Abstract
The Ohr (organic hydroperoxide resistance) family of 15-kDa Cys-based, thiol-dependent peroxidases is central to the bacterial response to stress induced by organic hydroperoxides but not by hydrogen peroxide. Ohr has a unique three-dimensional structure and requires dithiols, but not monothiols, to support its activity. However, the physiological reducing system of Ohr has not yet been identified. Here we show that lipoylated enzymes present in the bacterial extracts of Xylella fastidiosa interacted physically and functionally with this Cys-based peroxidase, whereas thioredoxin and glutathione systems failed to support Ohr peroxidase activity. Furthermore, we could reconstitute in vitro three lipoyl-dependent systems as the Ohr physiological reducing systems. We also showed that OsmC from Escherichia coli, an orthologue of Ohr from Xylella fastidiosa, is specifically reduced by lipoyl-dependent systems. These results represent the first description of a Cys-based peroxidase that is directly reduced by lipoylated enzymes.
Keywords: Antioxidant, Bacterial Metabolism, Oxidative Stress, Peroxidase, Thiol, Antioxidant Defense, Cys-based Thiol-dependent Peroxidase, Lipoic Acid, Organic Hydroperoxide, Phytopathogenic Bacteria
Introduction
Release of oxidants is one of the main responses of a host against pathogens. During host-pathogen interactions, in an attempt to kill the pathogen, plant or animal hosts generate various oxidants by enzymatic systems (1–3). For instance, organic hydroperoxides are oxidants that can be formed by the reaction of molecular oxygen with unsaturated fatty acids catalyzed by lipoxygenases in response to pathogen infection (4–7). To counteract this oxidative stress, pathogens have developed various antioxidant pathways (8, 9). Ohr (organic hydroperoxide resistance) protein is a Cys-based, thiol-dependent peroxidase that plays a central role in the response of bacteria against organic peroxide-induced insult (10). Ohr is exclusively present in bacteria (most of them pathogenic) and possesses a unique α/β fold that is not observed in the structures of peroxiredoxins and glutathione peroxidases (11, 12). Ohr peroxidase activity requires reduction of the enzyme's disulfide group formed upon catalytic reduction of the organic hydroperoxide (13). However, the enzyme's physiological reducing agent has not yet been identified.
Ohr belongs to a family of proteins that also comprise osmotically inducible protein (OsmC). These proteins were initially identified as part of the bacterial response to osmotic stress (14). On the basis of sequence analysis, Atichartpongkul et al. (10) proposed Ohr and OsmC homologues as two protein subfamilies. Later, it was demonstrated that OsmC enzymes also possess thiol-dependent peroxidase activity and share the same structural fold with Ohr proteins (15, 16).
Because only dithiols, and not monothiols, can be utilized by Ohr, we and others hypothesized that lipoate may act as a natural reducing agent of Ohr (11, 13, 17). Lipoate is a dithiol/disulfide redox compound well known to play important roles in metabolic pathways due to its capacity to serve as a cofactor in the multienzyme complexes that catalyze the oxidative decarboxylation of α-keto acids (18). Besides lipoylated E2 subunits, 2-oxo acid dehydrogenase multienzyme complexes also contain Lpd (dihydrolipoamide dehydrogenase, subunit E3 from α-ketoglutarate dehydrogenase complex) activity that can catalyze the redox processes between lipoylated E2 enzymes and NADH/NAD+ in both reductive and oxidative directions (19). Free dihydrolipoate, the reduced form of lipoate, has also been reported to scavenge free radicals and other oxidants (20). However, the cellular availability of free lipoate is significantly lower than that of other endogenous low molecular weight antioxidants, such as GSH and ascorbic acid (21).
Xylella fastidiosa is a Gram-negative, xylem-limited phytopathogenic bacterium that is the causative agent of several economically important diseases, such as citrus variegated chlorosis and Pierce disease in citrus and grapes, among other plants (22). Here, we show that peroxidase activity of Ohr from X. fastidiosa is supported by lipoylated proteins but not by thioredoxin and glutathione systems. Therefore, these data represent a previously undescribed enzymatic activity. We also showed that OsmC from Escherichia coli is specifically reduced by lipoyl-dependent systems.
EXPERIMENTAL PROCEDURES
Reagents
All reagents purchased had the highest degree of purity. Lipoic acid was purchased from Sigma in its oxidized form. It was solubilized in a phosphate-buffered solution through the high pH method (23). Lipoamide was purchased from Sigma in its oxidized form. A stock solution was prepared by diluting lipoamide to 100 mm in absolute methanol. Lipoamide (purchased from Sigma; catalog no. T5875) was reduced through the addition of sodium borohydride (purchased from Sigma; catalog no. S9125) as described previously (24). All commercial enzymes utilized in this paper were purchased from Sigma (glutaredoxin 1 (catalog no. G3531), glutathione reductase (catalog no. G3664), glutathione peroxidase (catalog no. G6137), thioredoxin (catalog no. T0910), and thioredoxin reductase (catalog no. T7915)). Reduced lipoamide was quantified using Ellman's reagent (5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB)2 (25). Nitrocellulose membrane was obtained from Bio-Rad. Protein A-Sepharose was purchased from GE Healthcare. The anti-lipoic antibody was obtained in the laboratory of Dr. Luke Szweda (Oklahoma Medical Research Foundation) as described previously (26). The anti-Ohr antibody was obtained by immunization of rabbits with recombinant Ohr. Briefly, in the first immunization, an Ohr solution (1 mg/ml) in phosphate-buffered saline (PBS) was mixed with Freund's adjuvant (Sigma, catalog no. F5506) (1:1, v/v). The Bench MarkTM protein ladder was purchased from Invitrogen. The rabbits were immunized three times, with 2-week intervals between immunizations. After the third immunization, the rabbits were sedated, and the blood was collected with a syringe. The serum from the samples was recovered and stored at −80 °C.
Nucleic Acid Extraction, Cloning, and Nucleotide Sequences
The lpd, lpdA (dihydrolipoamide dehydrogenase, subunit E3 from pyruvate dehydrogenase complex), PDHB (dihydrolipoamide acyltransferase, subunit E2 from pyruvate dehydrogenase complex), and sucB (dihydrolipoamide acyltransferase, subunit E2 from α-ketoglutarate dehydrogenase complex) genes were PCR-amplified from X. fastidiosa 9a5c strain genomic DNA. The following primers were used: lpdforw (5′-GGGAATTCCATATGACGTTGC-3′), lpdrev (5′-CCGCTCGAGTCAGTTTGCTTTATG-3′), lpdAforw (5′-GGGAATTCCATATGACGGTGATTG-3′), and lpdArev (5′-CCGCTCGAGTTACTTTGCTGAAG-3′) (the underlined bases denote the NdeI and XhoI restriction sites, respectively, for Lpd and LpdA genes); sucBforw (5′-CTAGCTAGCATGACCATCGAAG-3′) and sucBrev (5′-CGCGGATCCTTATAAGCC-3′) (the underlined bases represent the NheI and BamHI restriction sites, relative to the forward and reverse primers, respectively); PDHBforw (5′-GGGAATTCCATATGACCGAAATCAAGGAAG-3′) and PDHBrev (5′-CGGGATCCTCAACGGGCCTTAC-3′) (the underlined bases represent the NdeI and BamHI restriction sites, relative to the forward and reverse primers, respectively). The osmC gene was PCR-amplified from Escherichia coli DH5-α strain genomic DNA. The following primers were used: osmCforw (5′-CGATCCATATGACAATCCATAAGAAAGG-3′ and osmCrev (5′-CGCGGATCCTTACGATTTCAACTGGTAATCC-3′) (the underlined bases represent the NdeI and BamHI restriction sites, relative to the forward and reverse primers, respectively). PCR products were double-digested with the appropriate restriction enzymes and cloned into digested pET15b (for Lpd, LpdA, and PDHB), pET28a (for sucB), and pPROEX (for osmC) expression vectors. The sequences of the resulting pET15b/Lpd, PET15b/LpdA, PET15b/PDHB, pET28a/sucB, and pPROEX/osmC plasmids were confirmed by automated DNA sequencing. E. coli DH5-α strain was transformed by electroporation (Gene Pulser® II, Bio-Rad) with the expression vectors and cultured to increase plasmid production. Plasmid extraction was performed using Perfectprep® plasmid mini (Eppendorf). BL21(DE3) and AD494(DE3) strains were transformed by electroporation. sucB and PDHB genes were transformed into Bl21(DE3) cells, and lpd and lpdA genes were transformed into AD494(DE3) cells. The resulting strains were used for expression and purification of the Lpd, LpdA, SucB, and PDHB recombinant proteins. The osmC gene was expressed in the E. coli DH5-α strain. Cloning, expression, and purification of Ohr were performed as described previously (13).
Protein Expression and Purification
Bl21(DE3) strains transformed with pET15b/PDHB and pET28a/sucB, AD494 strains transformed with pET15b/lpd and pET15b/lpdA were cultured (50 ml) overnight in LB medium containing the appropriate antibiotics. Each culture was transferred to 1 liter of fresh LB plus antibiotics and grown until the OD reached 0.6–0.8. Isopropyl-1-thio-β-d-galactopyranoside was then added to a final concentration of 0.1 mm. The cells were grown overnight at 20 °C and were then harvested by centrifugation. For the expression of lipoylated enzymes from the strains containing pET15b/lpdA, pET15b/PDHB, and pET15b/sucB, 300 μg/ml lipoic acid was added to the culture medium to ensure maximum lipoylation of these proteins. For the DH5-α strain transformed with pPROEX/osmC, isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm, and cells were grown for 3 h at 37 °C and were then harvested by centrifugation. The pellets were washed and resuspended in start buffer (phosphate buffer, 20 mm, pH 7.4). Eight cycles of 25 s of sonication (40% amplitude) followed by 50 s on ice were applied to cell suspensions. Cell extracts were kept on ice during 1% streptomycin sulfate treatment for 20 min. The suspension was centrifuged at 31,500 × g for 40 min to remove nucleic acid precipitates. Finally, the cell extracts were each applied to a nickel affinity column (Hi-Trap from GE Healthcare). The conditions of protein purification were optimized using the gradient procedure for imidazole concentration described by the manufacturer. All recombinant proteins were quantified by A280 through in silico prediction of extinction coefficients (ExPASy; available on the World Wide Web). Lipoylated enzymes SucB and LpdA possess 1 mol of lipoate per 1 mol of enzyme, and PDHB possesses 2 mol of lipoate per 1 mol of enzyme.
Preparation of Crude Lysates from E. coli and X. fastidiosa for Immunodetection Assays
Cell pellets from X. fastidiosa J1A12 and E. coli DH5α (OD ∼0.6–1.0) were resuspended in phosphate buffer (50 mm, pH 7.4) containing NaCl (50 mm) and disrupted by sonication. Cell extracts were kept on ice during 1% streptomycin sulfate treatment for 20 min. The suspension was centrifuged at 31,500 × g for 40 min to remove nucleic acid precipitates. Total protein concentration was determined by the Bradford method (27).
Immunodetection of Lipoylated Proteins and Ohr
For immunodetection, the samples were run on SDS-polyacrylamide gels, and the gels were electroblotted onto a 0.45-μm nitrocellulose membrane in Tris-glycine buffer at 50 V for 90 min. Blotted membranes were blocked in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T) with 5% skim milk (Bio-Rad) overnight at 4 °C. The membrane was washed three times for 5 min with PBS-T and incubated with an anti-lipoic acid (1:5000 in PBS-T) or anti-Ohr (1:1000 in PBS-T) antibody for 1 h at room temperature, followed by washing. The membrane was subsequently incubated with an anti-phosphatase rabbit antibody (1:1000) for 1 h (purchased from the Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). After washing, the proteins were detected following incubation with the 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphatase substrate. The levels of lipoylated proteins were quantitated by densitometry using ImageQuant TL software (GE Healthcare).
Immunoprecipitation of Lipoic Acid
Immunoprecipitation was performed as follows. 1 mg of X. fastidiosa lysate (previously incubated for 30 min with Protein A-Sepharose) was incubated with anti-lipoic acid (1:50) overnight at 4 °C. Immune complexes were then incubated with Protein A-Sepharose for 4 h at 4 °C, followed by centrifugation. The immunoprecipitate was washed five times with Tris-HCl buffer (20 mm), NaCl (300 mm) and boiled with 40 μl of sample buffer containing dithiothreitol (10 mm). Electrophoresis and transfer were performed. Blotted membranes were incubated with an anti-Ohr antibody (1:1000 in PBS-T) overnight at 4 °C. The membrane was subsequently incubated with an anti-phosphatase rabbit antibody for one hour and proteins were detected following incubation with the 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphatase substrate (Kirkegaard and Perry Laboratories).
Isolation of Extracellular Protein Fraction
To isolate secreted proteins as well as proteins weakly attached to the cell surface, we use a modified protocol from Smolka et al. (28). Briefly, 50 ml of X. fastidiosa was centrifuged, and the pellet was washed twice with buffer containing 10 mm Tris-HCl (pH 8.8), 3 mm KCl, 50 mm NaCl, 5 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride. The supernatant from each washing step was precipitated with a cold ethanol/acetone/acetic acid (50:50:0.1, v/v/v) solution on ice for 30 min and solubilized in 50 μl of 10 mm Tris (pH 8.8), 0.5% SDS, 5 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride. After the washing steps, the pelleted cells were then lysed with 200 μl of the following solution: 10 mm Tris (pH 8.8), 0.5% (w/v) SDS, 5 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride. After the addition of 100 mm dithiothreitol, the proteins (from the supernatant and cellular fraction) were boiled for 3 min and loaded onto a 14% SDS-polyacrylamide gel under reducing conditions. Following electrophoresis, the gel was transferred to a nitrocellulose membrane and incubated with an anti-Ohr antibody (1:1000) or an anti-lipoic acid antibody (1:5000) for 1 h at room temperature.
Disulfide Reductase Activity Assay
The disulfide reductase activities of the reducing systems were determined using the DTNB (Ellman's reagent) assay (29). The reaction mixtures contained 50 mm sodium phosphate (pH 7.4), 1 mm diethylenetriamine pentaacetic acid (DTPA), 0.5 mm DTNB, and 0.2 mm NAD(P)H and were carried out at 37 °C and were initiated by the addition of NADH.
Peroxidase-coupled Lipoamide System Assay
The peroxidase activities of Cys-based, thiol peroxidases were measured using recombinant Lpd or LpdA from X. fastidiosa and lipoamide or recombinant E2 enzymes from X. fastidiosa (PDHB or SucB). Decay due to NADH consumption was measured by A340 (ϵ = 6290 m−1.cm−1). The reaction mixtures contained 50 mm sodium phosphate (pH 7.4), 50 mm NaCl, 1 mm DTPA (pH 7.4), and 0.2 mm NADH. All reactions were performed at 37 °C and were initiated by the addition of peroxide at various concentrations.
Peroxidase-coupled Thioredoxin System Assay
The peroxidase activities of Cys-based thiol peroxidases were measured using recombinant thioredoxin and thioredoxin reductase (TSNC and TRR, respectively) from X. fastidiosa or commercial thioredoxin and thioredoxin reductase from E. coli. Decay due to NADPH consumption was measured by A340 (ϵ = 6220 m−1·cm−1). The reaction mixtures contained 50 mm sodium phosphate (pH 7.4), 50 mm NaCl, 1 mm DTPA (pH 7.4), and 0.2 mm NADPH. All reactions were performed at 37 °C and were initiated by the addition of t-BHP at various concentrations.
Peroxidase-coupled Glutathione System Assay
The peroxidase activity of Ohr from X. fastidiosa, OsmC from E. coli, and human glutathione peroxidase were measured using reduced glutathione and commercial glutathione reductase from bakers' yeast. Decay due to NADPH consumption was measured by A340 (ϵ = 6220 m−1·cm−1). The reaction mixtures contained 50 mm sodium phosphate (pH 7.4), 50 mm NaCl, 1 mm DTPA (pH 7.4), and 0.2 mm NADPH. All reactions were performed at 37 °C and were initiated by the addition of t-BHP at various concentrations.
RESULTS
Ohr Interacts with Lipoylated Enzymes in Vivo
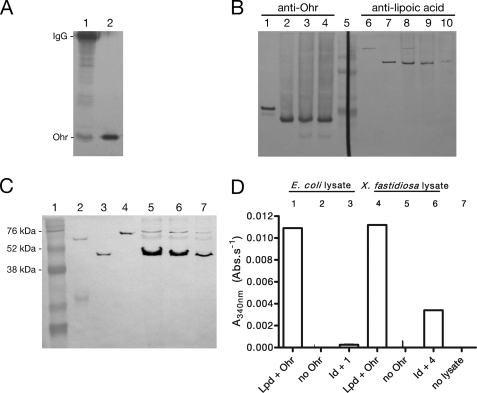
Lipoylated proteins from X. fastidiosa, a bacterial phytopathogen, were immunoprecipitated utilizing a polyclonal antibody specific for lipoic acid (anti-lipoic acid). Consistent with our hypothesis, we observed that Ohr co-precipitated with lipoylated enzymes (Fig. 1A). Moreover, we observed that Ohr is present in the extracellular fraction (or weakly attached to the cell surface) along with a protein reactive to anti-lipoic acid that co-migrated with SucB, the E2 subunit of the α-ketoglutarate dehydrogenase complex (Fig. 1B, lanes 3, 4, 9, and 10). Furthermore, another study detected Ohr and LpdA in the extracellular environment through proteomic analyses of X. fastidiosa (28). LpdA is an enzyme harboring its own substrate, a lipoyl group (30).
FIGURE 1.
In vivo interaction of Ohr with lipoylated proteins. A, Ohr co-immunoprecipitated with lipoylated enzymes. Proteins were precipitated with anti-lipoic acid and were analyzed by Western blot with anti-Ohr. Lane 1, immunoprecipitated proteins on beads (30 μl); lane 2, recombinant Ohr (0.1 μg). The His tag from recombinant Ohr (lane 2) was previously digested by thrombin treatment. B, fractions containing extracellular proteins were obtained by washing cells twice with Tris buffer. Lanes 1–4, anti-Ohr (1:1000); lanes 6–10, anti-lipoic acid (1:5000). Lane 1, recombinant Ohr (0.1 μg); lane 2, X. fastidiosa lysate; lane 3, supernatant first wash; lane 4, supernatant second wash; lane 5, RainbowTM markers; lane 6, recombinant LpdA (0.1 μg); lane 7, recombinant SucB (0.1 μg); lane 8, X. fastidiosa lysate; lane 9, supernatant first wash; lane 10, supernatant second wash. All recombinant proteins possess a His tag. C, immunodetection of lipoate enzymes in X. fastidiosa lysates. Lane 1, RainbowTM markers; lane 2, PDHB (0.1 μg); lane 3, SucB (0.1 μg); lane 4, LpdA (0.1 μg); lanes 5–7, X. fastidiosa lysates (15, 10, and 5 μg, respectively). D, ability of immunodepleted (Id) E. coli (Ec) and X. fastidiosa (Xf) lysates to support Ohr peroxidase activity. Bacterial lysates (0.5 mg/ml) were incubated with NADH (0.2 mm), Ohr (1 μm), Lpd (2.5 μm), and t-BHP (0.2 mm). For more details on the experimental procedures employed here, see the supplemental material.
Ohr was also found in the intracellular fraction of X. fastidiosa (Fig. 1B, lane 2). Therefore, we investigated whether any other lipoylated proteins were present in this compartment. Immunochemical evidence indicated that all three lipoylated enzymes (LpdA, PDHB, and SucB) predicted by the annotation of the X. fastidiosa genome (see the X. fastidiosa Comparative Genome Project Web site from Laboratório Nacional de Computação Científica) are present in the cell lysates (Fig. 1C). Thus, in principle, all of these proteins could support Ohr peroxidase activity in the intracellular compartment. If this is the case, then the ability of X. fastidiosa lysates to support Ohr activity should be decreased if lipoylated proteins were immunodepleted. In fact, the removal of lipoylated proteins from X. fastidiosa and E. coli lysates reduced the rate of NADH oxidation compared with the same preparation that was not immunodepleted with anti-lipoic acid (Fig. 1D). The residual activity detected in the immunodepleted lysates correlated with the presence of lipoylated proteins that were not completely removed during the immunoprecipitation step (supplemental Fig. 1).
Lipoyl-dependent Peroxidase Activity of Ohr
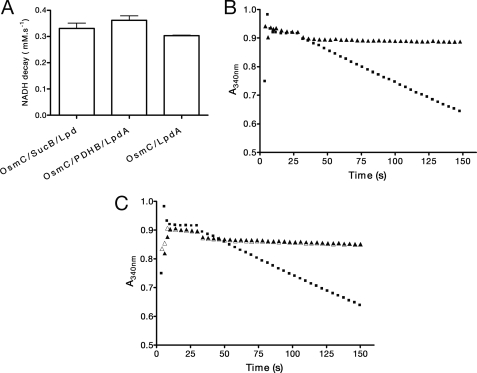
Next, we investigated the ability of Lpd from X. fastidiosa to alternatively function as a physiological reductant of Ohr instead of their classical function as oxidants during α-keto acid oxidative decarboxylation. Initially, the disulfide reductase activity of recombinant Lpd from X. fastidiosa in the presence of free lipoamide was demonstrated (Fig. 2A). We then showed that this lipoamide-dependent system supported the peroxidase activity of Ohr (Fig. 2B). Interestingly, only Ohr and not the other two Cys-based peroxidases from X. fastidiosa (PrxQ (peroxiredoxin Q) and AhpC (alkyl hydroperoxide reductase, subunit C)) presented lipoyl-dependent peroxidase activity (Fig. 2B). Parallel lines were obtained in Lineweaver-Burk plots, indicating a ping-pong reaction mechanism (Fig. 2C), as expected for other Cys-based peroxidases (31, 32). Using a bisubstrate kinetic approach (33), Km values in the micromolar range for lipoamide and t-BHP were obtained (Table 1, first lane, and supplemental Fig. 2). In contrast, millimolar amounts of H2O2 are required to saturate Ohr (Fig. 2D) under non-limiting amounts of lipoamide. Therefore, Ohr reduced t-BHP at least 10,000 times more efficiently than H2O2 (Table 1). The Ohr active site pocket is surrounded by several hydrophobic residues (11, 12), which may explain the very high Km value for H2O2. This large difference in affinities for hydroperoxides has never been described before for other Cys-based peroxidases. Furthermore, the catalytic efficiency of Ohr to detoxify t-BHP (106 m−1·s−1) is similar or even higher than other Cys-based thiol peroxidases (9, 34, 35). This is the first detailed enzymatic characterization of a Cys-based peroxidase belonging to the Ohr family of proteins.
FIGURE 2.
Characterization of lipoyl-dependent peroxidase activity. A, disulfide reductase activity of lipoamide systems. ●, 1 μm LpdA plus 0.5 μm PDHB; ♦, 1 μm Lpd plus 1 μm SucB; ■, 1 μm lipoamide plus 1 μm Lpd; ▴, 1 μm LpdA; □, 1 μm LpdA−LA; ▵, 1 μm Lpd plus 1 μm SucB−LA. −LA denotes proteins whose expression was not enriched with lipoic acid. B, lipoamide-dependent peroxidase activity of Cys-based thiol-dependent peroxidases (0.1 μm) in the presence of free lipoamide (0.2 mm), Lpd (0.5 μm), and t-BHP (0.2 mm). ●, Ohr; ▴, AhpC; ■, PrxQ; ♦, no further addition. C, Lineweaver-Burk plot of the lipoamide-dependent peroxidase activity of Ohr. Lipoamide concentrations were as follows: 7.5 μm (■), 10 μm (▴), and 15 μm (●). D, Henri-Michaelis-Menten plot for Ohr catalysis in reactions containing Ohr (1 μm), Lpd (5 μm), and lipoamide (0.1 mm). The result is the average of three independent experiments. Data are means ± S.D.
TABLE 1.
Kinetic constant for different Ohr substrates
Parameters for H2O2 were obtained through a non-linear regression fit using saturating concentrations of lipoamide. Parameters for all other substrates were obtained with the secondary plots of 1/Vmax(app) and 1/Km(app) versus 1/[LA]. In the case of lipoylated enzymes SucB and PDHB, LA represents the lipoic acid covalently attached to the enzyme (1 mol of LA per 1 mol of SucB and 2 mol of LA per 1 mol of PDHB).
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | m−1s−1 | |
| t-BHPa | 14.51 ± 0.15 | 17.95 ± 7.25 | (2.06 ± 0.08) × 106 |
| H2O2a | 41,720.00 ± 6530.00 | 9.53 ± 0.56 | (2.30 ± 0.20) × 102 |
| Lipoamide | 44.46 ± 0.98 | 90.9 ± 4.70 | (2.06 ± 0.08) × 106 |
| PDHB | 10.93 ± 0.15 | 9.5 ± 0.20 | (0.87 ± 0.01) × 106 |
| SucB | 38.51 ± 14.12 | 17.95 ± 7.25 | (0.46 ± 0.02) × 106 |
a Parameters for t-BHP and H2O2 were calculated using free lipoamide. The results are the average of three independent experiments and represent mean ± S.D.
Reconstitution of the Ohr Physiological Reducing System
Initially, we cloned and expressed enzymes from X. fastidiosa predicted to have lipoyl binding domains (supplemental Fig. 3), which include SucB (the E2 component of 2-oxoglutarate dehydrogenase complex), PDHB (the E2 component of pyruvate dehydrogenase complex), and LpdA. The physiological role of LpdA is still unclear. Because its gene is located close to genes encoding proteins of the pyruvate dehydrogenase complex, it was annotated as its E3 component. However, LpdA in Streptococcus pneumonia is not associated with any two oxo-acid dehydrogenase complexes (30). In any case, recombinant SucB, PDHB, and LpdA presented disulfide reductase activity (Fig. 2A).
Then we tested whether these lipoyl-dependent reducing systems were capable of supporting Ohr peroxidase activity. Both SucB and PDHB promoted Ohr-dependent peroxide removal in the presence of Lpd. Besides that, LpdA alone or in combination with SucB or PDHB also supported Ohr peroxidase activity (Fig. 3A). Therefore, we have reconstituted three possible pathways involved in the decomposition of organic hydroperoxides.
FIGURE 3.
Reconstitution of the Ohr physiological reducing system. A, Ohr peroxidase activity measured in the presence of lipoylated recombinant proteins (LpdA (2 μm), SucB (2 μm), and PDHB (1 μm)) or free lipoamide (2 μm). Reaction mixtures contained NADH (0.2 mm), Lpd (2 μm), and Ohr (0.1 μm). B, disulfide reductase activity of the thioredoxin system from X. fastidiosa. ●, 5 μm TSNC; ♦, 5 μm TRR; ▴, 5 μm TRR plus 5 μm TSNC. C, peroxidase activity of Ohr and PrxQ in the presence of the thioredoxin system from X. fastidiosa. ▾, 5 μm TRR plus 5 μm TSNC and 5 μm PrxQ; ♦, 5 μm TRR plus 5 μm TSNC and 10 μm Ohr. D, dependence of Ohr activity on LpdA concentration. Reaction mixtures as described under “Experimental Procedures” contained Ohr (0.1 μm). The results presented in a and d are the average of three independent experiments. Data are means ± S.D. (error bars).
We also investigated whether other classical thiol-containing systems, well known reductants for other thiol peroxidases (peroxiredoxins and glutathione peroxidases), could act as alternative electron donors to Ohr. Recombinant TSNC and TRR from X. fastidiosa were obtained, and they presented disulfide reductase activity (Fig. 3B). However, Ohr presented no peroxidase activity, even with the use of high concentrations of thioredoxin and thioredoxin reductase (Fig. 3C). Under the same conditions, PrxQ from X. fastidiosa was active (Fig. 3C). In fact, taking into account data from the literature, it is evident that the concentrations of thioredoxin and thioredoxin reductase employed here were high enough to support the enzymatic activity of other Cys-based peroxidases (36, 37). Some Cys-based peroxidases display peroxidase activity supported by glutathione or glutaredoxin (38, 39). Therefore, it was relevant to investigate whether Ohr would present such activities. We previously observed that GSH does not support Ohr activity, even at very high concentrations (13). We have now verified that glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae and glutaredoxin 1 from E. coli in the presence of GSH and glutathione reductase did not support Ohr peroxidase activity. We did not observe any NADPH consumption even using millimolar concentrations of GSH. Given the fact that 2 μm lipoamide is sufficient to support Ohr peroxidase activity (Fig. 3A), the glutathione system is probably not its physiological reductant. In conclusion, Ohr is highly specific for lipoylated enzymes among other biological reducing systems.
To gain further insight into the physiological relevance of the interactions of Ohr with lipoylated enzymes, we performed bisubstrate kinetic analysis (supplemental Fig. 2). The catalytic parameters of Ohr that were determined relative to SucB and PDHB were similar in comparison with free lipoamide (Table 1 and supplemental Fig. 2). Moreover, using saturating concentrations of t-BHP, we were able to analyze the dependence of the reaction on LpdA concentration (Fig. 3D). The Km value for LpdA (13.5 ± 1.9 μm) was also very similar to that obtained for SucB and PDHB (Table 1). Comparison of Km values is difficult because the parameters for SucB and PDHB (Table 1) were calculated in a fixed concentration of flavoenzyme (Lpd = 1 μm), whereas for LpdA, the concentration of flavoenzyme present in the reaction mixture varies according to the value indicated on the x axis (Fig. 2D) because this enzyme possesses both an Lpd and a lipoyl binding domain (30). To test whether the increase in peroxidase activity could also be influenced by an increase in Lpd concentration, we performed the assay in the presence of SucB and various concentrations of Lpd (supplemental Fig. 4). Increasing concentrations of Lpd did not influence the activity of the system, showing that the concentration of Lpd used in these assays was not rate-limiting.
Moreover, to determine whether lipoylated enzymes are present in the cell at concentrations that make Ohr-dependent hydroperoxide reduction kinetically favorable, we employed semiquantitative Western blot analysis using anti-lipoic acid. SucB was the most abundant lipoyl protein, present at micromolar concentrations (0.9 μm). LpdA and PDHB could also be detected although at lower concentrations (0.07 and 0.13 μm, respectively) (Fig. 1C).
Finally, we showed that recombinant OsmC (osmotically inducible protein) from E. coli, another Cys-based, thiol peroxidase belonging to the OsmC/Ohr family of proteins (10, 14, 15, 16), also had its activity supported by lipoylated proteins from X. fastidiosa (Fig. 4A). Again, classic reducing systems, such as thioredoxin and glutathione, failed to support OsmC activity, even at high concentrations (Fig. 4, B and C, respectively). Furthermore, the addition of glutaredoxin 1 from E. coli to the glutathione system did not lead to NADH consumption dependent on OsmC activity (data not shown). These data suggested that the specificity for lipoyl-dependent systems is a common feature for proteins belonging to the Ohr/OsmC family.
FIGURE 4.
Lipoyl-dependent peroxidase activity of OsmC from E. coli. A, OsmC peroxidase activity in the presence of lipoylated recombinant proteins from X. fastidiosa (LpdA (2 μm), PDHB (1 μm), and SucB (2 μm)). Reaction mixtures contained NADH (0.2 mm), Lpd (2 μm), and OsmC (0.5 μm). B, peroxidase activity of OsmC (10 μm, ▴) and PrxQ (5 μm, ■) from X. fastidiosa in the presence of the thioredoxin system from E. coli (thioredoxin (1 μm) and thioredoxin reductase (0.5 μm)). C, peroxidase activity of Ohr (▵, 10 μm), OsmC (▴, 10 μm), and human glutathione peroxidase (■, 1 μm glutathione peroxidase) in the presence of a heterologous glutathione system (6 μg/μl glutathione reductase, 1 mm GSH). Reaction mixtures were added as described under “Experimental Procedures.” The results presented in a are the average of three independent experiments. Data are means ± S.D. (error bars).
DISCUSSION
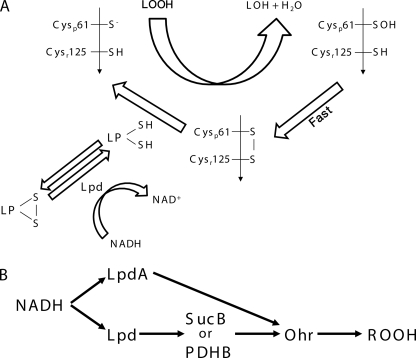
The results presented here indicate that lipoylated proteins (SucB, PDHB, and/or LpdA) are the biological reducing agents for Ohr (Fig. 5). Traditionally, protein-associated lipoyl groups are viewed as electron acceptors in oxidative pathways, but they can also act in reductive processes (19). For instance, lipoylated E2 from Mycobacterium tuberculosis reduces a thioredoxin-like protein (AhpD), thereby indirectly supporting the peroxidase activity of a peroxiredoxin (AhpC) (26). Besides, it was also proposed that the reduced form of lipoylated enzymes could donate electrons to ribonucleotide reductase via Grx1 (40). Interestingly, the redox state of the complex-bound lipoate is an indicator of the availability of the reaction substrates (2-oxo acid, CoA, and NAD+) and thiol-disulfide status of the medium (19).
FIGURE 5.
Proposed scheme for the Ohr reduction pathways. A, the Ohr disulfide is reduced by a lipoylated protein (LP). B, Ohr reduction by lipoylated proteins. The arrows represent the flow of electrons coming from NADH.
Importantly, levels of components belonging to the lipoamide system are probably present at concentrations capable of efficiently supporting Ohr peroxidase activity in vivo because these concentrations were in the range of the Km values determined by bisubstrate kinetic analyses. In fact, in another bacteria (E. coli), whose cells were grown in minimal medium, E1, E2, and E3 were identified as three of the 19 proteins most induced by aerobic growth after anaerobiosis (41, 42). Levels of lipoic acid in bacterial cytosols are also abundant (43, 44), making their ability to support Ohr peroxidase activity feasible. Remarkably, we did not find an orthologous of LplA (lipoate ligase), in the X. fastidiosa genome. This enzyme is responsible for lipoylation of enzymes using exogenous lipoic acid as substrate (45). However, we found all of the three genes involved in the biosynthetic pathway of lipoic acid: LipA (lipoyl synthase, XF1050), LipB (lipoyltranferase, XF1051), and ACP (acyl carrier protein, XF0672) (46, 47). This finding is in line with the observation that the plant pathogen X. fastidiosa colonizes an environment with poor availability of nutrients.
Interestingly, OsmC from E. coli was also reduced by lipoylated proteins from X. fastidiosa (Fig. 4). This result indicates that lipoylated proteins may also be the physiological reductant for other members of the Ohr/OsmC family of proteins.
It is important to mention that a search in the genome data bank of X. fastidiosa revealed that there are at least three Grxs with variable lengths and active sites (XF2066, XF2263, and XF2323). Also, X. fastidiosa possesses at least another three thioredoxin-like proteins (XF0992, XF1569, and XF1714) besides TSNC (XF2343). Therefore, we cannot exclude the possibility that one of these Trxs and/or Grxs donate electrons to Ohr.
This work is the first description of a protein endowed with lipoyl-dependent peroxidase activity. The ability of Ohr to utilize reducing equivalents from lipoyl sulfhydryls, its unique structure, and the fact that it is exclusively present in many types of pathogenic bacteria might represent an unexplored microbial niche for new molecular scaffolds. This finding is relevant because antibiotic-resistant strains of pathogenic bacteria are increasingly prevalent (48), and several major classes of bactericidal antibiotics operate through the production of highly deleterious hydroxyl radicals in both Gram-negative and Gram-positive bacteria (49). Remarkably, lipoyl groups are essential for parasite survival in host cells (50, 51) and for virulence (52). Indeed, the utilization of lipoylated proteins for Ohr reduction represents an emerging area of investigation (19) into the likelihood that specific proteins perform multiple catalytic functions to provide a means to regulate divergent processes, with a single molecular switch.
Supplementary Material
Acknowledgments
We are grateful to A. M. Silva (Instituto de Química, Universidade de São Paulo); José FS Netto (Instituto de Ciências Biomédicas, Universidade de São Paulo); B. B. Horta, K. F. Discola, and Dr. M. Vainzof (Instituto de Biociencias, Universidade de Sao Paulo); and Dr. J. R. K. Júnior (Faculdade de Medicina Veterinária e Zootecnia, Universidade de São Paulo) for providing material for this work.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo Grant 07/58147-6 and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico under the National Institute of Science and Technology for Redox Processes in Biomedicine (Redoxome).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- DTNB
- 5, 5′-dithiobis-(2-nitrobenzoic acid)
- t-BHP
- tert-butyl hydroperoxide
- PBS
- phosphate-buffered saline
- DTPA
- diethylenetriamine pentaacetic acid.
REFERENCES
- 1.Tenhaken R., Levine A., Brisson L. F., Dixon R. A., Lamb C. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen R., van der Straaten T., van Diepen A., van Dissel J. T. (2003) Microbes Infect. 5, 527–534 [DOI] [PubMed] [Google Scholar]
- 3.Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., Brumell J. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalloul A., Montillet J. L., Assigbetsé K., Agnel J. P., Delannoy E., Triantaphylidès C., Daniel J. F., Marmey P., Geiger J. P., Nicole M. (2002) Plant J. 32, 1–12 [DOI] [PubMed] [Google Scholar]
- 5.Gobel C., Feussner I., Rosahl S. (2003) J. Biol. Chem. 278, 52834–52840 [DOI] [PubMed] [Google Scholar]
- 6.Montillet J. L., Agnel J. P., Ponchet M., Vailleau F., Roby D., Triantaphylidès C. (2002) Plant Physiol. Biochem. 40, 633–639 [Google Scholar]
- 7.Maeng J. H., Sakai Y., Tani Y., Kato N. (1996) J. Bacteriol. 178, 3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G., Alamuri P., Maier R. J. (2006) Mol. Microbiol. 61, 847–860 [DOI] [PubMed] [Google Scholar]
- 9.Poole L. B. (2005) Arch. Biochem. Biophys. 433, 240–254 [DOI] [PubMed] [Google Scholar]
- 10.Atichartpongkul S., Loprasert S., Vattanaviboon P., Whangsuk W., Helmann J. D., Mongkolsuk S. (2001) Microbiology 147, 1775–1782 [DOI] [PubMed] [Google Scholar]
- 11.Oliveira M. A., Guimarães B. G., Cussiol J. R., Medrano F. J., Gozzo F. C., Netto L. E. (2006) J. Mol. Biol. 359, 433–445 [DOI] [PubMed] [Google Scholar]
- 12.Lesniak J., Barton W. A., Nikolov D. B. (2002) EMBO J. 21, 6649–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cussiol J. R., Alves S. V., de Oliveira M. A., Netto L. E. (2003) J. Biol. Chem. 278, 11570–11578 [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez C., Devedjian J. C. (1991) J. Mol. Biol. 220, 959–973 [DOI] [PubMed] [Google Scholar]
- 15.Lesniak J., Barton W. A., Nikolov D. B. (2003) Protein Sci. 12, 2838–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin D. H., Choi I. G., Busso D., Jancarik J., Yokota H., Kim R., Kim S. H. (2004) Acta Crystallogr. D 60, 903–911 [DOI] [PubMed] [Google Scholar]
- 17.Meunier-Jamin C., Kapp U., Leonard G. A., McSweeney S. (2004) J. Biol. Chem. 279, 25830–25837 [DOI] [PubMed] [Google Scholar]
- 18.Reed L. J. (2001) J. Biol. Chem. 276, 38329–38336 [DOI] [PubMed] [Google Scholar]
- 19.Bunik V. I. (2003) Eur. J. Biochem. 270, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 20.Moini H., Packer L., Saris N. E. (2002) Toxicol. Appl. Pharmacol. 182, 84–90 [DOI] [PubMed] [Google Scholar]
- 21.Schupke H., Hempel R., Peter G., Hermann R., Wessel K., Engel J., Kronbach T. (2001) Drug Metab. Dispos. 29, 855–862 [PubMed] [Google Scholar]
- 22.Purcell A. H., Hopkins D. L. (1996) Annu. Rev. Phytopathol. 34, 131–151 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki Y. J., Tsuchiya M., Packer L. (1994) Methods Enzymol. 234, 454–461 [DOI] [PubMed] [Google Scholar]
- 24.Reed L. J., Koike M., Levitch M. E., Leach F. R. (1958) J. Biol. Chem. 232, 143–158 [PubMed] [Google Scholar]
- 25.Ellman G., Lysko H. (1979) Anal. Biochem. 93, 98–102 [PubMed] [Google Scholar]
- 26.Bryk R., Lima C. D., Erdjument-Bromage H., Tempst P., Nathan C. (2002) Science 295, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 27.Kruger N. J. (1994) Methods Mol. Biol. 32, 9–15 [DOI] [PubMed] [Google Scholar]
- 28.Smolka M. B., Martins D., Winck F. V., Santoro C. E., Castellari R. R., Ferrari F., Brum I. J., Galembeck E., Della Coletta Filho H., Machado M. A., Marangoni S., Novello J. C. (2003) Proteomics 3, 224–237 [DOI] [PubMed] [Google Scholar]
- 29.Luthman M., Holmgren A. (1982) Biochemistry 21, 6628–6633 [DOI] [PubMed] [Google Scholar]
- 30.Håkansson A. P., Smith A. W. (2007) J. Biol. Chem. 282, 29521–29530 [DOI] [PubMed] [Google Scholar]
- 31.Trujillo M., Ferrer-Sueta G., Thomson L., Flohé L., Radi R. (2007) Subcell. Biochem. 44, 83–113 [DOI] [PubMed] [Google Scholar]
- 32.Tosatto S. C., Bosello V., Fogolari F., Mauri P., Roveri A., Toppo S., Flohé L., Ursini F., Maiorino M. (2008) Antioxid. Redox Signal. 10, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 33.Segel H. I. (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 606–612, Wiley Interscience, New York [Google Scholar]
- 34.Fourquet S., Huang M. E., D'Autreaux B., Toledano M. B. (2008) Antioxid. Redox Signal. 10, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 35.Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 36.Munhoz D. C., Netto L. E. (2004) J. Biol. Chem. 279, 35219–35227 [DOI] [PubMed] [Google Scholar]
- 37.Rhee S. G., Chae H. Z., Kim K. (2005) Free Radic. Biol. Med. 38, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 38.Rouhier N., Gelhaye E., Jacquot J. P. (2002) J. Biol. Chem. 277, 13609–13614 [DOI] [PubMed] [Google Scholar]
- 39.Bréhélin C., Meyer E. H., de Souris J. P., Bonnard G., Meyer Y. (2003) Plant Physiol. 132, 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beckwith J. (2009) J. Biol. Chem. 284, 12585–12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. (1978) Cell 14, 179–190 [DOI] [PubMed] [Google Scholar]
- 42.Smith M. W., Neidhardt F. C. (1983) J. Bacteriol. 154, 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noll K. M., Barber T. S. (1988) J. Bacteriol. 170, 4315–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed K. E., Cronan J. E., Jr. (1993) J. Bacteriol. 175, 1325–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris T. W., Reed K. E., Cronan J. E., Jr. (1995) J. Bacteriol. 177, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jordan S. W., Cronan J. E., Jr. (1997) J. Biol. Chem. 272, 17903–17906 [DOI] [PubMed] [Google Scholar]
- 47.Ma Q., Zhao X., Nasser Eddine A., Geerlof A., Li X., Cronan J. E., Kaufmann S. H., Wilmanns M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8662–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischbach M. A., Walsh C. T. (2009) Science 325, 1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. (2007) Cell 130, 797–810 [DOI] [PubMed] [Google Scholar]
- 50.O'Riordan M., Moors M. A., Portnoy D. A. (2003) Science 302, 462–464 [DOI] [PubMed] [Google Scholar]
- 51.Allary M., Lu J. Z., Zhu L., Prigge S. T. (2007) Mol. Microbiol. 63, 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith A. W., Roche H., Trombe M. C., Briles D. E., Håkansson A. (2002) Mol. Microbiol. 44, 431–448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.