Abstract
Although sphingomyelin is an important cellular lipid, its subcellular distribution is not precisely known. Here we use a sea anemone cytolysin, equinatoxin II (EqtII), which specifically binds sphingomyelin, as a new marker to detect cellular sphingomyelin. A purified fusion protein composed of EqtII and green fluorescent protein (EqtII-GFP) binds to the SM rich apical membrane of Madin-Darby canine kidney (MDCK) II cells when added exogenously, but not to the SM-free basolateral membrane. When expressed intracellularly within MDCK II cells, EqtII-GFP colocalizes with markers for Golgi apparatus and not with those for nucleus, mitochondria, endoplasmic reticulum or plasma membrane. Colocalization with the Golgi apparatus was confirmed by also using NIH 3T3 fibroblasts. Moreover, EqtII-GFP was enriched in cis-Golgi compartments isolated by gradient ultracentrifugation. The data reveal that EqtII-GFP is a sensitive probe for membrane sphingomyelin, which provides new information on cytosolic exposure, essential to understand its diverse physiological roles.
Keywords: Golgi, Lipid, Membrane Bilayer, Membrane Lipids, Plasma Membrane
Introduction
Sphingomyelin (SM)2 plays an important structural role in the formation of so-called lipid rafts, plasma membrane domains where it is enriched along with cholesterol (CHO) (1). Membrane microdomains take part in many eukaryotic cellular processes, i.e. signal transduction, and are important sites for entrance or exit of bacteria, viruses, and other ligands (2, 3). However, the role of SM in the cell extends much further. It also serves as a precursor reservoir for many important lipid signaling molecules, such as ceramide, sphingosine-1-phosphate, sphingosine, ceramide-1-phosphate, etc. These lipids are critical regulators of cell proliferation, differentiation and apoptosis (4–6), while ceramide and sphingosine 1-phosphate are also important in cancer pathogenesis and treatment (7). SM synthesis is also an important source of diacylglycerol (DAG), which is produced from phosphatidylcholine following the transfer of phosphocholine to ceramide by SM synthase 1 (SMS1) (8). The participation of SM in these diverse processes confirms the important role it has in cell function.
SM subcellular distribution is, however, not well understood largely because of the lack of a suitable probe, which specifically binds SM. For example, of the many lipid-binding proteins only a handful have been shown to selectively bind SM. These include enzymes that degrade SM to ceramides (9, 10) and earthworm protein lysenin (11) for which the molecular details of SM recognition are not known. One further example is the actinoporin family, ≈20 kDa pore-forming toxins from sea anemones (12). The most studied representative, EqtII from Actinia equina, is a globular protein with a tightly folded 12 strand β-sandwich core flanked on two sides by single α-helices. The first of these, situated within the 30 N-terminal amino acids, is the only part of the molecule that can move freely without disrupting the general fold of the protein and is essential for formation of transmembrane pores (13, 14). However, pore formation by EqtII is a multistep process involving, respectively, membrane binding, transfer of the amphipathic N-terminal segment to the lipid-water interface, oligomerization on the membrane surface and formation of the final pore by the N-terminal α-helices (13–17). Regarding this study, the most important characteristic of EqtII is its remarkable SM specificity. EqtII recognizes SM during membrane binding via a phosphocholine (POC) binding site and an adjacent aromatic amino acid cluster (13, 16, 18). Because PC and SM possess the same POC headgroup and EqtII binds isolated SM analogues (16), the toxin therefore is sensitive to the characteristic SM chemistry below the headgroup rather than its headgroup or bulk phase behavior. Residues Trp-112 and Tyr-113, both located at the membrane interface and required for the recognition of SM (16), probably lie within hydrogen bonding distance of the distinct SM hydroxyl and amide groups. This protein architecture is unique and allows exclusive recognition of SM and not other sphingolipids, such as ceramide, glucosyl-ceramide, galactosyl-ceramide, lactosyl-ceramide, or ganglioside GM1 (16). Interestingly, strict SM dependence has a clear biological role because sea anemones possess a modified SM to which actinoporins do not bind (19).
In this work we demonstrate the feasibility of using EqtII-GFP fusions as new markers for cellular SM. Recombinant EqtII-GFP fusion protein has the same SM-dependent membrane-binding capacity as the wild-type EqtII and using this new SM marker, we demonstrate distribution of SM within the MDCKII cells and NIH 3T3 fibroblasts.
EXPERIMENTAL PROCEDURES
Materials
Rabbit polyclonal anti-GFP and anti-TGN38 were obtained from Calbiochem and Abcam, respectively. Rabbit polyclonal anti-calnexin, anti-flotillin-2, and mouse monoclonal anti-GM130 were purchased from Santa Cruz Biotechnology. 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), porcine brain SM, CHO, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dipalmitoyl-sn-glycero-3-phospho-l-serine (DPPS), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol) (DOPI) were from Avanti Polar Lipids (Alabaster, AL). Tricyclodecan-9-yl xanthogenate (D609) was from Biomol-Enzo Life Sciences. All other chemicals were from Sigma unless stated otherwise. Recombinant EqtII, mutant EqtII 8–69, and histidine-tagged variant were prepared in Escherichia coli as described previously (20, 21). The fusion protein composed of EqtII and GFP (EqtII-GFP) was expressed as a His6 fusion protein in an E. coli Rosetta (DE3) pLysS strain as detailed in supplemental data.
Liposome Preparations
Large unilamellar vesicles (LUV) of 100 nm diameter were prepared by extrusion of multilamellar vesicles as described previously (22). When using calcein, excess dye was removed by gel filtration through a small G-50 column. DOPC concentration was measured by using a Phospholipids B kit (Wako Chemicals GmbH).
Permeabilization Assay
For calcein release experiments, calcein-loaded liposomes were used, and changes in the fluorescence intensity were followed on a Jasco FP-750 spectrofluorimeter. Liposomes were stirred in a 1.5-ml cuvette in vesicle buffer at a lipid concentration 20 μm and at 25 °C. Protein was added to achieve desired lipid/protein ratio. Excitation and emission wavelengths were set to 485 and 520 nm, respectively. Excitation and emission slits were set to 5 nm. The permeabilization induced by the proteins was expressed in percentage, as compared with the maximal permeabilization obtained at the end of the assay by the addition of Triton X-100 at a final concentration of 2 mm.
Surface Plasmon Resonance Assay
Surface plasmon resonance (SPR) measurements were performed on a Biacore X (GE Healthcare, Biacore) at 25 °C. An L1 sensor chip was equilibrated with vesicle buffer (140 mm NaCl, 20 mm Tris-HCl, 1 mm EDTA, pH 8,5) and a liposome-coated chip surface was prepared as described previously (23). Proteins were injected over immobilized LUV at a final concentration of 100 nm for 2 min at 5 μl/min, and the dissociation was followed for 5 min.
Lipid Monolayer Assay
Increase in the surface pressure of lipid monolayers of different compositions was measured with a MicroTrough-S system from Kibron (Helsinki, Finland) at room temperature. The aqueous subphase consisted of 500 μl of 50 mm NaH2PO4, 300 mm NaCl, pH 8.5. Appropriate lipid mixtures, dissolved in chloroform: methanol (2:1, v:v), were gently spread over the subphase to create a monolayer with the 20 mN/m initial surface pressure. The protein was injected through a hole connected to the subphase and the final concentration of EqtII-GFP and EqtII in the subphase was 0.2 μm. The increment in surface pressure against time was recorded until a stable signal was obtained.
Cell Culture
MDCK II (Madin-Darby canine kidney II) cells were maintained in Minimum Essential Medium (MEM) supplemented with 10% (v/v) fetal calf serum, l-glutamine, non-essential amino acids and kanamycin. 3T3 fibroblasts were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, glutamine and 1% penicillin-streptomycin. Both types of cells were cultured at 37 °C with 5% CO2.
Binding of EqtII-GFP to MDCK II Cells
MDCK II cells grown on coverslips were washed three times with phosphate-buffered saline (50 mm sodium phosphate, 150 mm NaCl, pH 7.4; PBS) and then incubated for 15 min with 0.3 μg of EqtII-GFP or 0.3 μg of GFP in PBS at 4 °C. Cells were washed again with PBS after incubation, fixed by 3% paraformaldehyde and washed once more with PBS. Plasma membrane was stained with rhodamine-conjugated wheat germ agglutinin (R-WGA) (Vector Laboratories), and cells were prepared for confocal microscopy as described below. To bind EqtII-GFP separately to apical and basolateral membrane, 5 × 105 MCDK II cells were plated onto 12-mm clear Transwell filters (Costar) and grown for 3 days. Both sides of the membrane were incubated with 0.5 μg of the EqtII-GFP in PBS for 15 min at 4 °C. After washing with PBS, fixing with paraformaldehyde and staining with R-WGA, filters were cut out and imaged with confocal microscopy as described below.
Plasmid Construction and Cell Transfection
PCR-amplified sequences of wild-type EqtII, EqtII mutants, and Dr1 were cloned N-terminal to GFP into mammalian expression vector, pcDNA3.1/CT-GFP (Invitrogen). The nucleotide sequence of all constructs was verified by sequencing. Transfection was carried out using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Cells were fixed 24–48 h after transfection to prepare samples for confocal microscopy or harvested for subcellular centrifugation.
Confocal Microscopy
Cells were seeded on coverslips and 24–48 h after transfection they were washed three times with PBS and then fixed in 3% paraformaldehyde diluted in PBS for 20 min at room temperature. Following a further three washes with PBS, the cells were incubated for 30 s with R-WGA diluted (1:50) in PBS. After washing three times in PBS, cells were mounted with Vectashield mounting medium and visualized with a confocal microscope (TCS-NT, Leica with Kr-Ar laser). To stain nuclei, cells were fixed with methanol (after washing in PBS) and after washing incubated with propidium iodide diluted in PBS (25 μg/ml) for 10 min at room temperature. Cells were washed with PBS and mounted as described above. The Golgi apparatus (GA) was visualized by staining with Bodipy-TR-ceramide complexed to bovine serum albumin (BSA) (Molecular Probes) as described previously (24) with few changes. Briefly, after removal of the medium cells on coverslips in 12-well plate were washed twice in serum free MEM, 25 mm HEPES (MEM-H) and incubated in 100 μl of 5 μm Bodipy-TR ceramide for 30 min at 4 °C. Cells were then washed with MEM-H for 30 min at 37 °C. After removal of the medium and washing three times with PBS, cells were fixed with 3% paraformaldehyde and mounted as described above. Cotransfection with mammalian expression vector pDsRed2-ER (BD Biosciences, targeted to endoplasmic reticulum (ER) with a calreticulin ER-retention sequence KDEL) allowed identification of ER. To visualize mitochondria, cotransfection using pDsRed2-Mito was performed (BD Biosciences, incorporating the mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase). A ratio of 25:1 (μg DNA, EqtII-GFP:pDsRed) was used for cotransfection.
Colocalization Analysis
Images used for EqtII-GFP and Bodipy-TR-ceramide colocalization analyses were acquired with a laser-scanning confocal microscope (Leica) in sequential mode for acquisition of both channels as Z-stacks. GFP and Bodipy fluorescence were recorded using 488 nm and 568 nm excitation, respectively. Image analysis was performed with MBF ImageJ bundle (ImageJ version 1.43d; Rasband, 1997–2009). The background resulting from dark noise and/or offset was subtracted with “BG Subtraction from ROI” plugin. The colocalization amount was measured as thresholded Mander's coefficients (25) that are a measure of colocalization of signal in one channel with signal in the other channel using the “Colocalisation Threshold” plugin developed by T. Collins (McMaster Biophotonics Facility, Hamilton, Ontario, Canada) using a defined ROI surrounding the cell of interest and excluding zero-zero values in automatic threshold computation. The plugin calculates a threshold value for each channel, so that for the pixel intensity values below the threshold for both channels, the Pearson's correlation coefficient is near zero (25).
Statistical Analysis
The differences in thresholded Mander's coefficients (tM) distribution of different data sets were tested with t test using GraphPad Prism version 5.01 for Windows (GraphPad Software, San Diego CA). The true significance of colocalization coefficients was tested using “Colocalisation Test” plugin that uses the approach by Costes et al. (25). For all channel pairs that were analyzed the colocalization coefficients were confirmed as statistically significant, e.g. colocalization was higher than in randomly scrambled images.
Subcellular Fractionation
Subcellular fractionation was performed using OptiPrep gradient (Axis-Shield) and ultracentrifugation according to the manufacturer's recommendations. The cells in five confluent 75-cm2 culture flasks (total number of cells ∼17 ± 5 × 106) were washed twice with ice-cold PBS and once with ice-cold homogenization buffer with protease inhibitor mixture (Roche) (0.25 m sucrose, 1 mm EDTA, 10 mm Hepes-NaOH, pH 7.4). The cells were harvested by scraping in ice-cold homogenization buffer and broken open using 15 strokes in a Dounce tissue homogenizer to obtain 80–85% cell lysis. Cell debris and nuclei were pelleted by centrifugation at 1000 × g for 10 min. The postnuclear supernatant (1 ml) was overlaid onto prepared discontinuous gradient (2.5/5/7.5/10/12.5/15/17.5/20/30% (w/v) iodixanol solution) and centrifuged at 200,000 × g for 3 h at 4 °C in a SW40Ti rotor (ultracentrifuge Beckman Optima L-100×). The gradient was removed in 13 equal fractions collected from the top of the gradient and stored at −80 °C.
Western Blotting
100 μl of each fraction was precipitated with 50% trichloroacetic acid, and the pellet was resuspended in 20 μl of SDS-PAGE sample loading buffer (120.5 mm Tris-HCl, 4% SDS (w/v), 10% glycerol (v/v), and 0.002% (w/v) bromphenol blue) and resolved on 12% acrylamide gel. The proteins were transferred to ImmobilonTR-P polyvinylidine difluoride membranes with pore size 0.45 μm (polyvinylidene difluoride membrane, Millipore) and blocked with 4% BSA in Tris-buffered saline (50 mm NaCl, 10 mm Tris-HCl, 1 mm EDTA, pH 7.4) overnight at room temperature. Next, the membrane was incubated with primary antibodies in the Tris-buffered saline buffer with 4% BSA for 2 h at room temperature. After incubation of the membrane with primary antibodies, washing (5 × 5 min) with 1% of the BSA in 20 ml of Tris-buffered saline buffer followed and incubation with secondary antibodies conjugated to horseradish peroxidase in Tris-buffered saline buffer with 4% BSA for 2 h at room temperature. The blots were developed by using 4-chloro-1-naphthol, methanol, and H2O2 as a substrate. The following primary antibodies were used: anti-TGN38 (for trans-Golgi), anti-GM130 (for cis-Golgi), anti-calnexin (for ER), anti-flotillin-2 (for plasma membrane), and anti-GFP (for EqtII-GFP).
Determination of Protein Content and Enzyme Activities in Fractions from Subcellular Centrifugation
All assays were performed in 96-well plates. Marker for the middle-Golgi compartments (mannosidase II) (26) was assayed as described previously. Briefly, hydrolysis of p-nitrophenyl α-d-mannopyranoside in a 50-μl standard incubation mixture, containing 2 mm substrate, 0.1 m sodium acetate buffer, and 0.1% Triton X-100 was measured. After incubation for 1 h at 37 °C, the reaction was stopped by 30 mm NaOH, and absorbance at 400 nm was recorded. K+-(p-nitrophenyl phosphatase) activity, as a marker for the plasma membrane, was assayed by the method of Marcheselli et al. (27). Hydrolysis of p-nitrophenyl phosphate was measured in a 250-μl assay mixture (50 mm Tris-HCl, 3 mm MgCl2, 10 mm KCl, pH 7.5), containing 3 mm substrate for 1 h at 37 °C. After incubation the absorbance at 405 nm was recorded. Cytochrome c reductase activity, as a marker for ER, was measured by cytochrome c reductase assay kit following the manufacturer's instructions (Sigma). Proteins were determined by the assay with Bradford reagent (Sigma). Refractive index of the fractions was determined by refractometer.
RESULTS
SM-dependent Membrane Association of EqtII-GFP
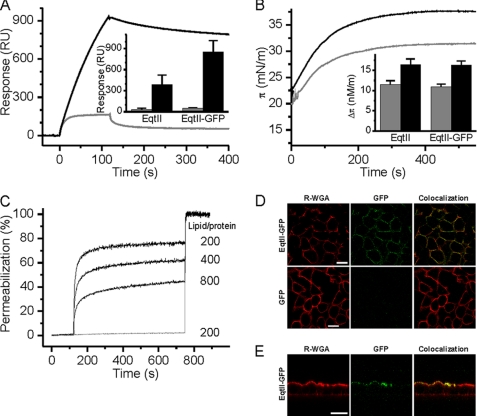
We first ensured that a fusion protein composed of EqtII and C-terminal GFP (EqtII-GFP) retains an SM-dependent membrane binding capacity. A hexahistidine-tagged EqtII-GFP was expressed in and purified from E. coli cytoplasm (supplemental Fig. S1A). His-tagged EqtII-GFP is less hemolytically active than the wild-type EqtII, but similar to EqtII that is simply His-tagged (supplemental Fig. S1B), in agreement with the published data (21). Surface plasmon resonance (SPR) experiments showed much greater binding of EqtII-GFP to membranes composed of porcine brain SM and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) as compared with membranes containing only DOPC (Fig. 1A). Again in comparison with pure DOPC, there was greater insertion of EqtII-GFP into mixed SM-DOPC monolayers at the air-water interface (Fig. 1B). Finally, EqtII-GFP was able to permeabilize LUVs composed of equimolar ratios of SM and DOPC, but not DOPC (Fig. 1C). The data are qualitatively similar to the properties of the wild-type EqtII (16) and confirm that the addition of GFP to the C terminus of EqtII does not affect its SM-specific membrane association.
FIGURE 1.
SM-dependent activity of EqtII-GFP. A, SPR analysis of EqtII-GFP binding to SM:DOPC liposomes (black) and DOPC liposomes (gray) immobilized on the surface of L1 Biacore sensor chip to ∼10000 RU. The concentration of EqtII-GFP was 100 nm. The inset shows the amount of stably bound EqtII-GFP in comparison to the EqtII after 5 min of dissociation, as determined from the sensorgrams. Black columns, SM:DOPC liposomes; gray columns, DOPC liposomes. n = 3–10; average ± S.D. B, insertion of the EqtII-GFP in lipid monolayers composed of DOPC:SM (black) and DOPC (gray). The concentration of EqtII-GFP was 0.2 μm. The inset shows the increase in the surface pressure at the equilibrium. Black columns, SM:DOPC monolayer; gray columns, DOPC monolayer. n = 2; average ± S.D. C, permeabilization of calcein-loaded LUVs by EqtII-GFP. LUVs were composed of DOPC:SM (black) and DOPC (gray). LUVs were at 20 μm lipid concentration. The lipid/toxin ratio used is indicated. D, binding of EqtII-GFP to the MDCK II cells. Cells, seeded on coverslips, were incubated with 0.3 μg of the proteins in PBS for 15 min at 4 °C. After washing, cells were fixed with 3% paraformaldehyde and plasma membrane was stained with R-WGA. E, binding of the EqtII-GFP to MDCK II cells plated on 12-mm clear Transwell filters (Costar). EqtII-GFP (0.5 μg in PBS) was added separately to the apical and to the basolateral side of the membrane. Plasma membrane was stained with R-WGA. Scale bar is 20 μm.
We then tested the binding of EqtII-GFP to MDCK II epithelial cells. The plasma membrane was intensively stained when EqtII-GFP was added to the medium, but not upon GFP addition (Fig. 1D). It is known that polarized MDCK II cells have an asymmetric distribution of sphingolipids, where SM is present in the apical and absent from the basolateral plasma membrane (28). Thus EqtII-GFP SM selectivity in vivo was confirmed when we added it to both sides of MDCK II cells, and upon imaging of cells in cross-section we observed GFP staining of only the apical side (Fig. 1E). From these data, we concluded that EqtII-GFP could be used to stain SM-containing membranes and hence we used it to explore the intracellular exposure of SM.
Cellular Distribution of EqtII-GFP
We used transiently expressed EqtII-GFP in the cytoplasm of MDCK II cells to assess intracellular distribution of SM. The cells were not affected by the intracellular expression of EqtII-GFP or other constructs used for controls (see below) and were indistinguishable from the non-transfected cells (Fig. 2). EqtII-GFP was found to have an inhomogeneous distribution characterized by a distinct punctuate, perinuclear distribution. Importantly no staining of the inner plasma membrane was observed (Fig. 2). When GFP alone was similarly expressed as a control, it was distributed homogeneously throughout the cytoplasm and nucleus. The distribution of the fusion protein fluorescence is thus driven by EqtII.
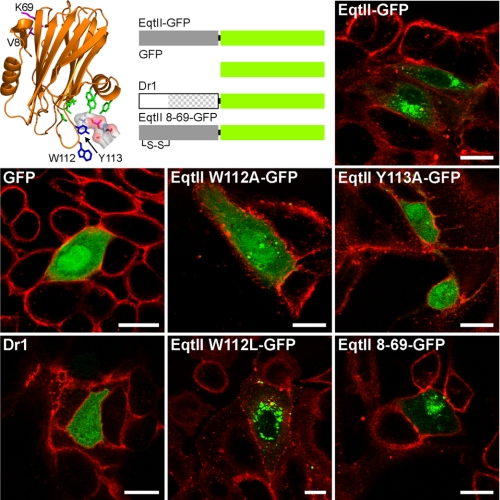
FIGURE 2.
Subcellular distribution of EqtII-GFP and its mutants or homologue protein. MDCK II cells were transfected with DNA constructs encoding EqtII, its mutants or a homologue from zebrafish, Dr1. Proteins used in this experiment are presented schematically in the top row. The structure of EqtII shows the position of the residues that have been changed in mutant proteins. The residues that participate in POC binding are labeled green (17), whereas residues responsible for SM specificity, Trp-112 and Tyr-113 (16), are shown in blue. The position of SM head group is denoted by sticks and surface presentation. Residues at position 8 and 69, which were replaced with cysteines, are labeled pink. The Dr1 region homologous to EqtII is shown by dots. The plasma membrane is stained with R-WGA. Scale bar is 20 μm.
To ensure that EqtII does not associate with other intracellular molecules we used specific control proteins, based upon our previous studies, consisting of point mutants that do not show SM specific association. Position 113 (Tyr) is very fastidious with the aromatic ring absolutely required for SM recognition, while at position 112 (Trp) any bulky hydrophobic amino acid, such as phenylalanine or leucine can maintain a wild-type phenotype (16). Mutations at these two sites to alanine diminish specific binding of SM, and prevent insertion and binding into SM containing lipid monolayers and liposomes, respectively (16). In particular EqtII W112A retains weak binding to DOPC but loses its SM specificity. The mutant proteins EqtII W112A-GFP and EqtII Y113A-GFP did not show membrane association, when expressed intracellularly and their pattern was similar to that of GFP (Fig. 2). Thus the distribution of EqtII-GFP is driven by the SM association and not by the much weaker phosphatidylcholine membrane binding. As a final confirmation EqtII W112L-GFP exhibited spotted perinuclear distribution, similar to EqtII-GFP.
We have further used a mutant with inserted cysteines at positions 8 and 69 that are precisely placed to form a disulfide bridge upon the synthesis of the protein. The disulfide bond prevents the dislocation of the N-terminal helix, and consequently the formation of pores, but SM binding is unaffected (13, 14). Disulfide bond formation is inhibited in the reducing cytoplasm but not entirely prevented and the favorable placing of the thiols in the native fold removes the entropic penalty of binding. The disulfide of EqtII 8–69 mutant is not reduced when preincubated in cytosolic concentrations of reduced glutathione (29) (supplemental Fig. S2). This mutant (EqtII 8-69-GFP) showed the same distribution as the EqtII-GFP (Fig. 2), and coupled with the unaltered GA morphology in the wild-type EqtII-GFP (see below) suggests that neither membrane insertion of the N-terminal helix nor membrane permeabilization are significant elements of the cellular SM labeling.
Finally, we used an EqtII homologue Dr1 of unknown function from zebrafish (Danio rerio) as a negative control. It displayed even staining of cells with no distinct membrane association. This agrees with its lack of SM specificity, non-hemolytic activity and weak in vitro membrane binding capacity (30) (Fig. 2). The control experiments thus indicate that perinuclear distribution of EqtII-GFP is due to a specific recognition of membrane SM by EqtII.
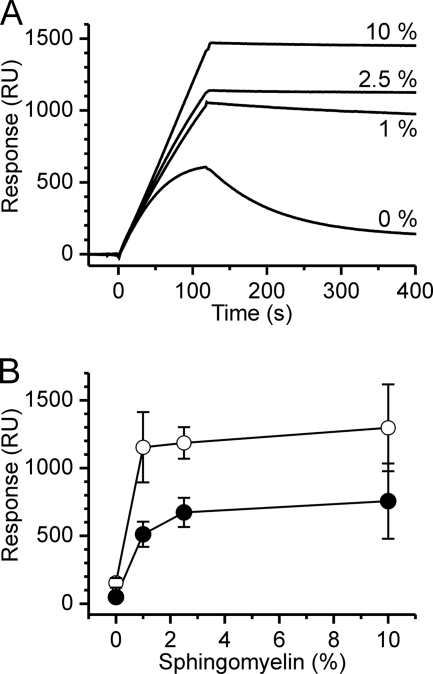
We showed above that EqtII-GFP binds to the apical membrane of the MDCK II cells when added from outside (Fig. 1, D and E). However, when expressed intracellularly no staining of plasma membrane was observed (Fig. 2), strongly indicating that inner leaflet of the plasma membrane contains too little SM to be visualized by intracellular EqtII-GFP. To assess the sensitivity of EqtII-GFP to SM in membranes we tested the binding to liposomes composed of an “inner leaflet lipid mixture” (31) (DOPC:DPPE:DPPS:CHO:DOPI in molar ratio of 30.5:13.5:19.5:35.5:1) immobilized on an L1 sensor chip. The mixture did not allow the stable binding as shown in Fig. 3. However, addition of only 1% mole fraction of SM to the inner leaflet mixture results in stable binding with low dissociation of the protein from the membrane (Fig. 3). The increase of the SM content to 2.5% or 10% increased the binding and further reduced dissociation. These results indicate that EqtII-GFP is highly sensitive to SM content.
FIGURE 3.
Binding of EqtII-GFP and EqtII to liposomes composed of the inner leaflet mixture. Experimental conditions were as described in Fig. 1A. A, binding of EqtII-GFP to liposomes composed from inner leaflet mixture (DOPC:DPPE:DPPS:CHO:DOPI in the molar ratio 30.5:13.5:19.5:35.5:1) and various % of SM as indicated. B, amount of stably bound protein after 4 min of dissociation. Open circles, EqtII-GFP; solid circles, native EqtII. n = 2–7; average ± S.D.
Subcellular Localization of EqtII-GFP
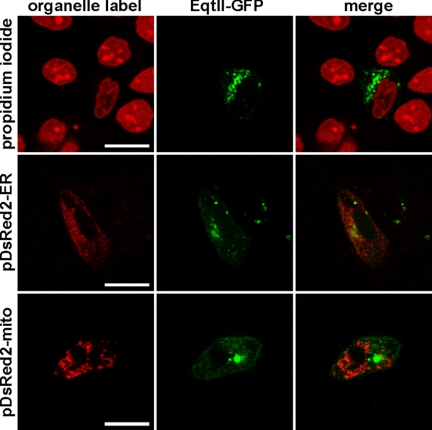
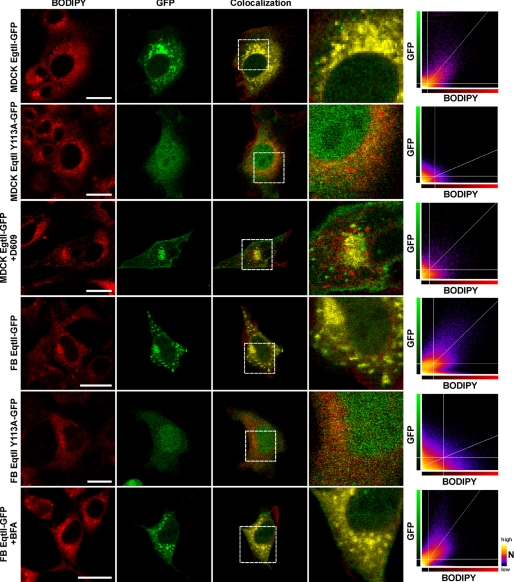
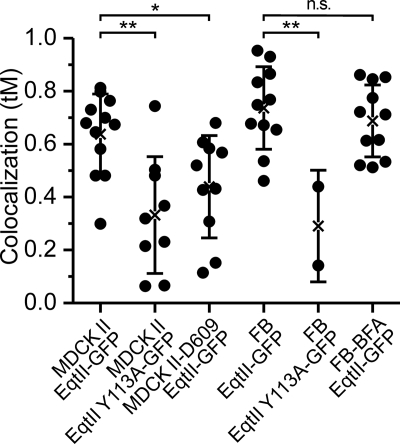
However, EqtII-GFP does bind to specific subcellular structures and the accumulated evidence indicates that these must be membranous. To discover to which compartments EqtII-GFP binds, we used MDCK II cells transfected with plasmid encoding EqtII-GFP and co-stained them with markers specific for either nucleus (propidium iodide) or GA (Bodipy-TR-ceramide). In a separate approach we co-transfected the EqtII-GFP expressing cells with pDsRed2 plasmids encoding mitochondrial or ER targeting sequences. EqtII-GFP did not co-localize with markers for nucleus, ER or mitochondria (Fig. 4), but showed strong colocalization with GA (Fig. 5). The extent of the colocalization was quantified by pixel analysis. The average Manders coefficient (tM) was 0.64 ± 0.15 (Fig. 6). In contrast, the mutant EqtII Y113A-GFP did not show colocalization and consequently tM was low (0.33 ± 0.22) (Fig. 5). The strong colocalization was revealed also for EqtII W112L-GFP, but not for EqtII W112A-GFP (supplemental Fig. S3). We further confirmed this strong colocalization of EqtII-GFP and GA-marker by using NIH 3T3 fibroblasts. Here EqtII-GFP was also co-localized with Bodipy-TR-ceramide (tM = 0.74 ± 0.16), whereas EqtII Y113A-GFP was not (tM = 0.29 ± 0.21) (Fig. 6).
FIGURE 4.
Subcellular localization of EqtII-GFP. MDCK II cells were transfected with plasmid encoding EqtII-GFP and co-stained with propidium iodide (marker for nucleus) or co-transfected with pDsRed2-ER (marker for endoplasmic reticulum), or pDsRed2-Mito (marker for mitochondria). Scale bar is 20 μm.
FIGURE 5.
Colocalization of EqtII-GFP with BODIPY-TR ceramide. MDCK II cells and NIH 3T3 fibroblasts (FB) transfected with EqtII-GFP or EqtII Y113A-GFP and co-stained with Golgi-specific marker, Bodipy-TR-ceramide (BODIPY). MDCK cells expressing the wild-type EqtII were also treated with 25 μg/ml of D609, while fibroblasts expressing the wild-type EqtII were also treated with BFA at final 50 μg/ml. Scale bar is 20 μm. The colocalization panels show in yellow color the sites where the pixel intensities in both channels are above the calculated threshold values. The framed area is also shown enlarged to reveal details. The BODIPY-GFP scatter-graphs represent the relationship between the intensity of fluorescence in both channels at the same pixel locations in the two channel images. The x axis represents the intensity values 0–255 in the red channel, and the y axis represents the intensity values in the green channel, while the color of the dots represents the frequency of pixels with a given combination of intensities in both channels. The straight white lines represent the automatically calculated threshold values by the method of Costes et al. (25) with the “Colocalisation Threshold” plugin for ImageJ.
FIGURE 6.
Distribution of thresholded Mander's coefficients (tM) of colocalization of EqtII-GFP or EqtII Y113A-GFP and Bodipy-TR-ceramide. The MDCK cells were also analyzed in the presence of 25 μg/ml of D609, while the NIH 3T3 fibroblasts (FB) were also analyzed in the presence of 50 μg/ml BFA. 0 means no colocalization, 1 is perfect colocalization. The average (shown by cross) ± S.D. is also presented. The differences between groups were tested using t test. *, p < 0.05; **, p < 0.01; n.s., not significant.
We also checked the intracellular distribution of EqtII-GFP and colocalization with GA in the presence of 25 μg/ml D609 (Figs. 5 and 6). D609 is an inhibitor of SMS, which inhibits transfer of POC from phosphatidylcholine to ceramide (32). The staining clearly differs from that in the absence of D609 (Fig. 5). Although there was still some colocalization with GA probe, we notably observed more dispersed staining of EqtII-GFP throughout the cells and, consequently, tM was significantly lower than in controls (tM = 0.44 ± 0.19) (Fig. 6). This is in good agreement with the reduced, but not abrogated, synthesis of SM at sublethal concentrations of the used SMS inhibitor (32–34).
To check if artificial redistribution of GA could also change redistribution of EqtII-GFP, we treated transfected NIH 3T3 fibroblasts with brefeldin A (BFA). BFA is a fungal macrocyclic lactone, which disrupts the GA and redistributes both resident and itinerant Golgi proteins and lipids into the ER (35, 36). Effect of the BFA is to disperse both the Bodipy-TR-ceramide and EqtII-GFP staining of the GA (Fig. 5). The quantified co-localization remained as strong (tM = 0.69 ± 0.14) as that observed in the absence of BFA (Fig. 6).
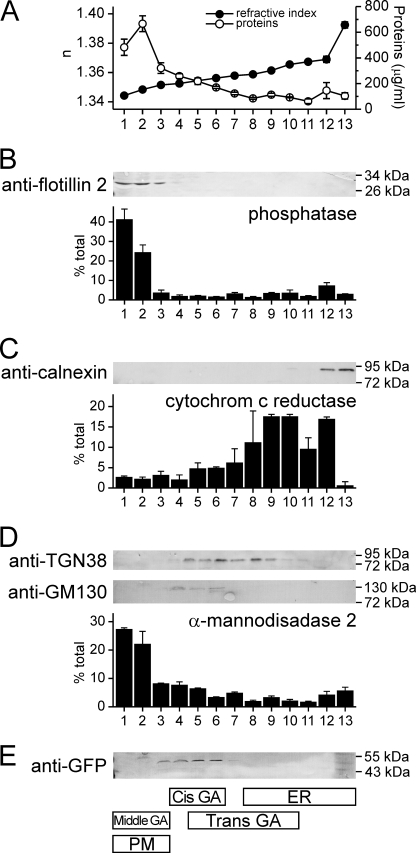
For an independent assay of co-localization of EqtII-GFP with GA, we carried out subcellular fractionation of NIH 3T3 fibroblasts using an Optiprep density gradient (Fig. 7). Thirteen fractions were collected and probed by Western blot or enzyme assay for various markers of plasma membrane, ER and GA. GM130, a cis-GA protein (37), was found in fractions 4–6 and TGN38, a marker for trans-GA (38), in fractions 5–10. The enzyme activity of mannosidase II, a marker for middle GA (26), was found to be mainly in fractions 1 and 2, but also in fractions 3–5. Markers for plasma membrane (flotillin-2 and K+-(p-nitrophenyl phosphatase)) (27) were found in the low density fractions 1–3 while ER markers (cytochrome c reductase and calnexin) were found in the highest density fractions 9–13. EqtII-GFP was found in fractions 3–6, thus clearly co-localized with GA and specifically with GM130.
FIGURE 7.
Subcellular distribution of EqtII-GFP. Cell lysate of NIH 3T3 fibroblasts expressing EqtII-GFP was fractionated by centrifugation on a discontinuous Optiprep gradient and the resulting fractions were immunoblotted and assayed for enzyme activities as indicated. The value in a particular fraction is expressed as percentage to the sum of the enzyme activity in all fractions. Average ± S.D. is shown of two independent fractionations. A, refraction index (solid circles) was measured to check the gradient. Protein content (open circles) was measured by using Bradford reagent. B, markers for plasma membrane (flotillin-2 and K+-(p-nitrophenyl phosphatase)). C, markers for ER (calnexin and cytochrome c reductase). D, markers for GA (TGN38, GM130 and mannosidase II). E, Eqt-GFP. The summary of distribution of various markers presented in B–D is shown below schematically.
DISCUSSION
In the plasma membrane, SM mainly resides in the outer leaflet, especially in cholesterol-rich microdomains termed lipid rafts (1, 39). However, it is mainly synthesized in the lumen of GA (40–43) by the transfer of phosphocholine from PC to ceramide (44). Ceramide, synthesized in ER, is delivered to GA mainly by a ceramide transport protein CERT (43). The SM synthesis is then catalyzed in the lumen of GA by SMS1 (42, 45) and not significantly by SM synthase 2 (SMS2), which resides in the plasma membrane (42). Nevertheless, both enzymes, SMS1 and SMS2, are responsible for controlling SM levels within the cells and on the plasma membrane (34, 46). The distribution of SM in various subcellular compartments has been determined but the amounts and extent of lipid asymmetry across the bilayers is not yet precisely known. SM is absent from ER and mitochondria, but present in the plasma membrane, GA, and endosomes (47).
To improve our understanding of subcellular SM distribution, specific probes are required. In order to avoid the need for intracellular injection of probe, the ability to use transiently expressed GFP fusions is a clear benefit and, therefore, a recombinant probe is preferred. Recently, we have shown that EqtII specifically employs SM as its membrane receptor and have described the molecular mechanism of SM recognition (16). It has a phosphocholine binding site, which might bind PC but recognizes SM in the membrane phase by inserting at the interface deeply enough to detect the difference between sphingomyelin and glycerophosphocholine backbones. This ability to bind to SM in the membrane phase without disrupting the bilayer structure is critical to its use as a reporter of SM membrane distribution. Its utility in discriminating between cellular membranes with different levels of SM was recently shown in a study of HIV-1. EqtII was able to inhibit infectivity of the virus produced in MT-4 cell line in a dose-dependent manner. HIV-1 derived from 293T cells, which possess significantly less SM in the plasma membrane, was much more resistant to EqtII treatment (48). In this report, we explore the possibility of using cytosolic EqtII-GFP as a probe for cellular SM. We first showed that the fusion to GFP does not affect membrane binding properties in vitro; SM enabled EqtII-GFP binding to liposomes and increased its insertion into monolayers (Fig. 1, A–C), thus faithfully reproducing the behavior of the wild-type EqtII (16).
Subcellular distribution of EqtII-GFP transiently expressed in MDCK II cells is perinuclear and punctate (Fig. 2). However EqtII point mutants, in which SM specificity is specifically disrupted, and the non SM-selective EqtII homologue Dr1, display a diffuse distribution, similar to GFP. By using a range of markers for specific organelles we show that EqtII-GFP colocalizes with markers for GA (Figs. 5 and 6) in MDCK II cells and NIH 3T3 fibroblasts. Moreover, control experiments, treatment with SMS inhibitor D609 or with GA-disrupting BFA, revealed that the colocalization with GA is persistent despite SM depletion or GA diffusion, respectively (Figs. 5 and 6). Results obtained by subcellular fractionation (Fig. 7) confirmed these results and indicate that EqtII-GFP mainly colocalizes with the cis-Golgi, in agreement with the literature data that shows that most of the SM is synthesized in the lumen of the cis/medial Golgi (40, 49). However, this sensitive and selective probe provides two important clarifications of trans bilayer SM distribution in the cell. The size and water solubility of the fusion protein means that it is extremely unlikely to cross the lipid bilayer and so any SM-dependent localization must reflect the exposure of SM on the cytosolic leaflet of the membrane bilayer.
It is known that plasma membranes are asymmetric with respect to lipid distributions across the bilayer (39, 50). The outer leaflet is enriched with PC and SM, while the inner leaflet is rich in PS, PE and PI. The presence or absence of SM in the inner leaflet is still an unsolved question. In contrast to some reports (51–53), no SM is detected by intracellularly expressed EqtII-GFP in the inner leaflet of the plasma membrane in either MDCK II cells or NIH 3T3 fibroblasts (Figs. 2 and 7). It binds the plasma membrane of MDCK II cells only when added from outside of the cells (Fig. 1D) and, moreover, it binds only to apical and not to basolateral membranes (Fig. 1E), in agreement with known distribution of SM in polarized epithelial cells (28). There is reported to be less than 5% of SM in the inner leaflet of mammalian plasma membranes (54) and our SPR results (Fig. 3) show that EqtII-GFP binds stably to the inner leaflet lipid mixture when there is only 1 mole % of SM present. It is therefore likely that even if the inner leaflet of the MDCK II or NIH 3T3 plasma membranes would contain a few mol% of SM, then colocalization with membrane markers should be observed. Thus only a lack, or very low amount, of SM in the plasma membrane can explain the lack of association of EqtII-GFP observed by confocal microscopy (Fig. 2) or cell fractionation (Fig. 7).
Thus the localization by microscopy and sub-cellular fractionation confirms that SM is exposed on the cytosolic side of the cis-Golgi. Membrane disruption by EqtII, which may also cause transbilayer redistribution of SM in GA membrane via a toroidal pore (14, 15, 55, 56), is unlikely because EqtII-GFP is probably expressed in too low amounts to allow formation of pores in the membranes of GA. Furthermore, the viability and morphology of GA was not affected in cells that transiently expressed EqtII-GFP as compared with untransfected cells. And even, if EqtII-GFP does alter the transbilayer distribution of SM across the GA bilayer, membranes of GA must contain some amount of SM exposed to the cytosol to allow initial stable EqtII-GFP association as shown in in vitro assays (Figs. 1A and 3A). Notably, EqtII-GFP-induced redistribution of SM was not observed in plasma membrane. This means there must be effective flip-flop across the GA membrane that is catalyzed by specific enzymes (57) and which exposes SM in sufficient amounts for significant EqtII-GFP labeling of the cytosolic face of Golgi.
Such staining pattern is at odds with the luminal synthesis of SM, but in agreement with the notion that a small pool of SM should be made available, via exposure to the cytosol, for ceramide signaling by neutral SMase 2 (nSMase2) (58). SMase-mediated hydrolysis of SM is recognized as the most important pathway of ceramide production induced by stress (4). It was proposed that the pool of SM for ceramide generation is located in the cytosolic leaflet of the plasma membrane or in GA (51–53, 59). Our finding also correlates with cellular distribution of nSMase2, which was found to be localized on the plasma membrane and GA (60–62), with its catalytic side exposed to the cytosol (62).
In summary, we have used a novel SM-specific probe to label intracellular distribution of SM and found it to be exposed in considerable amounts to the cytosolic side of the GA but absent from the inner leaflet of the plasma membrane. Our results indicate that there is a significant pool of SM on the cytosolic face of GA and that EqtII-GFP could be useful marker for studies that may clarify mechanisms of ceramide signaling in cells.
Supplementary Material
Acknowledgments
We would like to acknowledge Nick Morris for help with gradient ultracentrifugation and Helen Ridley for excellent technical help.
This work was supported in part by the Slovenian Research Agency (P1-0207) and a fellowship from the Slovene Human Resources Development and Scholarship Fund (to B. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- SM
- sphingomyelin
- BFA
- brefeldin A
- BSA
- bovine serum albumin
- CHO
- cholesterol
- D609
- tricyclodecan-9-yl xanthogenate
- DAG
- diacylglycerol
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPI
- 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol)
- DPPE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine
- DPPS
- 1,2-dipalmitoyl-sn-glycero-3-phospho-l-serine
- ER
- endoplasmic reticulum
- EqtII
- equinatoxin II
- GFP
- green fluorescent protein
- GA
- Golgi apparatus
- LUV
- large unilamellar vesicles
- MDCK II
- Madin-Darby canine kidney II
- nSMase2
- neutral SMase 2
- PBS
- phosphate-buffered saline
- POC
- phosphocholine
- R-WGA
- rhodamine-conjugated wheat germ agglutinin
- SMS
- SM synthase
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Simons K., Ikonen E. (1997) Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 2.Edidin M. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 257–283 [DOI] [PubMed] [Google Scholar]
- 3.Mañes S., del Real G., Martinez A. C. (2003) Nat. Rev. Immunol. 3, 557–568 [DOI] [PubMed] [Google Scholar]
- 4.Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, (suppl.), S91–S96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wymann M. P., Schneiter R. (2008) Nat. Rev. Mol. Cell Biol. 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 6.Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 7.Ogretmen B., Hannun Y. A. (2004) Nat. Rev. Cancer 4, 604–616 [DOI] [PubMed] [Google Scholar]
- 8.Tafesse F. G., Ternes P., Holthuis J. C. (2006) J. Biol. Chem. 281, 29421–29425 [DOI] [PubMed] [Google Scholar]
- 9.Openshaw A. E., Race P. R., Monzó H. J., Vázquez-Boland J. A., Banfield M. J. (2005) J. Biol. Chem. 280, 35011–35017 [DOI] [PubMed] [Google Scholar]
- 10.Murakami M. T., Fernandes-Pedrosa M. F., Tambourgi D. V., Arni R. K. (2005) J. Biol. Chem. 280, 13658–13664 [DOI] [PubMed] [Google Scholar]
- 11.Shogomori H., Kobayashi T. (2008) Biochim. Biophys. Acta 1780, 612–618 [DOI] [PubMed] [Google Scholar]
- 12.Anderluh G., Maček P. (2002) Toxicon 40, 111–124 [DOI] [PubMed] [Google Scholar]
- 13.Hong Q., Gutierrez-Aguirre I., Barlič A., Malovrh P., Kristan K., Podlesek Z., Maček P., Turk D., Gonzalez-Manas J. M., Lakey J. H., Anderluh G. (2002) J. Biol. Chem. 277, 41916–41924 [DOI] [PubMed] [Google Scholar]
- 14.Kristan K., Podlesek Z., Hojnik V., Gutiérrez-Aguirre I., Gunčar G., Turk D., González-Mañas J. M., Lakey J. H., Maček P., Anderluh G. (2004) J. Biol. Chem. 279, 46509–46517 [DOI] [PubMed] [Google Scholar]
- 15.Malovrh P., Viero G., Serra M. D., Podlesek Z., Lakey J. H., Maček P., Menestrina G., Anderluh G. (2003) J. Biol. Chem. 278, 22678–22685 [DOI] [PubMed] [Google Scholar]
- 16.Bakrač B., Gutiérrez-Aguirre I., Podlesek Z., Sonnen A. F., Gilbert R. J., Maček P., Lakey J. H., Anderluh G. (2008) J. Biol. Chem. 283, 18665–18677 [DOI] [PubMed] [Google Scholar]
- 17.Mancheño J. M., Martín-Benito J., Martínez-Ripoll M., Gavilanes J. G., Hermoso J. A. (2003) Structure 11, 1319–1328 [DOI] [PubMed] [Google Scholar]
- 18.Alegre-Cebollada J., Lacadena V., Oñaderra M., Mancheño J. M., Gavilanes J. G., del Pozo A. M. (2004) FEBS Lett. 575, 14–18 [DOI] [PubMed] [Google Scholar]
- 19.Meinardi E., Florin-Christensen M., Paratcha G., Azcurra J. M., Florin-Christensen J. (1995) Biochem. Biophys. Res. Commun. 216, 348–354 [DOI] [PubMed] [Google Scholar]
- 20.Anderluh G., Pungerčar J., Štrukelj B., Maček P., Gubenšek F. (1996) Biochem. Biophys. Res. Commun. 220, 437–442 [DOI] [PubMed] [Google Scholar]
- 21.Kristan K., Viero G., Maček P., Dalla Serra M., Anderluh G. (2007) FEBS J. 274, 539–550 [DOI] [PubMed] [Google Scholar]
- 22.Anderluh G., Barlič A., Podlesek Z., Maček P., Pungerčar J., Gubenšek F., Zecchini M. L., Serra M. D., Menestrina G. (1999) Eur. J. Biochem. 263, 128–136 [DOI] [PubMed] [Google Scholar]
- 23.Anderluh G., Beseničar M., Kladnik A., Lakey J. H., Maček P. (2005) Anal. Biochem. 344, 43–52 [DOI] [PubMed] [Google Scholar]
- 24.Marchetti B., Ashrafi G. H., Tsirimonaki E., O'Brien P. M., Campo M. S. (2002) Oncogene 21, 7808–7816 [DOI] [PubMed] [Google Scholar]
- 25.Costes S. V., Daelemans D., Cho E. H., Dobbin Z., Pavlakis G., Lockett S. (2004) Biophys. J. 86, 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canty E. G., Kadler K. E. (2005) J. Cell Sci. 118, 1341–1353 [DOI] [PubMed] [Google Scholar]
- 27.Marcheselli V. L., Rossowska M. J., Domingo M. T., Braquet P., Bazan N. G. (1990) J. Biol. Chem. 265, 9140–9145 [PubMed] [Google Scholar]
- 28.van Meer G., Simons K. (1988) J. Cell Biochem. 36, 51–58 [DOI] [PubMed] [Google Scholar]
- 29.Hansen R. E., Roth D., Winther J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutiérrez-Aguirre I., Trontelj P., Maček P., Lakey J. H., Anderluh G. (2006) Biochem. J. 398, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins M. D., Keller S. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luberto C., Hannun Y. A. (1998) J. Biol. Chem. 273, 14550–14559 [DOI] [PubMed] [Google Scholar]
- 33.Meng A., Luberto C., Meier P., Bai A., Yang X., Hannun Y. A., Zhou D. (2004) Exp. Cell Res. 292, 385–392 [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Hailemariam T. K., Zhou H., Li Y., Duckworth D. C., Peake D. A., Zhang Y., Kuo M. S., Cao G., Jiang X. C. (2007) Biochim. Biophys. Acta 1771, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiwara T., Oda K., Yokota S., Takatsuki A., Ikehara Y. (1988) J. Biol. Chem. 263, 18545–18552 [PubMed] [Google Scholar]
- 36.Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. (1989) Cell 56, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luzio J. P., Brake B., Banting G., Howell K. E., Braghetta P., Stanley K. K. (1990) Biochem. J. 270, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Meer G. (2005) EMBO J. 24, 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Futerman A. H., Stieger B., Hubbard A. L., Pagano R. E. (1990) J. Biol. Chem. 265, 8650–8657 [PubMed] [Google Scholar]
- 41.Jeckel D., Karrenbauer A., Burger K. N., van Meer G., Wieland F. (1992) J. Cell Biol. 117, 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. (2004) EMBO J. 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Futerman A. H., Riezman H. (2005) Trends Cell Biol. 15, 312–318 [DOI] [PubMed] [Google Scholar]
- 44.Ullman M. D., Radin N. S. (1974) J. Biol. Chem. 249, 1506–1512 [PubMed] [Google Scholar]
- 45.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. (2004) J. Biol. Chem. 279, 18688–18693 [DOI] [PubMed] [Google Scholar]
- 46.Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. (2007) J. Biol. Chem. 282, 17537–17547 [DOI] [PubMed] [Google Scholar]
- 47.van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lorizate M., Brügger B., Akiyama H., Glass B., Müller B., Anderluh G., Wieland F. T., Kräusslich H. G. (2009) J. Biol. Chem. 284, 22238–22247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeckel D., Karrenbauer A., Birk R., Schmidt R. R., Wieland F. (1990) FEBS Lett. 261, 155–157 [DOI] [PubMed] [Google Scholar]
- 50.Koval M., Pagano R. E. (1991) Biochim. Biophys. Acta 1082, 113–125 [DOI] [PubMed] [Google Scholar]
- 51.Linardic C. M., Hannun Y. A. (1994) J. Biol. Chem. 269, 23530–23537 [PubMed] [Google Scholar]
- 52.Andrieu N., Salvayre R., Levade T. (1996) Eur. J. Biochem. 236, 738–745 [DOI] [PubMed] [Google Scholar]
- 53.Nagata Y., Partridge T. A., Matsuda R., Zammit P. S. (2006) J. Cell Biol. 174, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiessling V., Wan C., Tamm L. K. (2009) Biochim. Biophys. Acta 1788, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderluh G., Dalla Serra M., Viero G., Guella G., Macek P., Menestrina G. (2003) J. Biol. Chem. 278, 45216–45223 [DOI] [PubMed] [Google Scholar]
- 56.Valcarcel C. A., Dalla Serra M., Potrich C., Bernhart I., Tejuca M., Martinez D., Pazos F., Lanio M. E., Menestrina G. (2001) Biophys. J. 80, 2761–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aye I. L., Singh A. T., Keelan J. A. (2009) Chem. Biol. Interact. 180, 327–339 [DOI] [PubMed] [Google Scholar]
- 58.Clarke C. J., Snook C. F., Tani M., Matmati N., Marchesini N., Hannun Y. A. (2006) Biochemistry 45, 11247–11256 [DOI] [PubMed] [Google Scholar]
- 59.Zhang P., Liu B., Jenkins G. M., Hannun Y. A., Obeid L. M. (1997) J. Biol. Chem. 272, 9609–9612 [DOI] [PubMed] [Google Scholar]
- 60.Hofmann K., Tomiuk S., Wolff G., Stoffel W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karakashian A. A., Giltiay N. V., Smith G. M., Nikolova-Karakashian M. N. (2004) FASEB J. 18, 968–970 [DOI] [PubMed] [Google Scholar]
- 62.Tani M., Hannun Y. A. (2007) FEBS Lett. 581, 1323–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.