Abstract
The murine KRAS promoter contains a G-rich nuclease hypersensitive element (GA-element) upstream of the transcription start site that is essential for transcription. Pulldown and chromatin immunoprecipitation assays demonstrate that this GA-element is bound by the Myc-associated zinc finger (MAZ) and poly(ADP-ribose) polymerase 1 (PARP-1) proteins. These proteins are crucial for transcription, because when they are knocked down by short hairpin RNA, transcription is down-regulated. This is also the case when the poly(ADP-ribosyl)ation activity of PARP-1 is inhibited by 3,4-dihydro-5-[4-(1-piperidinyl) butoxyl]-1(2H) isoquinolinone. We found that MAZ specifically binds to the duplex and quadruplex conformations of the GA-element, whereas PARP-1 shows specificity only for the G-quadruplex. On the basis of fluorescence resonance energy transfer melting and polymerase stop assays we saw that MAZ stabilizes the KRAS quadruplex. When the capacity of folding in the GA-element is abrogated by specific G → T or G → A point mutations, KRAS transcription is down-regulated. Conversely, guanidine-modified phthalocyanines, which specifically interact with and stabilize the KRAS G-quadruplex, push the promoter activity up to more than double. Collectively, our data support a transcription mechanism for murine KRAS that involves MAZ, PARP-1 and duplex-quadruplex conformational changes in the promoter GA-element.
Keywords: DNA-Protein Interaction, DNA Structure, DNA Transcription, Gene Regulation, General Transcription Factors, G-rich Sequences, Promoters, Quadruplex DNA
Introduction
Guanine-rich sequences have the potential to fold into intramolecular G-quadruplex (or G4-DNA) structures that are stabilized by planar arrays of four guanines paired by Hoogsteen hydrogen bonds (G-tetrad) (1). Quadruplex-forming sequences (QFS)2 are present in prokaryotic and eukaryotic genomes, promoter regions, micro- and mini-satellite repeats, telomeres, rDNA, and the vertebrate immunoglobulin heavy chain switch regions (2). Recent bioinformatic search analyses have shown a surprisingly high presence in the human genome of QFS, on the order of 3–4 × 105 (3, 4). The gene distribution of QFS is highly skewed because tumor suppressor genes have a very low level of QFS, whereas proto-oncogenes have a high level of such sequences (5). There seems to be a correlation between QFS and genomic instability; a low level of QFS in tumor suppressor genes is associated with genomic stability, and a high level is associated with genomic instability (5). Furthermore, the observation that QFS are often located in the region surrounding the transcription start sites of the genes and within cis-elements suggests that they may be involved in transcription regulation. This hypothesis has been formulated for a number of genes including CMYC, KRAS, C-MYB, VEGF, PDGFA, CKIT, and human insulin (6–13). The best studied G-rich sequence folding into a G-quadruplex, whose function has been correlated with a mechanism of transcription regulation, is the one found in the promoter of the CMYC gene (6). Upstream of the P1 promoter, controlling about 80% of transcription, there is a QFS that can fold into a G-quadruplex. When this quadruplex is destabilized by a G-to-A mutation, the basal transcription increases 3-fold, but when the G-quadruplex is stabilized by cationic porphyrins, transcription is repressed. From these data it has been suggested that the G-quadruplex formed in the CMYC promoter may function as a transcription repressor. A comprehensive review on the mechanism controlling transcription in CMYC has been recently reported (14). Interestingly, G-rich elements do not seem to be a unique feature of mammalian genomes, as they are also overrepresented in Saccharomyces cerevisiae. For instance, it has been reported that the gene promoters in this organism contain a level of G-rich sequences, which is 14-fold higher than the entire genome (15). A mutant of S. cerevisiae lacking the SGS1 gene, encoding for a helicase specific for quadruplex DNA, was characterized by a reduced expression of the genes in which open reading frames have a high potential to form quadruplexes (16). In addition, the human pathogen Neisseria gonorrhoeae contains a quadruplex-forming 16-base pair G-rich sequence that is required to promote pilin antigenic variation of surface structures to avoid immune detection (17). Evidence that G4-DNA formed within the regulatory elements of promoters may be involved in transcription regulation has also been produced by other research groups (6–13). Nevertheless, whether quadruplex DNA is formed inside the cell or whether it has any biological function in cellular events such as transcription or recombination is still a matter of debate. The role of G-quadruplex DNA on transcription is essentially based on the observation that G-quadruplex destabilizing point mutations or deletions bring about a significant enhancement of transcription, whereas G4-DNA ligands repress transcription (6–13). Although these findings support the notion that G4-DNA may be a transcription repressor, they do not provide a conclusive answer about the cellular function of quadruplex DNA. Indeed, it can be argued that the mutations introduced into the guanine element of the promoter could abrogate the binding of proteins to DNA and consequently alter the level of transcription. It could also be that the quadruplex-stabilizing ligands affect transcription simply because they compete with the binding of the proteins to the promoter. To provide an answer to these questions, we have in the present paper extended our previous work (7) and analyzed the role played in transcription by a critical GA-element located in the promoter of the murine KRAS proto-oncogene (18). This promoter contains a 34-bp G-rich sequence between nucleotides −322 and −288 (−1 is the 3′ boundary of the exon 0), which is sensitive to nuclease S1 and able to fold into a parallel G-quadruplex conformation. This intramolecular quadruplex was characterized by circular dichroism and UV spectroscopy, dimethyl sulfate footprinting, polymerase stop assays, and electrophoresis (7). The GA-element contains two perfect copies of the consensus sequence for the myc-associated zinc finger (MAZ) transcription factor. By pulldown and chromatin immunoprecipitation (ChIP) assays, we demonstrated that not only MAZ but also PARP-1 is associated to the GA-element. Although MAZ binds to both duplex and quadruplex conformations of the GA-element, PARP-1 binds specifically only to the G-quadruplex. We provide here compelling evidence that MAZ and PARP-1 are transcription activators and propose that these proteins are recruited to the KRAS promoter by the G-quadruplex structure formed within the GA-element.
EXPERIMENTAL PROCEDURES
Site-specific Mutagenesis; Wild-type and Mutant Plasmid Construction
Plasmid pKRS413 harboring the CAT gene driven by the murine KRAS promoter was subjected to PCR using primers 5′-TGCAGCCGCTCCCTCTCTCTCTCCTTCTCTCTCTCCCGCGCG and 5′-GAGGGAGCGGCTGCAGCGCTGGGAG (to introduce four G → A point mutations) and 5′-TGCAGCCGCTCCCTCACTCACTCCTTCCCT and 5′-GAGGGAGCGGCTGCAGCGCTGGGAG (to introduce two G → T point mutations). The mutant plasmid with two T residues was amplified with 5′-CTCACTCACTCCTTCACTCACTCCCGCGCG and 5′-GAAGGAGTGAGTGAGGGAGCGGCTGCAG to obtain a mutant with four T residues. PCR was performed with 3 ng/μl DNA template, 0.1 μm each primer, 0.05 units/μl AccuPrime pfx DNA polymerase (Invitrogen) in 1× AccuPrime pfx reaction mix for 3 min at 95 °C, 30 cycles 1 min at 95 °C, 30 s at 65 °C, and 5 min at 68 °C. Bacteria DIH 101 were transformed with PCR product, and DNA was extracted and sequenced (primer 5′-CCTCTCGGCACCACCCTC, accession number U49448, Entrez Nucleotide database (NCBI), bases 403–420). A 300-bp XmaI-AvaII fragment containing the KRAS murine promoter from the wild-type and mutant plasmids was subcloned in pGL3–1B basic in XmaI-HindIII blunted site in order to have the reporter firefly luciferase gene driven by wild-type and mutant KRAS promoters. The inserts were sequenced with pGL3for primer (5′-CTAGCAAAATAGGCTGTCCC). The plasmids constructed in this way were pKRS413-luc, pKRS413-luc-2T, pKRS413-luc-4T, and pKRS413-luc-4A.
Cell Culture and Proliferation Assay
NIH 3T3 cells were maintained in exponential growth in Dulbecco's modified Eagle's medium containing 100 units/ml penicillin, 100 mg/ml streptomycin, 20 mm l-glutamine, and 10% fetal bovine serum (Euroclone, Milano, Italy). Cell viability was measured by resazurin assays following standard procedures.
Dual Luciferase Assay
NIH 3T3 cells were seeded the day before transfection at 8000 cells/well in a 96-well plate. When the experiment involved a phthalocyanine treatment, this was done on cells let for adhesion for 16 h. Transfection was performed by mixing each vector 250 ng/well with control plasmid pRL-CMV expressing Renilla luciferase under control of the CMV promoter 10 ng/well using JetPEI transfection reagent (Polyplus transfection) following the manufacturer's instructions. For cotransfection with pCMV-MAZ (19), 150 ng of pKRS413-luc or mutant with 10 ng of pRL-CMV were transfected with either 150 ng of pCMV-MAZ or pcDNA3 plasmid (empty vector) as the mass for control transfections. Each transfection was performed in triplicate. Luciferase assays were performed 48 h after transfection with the Dual-Glo Luciferase assay system (Promega, Milan, Italy) following instructions. Samples were read with Turner Luminometer and expressed as relative luciferase, i.e. RT/RC × 100, where RT and RC are (firefly luciferase)/(Renilla luciferase) in phthalocyanine-treated and untreated cells.
Recombinant MAZ Expression and Electrophoretic Mobility Shift Assays (EMSA)
Recombinant MAZ protein tagged to GST was expressed in Escherichia coli BL21 using plasmid pGEX-hMAZ (19). The bacteria were grown for 1–2 h at 37 °C to an A600 of 0.5–2.0 before induction with isopropyl 1-thio-β-d-galactopyranoside (1.5 mm final concentration). Cells were allowed to grow for 7 h before harvesting. The cells were centrifuged at 5000 rpm, 4 °C, the supernatant was removed, and the cells washed twice with PBS. The pellet was resuspended in a solution of PBS with 100 mm phenylmethylsulfonyl fluoride, 1 m DTT, and protease inhibitor cocktails (for general use and for purification of histidine-tagged protein (Sigma) according to the manufacturer's protocols). The bacteria were lysed by sonication, added with Triton X-100 (1% final concentration), and incubated for 30 min on a shaker at 4 °C. The lysate was then centrifuged for 1 h at 4 °C at 20,000 rpm. Glutathione-Sepharose 4B (GE Healthcare) (50% slurry in PBS) was added to the supernatant from the previous step and incubated for 30 min at 4 °C on a shaker. The mix was centrifuged for 5 min at 500 × g, and the pellet was washed 5 times with PBS. The protein was eluted from the pellet with elution buffer containing 50 mm Tris-HCl, pH 8, and 10 mm reduced glutathione by centrifugation for 5 min at 500 g, 4 °C, and the tagged proteins collected from the supernatant were checked by SDS-PAGE.
Protein-DNA interactions were analyzed by EMSA. 5 nm radiolabeled GA-duplex, GA-duplex (2T), GA-duplex (4T), or quadruplex 28R (see Table 1) were incubated with MAZ protein (or NIH 3T3 nuclear extract) as indicated in legends for Figs. 4, 5, and 9 for 30 min at room temperature in 20 mm Tris HCl, pH 8, 30 mm KCl, 1.5 nm MgCl2, 1 mm DTT, 8% glycerol, 1% phosphatase Inhibitor Cocktail I (Sigma), 5 mm NaF, 1 mm Na3VO4, 2.5 ng/μl poly(dI-dC). The analyses were carried out in 5% polyacrylamide gels in 1× TBE at 20 °C.
TABLE 1.
Oligonucleotides used in this study
B, biotin.
| 5′-GGGAGGGAGGGAAGGAGGGAGGGAGGGA | 28R |
| FAM-GGGAGGGAGGGAAGGAGGGAGGGAGGGA-TAMRA | F-28R-T |
| 5′-GGGAGGGAGGGAAGGAGTGAGTGAGGGA | 28R(2T) |
| 5′-GGGAGTGAGTGAAGGAGTGAGTGAGGGA | 28R(4T) |
| 5′-TCCCTCCCTCCCTCCTTCCCTCCCTCCC | 28Y |
| 5′-TCCCTCACTCACTCCTTCCCTCCCTCCC | 28Y(2A) |
| 5′-TCCCTCACTCACTCCTTCACTCACTCCC | 28Y(4A) |
| B-5′-CGCGCGGGAGGGAGGGAAGGAGGGAGGGAGGGAGCGGCT (accession number U49448, bases 490–528) | G4-biotin |
| B-5′-TACCACCGAAGAGTTAAACAGGGAGGCAAATAAGCGTGCG (accession number U49448, bases 11–50) | SRC-biotin |
| 5′-ACCTTGATGAATCCAGGGAGGGAGGGAAGGAGGGAGGGAGGGACATCG-CTACCGTTAAAGATCATATCAAGTGATAGTAC | wtR-Mur80 |
| 5′-GTACTATCACTTGATATG | pMur80 |
| 5′-GTACTATCACTTGATATGATGCTTAACCGTAGCGATCTCCCTCCCTCC-CTCCTTCCCTCCCTCCCTGGATTCATCAAGGT | wtY-Mur80 |
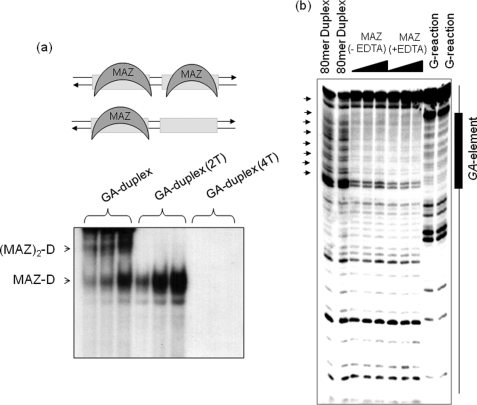
FIGURE 4.
Binding of MAZ to duplex of the GA-element. a, an EMSA shows the formation of two DNA·MAZ complexes with a 1:1 and 1:2 stoichiometry. The targets used are the GA-duplex, the mutant duplexes with 2 and 4 G → T mutations (GA-duplex (2T) and GA-duplex (4T) were obtained annealing 28R(2T) and 28R(4T) (Table 1) to their complementary oligonucleotides). MAZ amounts of 1, 2, and 4 μg were used, whereas the target 32P duplex was 20 nm. The experiment was performed with 5% PAGE in TBE. b, DNase I footprinting of an 80-mer promoter fragment containing the GA-element is shown. From left, first and second lanes, duplex digested with DNase I; third-fifth lanes and sixth-eighth lanes, DNase I digestion in the absence and presence of EDTA in the presence of 0.5, 1 and 2 μm MAZ-GST.
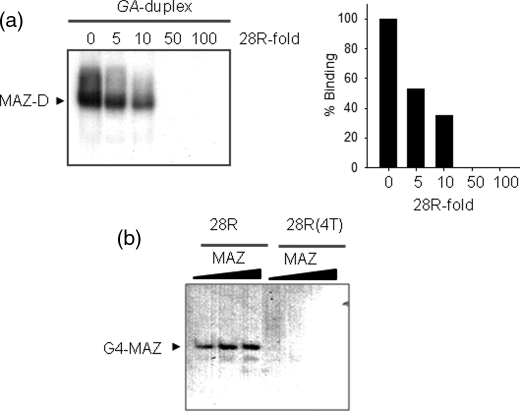
FIGURE 5.
EMSA competition experiments and binding of MAZ to G4-DNA. a, a competition assay shows that the DNA-MAZ complex is competed away by 5-, 10-, 50-, and 100-fold of cold quadruplex 28R. Experiments performed with 20 nm GA-duplex and 3 μg of MAZ-GST. Histograms show the densitometric analysis of the gel. b, an EMSA shows the binding of quadruplex 28R to the MAZ protein. Radiolabeled quadruplex 28R and mutant 28R(4T) (20 nm) have been incubated with 2, 3, and 5 μg MAZ. 5% PAGE in TBE.
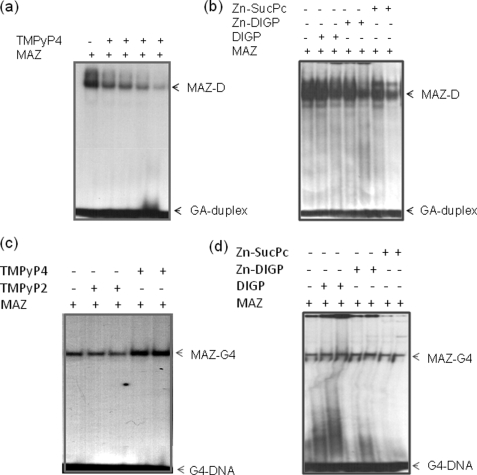
FIGURE 9.
Effect of G4-DNA ligands on KRAS transcription. a, an EMSA shows that the complex between GA-duplex and MAZ-GST is competed by increasing amounts of TMPyP4 (0.5, 1, 2, and 5 μm). Duplex concentration is 20 nm, and MAZ is 4 μg (5% PAGE in TBE). b, shown is the same experiment as in a but with phthalocyanines DIGP, Zn-DIGP, and Zn-SucPc at concentrations 5 and 10 μm. c, an EMSA show that the KRAS G-quadruplex binds to MAZ-GST in the absence and presence of TMPyP2 or TMPyP4 (2.5 and 10 μm), 10% PAGE in TBE. d, shown is the same experiment as in c but phthalocyanines DIGP, Zn-DIGP, and Zn-SucPc at the concentrations of 5 and 10 μm. The streaking in the second, third, and fifth lanes from the left is due to the effect of phthalocyanines binding to the quadruplex.
Pulldown and Western Blotting Assays
One milligram of nuclear protein extract (0.25 mg/ml), prepared as described (8), was incubated for 1 h at 37 °C with 60 nm biotinylated G4-DNA (G4-biotin, prepared in 100 mm KCl) or biotinylated duplex (G4-biotin annealed with its complementary in 100 mm NaCl) (Table 1) in a solution containing 20 mm Tris-HCl, pH 8, 8% glycerol, 150 mm KCl, 25 ng/ml poly(dI-dC), 1 mm Na3VO4, 5 mm NaF, 1 mm DTT, 0.2 mm phenylmethylsulfonyl fluoride, 0.1 mm zinc acetate. Then 250 μg of Streptavidin MagneSphere Paramagnetic Particles (Promega) pretreated for 30 min with 0.25 mg/ml BSA were added and incubated for 30 min at 37 °C. Particles were captured with a magnet, and the proteins were eluted with the buffer containing 0.5 and 1 m NaCl. The eluted proteins were concentrated and desalted with Microcon YM-3 filters (Millipore, Billerica, MA) for further analyses. The eluted proteins were separated by 10% SDS-PAGE and blotted overnight in 25 mm Tris, 192 mm glycine, and 20% methanol at 4 °C on a nitrocellulose membrane. The membrane was incubated with different antibodies: MAZ H-50 and PARP-1 H-300 diluted 1:200 (Santa Cruz). The secondary antibody used was rabbit IgG peroxidase conjugate (1:10,000) (Calbiochem). The antibodies were diluted in 10 mm Tris, pH 7.9, 150 mm NaCl, 0.05% Tween, and 5% BSA. The signal was developed with SuperSignal West Pico or Femto (Pierce) and detected with ChemiDOC XRS, Quantity One 4.6.5 software (Bio-Rad).
ChIP Assay
NIH 3T3 cells were plated in 2 × 15-cm diameter plates, grown to 80% confluence (about 1.5 × 107 cells), and fixed in formaldehyde 1% in PBS for 2 or 5 min. Nuclei were isolated following the ChIP-ITTM Express kit (Active Motif, Rixensart, Belgium) and resuspended in radioimmune precipitation assay buffer. Sonication was performed with a BioruptorTM sonicator (Diagenode, Liege, Belgium) following the manufacturer's instructions to obtain 200–500-bp fragments. Chromatin immunoprecipitation, washes, elution, reverse cross-linking, and proteinase K treatment were performed following the ChIP-ITTM Express kit manual. Antibodies used were MAZ H-50x and PARP-1 H-300 (Santa Cruz), 100 and 20 ng/μl, respectively. Control antibodies were RNA polymerase II mouse monoclonal antibody and negative control mouse IgG (ChIP-ITTM Control kit-mouse, Active Motif). About 15 μg of chromatin was used for each sample and fixed for 2 min for PARP-1 and 5 min for MAZ. The PCR reaction mixture was 1× PCR buffer (from the kit), 0.2 mm dNTPs, 0.4 μm primers, 0.04 units of Taq polymerase (EuroTaq Euroclone), and of the ChIP samples. Primers used were EF1-α control primers (from control kit), which give a 233-bp product, and 5′-CCTCTCGGCACCACCCTC and 5′-GATGCGCTCGGTGCTC (respectively, 403–420 and 561–546, sequence accession number U49448), which give a 160-bp product. PCR was performed as follows: 3 min at 94 °C, 40 cycles of 1 min at 94 °C, 30 s at 59 °C for EF1-α amplification and 61 °C for KRAS promoter amplification, and 30 s at 72 °C, with a final elongation 10 min at 72 °C. Amplification products were separated by a 10% acrylamide gel in TBE and visualized with a Gel-DOC apparatus (Bio-Rad).
shRNA Transfections and 3,4-Dihydro-5-[4-(1-piperidinyl) Butoxyl]-1(2H)-isoquinolinone (DPQ) Treatment
Cells were seeded 20–50,000/well in a 24-well plate. The day after plating they were either treated with DPQ at the concentrations indicated, or shRNA plasmids were transfected 0.5 μg/well. Plasmids used are Control shRNA Plasmid-A, PARP-1 shRNA Plasmid (m), and MAZ shRNA Plasmid (m) (Santa Cruz Biotechnology). Cells were collected 48 or 72 h after transfection.
RNA Extraction, Reverse Transcription, and Real-time PCR
RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer's instructions. For cDNA synthesis 5 μl of RNA in diethyl pyrocarbonate-treated water (extracted from about 25,000 cells) was heated at 55 °C and placed in ice. The solution was added to 7.5 μl of mix containing (final concentrations) 1× buffer, 0.01 m DTT (Invitrogen), 1.6 μm primer dT (MWG Biotech, Ebersberg, Germany; d(T)16), 1.6 μm Random primers (Promega), 0.4 mm dNTPs solution containing equimolar amounts of dATP, dCTP, dGTP, and dTTP (Euroclone, Pavia, Italy), 0.8 units/μl RNase OUT, and 8 units/μl of Maloney murine leukemia virus reverse transcriptase (Invitrogen). The reactions were incubated for 1 h at 37 °C and stopped with heating at 95 °C for 5 min. As a negative control the reverse transcription reaction was performed with 5 μl of diethyl pyrocarbonate water. Real-time PCR reactions were performed with 1x iQTM SYBR Green Supermix (Bio-Rad), 300 nm of each primer, 1 μl of reverse transcription reaction. The sequences of the primers used for amplifications are reported below. The PCR cycle was: 3 min at 95 °C, 40 cycles 10 s at 95 °C, 30 s at 60 °C with an iQ5 real-time PCR controlled by an Optical System software Version 2.0 or with CFX 96 controlled by Bio-Rad CFX Manager V1.5 (Bio-Rad). KRAS, MAZ, and PARP-1 expression are normalized with glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine-guanine phosphoribosyltransferase, and β2-microglobulin. The PCR primers used are reported in supplemental Table S1).
Polymerase-stop Assay
A linear DNA sequence of 80 nucleotides, named wtR-Mur80 (Table 1), containing the G-rich element of murine KRAS, was used as a template for Taq polymerase primer-extension reactions. The primer used was an 18-mer sequence named pMur80 (Table 1). This DNA sequence was purified by PAGE. The template (50 nm) was mixed with the labeled primer (25 nm) in 25 mm KCl, 1× Taq buffer and incubated overnight at 37 °C. The primer extension reactions were carried out for 1 h by adding 10 mm DTT, 100 μm dATP, dGTP, dTTP, dCTP, and 3.75 units of Taq polymerase (EuroTaq, Euroclone, Milan). The reactions were stopped by adding an equal volume of stop buffer (95% formamide, 10 mm EDTA, 10 mm NaOH, 0.1% xylene cyanol, 0.1% bromphenol blue). The products were separated on a 12% polyacrylamide sequencing gel prepared in 1× TBE, 8 m urea. The gel was dried and exposed to autoradiography. Standard dideoxy sequencing reactions were performed to detect the points where DNA polymerase I was arrested.
DNase I Footprinting
DNase I footprint was performed with the duplex obtained by annealing wtR-Mur80 with its complementary wtY-Mur80. The purine strand was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. The labeled duplex was preincubated at different ratios of MAZ-GST for 30 min in 20 mm Tris-HCl, pH 8, 30 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 8% glycerol, 50 μm zinc acetate, either in the presence or absence of 0.5 mm EDTA, and digested with DNase I (1 μl of a solution containing 0.002 unit of DNase I (Takara Biomedicals, Japan), 50 mm Tris-HCl, pH 7.4, 0.1 mg/ml BSA, 30 mm MnCl2). The reaction was conducted for 1 min at room temperature and stopped by adding to the reaction mixture 10 μl of stop solution (90% formamide, 50 mm EDTA, bromphenol blue). The analyses were carried out in 15% polyacrylamide sequencing gel prepared in 1× TBE, 8 m urea. After running, the gel was fixed and exposed to autoradiography (Hyperfilm, GE Healthcare) at −80 °C for few hours. A standard dimethyl sulfate G-reaction was performed with wtR-Mur80 purine strand to locate the binding of MAZ within murine duplex.
Fluorescence Resonance Energy Transfer (FRET) Experiments
Fluorescence measurements were carried out with a Microplate Spectrofluorometer System (Molecular Devices, Concorde, Canada) using a 96-well black plate in which each well contained 50 μl of 200-nm dual-labeled 28R in 50 mm Tris-HCl, pH 7.4, KCl as specified in legends for Figs. 2 and 6. The samples were incubated overnight at 37 °C before measurements. The emission spectra were obtained setting the excitation wavelength at 475 nm, the cut-off was at 515 nm, and recording the emission was from 500 to 650 nm. FRET melting experiments were performed on a real-time PCR apparatus (CFX 96, Bio-Rad) using a 96-well plate filled with 50-μl solutions of dual-labeled 28R, called F-28R-T (Table 1) in 50 mm Tris-HCl, pH 7.4, and potassium chloride at different concentrations as specified on the figure. The protocol used for the melting experiments was (i) an equilibration step of 5 min at low temperature (20 °C) and (ii) a stepwise increase of the temperature of 1 °C/min for 76 cycles to reach 95 °C. All the samples in the wells were melted in 76 min. After melting, the samples were re-annealed then melted again to give the same melting curves.
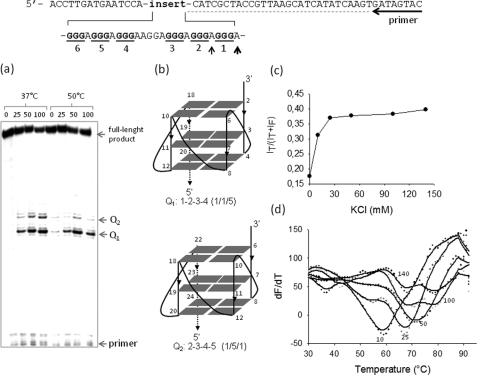
FIGURE 2.
Polymerase stop and FRET-melting assays. The DNA template used for the polymerase stop assay is shown. a, the G-runs of the GA-element are underlined. Primer-extension reactions were performed at 37 and 50 °C. Two polymerase pauses were observed at the adenines (determined by Sanger sequencing, see Ref. 7 and supplemental Fig. S1) before the first and second G-run at the 3′ end of the GA-element. The numbers above the gel indicate the KCl concentrations in the samples. b, shown are structures of the putative G-quadruplexes determined by dimethyl sulfate footprinting and circular dichroism (see Ref. 7). c, energy transfer, p = I580/(I525 + I580)-associated to the folding of F-28R-T as a function of KCl concentration, is shown. The experiment was performed in 50 mm Tris-HCl, pH 7.4, KCl as indicated in the figure (10, 25, 50, 100, and 140 mm). d, shown are FRET-melting curves obtained by measuring the increase of fluorescence at 525 nm emitted by F-28R-T as a function of temperature (excitation, 485 nm). The numbers indicate the KCl concentration. F-28R-T melts with a biphasic profile in the presence of 100 and 140 mm KCl.
FIGURE 6.
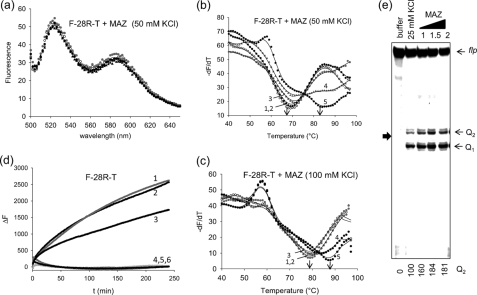
MAZ stabilizes the G-quadruplex. a, emission spectra are shown of quadruplex F-28R-T treated with BSA (r ([protein])/[quadruplex]) = 5) or increasing amounts of recombinant MAZ (r = 0, 1, 2. 5, 5). Excitation 475 nm. The experiment was carried out in 50 mm Tris-HCl, pH 7.4, 50 mm KCl, 50 μm zinc acetate; spectra were recorded using a fluorometer. b, shown is FRET melting of the same samples described in a. Curves 1 and 2, F-28R-T with and without BSA; curves 3, 4, and 5, F-28R-T with MAZ at r = 1, 2.5, and 5; the experiment was carried out in 50 mm Tris-HCl, pH 7.4, 50 mm KCl, 50 μm zinc acetate. c, the experiment was as in b but with 100 mm KCl. d, shown is the rate of assembly between quadruplex F-28R-T and 28Y. Curve 1, F-28R-T + 28Y (1:1); curve 2, F-28R-T + 28Y (1:1) + BSA (r = 2); curve 3, F-28R-T + 28Y (1:1) + MAZ (r = 1); curve 4, F-28R-T + 28Y (1:1) + MAZ (r = 2); curve 5, F-28R-T + BSA (r = 2); curve 6, F-28R-T MAZ (r = 2). The experiment was carried out in 50 mm Tris-HCl, pH 7.4, 50 mm KCl, 50 μm zinc acetate at 37 °C. The data shown in panels b–d have been collected by a real time apparatus (CFX96 Bio-Rad) measuring the FAM emission at 525 nm. e, shown is a polymerase-stop assay using the wtR-Mur80 template, 18-mer primer pMur80, and increasing amounts of MAZ (1, 1.5, and 2 μg) as indicated in the figure. The experiment was carried out in 50 mm Tris-HCl, pH 7.4, 25 mm KCl, 50 μm zinc acetate, 12% polyacrylamide gel in 1× TBE. flp, full-length product. Numbers at the bottom indicate the intensity of the Q2 band.
RESULTS
The GA-element of the Murine KRAS Promoter Folds in One of Two Topologically Distinct G4-DNA Conformations
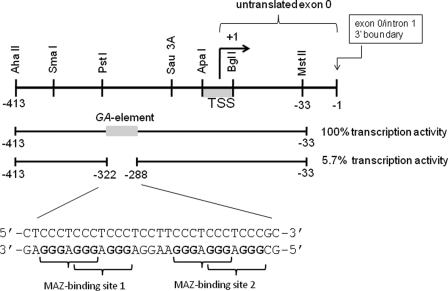
The promoter activity of murine KRAS resides within a 380-bp DNA fragment including a guanine-rich element (GA-element) between −322 and −288 (positions relative to exon 0/intron 1 boundary (accession number M16708)). The deletion of the GA-element drops the promoter activity to 5.7% that of the control (18) (Fig. 1). As it exhibits S1 nuclease sensitivity (18), the GA-element is likely to assume an unusual DNA structure under superhelical stress. A previous study has shown that the GA-element formed an intramolecular H-DNA structure, but under acidic conditions (20). We recently discovered that the polypurine strand of the GA-element (called 28R, Table 1), composed of six runs of guanines (G-runs), folds into an intramolecular G-quadruplex under physiological conditions (7). This structure is rather stable because it arrests the progression of Taq polymerase even at 50 °C, as demonstrated by polymerase stop assays with an 80-mer duplex containing the GA-element (Fig. 2a). Two polymerase arrests are observed at the adenines before the first and second G-run, at the 3′ end of the GA-element (supplemental Fig. S1). This suggests the formation of two G-quadruplexes, Q1 or Q2, the former involving G-runs 1–2-3–4 and the latter involving G-runs 2–3-4–5 (Fig. 2b). To provide further support that the GA-element folds in two topologically distinct G-quadruplex conformations, we performed FRET experiments. The polypurine strand of the GA-element (28R) was labeled at the 5′ and 3′ ends with a donor (6-carboxyfluorescein (FAM)) and an acceptor (tetramethylrodhamine) fluorophore (F-28R-T). In the presence of increasing amounts of KCl, F-28R-T folds into a G-quadruplex that upon FAM excitation at 475 nm gives rise to a FRET signal at 580 nm due to tetramethylrodhamine emission (21) (supplemental Fig. S2). The energy transfer between the two dyes is empirically measured by the p value, p = IT/(IT + IF), where IF and IT are the fluorescence intensities of the donor and acceptor at 525 and 580 nm, respectively. Fig. 2c shows that the p value increases with KCl concentration from 0.175 (open sequence) to 0.39 (folded sequence); in 140 mm KCl, the p value is not higher than 0.39 because F-28R-T forms a parallel G-quadruplex where the two fluorophores are located at the opposite ends of the structure and, therefore, not very close one to the other. When the G-quadruplex is melted, the two fluorophores increase their distance, and the FAM fluorescence at 525 nm (F525) raises with temperature. Plotting dF525/dt versus t, one obtains well resolved melting curves. In keeping with the polymerase stop assay, FRET-melting curves show that F-28R-T melts with a biphasic profile, confirming the formation of two G-quadruplexes with TMs of 68 and 80 °C in 100 mm KCl (Fig. 2d).
FIGURE 1.
The murine KRAS promoter contains a nuclease hypersensitive G-rich element (GA-element) that is essential for transcription. The GA-element is characterized by six runs of guanines that can fold into an intramolecular G-quadruplex structure. The GA-element contains two perfect binding sites for the MAZ transcription factor. TSS means transcription start site. The sequence is numbered with the exon 0/intron 1 boundary taken as −1.
Functional Characterization of the GA-element
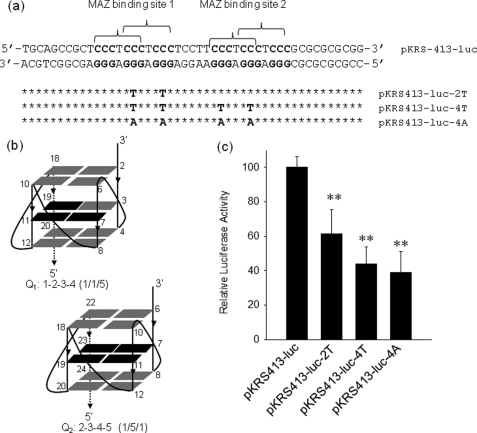
To functionally characterize how the GA-element regulates transcription, we first studied if the introduction of quadruplex-destabilizing point mutations affects the level of KRAS transcription. We constructed plasmid pKRS413-luc bearing the murine KRAS promoter driving firefly luciferase. From pKRS413-luc we obtained by site-directed mutagenesis three mutant plasmids, pKRS413-luc-2T, pKRS413-luc-4T, and pKRS413-luc-4A, in which two or four guanines located in the mid-G-tetrad of Q1 or Q2 have been replaced with thymines or adenines (Fig. 3, a and b). To explore whether the quadruplex-destabilizing point mutations change the level of transcription, wild-type or mutant plasmids were co-transfected with pRL-CMV, a vector encoding for Renilla luciferase, in NIH 3T3 cells. The levels of firefly and Renilla luciferases were measured with a luminometer 48 h after transfection. When firefly/Renilla ratio obtained with pKRS413-luc and pRL-CMV was set to 100, the same ratio given by the mutant plasmids pKRS413-luc-2T, pKRS413-luc-4T, and pKRS413-luc-4A turned out to be 60 ± 14, 44 ± 10, and 39 ± 12, respectively (Fig. 3c). This result shows that the abrogation of the capacity of quadruplex formation by the GA-element significantly decreases the expression of firefly luciferase, suggesting that the G-quadruplex could behave as a structural element favoring KRAS transcription.
FIGURE 3.
Transcription activity of wild-type and mutant KRAS promoter. a, shown is the sequence of the KRAS promoter in the region spanning over the GA-element. The two MAZ-binding sites are indicated by brackets. By site-directed mutagenesis the promoter sequence was modified in the GA-element either in one MAZ-binding site or in both MAZ-binding sites. The point mutations abrogated the capacity of the sequence to fold into a G-quadruplex. b, the structure of the putative KRAS G-quadruplexes shows that the point mutations fall in the mid G-tetrad of the structure (depicted in black), c, results of dual luciferase assay with wild-type and mutant plasmids show that the activity of the KRAS promoter is reduced by the introduction in the GA-element of the point mutations that destabilize G-quadruplex formation. All mutant expressions are different than wild-type expression by Student's t test; p < 0,01 (two asterisks).
MAZ Binds to the Duplex and G4-DNA Conformations of the GA-element
Considering that the GA-element is essential for the activation of transcription, we searched for putative transcription factors binding to this critical sequence by using the MatInspector software. We found that the protein with the highest binding prediction is MAZ, whose consensus sequence is GGGAGGG. A tandem of two perfect MAZ-binding sites is indeed present in the GA-element. To find out whether MAZ actually binds to the GA-element, we expressed MAZ fused to GST in E. coli and purified the chimeric protein by affinity chromatography with glutathione-Sepharose 4B. The wild-type GA-element in duplex (28R:28Y, namely GA-duplex, Table 1) gave two retarded bands with MAZ-GST, in keeping with the binding of one protein to each of the MAZ-binding sites (Fig. 4a). This is consistent with the fact that (i) the introduction in the GA-duplex of two G → T point mutations in only one of the two MAZ-binding sites resulted in the abrogation of the DNA-protein complex of lower mobility, i.e. the complex where both protein sites are occupied by MAZ (slower band), and (ii) the introduction of four G → T point mutations, two in each MAZ-binding site, resulted in the abrogation of both DNA-protein complexes, as expected. This clearly demonstrates that the interaction of MAZ with the KRAS promoter is highly sequence-specific and that two MAZ molecules bind to the GA-element.
According to the luciferase transfection experiments, we cautiously hypothesized that a G-quadruplex in the GA-element could function as a transcription activator. However, it is also possible that the mutant plasmids expressed less luciferase because the G → T and G → A point mutations introduced in the GA-element abolished the binding of MAZ to the promoter. Therefore, mutational and EMSA studies did not allow us to understand whether repression of KRAS transcription was due to the loss of quadruplex formation by the GA-element or to the loss of the MAZ-binding sites. This suggests that the use of mutagenesis to investigate whether non-orthodox DNA secondary structures influence transcription should be considered with great caution. There is a correlation between the level of transcription and the number of mutations introduced in the GA-element; pKRS413-luc-4T and pKRS413-luc-4A with both MAZ-binding sites abrogated show a lower level of residual transcription (∼40%) than pKRS413-luc-2T having only one MAZ-binding site abrogated (∼60%). The fact that a residual transcription of ∼40% is observed when the MAZ binding is abrogated suggests that KRAS transcription also depends on other nuclear proteins.
To confirm that recombinant MAZ binds to the KRAS promoter in a sequence-specific manner, we performed a DNase I footprinting assay on an 80-mer promoter fragment spanning over the GA-element (Fig. 4b). MAZ was found to protect the DNA portion containing the cluster of G-runs, confirming that both MAZ sites are indeed occupied by the protein.
As MAZ binds to a promoter sequence that can extrude a G-quadruplex, we asked whether it also recognizes the folded conformation of the GA-element. Previous studies have shown that MAZ binds to the G-quadruplexes formed by the G-rich region of the diabetes susceptibility locus IDDM2 (13) and the GGA repeat region in the CMYB promoter (9). Fig. 5a shows that quadruplex 28R efficiently competed away the DNA-protein complexes between MAZ and the GA-duplex; a 5-fold excess of cold quadruplex 28R reduces the MAZ-duplex complexes to ∼50%, whereas a 50-fold excess completely abrogates the complexes. From these data we roughly estimated for the interaction of MAZ with the G-quadruplex a KD of about 0.5 × 10−9 m. Additionally, when MAZ was incubated with radiolabeled quadruplex 28R, a clear DNA-protein complex was observed by EMSA. Instead, the mutant oligonucleotide with four G → T mutations, which is unable to fold into a G-quadruplex, did not form any DNA-protein complex (Fig. 5b).
MAZ Stabilizes the KRAS Quadruplex
The interaction between recombinant MAZ and the KRAS G-quadruplex has been investigated by FRET experiments, measuring the increase of FAM emission at 525 nm as a function of temperature (excitation, 485 nm). At r ([protein]/[DNA]) ratios of 0, 1, 2.5, and 5, MAZ does not change the emission spectrum of F-28R-T, suggesting that in the DNA-protein complex the G-quadruplex maintains its characteristic p value of 0.39 (Fig. 6a). To explore whether the binding to MAZ results in the stabilization of the G- quadruplex, we performed FRET-melting experiments. In 50 mm KCl, the folded DNA melts cooperatively, with a TM of 70 °C (Fig. 6b). When an unrelated protein such as BSA is added to the G-quadruplex solution (r = 5), the TM does not change, as expected. In contrast, when increasing aliquots of MAZ are added (r = 1, 2.5, and 5), the TM increases up to 85 °C, indicating that the protein strongly stabilizes the DNA structure. The experiment was repeated in 100 mm KCl, and in this case the TM increased from 80 to 90 °C (Fig. 6c). To provide further evidence about the stabilizing effect of MAZ on the G-quadruplex, we used a polymerase stop assay. In the presence of 25 mm KCl, Taq polymerase was arrested at the 3′ end of GA-element, before the first and second runs of guanines. The blocking of the polymerase increases in the presence of MAZ; Q2 increases by 80% and Q1 by 25%, confirming that MAZ facilitates the folding (Fig. 6e). Moreover, we expected that the stabilizing activity of MAZ should slow down the assembly rate between quadruplex F-28R-T and its complementary 28Y strand. When quadruplex F-28R-T in 50 mm KCl (TM = 70 °C) was mixed with 28Y at 37 °C, the G-quadruplex was transformed into the more stable duplex, and the fluorescence of FAM at 525 nm increased (Fig. 6d). The assembly process could be monitored by measuring the increase of fluorescence, ΔF, as a function of time (ΔF = F - F0, where F0 is the FAM fluorescence at 525 nm at t = 0, and F is the fluorescence at time t). The ΔF versus t curves showed an exponential shape that was best-fitted to a double-exponential equation (22). For the slow phase, a kinetic constant k of 5.1 × 10−3 ± 7 × 10−6 s−1 was obtained. But when the hybridization was performed in the presence of MAZ at r = 1, the constant dropped to 1.7 × 10−3 ± 4 × 10−5 s−1. When a higher amount of MAZ was used (r = 2), the hybridization was completely inhibited. This demonstrates that MAZ, although stabilizing the G-quadruplex, hinders the quadruplex-to-duplex transformation in the GA-element.
Selective Enrichment Strategy and ChIP Assay
To see if the GA-element is bound by endogenous MAZ present in NIH 3T3 nuclear extract, we used a selective enrichment strategy (Fig. 7a). Biotinylated oligonucleotides (Table 1) mimicking the KRAS GA-element (G4-biotin annealed to its complementary) or the KRAS quadruplex (folded G4-biotin) (Table 1) were used as bait in pulldown biotin-streptavidin assays. The pulled-down samples were separated by SDS-PAGE and analyzed by Western blot with specific antibodies. As shown in Fig. 7a, the GA-element enriched the pulled-down sample with MAZ, whereas a scramble duplex (scr-biotin annealed to its complementary) did not, indicating that this transcription factor binds to the KRAS GA-element with good affinity and specificity. In keeping with EMSA (Fig. 5), endogenous MAZ was efficiently pulled down by G4-biotin in the quadruplex conformation. Instead, the scramble oligonucleotide (scr-biotin) showed a much lower MAZ affinity than quadruplex G4-biotin.
FIGURE 7.
Pulldown and ChIP assays. a, biotinylated GA-element in duplex (G4-biotin hybridized to its complementary) or in quadruplex (G4-biotin) were used as bait in pulldown experiments with NIH 3T3 extract. The concentrations of NaCl in the elution buffer used to elute the protein fractions were 0.5 and 1 m. The panels show the Western blots of the pulled down fractions obtained with anti MAZ (top panel) and anti-PARP-1 (bottom panel) antibodies. The band intensities have been measured with ChemiDOC XRS apparatus (Bio-Rad). b, a chromatin immunoprecipitation assay was performed with anti-MAZ, anti-PARP-1, anti- RNA polymerase II (positive control), and IgG (negative control) antibodies. PCR analysis was performed on DNA isolated from ChIP reactions using controls, anti-PARP-1, and anti MAZ antibodies. PCR was performed with KRAS primers (see “Experimental Procedures”) and EF1-α control primers (EF1-α primers provided by the kit amplify a 233-bp fragment from the DNA immunoprecipitated with anti-RNA polymerase II, used as a positive control). The KRAS PCR amplification product obtained with anti MAZ and anti-PARP-1antibodies show that under in vivo conditions the GA-element is bound by PARP-1 and MAZ.
In addition to MAZ, we focused our efforts on PARP-1, having found in a previous study that this protein binds to the human KRAS promoter at a sequence homologous to the GA-element (8) (supplemental Fig. S3). We found that PARP-1 interacts non-specifically with the GA-element, as it was pulled down by the GA-element and also by a scramble duplex (Fig. 7a). This is probably due to the fact that the protein recognizes the ends of the duplex (23). By contrast, PARP-1 showed binding specificity for quadruplex G4-biotin, as the unstructured oligonucleotide scr-biotin is weakly bound. The binding of PARP-1 to the G-quadruplex was confirmed by EMSA (supplemental Fig. S4).
To investigate whether MAZ and PARP-1 bind to the GA-element under in vivo conditions, we performed ChIP experiments. ChIP is a powerful tool for studying protein-DNA interactions under real physiological conditions. Living NIH 3T3 cells were treated with formaldehyde to cross-link and fix the protein-DNA complexes at the chromatin level. The chromatin was sheared into short fragments, and the DNA·MAZ and DNA·PARP-1 complexes were immunoprecipitated with specific anti-MAZ and anti-PARP-1 antibodies. The DNA present in the immunoprecipitated complexes was recovered and amplified by PCR. Fig. 7b shows that the amplification of sheared chromatin produced the expected band from the KRAS promoter region. It can be seen that although anti-RNA polymerase II and anti-IgG antibodies did not immunoprecipitate any DNA-protein complex involving the KRAS DNA (negative control), anti-MAZ and anti-PARP-1 antibodies effectively immunoprecipitated the MAZ·DNA and PARP-1·DNA complexes. The finding that antibody anti-MAZ was more efficient than antibody anti-PARP-1 may be explained by a different binding strength of the two antibodies or by the fact that the GA-element is prevalently occupied by MAZ. Together, the data demonstrate that under physiological conditions MAZ and, to a lower extent, PARP-1, is associated to the GA-element of murine KRAS promoter.
Role of MAZ and PARP-1 on KRAS Transcription
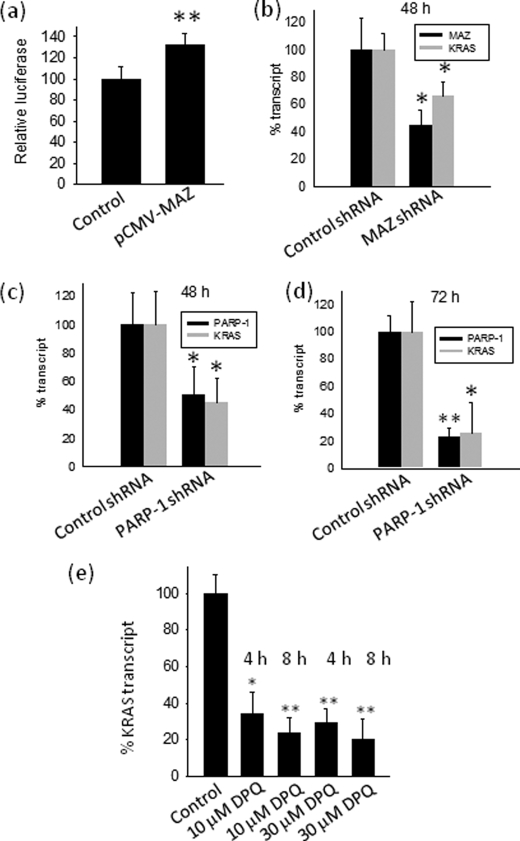
To investigate the role of MAZ in the activation of the murine KRAS promoter, NIH 3T3 cells were co-transfected with pKRS413-luc and pCMV-MAZ, a plasmid encoding for MAZ. The overexpression of MAZ increased luciferase by ∼35%, suggesting that it activates the KRAS promoter (Fig. 8a). The fact that MAZ did not strongly stimulate the KRAS promoter may be due to a relatively high level of endogenous MAZ in NIH 3T3.
FIGURE 8.
Effect of MAZ and PARP-1 on transcription. a, transient transfection experiments show the effect of MAZ overexpression on firefly luciferase driven by the wild-type KRAS promoter. Left bar (control), cells transfected with pKRS413-luc, pcDNA3 (empty vector), and pRL-CMV; right bar, cells transfected with pKRS413-luc, pCMV-MAZ, and pRL-CMV. The ordinate reports firefly luciferase normalized by Renilla luciferase; b–d, real-time PCR shows the effect on KRAS transcription of knocking down MAZ or PARP-1 with specific shRNAs at 48 and 72 h. The ordinate reports the % transcript (KRAS or MAZ or PARP-1), i.e. RT/RC × 100, where RT is (transcript)/(average transcripts from β2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, and hypoxanthine-guanine phosphoribosyltransferase) in shRNA-treated cells, and RC is (transcript)/(average transcripts from β2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, and hypoxanthine-guanine phosphoribosyltransferase) in cells treated with unrelated shRNA. e, real time PCR shows the effect on KRAS transcription of 10 and 30 μm DPQ, an inhibitor of PARP-1 poly (ADP-ribose) activity. The ordinate reports the % KRAS transcript, i.e. RT/RC × 100, where RT is (KRAS transcript)/(β2microglobulin and hypoxanthine-guanine phosphoribosyltransferase transcripts) in untreated cells, and RC is (KRAS transcript)/(β2-microglobulin and hypoxanthine-guanine phosphoribosyltransferase transcripts) in cells treated with DPQ. Differences from the control are supported by Student's t test, p < 0.05 (one asterisk), p < 0.01 (two asterisks).
To further evaluate the impact of MAZ on KRAS transcription, we carried out shRNA knockdown experiments. NIH 3T3 cells were treated with a validated anti-MAZ shRNA, and the level of both MAZ and KRAS transcripts was measured by real-time PCR 48 h after shRNA treatment. As a control, we transfected the cells with an unrelated shRNA. An inhibition of KRAS transcription (∼40%) was observed upon MAZ silencing even if this was only partial (Fig. 8b). This is in keeping with the dual luciferase assays showing that KRAS transcription is down-regulated by 40% when the MAZ-binding sites are abrogated. From these data we can conclude that MAZ is an essential factor for activating KRAS transcription.
Similarly, the functional role of PARP-1 at the KRAS promoter was determined by using a validated shRNA. Fig. 8, c and d, shows that the level of KRAS transcripts parallels the inhibition of PARP-1 at both 48 and 72 h post-transfection, showing a dependence of KRAS transcription from PARP-1. Considering that PARP-1 is a protein that catalyzes the poly(ADP-ribosyl)ation of target proteins (heteromodification) and itself (automodification) (24), we tested whether this activity is important for KRAS transcription by treating NIH 3T3 cells for 4 and 8 h with 10 and 30 μm DPQ, an inhibitor of PARP-1 activity (25). Fig. 8e, which reports the levels of KRAS transcript determined by real-time PCR in NIH 3T3 cells treated with DPQ for 4 and 8 h, shows that the inhibitor suppresses the transcription to ∼30% of the control. This suggests that PARP-1 acts on KRAS in a complex way and at two distinct levels; the first through its poly(ADP-ribosyl)ation activity and the second by binding directly to the promoter like a transcription factor (24, 26) (see “Discussion”).
Effect of G4-DNA Ligands on KRAS Transcription
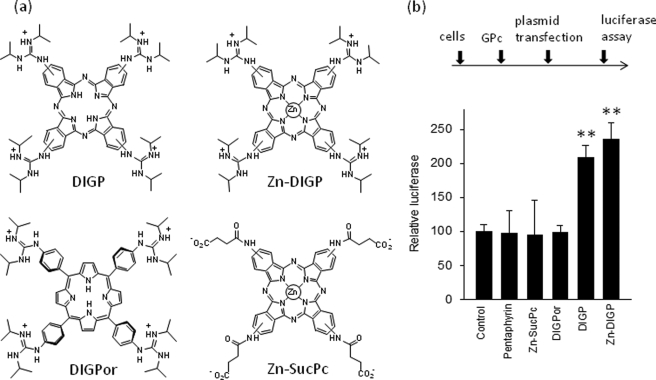
An approach adopted by several authors to investigate the possible role played by G4-DNA in transcription regulation is to treat the cells with G4-DNA ligands and then measure the expression of the target gene. This approach assumes that the ligands have a higher affinity for G4-DNA than for duplex DNA and that they do not compete with the binding of proteins to the duplex or quadruplex forms of the promoter. The cationic porphyrin TMPyP4 has been widely used as a G4-DNA ligand, whereas its positional isomer TMPyP2, which does not bind to G4-DNA, has been normally used as a control. However, TMPyP4 is characterized by poor binding specificity and similar affinities for quadruplex and duplex DNAs (27, 28). Fig. 9, a and c, shows that TMPyP4 competes with the binding of MAZ to the GA-duplex (at 5 μm the porphyrin practically abrogates the DNA-protein complex) but not to the G-quadruplex (TMPyP4 and, to a lower extent, DIGP/Zn-DIGP, seem to enhance the binding of MAZ to the G-quadruplex). A similar result was obtained by using a NIH 3T3 nuclear extract (supplemental Fig. S5). In light of these findings we used guanidine-modified phthalocyanines (GPc), DIGP and Zn-DIGP, a new class of G4-DNA ligands that specifically binds to G4-DNA and poorly competes with the binding of proteins to duplex DNA (21, 29). Indeed, Fig. 9b shows that 10 μm DIGP and Zn-DIGP weakly compete with the binding of MAZ to the GA-duplex. In a recent study we found that DIGP and Zn-DIGP interact with the murine KRAS quadruplex with a KD between 10−7 and 10−6 m (21). Polymerase stop assays showed that these ligands efficiently stabilize the G-quadruplex formed by the GA-element (21). Indeed, circular dichroism spectra obtained at various temperatures (20–90 °C) showed that the TM of the G-quadruplex in the presence of DIGP (r = 4) increased from 68 to >90 °C in 50 mm KCl (supplemental Fig. S6). Furthermore, the assembly between F-28R-T and 28Y was practically inhibited in the presence of DIGP at r = 5 (supplemental Fig. S7). Because MAZ binds to the KRAS quadruplex, we analyzed if this also occurs in the presence of phthalocyanines and found that 10 μm DIGP and Zn-DIGP promoted only a weak competition of the MAZ-quadruplex complex (Fig. 9d). To test the effect of the guanidine phthalocyanines on KRAS transcription, we performed dual luciferase assays. We used ligand concentrations within the window where the molecules showed a relatively low cytotoxicity, i.e. at concentrations lower than IC50. NIH 3T3 cells were first treated with 10 μm GPc for 16 h, then transfected with a mixture of pKRS413-luc and pRL-CMV. They were grown for a further 48 h before firefly and Renilla luciferase were measured with a luminometer. As a control, we treated the cells with porphyrins or phthalocyanines that do not have affinity for G4-DNA (Zn-SucPc, and pentaphyrin; see supplemental Fig. S8) (21). The results show that DIGP and Zn-DIGP stimulate KRAS transcription by more than 2-fold, suggesting that the G-quadruplex behaves as a transcription activator element (Fig. 10). In contrast, the control molecules Zn-SucPc, DIGPor (tetrakis-(diisopropylguanidine) porphine), and pentaphyrin, which do not stabilize G4-DNA, have no effect on transcription. This result strengthens our hypothesis that quadruplex DNA in the GA-element is involved in the activation of transcription. Taken together, our data indicate that G4-DNA mediates the activation of KRAS transcription probably by recruiting MAZ to the KRAS promoter.
FIGURE 10.
Transcription assays. a, structures of guanidine phthalocyanines used are shown. b, a dual luciferase assay was performed in NIH 3T3 cells treated with 10 μm DIGP, Zn-DIGP, Zn-SucPc, DIGPor, or pentaphyrin for 16 h and subsequently transfected with a mixture of pKRS413-luc and pRL-CMV. The signal was normalized to Renilla luciferase (control). Luciferase expressions in the presence of DIGP and Zn-DIGP are different from control, by Student's t test, p < 0.01 (two asterisks).
DISCUSSION
The GA-element of the murine KRAS gene promoter between −322 to −288, which contains six runs of guanines separated by adenine nucleotides, is essential for transcription; its deletion reduces transcription to 5.7% (18). This critical GA-element is structurally polymorphic, as in the presence of potassium and under physiological conditions it folds into one of two topologically distinct G-quadruplexes. The stability of the G-quadruplex is sufficiently high to arrest Taq polymerase even at 50 °C. We report here for the first time that the murine KRAS GA-element interacts with zinc finger proteins MAZ and PARP-1. These interactions are complex as both proteins recognize the duplex and/or folded conformations of the GA-element. This has been discovered by employing two independent techniques: a selective enrichment strategy (pull-down assay with biotinylated oligonucleotides) and ChIP assays.
MAZ was predicted to bind to the GA-element by using the MAT-Inspector bioinformatic software, which indicated the presence in the GA-element of two MAZ consensus sequences, GGGAGGG. The binding of this nuclear factor to the GA-element was analyzed by recombinant MAZ-GST. As the GA- element contains two sites for MAZ, it forms two DNA-protein complexes, one in which only one site is occupied by MAZ, the other in which both sites are occupied. The interaction of MAZ to the GA-element is highly sequence-specific, as it is sufficient to introduce in one binding site two point mutations to completely abrogate the interaction.
MAZ was first identified as a GA-box binding transcription factor in the CMYC promoter controlling transcription initiation and termination (30, 31). This nuclear protein is expressed in many tissues (32) and its functional role in the context of transcription seems to be complex, as some genes are activated by MAZ, whereas others are repressed. MAZ was reported to activate the serotonin 1a receptor gene promoter (33), the adenovirus major later promoter (34), the parathyroid hormone PTH/PTHrP promoter (35), the PNMT promoter (36), the insulin promoter (13) and the muscle creatine kinase promoter (37). Instead, MAZ was reported to repress the endothelial nitric-oxide synthase promoter (38) and the CMYB promoter (9). Here, we provide sound evidence that MAZ activates transcription of the murine KRAS gene. When MAZ was partially knocked-down with a specific shRNA, transcription dropped to ∼60% of control. In contrast, when MAZ was overexpressed, transcription increased by 35%. These values are low probably because (i) endogenous MAZ is not limiting in NIH 3T3 cells; (ii) MAZ may require post-translational modifications, for instance phosphorylation, to become active and this cannot occur in all overexpressed MAZ molecules (39); (iii) MAZ may need to interact with other proteins to activate KRAS transcription.
An important finding of this study is that MAZ stabilizes the quadruplex structure formed by the GA-element. By means of FRET experiments we have discovered that in 50 mm KCl, the TM of the KRAS quadruplex increased from 70 to 85 °C, in the presence of MAZ at r = 5. The ability of MAZ to recognize non B-DNA structures was previously reported. Indeed, MAZ binds to the G-quadruplex structures formed by the IDDM2 locus (13) and the GGA repeat sequence in the CMYB promoter (9). Because MAZ shows binding activities to both duplex and quadruplex DNA and is involved in the trans-activation of several genes, the activation of KRAS is likely to involve other cofactors. Several proteins have been reported to interact with MAZ. For instance, autoregulation of MAZ occurs through its interaction with histone deacetylase (40), whereas FAC1 has been shown to repress MAZ-mediated activation of the SV40 promoter and to co-localize with MAZ in plaque-like structures in the brains of Alzheimer's patients (41). Furthermore, DCC, a type I membrane protein and putative tumor suppressor, has been shown to associate with MAZ during neuronal differentiation of P19 cells, which correlates with the loss of CMYC expression (42).
The other zinc finger protein that was found to influence the KRAS transcription is PARP-1. In addition to its capacity of direct binding to DNA, it shows catalytic activity by attaching poly(ADP-ribose) units to target proteins including itself (22). The addition of negative charges on the modified proteins is expected to affect their interactions with DNA and other proteins. The intracellular level of PARP-1 and the removal of the ribose chains on the target proteins by specific glycohydrolases are tightly controlled (43). Data accumulated in the years suggest that the role of PARP-1 can be divided into emergency and housekeeping functions. The emergency role occurs after DNA damage, whereas the housekeeping role is the modulation of gene expression (44). Genome-wide localization studies (Chip-on-chip) have revealed that PARP-1 is associated with upstream of transcription start sites of actively transcribed genes (45). A significant number of genes (about 3.5% of the transcriptome) involve PARP-1 in transcription regulation (46). It occurs by heteromodification of histones, which promotes the decondensation of high order chromatin, and by binding to enhancer/promoter regulatory cis-elements (44). Previous studies have reported that PARP-1 recognizes unusual DNA conformations such as hairpins/cruciforms (47–49) and the G-quadruplex formed by a cis-element in the human KRAS promoter (8). Interestingly, Ladame and co-workers (50) have shown that PARP-1 undergoes automodification after binding to the c-kit quadruplex. For a review on the role of PARP-1 in transcription, we refer to Kraus and Lis (44). Many studies have shown that PARP-1 is implicated in the activation or repression of transcription (44). Our data show that PARP-1 acts as an activator because KRAS transcription decreases to 20% that of control when PARP-1 is knocked down by specific shRNA. Furthermore, as PARP-1 is an enzyme with poly(ADP-ribosyl)ation activity, we addressed the question of whether the inhibition of its catalytic activity affected KRAS transcription. We found that DPQ, a PARP-1 inhibitor, strongly down-regulates KRAS transcription. Combining these data, one can conclude that the action of PARP-1 on the KRAS promoter requires that the protein is catalytically active on other proteins and probably itself.
One approach to investigate the role of G4-DNA on transcription is to introduce point mutations in the quadruplex-forming sequence of the promoter and study the effect on transcription (6, 9, 51). It is assumed that the point mutations do not alter the binding of the proteins to DNA. Because we found that the insertion in the GA-element of two-point mutations completely abrogated the MAZ binding, we could not draw any conclusion about the role of G4-DNA on transcription by promoter mutational analysis. We, therefore, adopted another strategy to ascertain the role of G4-DNA within the GA-element; we used ligands that specifically interact with G4-DNA. If the ligands stabilize the quadruplex and shift the duplex-quadruplex equilibrium, their effects on transcription can give information about the functional role of G4-DNA. However, this rationale is valid if the ligands do not compete with the proteins binding to the duplex or quadruplex conformations of the GA-element. We employed for our functional studies a class of G4-DNA ligands showing a higher specificity for G4-DNA than TMPyP4. TMPyP4 is a well known cationic porphyrin that has been extensively used as G4-DNA ligand in transcription assays, although it shows a poor binding specificity for quadruplex DNA (27, 28). We found that the complex between MAZ and GA-duplex is strongly competed by TMPyP4 but not by GPcs. Furthermore, GPcs and also TMPyP4 did not compete the complex between MAZ and quadruplex 28R. By means of luciferase assays we found that GPcs enhanced the activity of the KRAS promoter by more than 2-fold, whereas the control ligands did not. This result is apparently in conflict with a previous study in which we showed that TMPyP4 down-regulated the promoter activity of murine KRAS (7). In that study we cotransfected TMPyP4 and pKRS413-CAT, a plasmid-bearing CAT driven by the murine KRAS promoter. Most likely, in the transfection mixture TMPyP4 saturated the GA-element, so that the observed down-regulation of transcription was due to the inability of MAZ (and other proteins) to bind to the promoter. Thus, our previous hypothesis that G4-DNA might be a repressor is not supported by the present study. On the contrary, the data here reported provide strong evidence that G4-DNA should be an activator element. Similarly to our study, Lew et al., (13) reported that the insulin-linked polymorphic region located in the promoter of the insulin gene forms intra- and inter-molecular G-quartets that bind to Pur-1 (MAZ) and stimulate transcription activity.
In light of these new discoveries, how can MAZ, PARP-1 and G4-DNA regulate transcription of KRAS? A possible model that rationalizes the results of this study is the following. PARP-1 is likely to act primarily at chromatin level by promoting, through heteromodification of histones, a local decondensation of chromatin that increases the DNA accessibility (44). This is in keeping with the fact that when heteromodification is inhibited by DPQ, KRAS transcription is repressed. Under negative supercoiling, the tract of promoter containing the GA-element unwinds (this is a nuclease hypersensitive site (18)), and the unpaired G-rich strand is expected to fold into a G-quadruplex (52, 53). As MAZ stabilizes the G-quadruplex, the protein should shift the duplex-folded equilibrium toward the folded form. This may also be favored by the binding to the complementary polypyrimidine strand of proteins recognizing C-rich sequences such as heterogeneous nuclear ribonucleoprotein K (54, 55) and poly(C)-binding proteins PCBP1–4 (56). The binding of MAZ to the promoter results in the activation of transcription, as indicated by the fact that the overexpression of MAZ in NIH 3T3 cells up-regulates transcription, whereas the depletion of MAZ by shRNA down-regulates transcription. Our data indicate that the murine KRAS transcription is favored by the folded form of the GA-element as (i) KRAS transcription is reduced to 50% of control when point mutations abrogating DNA folding are introduced in the GA-element, (ii) transcription is 2-fold up-regulated by guanidine phthalocyanines that stabilize the murine KRAS quadruplex (21), (iii) MAZ stabilizes the G-quadruplex and activates transcription, and (iv) hydridization of the G-quadruplex to its complementary strand is inhibited by MAZ. It is likely that the unwinding of the GA-element serves to recruit at the promoter the proteins forming a multiprotein complex that activates transcription. Therefore, the G-quadruplex should function as an attraction point to recruit MAZ and the other proteins of the transcription machinery.
In summary, in this study we show that the critical GA-element of murine KRAS interacts with MAZ and PARP-1 in a very complex way involving DNA conformation changes from duplex to quadruplex DNA. We provide compelling evidence that both MAZ and PARP-1 are activators of the KRAS promoter. Both proteins recognize the parallel quadruplex conformation adopted by the GA-element, which probably has the function of recruiting these proteins to the promoter. Our study provides the basis for the rationale design of anticancer drugs, for example, G-rich G4-aptamers specific for MAZ and PARP-1 to down-regulate the expression of oncogenic KRAS in cancer cells (57, 58).
Acknowledgments
Plasmids pGEX-hMAZ and pCMV-MAZ were provided by RIKEN BioResource Center, which participates in the National Bio-Resources Project of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We thank Nathan Luedtke (Zurich University) for the gift of the guanidine-modified phthalocyanines.
This work was supported by Associazione Italiana per la Ricerca sul Cancro 2008 and Italian Ministry of Scientific Research (PRIN 2007).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
- QFS
- quadruplex-forming sequence
- MAZ
- myc-associated zinc finger protein
- PARP-1
- poly(ADP-ribose) polymerase
- DPQ
- (3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone)
- DIGP
- tetrakis-(diisopropyl-guanidine) phthalocyanine
- shRNA
- short hairpin RNA
- FRET
- fluorescence resonance energy transfer
- CAT
- chloramphenicol acetyltransferase
- FAM
- 6-carboxyfluorescein
- GPc
- guanidine-modified phthalocyanine
- ChIP
- chromatin immunoprecipitation
- CMV
- cytomegalovirus
- EMSA
- electrophoretic mobility shift assay
- GST
- glutathione S-transferase
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- TBE
- Tris borate EDTA
- BSA
- bovine serum albumin
- SucPc
- tetrakis-(succinyl)phthalocyanines.
REFERENCES
- 1.Sen D., Gilbert W. (1988) Nature 334, 364–366 [DOI] [PubMed] [Google Scholar]
- 2.Maizels N. (2006) Nat. Struct. Mol. Biol. 13, 1055–1059 [DOI] [PubMed] [Google Scholar]
- 3.Huppert J. L., Balasubramanian S. (2007) Nucleic Acids Res. 35, 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd A. K., Johnston M., Neidle S. (2005) Nucleic Acids Res. 33, 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddy J., Maizels N. (2006) Nucleic Acids Res. 34, 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogoi S., Xodo L. E. (2006) Nucleic Acids Res. 34, 2536–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogoi S., Paramasivam M., Spolaore B., Xodo L. E. (2008) Nucleic Acids Res. 36, 3765–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palumbo S. L., Memmott R. M., Uribe D. J., Krotova-Khan Y., Hurley L. H., Ebbinghaus S. W. (2008) Nucleic Acids Res. 36, 1755–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D., Liu W. J., Guo K., Rusche J. J., Ebbinghaus S., Gokhale V., Hurley L. H. (2008) Mol. Cancer Ther. 7, 880–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin Y., Rezler E. M., Gokhale V., Sun D., Hurley L. H. (2007) Nucleic Acids Res. 35, 7698–7713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rankin S., Reszka A. P., Huppert J., Zloh M., Parkinson G. N., Todd A. K., Ladame S., Balasubramanian S., Neidle S. (2005) J. Am Chem. Soc. 127, 10584–11059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew A., Rutter W. J., Kennedy G. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12508–12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks T. A., Hurley L. H. (2009) Nat. Rev. Cancer 9, 849–861 [DOI] [PubMed] [Google Scholar]
- 15.Hershman S. G., Chen Q., Lee J. Y., Kozak M. L., Yue P., Wang L. S., Johnson F. B. (2008) Nucleic Acids Res. 36, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S. K., Johnson R. E., Yu S. L., Prakash L., Prakash S. (1999) Science 286, 2339–2342 [DOI] [PubMed] [Google Scholar]
- 17.Cahoon L. A., Seifert H. S. (2009) Science 325, 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman E. K., Trusko S. P., Murphy M., George D. L. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2705–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsutsui H., Sakatsume O., Itakura K., Yokoyama K. K. (1996) Biochem. Biophys. Res. Commun. 226, 801–809 [DOI] [PubMed] [Google Scholar]
- 20.Pestov D. G., Dayn A., Siyanova E. Yu., George D. L., Mirkin S. M. (1991) Nucleic Acids Res. 19, 6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Membrino A., Paramasivam M., Cogoi S., Alzeer J., Luedtke N. W., Xodo L. E. (2010) Chem. Comm. 46, 625–627 [DOI] [PubMed] [Google Scholar]
- 22.Green J. J., Ying L., Klenerman D., Balasubramanian S. (2003) J. Am. Chem. Soc. 125, 3763–3767 [DOI] [PubMed] [Google Scholar]
- 23.de Murcia G., Ménissier de Murcia J. (1994) Trends Biochem. Sci. 19, 172–176 [DOI] [PubMed] [Google Scholar]
- 24.Caiafa P., Zlatanova J. (2009) J. Cell. Physiol. 219, 265–270 [DOI] [PubMed] [Google Scholar]
- 25.Martin-Oliva D., Aguilar-Quesada R., O'valle F., Muñoz-Gámez J. A., Martínez-Romero R., García Del Moral R., Ruiz de Almodóvar J. M., Villuendas R., Piris M. A., Oliver F. J. (2006) Cancer Res. 66, 5744–5756 [DOI] [PubMed] [Google Scholar]
- 26.Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren J., Chaires J. B. (1999) Biochemistry 38, 16067–16075 [DOI] [PubMed] [Google Scholar]
- 28.Freyer M. W., Buscaglia R., Kaplan K., Cashman D., Hurley L. H., Lewis E. A. (2007) Biophys. J. 92, 2007–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alzeer J., Roth P. J., Luedtke N. W. (2009) Chem. Comm. 15, 1970–1971 [DOI] [PubMed] [Google Scholar]
- 30.Bossone S. A., Asselin C., Patel A. J., Marcu K. B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7452–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izzo M. W., Strachan G. D., Stubbs M. C., Hall D. J. (1999) J. Biol. Chem. 274, 19498–19506 [DOI] [PubMed] [Google Scholar]
- 32.Song J., Murakami H., Tsutsui H., Tang X., Matsumura M., Itakura K., Kanazawa I., Sun K., Yokoyama K. K. (1998) J. Biol. Chem. 273, 20603–20614 [DOI] [PubMed] [Google Scholar]
- 33.Parks C. L., Shenk T. (1996) J. Biol. Chem. 271, 4417–4430 [DOI] [PubMed] [Google Scholar]
- 34.Parks C. L., Shenk T. (1997) J. Virol. 71, 9600–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leroy C., Manen D., Rizzoli R., Lombès M., Silve C. (2004) J. Mol. Endocrinol. 32, 99–113 [DOI] [PubMed] [Google Scholar]
- 36.Her S., Claycomb R., Tai T. C., Wong D. L. (2003) Mol. Pharmacol. 64, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 37.Himeda C. L., Ranish J. A., Hauschka S. D. (2008) Mol. Cell. Biol. 28, 6521–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karantzoulis-Fegaras F., Antoniou H., Lai S. L., Kulkarni G., D'Abreo C., Wong G. K., Miller T. L., Chan Y., Atkins J., Wang Y., Marsden P. A. (1999) J. Biol. Chem. 274, 3076–3093 [DOI] [PubMed] [Google Scholar]
- 39.Tsutsui H., Geltinger C., Murata T., Itakura K., Wada T., Handa H., Yokoyama K. K. (1999) Biochem. Biophys. Res. Commun. 262, 198–205 [DOI] [PubMed] [Google Scholar]
- 40.Song J., Ugai H., Kanazawa I., Sun K., Yokoyama K. K. (2001) J. Biol. Chem. 276, 19897–19904 [DOI] [PubMed] [Google Scholar]
- 41.Jordan-Sciutto K. L., Dragich J. M., Caltagarone J., Hall D. J., Bowser R. (2000) Biochemistry 39, 3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugai H., Li H. O., Komatsu M., Tsutsui H., Song J., Shiga T., Fearon E., Murata T., Yokoyama K. K. (2001) Biochem. Biophys. Res. Commun. 286, 1087–1097 [DOI] [PubMed] [Google Scholar]
- 43.Bonicalzi M. E., Haince J. F., Droit A., Poirier G. G. (2005) Cell. Mol. Life Sci. 62, 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraus W. L., Lis J. T. (2003) Cell 113, 677–683 [DOI] [PubMed] [Google Scholar]
- 45.Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 46.Ogino H., Nozaki T., Gunji A., Maeda M., Suzuki H., Ohta T., Murakami Y., Nakagama H., Sugimura T., Masutani M. (2007) BMC Genomics 8, 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Mol. Biol. 348, 609–615 [DOI] [PubMed] [Google Scholar]
- 48.Chasovskikh S., Dimtchev A., Smulson M., Dritschilo A. (2005) Cytometry A. 68, 21–27 [DOI] [PubMed] [Google Scholar]
- 49.Lonskaya I., Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Biol. Chem. 280, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 50.Soldatenkov V. A., Vetcher A. A., Duka T., Ladame S. (2008) ACS Chem. Biol. 3, 214–219 [DOI] [PubMed] [Google Scholar]
- 51.Thakur R. K., Kumar P., Halder K., Verma A., Kar A., Parent J. L., Basundra R., Kumar A., Chowdhury S. (2009) Nucleic Acids Res. 37, 172–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun D., Hurley L. H. (2009) J. Med. Chem. 52, 2863–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. (2008) Nat. Struct. Mol. Biol. 15, 146–154 [DOI] [PubMed] [Google Scholar]
- 54.Michelotti E. F., Michelotti G. A., Aronsohn A. I., Levens D. (1996) Mol. Cell. Biol. 16, 2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomonaga T., Levens D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi H. S., Hwang C. K., Song K. Y., Law P. Y., Wei L. N., Loh H. H. (2009) Biochem. Biophys. Res. Commun. 380, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi E. W., Nayak L. V., Bates P. J. (2010) Nucleic Acids Res. 38, 1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teng Y., Girvan A. C., Casson L. K., Pierce W. M., Jr., Qian M., Thomas S. D., Bates P. J. (2007) Cancer Res. 67, 10491–10500 [DOI] [PubMed] [Google Scholar]