Abstract
MADD plays an essential role in cancer cell survival. Abrogation of endogenous MADD expression results in significant spontaneous apoptosis and enhanced susceptibility to tumor necrosis factor α-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. However, the regulation of MADD function is largely unknown. Here, we demonstrate that endogenous MADD is phosphorylated at three highly conserved sites by Akt, and only the phosphorylated MADD can directly interact with the TRAIL receptor DR4 thereby preventing Fas-associated death domain recruitment. However, in cells susceptible to TRAIL treatment, TRAIL induces a reduction in MADD phosphorylation levels resulting in MADD dissociation from, and Fas-associated death domain association with DR4, which allows death-inducing signaling complex (DISC) formation leading to apoptosis. Thus, the pro-survival function of MADD is dependent upon its phosphorylation by Akt. Because Akt is active in most cancer cells and phosphorylated MADD confers resistance to TRAIL-induced apoptosis, co-targeting Akt-MADD axis is likely to increase efficacy of TRAIL-based therapies.
Keywords: Akt PKB, Apoptosis, Cancer Therapy, Caspase, Cell Death, Confocal Microscopy, PI 3-Kinase, Tumor Necrosis Factor (TNF)
Introduction
A fine balance between opposing signaling events that regulate survival and death appears to determine the outcome of cell fate. A complex array of genes regulates the above process, and one such gene we previously identified is IG20 (insulinoma-glucagonoma 20) (1). The IG20 can profoundly affect cancer cell survival and death through alternative splicing (2, 3). The IG20 gene encodes six different splice variants (SVs).3 The KIAA and IG20-SV4 isoforms are selectively expressed only in neuronal tissues (4). The KIAA is analogous to rat Rab3a GEP (also referred to as MADD/DENN) and plays an important role in neuronal vesicular trafficking and is required for animal survival (5–7). The IG20pa, MADD, IG20-SV2, and DENN-SV are more ubiquitously expressed. Of these, MADD is physiologically the most important isoform. MADD is expressed at very low levels in a variety of healthy tissues; however, its expression levels are much higher in many types of human tumors and tumor cell lines (1, 8). Knockdown of endogenous MADD or all IG20 SVs results in enhanced spontaneous as well as tumor necrosis factor α-related apoptosis-inducing ligand (TRAIL)-induced apoptosis (9–11). Interestingly, expression of exogenous MADD, and not other SVs, in the absence of endogenous IG20 SVs can rescue the cells from undergoing apoptosis and indicates that only the MADD isoform is required and sufficient to promote cancer cell survival (10, 11).
The extrinsic apoptotic pathway is initiated by death ligands such as Fas ligand, TRAIL, or tumor necrosis factor α (12–15). Unlike Fas ligand, TRAIL can induce cancer cell death with little or no effect on most normal cells (16). However, a number of different factors can confer resistance to TRAIL-induced apoptosis in different cells (17). TRAIL binding to death receptors 4 and 5 (DR4/DR5) induces receptor trimerization and recruitment of FADD (16, 18–20). This facilitates recruitment of procaspase-8 and DISC formation, caspase-8 and -3 activation, and cell death. MADD can bind to the cytoplasmic tail of DR4 and DR5, thereby inhibiting DISC formation and/or activation of caspase-8 (1, 10, 21). Although these earlier studies revealed a critical role for MADD in cancer cell survival, they failed to provide insight into the regulation of endogenous MADD function.
Important cellular functions, including apoptosis, are regulated through post-translational modification of key molecules. Protein kinase B (Akt) plays a key role in promoting cell survival by regulating the function of a variety of apoptosis-related proteins by phosphorylating the consensus sequence RXRXX(S/T) (22, 23). Akt phosphorylates mouse double minute 2 (mdm2) and enhances its ability to degrade p53 (23, 24). It targets caspase-9, Bad, IκB kinase α, Forkhead transcription factor, and Yap and plays a critical role in TSC1/2 and Rheb/mTOR signaling pathway (25, 26). We analyzed the sequences of human, mouse, and rat MADD proteins and found that they all contained three Akt consensus sites. Accordingly, we investigated whether MADD is an Akt substrate and its function is regulated by Akt-mediated phosphorylation.
In the present study we demonstrate that MADD is an endogenous substrate for Akt and only the phosphorylated MADD (pMADD) can interact with DR4 and confers resistance to apoptosis. In susceptible cells TRAIL treatment causes a reduction in the levels of pMADD. This results in MADD dissociation from, and FADD association with, DR4 leading to DISC formation and cell death.
EXPERIMENTAL PROCEDURES
Cell Culture and Viability Assay
HEK293 and HeLa cells were cultured as described elsewhere (10). Cell death was determined by trypan blue exclusion and the numbers of trypan blue-positive and -negative cells were counted using a hemocytometer. For immunoprecipitation of endogenous proteins and overexpressed proteins, we used 4 × 106 and 2 × 106 cells, respectively. For immunoblotting of endogenous and overexpressed proteins, we used 3 × 106 and 2 × 106 cells, respectively.
Construction of MADD Mutants, Small Interfering RNAs, Virus Infection, and Transfection of Cells
The serine or threonine residue at the consensus Akt phosphorylation sites of MADD were mutated to an alanine residue using the QuikChange II XL site-directed mutagenesis kit (catalogue #200521, Stratagene) according to the manufacturer's instructions. This kit was also used to produce MADD mutant devoid of the death domain (i.e. deletion of amino acids 1278–1588). All constructs were sequenced to ensure that only the desired mutations had been introduced. Construction of shRNAs expressing lentiviruses and MADD knockdown are described elsewhere (10, 27). The transfection was performed using an Effectene Transfection kit (Qiagen catalogue #301425).
Phosphorylation of MADD in Vitro
A kinase assay was performed as described previously (28). In brief, 2 μg of purified glutathione S-transferase (GST)-tagged wtMADD or MADD mutants were incubated for 1 h at 30 °C with 0.2 μg of rAkt and 5 μCi of γATP in a kinase buffer. The reaction products were separated by SDS-PAGE, and 32P-labeled proteins were visualized by autoradiography. Loaded GST-MADD was visualized by probing the same membrane with an anti-MADD antibody. To prepare pMADD for in vitro protein binding assays, recombinant wtMADD or MADD mutants were treated with recombinant Akt as described above except that cold ATP was added.
Metabolic Labeling
HEK293 cells were washed twice with phosphate-free Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal bovine serum and subsequently incubated with 0.3 mCi/ml [32P]orthophosphate in the same medium for 6 h. MADD was immunoprecipitated with an anti-MADD antibody. The precipitated MADD was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to autoradiography. Loaded MADD was visualized by immunoblotting the same membrane using an anti-MADD antibody.
Generation of the Anti-phospho-Ser-70, -Thr-173, and –Thr-1041 Antibodies
The anti-phospho-MADD antibodies and the anti-MADD antibody were produced by Eurogentec. The phosphopeptides CRQRRMpSLRDDTS (S-70), GSRSRNSpTLTSL (T-173), and KRKRSPpTESVNTP (T-1041) were used to immunize the rabbits. Antisera were depleted of antibodies that recognized non-phosphorylated MADD.
Membrane Preparations
Membrane and cytosolic fractions were prepared as previously described (29). Cells were washed in an ice-cold buffer (160 mm NaCl, 38 mm HEPES, pH 7.4, 1 mm MgCl2, 1 mm EGTA) containing a protease inhibitor mixture (Roche Applied Science). Cells were sonicated on ice and subjected to centrifugation at 400 × g for 10 min at 4 °C to remove unbroken cells, debris, and nuclei. Lysates were separated by ultracentrifugation at 117,000 × g for 60 min at 4 °C into cytosol and membrane fractions. Membrane pellets were re-suspended in 2x Laemmli sample buffer and boiled for 5 min. All samples were frozen at −80 °C. Equal loading for membrane and cytosolic fractions were monitored by re-probing the membrane with anti-caveolin-1 antibody or anti-β-actin antibody, respectively.
Immunoprecipitation and in Vitro Protein Binding Assay
The samples were precleared with 10% (v/v) Protein A-agarose (Roche Applied Science) for 1 h on a rocking platform (10). The anti-DR4 (catalogue #06-744, Millipore) antibody or the anti-DR5 (catalogue #sc-7191, Santa Cruz Biotechnology) was added and incubated overnight at 4 °C. Immunoprecipitates were captured by incubating with 10% (v/v) Protein A/G-agarose for 1 h. The agarose beads were spun down and washed 3 times with NET/Nonidet P-40 buffer (150 mm NaCl, 2 mm EDTA, 50 mm Tris-HCl, pH 7.5, 0.1% Nonidet P-40). Samples were resolved by SDS-PAGE, and immunoblots were prepared and analyzed as described above.
In vitro protein binding assay was performed as described elsewhere (30, 31). In brief, recombinant proteins were incubated in 50 μl of binding buffer (142 mm KCl, 5 mm MgCl2, 10 mm HEPES (pH 7.4), 0.5 mm dithiothreitol, 1 mm EGTA, 0.1% Nonidet P-40, and a protease inhibitor mixture) at 4 °C for 2 h and immunoprecipitated as described above.
Immunoblot Analysis
Cells were lysed for 1 h at 4 °C in a lysis buffer (20 mm Tris pH 7.5, 2 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, 250 mm sucrose, 0.1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100) containing a protease inhibitor mixture (Sigma). Equal protein loading was controlled by Ponceau Red staining of membranes. For immunoblotting, anti-YFP (catalogue #632380, Clontech), anti-caspase-8 (catalogue #9746, clone 1C12, Cell Signaling), anti-caveolin-1 (N20) (catalogue #sc-894, Santa Cruz Biotechnology), anti-pAkt (catalogue #9271, Cell Signaling), anti-Akt (catalogue #9272, Cell signaling), anti-β actin (Abcam), anti-Foxo3a and anti-phospho-Foxo3a (T32, Santa Cruz Biotechnology), anti-GST (GE Healthcare), anti-tubulin (catalogue #sc-8035, Santa Cruz Biotechnology), and anti-FADD (catalogue #06-711, Millipore) antibodies were used as primary antibodies followed by the addition of horseradish peroxidase-conjugated secondary antibodies. To avoid the hindrance by immunoprecipitating immunoglobulin heavy and light chains, the TrueBlot (eBioscience) was employed when necessary. The protein bands were visualized by enhanced chemiluminescence. When appropriate, the intensities of the protein bands were determined using densitometry, the ratios were calculated, and the ratios of controls were assigned a value of 1.
DNA Fragmentation and Caspase Activity Assays
Histone-associated DNA fragments were analyzed using a Cell Death Detection ELISA Kit (Roche Applied Science catalogue #11544675001). Caspase-3 and caspase-8 activities were quantitatively analyzed using the commercial assay kits (R&D Systems; for caspase-3 catalogue, #BF3100; for caspase-8, catalogue #BF4100).
Co-localization of DR4 and MADD
Immunofluorescence microscopy was carried out as described earlier (21). Briefly, HeLa cells were co-transfected with the plasmid constructs of wtMADD-YFP or MADD3A-YFP along with DR4-CFP using Lipofectamine 2000 (Invitrogen). Cells were imaged using a laser scanning confocal microscope (Zeiss LSM 510 META). The excitation/emission for CFP and YFP detection was at 458/475 nm and 488/509 nm, respectively. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole and imaged with a two-photon excitation at 790 nm.
Statistical Analysis
All results are expressed as the means ± S.E. of at least three independent experiments. Comparisons between means were evaluated by Student's t test. A one-way analysis of variance was used for multiple comparisons. A value of p < 0.05 was considered significant.
RESULTS
MADD Can Be Phosphorylated by Akt in Vitro and Metabolically Labeled in Vivo
Cancer cells express higher levels of MADD, which confers resistance against apoptosis, and yet are often susceptible to TRAIL-induced apoptosis. Therefore, we tested whether TRAIL can alter the levels of MADD expression. TRAIL treatment induced caspase-3 activation and cell death (Fig. 1A) without significantly altering the levels of MADD expression up to 4 h after treatment (Fig. 1B). However, MADD knockdown alone was sufficient to induce cell death, which was further enhanced upon treatment with a suboptimal dose of TRAIL (5 ng/ml) that was insufficient to cause apoptosis in cells expressing MADD (Fig. 1C). These data suggested that MADD contributes to cell survival and TRAIL resistance, and TRAIL can partially overcome this resistance without reducing the levels of MADD expression.
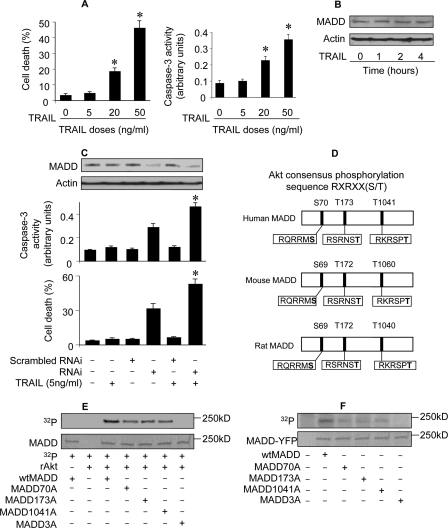
FIGURE 1.
MADD can be phosphorylated by Akt in vitro and metabolically labeled in vivo. A, TRAIL-induces dose-dependent cell death. HeLa cells were treated with indicated doses of TRAIL, and 12 h later cell death was analyzed. Caspase-3 activity was analyzed 6 h after treatment. *, p < 0.05 versus control. Data are expressed as the mean ± S.E. of three independent experiments. B, TRAIL treatment does not alter MADD protein expression. MADD levels were determined by immunoblotting upon TRAIL treatment. A representative result from three independent experiments is shown. C, knockdown of endogenous MADD enhances sensitivity to TRAIL-induced cell death. HeLa cells were infected with the lentivirus harboring MADD shRNAi (RNAi) or its scrambled form (Scrambled RNAi), and 48 h later the cells were treated with 5 ng/ml TRAIL. Cells were harvested 12 h after TRAIL treatment and analyzed for MADD expression (upper panel immunoblot) and cell death (lower panel). Caspase-3 activity was analyzed 6 h after treatment (middle panel). Data are expressed as the mean ± S.E. of three independent experiments. D, Akt consensus phosphorylation sequences on MADD are shown. The three potential phosphorylation sites of Akt in MADD are conserved in human, mouse, and rat. E, in vitro phosphorylation of recombinant MADD by Akt is shown. GST fused-wtMADD or GST fused-MADD mutants were expressed in bacteria. They were then purified and incubated with recombinant Akt (rAkt) and γ[32P]ATP for 1 h at 30 °C. The products were subjected to SDS-PAGE, and 32P-labeled proteins were visualized by autoradiography. F, MADD is labeled by [32P]orthophosphate in vivo. HEK293 cells were transfected with wtMADD or MADD mutant constructs, and 24 h later the transfected cells were incubated with 0.3 mCi/ml [32P]orthophosphate for 6 h. To obtain relative pure MADD, YFP-tagged MADD was immunoprecipitated with an anti-YFP antibody and then separated by SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to autoradiography. MADD-YFP was visualized by staining the same membrane with an anti-YFP antibody. The molecular weight marker is indicated.
Because protein function is often regulated by its phosphorylation state, we analyzed the MADD sequence and found three conserved Akt phosphorylation sites (Fig. 1D). To test whether MADD is an Akt substrate, we treated bacterial recombinant GST-MADD with recombinant Akt in vitro and found that the MADD (wtMADD) was phosphorylated (Fig. 1E). Next, we generated and tested MADD mutants MADD70A (S70A), MADD173A (T173A), MADD1041A (T1041A), and MADD3A (triple mutant with S70A,T173A,T1041A). Phosphorylation was considerably reduced in MADD single mutants and absent in the triple mutant (Fig. 1E). We subjected MADD protein to metabolic labeling and found that WtMADD, but not MADD3A, could be labeled with [32P]orthophosphate (Fig. 1F). HEK293 cells express a lower level of MADD in comparison with HeLa cells as revealed by immunoblotting (data not shown). Therefore, to minimize the influence of endogenous MADD, HEK293 cells were used in this set of experiments. A reduction in the levels of [32P]orthophosphate labeling was observed in each of the single mutants and absent in the triple mutant. These results suggested that all three sites identified in MADD can be phosphorylated by Akt in vitro and metabolically labeled in vivo.
MADD Can Be Phosphorylated by Akt in Vivo
To more definitively demonstrate the phosphorylation of each of the three sites by Akt, we raised three different antibodies that reacted specifically with either phosphorylated Ser-70, Thr-173, or Thr-1041 (anti-phospho-Ser-70, -Thr-173, and Thr-1041) MADD peptides and used them for in vivo studies. Although wtMADD was recognized by each of the three antibodies, MADD70A, MADD173A and MADD1041A were not recognized by their corresponding anti-phospho-specific antibodies (Fig. 2A). These results further demonstrated that all three sites are phosphorylated.
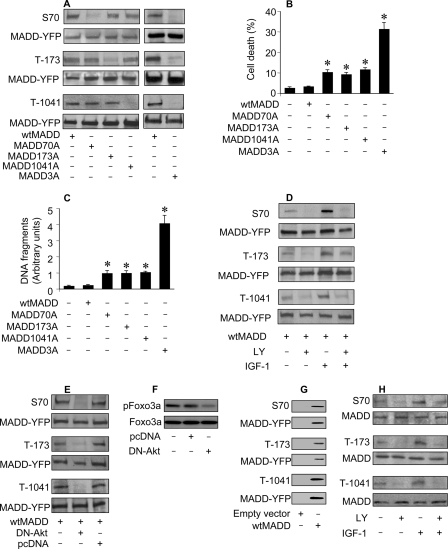
FIGURE 2.
In vivo phosphorylation of MADD. A, detection of MADD phosphorylation using specific anti-phospho antibodies is shown. HEK293 cells were transfected with various cDNA constructs, and 36 h later the cells were harvested for immunoblotting using the anti-phospho-Ser-70, anti-phospho-Thr-173, or anti-phospho-Thr-1041 antibodies or an anti-YFP antibody to detect exogenous MADD. B, shown is an analysis of cell fate. HEK293 cells were transfected with various MADD constructs. Cell death was determined 48 h after transfection by trypan blue exclusion. *, p < 0.05 versus control. C, measurement of apoptosis using Cell death detection enzyme-linked immunosorbent assay is shown. Cells were treated as described for B. *, p < 0.05 versus control. D, induction, or inhibition of MADD phosphorylation is shown. HEK293 cells were transfected with wtMADD, and 24 h later they were cultured in serum-free medium for 20 h. Subsequently, they were treated with LY294002 (LY, 10 μm) for 1 h and/or IGF-1 (150 ng/ml) for 20 min. The cell lysates were immunoblotted and probed with anti-phospho-specific antibodies or an anti-YFP antibody. E, DN-Akt inhibits MADD phosphorylation. HEK293 cells were co-transfected with wtMADD along with dominant negative Akt (DN-Akt) or an empty vector pcDNA3.1 (pcDNA). Immunoblots were probed with indicated antibodies. F, DN-Akt could reduce Foxo3a phosphorylation levels. HeLa cells were transfected with DN-Akt or an empty vector pcDNA3.1 (pcDNA). Immunoblots were probed with the anti-Foxo3a antibody or the anti-phospho Foxo3a (T32) antibody. G, exogenous MADD is expressed and phosphorylated. HEK293 cells were transfected with either a empty vector is a vector containing YFP-fused MADD. Expression and phosphorylation of exogenous MADD were analyzed by immunoblotting. H, phosphorylation of endogenous MADD is shown. HeLa cells were serum-starved for 20 h and treated with LY (10 μm) for 1 h and/or IGF-1 (150 ng/ml) for 20 min. Immunoblot was probed with the indicated antibodies.
To test whether the function of MADD is related to its phosphorylation status, we detected the effect of phosphorylate-able wtMADD and non-phosphorylate-able MADD3A on apoptosis. Expression of wtMADD alone did not induce apoptosis. Surprisingly, although expression of MADD70A, MADD173A, or MADD1041A modestly enhanced apoptosis, expression of MADD3A substantially enhanced apoptosis (Fig. 2B). Apoptotic cell death was confirmed by estimating histone-associated DNA fragments (Fig. 2C). These data suggested that the cell protective function of MADD is dependent upon its ability to undergo phosphorylation.
Akt activation is dependent upon phosphatidylinositol 3-kinase (PI3K) activity and together they form a signaling axis (PI3K-Akt axis) (26). To understand whether MADD phosphorylation in vivo is controlled by PI3K-Akt axis, we stimulated or repressed the PI3K activity. Although the Ser-70, Thr-173, and Thr-1041 phosphorylation levels in MADD were increased upon PI3K activation with IGF-1, they were decreased in the presence of the PI3K inhibitor LY294002 (Fig. 2D). These data suggested that PI3K is involved in regulating MADD phosphorylation. To directly demonstrate the specific requirement of Akt, we determined pMADD levels in cells expressing a dominant negative form of Akt (DN-Akt). This DN-Akt is deficient in the kinase activity due to a point mutation in the ATP binding domain K179M but can bind to the substrate (32). Our results showed that pMADD levels were dramatically decreased (Fig. 2E). To determine the efficiency and specificity of DN-Akt, we used Foxo3a, a well characterized substrate of Akt (33), as a positive control. DN-Akt could reduce the phosphorylation levels of Foxo3a (Fig. 2F). As a control for the specificity of exogenous MADD expression and phosphorylation, we transfected cells with an empty vector, and a protein band comparable with that noted in cells transfected with wtMADD could not be observed (Fig. 2G). These data suggested that MADD is likely an Akt substrate.
Next, we confirmed endogenous MADD phosphorylation by Akt using the specific anti-phospho-peptide antibodies. Our results showed that endogenous MADD was phosphorylated at all three sites, which increased upon IGF-1 treatment. In contrast, phosphorylation levels were decreased considerably in the presence of LY294002 (Fig. 2H). Taken together, these data demonstrated that endogenous MADD is an Akt substrate.
TRAIL Initiates Cell Death by Reducing MADD Phosphorylation Levels
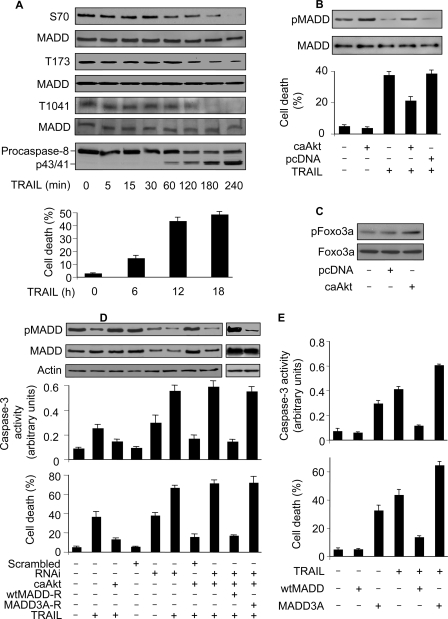
Because MADD function is dependent upon its phosphorylation status, we wondered whether TRAIL treatment can influence MADD phosphorylation. In cells treated with TRAIL, we found a considerable reduction in the levels of pMADD with a concomitant activation of caspase-8 that positively correlated with cell death (Fig. 3A). We employed the constitutively activated form of Akt (caAkt), which is composed of the entire coding sequence of Akt fused in-frame to the myristoylation signal, which facilitates translocation of Akt to the plasma membrane. This fusion protein is constitutively active, and its activity is independent of growth factor signaling (32). Expression of caAkt attenuated decrease in pMADD (upper panel, Fig. 3B) as well as cell death (lower panel, Fig. 3B) seen upon TRAIL treatment. To determine the efficiency of caAkt, we used Foxo3a, which is a very well characterized Akt substrate, as a positive control (33). The caAkt could augment the phosphorylation levels of Foxo3a (Fig. 3C). Because Akt can phosphorylate a variety of proteins (22, 33), we set out to determine whether the protective effect of caAkt on TRAIL-induced apoptosis is related to the levels of pMADD. To this end, we generated and used shRNA resistant wtMADD (wtMADD-R) and MADD3A (MADD3A-R). The caAkt was unable to protect cells from death upon MADD knockdown. However, expression of wtMADD-R, but not the MADD3A-R, restored the protective effect of caAkt against TRAIL-induced apoptosis in cells devoid of endogenous MADD (Fig. 3D). To further understand whether TRAIL-induced cell death is related to MADD phosphorylation, we tested whether the Akt phosphorylate-able wild type MADD (wtMADD) or the Akt non-phosphorylate-able MADD3A can influence TRAIL-induced cell death. WtMADD, and not the MADD3A, was able to inhibit TRAIL-induced caspase-3 activation and cell death (Fig. 3E). Taken together, these data suggested that MADD-mediated regulation of TRAIL-induced apoptosis is dependent upon its phosphorylation status.
FIGURE 3.
TRAIL initiates cell death by reducing MADD phosphorylation levels. A, TRAIL treatment reduces the levels of pMADD. HeLa cells were treated with 50 ng/ml TRAIL. MADD phosphorylation was analyzed by immunoblotting using the anti-phospho antibodies. Total MADD was detected using an anti-MADD antibody. Caspase-8 activation was analyzed by immunoblotting using an anti-caspase-8 antibody. Cell death was analyzed by trypan blue exclusion at the indicated time. B, caAkt attenuates TRAIL-induced decrease in MADD phosphorylation. HeLa cells were transfected with caAkt or the empty vector pcDNA3.1 (pcDNA), and 36 h later the cells were treated with 50 ng/ml TRAIL. Two hours later cells were harvested and analyzed for MADD phosphorylation. Cell death was analyzed 12 h after the TRAIL treatment. C, caAkt could augment Foxo3a phosphorylation levels. HeLa cells were transfected with caAkt or an empty vector pcDNA3.1 (pcDNA). Immunoblots were probed with the anti-Foxo3a antibody or the anti-phospho Foxo3a (T32) antibody. D, MADD knockdown attenuates the protective effect of caAkt. HeLa cells were subjected to indicated treatment. Cells were harvested 2 h after TRAIL treatment (50 ng/ml) and analyzed for MADD phosphorylation (upper panel). In a parallel experiment cells were harvested 12 h after TRAIL treatment and analyzed for cell death (lower panel). E, wtMADD but not MADD3A can inhibit apoptosis induced by TRAIL. HeLa cells were transfected with the constructs of wtMADD or MADD3A and then treated with TRAIL. Caspase-3 activity (upper panel) and cell death (lower panel) were analyzed. Data are expressed as the mean ± S.E. of three independent experiments.
Akt-phosphorylated MADD Associates with DR4
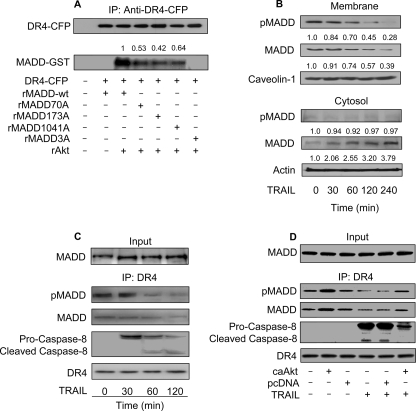
Earlier we showed that MADD can prevent caspase-8 activation and the ensuing cell death (10), perhaps due to its interaction with DR4 (17). To test whether phosphorylation of MADD affected its cellular localization, we carried out cell fractionation studies and found segregation of wtMADD and MADD3A mutant predominantly with the membrane and the cytosol, respectively (Fig. 4A). Immunofluorescence microscopy showed co-localization of wtMADD with DR4 predominantly in the membrane. In contrast, the pattern of distribution of MADD3A was distinct from that of DR4, and the protein was distributed more diffusely throughout the cytoplasm (Fig. 4B). Additionally, although wtMADD readily co-precipitated with DR4, the interaction of MADD single mutants with DR4 was decreased, whereas that of MADD3A was almost non-existent (Fig. 4C), suggesting that Akt phosphorylation of MADD is required for its interaction with DR4.
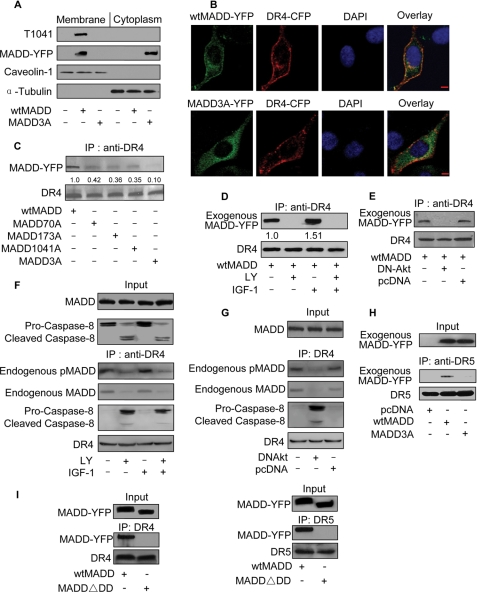
FIGURE 4.
Phosphorylation of MADD by Akt influences its binding to DR4. A, cellular distribution of wtMADD and MADD3A is shown. HEK293 cells were transfected with wtMADD or MADD3A. 36 h after transfection the cells were harvested, and membrane and cytosolic fractions were prepared and subjected to immunoblotting using an anti-YFP antibody. Tubulin and caveolin-1 served as loading controls. B, cellular distribution of wtMADD and MADD3A was detected by immunofluorescence staining. HeLa cells were co-transfected with wtMADD-YFP or MADD3A-YFP plasmid along with DR4-CFP plasmid. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). For the overlay, DR4-CFP is shown in red. Bar = 10 μm. The amount of overlap between DR4-CFP and MADD-YFP was determined using the computer program available in the laser scanning confocal microscope (Zeiss LSM 510 META), and the correlation indicating the degree of overlap in wtMADD was 0.7, the correlation in MADD3A is 0.26. C, phosphorylation status of MADD influences its binding to DR4. HEK293 cells were transfected with wtMADD or MADD mutants, and 36 h after transfection the cell lysates were immunoprecipitated (IP) using an anti-DR4 antibody followed by immunoblotting using an anti-YFP antibody to detect exogenous MADD. The membrane was re-probed with an anti-DR4 antibody to show the protein loading. D, PI3K can influence exogenous MADD association with DR4. HEK293 cells were transfected with wtMADD, and 24 h after transfection the cells were serum-starved for 20 h and treated with LY294002 (LY, 10 μm) for 1 h and/or IGF-1 (150 ng/ml) for 20 min. Immunoprecipitation using an anti-DR4 antibody was followed by immunoblotting. The numbers show the ratio of MADD to DR4. E, Akt can influence exogenous MADD association with DR4. HEK293 cells were co-transfected with wtMADD along with DN-Akt or the empty vector pcDNA3.1 (pcDNA), and 36 h later the cells were harvested for immunoprecipitation using an anti-DR4 antibody followed by immunoblotting. F, PI3K can influence the association of endogenous MADD with DR4. HeLa cells were serum-starved for 20 h, then treated with LY294002 (10 μm) for 1 h and/or IGF-1 (150 ng/ml) for 20 min. Immunoprecipitation using an anti-DR4 antibody was followed by immunoblotting. G, Akt can influence endogenous MADD association with DR4. HeLa cells were transfected with DN-Akt or pcDNA, and 36 h later the cells were collected for immunoprecipitation using an anti-DR4 antibody followed by immunoblotting. H, wtMADD but not MADD3A is able to bind to DR5. HeLa cells were transfected with wtMADD, MADD3A, or the empty vector pcDNA3.1 (pcDNA), and 36 h after transfection the cell lysates were immunoprecipitated using an anti-DR5 antibody followed by immunoblotting using an anti-YFP antibody to detect MADD and an anti-DR5 antibody to detect DR5. I, the death domain is required for MADD interaction with DR4 or DR5. HEK293 cells were transfected with wtMADD or a MADD mutant (MADD▵DD) in which the death domain is deleted. Immunoprecipitation using an anti-YFP antibody was followed by immunoblotting. Immunoprecipitation using an anti-DR4 antibody (left panel) or DR5 antibody (right panel) was followed by immunoblotting.
Subsequently, we determined whether the modulation of PI3K-Akt axis could influence the ability of wtMADD to interact with DR4. Treating cells with IGF-1 resulted in enhanced association of wtMADD with DR4, whereas treatment with a PI3K inhibitor, LY294002, resulted in a loss of interaction between the two (Fig. 4D). Similarly, expression of the DN-Akt also resulted in the loss of wtMADD binding to DR4 (Fig. 4E), indicating the specific and critical requirement of Akt. To understand whether the association of pMADD with DR4 plays a role in regulating the DR4-mediated signaling, we analyzed the components of DISC. Caspase-8 could be observed in the absence, but not in the presence, of MADD association with DR4, suggesting that MADD interaction with DR4 could inhibit DR4-DISC formation (Fig. 4F). In addition, although the endogenous pMADD-DR4 complex was drastically reduced, the procaspase-8 recruitment (DISC formation) was increased upon expression of DN-Akt (Fig. 4G). We tested to see whether MADD can bind to DR5 and found that that wtMADD, but not MADD3A, could associate with DR5 (Fig. 4H). Because MADD contains a death domain (DD) at its C terminus, we analyzed to see whether the DD was required for MADD interaction with the DR4 and the DR5. We generated and used a MADD mutant (MADD▵DD) in which the DD domain was deleted (i.e. deletion of amino acids 1278–1588). MADD without DD lost its ability to bind to DR4 and DR5, suggesting that MADD interacts with DR4 and DR5 through its DD (Fig. 4I). These results suggest that the Akt-phosphorylated MADD is tethered to DR4, and this association can be increased or decreased by either activating or suppressing PI3K/Akt signaling, respectively.
To determine whether phosphorylation is essential for direct binding of MADD to DR4 we carried out in vitro studies. Although the non-phosphorylated MADD failed to associate with DR4, the recombinant Akt-phosphorylated wtMADD, but not the MADD3A mutant, interacted with DR4 (Fig. 5A). In addition, MADD single mutants interacted much less efficiently with the DR4 as compared with recombinant wtMADD. Thus, it appears that phosphorylation of MADD is essential for its direct biding to DR4.
FIGURE 5.
MADD phosphorylation is required for its binding to DR4. A, the recombinant pMADD binds to DR4. The recombinant wt-MADD or MADD mutants were phosphorylated in vitro by Akt (recombinant Akt (rAkt)) and purified using glutathione-Sepharose columns (Roche Applied Science). The DR4-CFP was expressed in HEK 293 cells and immunoprecipitated (IP) using an anti-CFP antibody and protein A-agarose. To this, purified MADD preparations were added for 2 h at 4 °C, washed, and centrifuged. Pellets were analyzed by immunoblotting using the indicated antibodies. The numbers show the ratio of MADD-GST to DR4-CFP. B, TRAIL induces MADD relocation. HeLa cells were treated with TRAIL (50 ng/ml). The localizations of phosphorylated and nonphosphorylated MADD in the membrane and cytosolic fractions were analyzed by immunoblotting using the anti-MADD antibody or the anti-phospho-Thr-1041 antibody. The numbers in the upper panel show the ratio of pMADD and MADD to caveolin-1 and in the lower panel the ratio of pMADD and MADD to the actin. C, TRAIL induces a decrease in endogenous MADD association with DR4. HeLa cells were treated with 50 ng/ml TRAIL. Cells were harvested at indicated times after treatment, immunoprecipitated using an anti-DR4 antibody, and subjected to immunoblotting with the indicated antibodies. D, caAkt can alter MADD association with DR4. HeLa cells were transfected with caAkt or the empty vector pcDNA3.1 (pcDNA), and 36 h later the cells were treated with 50 ng/ml TRAIL. Cells were harvested 2 h after treatment and analyzed for MADD association with DR4 as described under C. A representative result of three independent experiments is shown.
Because cancer cells express higher levels of pMADD and yet are often susceptible to TRAIL-induced apoptosis, we asked whether TRAIL affects MADD phosphorylation with a consequent effect on its ability to interact with DR4. TRAIL treatment reduced pMADD levels in the membrane while elevating non-phosphorylated MADD levels in the cytoplasm (Fig. 5B). Furthermore, we observed a progressive decrease in the levels of MADD that co-precipitated with DR4 upon TRAIL treatment (Fig. 5C). Interestingly, expression of caAkt alone was able to increase pMADD association with DR4. Although TRAIL treatment reduced the level of pMADD association with DR4, this reduction was attenuated upon caAkt expression (Fig. 5D). These results combined with those shown in Fig. 4 demonstrated that pMADD interacts with DR4, and the dissociation of MADD from DR4 upon TRAIL treatment is likely a consequence of reduced levels of pMADD.
MADD Phosphorylation Status Regulates FADD Binding to DR4
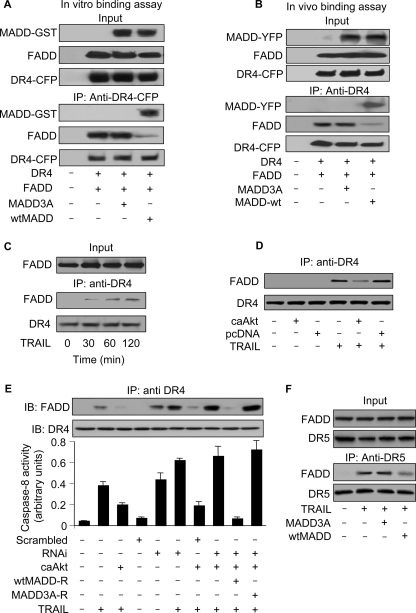
Ligation of DR4 with TRAIL is known to trigger the DISC formation that is characterized by the recruitment of both FADD and procaspase-8 (16). It is, therefore, possible that pMADD, which preferentially binds to DR4, can inhibit FADD recruitment. To test this hypothesis we carried out in vitro binding assays using recombinant FADD and DR4 in the presence of recombinant Akt-phosphorylate-able MADD or Akt non-phosphorylate-able MADD3A. As shown in Fig. 6A, FADD binds to DR4 only in the presence of MADD3A but not in the presence of wtMADD. This observation was confirmed when we observed that wtMADD, but not MADD3A, was able to suppress the association of DR4 with FADD in vivo (Fig. 6B). The time-dependent recruitment of FADD to DR4 in TRAIL-treated cells (Fig. 6C) was dramatically reduced when caAkt was expressed (Fig. 6D), suggesting that the pMADD prevented DR4-FADD interaction. To test whether the protective effect of caAkt is mediated through MADD, we performed a rescue experiment and found that caAkt reduced both TRAIL-induced association of DR4 with FADD (upper panel, Fig. 6E) and caspase-8 activation (lower panel, Fig. 6E) in cells expressing RNAi-resistant wtMADD-R and not the RNAi-resistant MADD3A-R in the absence of endogenous MADD expression. Finally, we tested whether wtMADD and MADD3A can influence DR5 and FADD association. wtMADD but not MADD3A was able to significantly inhibit the association of DR5 with FADD (Fig. 6F). Taken together, these results demonstrated that pMADD binds to DR4/DR5 and prevents FADD recruitment. However, upon TRAIL treatment, MADD phosphorylation levels were reduced, thereby resulting in its loss of DR4 binding, which allowed FADD recruitment and DISC formation leading to apoptosis.
FIGURE 6.
The association of DR4 and FADD depends on MADD phosphorylation status. A, in vitro binding to DR4 is shown. FADD was incubated with DR4 in the presence of Akt-phosphorylate-able recombinant wtMADD or Akt non-phosphorylate-able MADD3A. Immunoprecipitation with anti-CFP antibody was followed by immunoblotting with anti-GST or anti-FADD antibody. B, in vivo binding to DR4 is shown. HEK293 cells were transfected with the DR4-CFP, FADD, wtMADD-YFP, or MADD3A-YFP constructs, and 24 h later the cells were harvested and analyzed. Immunoprecipitation (IP) with anti-DR4 antibody was followed by immunoblotting with anti-FADD antibody or anti-CFP antibody. C, TRAIL treatment results in DR4 association with FADD. HeLa cells were treated with TRAIL (50 ng/ml). Immunoprecipitation with anti-DR4 antibody was followed by immunoblotting with anti-FADD antibody. D, caAkt attenuates FADD association with DR4. HeLa cells were transfected with caAkt or an empty vector pcDNA3.1 (pcDNA), and 36 h later the cells were treated with 50 ng/ml TRAIL. Two hours later the cells were harvested, and the cell lysates were immunoprecipitated using an anti-DR4 antibody followed by immunoblotting with anti-FADD antibody. E, MADD knockdown attenuates the inhibitory effect of caAkt on DISC formation and caspase-8 activation. HeLa cells were transfected with the indicated constructs and treated with 50 ng/ml TRAIL for 2 h. Cell lysates were immunoprecipitated with an anti-DR4 antibody followed by immunoblotting (IB) with anti-FADD antibody (upper panel). Caspase-8 activity is shown in the lower panel. Data are expressed as the mean ± S.E. of three independent experiments. F, wtMADD but not MADD3A can significantly inhibit the association of DR5 with FADD. HeLa cells were transfected with wtMADD or MADD3A construct and treated with 50 ng/ml TRAIL. Cell lysates were immunoprecipitated with an anti-DR5 antibody followed by immunoblotting with anti-FADD antibody or anti-DR5 antibody.
DISCUSSION
MADD was originally discovered as a tumor necrosis factor αR1 cytoplasmic tail interacting protein that when overexpressed enhanced tumor necrosis factor α-induced activation of mitogen-activated protein kinase (MAPK) and c-Jun N-terminal kinase (JNK) (34). These pathways play crucial roles in cell growth and proliferation (34, 35). Subsequently, we showed that the IG20 gene can encode six different isoforms. Expression of two of these isoforms namely, KIAA and IG20-SV4, is restricted to certain neuronal cells (4). The KIAA (often referred to as MADD), which was isolated from a brain cDNA library, is homologous to rat Rab3a-GEP, which plays a critical role in vesicle trafficking and is required for the survival of the animal (5–7). Of the other four isoforms, MADD is the most physiologically relevant isoform, as it plays a critical role in cancer cell survival (11). Using specific shRNAs, we have demonstrated that abrogation of endogenous MADD results in enhanced spontaneous and TRAIL-induced apoptosis, and the cells can be rescued only when an shRNA-resistant MADD cDNA, and not cDNAs encoding the other three isoforms, is expressed in the absence of endogenous IG20 expression (10, 11). The novel finding of our present work reveals that MADD-mediated resistance to TRAIL-induced apoptosis is regulated through Akt-dependent phosphorylation. Furthermore, we demonstrate that it is the pMADD that interacts specifically with the TRAIL receptor DR4 and DR5 and prevents DISC formation by precluding FADD binding to DRs. TRAIL treatment on the other hand triggered a reduction in phosphorylation of MADD, thereby facilitating its release from DR4 and allowing for increased recruitment of FADD, resulting in enhanced DISC formation leading to apoptosis. However, we do not yet know whether TRAIL-induced reduction in MADD phosphorylation is due to the activation of a lipid or a protein phosphatase or suppression of kinase activity and is currently under investigation.
The resistance offered by endogenous MADD to TRAIL-induced apoptosis can be appreciated from results shown in Fig. 1. Although TRAIL is capable of inducing apoptosis in HeLa cells (Fig. 1A), at a suboptimal concentration of 5 ng/ml it hardly has any effect in cells that express significant levels of MADD (Fig. 1C). However, upon MADD knockdown, the same cells not only undergo significant spontaneous cell death, but they become more susceptible to treatment with a suboptimal dose of TRAIL (Fig. 1C). These results show MADD as a key regulator of TRAIL-induced apoptosis. As shown previously, the above function of MADD cannot be mimicked by other related isoforms encoded by the IG20 gene, such as DENN-SV, IG20pa, or SV-2 (10, 11).
The three Akt phosphorylation sites in MADD are evolutionarily conserved, thereby suggesting the importance of MADD in some of the key cellular functions. Akt is a key player in tumor growth and metastasis, and its basal activity is likely to be relatively high in a majority of cancers, and by corollary the levels of pMADD are likely to be increased (25, 28, 36, 37). It will be interesting to determine whether differential susceptibility of cancer cells to TRAIL-induced apoptosis is related to the levels of Akt activity and MADD phosphorylation. Several Akt substrates are known to play a critical role in apoptosis. For instance, in a healthy cell the pro-apoptotic molecules BAD and BAX are phosphorylated by Akt, resulting in their sequestration in cytosol by 14-3-3. Dephosphorylation results in their translocation to mitochondria leading to a loss of mitochondrial integrity and apoptotic cell death (22, 38–43). In contrast, pMADD is sequestered to the membrane and only upon TRAIL treatment MADD is released from the DR4 and accumulates in the cytosol. Similarly, the non-phosphorylate-able MADD3A is pro-apoptotic and is largely localized to cytosol. These findings indicate that the phosphorylation status of MADD likely determines its cellular localization and physiological function.
Death receptor-induced apoptosis is negatively regulated by the cellular form of FLICE inhibitory protein, Inhibition of Apoptosis, and caspase 8 and FADD-like apoptosis regulator that interact with either caspases or FADD and inhibit their function (16, 44–48). However, pMADD appears to act further upstream by blocking FADD recruitment to the death receptors and DISC formation. Unlike pMADD, the non-phosphorylated MADD cannot bind to DR4 and, thus, allows for FADD recruitment. Recent reports have revealed that cell surface TRAIL receptors move to lipid rafts upon TRAIL engagement (49, 50), which appears to be required for FADD binding and DISC formation leading to apoptosis. Whether or not pMADD prevents DR4 movement into lipid rafts or it physically interferes with FADD recruitment and thereby inhibits DISC formation is not yet known and is under investigation.
Unlike Fas ligand, the TRAIL appears to be an excellent choice of treatment for a variety of cancers as it has no significant deleterious effects on physiologically normal cells (16, 51–53). However, the possibility that the treatment itself can quickly induce resistance and promote metastasis could limit its potential utility (17). Akt is overexpressed in 20% of gastric adenocarcinomas, 12% of pancreatic cancers, and 15% of ovarian carcinomas (54). Enhanced Akt expression or loss-of-function mutation in PTEN could lead to increased Akt activity and, thus, increased phosphorylation of MADD resulting in enhanced resistance to TRAIL. Therefore, a combinatorial therapy with TRAIL and an inhibitor of the PI3K/Akt pathway might be an attractive alternative. However, it is fraught with problems as the PI3K/Akt pathway plays an indispensable role in normal cellular functions, thereby raising the possibility of severe side effects. Instead, targeting MADD may be a better alternative as its anti-apoptotic and pro-survival functions appear to be limited to death receptor signaling (10). In addition, loss of MADD appears to have little or no deleterious effects on primary cells (55, 56). We, therefore, propose MADD as an adjuvant therapeutic target for developing TRAIL-based combinatorial therapies.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01CA107506.
- SV
- splice variant
- caAkt
- constitutively activated form of Akt
- DISC
- death-inducing signaling complex
- DN-Akt
- dominant negative form of Akt
- DR4
- death receptor 4
- FADD
- Fas-associated death domain
- IGF-1
- insulin-like growth factor-1
- MADD
- mitogen-activated kinase activating death domain protein
- pMADD
- phosphorylated MADD
- shRNA
- small hairpin RNA
- PI3K
- phosphatidylinositol 3-kinase
- TRAIL
- tumor necrosis factor α-related apoptosis-inducing ligand
- GST
- glutathione S-transferase
- wt
- wild type
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein.
REFERENCES
- 1.Al-Zoubi A. M., Efimova E. V., Kaithamana S., Martinez O., El-Idrissi Mel-A., Dogan R. E., Prabhakar B. S. (2001) J. Biol. Chem. 276, 47202–47211 [DOI] [PubMed] [Google Scholar]
- 2.Efimova E., Martinez O., Lokshin A., Arima T., Prabhakar B. S. (2003) Cancer Res. 63, 8768–8776 [PubMed] [Google Scholar]
- 3.Efimova E. V., Al-Zoubi A. M., Martinez O., Kaithamana S., Lu S., Arima T., Prabhakar B. S. (2004) Oncogene 23, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 4.Li L. C., Sheng J. R., Mulherkar N., Prabhakar B. S., Meriggioli M. N. (2008) Cancer Res. 68, 7352–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka M., Miyoshi J., Ishizaki H., Togawa A., Ohnishi K., Endo K., Matsubara K., Mizoguchi A., Nagano T., Sato M., Sasaki T., Takai Y. (2001) Mol. Biol. Cell 12, 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa S., Tanaka Y., Hirokawa N. (2008) Nat. Cell Biol. 10, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 7.Coppola T., Perret-Menoud V., Gattesco S., Magnin S., Pombo I., Blank U., Regazzi R. (2002) Biochem. J. 362, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow V. T., Lee S. S. (1996) DNA Seq. 6, 263–273 [DOI] [PubMed] [Google Scholar]
- 9.Subramanian M., Pilli T., Bhattacharya P., Pacini F., Nikiforov Y. E., Kanteti P. V., Prabhakar B. S. (2009) J. Clin. Endocrinol. Metab. 94, 1467–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulherkar N., Prasad K. V., Prabhakar B. S. (2007) J. Biol. Chem. 282, 11715–11721 [DOI] [PubMed] [Google Scholar]
- 11.Mulherkar N., Ramaswamy M., Mordi D. C., Prabhakar B. S. (2006) Oncogene 25, 6252–6261 [DOI] [PubMed] [Google Scholar]
- 12.Bennett M., Macdonald K., Chan S. W., Luzio J. P., Simari R., Weissberg P. (1998) Science 282, 290–293 [DOI] [PubMed] [Google Scholar]
- 13.Desbarats J., Birge R. B., Mimouni-Rongy M., Weinstein D. E., Palerme J. S., Newell M. K. (2003) Nat. Cell Biol. 5, 118–125 [DOI] [PubMed] [Google Scholar]
- 14.Itoh N., Yonehara S., Ishii A., Yonehara M., Mizushima S., Sameshima M., Hase A., Seto Y., Nagata S. (1991) Cell 66, 233–243 [DOI] [PubMed] [Google Scholar]
- 15.Kurada B. R., Li L. C., Mulherkar N., Subramanian M., Prasad K. V., Prabhakar B. S. (2009) J. Biol. Chem. 284, 13533–13541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnstone R. W., Frew A. J., Smyth M. J. (2008) Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 17.Prabhakar B. S., Mulherkar N., Prasad K. V. (2008) Clin. Cancer Res. 14, 347–351 [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Thakkar H., Tyan F., Gim S., Robinson H., Lee C., Pandey S. K., Nwokorie C., Onwudiwe N., Srivastava R. K. (2001) Oncogene 20, 6073–6083 [DOI] [PubMed] [Google Scholar]
- 19.Pan G., Ni J., Wei Y. F., Yu G., Gentz R., Dixit V. M. (1997) Science 277, 815–818 [DOI] [PubMed] [Google Scholar]
- 20.Pan G., O'Rourke K., Chinnaiyan A. M., Gentz R., Ebner R., Ni J., Dixit V. M. (1997) Science 276, 111–113 [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy M., Efimova E. V., Martinez O., Mulherkar N. U., Singh S. P., Prabhakar B. S. (2004) Oncogene 23, 6083–6094 [DOI] [PubMed] [Google Scholar]
- 22.Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. (1997) Cell 91, 231–241 [DOI] [PubMed] [Google Scholar]
- 23.Mayo L. D., Donner D. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou B. P., Liao Y., Xia W., Zou Y., Spohn B., Hung M. C. (2001) Nat. Cell Biol. 3, 973–982 [DOI] [PubMed] [Google Scholar]
- 25.Bhaskar P. T., Hay N. (2007) Dev. Cell 12, 487–502 [DOI] [PubMed] [Google Scholar]
- 26.Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P. F., Dietz R., von Harsdorf R. (1999) EMBO J. 18, 6027–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudek H., Datta S. R., Franke T. F., Birnbaum M. J., Yao R., Cooper G. M., Segal R. A., Kaplan D. R., Greenberg M. E. (1997) Science 275, 661–665 [DOI] [PubMed] [Google Scholar]
- 29.Barber M. A., Donald S., Thelen S., Anderson K. E., Thelen M., Welch H. C. (2007) J. Biol. Chem. 282, 29967–29976 [DOI] [PubMed] [Google Scholar]
- 30.Moreau C., Cartron P. F., Hunt A., Meflah K., Green D. R., Evan G., Vallette F. M., Juin P. (2003) J. Biol. Chem. 278, 19426–19435 [DOI] [PubMed] [Google Scholar]
- 31.Cartron P. F., Gallenne T., Bougras G., Gautier F., Manero F., Vusio P., Meflah K., Vallette F. M., Juin P. (2004) Mol. Cell 16, 807–818 [DOI] [PubMed] [Google Scholar]
- 32.Gingras A. C., Kennedy S. G., O'Leary M. A., Sonenberg N., Hay N. (1998) Genes Dev. 12, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 34.Schievella A. R., Chen J. H., Graham J. R., Lin L. L. (1997) J. Biol. Chem. 272, 12069–12075 [DOI] [PubMed] [Google Scholar]
- 35.Dhillon A. S., Hagan S., Rath O., Kolch W. (2007) Oncogene 26, 3279–3290 [DOI] [PubMed] [Google Scholar]
- 36.Okano J., Gaslightwala I., Birnbaum M. J., Rustgi A. K., Nakagawa H. (2000) J. Biol. Chem. 275, 30934–30942 [DOI] [PubMed] [Google Scholar]
- 37.Way T. D., Kao M. C., Lin J. K. (2004) J. Biol. Chem. 279, 4479–4489 [DOI] [PubMed] [Google Scholar]
- 38.Gardai S. J., Hildeman D. A., Frankel S. K., Whitlock B. B., Frasch S. C., Borregaard N., Marrack P., Bratton D. L., Henson P. M. (2004) J. Biol. Chem. 279, 21085–21095 [DOI] [PubMed] [Google Scholar]
- 39.Wang H. G., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., Shibasaki F., McKeon F., Bobo T., Franke T. F., Reed J. C. (1999) Science 284, 339–343 [DOI] [PubMed] [Google Scholar]
- 40.Zha J., Harada H., Yang E., Jockel J., Korsmeyer S. J. (1996) Cell 87, 619–628 [DOI] [PubMed] [Google Scholar]
- 41.Antignani A., Youle R. J. (2006) Curr. Opin. Cell Biol. 18, 685–689 [DOI] [PubMed] [Google Scholar]
- 42.Tsuruta F., Sunayama J., Mori Y., Hattori S., Shimizu S., Tsujimoto Y., Yoshioka K., Masuyama N., Gotoh Y. (2004) EMBO J. 23, 1889–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi H., Wang H. G. (2001) Oncogene 20, 7779–7786 [DOI] [PubMed] [Google Scholar]
- 44.Chu Z. L., McKinsey T. A., Liu L., Gentry J. J., Malim M. H., Ballard D. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10057–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deveraux Q. L., Reed J. C. (1999) Genes Dev. 13, 239–252 [DOI] [PubMed] [Google Scholar]
- 46.Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. (1997) Nature 388, 300–304 [DOI] [PubMed] [Google Scholar]
- 47.Dohi T., Xia F., Altieri D. C. (2007) Mol. Cell 27, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., Koentgen F., Vaux D. L., Silke J. (2007) Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 49.Delmas D., Rébé C., Micheau O., Athias A., Gambert P., Grazide S., Laurent G., Latruffe N., Solary E. (2004) Oncogene 23, 8979–8986 [DOI] [PubMed] [Google Scholar]
- 50.Song J. H., Tse M. C., Bellail A., Phuphanich S., Khuri F., Kneteman N. M., Hao C. (2007) Cancer Res. 67, 6946–6955 [DOI] [PubMed] [Google Scholar]
- 51.Zhang L., Fang B. (2005) Cancer Gene Ther. 12, 228–237 [DOI] [PubMed] [Google Scholar]
- 52.Guo F., Nimmanapalli R., Paranawithana S., Wittman S., Griffin D., Bali P., O'Bryan E., Fumero C., Wang H. G., Bhalla K. (2002) Blood 99, 3419–3426 [DOI] [PubMed] [Google Scholar]
- 53.Deng Y., Lin Y., Wu X. (2002) Genes Dev. 16, 33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan S. M., Weng A. P., Tibshirani R., Aster J. C., Utz P. J. (2007) Blood 110, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim K. M., Chow V. T. (2002) Mol. Carcinog. 35, 110–126 [DOI] [PubMed] [Google Scholar]
- 56.Lim K. M., Yeo W. S., Chow V. T. (2004) Int. J. Cancer 109, 24–37 [DOI] [PubMed] [Google Scholar]