Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a Cl− channel physiologically important in fluid-transporting epithelia and pathologically relevant in several human diseases. Here, we show that mutations in the C terminus of the first nucleotide binding domain comprising the latest β strands (βc5 and βc6) influence the trafficking, channel activity, and pharmacology of CFTR. We mutated CFTR amino acids located in the βc5-βc6 hairpin, within the βc5 strand (H620Q), within the β-turn linking the two β strands (E621G, G622D), as well as within (S623A, S624A) and at the extremity (G628R) of the βc6 strand. Functional analysis reveals that the current density was largely reduced for G622D and G628R channels compared with wt CFTR, similar for E621G and S624A, but increased for H620Q and S623A. For G622D and G628R, the abnormal activity is likely due to a defective maturation process, as assessed by the augmented activity and mature C-band observed in the presence of the trafficking corrector miglustat. In addition, in presence of the CFTR activator benzo[c]quinolizinium, the CFTR current density compared with that of wt CFTR was abolished for G622D and G628R channels, but similar for H620Q, S623A, and S624A or slightly increased for E621G. Finally, G622D and G628R were activated by the CFTR agonists genistein, RP-107, and isobutylmethylxanthine. Our results identify the C terminus of the CFTR first nucleotide binding domain as an important molecular site for the trafficking of CFTR protein, for the control of CFTR channel gating, and for the pharmacological effect of a dual activity agent.
Keywords: ABC Transporter, Chloride Channels, Cystic Fibrosis, Protein Folding, Protein Structure, CFTR, NBD1, Pharmacology
Introduction
Mapping ligand binding sites within ionic channel structures is important to understand the mechanism of action of channel modulators (i.e. activators and inhibitors), to identify regions required for selective interaction with modulators, and to design more selective and more potent agents. Such mapping, called pharmaco-topology, has been performed for the sulfonylurea receptor (SUR) regulatory subunits of KATP channels (reviewed in Ref. 1), which are members of the ATP-binding cassette (ABC)3 transporter superfamily (2). The drug binding sites for several modulators (benzopyran, cromakalim; pyridine, pinacidil; sulfonylurea, glibenclamide, tolbutamide; benzothiadiazine, diazoxide) have been mapped to different predicted transmembrane segments within SUR1 and SUR2A (3). More recently, the analysis of the experimental three-dimensional structure of P-glycoprotein, another ABC transporter, in an inward-facing conformation revealed a large internal cavity formed from two bundles of six transmembrane helices, which may accommodate several compounds simultaneously (4).
For chloride (Cl−) channels in general and for the cystic fibrosis transmembrane conductance regulator (CFTR) protein in particular, such pharmaco-topology is missing. CFTR (ABCC7) also belong to the ABC transporter superfamily (2) and forms a Cl− channel whose gating depends on the intracellular equilibrium of cAMP and ATP amounts (5–8). Furthermore, there is a complex and fine tuned regulation between the phosphorylation/dephosphorylation of the unique regulatory (R)-domain and the fundamental ABC regulation mode by ATP binding/hydrolysis occurring on NBDs (nucleotide binding domains 1 and 2). Because mutations of the CFTR gene caused cystic fibrosis (CF), several studies have been conducted aiming at identifying original, selective, potent, and therapeutically active small molecules. Despite the fact that numerous and diverse chemical scaffolds are now available, no pharmacophore has been identified, and surprisingly, very few drug binding sites have been mapped on the CFTR protein. However, studies with the CFTR activator isoflavonoid genistein highlighted the contribution of glycine residues Gly551 and Gly1349 of NBD1 and NBD2 domains, respectively, to the pharmacological modulation of CFTR by genistein (4, 9). Mechanism of inhibition of CFTR activity by glibenclamide or CFTRinh-172 are associated with a positive charge of amino acids located on the third intracellular loop (Lys918) and pore-forming transmembrane domains (Arg347) respectively (4, 10).
Among the modulators of CFTR, some benzo[c]quinolizinium compounds (MPBs), such as the 5-butyl-6-hydroxy-10-chlorobenzo[c]quinolizinium chloride (MPB-91), are dual activity compounds able to rescue misprocessed CFTR mutants at the apical membrane (11) and to stimulate the Cl− channel activity of CFTR selectively (12). MPB-91 prevented the degradation of a polypeptide comprising NBD1 and the R-domain of F508del CFTR (13). These findings indicate that MPB-91 influences protein trafficking and stimulates the opening of CFTR Cl− channels. Although a direct interaction between the two has not yet been demonstrated, these findings suggest that the mechanism of action is associated with some putative sites or amino acids located most probably on the cytoplasmic domains of CFTR.
The alignment of the NBD sequences of human CFTR with those of various ABC transporters revealed conserved amino acids in this region, outside those belonging to motifs involved in ATP binding and hydrolysis (14). Among them there are two glycine residues in position 622 and 628 located at the C terminus of NBD1, within a β hairpin formed by the two latest β strands (βc5 and βc6, c indicating the participation of these two β strands into the NBD1 β sheet core) of the domain (Fig. 1). Interestingly, when individually mutated, these amino acids cause CF (15). We have mutated these two glycine residues in aspartic acid (G622D) and arginine (G628R) and considered other mutants in their neighborhood (H620Q, E621G, S623A, and S624A). To understand better the importance of this region, the activity and pharmacology of wild-type (wt) and mutated CFTR proteins were explored.
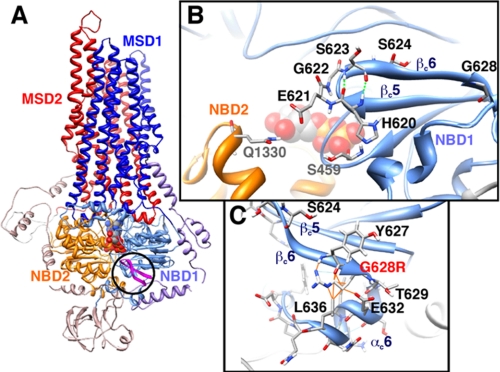
FIGURE 1.
β5c/β6c hairpin within the model of the three-dimensional structure of human CFTR. A, global view of the model of human CFTR in an outward-facing conformation (open channel) in which the β5c/β6c hairpin (in pink) discussed here is circled. B, focus on the β5c/β6c hairpin and on the amino acids, which are mutated in this study. His620, Glu621, Gly622, and Ser623 are the four residues (i to i+3) of the β turn, with His620 and Ser623 making two hydrogen bonds between their NH and O atoms (dashed green lines). Neighbors of His620 (Ser459) and Glu621 (Gln1330) are also shown. C, focus on the Gly628 position, which was mutated in the three-dimensional model in an arginine residue (G628R), highlighting the steric clashes (orange lines), which would be associated with this mutation.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293 cells were cultured in Dulbecco's modified Eagle's medium + GlutaMAX (Invitrogen) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. BHK-21 cells were cultured in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) with 5% SVF and 1% penicillin/streptomycin. Both cell lines were grown in standard conditions (37 °C, 5% CO2).
CFTR Mutagenesis
Mutations were introduced by PCR using the oligonucleotide-directed mutagenesis system (QuikChangeXL Site-directed Mutagenesis kit; Stratagene, La Jolla, CA) into the pEGFP CFTRwt (human cDNA) (16) as described previously (9). Mutagenesis primers are depicted in supplemental Table 1. Mutations in individual clones were verified through plasmid sequencing on both strands using the ABI PRISM Big Dye TerminatorTM Cycle sequencing Ready Reaction kit (Applied Biosystems, Courtaboeuf, France). Reactions were run on an ABI PRISM 310 automatic sequencer.
Transient Transfection of Mutant CFTR
HEK293 cells (for patch clamp and Western blot analysis) and BHK-21 cells (for iodide efflux experiments) were transfected with the corresponding cDNA construct using cationic lipids (JetPEI; QBiogene Illkirch, France) with 0.2 μg/ml plasmid for the patch clamp method, 0.5 μg/well for iodide efflux, and 1 μg/ml plasmid for Western blot analysis. Media were replaced 24 h after transfection.
Western Blot Analysis
HEK293 cell lysates (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 100 mm NaCl, 1% Triton X-100, 20 μm leupeptin, 10 μm pepstatin, 0.8 μm aprotinin, 2.1 mm 4-(2-aminoethyl) benzensulfonyl fluoride hydrochloride (AEBSF) were separated 48 h after transfection by 5% SDS-PAGE (50 μg of protein/lane). Membrane was incubated overnight at 4 °C in phosphate-buffered saline 0.1% Tween 20 with 4 μg/ml mouse anti-CFTR monoclonal antibody (clone MAB3480; Chemicon International, Millipore Bioscience Research Reagents, Temecula, CA). Horseradish peroxidase-conjugated sheep anti-mouse IgG (1:10,000; Amersham Biosciences) was used as secondary antibody and revealed with ECL Western blotting detection reagent (Millipore).
Patch Clamp Experiments
Ionic currents were measured in the broken-patch, whole cell configuration of the patch clamp method using an EPC-7 amplifier (List Electronic, Darmstadt, Germany). The holding potential was −40 mV in all whole cell experiments. Current/voltage (I/V) relationships were built by clamping the membrane potential to −40 mV and by pulses from −100 to +100 mV in 20 mV increments. Pipettes were prepared by pulling borosilicate glass capillary tubes (GC150-TF10; Clark Electromedical Inc., Reading, UK) using a two-step vertical puller (Narishige Tokyo, Japan). They were connected to the head stage of the patch clamp amplifier through an Ag-AgCl pellet (pipette resistance of 3–5 MΩ). Pipette capacitance was electronically compensated in cell-attached mode. Membrane capacitance and series resistances were measured in the whole cell mode by fitting capacitance currents, obtained in response to a hyperpolarization of 40 mV, with a first-order exponential and by integrating the surface of the capacitance current. Voltage clamp signals, allowing the membrane potential to be held at different values, were recorded via a microcomputer equipped with an analog/digital-digital/analog conversion board (Digidata 1200 interface; Axon Instruments, Burlingame, CA). Results were analyzed with pCLAMP version 9 software (Axon Instruments). The external bath solution contained 145 mm NaCl, 4 mm CsCl, 1 mm CaCl2, 1 mm MgCl2, 10 mm glucose, and 10 mm tetradecyl sulfate (titrated with NaOH to pH 7.4). The osmolarity was 315 ± 5 mOsmol. The intrapipette solution contained 113 mm l-aspartic acid, 113 mm CsOH, 27 mm CsCl, 1 mm NaCl, 1 mm MgCl2, 1 mm ethyleneglycoltetraacetic acid, 1 mm tetradecyl sulfate, and 3 mm MgATP (ex temporane) (titrated with CsOH to pH 7.2). The osmolarity was 285 ± 5 mOsmol. The pipette solution was always hypotonic (with respect to the bath solution) to prevent cell swelling and activation of the volume-sensitive chloride channels. All experiments were conducted at room temperature (20–25 °C). The calculated chemical equilibrium potential for chloride (ECl−) is −42 mV. Cells expressing GFP-CFTR proteins were visually identified on the patch clamp setup by green fluorescence detected by the CKX41 microscope supplied by the Olympus (Tokyo, Japan) Reflected Fluorescence system.
Iodide Efflux Experiments
CFTR chloride channel activity was assayed by measuring the rate of iodide (125I) efflux versus time from BHK-21 cells at 72 h after transfection. All experiments were performed and analyzed as described previously (17). Iodide efflux curves were constructed by plotting rate (k, min−1) of 125I versus time. All comparisons were based on maximal values for the time-dependent rate (k = peak rates, min−1), excluding the point used to establish the baseline (kpeak − kbasal, min−1).
Chemicals
All MPB compounds were synthesized in our laboratory as described previously (12, 18). We used MPB-91 (5-butyl-10-chloro-6-hydroxybenzo[c]quinolizinium chloride), MPB-95 (10-chloro-6-hydroxy-5-isobutylbenzo[c]quinolizinium chloride), and MPB-97 (10-chloro-6-hydroxy-5-pentylbenzo[c]quinolizinium chloride). Forskolin (Fsk) was purchased from LC Laboratories (PKC Pharmaceuticals, Woburn, MA). CFTRinh-172 (19) was from Calbiochem. All other products were from Sigma. Stock solutions of MPB and Fsk (100 mm), CFTRinh-172 (10 mm) were prepared in dimethyl sulfoxide. Miglustat was from Toronto Chemical Research and was dissolved in water (20).
Statistics
Results are expressed as means ± S.E. of n observations. To compare activation rate of channels, the slope of each I/V relationship was calculated. Data were compared using Student's t test. Differences were considered statistically significant at p < 0.05. All statistical tests were performed using GraphPad Prism version 4.0 for Windows (GraphPad Software).
RESULTS
Expression of the NBD1 C-terminal CFTR Mutants
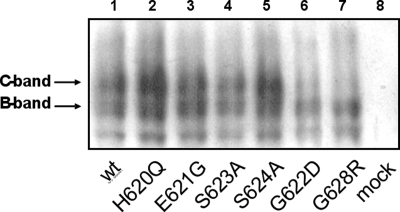
We have introduced EGFP-tagged CFTR proteins into HEK293 cells, wt CFTR and six CFTR mutants, i.e. H620Q, E621G, G622D, S623A, S624A, and G628R. EGFP-CFTR proteins were detected by Western blot analysis using antibodies directed against the CFTR NBD2 C terminus (Fig. 2, lanes 1–7). Control experiments were performed using nontransfected HEK293 cells (Fig. 2, lane 8). For the expression of CFTR mutants H620Q, E621G, S623A, and S624A, the profiles of core-glycosylated and mature-glycosylated forms were similar to that of the wt protein. However, no mature-glycosylated C-band form for G622D and G628R was detected (Fig. 2, lanes 6 and 7).
FIGURE 2.
Western blot analysis of wt and mutant CFTR channels. Western blot shows the EGFP-CFTR expression detected with CFTR NBD2 C-terminal antibody. B-band, core-glycosylated EGFP-CFTR; C-band, mature-glycosylated EGFP-CFTR.
Functional Analysis of the NBD1 C Terminus CFTR Mutants
We explored the chloride (Cl−) channel function for each mutated CFTR protein using the whole cell configuration of the patch clamp technique. Fig. 3A shows representative whole cell current traces recorded in HEK293 cells expressing either wt or CFTR mutants as indicated. Voltage pulse protocols were applied in the basal conditions, in presence of 10 μm Fsk or 10 μm Fsk + 10 μm CFTRinh-172. The thiazolidinone blocker CFTRinh-172 was used systematically to selectively inhibit CFTR channels (19).
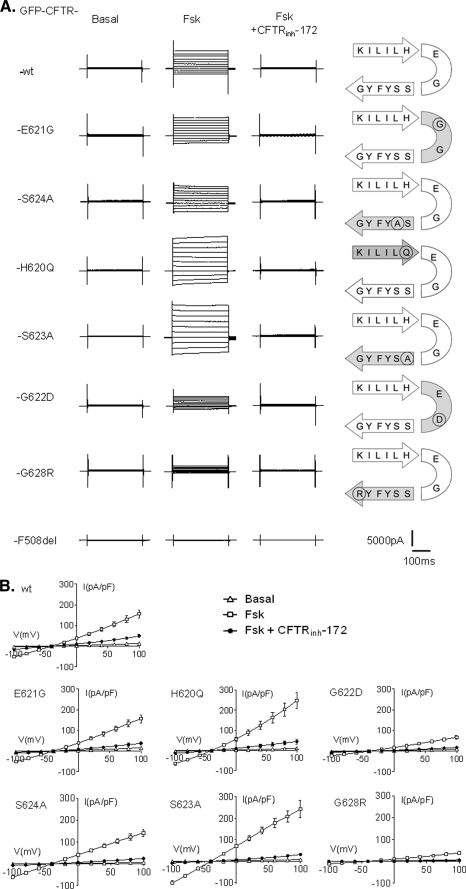
FIGURE 3.
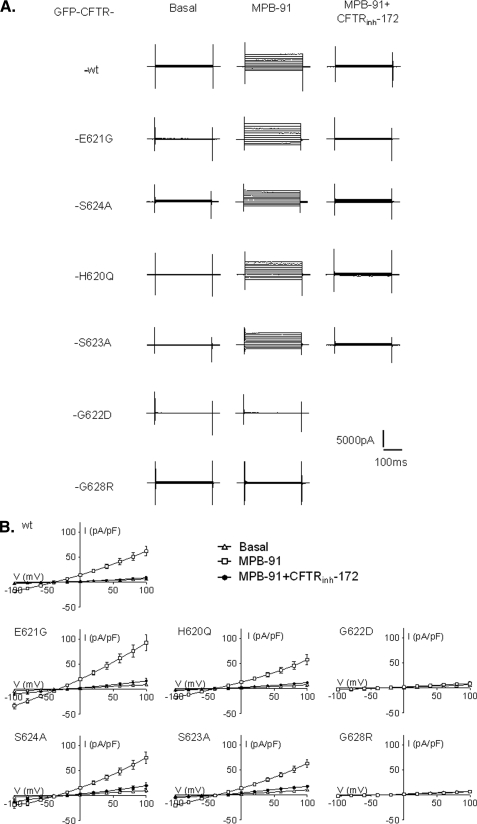
Effect of Fsk and CFTRinh-172 on the whole cell Cl− currents recorded for each CFTR mutant. A, representatives traces of whole cell Cl− currents elicited by stepping from holding potential of −40 mV to a series test potentials from −100 to +100 mV in 20-mV increments. B, corresponding current densities (pA/pF) obtained by current-voltage relationships normalized by cell capacitance (▵, basal; □, 10 μm Fsk; ●, 10 μm Fsk + 10 μm CFTRinh-172). Schemes on the right side indicate the position of each amino acid and its substitution on the β5c/β6c hairpin. Error bars, S.E. The bottom traces were recorded in F508del-expressing cells.
Except for F508del (Fig. 3A, bottom traces), 10 μm Fsk elicited a time- and voltage-independent current for all other mutated proteins and was fully blocked by CFTRinh-172 (Fig. 3). However some differences could be observed, and the comparison of current densities highlights three types of responses. First, Cl− currents recorded for wt CFTR, E621G, and S624A CFTR mutants were not significantly different (mean current densities for each construct are reported in supplemental Table 2). Second, with the two mutations introduced on both sides of the β turn, H620Q and S623A, we recorded an increased (p < 0.001) Cl− current (supplemental Table 2) compared with wt CFTR. Third, the current densities for G622D and G628R, although not abolished, are both significantly reduced (p < 0.001) compared with wt (supplemental Table 2).
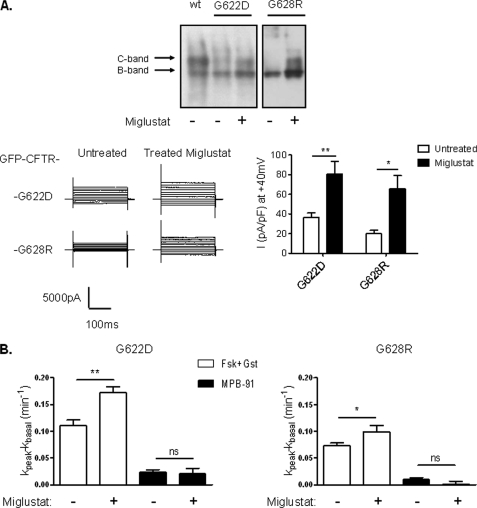
Some mutations of CFTR generated proteins with abnormal trafficking along the biosynthesis pathway. This is the case for F508del which trafficking is arrested in the endoplasmic reticulum, and subsequently the abnormal protein is conducted in the ubiquitin/proteasome degradation pathway (21). To overcome this traffic jam, correctors are in development (22). Among them, miglustat partially restored the abnormal location of F508del CFTR in epithelial cells (20, 23). Because we observed a pronounced reduction of the current density for the mutants G622D and G628R, we incubated transfected HEK293 and BHK-21 cells with this corrector and analyzed the corresponding CFTR activity. For both mutants, the Cl− current density measured in HEK293 cells was significantly increased by a treatment with the corrector (Fig. 4A). Similarly in BHK-21 cells the iodide efflux responses stimulated by Fsk and genistein was significantly increased for G622D and G628R after treatment with the corrector (Fig. 4B). This observation indicates that the diminution of cAMP-induced Cl− current is probably due to the diminution or to the absence of a mature form of G622D and G628R mutants. In support of this hypothesis, Western blot analysis of cells treated with miglustat shows enhanced mature C-band for G622D and G628R mutants (Fig. 4A).
FIGURE 4.
Effect of the CFTR corrector miglustat on G622D and G628R. A, upper, Western blot showing wt-CFTR, G622D-, and G628R-CFTR expression with or without pretreatment with miglustat (100 μm) and detected with CFTR NBD2 C-terminal antibody. Band B, core-glycosylated and band C, mature-glycosylated. Lower, representative traces of whole cell Cl− currents elicited by stepping from holding potential of −40 mV to a series test potential from −100 to +100 mV in 20 mV increments (left) and histograms of the corresponding current densities at +40 mV (right). Error bars, S.E. B, bar chart showing 10 μm Fsk + 30 μm Gst and 100 μm MPB-91 dependent iodide efflux in BHK transfected cells treated or not during 4 hours with 100 μm miglustat. n = 4 for each. ns, nonsignificant difference; *, p < 0.05; **, p < 0.01. Error bars, S.E.
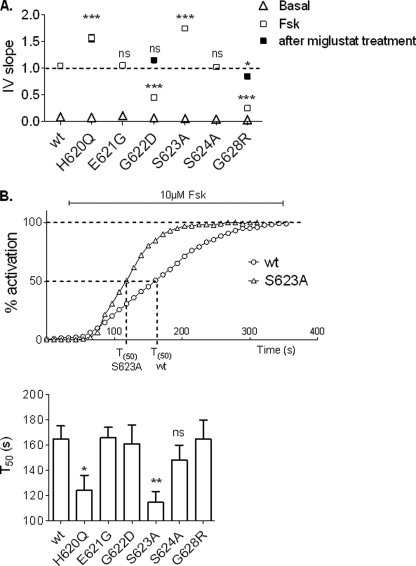
For each CFTR construct we calculated the I/V slope, which reflects the global conductance. Results confirmed the difference of Cl− current densities observed previously (Fig. 5A). Concerning the activation kinetics, only the two CFTR mutants with an increased Cl− transport activity (H620Q and S623A) also present a faster time course of activation (Fig. 5B). We calculated the time to obtain 50% of the maximum activation level (T50) after addition of 10 μm Fsk. T50 of the two mutated channels (T50 = 124.45 ± 8.8 s for H620Q, n = 8 and T50 = 114.63 ± 8.8 s for S623A, n = 8) were significantly different compared with wt (T50 = 164.85 ± 10.2 s, n = 8). Our results indicated that H620Q and S623A channels could be activated more rapidly than the other CFTR mutant channels studied.
FIGURE 5.
Analysis of the global Fsk-activated Cl− currents recorded for each CFTR mutant. A, I/V slope of current/voltage relationships. Dotted line corresponds to the wt level. B, upper, representatives time course of wt and S623A mutant in the presence of 10 μm Fsk. Lower, histograms representing mean of time (s) to reach 50% activation for each mutant. ns, nonsignificant difference; *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with wt. Error bars, S.E.
Pharmacological Analysis of the NBD1 C-terminus CFTR Mutants
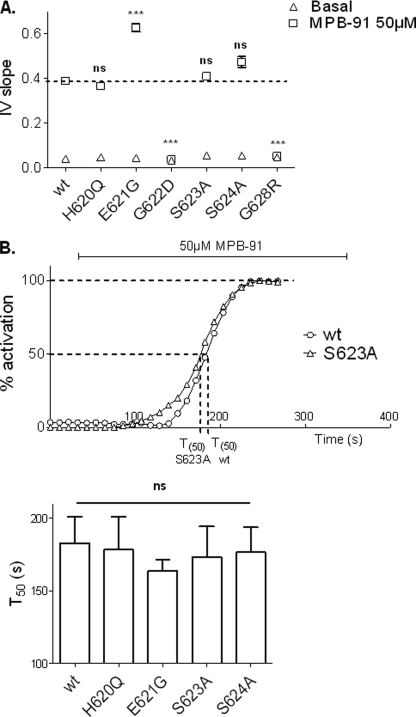
In a second series of experiments, we tested MPB-91 on wt protein and on each individual CFTR mutant. Representative whole cell current traces recorded in basal condition, in the presence of 50 μm MPB-91, or 50 μm MPB-91 + 10 μm CFTRinh-172, and the corresponding I/V curves are presented Fig. 6. As expected, wt CFTR channels activated by MPB elicited a linear and CFTRinh-172-sensitive Cl− current. However, in the presence of MPB the current density of wt CFTR (31.54 ± 4.69 pA/pF at +40 mV, n = 6; supplemental Table 3) was 2.5-fold less than the cAMP-activated Cl− current (see Fig. 3 and supplemental Table 2). Interestingly, although the level of current for H620Q and S623A was greater than the response of wt channels (Fig. 3), both mutated CFTRs were activated by MPB-91 to a level similar to that of wt CFTR (Fig. 6) (at 40 mV, current densities were: H620Q, 27.86 ± 4.2 pA/pF, n = 5; S623A, 30.83 ± 3.4 pA/pF, n = 6). On the contrary, a significant increase of I/V slope (Fig. 7A) in presence of MPB-91 was observed for E621G CFTR channels compared with wt (I/V slope of wt, 0.390 ± 0.014, n = 6; E621G, 0.628 ± 0.018, n = 8). No statistically significant difference was observed concerning the activation kinetics (Fig. 7B). MPB-91 stimulated the Cl− current for each CFTR mutants studied except the glycine mutants G622D and G628R (Fig. 6 and supplemental Table 3). For them, comparison of the corresponding IV slope (Fig. 7A) did not reveal significant differences between basal condition (I/V slope of G622D, 0.032 ± 0.001, n = 7; G628R, 0.046 ± 0.002, n = 5) and in presence of 50 μm MPB-91 (I/V slope of G622D, 0.037 ± 0.001, n = 7; G628R, 0.053 ± 0.003, n = 5) indicating the absence of a response of the two mutated channels to that agent.
FIGURE 6.
Effect of MPB-91 and CFTRinh-172 on the whole cell Cl− currents for each CFTR mutant. A, representatives traces of whole cell Cl− currents elicited by stepping from holding potential of −40 mV to a series test potentials from −100 to +100 mV in 20-mV increments. B, corresponding current densities (pA/pF) obtained by I/V relationships normalized by cell capacitance (▵, basal; □, 50 μm MPB-91; ●, 50 μm MPB-91 + 10 μm CFTRinh-172). Error bars, S.E.
FIGURE 7.
Analysis of the global MPB-activated Cl− currents recorded for each CFTR mutant. A, I/V slope of current/voltage relationships. Dotted line corresponds to the wt level. B, upper, representatives time course of wt and S623A mutant in presence of 50 μm MPB-91. Lower, histograms representing the mean of time corresponding to 50% of activation for each mutant. ns, nonsignificant difference. ***, p < 0.001 compared with wt. Error bars, S.E.
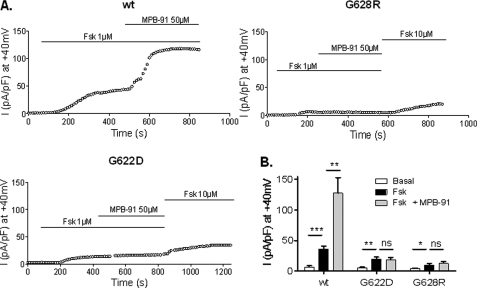
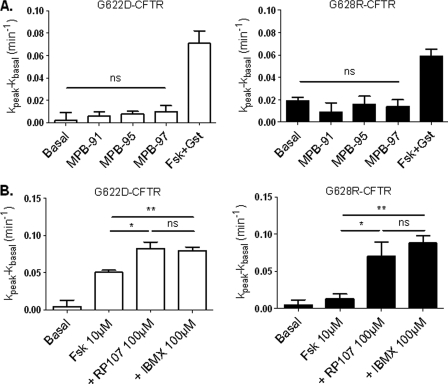
We have also tested MPB-91 on CFTR channels after phosphorylation of the R-domain promoted by 1 μm Fsk. Results in Fig. 8 show a potentiation of the cAMP-dependent Cl− current by MPB-91 for wt channels but neither for G622D nor G628R mutants. These results were confirmed by the determination of the mean current densities at 40 mV (Fig. 8B) showing no significant differences before or after addition of MPB-91 to cells expressing the two mutant proteins in the presence of Fsk. We also tested analogues of MPB-91 (12), but again for the mutant G622D or G628R, the Cl− channel function of CFTR was not stimulated by MPB-95 and MPB-97 (Fig. 9A). Finally, we tested three other compounds with a chemical structure unrelated to that of MPB, i.e. the isoflavone genistein (24), the pyrrolo[2,3-b]pyrazine RP-107 (25), and the xanthine derivative isobutylmethylxanthine (26). Importantly, all of these CFTR activators stimulated the Cl− channel activity of G622D and G628R CFTR, suggesting the relative specificity of the effect observed in the presence of the benzoquinolizinium drugs (Fig. 9B).
FIGURE 8.
Kinetic of activation of whole cell Cl− currents for wt, G622D, and G628R CFTR. A, representative time course of whole cell Cl− current densities at 40 mV recorded in presence of 1 μm Fsk and 1 μm +50 μm MPB-91. B, histograms of the corresponding means of global current densities. ns, nonsignificant difference; *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E.
FIGURE 9.
Iodide efflux of G622D and G628R CFTR-expressing cells in the presence of different MPB compounds or different activators. A, bar chart showing iodide efflux in BHK-21-transfected cells stimulated by 10 μm Fsk + 30 μm genistein (Gst) or 100 μm MPBs. n = 4 for each. B, bar chart showing iodide efflux in BHK-21-transfected cells stimulated by 10 μm Fsk, 10 μm Fsk + 100 μm RP-107, or 10 μm Fsk + 100 μm isobutylmethylxanthine (IBMX). n = 4 for each. ns, nonsignificant difference; *, p < 0.05; **, p < 0.01. Error bars, S.E.
DISCUSSION
In the present work we have identified the C terminus of NBD1 as an important region with structural features that are essential for CFTR trafficking and amino acids that are involved in the mechanism of activation of CFTR channels as well as in the pharmacological effect of the CFTR agonists and MPB dual activity compounds. The C-terminal region of NBD1 and in particular the fragment comprising amino acids 622–634 have already been shown to be crucial for the maturation process of CFTR proteins (27). Using Western blot analysis, we now observed a strong decrease of the mature-glycosylated form for the two mutants G622D and G628R. This suggests a defective maturation process due to impairment of the folding process or to a lower stability of the folded protein and highlights the importance of these two amino acids for this process. However, despite the defective maturation, we recorded a cAMP-dependent Cl− current, indicating a plasma membrane expression of these mutants.
Stabilization of C-terminal NBD1 Region and CFTR Maturation
We considered the structural information provided by the models of the three-dimensional structure of CFTR (14, 28, 29) to evaluate the possible impact of the mutations studied herein (Fig. 1A). All of the mutated amino acids belong to a β hairpin, formed by two adjacent β strands (strands βc5 and βc6) (Fig. 1B). The two antiparallel β strands are linked by a type I′ β turn (defined as four consecutive residues allowing the polypeptide chain folding back on itself; Fig. 1B). Because of its unique ϕ-ψ torsion angles, located in a characteristic region on the Ramachandran plot, the third residue of a type I′ β turn is always a glycine; substitution of this glycine by any other residue would cause steric hindrance. In CFTR, substitution of this third residue, Gly622 (ϕ = 70°, ψ = 4°) by an aspartic acid would thus perturb the local structure or even affect the folding process itself.
Although Gly622 is located in a turn participating in the interface between the two NBDs, Gly628 is located on the other side of the domain, at the extremity of the last β strand (βc6), which is immediately followed by the last helix of the domain (Fig. 1C). Gly628 is buried within the NBD1 structure, and substitution of this amino acid by an arginine, possessing a long side chain, would lead to steric hindrance with the NBD1 last helix. Consistently, the defect in the maturation process induced by the G622D and G628R mutations, which causes the retention of the mutated CFTR protein, can be clearly explained by the key role of these glycine residues at the structure level. However, mutation of these glycine residues also affects the Cl− function of the channels. Although the trafficking of the mutated proteins was improved by the corrector miglustat, the level of cAMP-dependent Cl− current remained clearly below that of wt channels under the stimulation by Fsk. Therefore, stabilization of this region encompassing the Gly622 and Gly628 seems to be required for correct trafficking and chloride channel functions.
On the contrary, mutations of the amino acids located on both sides of the β hairpin type I′ turn should not affect the local structure or folding process, as assessed by the mature glycosylated forms of the mutated proteins. Instead and interestingly, some of these mutations impacted on the channel gating. In particular, the cAMP-activated Cl− current densities elicited by H620Q and S623A channels were increased compared with wt. Moreover, the variation of the time to reach the maximum activation level indicates that the mutated channels could be activated more rapidly than wt. For H620Q mutant, this increase could be explained by the reported increased of open probability (15). These two amino acids correspond to the first (i) and fourth (i+3) residues of the type I′ turn and form through their main chain N and O atoms the two first hydrogen bonds of the antiparallel β strands (Fig. 1B). The change of the lateral chain of one of them, resulting from their substitution by another amino acid, might not, in principle, disturb these bonds and should thus not destabilize the local structure of the β hairpin. According to the model of the CFTR three-dimensional structure, the side chain of His620, however, is in tight contact with Ser459, located in the walker A motif (P-loop) of NBD1, participating in the noncanonical ATP binding site (Fig. 1B). The H620Q mutation might thus disturb the contacts existing between the β hairpin and the P-loop of NBD1 and thus might result in a perturbation of the channel gating through an effect on the ATP binding and/or hydrolysis in the noncanonical site. One could hypothesize that an enhancement of ATP binding and/or decrease of ATP hydrolysis at this site might favor the open state of the channel. On the contrary, mutation of the neighbor amino acids (Glu621 and Ser624) did not modify the kinetic parameters of the mutated channels. Their side chains do not establish any particular contacts with critical parts of the molecule, and consequently, their mutation in another amino acid should not alter CFTR function. Indeed, our recordings of Cl currents supported by CFTR mutants E621G and S624A were not different from the non-mutated channels stimulated either by Fsk or by MPB agents.
NBD1 C Terminus and Drug Interacting Sites
After having explored the functionality of the mutated channel, we studied the effect of MPB-91 on these mutants. As expected, MPB-91 was able to activate wt CFTR in presence or in absence of Fsk. In addition, MPB-91 was also able to activate H620Q, E621G, S623A, and S624A mutants. The Cl− current densities elicited by these mutants, except for E621G, were similar to wt. Contrary to the activation by Fsk, no difference of kinetic parameters was detected with the two mutants H620Q and S623A. This suggests an activation pathway independent of the cAMP-activating pathway. In contrast, for the two mutants G622D and G628R, no activation was recorded in the presence of MPB-91 or other MPB compounds. One could hypothesize that the absence of response might be due to a folding or stability defect, according to the structural key role of Gly622 and Gly628. Thus, the lack of sensitivity of the mutated channels to MPB would be unspecific, and in this case, we should therefore have also observed a similar lack of effect with other CFTR modulators. However, this is not the case since xanthine (isobutylmethylxanthine), RP-107, or isoflavonoide (genistein) successfully stimulated the channel activity of G622D and G628R CFTR channels. Therefore, one could hypothesize that the mechanism of activation of CFTR by MPB was itself affected by the mutations G622D and G628R and that the binding site of MPB might be located, on the folded protein, in the vicinity of the last β hairpin of CFTR NBD1. However, the perturbation induced by the G622D and G628R mutations on the overall NBD1 structure might be felt at long range and thus influence potential binding sites, which may be distant at the three-dimensional level.
For the CFTR E621G mutant, Cl− current activated by MPB-91 was increased compared with wt. Glu621 corresponds to the second residue of the type I′ β turn and participates in the NBD heterodimer interface (Fig. 1B). The head-to-tail conformation of the two NBDs implies that, in the open form of the CFTR channel (outward-facing conformation) (14, 28), the ATP binding sites are formed by the Walker A and Walker B motifs of one NBD and the ABC signature sequence of the other NBD. A sliding of the two NBDs has been predicted to occur when CFTR evolves toward a closed form of the channel (inward-facing conformation) (29) in which, according to the conformation observed of a closed-apo conformer in MsbA, the two Walker A/Walker B motifs face each other. It is interesting to note that Glu621 might interact with an amino acid located in the NBD1 α subdomain (Gln1330; open channel, Fig. 1B) or with a NBD2 P-loop residue (Arg1245; closed channel, data not shown). Thus, one might hypothesize that the E621G mutation may reinforce the effect of MPB-91 by acting on critical features of the NBD interface, for example by contributing to reinforce it.
Dual Activity Pharmacological Agents
Development of dual activity compounds is an attractive option for a pharmacological therapy in CF. Indeed a single molecule could be used for rescuing CFTR misprocessed mutants at the cellular membrane and for restoring its Cl− channel function once located at the apical plasma membrane. The molecular mechanism of action of dual activity compounds is unknown despite the fact it remains a major challenge for development of more active and less toxic lead compounds. We showed that the dual activity MPB compounds do not alter directly the intracellular levels of cAMP or ATP (18) or modulate the ATPase activity of NBD domains (30), findings suggesting that the effect of MPB could be explained by a direct interaction with CFTR channel. We also demonstrated that MPB-91 in an in vitro assay prevented the degradation of proteins comprising NBD1 and R-domain of F508del CFTR (13). Contacts between the different domains of CFTR are essential to the maturation and activity of CFTR. For instance, the contacts established between the intracellular loops of the membrane spanning domains and the NBDs (28, 31), in which Phe508 participates, are critical for the interdomain assembly and for the regulation of channel gating (31–33). The dimeric organization of the two NBDs is also a key feature for an optimal catalytic activity of CFTR (34). In view of the present results concerning MPB and CFTR, it is tempting to hypothesize that MPB might promote or stabilize these crucial interdomain interactions. Altogether our findings that the β hairpin, formed by two adjacent β strands (strands βc5 and βc6) influences the trafficking and the channel activity of CFTR and that the pharmacological effects of MPB are affected by mutating the β hairpin support the dual activity nature of these compounds. This region of CFTR could thus be targeted to design more selective and more potent dual activity CFTR modulators.
Supplementary Material
Acknowledgment
We thank Sandra Mirval for technical assistance.
This work was supported by grants from Vaincre La Mucoviscidose, MucoVie, and ABCF2.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3.
- ABC
- ATP-binding cassette
- R-domain
- regulator domain
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- NBD
- nucleotide binding domain
- MPB
- benzo[c]quinolizinium compound
- wt
- wild type
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- Fsk
- forskolin
- T50
- time to obtain 50% of the maximum activation level
- pF
- picofarad.
REFERENCES
- 1.Babenko A. P., Aguilar-Bryan L., Bryan J. (1998) Annu. Rev. Physiol. 60, 667–687 [DOI] [PubMed] [Google Scholar]
- 2.Dean M., Rzhetsky A., Allikmets R. (2001) Genome Res. 11, 1156–1166 [DOI] [PubMed] [Google Scholar]
- 3.Babenko A. P., Gonzalez G., Bryan J. (2000) J. Biol. Chem. 275, 717–720 [DOI] [PubMed] [Google Scholar]
- 4.Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., Chang G. (2009) Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aleksandrov A. A., Aleksandrov L. A., Riordan J. R. (2007) Pflugers Arch. 453, 693–702 [DOI] [PubMed] [Google Scholar]
- 6.Anderson M. P., Gregory R. J., Thompson S., Souza D. W., Paul S., Mulligan R. C., Smith A. E., Welsh M. J. (1991) Science 253, 202–205 [DOI] [PubMed] [Google Scholar]
- 7.Gadsby D. C., Nairn A. C. (1999) Physiol. Rev. 79, S77–S107 [DOI] [PubMed] [Google Scholar]
- 8.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 9.Melin P., Thoreau V., Norez C., Bilan F., Kitzis A., Becq F. (2004) Biochem. Pharmacol. 67, 2187–2196 [DOI] [PubMed] [Google Scholar]
- 10.Melin P., Hosy E., Vivaudou M., Becq F. (2007) Biochim. Biophys. Acta 1768, 2438–2446 [DOI] [PubMed] [Google Scholar]
- 11.Dormer R. L., Dérand R., McNeilly C. M., Mettey Y., Bulteau-Pignoux L., Métayé T., Vierfond J. M., Gray M. A., Galietta L. J., Morris M. R., Pereira M. M., Doull I. J., Becq F., McPherson M. A. (2001) J. Cell Sci. 114, 4073–4081 [DOI] [PubMed] [Google Scholar]
- 12.Marivingt-Mounir C., Norez C., Dérand R., Bulteau-Pignoux L., Nguyen-Huy D., Viossat B., Morgant G., Becq F., Vierfond J. M., Mettey Y. (2004) J. Med. Chem. 47, 962–972 [DOI] [PubMed] [Google Scholar]
- 13.Stratford F. L., Pereira M. M., Becq F., McPherson M. A., Dormer R. L. (2003) Biochem. Biophys. Res. Commun. 300, 524–530 [DOI] [PubMed] [Google Scholar]
- 14.Callebaut I., Eudes R., Mornon J. P., Lehn P. (2004) Cell Mol. Life Sci. 61, 230–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vankeerberghen A., Wei L., Jaspers M., Cassiman J. J., Nilius B., Cuppens H. (1998) Hum. Mol. Genet. 7, 1761–1769 [DOI] [PubMed] [Google Scholar]
- 16.Moyer B. D., Loffing J., Schwiebert E. M., Loffing-Cueni D., Halpin P. A., Karlson K. H., Ismailov I. I., Guggino W. B., Langford G. M., Stanton B. A. (1998) J. Biol. Chem. 273, 21759–21768 [DOI] [PubMed] [Google Scholar]
- 17.Derand R., Bulteau-Pignoux L., Becq F. (2002) J. Biol. Chem. 277, 35999–36004 [DOI] [PubMed] [Google Scholar]
- 18.Becq F., Mettey Y., Gray M. A., Galietta L. J., Dormer R. L., Merten M., Métayé T., Chappe V., Marvingt-Mounir C., Zegarra-Moran O., Tarran R., Bulteau L., Dérand R., Pereira M. M., McPherson M. A., Rogier C., Joffre M., Argent B. E., Sarrouilhe D., Kammouni W., Figarella C., Verrier B., Gola M., Vierfond J. M. (1999) J. Biol. Chem. 274, 27415–27425 [DOI] [PubMed] [Google Scholar]
- 19.Ma T., Thiagarajah J. R., Yang H., Sonawane N. D., Folli C., Galietta L. J., Verkman A. S. (2002) J. Clin. Invest. 110, 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norez C., Noel S., Wilke M., Bijvelds M., Jorna H., Melin P., DeJonge H., Becq F. (2006) FEBS Lett. 580, 2081–2086 [DOI] [PubMed] [Google Scholar]
- 21.Ward C. L., Omura S., Kopito R. R. (1995) Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 22.Becq F. (2010) Drugs 70, 241–259 [DOI] [PubMed] [Google Scholar]
- 23.Norez C., Antigny F., Noel S., Vandebrouck C., Becq F. (2009) Am. J. Respir. Cell Mol. Biol. 41, 217–225 [DOI] [PubMed] [Google Scholar]
- 24.Illek B., Fischer H., Santos G. F., Widdicombe J. H., Machen T. E., Reenstra W. W. (1995) Am. J. Physiol. Cell Physiol. 268, C886–C893 [DOI] [PubMed] [Google Scholar]
- 25.Noel S., Faveau C., Norez C., Rogier C., Mettey Y., Becq F. (2006) J. Pharmacol. Exp. Ther. 319, 349–359 [DOI] [PubMed] [Google Scholar]
- 26.Chappe V., Mettey Y., Vierfond J. M., Hanrahan J. W., Gola M., Verrier B., Becq F. (1998) Br. J. Pharmacol. 123, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan K. W., Csanády L., Seto-Young D., Nairn A. C., Gadsby D. C. (2000) J. Gen. Physiol. 116, 163–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mornon J. P., Lehn P., Callebaut I. (2008) Cell Mol. Life Sci. 65, 2594–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mornon J. P., Lehn P., Callebaut I. (2009) Cell Mol. Life Sci. 66, 3469–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dérand R., Bulteau-Pignoux L., Mettey Y., Zegarra-Moran O., Howell L. D., Randak C., Galietta L. J., Cohn J. A., Norez C., Romio L., Vierfond J. M., Joffre M., Becq F. (2001) Am. J. Physiol. Cell Physiol. 281, C1657–C1666 [DOI] [PubMed] [Google Scholar]
- 31.Serohijos A. W., Hegedus T., Aleksandrov A. A., He L., Cui L., Dokholyan N. V., Riordan J. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du K., Sharma M., Lukacs G. L. (2005) Nat. Struct. Mol. Biol. 12, 17–25 [DOI] [PubMed] [Google Scholar]
- 33.He L., Aleksandrov A. A., Serohijos A. W., Hegedus T., Aleksandrov L. A., Cui L., Dokholyan N. V., Riordan J. R. (2008) J. Biol. Chem. 283, 26383–26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidd J. F., Ramjeesingh M., Stratford F., Huan L. J., Bear C. E. (2004) J. Biol. Chem. 279, 41664–41669 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.