-dependent unwinding mediated by eIF4F promotes the recruitment and/or scanning of the capped mRNA. The involvement of these interactions in selecting a start codon following uORF1 during re-initiation (Figs. 4 and 5) is also significant, because it not only shows that eIF4F promotes re-initiation in yeast, but also extends our previous proposal that eIF4F is an important part of the scanning PIC (18).
The Role of eIF4F and Cap-dependent mRNA Unwinding in mRNA Scanning, AUG Selection, and Re-initiation
The GCN4-lacZ reporter plasmid pM226 (Fig. 3A) has been used to detect a defect in AUG recognition cause by mutations altering eIF5 and eIF5B (1). The elongated uORF1′ in this plasmid effectively suppresses translation from GCN4, and only a fraction (∼2–5%) of the 40 S subunits loaded onto the 5′-cap can initiate at GCN4 (20). The mutations in the above-mentioned factors were interpreted to allow the loaded ribosomes to bypass the start codon of uORF1′, thereby increasing initiation from GCN4. Here we showed that eIF4G2 mutations altering the eIF4E- and eIF4A-binding sites display such a phenotype (Fig. 3). The suppression of the phenotypes caused by tif4632-430 and -1 by eIF4E and eIF4A overexpression, respectively, suggests that eIF4G interaction with the 5′-cap and eIF4A is important for the recognition of uORF1 start codon (Fig. 3C). The requirement for the interaction with 5′-cap can be interpreted as that for loading of the whole eIF4F complex, including eIF4A, onto the mRNA.
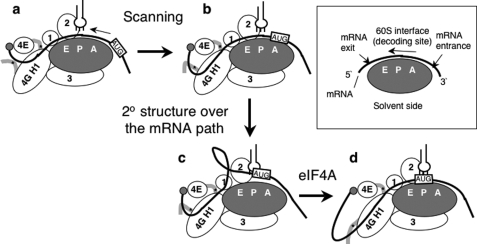
Then how does eIF4G binding to eIF4A promote the selection of start codons? In principle, eIF4A can promote the PIC scanning for start codon by unwinding mRNA structures at the leading edge before mRNA enters the PIC or at the trailing edge after mRNA exits it. Cryoelectron microscopy studies on the structure of mammalian eIF3/eIF4F and eIF3/40 S complexes proposed a three-dimensional model locating eIF4F (linked to the 40 S via eIF3) close to the mRNA exit of the 40 S (33) (Fig. 8), in agreement with previous footprinting studies suggesting that factors in the 48 S complex protects the 5′ but not 3′ side of the ribosome-bound mRNA (34). If this is the case, eIF4A would access the mRNA from the exit side, whereas the mRNA structure at the leading edge is unwound by the helicase activity of the ribosome itself (35) or additional mRNA helicases, such as DHX27 in mammals (36) or Ded1p and Dbp1p in yeast (37, 38). Mammalian eIF4G contains two HEAT domains, and the interaction of eIF4A with HEAT2 can potentially locate eIF4A at the leading edge (4). However, yeast eIF4G lacks HEAT2; thus the role of eIF4A in mRNA unwinding at the leading edge is less feasible in yeast. If distinct hypothetical mRNA helicase(s) unwind mRNA at the leading edge, secondary structures would develop by refolding over the mRNA path as mRNA enters the 48 S PIC and migrates along the path (Fig. 8b). eIF4A associated with eIF4G would be situated at the appropriate position to resolve the secondary structure and promote matching of codon-anticodon pairing (Fig. 8, c and d).
FIGURE 8.
Model depicting the roles of eIF4G HEAT domain 1 (4G H1) and eIF4A in scanning and AUG selection. The scanning starts with the 40 S subunit (gray oval) bound to the mRNA (thick line) near the 5′-cap (gray circle) (a) and continues as the mRNA migrates over the path on the 40 S subunit (b). The model hypothesizes that a secondary structure develops as mRNA migrates over the path and thereby prevents correct matching of tRNAiMet (plug) anticodon to start codon (boxed and labeled AUG) (c). eIF4A resolves the structure and promotes AUG recognition (d). In addition to eIF4A, RNA binding to eIF4G HEAT1 and/or its surrounding peptides (shown by thick gray line with asterisk indicating the RNA-binding site) prevents mRNA refolding, thereby promoting scanning and AUG recognition. The orientation of eIF4G HEAT1 relative to the 40 S is based on the speculative model proposed recently (4). eIF4G HEAT1 binding to eIF1 (circle labeled 1) located near the E-site (9, 43) is proposed to contribute to maintaining a scanning-competent conformation (18). The box describes the 40 S subunit structure with A-, P-, and E-site and the orientation of bound mRNA.
Here we also showed that the eIF4G interactions with both eIF4E and eIF4A stimulate (normal) re-initiation at GCN4 (Fig. 4B, columns 1–4) and at uORF4 (Fig. 5C), respectively, using different reporter plasmids termed pM199 and pG4. In the mammalian re-initiation that occurs following a short uORF, eIF4F that had apparently been loaded onto the mRNA via uORF translation was required for re-initiation at a downstream start codon (13). The eIF4F used in the assay could be replaced by recombinant forms of eIF4G lacking the N-terminal poly(A)-binding protein- and eIF4E-binding sites, but re-initiation with these proteins depended on eIF4A (13). Thus, the interaction with eIF4A and likely attendant ATP-dependent unwinding was necessary for re-initiation following a short uORF, in agreement with our results using yeast carrying pG4 (Fig. 5C). However, the link of post-termination PIC to eIF4E, and hence to the cap structure, was not necessary for re-initiation in the mammalian case (13), in contrast to the re-initiation of GCN4 found in pM199 (Fig. 4B). This difference in requirement for eIF4E could be accounted for by the well documented difference between the mechanisms whereby eIF4G is recruited to the PIC in yeast and mammals (1). In the case of mammals, eIF4G interacts with eIF3, thereby recruiting the 43 S PIC to eIF4F-primed mRNA (33) or linking eIF4F to eIF3-bound 40 S after translation of short uORFs (13). However, in the case of yeast, eIF5 is a major link of eIF4F to the 43 S PIC (18, 39). Because eIF5 would dissociate from the PIC in complex with eIF2/GDP on AUG selection at uORF1 (40), eIF3 that is preloaded by uORF1 translation probably lacks this component and hence eIF4G prior to re-initiation. Therefore, eIF4F might need to be re-acquired through the cap interaction with eIF4E (Fig. 1, f and k).
An eIF4A-independent Role of the eIF4G HEAT1 in mRNA Scanning, AUG Selection, and Re-initiation
Our finding that an increase in the intergenic distance between uORF1 and GCN4 partially restored the increase in the re-initiation efficiency caused by tif4632 mutants tested (Fig. 5B, panel 2) support the model that these mutations decrease the migration rate (scanning) of the PIC, thereby increasing the percentage of the migrating PIC to gain competency to re-initiate at GCN4. Because this increase in re-initiation with pM199 by tif4632-1 was not suppressed by eIF4A overexpression (Fig. 4C), we suggest that the intact HEAT1 itself also promotes mRNA scanning aside from its role in loading eIF4A. Biochemical studies showed that eIF4F, and perhaps a part of eIF3, forms an extended mRNA path 5′ to the mRNA path of the 40 S itself (3, 34). A hypothetical path of mRNA along the eIF4G HEAT domains was visualized in a three-dimensional model, based on the homology with the nuclear cap-binding protein CBP80 bound to mRNA (41). RNA-binding activities observed with the eIF4G HEAT1 (17) as well as their flanking segments (30, 31) (Fig. 7) might be used to antagonize refolding of mRNA, thereby promoting the PIC migration and recognition of the start codon (Fig. 8). If this is the case, the eIF4A-independent role of eIF4G HEAT1 would be to coordinate such RNA-binding activities and keep mRNA in an unwound state; tif4632-1, -6, and -8 directly altering the HEAT1 and tif4632-430 impairing cap-dependent eIF4G recruitment to the PIC in the GCN4 leader would slow down the scanning because of the disruption or lack, respectively, of the presumed regulation of RNA-binding activity carried by HEAT1.
The relatively minor difference in the increase in re-initiation between pM199 and pA77 (compare columns 1 with 2, 3 with 4, and 5 with 6 in Fig. 5B, panel 2) suggests that in order for all the PICs in the GCN4 leader to gain competency to re-initiate at GCN4, the distance between uORF1 and GCN4 has to be much wider than the distance of ∼500 bp found in pA77 (Fig. 5A). This is in contrast to re-initiation at uORF4, because the majority (∼80%) of the post-uORF1 ribosomes can re-initiate at this uORF even when it is located only 54 bp after uORF1 (Fig. 5A, pG4) (20). We suggest that this difference depends on the sequence context of the respective start codons, in particular the stability of local secondary structures surrounding them. In agreement with this idea, translation from a luciferase reporter construct with 5′-untranslated region of GCN4, but lacking all uORFs due to start codon mutations (L2-luc), was decreased 2-fold by ded1-Ts− or dbp1 deletion mutations, but was not decreased by eIF4A-Ts− or eIF4B deletion mutations (42). Thus, the helicases Ded1p and/or Dbp1p appear to contribute to the competency to initiate at GCN4. It would be important to examine whether start codons found in the GCN4 leader region require different helicase activities for their use in translation and whether this forms the molecular basis for the efficient GCN4 translational control by eIF2 phosphorylation.
Finally, how did the HEAT1 mutations tif4632-1 and -8 suppress the Sui− phenotype caused by SUI3-2 (Fig. 6)? We suggest that the 48 S complex containing the mutant eIF2, which has encountered the UUG codon, cannot tolerate the additional mutation(s) in the HEAT1 that destabilizes the eIF4G interaction with a 48 S complex component, such as eIF1 (18) and mRNA (Fig. 7). As a consequence, the 48 S complex on the UUG codon would be disrupted (rather than producing the 80 S IC), inhibiting translation of the mutant his4 allele. Alternatively, the slow scanning per se potentially caused by these mutations might somehow suppress the conformational change of the scanning PIC that would promote the release of eIF2 carrying SUI3-2. In support of the former scenario, we previously showed that tif4632-1 reduces the interaction with eIF1 and weakly increases translation from a UUG codon of the mutant his4-lacZ reporter in a manner suppressible by eIF1 overexpression (18) (also see Fig. 8 for the location of eIF1 close to eIF4G HEAT1). Apparently, this level of his4 expression was not sufficient to confer His+ phenotype to the tif4632-1 his4-303 strain (Fig. 6B, row 3) but was not increased further by the additional disruption of the 48 S PIC by SUI3-2 dislodging eIF2 from the PIC (Fig. 6B, row 4). In the case of tif4632-8, this alters the linker segment, which alone can also bind RNA as well as eIF1 and eIF5, although the three tif4632-8 amino acid substitutions altering this segment alone cannot eliminate the interaction with any of them (Fig. 7).7 Therefore, its Ssu− phenotype must be understood in the context of the entire eIF4G structure, including the HEAT domain. Further studies dissecting factor interactions within the scanning PIC will reveal how the PIC regulates translation initiation as well as the potential to re-initiate translation of certain mRNAs with uORF(s).
Supplementary Material
Acknowledgments
We are indebted to Ernie Hannig for critical reading of the manuscript and Alan Hinnebusch and Gerhard Wager for inspiring discussion and advice. We also thank Alan Sachs and Hui He for sharing results and materials prior to publication, Yuka Ikeda for technical assistance, and Tao You for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM64781 (to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
K. Asano, unpublished material.
K. Asano and M. J. Murai, unpublished material.
R. Watanabe and K. Asano, unpublished observations.
M. J. Murai and K. Asano, unpublished observations.
C. R. Singh, R. Watanabe, and K. Asano, unpublished observations.
- IC
- initiation complex
- PIC
- pre-IC
- eIF
- eukaryotic initiation factor
- uORF
- upstream open reading frame
- TC
- ternary complex
- GB
- B1 domain of streptococcal protein G
- GST
- glutathione S-transferase
- aa
- amino acid(s)
- nt
- nucleotide(s)
- hc
- high copy plasmid
- NTD
- N-terminal domain.
REFERENCES
- 1.Hinnebusch A. G., Dever T. E., Asano K. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 225–268 [Google Scholar]
- 2.Pestova T. V., Lorsch J. R., Hellen C. U. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. eds) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 87–128 [Google Scholar]
- 3.Pisarev A. V., Kolupaeva V. G., Yusupov M. M., Hellen C. U., Pestova T. V. (2008) EMBO J. 27, 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marintchev A., Edmonds K. A., Maritcheva B., Hendrickson E., Oberer M., Suzuki C., Herdy B., Sonenberg N., Wagner G. (2009) Cell 136, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano K., Sachs M. S. (2007) Genes Dev. 21, 1280–1287 [DOI] [PubMed] [Google Scholar]
- 6.Huang H. K., Yoon H., Hannig E. M., Donahue T. F. (1997) Genes Dev. 11, 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Algire M. A., Maag D., Lorsch J. R. (2005) Mol. Cell 20, 1–12 [DOI] [PubMed] [Google Scholar]
- 8.Cheung Y. N., Maag D., Mitchell S. F., Fekete C. A., Algire M. A., Takacs J. E., Shirokikh N., Pestova T., Lorsch J. R., Hinnebusch A. G. (2007) Genes Dev. 21, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Passmore L. A., Schmeing T. M., Maag D., Applefield D. J., Acker M. G., Algire M. A., Lorsch J. R., Ramakrishnan V. (2007) Mol. Cell 26, 41–50 [DOI] [PubMed] [Google Scholar]
- 10.Maag D., Fekete C. A., Gryczynski Z., Lorsch J. R. (2005) Mol. Cell 17, 265–275 [DOI] [PubMed] [Google Scholar]
- 11.Shin B. S., Maag D., Roll-Mecak A., Arefin M. S., Burley S. K., Lorsch J. R., Dever T. E. (2002) Cell 111, 1015–1025 [DOI] [PubMed] [Google Scholar]
- 12.Jackson R. J., Hellen C. U., Pestova T. V. (2010) Nat. Rev. Mol. Cell Biol. 10, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pöyry T. A., Kaminski A., Jackson R. J. (2004) Genes Dev. 18, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 15.Szamecz B., Rutkai E., Cuchalová L., Munzarová V., Herrmannová A., Nielsen K. H., Burela L., Hinnebusch A. G., Valásek L. (2008) Genes Dev. 22, 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross J. D., Moerke N. J., von der Haar T., Lugovskoy A. A., Sachs A. B., McCarthy J. E., Wagner G. (2003) Cell 115, 739–750 [DOI] [PubMed] [Google Scholar]
- 17.Marcotrigiano J., Lomakin I. B., Sonenberg N., Pestova T. V., Hellen C. U., Burley S. K. (2001) Mol. Cell 7, 193–203 [DOI] [PubMed] [Google Scholar]
- 18.He H., von der Haar T., Singh R. C., Ii M., Li B., Hinnebusch A. G., McCarthy J. E., Asano K. (2003) Mol. Cell. Biol. 23, 5441–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., Wagner G., Asano K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant C. M., Miller P. F., Hinnebusch A. G. (1994) Mol. Cell. Biol. 14, 2616–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neff C. L., Sachs A. B. (1999) Mol. Cell. Biol. 19, 5557–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarun S. Z., Jr., Sachs A. B. (1997) Mol. Cell. Biol. 17, 6876–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., Lugovskoy A. A., Wagner G. (2001) J. Biomol. NMR 20, 11–14 [DOI] [PubMed] [Google Scholar]
- 24.Singh C. R., Asano K. (2007) Methods Enzymol. 429, 139–161 [DOI] [PubMed] [Google Scholar]
- 25.Udagawa T., Nemoto N., Wilkinson C. R., Narashimhan J., Watt S., Jiang L., Zook A., Jones N., Wek R. C., Bähler J., Asano K. (2008) J. Biol. Chem. 283, 22063–22075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller P. P., Hinnebusch A. G. (1986) Cell 45, 201–207 [DOI] [PubMed] [Google Scholar]
- 27.Wei C. L., MacMillan S. E., Hershey J. W. B. (1995) J. Biol. Chem. 270, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 28.Asano K., Vornlocher H. P., Richter-Cook N. J., Merrick W. C., Hinnebusch A. G., Hershey J. W. (1997) J. Biol. Chem. 272, 27042–27052 [DOI] [PubMed] [Google Scholar]
- 29.Oberer M., Marintchev A., Wagner G. (2005) Genes Dev. 19, 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H. (2003) RNA 9, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prévôt D., Décimo D., Herbreteau C. H., Roux F., Garin J., Darlix J. L., Ohlmann T. (2003) EMBO J. 22, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurino J. P., Thompson G. M., Pacheco E., Castilho B. A. (1999) Mol. Cell. Biol. 19, 173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siridechadilok B., Fraser C. S., Hall R. J., Doudna J. A., Nogales E. (2005) Science 310, 1513–1515 [DOI] [PubMed] [Google Scholar]
- 34.Lazarowitz S. G., Robertson H. D. (1977) J. Biol. Chem. 252, 7842–7849 [PubMed] [Google Scholar]
- 35.Takyar S., Hickerson R. P., Noller H. F. (2005) Cell 120, 49–58 [DOI] [PubMed] [Google Scholar]
- 36.Pisareva V. P., Pisarev A. V., Komar A. A., Hellen C. U., Pestova T. V. (2008) Cell 135, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de la Cruz J., Iost I., Kressler D., Linder P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. (1997) Science 242, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 39.Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valásek L., Donahue T. F., Hinnebusch A. G. (2001) EMBO J. 20, 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh C. R., Lee B., Udagawa T., Mohammad-Quereshi S. S., Yamamoto Y., Pavitt G. D., Asano K. (2006) EMBO J. 25, 4537–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marintchev A., Wagner G. (2005) Biochemistry 44, 12265–12272 [DOI] [PubMed] [Google Scholar]
- 42.Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 43.Lomakin I. B., Kolupaeva V. G., Marintchev A., Wagner G., Pestova T. V. (2003) Genes Dev. 17, 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donahue T. F., Cigan A. M. (1988) Mol. Cell Biol. 8, 2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. (1991) Mol. Cell Biol. 11, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.