Abstract
Peroxisome proliferator-activated receptors (PPARs) are nuclear transcription factors that play central roles in metabolism and inflammation. Although a variety of compounds have been shown to activate PPARs, identification of physiologically relevant ligands has proven difficult. In silico studies of lipid derivatives reported here identify specific 5-lipoxygenase products as candidate physiologically relevant PPAR-α activators. Subsequent studies show both in vitro and in a murine model of inflammation that 5-lipoxygenase stimulation induces PPAR-α signaling and that this results specifically from production of the inflammatory mediator and chemoattractant leukotriene B4 (LTB4). Activation of PPAR-α is a direct effect of intracellularly generated LTB4 binding to the nuclear receptor and not of secreted LTB4 acting via its cell-surface receptors. Activation of PPAR-α reduces secretion of LTB4 by stimulating degradation of this fatty acid derivative. We also show that the LTB4 precursors leukotriene A4 (LTA4) and 5-hydroperoxyeicosatetrenoic acid (5-HPETE) activate PPAR-α but have no significant endogenous effect independent of conversion to LTB4. We conclude that LTB4 is a physiologically relevant PPAR-α activator in cells of the immune system. This, together with previous findings, demonstrates that different types of lipids serve as endogenous PPAR-α ligands, with the relevant ligand varying between functionally different cell types. Our results also support the suggestion that regulation of inflammation may involve balancing proinflammatory effects of LTB4, exerted through cell-surface receptors, and anti-inflammatory effects exerted through PPAR-α activation.
Keywords: Eicosanoid, Leukotriene, Lipid, Nuclear Receptors, Prostaglandins, Transcription Factors, PPAR
Introduction
Peroxisome proliferator-activated receptors (PPARs)2 are ligand-activated nuclear transcription factors that play central roles in regulation of both metabolism and inflammation (1). PPAR-α in particular regulates many aspects of lipid metabolism, including some aspects of plasma lipoprotein metabolism as well as fatty acid uptake, intracellular transport, and energy-generating metabolism. A further role for PPAR-α as a modulator of inflammation is supported by several lines of evidence (2), including the greater severity or duration of inflammation in PPAR-α knock-out mice (3, 4). These anti-inflammatory effects may reflect changes in intracellular redox status and decreased activity of the proinflammatory transcription factor NF-κB (5), although other mechanisms are likely as well.
Numerous synthetic agonists for the various PPARs have been developed. The PPAR-γ-activating thiazolidinediones are used clinically as insulin-sensitizing agents in type 2 diabetes, whereas the PPAR-α-activating fibrates are used as lipid-lowering agents. Endogenous compounds also bind to PPARs, yet unambiguous identification of physiologically important ligands has proven difficult. Most endogenous PPAR ligands identified to date either exhibit relatively low affinity or are present in low concentrations, and, in either case, appear unlikely to be important activators in vivo. For example, although 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) was found to be a high affinity ligand for PPAR-γ (6), further investigation has shown that concentrations are low and that 15d-PGJ2 is unlikely to play a significant physiological role (7). Likewise, a recent study has demonstrated that differentiating preadipocytes produce a PPAR-γ agonist essential for the differentiation process (8), thus providing unambiguous evidence for the existence of an endogenous ligand. Nevertheless, the investigators were unable to isolate and identify the ligand.
Endogenous lipid-derived compounds have likewise been identified as activating ligands for PPAR-α. As with early studies of 15d-PGJ2, however, until recently, all studies of these compounds have employed exogenous addition of the compound being investigated. The in vivo relevance of these ligands thus remains unclear, as their ability to activate PPAR-α under physiologically relevant conditions has never been demonstrated. The phospholipid 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine was shown recently to activate PPAR-α in liver but, as that study's authors note, its relevance to other tissues or cell types remains uncertain (9).
To determine the physiologically relevant PPAR-α ligand in cells of the immune system, we used in silico methods to identify lipid-derived compounds that bind strongly to the PPAR-α activation site. As we found that the most strongly binding compounds were products of 5-lipoxygenase action on arachidonic acid, we chose to further investigate this system. Involvement of 5-lipoxygenase in endogenous PPAR-α activation is plausible because this enzyme produces leukotrienes and related eicosanoids that play important roles as inflammatory mediators and chemoattractants (10) and because exogenously added eicosanoids have been shown to activate PPAR-α (3, 11–13).
The results we report here demonstrate that PPAR-α in cells of the immune system is activated in a physiologically relevant manner by a lipid quite different from the one active in hepatocytes. They also support the emerging concept that LTB4 plays a central role in the regulation of inflammation through its ability to exert both proinflammatory and anti-inflammatory effects via different receptors.
MATERIALS AND METHODS
Materials
Wy 14643, arachidonic acid, LTA4, LTB4, LTC4, LTD4, LTE4, 5-oxo-ETE, 5-HETE, 5-HPETE, 20-OH-LTB4, and zileuton were obtained from Cayman Chemicals (Ann Arbor, MI). SC22716, A23187, and lipopolysaccharide (LPS) from Escherichia coli (serotype 0111:B4) were obtained from Sigma Aldrich (St. Louis, MO). Cell culture medium and Lipofectamine 2000 were obtained from Invitrogen. Lipids supplied in ethanol were evaporated under a gentle stream of nitrogen and immediately resuspended with DMSO (purged with inert gas) at a concentration of 10 mm. Stocks were stored at −30 °C until use. Rabbit polyclonal 5-lipoxygenase antibody was from Abcam (Cambridge, MA); rabbit polyclonal PPAR-α (H-98), goat polyclonal LTA4 hydrolase (C-21), and β-actin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Gal4-PPAR plasmids were a kind gift from Ron Evans (Salk Institute), the aP2-luc plasmid was from Todd Leff (Wayne State University), and the CMX-PPAR-α plasmid was from Kirin Brewery Co., Ltd.
Animals
5-Lipoxygenase knock-out (129-Alox5tm1Fun) and strain-matched wild-type sv129 mice were bred in the University of Michigan Unit for Laboratory Animal Medicine from breeders obtained from Jackson Laboratories (Bar Harbor, ME). All experiments were conducted in accordance with protocols approved by the University of Michigan Committee on Use and Care of Animals.
Cell Culture and Transfection
RBL-2H3, CV-1, HeLa, and Jurkat T cells were obtained from American Type Culture Collection (Manassas, VA), grown in medium supplemented with 10% fetal bovine serum (Invitrogen) and incubated at 37 °C with 5% CO2. In all transfection experiments, cells were transiently cotransfected with pRL-SV40 and reporter plasmids with or without expression plasmids. In separate experiments, cells were cotransfected with pRL-SV40 plus a luciferase gene under the control of four Gal4 DNA-binding elements (UASG × 4 TK-luciferase) and a plasmid containing the ligand-binding domain for a PPAR fused to the Gal4 DNA-binding domain. All transfections were performed using Lipofectamine 2000 or Amaxa Nucleofector (Walkersville, MD) according to the manufacturer's instructions. Twenty-four h after transfection, cells were treated with test compounds and incubated for an additional 24 h in medium with 10% fetal bovine serum. The resulting luciferase activity was measured with reporter luciferase assay kits (Promega; Madison, WI) employing a Modulus 9201 luminometer (Turner Biosystems, Sunnydale, CA) and normalized by comparison to Renilla luciferase.
Isolation of PMNs
Human peripheral blood PMNs were obtained from heparinized venous blood of healthy volunteers by Ficoll-Hypaque gradient centrifugation and sedimentation in 5% dextran according to a protocol approved by the University of Michigan Institutional Review Board for Human Subject Research. This method yielded ∼2 × 106 cells/ml, with PMNs representing >95% of the cells isolated. Informed consent was obtained from all subjects.
LPS Injection and Peritoneal Lavage
Mice were injected intraperitoneally with 20 mg/kg body weight of E. coli LPS or vehicle. Four h after injection, the mice were euthanized, and the peritoneal cavity was opened and rinsed with 10 ml of phosphate-buffered saline. Total cell counts were performed, and cells were pelleted by centrifugation. Immediately following peritoneal lavage, liver tissue was excised and processed.
Nuclear Protein Preparation and PPAR-α DNA-binding Assay
Nuclear protein was isolated using a nuclear protein extraction kit (Cayman Chemicals). Protein concentrations were estimated using the Bio-Rad (Hercules, CA) DC protein assay. PPAR-α DNA-binding activity in the nuclear protein was detected by an ELISA-based PPAR-α transcription factor assay (Cayman Chemicals) that detects PPAR-α bound to PPAR response element-containing double-stranded DNA sequences.
RNA Interference and Transfection
SMARTpool siRNA reagents, along with nonspecific negative control pool siRNAs, were purchased from Dharmacon (Lafayette, CO). Transfection of these pooled 21-nucleotide siRNA duplexes was carried out using Lipofectamine 2000.
Zymosan Preparation and Opsonization with Human PMNs
Zymosan particles (Sigma Aldrich) were boiled in serum-free medium for 10 min and washed twice with the same medium. The particles were then resuspended in serum-free medium and counted with a Coulter counter. Isolated PMNs (2 × 106 in 250 μl culture medium) were incubated with 5 × 108 zymosan particles for 10 min at 37 °C.
LTB4 Measurement
Cells (2 × 106) were cultured in medium and treated with compounds. Supernatants were collected, and immunoreactive LTB4 was quantitated by ELISA (Cayman Chemical) following the manufacturer's instructions. LTB4 in peritoneal lavage fluid was quantitated similarly. The detection limit for LTB4 was 4 pg/ml; cross-reactivity for arachidonic acid, 5-HETE, LTC4, LTD4, or LTE4 was <0.01%.
Western Blot Analysis
Cells were lysed in ice-cold lysis buffer, mixed with a commercial sample buffer (Invitrogen), and heated at 95 °C for 5 min. Samples were then electrophoresed on 4–20% Tris-glycine Novex SDS-polyacrylamide gels (Invitrogen), after which the gels were transferred to polyvinylidene difluoride membranes (0.45 μm). Blots were incubated overnight at 4 °C with the primary antibody, followed by appropriate secondary antibody. They were then washed and signal detected using a SuperSignal chemiluminescent substrate Western blotting reagent (Pierce Biotechnology) with chemiluminescence-sensitive film.
In Silico Docking
The compounds of interest were sketched and converted into three-dimensional structures using the program Sybyl (Tripos; St. Louis, MO). Minimization for these compounds employed 1000 steps utilizing the steepest-descents method followed by conjugate gradients and finally BFGS method up to a gradient of 0.01 kcal/mol/Å. Docking of pterostilbene analogs was performed in the PPAR-α ligand-binding domain (Protein Data Bank code 1K7L). Protein preparation was done using the program Discovery Studio (version 2.1) (Accelrys; San Diego, CA), which involved addition of hydrogen atoms, minimization of the protein, and assigning of formal charges to the amino acid residues. The x-ray crystal bound pose of GW409544 was used to define the binding site during protein preparation. Docking of the compounds was performed using the program GOLD (Cambridge Crystallographic Data Centre; Cambridge, UK). The binding site for docking was defined using all amino acid residues within 12 Å. The “standard mode” settings were used, and no artificial restraints were defined during docking. The “early termination” criterion was set to off. For each compound, a total of 10 different docking poses were collected. The GOLD scoring function was used to identify docking poses and to rank these poses. The various docking poses of each compound were visually inspected for anticipated ligand-receptor interactions and unreasonable conformations.
Measurement of LTB4 Metabolites
Jurkat T cells were transfected with the human PPAR-α expression plasmid or control vector. Cells were then changed to Hanks' balanced salt solution (with calcium and magnesium) and stimulated with A23187 or control for 15 min at 37 °C. The reaction was stopped by addition of 1 ml chilled methanol to the cells followed by incubation for 1 h at 0 °C. Supernatants were collected by centrifugation at 2000 × g for 10 min and diluted with HPLC grade water to bring the methanol concentration to 15%. Extraction of LTB4 metabolites and their separation and quantification was done according to the method of Christmas et al. (14) with slight modifications. Briefly, diluted supernatants were passed through a solid phase extraction cartridge (C-18 Sep-Pak column, 1 cc, 100 mg, Waters), eluted with 1 ml of methanol, and evaporated under inert nitrogen gas at room temperature. The dried substance was immediately suspended in an HPLC mobile phase, and peaks were analyzed by LC-MS/MS.
Statistical Analysis
Data are represented as mean ± S.E. and were analyzed using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA). Comparisons among experimental groups were performed with one-way analysis of variance (ANOVA), followed by Dunnett's adjustment for multiple comparisons or Student's t test as applicable. Differences were considered significant if p < 0.05.
RESULTS
In Silico Docking Suggests 5-Lipoxygenase Produces Potential PPAR-α Ligands
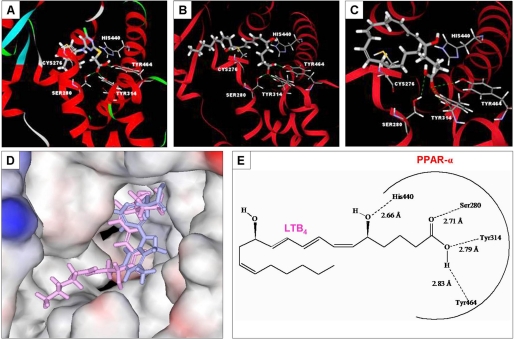
A number of lipid-derived compounds, especially arachidonic acid derivatives, have been shown to activate PPAR-α when added exogenously at high concentrations. Although these compounds may be present endogenously, previous studies have not established the physiological relevance of any outside hepatocytes. As a first step toward identifying physiologically relevant endogenous PPAR-α ligands, we performed molecular docking studies in silico. As shown in Table 1, strong binding was observed for LTB4, 20-hydroxy-LTB4 (20-OH-LTB4), LTE4, LTA4, 5-hydroperoxyeicosatetraenoic acid (5-HPETE), and to a lesser extent 5-oxoeicosatetraenoic acid (5-oxo-ETE). Other tested compounds demonstrated either weak or absent binding (Table 1 and data not shown). Previous studies have established that typical PPAR-α agonists form hydrogen bonds to Ser-280, Tyr-314, His-440, and Tyr-464 of the receptor (15, 16) and that these hydrogen bonds are necessary for receptor activation. Both LTB4 and the potent synthetic PPAR-α agonist Wy 14643 form hydrogen bonds with these four receptor residues (Table 1 and Fig. 1). LTB4 forms an additional hydrogen bond to Cys-276, which is not essential for receptor activation. These four important hydrogen bonds are also observed on docking of 20-OH-LTB4 and 5-HPETE, although the binding energies (docking scores) for these two compounds are somewhat less than those for LTB4 and Wy 14643 (Table 1). By contrast, the hydrogen bonds formed by LTE4 (Met-220, Leu-331, Glu-286, and Gly-335), LTA4 (Tyr-334, Glu-286, and Met-220) and 5-oxo-ETE (Met-220, Asn-219, and Glu-286) are not those recognized as important for receptor activation. Notably, in silico binding of LTB4 (Fig. 1B) is similar to that of Wy 14643 (Fig. 1A). Indeed, in silico docking poses of the two compounds are almost superimposable (Fig. 1D). 5-HPETE also binds in a very similar manner (Fig. 1C). Consequently, LTB4 and 5-HPETE may be viewed as plausible candidate endogenous activators.
TABLE 1.
Docking score and key interacting residues of PPAR-α with 5-lipoxygenase compounds
Compounds interacting with amino acid residues important for activation are in boldface type.
| Compound name | Docking score | Interactive residues |
|---|---|---|
| kcal/mol | ||
| Wy 14643 | −7.771 | Ser-280, Tyr-314, Tyr-464, and His-440 |
| LTB4 | −7.803 | Ser-280, Tyr-314, Tyr-464, His-440, and Cys-276 |
| 20-OH-LTB4 | −6.041 | Ser-280, Tyr-314, Tyr-464, His-440, and Cys-276 |
| 5-HPETE | −5.903 | Ser-280, Tyr-314, Tyr-464, and His-440 |
| LTE4 | −6.288 | Met-220, Leu-331, Glu-286, and Gly-335 |
| LTA4 | −6.240 | Tyr-334, Glu-286, and Met-220 |
| 5-oxo-ETE | −4.535 | Met-220, Asn-219, and Glu-286 |
| LTC4 | No pose | |
| LTD4 | No pose | |
| 5-HETE | No pose | |
| 12-oxo-LTB4 | No pose |
FIGURE 1.
In silico molecular docking with PPAR-α. A, docking of Wy 14643 (ribbon representation). B, docking of LTB4. C, docking of 5-HPETE. D, superimposed docking of Wy 14643 and LTB4 (space-filling representation). E, docking of LTB4 with PPAR-α (two-dimensional representation) showing key amino acid residues and interactions.
5-Lipoxygenase Generates a Physiologically Relevant PPAR-α Agonist
Studies of nuclear receptor activation traditionally involve addition of exogenous ligand either to cells or to isolated receptor molecules. Such studies provide no information about the amount of ligand present at the receptor site in vivo and whether it is adequate to induce effective activation. By contrast, generation of an activating ligand within the cell under physiological conditions clearly establishes its relevance. Since all of the compounds that demonstrated strong binding and relevant hydrogen bond formation in silico are products of the enzyme 5-lipoxygenase (17), which is activated by a variety of proinflammatory stimuli through mechanisms dependent on an influx of extracellular Ca2+ (18), we hypothesized that this enzyme might generate a physiologically relevant ligand.
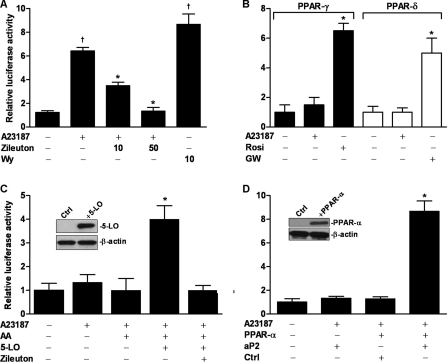
In initial studies, we activated 5-lipoxygenase through addition of the Ca2+ ionophore A23187. Activation of PPAR-α was assessed by a Gal4-luciferase reporter in conjunction with a chimeric protein having the PPAR-α ligand binding domain fused to the Gal4 DNA-binding domain. Plasmids coding for these proteins were transfected into Jurkat T cells (Fig. 2A), which constitutively express 5-lipoxygenase. PPAR-α was activated ∼7-fold by A23187 and to a similar level by the potent synthetic PPAR-α agonist Wy 14643 as a positive control. Similar studies with PPAR-γ or PPAR-β/δ reporters demonstrated no activation (Fig. 2B). Because A23187-stimulated Ca2+ influx can affect a number of different signaling pathways, we employed the 5-lipoxygenase inhibitor zileuton as a probe for the specific role of this enzyme in the observed increase in PPAR-α activity. As hypothesized, zileuton treatment produced a significant, dose-dependent reduction in PPAR-α activation (Fig. 2A).
FIGURE 2.
Activation of PPAR-α following stimulation of 5-lipoxygenase. A, Jurkat T cells were transiently transfected with the PPAR-α Gal4-luciferase reporter system. Transfected cells were treated with the 5-lipoxygenase inhibitor zileuton (10 and 50 μm) and stimulated with A23187 (5 μm). Twenty-four h later, cells were lysed and analyzed by Dual-Luciferase assay. Wy 14643 (Wy, 10 μm) was used as a positive control. *, p < 0.5 versus A23187 (one-way ANOVA); †, p < 0.5 versus DMSO (one-way ANOVA). B, Jurkat T cells were similarly transfected with PPAR-γ or PPAR-δ Gal4-luciferase reporter systems and treated as described in A. Rosiglitazone (Rosi, 5 μm) was used as a positive control for PPAR-γ and GW501516 (GW, 10 μm) as a positive control for PPAR-δ. *, p < 0.5 versus other groups (one-way ANOVA). C, CV-1 cells were transfected as in A and treated with the 5-lipoxygenase (5-LO) substrate arachidonic acid (AA, 10 μm) and A23187. Some cells were transfected with a 5-lipoxygenase expression plasmid in the presence or absence of zileuton (50 μm). Following treatment, cells were incubated for 24 h and then lysed and analyzed by Dual-Luciferase assay. Inset, shown is a Western blot 24 h after transfection with 5-lipoxygenase or control plasmid. *, p < 0.5 versus other groups (one-way ANOVA). D, RBL-2H3 cells were transfected with a PPAR-dependent luciferase reporter plasmid (aP2) with luciferase under control of PPAR response elements and either a PPAR-α expression plasmid (PPAR-α) or empty control (Ctrl). Transfected cells were stimulated with A23187 (5 μm) and then lysed and analyzed by Dual-Luciferase assay 24 h later. Inset, shown is a Western blot 24 h after transfection with PPAR-α or control plasmid. *, p < 0.5 versus other groups (one-way ANOVA). Data are mean values of three independent experiments performed in triplicate. Error bars represent S.E. Western blots were repeated at least three times.
To demonstrate conclusively that 5-lipoxygenase is essential for PPAR-α activation, we also carried out studies in cells (CV-1) that lack native 5-lipoxygenase (Fig. 2C). Stimulation with the Ca2+ ionophore did not activate PPAR-α in these cells unless they were transfected with a 5-lipoxygenase expression plasmid. To confirm that the 5-lipoxygenase product not only binds to but functionally activates PPAR-α, we studied binding of activated receptor to PPAR response elements. We transfected RBL-2H3 cells, which lack native PPAR-α, with plasmids containing luciferase under control of PPAR response elements. Stimulation with A23187 induced luciferase activity only in the presence of PPAR-α expression and not in native cells (Fig. 2D). In additional studies, PPAR-α-transfected cells were treated with zileuton then stimulated with A23187. Zileuton effectively inhibited A23187-induced PPAR-α activation (data not shown).
LTB4 Is the 5-Lipoxygenase Product That Activates PPAR-α in Vitro
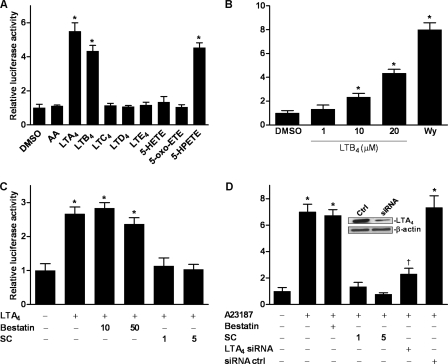
To determine the specific identity of the 5-lipoxygenase product that activates PPAR-α in vitro, we investigated the ability of candidate compounds to activate PPAR-α in CV-1 cells transfected with a Gal4-luciferase PPAR-α reporter system. PPAR-α activity was increased significantly by LTA4, LTB4, and 5-HPETE but not by arachidonic acid, LTC4, LTD4, LTE4, 5-HETE, or 5-oxo-ETE (Fig. 3A). 5-HPETE, LTA4, and LTB4 are thus activating ligands of PPAR-α, although these results with exogenously added compound do not establish physiological relevance. Most LTB4 is secreted into surrounding medium or tissues, where it activates cell-surface BLT1 or BLT2 receptors on adjacent cells. To eliminate the possibility that PPAR-α activation was an indirect result of such effects, studies were repeated using HeLa cells, which lack such cell-surface receptors (19, 20). Treatment of HeLa cells with LTB4 activated PPAR-α in a manner identical to that seen in CV-1 cells and was dose-dependent in the range of 1–20 μm (Fig. 3B).
FIGURE 3.
Identification of the specific PPAR-α-activating compound. In A, B, C, and D, cells were transfected with the PPAR-α Gal4-luciferase reporter system, treated, and then lysed and analyzed by Dual-Luciferase assay 24 h later. A, transfected CV-1 cells were treated with arachidonic acid (AA), LTA4, LTB4, LTC4, LTD4, LTE4, 5-HETE, 5-oxo-ETE, or 5-HPETE, all at 20 μm. *, p < 0.5 versus DMSO (one-way ANOVA). B, transfected HeLa cells were treated with LTB4 (1–20 μm) or with Wy 14643 (WY, 10 μm). *, p < 0.5 versus DMSO (one-way ANOVA). C, transfected CV-1 cells were treated with the LTA4 hydrolase inhibitors bestatin (10 and 50 μm) or SC22716 (SC; 1 and 5 μm) and LTA4 (10 μm). *, p < 0.5 versus DMSO (one-way ANOVA). D, transfected Jurkat T cells were treated with bestatin (50 μm) or SC22716 (1 and 5 μm) and stimulated with A23187 (5 μm). Some cells were transfected with LTA4 hydrolase siRNA (LTA4 siRNA) or scrambled siRNA control (siRNA ctrl) for 48 h prior to A23187 stimulation. Inset, shown is a Western blot 48 h after transfection with LTA4 hydrolase siRNA. *, p < 0.5 versus DMSO (one-way ANOVA); †, p < 0.5 versus siRNA control (one-way ANOVA). Data are mean values of three independent experiments performed in triplicate. Error bars represent S.E. Western blots were repeated at least three times.
Interestingly, activation of PPAR-α was also seen following treatment with LTA4, although this compound did not form the hydrogen bonds considered essential for the effect. Furthermore, the extent of activation was greater than that following treatment with an equimolar concentration of LTB4 (Fig. 3A). Because LTA4 is converted to LTB4 by LTA4 hydrolase, we tested the effect on PPAR-α activation of LTA4 hydrolase inhibitors. Surprisingly, the commonly employed LTA4 hydrolase inhibitor bestatin had no effect on PPAR-α activation by LTA4 (Fig. 3C). Further study showed that this reflected failure of bestatin to traverse the cellular membrane (data not shown). By contrast, LTA4-induced activation was completely inhibited by the cell-permeable inhibitor SC22716 (Fig. 3C).
We next sought to show that LTB4 is the specific endogenous product that activates PPAR-α following 5-lipoxygenase stimulation. We therefore tested the effects of SC22716 on Ca2+ ionophore-induced PPAR-α activation. Jurkat T cells transfected with the Gal4-luciferase PPAR-α reporter system were treated with SC22716 and stimulated with A23187. Inhibition of LTA4 hydrolase abolished the stimulatory effects of ionophore treatment (Fig. 3D), thus demonstrating that the 5-lipoxygenase product involved in PPAR-α activation is specifically LTB4. To eliminate the possibility that SC22716 was acting through off-target mechanisms, we silenced LTA4 hydrolase by transfection with targeted LTA4 siRNA. LTA4 hydrolase silencing, like enzyme inhibition, inhibited PPAR-α activation following stimulation with A23187 (Fig. 3D). These results reinforce the conclusion that LTB4 is the only effective activating ligand within the cell and that 5-HPETE, although capable of activating PPAR-α when added exogenously at high concentrations, has little effect under intracellular conditions.
Stimulation of LTB4 Production under Physiological Conditions Activates PPAR-α
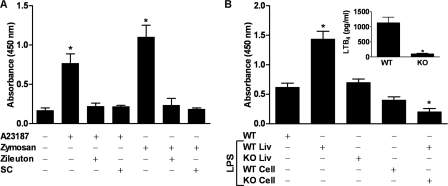
To demonstrate relevance in more physiological settings, studies were carried out using PMNs isolated from human blood. In some of these studies, cells were stimulated with opsonized zymosan (21) rather than A23187. Both stimuli induced PPAR-α activation in these cells (Fig. 4A), whereas both zileuton and SC22716 inhibited activation. These results confirm the identity of LTB4 as the relevant PPAR-α activator in primary cells exposed to physiological as well as pharmacological stimuli.
FIGURE 4.
5-Lipoxygenase stimulation of PPAR-α under physiologic conditions. A, human PMNs (pooled from three donors) were treated with either zileuton (50 μm), SC22716 (SC; 1 μm), or vehicle. Some PMNs were then stimulated with A23187 (5 μm), whereas other cells were stimulated with opsonized zymosan. After 4 h, nuclear protein was isolated for PPAR-α transcription factor assay. *, p < 0.5 versus DMSO (one-way ANOVA). B, wild-type (WT; n = 3) or 5-lipoxygenase knock-out (KO; n = 4) mice were injected intraperitoneally with 20 mg/kg body weight of LPS or vehicle (saline). Four h later, mice were euthanized, and the peritoneal cavity was opened and washed with 10 ml of phosphate-buffered saline. Liver tissue was then excised. Nuclear protein was isolated for PPAR-α transcription factor assay from both liver tissue (Liv), and cells pelleted by centrifugation from lavage fluid. *, p < 0.5 versus wild-type (one-way ANOVA). Inset, LTB4 in lavage fluid supernatant was quantitated by ELISA 4 h following LPS stimulation. *, p < 0.5 versus wild-type (Student's t test, two-sided). Data are mean values of three independent experiments performed in triplicate. Error bars represent S.E.
Because inflammation is the primary stimulus for LTB4 production in vivo, we hypothesized that inflammation would lead to PPAR-α activation in a 5-lipoxygenase-dependent manner. To test this hypothesis, we induced inflammation in wild-type and 5-lipoxygenase knock-out mice by intraperitoneal injection of bacterial lipopolysaccharide. Four hours later, the mice were euthanized, fluid from peritoneal lavage was collected, and liver tissue was excised. Lavage fluid from wild-type but not 5-lipoxygenase knock-out mice demonstrated increased LTB4 concentrations (Fig. 4B, inset). In both liver tissue and the cellular fraction of peritoneal lavage fluid, LPS treatment increased PPAR-α activity in wild-type but not 5-lipoxygenase knock-out mice (Fig. 4B). Thus, in vivo as in vitro, a 5-lipoxygenase product is essential for inflammation-induced increases in PPAR-α activity.
PPAR-α Decreases Secretion of LTB4 and Stimulates Its Breakdown
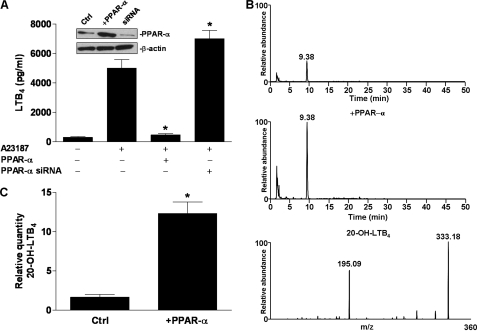
Because PPAR-α induces enzymes involved in lipid degradation, including that of LTB4 (22), it is plausible that PPAR-α activation may reduce net production of LTB4 following cell stimulation. To determine whether secretion of LTB4 is altered when PPAR-α is activated by endogenously produced LTB4, we transfected Jurkat T cells with PPAR-α expression plasmids and then stimulated them with A23187 as before. Secretion of LTB4 was significantly lower in the presence than in the absence of PPAR-α overexpression (Fig. 5A). To further confirm that this decrease in LTB4 secretion results from PPAR-α activity, we also knocked down endogenous PPAR-α using siRNA. PPAR-α knockdown significantly increased secretion of LTB4 (Fig. 5A). To confirm the hypothesis that the PPAR-α-induced decrease in LTB4 results at least in part from increased degradation, we transfected Jurkat T cells with PPAR-α, stimulated them with A23187, and measured the relative concentration of the first LTB4 degradation product, 20-OH-LTB4, using LC-MS/MS (14). PPAR-α transfection increased the relative concentration of 20-OH-LTB4 significantly (Fig. 5, B and C). The identity of 20-OH-LTB4 was confirmed by MS of the putative peak (Fig. 5B). It follows that in cells expressing PPAR-α, production of LTB4 is to some extent self-limiting due to enhanced degradation of the product.
FIGURE 5.
Effects of PPAR-α on LTB4 secretion and metabolism. A, Jurkat T cells were transfected either with a PPAR-α expression plasmid (PPAR-α) or PPAR-α-targeted siRNA. After 24 h, transfected cells were stimulated with A23187. After an additional 1 h of incubation, culture supernatant was collected, and LTB4 was quantified by ELISA. Inset, shown is a Western blot after transfection with PPAR-α plasmid or PPAR-α siRNA. *, p < 0.5 versus A23187 (one-way ANOVA). B, Jurkat T cells were transfected with PPAR-α expression plasmid (middle) or empty vector (top). After 24 h, cells were stimulated with A23187 for 1 h. The reaction was stopped by the addition of methanol, and medium was collected after centrifugation. Diluted supernatant was passed through a solid-phase extraction cartridge, eluted with methanol, evaporated under inert nitrogen gas at room temperature, and analyzed by HPLC followed by MS/MS (bottom panel). Mass spectrometric analyses were performed in the negative ion mode using multiple reaction monitoring and monitoring the transition m/z 351 → 195 for 20-OH-LTB4. C, shown are the relative expression levels of 20-OH-LTB4 in cells as treated in B. *, p < 0.5 versus control (Ctrl) (Student's t test, two-sided). Data are mean values of three independent experiments performed in triplicate. Error bars represent S.E. Western blots and mass spectrometric data were repeated at least three times.
DISCUSSION
Identification of physiologically relevant PPAR agonists has proven unexpectedly difficult. PPARs were initially identified as “orphan” receptors because no endogenous ligands were known. Improved screening methods have allowed identification of compounds that bind to and activate each of the three PPAR isoforms. Which, if any, of these compounds are important for activation of their target receptor(s) in vivo remains unclear, however. Many of the identified agonists either are low affinity ligands or exhibit intracellular concentrations that may be low or unknown. There has been only one previous instance in which any PPAR has been activated by an identified agonist generated specifically within the target cell. Here, we demonstrate conclusively that, in cells possessing the enzyme 5-lipoxygenase, stimulation of the activity of the enzyme leads to activation of PPAR-α via production of intracellular LTB4.
The ability of LTB4 to act as a PPAR-α agonist has been reported previously (3, 12, 13) but has also been disputed (11). Our results with CV-1 cells, however, directly contradict the report of Forman and colleagues (11) that LTB4 does not activate PPAR-α in these cells. Activation by a variety of other eicosanoids (11, 23), including a novel 15-lipoxygenase product (24), has also been reported. All of these studies, however, were carried out with exogenously added compounds and provide little evidence as to whether identified agonists are present intracellularly in concentrations and locations adequate for physiologically meaningful activation. As the example of the PPAR-γ ligand 15d-PGJ2 shows, studies with exogenous ligands can prove irrelevant physiologically. Our results with LTA4 and 5-HPETE also highlight how easily results with exogenously added putative ligands could be misinterpreted in the absence of rigorous investigation of their endogenous roles. Both of these compounds, when added exogenously, are highly effective PPAR-α activators. In the case of LTA4, this is due exclusively to its subsequent conversion to LTB4. In the case of 5-HPETE, the inability of 5-lipoxygenase stimulation to activate the receptor when LTA4 hydrolase is removed indicates that this 5-lipoxygenase metabolite is not present at sufficient intracellular concentrations or locations to play a significant physiological role.
By contrast, we demonstrate that LTB4 directly activates PPAR-α under conditions typically analogous to those encountered by cells of the immune system in vivo. We further extend and confirm these findings by showing that inflammatory conditions stimulating LTB4 production in vivo also up-regulate PPAR-α activity. Surprisingly, LTB4 does not appear to be the only physiologically relevant ligand in the body. Observations have demonstrated that liver PPAR-α is activated by the phospholipid 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (9). Concentrations of this compound vary with activity of fatty acid synthase in a way that is physiologically relevant to PPAR-α regulation of lipid metabolism in this organ. Although neither we nor Chakravarthy and colleagues (9) specifically identified the cell types involved, it is likely that the phospholipid activator is primarily relevant to hepatocytes, whereas our results may be dominated by Kupffer cells, mononuclear phagocytes found in the liver. In combination, these data suggest that functionally distinct cell types may have different PPAR-α ligands, each physiologically relevant to the specific function of the cell. One may speculate that the same will prove to be true of other PPARs as well. It should also not be assumed that LTB4 and 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine are the only physiologically relevant PPAR-α ligands. Chakravarthy and colleagues (9) not only raise the possibility of tissue-specific ligands regulating inflammation, as we have now shown to be the case, but also suggest that the specific phospholipid they have identified is unlikely to be physiologically relevant in brain. It is thus likely that there is at least one, and perhaps several, additional PPAR-α ligands that are physiologically relevant in different tissues.
In addition to showing that LTB4 is a physiologically relevant PPAR-α ligand, we also demonstrate that secretion of this compound is decreased by the presence of activated PPAR-α and that this effect is accompanied by increased levels of LTB4 metabolic breakdown products. Breakdown of LTB4 typically involves ω-oxidation by specific cytochrome P450s, followed by β-oxidation usual for long-chain fatty acids, with enzymes essential for both portions of this pathway subject to PPAR-α induction (22). These considerations led Devchand and colleagues (3) to propose that LTB4, via activation of PPAR-α, stimulated its own breakdown and hence contributed to the resolution of inflammation. Their finding that the duration of inflammation was increased in PPAR-α knock-out mice reinforces this suggestion, which draws further support from our observation. Fiedler and colleagues (25), however, reported that LTB4 did not stimulate LTB4 catabolism in isolated rat hepatocytes. This may be accounted for by the observed failure of LTB4 to up-regulate CYP4Fs, the cytochrome P450s primarily responsible for LTB4 metabolism in these cells (26). Because hepatocytes do not synthesize LTB4 and are therefore exposed only to the exogenous lipid (27), differences in cytochrome P450 induction mechanisms are not implausible. Youseff and Badr (2) also questioned the importance of LTB4 degradation in the anti-inflammatory properties of PPAR-α, as they observed that attenuation of rat paw edema by PPAR-α agonists was rapid and therefore unlikely to reflect transcription of oxidative enzymes. This in vivo system is complex, however; and it is likely that PPAR-α acts by several different mechanisms. The possible predominance of transcription-independent mechanisms in this model does not rule out the importance of transcriptional mechanisms in other settings.
Inflammation is a finely balanced process that must be stimulated when required to combat infection or other insults, yet limited and resolved when it is no longer needed. Given the importance of LTB4 in the inflammatory process (28), exerted mostly through activation of cell-surface receptors, it is fitting that in many of the cells where it is produced it also exerts anti-inflammatory effects, including limitation of its own activity, by activating the nuclear receptor PPAR-α. LTB4 may exert different effects in different immune and inflammatory cells capable of its synthesis. Notably, PPAR-α is absent from alveolar macrophages, often the first cells exposed in the lung to inhaled foreign antigens and therefore important for initiation of the inflammatory process. Given this crucial early role of macrophages, it is appropriate that there is no balancing anti-inflammatory LTB4 activity.
In summary, we show that LTB4, which exerts proinflammatory effects via cell-surface receptors, is also responsible for activating the anti-inflammatory nuclear transcription factor PPAR-α following stimulation of its intracellular production. As a quite different lipid has been shown to activate PPAR-α under conditions that are physiologically relevant to hepatocytes, this strongly suggests that PPARs in general may have alternative ligands in functionally different cell types. We also show that activation of PPAR-α results in reduced secretion of LTB4. This suggests an important homeostatic mechanism by which a crucial proinflammatory mediator ultimately limits its own activity and thus facilitates resolution of the inflammatory process.
Acknowledgments
We thank Todd Leff for comments on the manuscript and Kate Noon for technical assistance with the mass spectrometric analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant HL093196.
- PPAR
- peroxisome proliferator-activated receptor
- LTB4
- leukotriene B4
- LTA4
- leukotriene A4
- ANOVA
- analysis of variance
- DMSO
- dimethyl sulfoxide
- 15d-PGJ2
- 15-deoxy-Δ12,14-prostaglandin J2
- HETE
- hydroxyeicosatetraenoic acid
- ELISA
- enzyme-linked immunosorbent assay
- HPETE
- hydroperoxyeicosatetraenoic acid
- PMN
- polymorphonuclear leukocyte
- siRNA
- small interfering RNA
- LPS
- lipopolysaccharide
- HPLC
- high pressure liquid chromatography
- MS
- mass spectrometry
- LC-MS/MS
- liquid chromatography-tandem MS.
REFERENCES
- 1.Brown J. D., Plutzky J. (2007) Circulation 115, 518–533 [DOI] [PubMed] [Google Scholar]
- 2.Youssef J., Badr M. (2004) J. Biomed. Biotechnol. 2004, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devchand P. R., Keller H., Peters J. M., Vazquez M., Gonzalez F. J., Wahli W. (1996) Nature 384, 39–43 [DOI] [PubMed] [Google Scholar]
- 4.Woerly G., Honda K., Loyens M., Papin J. P., Auwerx J., Staels B., Capron M., Dombrowicz D. (2003) J. Exp. Med. 198, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poynter M. E., Daynes R. A. (1998) J. Biol. Chem. 273, 32833–32841 [DOI] [PubMed] [Google Scholar]
- 6.Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. (1995) Cell 83, 803–812 [DOI] [PubMed] [Google Scholar]
- 7.Bell-Parikh L. C., Ide T., Lawson J. A., McNamara P., Reilly M., FitzGerald G. A. (2003) J. Clin. Invest. 112, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzameli I., Fang H., Ollero M., Shi H., Hamm J. K., Kievit P., Hollenberg A. N., Flier J. S. (2004) J. Biol. Chem. 279, 36093–36102 [DOI] [PubMed] [Google Scholar]
- 9.Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. (2009) Cell 138, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters-Golden M., Henderson W. R., Jr. (2007) N. Engl. J. Med. 357, 1841–1854 [DOI] [PubMed] [Google Scholar]
- 11.Forman B. M., Chen J., Evans R. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. (1997) Mol. Endocrinol. 11, 779–791 [DOI] [PubMed] [Google Scholar]
- 13.Lin Q., Ruuska S. E., Shaw N. S., Dong D., Noy N. (1999) Biochemistry 38, 185–190 [DOI] [PubMed] [Google Scholar]
- 14.Christmas P., Tolentino K., Primo V., Berry K. Z., Murphy R. C., Chen M., Lee D. M., Soberman R. J. (2006) J. Biol. Chem. 281, 7189–7196 [DOI] [PubMed] [Google Scholar]
- 15.Cronet P., Petersen J. F., Folmer R., Blomberg N., Sjöblom K., Karlsson U., Lindstedt E. L., Bamberg K. (2001) Structure 9, 699–706 [DOI] [PubMed] [Google Scholar]
- 16.Zoete V., Grosdidier A., Michielin O. (2007) Biochim. Biophys. Acta 1771, 915–925 [DOI] [PubMed] [Google Scholar]
- 17.Murphy R. C., Gijón M. A. (2007) Biochem. J. 405, 379–395 [DOI] [PubMed] [Google Scholar]
- 18.Wong A., Cook M. N., Foley J. J., Sarau H. M., Marshall P., Hwang S. M. (1991) Biochemistry 30, 9346–9354 [DOI] [PubMed] [Google Scholar]
- 19.Pettersson A., Boketoft A., Sabirsh A., Nilsson N. E., Kotarsky K., Olde B., Owman C. (2000) Biochem. Biophys. Res. Commun. 279, 520–525 [DOI] [PubMed] [Google Scholar]
- 20.Sabirsh A., Bristulf J., Owman C. (2004) Eur. J. Pharmacol. 499, 53–65 [DOI] [PubMed] [Google Scholar]
- 21.Claesson H. E., Lundberg U., Malmsten C. (1981) Biochem. Biophys. Res. Commun. 99, 1230–1237 [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto T., Cook W. S., Qi C., Yeldandi A. V., Reddy J. K., Rao M. S. (2000) J. Biol. Chem. 275, 28918–28928 [DOI] [PubMed] [Google Scholar]
- 23.Ng V. Y., Huang Y., Reddy L. M., Falck J. R., Lin E. T., Kroetz D. L. (2007) Drug Metab. Dispos. 35, 1126–1134 [DOI] [PubMed] [Google Scholar]
- 24.Kozak K. R., Gupta R. A., Moody J. S., Ji C., Boeglin W. E., DuBois R. N., Brash A. R., Marnett L. J. (2002) J. Biol. Chem. 277, 23278–23286 [DOI] [PubMed] [Google Scholar]
- 25.Fiedler J., Simon F. R., Iwahashi M., Murphy R. C. (2001) J. Pharmacol. Exp. Ther. 299, 691–697 [PubMed] [Google Scholar]
- 26.Kalsotra A., Anakk S., Brommer C. L., Kikuta Y., Morgan E. T., Strobel H. W. (2007) Arch. Biochem. Biophys. 461, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leier I., Müller M., Jedlitschky G., Keppler D. (1992) Eur. J. Biochem. 209, 281–289 [DOI] [PubMed] [Google Scholar]
- 28.Crooks S. W., Stockley R. A. (1998) Int. J. Biochem. Cell Biol. 30, 173–178 [DOI] [PubMed] [Google Scholar]