Abstract
The cytoplasm and the nucleus have been identified as activity sites for granzyme B (GrB) following its delivery from cytotoxic lymphocyte granules into target cells. Here we report on the ability of exogenous GrB to insert into and function within a proteinase K-resistant mitochondrial compartment. We identified Hax-1 (HS-1-associated protein X-1), a mitochondrial protein involved in the maintenance of mitochondrial membrane potential, as a GrB substrate within the mitochondrion. GrB cleaves Hax-1 into two major fragments: an N-terminal fragment that localizes to mitochondria and a C-terminal fragment that localizes to the cytosol after being released from GrB-treated mitochondria. The N-terminal Hax-1 fragment major cellular impact is on the regulation of mitochondrial polarization. Overexpression of wild-type Hax-1 or its uncleavable mutant form protects the mitochondria against GrB or valinomycin-mediated depolarization. The N-terminal Hax-1 fragment functions as a dominant negative form of Hax-1, mediating mitochondrial depolarization in a cyclophilin D-dependent manner. Thus, induced expression of the N-terminal Hax-1 fragment results in mitochondrial depolarization and subsequent lysosomal degradation of such altered mitochondria. This study is the first to demonstrate GrB activity within the mitochondrion and to identify Hax-1 cleavage as a novel mechanism for GrB-mediated mitochondrial depolarization.
Keywords: Cell Death, Cytochrome c, Death Protease, Immunology, Mitochondria, Mitochondrial Apoptosis
Introduction
The granzyme family of serine proteases are pivotal mediators of apoptosis utilized by cytotoxic lymphocytes against targets such as virus-infected cells and tumor cells (1–6). Granzyme B (GrB)3 exerts its cytotoxic effect after perforin-dependent intracellular delivery, acting in both cytosolic and nuclear compartments. The capacity of GrB to initiate a mitochondrial apoptotic cascade that features permeabilization of the mitochondrial outer membrane and release of apoptogenic proteins has been tied directly to cleavage of Bid (7–10). GrB-generated tBid activates the downstream executioners of the mitochondrial apoptotic cascade, Bax and/or Bak (11). However, mitochondria of Bid-deficient cells continue to release cytochrome c in response to GrB, suggesting the existence of an alternative mechanism(s). Our recent studies demonstrated that GrB induces a mitochondrial apoptotic cascade by cleaving Mcl-1, an antiapoptotic Bcl-2 family member that resides on the mitochondrial outer membrane in a complex with a potent proapoptotic BH3-only protein, Bim. GrB-mediated degradation of Mcl-1 releases sequestered Bim, allowing the execution of a distinct apoptotic cascade (12, 13). The mitochondrial function of BH3-only proteins, including Bid and Bim, is strictly Bax/Bak-dependent (14). Accordingly, Bax/Bak-deficient cells are relatively resistant to GrB-mediated apoptosis, but mitochondria continue to undergo significant depolarization that cannot be attributed to currently described mechanisms of GrB-mediated apoptosis (10, 15, 16).
Mitochondrial polarization (i.e. maintenance of its inner membrane potential) is ultimately controlled by the mechanisms responsible for the opening of a multiprotein pore, the permeability transition pore (PTP), at the contact sites between the outer and the inner mitochondrial membranes (17). The PTP complex is putatively composed of a voltage-dependent anion channel at the outer mitochondrial membrane, adenine nucleotide translocase in the inner mitochondrial membrane, and cyclophilin D (Cyp-D), a peptidyl-prolyl isomerase in the mitochondrial matrix. Recent studies involving knock-out mice for each of these components provided evidence that only Cyp-D represents a necessary regulatory component for the PTP opening mechanism, whereas adenine nucleotide translocase and voltage-dependent anion channel participate in the process to a limited extent (18–22). Disruption in the mitochondrial membrane potential (MMP) due to the opening of the PTP has profound consequences for mitochondrial respiration, energy production, and hence, cell survival.
Recent studies have shown that Hax-1 is essential for maintenance of the MMP in myeloid and epithelial cells (23, 24). Hax-1 was originally identified by a yeast two-hybrid screen, associating with HS-1 (hematopoietic lineage cell-specific protein 1), a major substrate for tyrosine kinase activity (25). Hax-1 possesses two Bcl-2 homology domains BH1 and BH2, a PEST sequence, and a putative transmembrane domain. In addition, Hax-1 has limited sequence similarity to BNIP3, a BH3-only protein that is up-regulated in response to oxidative stress. Hax-1 is reportedly cleaved by the mitochondrial Omi/HtrA2, a serine protease that is released into the cytosol during apoptosis to antagonize inhibitors of apoptosis (24). Nevertheless, the biological significance of Omi/HtrA2-mediated Hax-1 cleavage is not fully understood. Hax-1 has also been identified as an endogenous inhibitor of caspase-9 in cardiac myocytes (26). Furthermore, an autosomal recessive severe congenital neutropenia resulting from a homozygous germ line mutation in Hax-1 is associated with increased apoptosis in myeloid cells (23, 27). Why a null mutation of a ubiquitously expressed gene causes a myeloid-specific phenotype is unclear. Current speculation suggests that the tissue-specific effect of Hax-1 may be due to intrinsic differences in the regulation of apoptosis in neutrophils. However, compared with fibroblasts from normal donors, stressed Hax-1-deficient fibroblasts from severe congenital neutropenia patients showed a more rapid loss of their membrane potential, suggesting that Hax-1-mediated stabilization of the MMP is not limited to neutrophils (23). Indeed, the likelihood that Hax-1 acts as a mitochondrial antiapoptotic protein in other cells has been verified by a Hax-1-null mouse model (28). Loss of Hax-1 resulted in postnatal lethality as well as lymphocyte and neuron apoptosis. Hax-1 interacts with mitochondrial proteases Parl and Omi/HtrA2. These interactions enable Hax-1 to present HtrA2 to Parl and thereby facilitate the processing of HtrA2 to an active protease localized in the mitochondrial intermembrane space. It has been further determined that in lymphocytes, the presence of processed HtrA2 delays the accumulation of mitochondrial outer membrane-associated activated Bax. Promotion of cell survival by Hax-1 was also elucidated in cardiomyocytes, where it functions in the regulation of calcium signaling through dynamic interactions with phospholamban and sarcoplasmic reticulum Ca-ATPase (SERCA2a) (29–31). Hax-1 has also been elucidated as a binding target for several viral proteins encoded by Epstein-Barr virus, Kaposi's sarcoma associated herpes virus, and human immunodeficiency virus, type 1 (32–35), but the biological implications of these interactions remain unclear.
In the current study, we show that GrB is targeted to mitochondria and that Hax-1 is processed by GrB within a proteinase K-resistant mitochondrial compartment. Hax-1 cleavage by GrB generates a two-pronged assault on the conservation of the MMP; it eliminates antiapoptotic full-length Hax-1 and produces an N-terminal fragment that functions to disrupt the MMP and to enhance the outer membrane permeability for the release of apoptogenic proteins.
EXPERIMENTAL PROCEDURES
Reagents and Cell Lines
Human GrB was purified from cytolytic granules of the human YT NK cell line, as described previously (36). Human replication-deficient adenovirus (Ad) was purified from infected 293 cells by the University of Pittsburgh Vector Core Facility. Anti-human Mcl-1 Abs were from Oncogene (Boston, MA) and from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); anti-β-actin monoclonal Ab was from Sigma; anti-Bim Ab was from ProSci (Poway, CA); Abs to apoptosis-inducing factor (AIF), histone 1, cytochrome c, SMAC, Hax-1, GrB (for confocal microscopy), LAMP2, and β-tubulin were from Santa Cruz Biotechnology, Inc.; anti-Xpress mouse monoclonal Ab for lacZ-encoded protein and anti-Cox IV monoclonal Ab were from Invitrogen; anti-GrB used for immunoblotting was from Serotec (Raleigh, NC); Abs to caspase-3 and Grp75 were from StressGen (Ann Arbor, MI); and Abs to prohibitin and Cyp-D were from Abcam (Cambridge, MA). MitoTracker Deep Red and goat anti-mouse Alexa Fluor 488 Ab were from Invitrogen; [35S]methionine, Protein A-Sepharose beads, and Protein G-Sepharose beads were from Amersham Biosciences. Z-VAD-fluoromethyl ketone and Z-VAD-aldehyde were from ICN (Aurora, OH). Valinomycin was from Cayman (Ann Arbor, Michigan), and proteinase K was from Sigma.
The Bax/Bak-deficient Jurkat clonal cell line was obtained from a wild-type (WT) Jurkat cell line, as described previously (37). Jurkat T leukemic cells were grown in RPMI 1640 medium containing 10% fetal calf serum, 20 mm HEPES, 2 mm l-glutamine, and 100 units/ml each of penicillin and streptomycin. Breast carcinoma MCF7, colon cancer Hct116, and T-REx-293 cells were grown in Dulbecco's modified Eagle's medium containing 15% fetal calf serum, 20 mm l-glutamine, and 100 units/ml each of penicillin and streptomycin. Transfected T-REx-293 clonal cell lines were maintained in the presence of blasticidin (5 μg/ml) and zeocin (20 μg/ml), as described previously (38).
RNAi
Hax-1 siRNA and a matched control siRNA were obtained from Dharmacon, as the siGENOME SMARTpool siRNA reagent and the siGENOME Non-targeting siRNA Pool 2, respectively. Each reagent consists of a pool of four distinct siRNAs. A Cyp-D siRNA based on the previously described siRNA-1 (39) was obtained from Ambion as a Custom Select siRNA along with the Silencer Select Negative Control 1. In addition, three distinct Cyp-D siRNAs and their negative control (with matching GC content) were obtained from Invitrogen as the Stealth Select 3 RNAi reagent (HSS115357-9) and the Stealth RNAi Negative Control Med GC, respectively. Transfection of HCT116 and T-REx-293 cells was carried out as described previously (38).
Sequence Analysis of GrB-generated Hax-1 Fragments
N-terminal (Edman) radiosequencing of [3H]leucine- and [35S]methionine-labeled Hax-1 fragments generated by GrB digestion of in vitro translated full-length Hax-1 and MALDI mass spectrometry of a tryptic digest of unlabeled Hax-1 also generated by in vitro translation were carried out by the W. M. Keck Foundation Biotechnology Resource Laboratory of Yale University.
Cellular Fractionation and Mitochondria Purification
To obtain an enriched mitochondrial fraction, Jurkat, MCF7, Hct116, or T-REx-293 cells were suspended in mitochondrial buffer (MIB) composed of 0.3 m sucrose, 10 mm MOPS, 1 mm EDTA, and 4 mm KH2PO4, pH 7.4, and lysed by Dounce homogenization, as described previously (12). Briefly, nuclei and debris were removed by a 10-min centrifugation at 650 × g, and a pellet containing mitochondria was obtained by two successive spins at 10,000 × g for 12 min. To obtain the cytosolic fraction, the postnuclear supernatant was further centrifuged at 20,000 × g for 1 h at 4 °C. To obtain the enriched mitochondrial fraction, the mitochondria-containing pellet was resuspended in MIB and layered on a Percoll gradient consisting of four layers of 10, 18, 30, and 70% Percoll in MIB. After centrifugation for 30 min at 15,000 × g, the mitochondrial fraction was collected at the 30/70 interface. Mitochondria were diluted in MIB containing 1 mg/ml bovine serum albumin (at least a 10-fold dilution required to remove Percoll). The mitochondrial pellet was obtained by a 40-min spin at 20,000 × g and used immediately. Purity was assessed by electron microscopy and by enzyme marker analysis, as described previously (38, 40).
Confocal Microscopy
Cells were grown and treated on Lab-Tek II chamber slides. After treatment with the MitoTracker Deep Red dye according to the manufacturer's instructions, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Images were captured by a confocal Olympus Fluoview 1000 system using a ×100 oil immersion objective with the companion FV10-ASW1.6 imaging software at 25 °C. The digital images were split and merged using ImageJ, and image contrast and brightness were adjusted with Adobe Photoshop.
Quantitation of colocalization was measured using Metamorph (Molecular Devices, Downingtown, PA). For this purpose, two-color confocal micrographs that contain defined positive regions of each protein were segmented using conventional thresholding tools. The thresholded images were compared by simple Boolean math. The numbers of positive pixels for each protein were determined using a logical AND operator, and the percentages of overlay were assessed.
Molecular Cloning of Hax-1 Plasmids
Isolation of total RNA from Jurkat T-cells, first strand cDNA synthesis, and PCR were carried out as described previously (38). A full-length Hax-1 amplicon that extended 9 nucleotides into the 5′- and 3′-untranslated regions, respectively, was generated with the following primer pair (forward and reverse): 5′-CGCGGATCCAGTACGGGAATGAGCCTC-3′ and 5′-ACGCGTCGACTTAACAAGGCTACCGGGA-3′. The putative Hax-1 amplicon was size-selected on a 1% agarose gel and restriction enzyme-digested with BamHI and SalI (New England Biolabs). The digested Hax-1 amplicon was ligated into either a Tet-inducible vector, pcDNA4/TO (Invitrogen), for stable expression in T-REx-293 cells or the constitutive expression vector pCR3.1 (Invitrogen) for in vitro translation experiments. Both plasmids had been previously digested with BamHI and XhoI. Following transformation (Escherichia coli Top10F′ (pcDNA4/TO); DH5α (pCR3.1)), randomly picked colonies underwent DNA sequence analysis (University of Pittsburgh DNA Sequencing Core Facility) to confirm sequence integrity.
Generation of Hax-1 Mutant Clones
Both single substitution Asp to Ala full-length Hax-1 clones, D148A and D159A, were produced from the WT Hax-1 clone (see above) by overlap extension using PCR. Single point mutations for D148A(GAT → GCT) and D159A(GAC → GCC) were introduced by mutated forward and reverse primers that overlap the specific Asp residue that was changed to Ala.
The mutated primer pairs (forward and reverse) for D148A were as follows: 5′-TTGGAGAGTGCTGCAAGAAGTGAATCCCCCCAA-3′ and 5′-ACTTCTTGCAGCACTCTCCAAGACCCCCCCAAA-3′; for D159A, 5′-CCAGCACCAGCCTGGGGCTCCCAGAGGCCATTT-3′ and 5′-GGAGCCCCAGGCTGGTGCTGGTTGGGGGGATTC-3′.
PCR, restriction enzyme digestion, ligation to pCR3.1, and sequence analysis to confirm both Asp to Ala conversions were carried out as above. The double mutant D148A/D159A Hax-1 clone was constructed from either single mutation clone (D148A or D159A) by overlap extension using PCR with the same mutated primers described above. PCR, restriction enzyme digestion, ligation to pCR3.1, and sequence analysis to confirm both Asp to Ala conversions were carried out as above.
Production of Hax-1 GrB N- and C-terminal Fragment-encoding Clones
The N- and C-terminal GrB Hax-1 fragment-encoding clones whose cDNA inserts correspond to fragments generated by GrB cleavage at Asp148 were produced from the WT Hax-1 clone. The N-terminal fragment was produced from a primer pair consisting of the forward primer for full-length Hax-1 (see above) and the reverse primer 5′-ACGCGTCGACCTAATCACTCTCCAAGACCCC-3′. The C-terminal fragment was made with the forward primer 5′-CGCGGATCCACGGGAATGGCAAGAAGTGAATCCCCCCAA-3′ and the reverse primer for full-length Hax-1 (see above). The C-terminal GrB fragment containing the D159A mutation was generated with C-terminal fragment primers and the full-length D159A clone (see above). PCR was carried out with the Expand High Fidelity PCR System kit (Roche Applied Science). Restriction enzyme digestion, ligation to pcDNA4/TO or pCR3.1, and sequence verification were as described above for full-length Hax-1.
Molecular Cloning of AIF
Isolation of total RNA from HeLa cells, and first strand cDNA synthesis using oligo (dT)12–18 and PCR were performed as described (38). A full-length AIF clone (which encodes the longest isoform of three variants) that extends 6 nucleotides into the 5′-untranslated region and 3 nucleotides into the 3′-untranslated region was produced with the following primer pair (forward and reverse): 5′- CGGGGTACCGCCGAAATGTTCCGGTGTG-3′ and 5′-CCGGAATTCGCTTCAGTCTTCATGAATGTTG-3′. The putative AIF full-length amplicon was size-selected on an agarose gel (0.9%) and digested with KpnI and EcoR I (New England Biolabs). It was ligated into pCR3.1 that had also been digested with KpnI and EcoRI. Following transformation (E. coli DH5α), randomly picked colonies underwent DNA sequence analysis as described above.
Stable Transfection of T-REx-293 Cells with Tet-inducible Plasmids
The cell line T-REx-293 (Invitrogen) (human embryonic kidney cells that stably express the Tet repressor) was stably transfected with the linearized Tet-inducible plasmids pcDNA4/TO, pcDNA4/TO/lacZ (Invitrogen), pcDNA4/ TO/Hax-1 (full-length), pcDNA4/TO/N-Hax-1 (Tet N-Hax-1), and pcDNA4/TO/C-Hax-1 (Tet C-Hax-1). All procedures involving linearization, transfection, and stable clone selection were carried out as described previously (38).
Cell Lysates and Cell Extracts
Cell lysates were prepared with 1% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 137 mm NaCl, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. To prepare cell extracts, cultured cells were washed twice with phosphate-buffered saline and then resuspended in ice-cold buffer (20 mm HEPES, pH 7.0, 10 mm KCl, 1.5 mm MgCl2, 1 mm sodium EDTA, 1 mm sodium EGTA, 1 mm dithiothreitol, 250 mm sucrose, and protease inhibitors). After incubation on ice for 20 min, cells (2.5 × 106/0.5 ml) were disrupted by Dounce homogenization. Cellular extracts were obtained as the supernatants resulting from centrifugation to remove nuclei at 650 × g for 10 min at 4 °C.
Mitochondrial Membrane Potential and Oxidative Stress Measurements
Flow cytometry was utilized to measure alterations in MMP and in production of reactive oxygen species (ROS). The cationic fluorescent dyes 1,1′,3,3′-tetraethylbenzamidazolocarbocyanin iodide (JC-1) and 3,3′-diethyloxacarbocyanine iodide (Invitrogen) were utilized for MMP measurement. 5-(and 6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA; Invitrogen) was utilized for measurement of ROS production. The staining was performed according to the manufacturer's procedures, and mitochondrial alterations were quantified by the Beckman Coulter Epics XL-MCL and analyzed with the EXPO32 software.
ATP Measurement
ATP levels were measured by the ATPLite kit (PerkinElmer Life Sciences) according to the manufacturer's procedure. This method is based on mono-oxygenation of luciferin, catalyzed by luciferase in the presence of Mg2+, ATP, and oxygen, resulting in a luminescent signal that is proportional to the ATP concentration.
In Vitro Transcription-Translation
Full-length Hax-1, D148A mutant Hax-1, D159A mutant Hax-1, double mutant D148A/D159A Hax-1, GrB N-terminal (Met1–Asp148) and C-terminal (Ala149–Arg279) Hax-1 fragments as well as the D159A mutant C-terminal Hax-1 fragment cDNAs were expressed in the TNT T7 transcription-translation reticulocyte lysate system (Promega). Each coupled transcription-translation reaction contained 1 μg of plasmid DNA in a final volume of 50 μl in a methionine-free reticulocyte lysate reaction mixture supplemented with 35S-labeled methionine according to the manufacturer's instructions. After incubation at 30 °C for 90 min, the reaction products were immediately used or stored at −78 °C.
Western Blot Analysis
Proteins in cell lysates, cell extracts, mitochondria, cytosol, or nuclei were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Following probing with a specific primary Ab and horseradish peroxidase-conjugated secondary Ab, the protein bands were detected by enhanced chemiluminescence (Pierce).
RESULTS
Mitochondrial Insertion of GrB
To investigate whether GrB has a direct effect on a protein(s) involved in controlling PTP function, we revisited its subcellular localization after intracellular delivery. Although the granzyme readily undergoes endocytic uptake, efficient induction of apoptosis requires the presence of perforin or an alternative endosomolytic reagent that act to ensure the release of the protease from endosomes (41–43). Based on multiple studies performed by our laboratory (12, 13, 44) as well as others (8, 42, 45, 46), replication-deficient Ad is considered an appropriate replacement for perforin. Using confocal microscopy, we observed a Bcl-2-independent association of GrB with mitochondria when GrB was exogenously delivered to HeLa cells together with Ad. Overexpression of Bcl-2 did not impact the mitochondrial localization of GrB because a similar pattern of co-localization was observed in vector or Bcl-2-transfected HeLa cells (Fig. 1A and supplemental Fig. S1). GrB was associated with mitochondria within 10 min of its exogenous application (Fig. 1, D, F, and H).
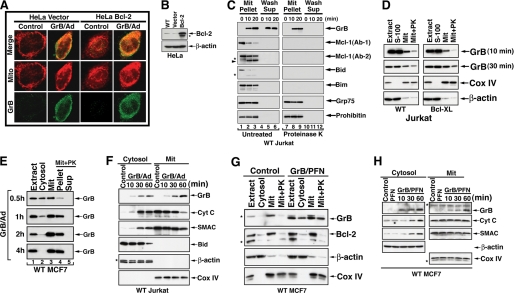
FIGURE 1.
Insertion of GrB into a proteinase K-resistant mitochondrial compartment. A and B, mitochondrial localization of GrB. HeLa cells stably transfected with vector control or Bcl-2 were treated with GrB/Ad (33 nm/10 pfu/ml, respectively) for 10 min. The cells were then stained with MitoTracker Deep Red and with anti-GrB Ab. Scale bar, 10 μm. Using the Metamorph colocalization method described under “Experimental Procedures,” it was determined that 52 and 61% of GrB colocalized with MitoTracker in control cells and Bcl-2-HeLa cells, respectively. A higher magnification of this staining is shown in supplemental Fig. S1. The expression level of Bcl-2 in the utilized cells is shown in B. No difference between vector or Bcl-2-transfected cells was detected with regard to the mitochondrial localization of GrB. C, insertion of GrB into purified mitochondria. Purified mitochondria from Jurkat cells were treated with GrB (160 nm) for the indicated time periods. Subsequently, the mitochondria were left untreated (left; lanes 1–6) or treated with proteinase K (30 μg/ml for 10 min followed by 1 μm phenylmethylsulfonyl fluoride to stop the reaction; right, lanes 7–12). The mitochondria were then spun to separate the pellets and their supernatants. All proteins in the supernatants of the proteinase K-treated mitochondria were digested (lanes 10–12). Like Grp75 and prohibitin, mitochondrial GrB remained proteinase K-resistant, whereas Mcl-1, Bid, and Bim were proteinase K-sensitive (lanes 7–9). The arrowhead indicates cleaved Mcl-1 fragments, and the asterisks show unidentified protein bands. The gel loading ratio between the mitochondrial pellet (Mit Pellet) and wash supernatant (Wash Supp) was 1:4, indicating that at 20 min following its application, ∼80% of the GrB was mitochondrially inserted. D, mitochondrial insertion of exogenously applied GrB onto WT or Bcl-XL-overexpressing Jurkat cells. The cells were treated with GrB/Ad (33 nm/10 pfu/ml) for the indicated time periods. The cells were then homogenized in the presence of a GrB inhibitor and subjected to cellular fractionation to obtain the indicated fractions. The mitochondrial fraction was further treated with proteinase K as described in C. The Cox IV and β-actin immunoblots correspond to the membrane GrB application for 30 min. No significant differences were detected between WT and Bcl-XL-overexpressing cells in the levels of mitochondrial association or insertion of GrB. E and F, kinetics of GrB mitochondrial localization following its cellular delivery with Ad. MCF7 (E) and WT Jurkat cells (F) were treated as described in D. A significant fraction of the cell-delivered GrB was associated with the mitochondria within 30 min of its exogenous application, and most of the delivered GrB was inserted into a proteinase K-resistant mitochondrial compartment within 1 h (E, lanes 3 and 4). In Jurkat cells (F), exogenously applied GrB was associated with mitochondria as early as 10 min after treatment, concomitant with the initial detection of the release of mitochondrial cytochrome c but prior to the detection of Bid processing. G and H, kinetics of GrB mitochondrial localization following its cellular delivery with perforin. MCF7 cells were treated with GrB (33 nm) and PFN titrated to kill less than 10% of the cells on its own. Results obtained with perforin (G and H) were similar to those obtained with Ad (E and F). In G, the cells were treated with GrB/PFN for 1 h, and following subcellular fractionation, the mitochondrial fraction was treated with proteinase K, as described in D. In H, GrB was associated with the mitochondria at the earliest time point tested (10 min), whereas cytosolic cytochrome c and SMAC were detected significantly later.
To investigate whether GrB translocates into the mitochondrion, we co-incubated isolated mitochondria with GrB for 10–20 min and then treated the mitochondria with proteinase K to remove excess as well as externally bound GrB. Based on the relative gel loading (1:4 pellet/supernatant), 80% of the applied GrB was detected in the mitochondrial pellet (20 min), and the rest was detected in the wash fraction (Fig. 1C). Subsequent treatment of the mitochondrial pellet with proteinase K showed that GrB remained confined to the pellet, whereas the Bcl-2 family members Mcl-1, Bid, and Bim were completely removed from the mitochondrial outer membrane. Similar to GrB, the mitochondrial matrix proteins, Grp75 and prohibitin, were detected exclusively in the pellet, and levels were unaffected by the proteinase K treatment.
To determine whether GrB enters mitochondria of intact cells, WT- or Bcl-XL-overexpressing Jurkat cells were treated with GrB/Ad, and following removal of excess GrB and the addition of a GrB pharmacological inhibitor, the cells were homogenized and subjected to subcellular fractionation. Within 10 min of its exogenous application, a significant portion of the detected GrB was associated with the mitochondria. To distinguish between mitochondrion-associated and mitochondrion-inserted GrB, the mitochondrial fraction was further treated with proteinase K. Again, a significant fraction of GrB was proteinase K-resistant (Fig. 1D). Furthermore, similar levels of GrB mitochondrial insertion were observed in WT or Bcl-XL-overexpressing Jurkat cells, suggesting that the insertion was not impacted by an increased expression of Bcl-XL. As confirmed by histone-1 immunoblotting, all of the cellular fractions were devoid of nuclei, whereas the reported nuclear localization of the granzyme was confirmed with highly purified nuclei (Fig. S2, A and B). Further kinetic studies were performed with MCF7 cells (which are caspase-3-deficient; Fig. 1E) and with MCF7 stably transfected with caspase-3 that demonstrated similar kinetics and insertion levels of GrB (not shown). In Jurkat cells (Fig. 1F), the mitochondrial translocation of GrB coincided with the initiation of cytochrome c release (10 min), yet marked processing of Bid was only observed significantly later (30–60 min), further suggesting that the mitochondrial translocation of GrB is not tBid-dependent. To further demonstrate the mitochondrial insertion of GrB following its intracellular delivery by perforin, a sublytic dose of perforin, which caused death in ∼10% of cells on its own, was used to deliver GrB into MCF7 cells. Again, GrB was detected in the mitochondrial fraction within 10 min of its intracellular delivery (Fig. 1G) and was further demonstrated to accumulate in a proteinase K-treated mitochondrial fraction (Fig. 1H). Taken together, the mitochondrion appears to be a major site for localization of GrB after its intracellular delivery.
Cleavage of Mitochondrial Proteins by GrB
To verify that mitochondria are indeed a biologically relevant site for GrB and to identify potential protein substrates, two-dimensional differential in-gel electrophoresis was performed on control and GrB-treated mitochondria. Among seven proteins that demonstrated a reduced level of at least 2-fold within 30 min of GrB treatment, Hax-1 (supplemental Fig. S3, A and B) and AIF (supplemental Fig. S4) were identified as potential substrates.
After granzyme delivery, we observed a time-dependent release of AIF into the cytosol and the presence of its putative cleavage products in the proteinase K-resistant mitochondrial pellet (supplemental Fig. S4A, lanes 3 and 4). In many models of apoptosis, mitochondrial AIF translocates to the nucleus, where it induces chromatin condensation and DNA degradation (47). It was therefore surprising to observe that GrB cleaves AIF within the mitochondrion. To confirm that granzyme processes AIF, we examined the rate of AIF cleavage product generation. Mitochondria were exposed to the granzyme for various times (0.5–2 h) and subsequently treated with proteinase K to remove outer membrane-associated proteins as well as bound GrB (supplemental Fig. S4B). Cleavage fragments of AIF were detected at the earliest time point tested (30 min; lanes 4–6). To learn whether GrB directly cleaves AIF, we examined the capacity of the protease to process in vitro translated AIF (supplemental Fig. S4C). Four possible cleavage products were observed, with two being also detected by immunoblotting. Although AIF is a putative death effector, the flavoprotein also functions as an NADH oxidase essential for oxidative phosphorylation and for efficient antioxidant defense (48). GrB may therefore cause cell death, in part, by disrupting AIF activity.
Mitochondrial Hax-1 was also identified as a potential GrB substrate. Hax-1 was detected in the mitochondrial compartment of multiple cell lines, including Jurkat, MCF7, HEK293 (not shown), and Bax/Bak-deficient Jurkat (Fig. 2A, lanes 1–4). In Bax/Bak-deficient Jurkat cells, Hax-1 co-localized with GrB within a proteinase K-resistant mitochondrial fraction. Interestingly, in these Bax/Bak-deficient Jurkat cells, GrB induced significant processing of Mcl-1 but no processing of Bid and low level processing of caspase-3 to its p20 subunit. Only low levels of intermembrane space apoptogenic proteins, such as cytochrome c, were detected in Bax/Bak-deficient cells (Fig. 2A), whereas increased levels were noted in WT Jurkat or MCF7 cells (Fig. 1, F and H). In isolated mitochondria sequentially exposed to GrB and proteinase K, we detected a Hax-1 cleavage product of ∼16 kDa. This fragment did not demonstrate time-dependent accumulation in the treated mitochondria (Fig. 2B); thus, the possibility that the Hax-1 fragment was released from the mitochondria was considered. Examining the liberated proteins from isolated mitochondria exposed to GrB, we observed, in addition to cytochrome c and AIF, a granzyme-mediated, time-dependent release of both full-length Hax-1 (31 kDa) and the cleavage fragment (16 kDa) (Fig. 2C). The mitochondrial release of Hax-1 and Hax-1 fragment(s) was specific to GrB-treated mitochondria because the BH3-only proteins, tBid or Bim, which induced the release of cytochrome c and SMAC, did not cause an efflux of mitochondrial Hax-1 (data not shown). It appears that two distinct efflux mechanisms are activated in mitochondria by GrB versus BH3-only proteins; although GrB can mediate the release of Hax-1 and intermitochondrial space proteins, BH3-only proteins do not impact the mitochondrial locality of Hax-1.
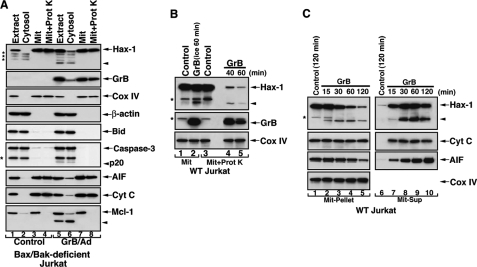
FIGURE 2.
GrB-mediated cleavage and release of mitochondrial Hax-1. A, mitochondrially localized Hax-1 is cleaved and released into the cytoplasm of GrB/Ad-treated cells. Bax/Bak-deficient Jurkat cells were treated with GrB/Ad (33 nm/10 pfu/ml, 2 h) as described in the legend to Fig. 1. The cells were then Dounce homogenized and subjected to subcellular fractionation to obtain mitochondrial and cytosolic fractions. The mitochondria were further treated with proteinase K, as described in the legend to Fig. 1. Subcellular fractions obtained from control or GrB/Ad-treated cells were assessed by immunoblotting for the expression of the indicated proteins. The low release levels of cytochrome c and AIF are probably due to the Bax/Bak-deficiency of these cells. Also, no significant processing of either Bid or caspase-3 was observed in these cells, despite marked processing of Mcl-1. B, GrB-mediated cleavage of Hax-1 within a proteinase K-resistant compartment of mitochondria. Purified mitochondria obtained from WT Jurkat cells were treated with GrB for the indicated time periods (66 nm; lane 2 on ice, lanes 4 and 5 at 37 °C). The mitochondria were then treated with proteinase K (lanes 3–5), as described in the legend to Fig. 1. Hax-1 cleavage product (arrowhead) is detected in mitochondria treated with GrB and subsequently with proteinase K (lanes 4 and 5). C, GrB mediates the release of Hax-1 and its cleaved fragment from mitochondria. Purified mitochondria obtained from WT Jurkat cells were treated with GrB, as described above. The mitochondrial pellet (Mit-Pellet) (30% of input, lanes 1–5) and supernatant (Mit-Sup) (90% of input, lanes 6–10) were separated and assessed by immunoblotting for the presence of the indicated proteins. Loss of mitochondrial full-length Hax-1 in the mitochondrial pellet is accompanied by the detection of the cleaved Hax-1 product (lanes 2–5). Release of Hax-1 and its cleavage fragment is accompanied by the release of cytochrome c and AIF (lanes 7–10). Expression of Cox IV in the mitochondrial pellet serves as a control for equal loading. The asterisks indicate unidentified protein bands.
Mapping of Hax-1 Cleavage Sites
Because GrB efficiently cleaves and activates cytosolic caspase-3, it was important to investigate whether Hax-1 cleavage is mediated by GrB-activated caspase-3. In cells treated with GrB/Ad and the broadly reactive caspase inhibitor, Z-VAD-fluoromethyl ketone, the level of Hax-1 was reduced despite the documented inhibition of caspase-3 activity (Fig. 3A). These results suggested that the reduction in the cellular level of Hax-1 was primarily due to GrB and not the activated caspase. We further demonstrated that GrB directly cleaves native Hax-1 immunoprecipitated from cellular extracts (Fig. 3B). Similarly, in vitro translated Hax-1 was processed by GrB, yielding the 16-kDa fragment that had a mobility similar to that of Hax-1 fragments obtained from GrB-treated mitochondria (Fig. 3C). We also utilized in vitro translated Hax-1 to investigate whether a 16-kDa Hax-1 fragment could be generated by caspase-3 (Fig. 3D). To this end, an immunoblot of isolated mitochondria or in vitro translated Hax-1 exposed to isolated GrB or recombinant caspase-3 showed that only GrB was able to generate the 16-kDa fragment. The activity of caspase-3 was verified by probing the same membrane for cleavage of mitochondria-associated Mcl-1, which is a substrate for both GrB and caspase-3. An autoradiographic approach showed that Hax-1 could be processed by caspase-3, but the cleavage products were distinct from the fragments generated by GrB.
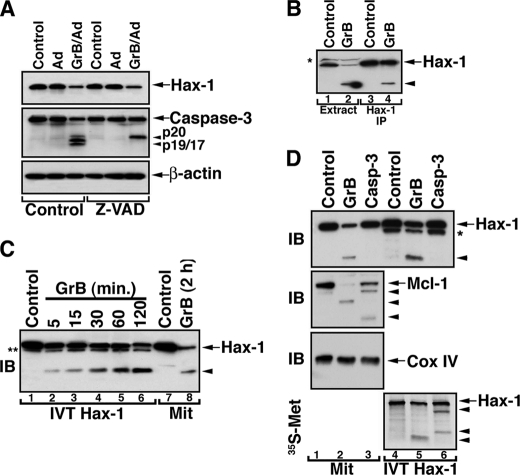
FIGURE 3.
Direct and preferential cleavage of Hax-1 by GrB. A, loss of Hax-1 expression in GrB/Ad-treated cells is not attenuated in the presence of the caspase inhibitor, Z-VAD-fluoromethyl ketone. Jurkat cells were treated with GrB/Ad (33 nm/10 pfu/ml) in the presence or absence of the pancaspase inhibitor, Z-VAD-fluoromethyl ketone (100 μm). The cells were lysed in the presence of GrB inhibitor, as described above, and assessed by immunoblotting for the expression of Hax-1, caspase-3, and β-actin. B, direct GrB cleavage of immunoprecipitated Hax-1. GrB (66 nm) was applied to the extract of Jurkat cells (lane 2) or a pellet of the protein A·anti-Hax-1·Hax-1 complex immunoprecipitated from the Jurkat cell extract (lane 4). Controls were the untreated extract or the Hax-1 immunoprecipitant (lane 1 and 3, respectively). Reaction products were resolved by SDS-PAGE and assessed by immunoblotting with anti-Hax-1 Ab. GrB acts directly on endogenous Hax-1 because immunoprecipitated (IP) Hax-1 is cleaved by GrB. C, direct GrB activity on in vitro translated Hax-1. In vitro translated Hax-1 was co-incubated with GrB (66 nm) for the indicated time periods (lanes 1–6). Lysates of control or GrB-treated mitochondria (lanes 7 and 8) were run on SDS-PAGE side by side with the in vitro translated Hax-1 to compare migration patterns. The asterisk indicates an unidentified protein band. D, mitochondrial Hax-1 is preferentially cleaved by GrB, although in vitro translated Hax-1 is also a caspase-3 substrate. Mitochondria purified from Jurkat cells (lanes 1–3) or 35S-labeled in vitro translated Hax-1 (lanes 4–6) were left untreated or treated with GrB (66 nm) or recombinant caspase-3 (100 nm). The reaction products were assessed by immunoblotting (IB) for the presence of the indicated proteins and by autoradiography for the 35S-labeled fragment(s) of in vitro translated Hax-1. Mitochondrial Hax-1 was cleaved by GrB but not by recombinant caspase-3 (top), and the GrB cleavage product had a migration pattern similar to that of GrB-treated in vitro translated (IVT) Hax-1 (lanes 2 and 5). The activity of GrB and caspase-3 was confirmed by their activity on mitochondrial Mcl-1 (second panel). Cox IV serves to demonstrate equal loading of mitochondrial proteins (third panel). Autoradiographic assessment of IVT Hax-1 cleavage products suggests that it is cleaved by either GrB or caspase-3, although different cleavage fragments are produced. The arrowheads indicate cleavage products. The asterisk indicates an unidentified protein band present in the in vitro translation mixture.
In experiments designed to map the GrB cleavage sites of Hax-1, we first performed a rate analysis of GrB-mediated cleavage of [35S]methionine-labeled in vitro translated Hax-1 (Fig. 4A). The results showed that a sufficient amount of the 16-kDa fragment would accumulate in a 3-h treatment for sequence analysis. Protein bands representing Hax-1 fragments were excised from an electroblotted membrane and subjected to Edman degradation and bioinformatic analysis (49). The major cleavage site for GrB was Asp148 (D148 in Fig. 4) with a minor cleavage site at Asp159 (D159 in Fig. 4). We then generated D148A and D159A mutant Hax-1-encoding plasmids that are suitable for in vitro translation. The WT Hax-1 and the two mutants were treated with GrB, and the reaction products were analyzed by immunoblotting and autoradiography (Fig. 4B). The D148A mutated Hax-1 has a pronounced resistance to GrB-mediated processing, whereas the D159A mutation resulted only in a partial resistance (Fig. 4B, top). The C-terminal fragment of Hax-1 (residues 149–279), which has a predicted size of ∼14 kDa (and includes a single [35S]methionine residue as compared with four such residues in the N-terminal fragment), was confirmed by tryptic digestion and MALDI mass spectrometry to be present in an excised gel band that turned out to encompass the two major Hax-1 fragments. To further confirm the identity of the cleavage sites, we generated the in vitro translated N-terminal Hax-1 fragment (residues 1–148), the corresponding C-terminal Hax-1 fragment (149–279), and the same C-terminal fragment with a D159A mutation. With these in vitro translated products, we observed that the C-terminal fragment is further cleaved at Asp159, as initially predicted (Fig. 4C). Once the two cleavage sites were mapped, we generated a double-mutant, D148A/D159A, Hax-1 construct whose corresponding in vitro translated protein demonstrated resistance to cleavage by GrB (Fig. 4D). These cleavage mapping results suggest that GrB converts full-length Hax-1 into a major N-terminal fragment (residues 1–148), which appears resistant to further degradation by GrB, and two C-terminal fragments: residues 149–279 (major) and residues 160–279 (minor).
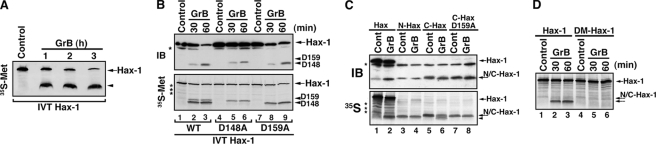
FIGURE 4.
Mapping of GrB cleavage sites in Hax-1. A, accumulation of a 16-kDa Hax-1 fragment during co-incubation with GrB. 35S-Labeled in vitro translated Hax-1 was co-incubated with GrB (100 nm) for 1–3 h. The reaction products were resolved by SDS-PAGE, and the 35S signal was assessed by autoradiography. B, GrB cleaves Hax-1 at Asp148 and Asp159. In vitro translated products of WT Hax-1 (lanes 1–3), D148A mutant Hax-1 (lanes 4–6), and D159A mutant Hax-1 (lanes 7–9) were treated with GrB, as indicated. The reaction products were resolved by SDS-PAGE and assessed by immunoblotting (IB) (top) or autoradiography (bottom). The two panels show the presence of two distinct bands that are generated by GrB cleavage: a major band that corresponds to the Asp148 cleavage site (lanes 2, 3, 8, and 9) and a minor band that corresponds to the Asp159 cleavage site (lanes 2, 3, 5, and 6). These results are in agreement with the predicted GrB Hax-1 cleavage sites by the GrB Hax-1 fragment analyses described under “Experimental Procedures.” D148 and D159 arrowheads correspond to bands generated by GrB cleavage at Asp148 and Asp159 (D148 and D159), respectively. The asterisks indicate unidentified protein bands. C, cleavage of in vitro translated C-terminal Hax-1 by GrB. 35S-Labeled in vitro translated full-length Hax-1 (lanes 1 and 2), N-terminal Hax-1 fragment (Met1–Asp148, lanes 3 and 4), C-terminal Hax-1 fragment (Ala149–Arg279, lanes 5 and 6), and D159A mutant C-terminal Hax-1 (lanes 7 and 8) were either untreated or treated with GrB (66 nm for 20 min) and subjected to SDS-PAGE followed by immunoblotting and autoradiography. The asterisks indicate nonspecific proteins. The arrowhead indicates the new C-terminal Hax-1 fragment (Trp160–Arg279) generated by GrB. Because of the close gel migration of the GrB Hax-1 fragments, they are not well distinguishable by immunoblotting or autoradiography. D, double mutant D148A/D159A Hax-1 protein is resistant to cleavage by GrB. 35S-Labeled, in vitro translated double mutant Hax-1 was treated with GrB, as described above. The reaction products of WT Hax-1 and double mutant Hax-1 were run side by side on SDS-PAGE.
Subcellular Localization of the GrB-generated Hax-1 Fragments
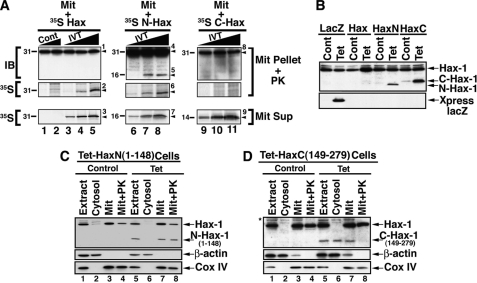
To determine whether the Hax-1 fragments had mitochondrial translocation potential, we examined the capacity of full-length Hax-1 and its N-terminal and C-terminal fragments to translocate into a proteinase K-resistant mitochondrial compartment (Fig. 5A). The full-length Hax-1 and the N-terminal Hax-1 fragment, but not the C-terminal fragment, were detected in proteinase K-treated mitochondria. To further investigate the subcellular localization of the Hax-1 fragments, we generated stably transfected T-REx-293 cells with Tet-inducible plasmids expressing either the lacZ encoded protein or the above described HAX-1-related proteins (Fig. 5B). Following Tet induction, the N-terminal Hax-1 fragment, but not the C-terminal fragment, translocated into a proteinase K-resistant mitochondrial compartment (Fig. 5, C and D).
FIGURE 5.
Subcellular localization of GrB Hax-1 cleavage products. A, translocation of in vitro translated full-length Hax-1 and the N-terminal Hax-1 fragment, but not the C-terminal Hax-1 fragment, into a proteinase K-resistant compartment of purified mitochondria. Jurkat cell-purified mitochondria were incubated with increasing doses of 35S-labeled IVT Hax-1 (lanes 3–5), 35S-labeled IVT N-terminal Hax-1 fragment (Met1–Asp148, lanes 6–8), 35S-labeled IVT C-terminal Hax-1 fragment (Ala149–Arg279, lanes 9–11), or control reaction lysate with no plasmids (lanes 1 and 2). The mitochondrial supernatant (Mit Sup) was separated, and the mitochondrial pellet (Mit Pellet) was treated with proteinase K. Following the addition of a proteinase K inhibitor and three washes, the mitochondrial pellet fractions were lysed and assessed for the presence of Hax-1-related proteins by immunoblotting and autoradiography. The identified proteins are indicated by numbered arrowheads as follows. 1, endogenous Hax-1 plus exogenous IVT Hax-1; 2 and 3, 35S-labeled IVT Hax-1; 4, endogenous Hax-1; 5–7, exogenous IVT N-terminal Hax-1 fragment; 8, endogenous Hax-1; 9, exogenous IVT C-terminal Hax-1 fragment. The molecular weight for each of the detected proteins is indicated on the left of each panel. B, inducible expression of N- and C-terminal fragments of Hax-1. T-REx-293 clonal cell lines stably transfected with Tet-inducible lacZ, Tet-inducible N-terminal Hax-1 fragment (residues 1–148), or Tet-inducible C-terminal Hax-1 fragment (residues 149–279) were treated with tetracycline (1 μg/ml, 16 h). The cells were lysed and assessed by immunoblotting for the expression of the Hax-1-related proteins or Xpress epitope (used as a tag for lacZ-encoded protein). Of note, The N-terminal Hax-1 fragment consistently demonstrated a faster migration pattern than C-terminal Hax-1 fragment, despite the lower predicted molecular mass of the latter. C and D, mitochondrial localization of the N-terminal, but not the C-terminal, GrB Hax-1 fragment. T-REx-293 clonal cell lines stably transfected with Tet-inducible N-terminal (C) or C-terminal Hax-1 fragment (D) were treated with tetracycline (1 μg/ml, 16 h). Control and treated cells were Dounce homogenized and subjected to subcellular fractionation to obtain cytosolic and mitochondrial fractions. The mitochondria were treated with proteinase K as described under “Experimental Procedures.” β-Actin and Cox IV serve as markers for the subcellular fractions and to demonstrate loading at a proportional cell ratio.
Mitochondrial Depolarization Function of the N-terminal Hax-1 Fragment
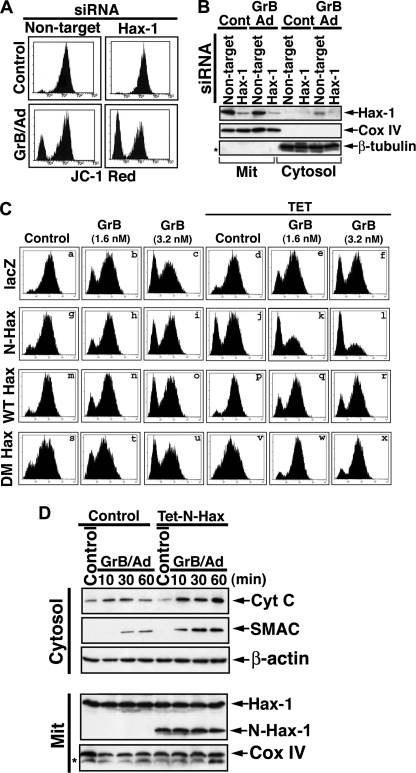
We then assessed the effects of the induced expression or knockdown of Hax-1 on the MMP of quiescent or GrB/Ad-treated cells. Hax-1 knockdown did not alter the steady-state MMP of quiescent cells, but it increased the sensitivity to GrB-mediated mitochondrial depolarization (Fig. 6, A and B). Induced expression of double mutant, uncleavable Hax-1 and also of WT Hax-1 protected the mitochondria against GrB-mediated depolarization (Fig. 6C). In contrast, the induced expression of the N-terminal fragment (Fig. 6C (j) and supplemental Fig. S5), but not the C-terminal fragment (not shown), resulted in significant mitochondrial depolarization. Furthermore, the presence of the N-terminal Hax-1 fragment markedly enhanced the mitochondrial depolarization induced by GrB (Fig. 6C, k and l). Thus, the N-terminal Hax-1 fragment appears to negate the positive effect of endogenous full-length Hax-1 on mitochondrial Δψm.
FIGURE 6.
The mitochondrial activity of GrB is enhanced by knockdown of Hax-1 and by the presence of the N-terminal Hax-1 fragment but inhibited by induced expression of WT or the uncleavable form of Hax-1. A and B, increased GrB-mediated mitochondrial depolarization following Hax-1 knockdown. Hct116 cells were treated with Hax-1 siRNA followed by treatment with GrB/Ad (33 nm/10 pfu/ml). Changes in Δψm were assessed by JC-1 flow cytometry (A), and the levels of expression of Hax-1 in the cells were assessed by immunoblotting (B). Because Hax-1 localizes mainly to mitochondria but is released from mitochondria treated with GrB, the mitochondrial and cytosolic fractions were separated to assess for the potential presence of Hax-1 in the cytosol. Cox-IV and β-tubulin serve as equal loading controls and purification markers for the mitochondrial and the cytosolic fractions, respectively. C, inhibition of GrB-mediated mitochondrial depolarization by induced expression of uncleavable double mutant Hax-1 and sensitization to such depolarization by induced expression of N-terminal Hax-1 fragment. T-REx-293 clonal cell lines stably transfected with Tet-inducible lacZ control, full-length Hax-1, double-mutant Hax-1, or the N-terminal Hax-1 fragment (residues 1–148) were treated with tetracycline (1 μg/ml, 16 h) and with GrB/Ad (1.6 or 3.2 nm/10 pfu/ml for 2 h). The cells were then assessed for changes in mitochondrial Δψm as indicated by JC-1 staining. The N-terminal Hax-1-mediated mitochondrial depolarization is shown in j; its sensitization to GrB-mediated depolarization is shown in k and l; and the double mutant (DM) Hax-1 inhibition of GrB-mediated depolarization is shown in w and x. Similar results were obtained in at least four independent experiments. D, GrB-mediated release of mitochondrial apoptogenic proteins is enhanced in cells overexpressing the N-terminal Hax-1 fragment. T-REx-293 cells stably transfected with Tet-inducible N-terminal Hax-1 fragment were kept as controls or treated with tetracycline and subsequently with GrB/Ad (33 nm/10 pfu/ml) for the indicated time periods. Following the addition of a GrB inhibitor, the cells were Dounce homogenized and fractionated to mitochondrial and cytosolic fractions. These subcellular fractions were assessed by immunoblotting for the expression of the indicated proteins.
To determine whether the N-terminal Hax-1 fragment enhances the mitochondrial apoptotic response to GrB, we evaluated the effect of the expressed fragment on GrB-mediated release of mitochondrial apoptogenic proteins, cytochrome c and SMAC. Control and Tet-treated cells stably transfected with N-terminal Hax-1 fragment were treated with GrB/Ad for 10–60 min, and the cytosolic levels of cytochrome c and SMAC were assessed. Expression of the N-terminal Hax-1 fragment did indeed enhance the level of GrB-mediated cytochrome c and SMAC release (Fig. 6D). Thus, the N-terminal Hax-1 fragment induces mitochondrial depolarization that further sensitizes the cells to GrB-mediated cell death.
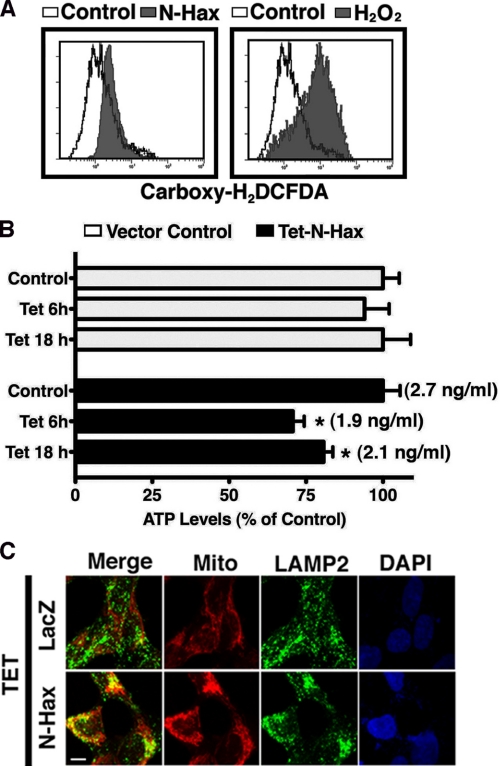
To further characterize the consequences of N-Hax-1-induced expression on mitochondrial physiology, changes in the levels of endogenous ROS and ATP were measured (Fig. 7, A and B). Flow cytometric analysis of carboxy-H2DCFDA, a ROS-sensitive membrane-permeant probe that becomes fluorescent upon oxidation, demonstrated a significant increase in oxidative stress in cells with Tet-induced expression of N-Hax-1 fragment. As a positive control for generation of oxidative stress, the cells were treated for 1 h with hydrogen peroxide. Induced expression of the N-Hax-1 fragment also led to a mild (∼30%) ATP depletion relative to the untreated control during the early period (6 h) of the tetracycline treatment, but the levels were slowly reduced (to approximately 20% of control during the following 12 h) (Fig. 7B).
FIGURE 7.
Mitochondrial alterations following Tet-induction of the N-terminal Hax-1 fragment. A, increased expression of the N-terminal Hax-1 fragment is associated with increased ROS production. T-REx-293 cells stably transfected with Tet-inducible N-terminal Hax-1 fragment were treated with tetracycline (1 μg/ml, 16 h, left) or with H2O2 (0.2 mm 1 h, right) and assessed by flow cytometry for carboxy-H2DCFDA fluorescence. B, increased expression of the N-terminal Hax-1 fragment is associated with ATP depletion. T-REx-293 cells stably transfected with Tet-inducible N-terminal Hax-1 fragment were treated with tetracycline (1 μg/ml) for the indicated time periods and assessed for cellular ATP concentration using the ATPLite Assay System (PerkinElmer Life Sciences). C, enhanced co-localization of mitochondria and lysosomes following Tet induction of the N-terminal Hax-1 fragment. T-REx-293 cells stably transfected with Tet-inducible lacZ or N-terminal Hax-1 fragment were treated with tetracycline (1 μg/ml, 16 h) and assessed by confocal microscopy for a merge between MitoTracker Deep Red and anti-LAMP2 Ab detected by a secondary green fluorescent Ab. Scale bar, 40 μm. Using the Metamorph colocalization method described under “Experimental Procedures,” it was determined that 18% of the MitoTracker labeled mitochondria colocalized with LAMP2 in tetracycline-treated lacZ control cells, and the colocalization level rose to 55% following Tet induction of the N-Hax-1 fragment. A higher magnification of this staining is shown in supplemental Fig. S6. Please note that the MitoTracker is utilized only for mitochondrial identification; this dye is not diagnostic for the level of mitochondrial polarization because its mitochondrial binding depends both on mitochondrial membrane potential and lipophilicity. Each of the panels represents results confirmed by at least three independent experiments. DAPI, 4′,6-diamidino-2-phenylindole.
Because induced expression of N-terminal Hax-1 did not mediate apoptosis on its own, we examined the cell response to the increased presence of depolarized mitochondria that was associated with enhanced ROS production and ATP depletion. Damaged mitochondria, particularly depolarized mitochondria, have been reported to be eliminated by lysosomic degradation (50, 51). Indeed, utilizing confocal microscopy, we observed in cells with induced expression of the N-Hax-1 fragment an increased co-localization of mitochondria and LAMP2, a lysosome-specific protein (Fig. 7C; higher magnification shown in supplemental Fig. S6). These findings suggest that the mere presence of the N-terminal Hax-1 fragment is not sufficient for the induction of cell death and that the consequent lysosomic elimination of damaged mitochondria potentially sustains cell survival.
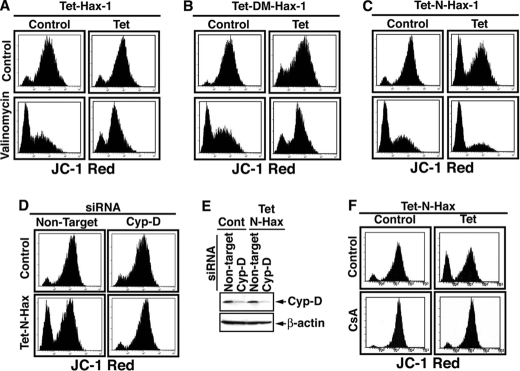
The depolarizing effect of the N-terminal Hax-1 fragment argues that this cleavage product acts as a dominant negative of endogenous full-length Hax-1. To test this possibility, we examined the effects of Tet-induced Hax-1, Tet-induced double mutant Hax-1, and Tet-induced N-terminal Hax-1 fragment on valinomycin-mediated mitochondrial depolarization. Valinomycin is a member of a group of natural neutral ionophores that disrupt the transmembrane ion concentration gradient. Induced expression of WT Hax-1 and of double mutant Hax-1 protected against valinomycin-mediated depolarization, whereas induced N-terminal Hax-1 fragment mediated depolarization on its own (Fig. 8, A–C). These results suggest that within mitochondria, Hax-1 functions to stabilize the membrane potential, and its cleavage by GrB generates an N-terminal fragment that is not only dysfunctional in stabilizing the mitochondrial ΔΨm, but also functions on its own to disrupt the membrane potential.
FIGURE 8.
N-terminal Hax-1 fragment functions as a dominant negative form of Hax-1 in the destabilization of MMP in a Cyp-D-dependent manner. A–C, WT Hax-1 and double mutant Hax-1, but not the N-terminal Hax-1(1–148) fragment, protects mitochondria against valinomycin-mediated depolarization. T-REx-293 cells stably transfected with Tet-inducible WT Hax-1 (A), DM-Hax-1 (B), or N-terminal Hax-1 fragment (C) were treated with tetracycline (1 μg/ml) for 16 h. The cells were then treated with valinomycin (100 nm, 2 h) and assessed by flow cytometry for mitochondrial membrane potential as measured by JC-1 staining. D–F, the mitochondrial depolarization effect of the N-terminal Hax-1 fragment is Cyp-D-dependent. D and E, T-REx-293 cells stably transfected with Tet-inducible N-terminal Hax-1 fragment were treated with Cyp-D or non-target siRNAs (Ambion) for 60 h. The cells were then treated with tetracycline (1 μg/ml) for 6 h and assessed by flow cytometry for JC-1 staining (D) and by immunoblotting for the expression level of Cyp-D (E). Similar results were obtained with three additional non-overlapping Cyp-D siRNAs from Invitrogen (not shown). F, T-REx-293 cells stably transfected with the Tet-inducible N-terminal Hax-1 fragment were treated with tetracycline (1 μg/ml) and CsA (1 μm) for 16 h and then assessed by flow cytometry for JC-1 staining.
Recent studies in knock-out mice have elucidated the function of Cyp-D in regulating the opening of the PTP (18, 20). Therefore, we examined whether Cyp-D is involved as an effector molecule downstream of Hax-1. To this end, we induced the expression of the N-terminal Hax-1 fragment while knocking down Cyp-D. Knockdown of Cyp-D alone did not have a significant effect on the steady-state MMP. However, the depolarizing effect of the induced N-terminal Hax-1 fragment was markedly inhibited in cells with reduced expression of Cyp-D (Fig. 8, D–E). The involvement of Cyp-D in the depolarizing effect of the N-terminal Hax-1 fragment was confirmed with four distinct non-overlapping Cyp-D siRNAs. Cyclosporine A (CsA), an inhibitor of mitochondrial depolarization, was recently demonstrated to specifically target Cyp-D because PTPs in mitochondria from Cyp-D knock-out mice lose their CsA sensitivity (18, 20). Thus, to verify that Cyp-D participated in the mitochondrial depolarization mediated by the N-terminal Hax-1 fragment, the effect of CsA was examined. Indeed, the presence of CsA completely inhibited the depolarizing activity of the Tet-induced N-terminal Hax-1 fragment (Fig. 8F). No changes in the MMP were observed in cells with induced expression of the C-terminal Hax-1 fragment in the presence or absence of CsA (not shown). Thus, inhibition of Cyp-D by knockdown or by CsA abolishes the depolarizing effect of induced N-terminal Hax-1 fragment.
DISCUSSION
The mitochondrial apoptotic events detected in GrB-treated cells have been linked to the function of substrates cleaved by GrB in the cytoplasm or at the outer mitochondrial membrane. The possibility that GrB might engage a cytotoxic pathway by cleaving intramitochondrial substrates has not been described. The current study has identified the mitochondrion as a unique locus for GrB activity following its translocation beyond the mitochondrial outer membrane. The biological relevance of these findings is indicated by the rapid intramitochondrial accumulation of GrB following its intracellular delivery within the estimated time frame required for the release of GrB from giant endosomes (43). Therefore, although GrB is delivered to and functions in the cytoplasm, its ultimate destination appears to be mitochondrial.
The identification of putative GrB substrates within the mitochondrion requires the cautious elimination of GrB itself and its potential substrates outside or at the mitochondrial outer membrane, as achieved in the current study by proteinase K treatment of mitochondria. To ensure that the identified substrates are cleaved by GrB and not by proteinase K, all experiments were performed with and without subsequent treatment of mitochondria with proteinase K. In addition, to exclude potential involvement of an intermediate cytosolic enzyme that might be activated when whole cells are subjected to GrB treatment, we utilized purified mitochondria that, following their direct GrB exposure, were treated with an effective concentration of proteinase K that eliminated mitochondrial outer membrane proteins as well as the GrB source. Because GrB was applied directly on mitochondria, the significance of all identified substrates needed to be further verified through their degradation by cell internalized GrB as well as by the kinetics of their degradation. Such corroborating experiments were performed for both Hax-1 and AIF.
Although mitochondrial insertion of GrB has not been previously reported, a recent seminal study (52) demonstrated the insertion of another granzyme, GrA, into the mitochondrial matrix, when it is applied directly on isolated mitochondria or together with perforin onto target cells. The resistance of Hax-1 to mitochondrial treatment with proteinase K in association with its release from GrB-treated mitochondria are in line with previous reports on the potential localization of Hax-1 on the mitochondrial inner membrane facing the intermembrane space (28). Thus, Hax-1 cross-talk with the mitochondrial matrix protein, Cyp-D, may take place on the mitochondrial inner membrane. In any event, the direct function of GrB on both Hax-1 and AIF lends support to the notion that GrB functions both within the mitochondrial intermembrane space (AIF locality) and on the inner membrane (Hax-1 locality).
Hax-1 has recently been reported to contribute to the stabilization of the MMP, but the underlying mechanism remains unknown (23). Overexpression of full-length Hax-1 and particularly of uncleavable Hax-1 protects the mitochondria from GrB-mediated depolarization. In contrast, reduction in the expression level of Hax-1, as achieved by RNA interference, increases the depolarization susceptibility of mitochondria to GrB. Furthermore, cleavage of Hax-1 by GrB does not only eliminate the presence of protective full-length Hax-1 but also generates an N-terminal Hax-1 fragment that possesses a functional activity opposing that of Hax-1. Specifically, this mitochondria-localized Hax-1 fragment does not possess the protective activity of full-length Hax-1 against depolarizing stimuli and on its own further dissipates the MMP and enhances ROS production and ATP depletion. The involvement of Cyp-D in the mitochondrial depolarizing activity of the N-terminal Hax-1 fragment serves as evidence that indeed the latter participates in regulation of the mitochondrial PTP. This finding is important not only in regard to GrB mitochondrial function but also in regard to the molecular mechanism involved in the function of Hax-1 itself. The underlying mechanism for the cross-talk between Hax-1 and Cyp-D is unknown and currently under investigation. The finding that the regulatory machinery of the mitochondrial PTP is an immediate but indirect attack target for GrB represents a novel concept in GrB biology. Not only does GrB put in motion a proteolytic cascade by activating cytosolic caspase-3, but it is also capable of targeting the ultimate cell survival core: the mitochondrial PTP.
Although Hax-1 has been considered as a prosurvival protein, its significance in GrB cellular function remains to be elucidated. Exogenous application of GrB with either perforin or Ad onto in vitro cell cultures leads to cell death associated with the degradation of multiple cellular targets of GrB. Indeed, the significance of GrB substrates as intermediate death effectors, including caspase-3, Bid, and Mcl-1, has been elucidated via the their contribution to the ultimate demise of the targeted cell. It is noteworthy that Hax-1 appears to be a highly sensitive target for GrB, yet its overexpression or elimination does not significantly impact the ultimate death following in vitro application of GrB. The protective effect of Hax-1 appears to be limited to the mitochondria, and mitochondria that are depolarized by the GrB-generated N-Hax-1 fragment appear to be targeted to lysosomes for degradation. Recently, an important aspect of GrB activity that is not associated with promoting target cell death has been recognized (1). Thus, GrB/perforin have been implicated in an inhibitory activity of CD8 lymphocytes against reactivation of HSV-1 in infected target cells. In particular, GrB was shown to degrade HSV-1 immediate early protein, ICP4, which is essential for further viral gene expression. In this experimental setting, the GrB-mediated degradation of ICP4 was not associated with the death of the infected cells. An equivalent mechanism of antipathogen activity with continuous survival of the target cell has been described for GrH (53). Future studies should investigate such scenarios for Hax-1, which has been identified as a binding target for proteins made by different viruses, including Epstein-Barr virus, Kaposi's sarcoma associated herpes virus, and human immunodeficiency virus, type 1 (32–35).
Although the significance of Hax-1 as a substrate in GrB antiviral activity remains to be determined, the ability of GrB to function within the mitochondria represents a novel concept in GrB research. With our current knowledge regarding the rationale behind GrB selectivity for its cellular substrates, the discovery that GrB can secure cellular demise by an immediate attack on the cell energy generator should not come as a surprise but rather as an expected finding.
Supplementary Material
Acknowledgment
We thank Dr. David Malehorn (University of Pittsburgh Proteomics Core) for help with the two-dimensional PAGE.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA109285 and RO1 CA111786 (to H. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- GrB
- granzyme B
- PTP
- permeability transition pore
- Cyp-D
- cyclophilin D
- MMP
- mitochondrial membrane potential
- Ad
- adenovirus
- Ab
- antibody
- Z-
- benzyloxycarbonyl-
- WT
- wild-type
- siRNA
- small interfering RNA
- MALDI
- matrix-assisted laser desorption ionization
- MIB
- mitochondrial buffer
- MOPS
- 4-morpholinepropanesulfonic acid
- ROS
- reactive oxygen species
- JC-1
- 1,1′,3,3′-tetraethylbenzamidazolocarbocyanin iodide
- carboxy-H2DCFDA
- 5-(and 6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate
- CsA
- cyclosporine A
- pfu
- plaque-forming units
- IVT
- in vitro translated
- RNAi
- RNA interference
- AIF
- apoptosis-inducing factor.
REFERENCES
- 1.Knickelbein J. E., Khanna K. M., Yee M. B., Baty C. J., Kinchington P. R., Hendricks R. L. (2008) Science 322, 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieberman J., Fan Z. (2003) Curr. Opin. Immunol. 15, 553–559 [DOI] [PubMed] [Google Scholar]
- 3.Packard B. Z., Telford W. G., Komoriya A., Henkart P. A. (2007) J. Immunol. 179, 3812–3820 [DOI] [PubMed] [Google Scholar]
- 4.Barry M., Bleackley R. C. (2002) Nat. Rev. Immunol. 2, 401–409 [DOI] [PubMed] [Google Scholar]
- 5.Bots M., Medema J. P. (2006) J. Cell Sci. 119, 5011–5014 [DOI] [PubMed] [Google Scholar]
- 6.Buzza M. S., Bird P. I. (2006) Biol. Chem. 387, 827–837 [DOI] [PubMed] [Google Scholar]
- 7.Casciola-Rosen L., Garcia-Calvo M., Bull H. G., Becker J. W., Hines T., Thornberry N. A., Rosen A. (2007) J. Biol. Chem. 282, 4545–4552 [DOI] [PubMed] [Google Scholar]
- 8.Heibein J. A., Goping I. S., Barry M., Pinkoski M. J., Shore G. C., Green D. R., Bleackley R. C. (2000) J. Exp. Med. 192, 1391–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waterhouse N. J., Sedelies K. A., Trapani J. A. (2006) Immunol. Cell Biol. 84, 72–78 [DOI] [PubMed] [Google Scholar]
- 10.Pardo J., Wallich R., Martin P., Urban C., Rongvaux A., Flavell R. A., Müllbacher A., Borner C., Simon M. M. (2008) Cell Death Differ. 15, 567–579 [DOI] [PubMed] [Google Scholar]
- 11.Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H. (2000) Cell Death Differ. 7, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 12.Han J., Goldstein L. A., Gastman B. R., Froelich C. J., Yin X. M., Rabinowich H. (2004) J. Biol. Chem. 279, 22020–22029 [DOI] [PubMed] [Google Scholar]
- 13.Han J., Goldstein L. A., Gastman B. R., Rabinovitz A., Rabinowich H. (2005) J. Biol. Chem. 280, 16383–16392 [DOI] [PubMed] [Google Scholar]
- 14.Zong W. X., Lindsten T., Ross A. J., MacGregor G. R., Thompson C. B. (2001) Genes. Dev. 15, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas D. A., Scorrano L., Putcha G. V., Korsmeyer S. J., Ley T. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14985–14990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goping I. S., Sawchuk T., Rieger A., Shostak I., Bleackley R. C. (2008) Blood 111, 2142–2151 [DOI] [PubMed] [Google Scholar]
- 17.Tsujimoto Y., Nakagawa T., Shimizu S. (2006) Biochim. Biophys. Acta. 1757, 1297–1300 [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa T., Shimizu S., Watanabe T., Yamaguchi O., Otsu K., Yamagata H., Inohara H., Kubo T., Tsujimoto Y. (2005) Nature 434, 652–658 [DOI] [PubMed] [Google Scholar]
- 19.Kokoszka J. E., Waymire K. G., Levy S. E., Sligh J. E., Cai J., Jones D. P., MacGregor G. R., Wallace D. C. (2004) Nature 427, 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baines C. P., Kaiser R. A., Purcell N. H., Blair N. S., Osinska H., Hambleton M. A., Brunskill E. W., Sayen M. R., Gottlieb R. A., Dorn G. W., Robbins J., Molkentin J. D. (2005) Nature 434, 658–662 [DOI] [PubMed] [Google Scholar]
- 21.Baines C. P., Kaiser R. A., Sheiko T., Craigen W. J., Molkentin J. D. (2007) Nat. Cell Biol. 9, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galluzzi L., Kroemer G. (2007) Nat. Cell Biol. 9, 487–489 [DOI] [PubMed] [Google Scholar]
- 23.Klein C., Grudzien M., Appaswamy G., Germeshausen M., Sandrock I., Schäffer A. A., Rathinam C., Boztug K., Schwinzer B., Rezaei N., Bohn G., Melin M., Carlsson G., Fadeel B., Dahl N., Palmblad J., Henter J. I., Zeidler C., Grimbacher B., Welte K. (2007) Nat. Genet. 39, 86–92 [DOI] [PubMed] [Google Scholar]
- 24.Cilenti L., Soundarapandian M. M., Kyriazis G. A., Stratico V., Singh S., Gupta S., Bonventre J. V., Alnemri E. S., Zervos A. S. (2004) J. Biol. Chem. 279, 50295–50301 [DOI] [PubMed] [Google Scholar]
- 25.Suzuki Y., Demoliere C., Kitamura D., Takeshita H., Deuschle U., Watanabe T. (1997) J. Immunol. 158, 2736–2744 [PubMed] [Google Scholar]
- 26.Han Y., Chen Y. S., Liu Z., Bodyak N., Rigor D., Bisping E., Pu W. T., Kang P. M. (2006) Circ. Res. 99, 415–423 [DOI] [PubMed] [Google Scholar]
- 27.Kostmann R. (1956) Acta. Paediatr. Suppl. 45, 1–78 [PubMed] [Google Scholar]
- 28.Chao J. R., Parganas E., Boyd K., Hong C. Y., Opferman J. T., Ihle J. N. (2008) Nature 452, 98–102 [DOI] [PubMed] [Google Scholar]
- 29.Vafiadaki E., Arvanitis D. A., Pagakis S. N., Papalouka V., Sanoudou D., Kontrogianni-Konstantopoulos A., Kranias E. G. (2009) Mol. Biol. Cell 20, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vafiadaki E., Sanoudou D., Arvanitis D. A., Catino D. H., Kranias E. G., Kontrogianni-Konstantopoulos A. (2007) J. Mol. Biol. 367, 65–79 [DOI] [PubMed] [Google Scholar]
- 31.Zhao W., Waggoner J. R., Zhang Z. G., Lam C. K., Han P., Qian J., Schroder P. M., Mitton B., Kontrogianni-Konstantopoulos A., Robia S. L., Kranias E. G. (2010) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi Y., Nakajima K., Igarashi M., Morita T., Tanaka M., Suzuki M., Yokoyama A., Matsuda G., Kato K., Kanamori M., Hirai K. (2000) J. Virol. 74, 10104–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp T. V., Wang H. W., Koumi A., Hollyman D., Endo Y., Ye H., Du M. Q., Boshoff C. (2002) J. Virol. 76, 802–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yedavalli V. S., Shih H. M., Chiang Y. P., Lu C. Y., Chang L. Y., Chen M. Y., Chuang C. Y., Dayton A. I., Jeang K. T., Huang L. M. (2005) J. Virol. 79, 13735–13746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda G., Nakajima K., Kawaguchi Y., Yamanashi Y., Hirai K. (2003) Microbiol. Immunol. 47, 91–99 [DOI] [PubMed] [Google Scholar]
- 36.Hanna W. L., Zhang X., Turbov J., Winkler U., Hudig D., Froelich C. J. (1993) Protein Expr. Purif. 4, 398–404 [DOI] [PubMed] [Google Scholar]
- 37.Han J., Goldstein L. A., Gastman B. R., Rabinovitz A., Wang G. Q., Fang B., Rabinowich H. (2004) Leukemia 18, 1671–1680 [DOI] [PubMed] [Google Scholar]
- 38.Han J., Goldstein L. A., Hou W., Rabinowich H. (2007) J. Biol. Chem. 282, 16223–16231 [DOI] [PubMed] [Google Scholar]
- 39.Machida K., Ohta Y., Osada H. (2006) J. Biol. Chem. 281, 14314–14320 [DOI] [PubMed] [Google Scholar]
- 40.Han J., Goldstein L. A., Gastman B. R., Rabinowich H. (2006) J. Biol. Chem. 281, 10153–10163 [DOI] [PubMed] [Google Scholar]
- 41.Browne K. A., Blink E., Sutton V. R., Froelich C. J., Jans D. A., Trapani J. A. (1999) Mol. Cell. Biol. 19, 8604–8615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Froelich C. J., Orth K., Turbov J., Seth P., Gottlieb R., Babior B., Shah G. M., Bleackley R. C., Dixit V. M., Hanna W. (1996) J. Biol. Chem. 271, 29073–29079 [DOI] [PubMed] [Google Scholar]
- 43.Pipkin M. E., Lieberman J. (2007) Curr. Opin. Immunol. 19, 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G. Q., Wieckowski E., Goldstein L. A., Gastman B. R., Rabinovitz A., Gambotto A., Li S., Fang B., Yin X. M., Rabinowich H. (2001) J. Exp. Med. 194, 1325–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talanian R. V., Yang X., Turbov J., Seth P., Ghayur T., Casiano C. A., Orth K., Froelich C. J. (1997) J. Exp. Med. 186, 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besenicar M. P., Metkar S., Wang B., Froelich C. J., Anderluh G. (2008) Biochem. Biophys. Res. Commun. 371, 391–394 [DOI] [PubMed] [Google Scholar]
- 47.Susin S. A., Lorenzo H. K., Zamzami N., Marzo I., Snow B. E., Brothers G. M., Mangion J., Jacotot E., Costantini P., Loeffler M., Larochette N., Goodlett D. R., Aebersold R., Siderovski D. P., Penninger J. M., Kroemer G. (1999) Nature 397, 441–446 [DOI] [PubMed] [Google Scholar]
- 48.Pospisilik J. A., Knauf C., Joza N., Benit P., Orthofer M., Cani P. D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C. R., Kroemer G., Rustin P., Burcelin R., Penninger J. M. (2007) Cell 131, 476–491 [DOI] [PubMed] [Google Scholar]
- 49.Backes C., Kuentzer J., Lenhof H. P., Comtesse N., Meese E. (2005) Nucleic Acids. Res. 33, W208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang H. T., Hwang E. S. (2009) Aging Cell 8, 426–438 [DOI] [PubMed] [Google Scholar]
- 51.Kim I., Rodriguez-Enriquez S., Lemasters J. J. (2007) Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinvalet D., Dykxhoorn D. M., Ferrini R., Lieberman J. (2008) Cell 133, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrade F., Fellows E., Jenne D. E., Rosen A., Young C. S. (2007) EMBO J. 26, 2148–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.