Abstract
Interleukin-1 (IL-1) induces extracellular matrix degradation as a result of increased expression of matrix metalloproteinases (MMPs). We examined adhesion-restricted signaling pathways that enable IL-1-induced MMP release in human gingival and murine fibroblasts. Of the seven MMPs and three tissue inhibitors of MMPs screened, IL-1 enhanced release only of MMP3 when cells formed focal adhesions. Inhibition of protein-tyrosine phosphatases (PTPs), which are enriched in focal adhesions, blocked IL-1-induced MMP3 release. Accordingly, in contrast to wild-type cells, fibroblasts null for PTPα did not exhibit IL-1-induced MMP3 release. IL-1 treatment enhanced the recruitment of SHP-2 and PTPα to focal adhesions and the association of PTPα with SHP-2. Pulldown assays confirmed a direct interaction between PTPα and SHP-2, which was dependent on the intact, membrane-proximal phosphatase domain of PTPα. Interactions between SHP-2 and PTPα, recruitment of SHP-2 to focal adhesions, IL-1-induced ERK activation, and MMP3 expression were all blocked by point mutations in the phosphatase domains of PTPα. These data indicate that IL-1-induced signaling through focal adhesions leading to MMP3 release and interactions between SHP-2 and PTPα are dependent on the integrity of the catalytic domains of PTPα.
Keywords: Integrin, Interleukin, Microarray, Protein-Protein Interactions, Tyrosine Protein Phosphatase (Tyrosine Phosphatase), Collagen, MMP3, Matrix
Introduction
Reversible phosphorylation of proteins on tyrosine residues is a pivotal, post-translational modification in many signal transduction pathways. The extent of these modifications is determined by the balance between the activities of protein-tyrosine kinases and phosphatases (1, 2). Whereas protein-tyrosine kinases are thought to regulate the amplitude of responses to extracellular signals, protein-tyrosine phosphatases (PTPs)2 may determine the rate and duration of these responses (3). For IL-1 signaling in adherent cells, tyrosine phosphorylation of focal adhesion proteins such as the focal adhesion kinase is a critical, rate-limiting process (4). Tyrosine phosphorylated proteins, such as focal adhesion kinase, paxillin, and Src family kinases, which are enriched in focal adhesions (5), influence the assembly, maturation, and disassembly of these adhesive structures (6–9) and also impact signaling through focal adhesions (10).
The dynamic and reversible nature of tyrosine phosphorylation of focal adhesion proteins suggests an important role for protein-tyrosine kinases and PTPs in focal adhesion-dependent signaling (11). There are numerous PTPs in focal adhesions, and, in particular, SHP-2 is recruited to focal adhesions upon integrin engagement (12–14). In the absence of SHP-2, the number of focal adhesions and actin stress fibers increases, which is associated with diminished spreading and motility (15, 16). Expression of a dominant-negative SHP-2 enhances the formation of focal adhesions and stress fibers (17). The dynamics of focal adhesion assembly and IL-1-induced signaling pathways, including Ca2+ release and ERK phosphorylation, are dependent on phosphorylation of Tyr542 of SHP-2 (12, 18, 19), which can in turn affect the phosphatase activity of SHP-2 (20).
We recently reported that another PTP, namely PTPα, plays a prominent role in the regulation of focal adhesions during cell adhesion, spreading, and motility (21). PTPα is a receptor-like PTP that can activate Src family kinases (e.g. Src and Fyn) via dephosphorylation of an inhibitory C-terminal tyrosine residue (22–27). We have also shown that through its interactions with Src, PTPα controls IL-1 induced phosphorylation of the inositol 1,4,5-phosphate receptor and consequently, Ca2+ release (28). Further, PTPα regulates formation of focal adhesions in response to mechanical force, strengthens connections between integrins and the cytoskeleton, and modulates cytoskeletal reorganization in response to integrin ligation (26, 27). In the absence of PTPα, fibroblasts demonstrated reduced spreading, increased numbers of abnormal focal adhesions, decreased tyrosine phosphorylation of focal adhesion kinase and p130Cas, and attenuated ERK activation during adhesion and spreading (24).
Although the individual roles of SHP-2 and PTPα in focal adhesion-mediated IL-1 signaling have been partially characterized independently of each of these PTPs (12, 21, 28, 29), whether PTPα and SHP-2 act cooperatively to regulate IL-1 signaling is not known. Consequently we examined the functional importance of the catalytic activity of PTPα in influencing the interactions between SHP-2 and PTPα, as well as IL-1-induced signaling that leads to ERK activation and MMP release.
EXPERIMENTAL PROCEDURES
Materials
Fibronectin, poly-l-lysine, BSA, puromycin, doxycycline, and mouse monoclonal antibodies to vinculin, phosphotyrosine (clone PY-20), and β-actin were from Sigma. Antibody microarrays to human MMPs and tissue inhibitors of MMPs were purchased from RayBiotech, Inc. (Norcross, GA). Rabbit polyclonal and mouse monoclonal anti-SHP-2 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibody directed against the membrane distal catalytic domain of PTPα and mouse monoclonal anti-Src (clone GD11) antibody against pp60Src were obtained from Millipore/Upstate (Lake Placid, NY). Rabbit polyclonal antibodies against phospho-PTPα (Tyr789), phospho-SHP-2 (Tyr542), phospho-MEK1/2, MEK1/2, phospho-ERK1/2 and ERK1/2 were purchased from Cell Signaling (Beverly, MA). Mouse monoclonal anti-MMP3 antibody and recombinant human IL-1β were obtained from R&D Systems (Minneapolis, MN). Mouse monoclonal antibody against calnexin was obtained from BD Biosciences (Mississauga, Canada). Goat anti-α5β1 integrin and mouse anti-GAPDH (clone 6C5) were bought from Millipore/Chemicon (Temecula, CA). The PTP inhibitor (bis(4-trifluoromethylsulfon-amidephenyl)-1,4-diisopropylbenzene; PTP inhibitor IV), inhibitors of IP3-mediated Ca2+ release (Xestospongin C and 2-APB) and MEK inhibitor (U0126) were purchased from Calbiochem. Latrunculin B was obtained from Invitrogen. FuGENE 6 transfection reagent and Src and Fyn kinases were purchased from Roche Applied Science (Indianapolis, IN). Glutathione-Sepharose 4B, thrombin protease, GSTrap 4B, GSTrap FF, and HiTrap benzamidine FF were purchased from GE Healthcare. Purified bovine collagen (PureCol) was purchased from Advanced BioMatrix (San Diego, CA).
Cell Culture
Human gingival fibroblasts were cultured in minimal essential medium containing 10% fetal bovine serum. Rat-2 cells were cultured in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum. All culture media also contained 0.17% (w/v) penicillin V, 0.01 g/ml amphotericin B, and 0.1% gentamycin sulfate, unless specified otherwise. Cells between the 5th and 12th passages were used (19, 29). Wild-type (PTPα+/+) and PTPα-null (PTPα−/−) mouse embryonic fibroblasts were provided by Jan Sap (University of Copenhagen, Denmark) (24). In some experiments, PTPα−/− mouse embryonic fibroblasts were transfected with wild-type PTPα (referred to as PTPαRescue). These cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Genetically modified NIH3T3 fibroblasts that express hemagglutinin-tagged wild-type PTPα (designated as NIH3T3PTPα) and PTPα C433S/C723S mutant (designated as NIH3T3CCSS; essential cysteine in catalytic site of both phosphatase domains mutated to serine) under control of a doxycycline sensitive repressor, were obtained from David Shalloway (Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY) and were generated as described (30). These modified NIH3T3 cells were grown in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum and 5 ng/ml doxycycline. Prior to experiments (14–16 h) doxycycline was removed to allow expression of recombinant PTPα.
Plasmid Constructs and Transient Transfection
Hemagglutinin-tagged wild-type PTPα, PTPα lacking the D2 domain (PTPαΔD2), and PTPα lacking the D1 and D2 domains (PTPαΔD1/D2) were a kind gift from J. den Hertog (Hubrecht Laboratory, Netherlands Institute for Developmental Biology, Utrecht, Netherlands). Cells were seeded in six-well plates at a density of 1 × 105/well for 24 h before transfection to yield a 30–40% confluent culture on the day of transfection. Transient transfections were performed using FuGENE 6 transfection reagent (Roche Applied Science), according to the manufacturer's protocol. Briefly, cells were incubated with DNA-Fugene 6 reagent (1:3) complexes for 5–7 h. Within 48 h after transfection, cells were subjected to further experiments.
Short Interfering RNA (siRNA)
Specific knockdown of PTPα expression was achieved by transfecting human gingival fibroblasts with commercially available PTPα-siRNA (Qiagen, Missisauga, ON) or GFP-siRNA (control) utilizing X-tremeGENE siRNA transfection reagent (Roche Applied Science) following the manufacturer's protocol, About 24–72 h after transfection, cells were washed with PBS, and lysates were collected with SDS lysis buffer to determine the degree of gene knockdown by Western blotting.
Quantitative Real-time PCR
Total RNA was isolated from cells using the RNeasy mini kit (Qiagen) following the manufacture's instructions. The integrity of isolated RNA was confirmed by an Agilent Bioanalyzer 2100 (Santa Clara, CA) prior to analysis. Total RNA samples (1 μg) were reverse transcribed using iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Real-time quantitative PCR was performed on Bio-Rad's CFX96 Real-time PCR system using SsoFastTM EvaGreen® Supermix (Bio-Rad, Hercules, CA) with validated mouse GAPDH primers (forward, 5′-CACACCGACCTTCACCATTTT-3′; reverse, 5′-AGACAGCCGCATCTTCTTGT-3′) and mouse MMP3 primers (forward, 5′-TGGAACAGTCTTGGCTCATGCCTA-3′; reverse, 5′-TGAGAGAGATGGAAACGGGACAAGT-3′). Relative quantification was done using the ΔΔCt method, in which the target gene (MMP3) was normalized to a reference gene (GAPDH), and the fold differences were calculated relative to the nontreatment controls of the NIH3T3CCSS or PTPα−/− cells.
Fluorescence Microscopy
Chamber slides (Lab-Tek) were coated with poly-l-lysine (100 μg/ml in PBS) and fibronectin- or BSA-coated latex microbeads. Cells were plated for 3–4 h at 37 °C. Prior to immunostaining, cells were stimulated with IL-1 (20 ng/ml for 20 min), fixed with 3.7% paraformaldehyde in PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and blocked with 0.2% BSA in PBS for 15 min at room temperature. Staining was performed with rabbit SHP-2 antibody (1:100 in PBS with 1% BSA) for 1 h at room temperature. Slides were washed with PBS, incubated with goat anti-rabbit fluorescein isothiocyanate-conjugated antibody or goat anti-mouse Texas Red conjugate antibody for 1 h, washed, sealed with coverslips, and viewed by total internal reflection or fluorescence microscopy.
Isolation of Focal Adhesions
Cells were grown to 80–90% confluence on 60 mm tissue culture dishes and were cooled to 4 °C prior to the addition of collagen or BSA-coated magnetite beads. Focal adhesion-associated proteins were isolated from cells after specific incubation time periods as described (29, 31). In brief, cells were washed three times with ice-cold PBS to remove unbound beads and scraped into ice-cold cytoskeleton extraction buffer (CKSB: 0.5% Triton X-100, 50 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 20 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm phenylmethylsulfonyl fluoride, 10 mm PIPES, pH 6.8). The cell bead suspension was sonicated, and beads were isolated from the lysate using a magnetic separator. The remainder of the lysates was used to assess the nonfocal adhesion fraction of cells. Beads were resuspended in fresh ice-cold CKSB, homogenized with a Dounce homogenizer (20 strokes), and reisolated magnetically. The beads were washed in CSKB, sedimented, resuspended in Laemmli sample buffer, and boiled for 3–5 min to allow collagen-associated complexes to dissociate from the beads. The beads were sedimented, and lysates were collected for analysis.
Immunoblotting and Immunoprecipitation
The protein concentrations of cell lysates were determined by Bradford assay. Equal amounts of protein were loaded onto 8–10% SDS-polyacrylamide gels, resolved by electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked for 1 h at room temperature in Tris-buffered saline solution with 0.1% Tween 20 and 5% milk or 0.2% BSA. Membranes were incubated with primary antibodies overnight at 4 °C in Tris-buffered saline with 0.1% Tween 20 and 5% milk or 0.2% BSA. Horseradish peroxidase-labeled secondary antibodies were incubated for 1 h at room temperature in Tris-buffered saline with 0.1% Tween 20. Labeled proteins were visualized by chemiluminescence as per the manufacturer's instructions (Amersham Biosciences).
For immunoprecipitations, cells were lysed in radioimmune precipitation assay buffer (50 mm HEPES, pH 7.4, 1% deoxycholate, 1% Triton X-100, 0.1% SDS, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4) containing 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. Equal amounts of protein from cleared extracts were immunoprecipitated and immunoblotted with standard procedures.
GST Pulldown Experiments
GST conjugated full cytoplasmic domain, GST-cPTPα (residues 167–793), GST-PTPαD1 (residues 167–503, domain 1) and GST-PTPαD2 (residues 504–793, domain 2) were kindly provided by J. den Hertog. Fibroblasts were scraped and lysed for 10 min on ice in radioimmune precipitation assay buffer. The lysed cells were centrifuged at 900 × g for 3 min to remove insoluble debris. Supernatants were removed and stored at −80 °C until use. Cell lysates were precleared with 50 μl of 50% slurry of glutathione-Sepharose 4B (1× PBS) and 25 μg of GST for 2 h at 4 °C. The Sepharose matrix was removed by centrifugation at 500 × g for 5 min, and supernatants were subsequently incubated with 50 μl of glutathione-Sepharose 4B, 5 μg of GST protein in PBS + 1% Triton X-100 with gentle agitation at room temperature for 30 min. The matrix was recovered by centrifugation at 500 × g for 5 min. The glutathione Sepharose 4B pellet was washed 4 times with PBS. GST proteins were eluted from the glutathione-Sepharose 4B matrix by incubating twice with 50 μl of elution buffer (10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0) for 10 min at room temperature and isolated by centrifugation at 500 × g for 5 min and pooling the supernatants. The samples were boiled for 5 min and analyzed by immunoblotting.
In Vitro Phosphorylation
For in vitro phosphorylation, bacterially expressed purified SHP-2 was incubated for 10 min at room temperature in 20 μl of kinase buffer (25 mm HEPES, pH 7.1, 10 mm MgCl2, 5 mm MnCl2, 0.5 mm EGTA, 1 mm Na3VO4, 1 mm dithiothreitol, 100 μm Mg-ATP) with or without 5 units of active Src. The reaction products were then incubated with GST-cPTPα in buffer (50 mm Tris, 1 mm EDTA, pH 6.8) for 30 min at 37 °C. The GST pulldown assay was performed, and bound proteins were eluted and immunoblotted using antibodies against phosphotyrosine and SHP-2.
Experimental Design and Analysis
All experiments were repeated at least three times on different days using different batches of cells. Data shown are representative examples of these experiments. For numerical data, means and S.E. were calculated and, where appropriate, Student's t test was performed. Statistical significance was set at p < 0.05.
RESULTS
PTPs in IL-1 Signaling
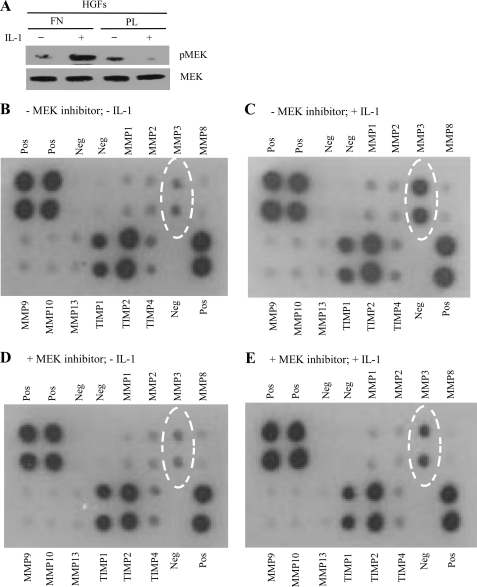
IL-1 induces focal adhesion-restricted ERK phosphorylation and Ca2+ signaling in fibroblasts (4, 12, 19, 29, 32, 33), but it is unknown whether these signals selectively lead to expression of matrix-destructive genes. We examined this possibility by first ensuring that under the experimental conditions used here, IL-1 induction of MEK phosphorylation was dependent on focal adhesions. Human gingival fibroblasts, which express IL-1 receptors at high levels (34), were plated on fibronectin (a condition that facilitates focal adhesion formation) or on poly-l-lysine (no focal adhesion formation) and were treated for 45 min with vehicle alone or with IL-1. Under these conditions, MEK phosphorylation in response to IL-1 required focal adhesions (Fig. 1A). In four separate experiments, we screened for human matrix metalloproteinases using antibody arrays (RayBiotech; catalog no. AAH-MMP-1) in conditioned media obtained from control or IL-1-treated human gingival fibroblasts, which had been plated previously on fibronectin to enable focal adhesion formation. The cell culture medium was examined undiluted for expression of MMPs and TIMPs following the manufacturer's instructions. IL-1 markedly and selectively increased MMP3 released into the medium; the other MMPs and TIMPs in the array were not detectably affected (Fig. 1, B and C). Duplicate experiments in which cells were pretreated with the MEK inhibitor U1026 (30 μm) (35) followed by overnight treatment with or without IL-1, indicated that MMP3 release in response to IL-1 is dependent on the MEK/ERK pathway (Fig. 1, D and E), which is consistent with earlier data showing that IL-1 induces ERK activation (32).
FIGURE 1.
IL-1-induced MMP3 expression depends on the ERK-mediated pathway. A, human gingival fibroblasts were plated on fibronectin (FN) or poly-l-lysine (PL) and treated with vehicle or IL-1 (40 ng/ml) overnight. Cell lysates were immunoblotted (WB) for MEK or phospho-MEK. B–E, human gingival fibroblasts grown on fibronectin were pretreated with vehicle (B and C) or MEK inhibitor, U0126 (30 μm; 30 min; D and E), which blocks ERK activation, and were stimulated with vehicle (B and D) or with IL-1 (40 ng/ml; C and E) overnight. Conditioned media were screened with human MMP/TIMP antibody arrays. Negative (Neg) and positive (Pos) controls were also screened.
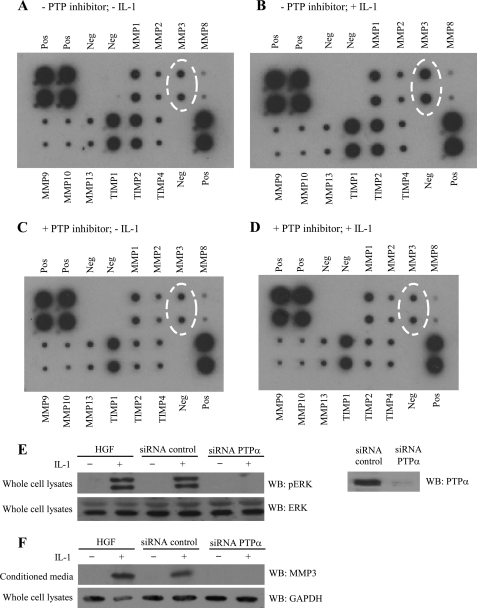
Several tyrosine phosphorylated proteins are crucial for the development and remodeling of focal adhesions (7, 8, 36), which, in turn are required for IL-1 signaling in fibroblasts (32) and chondrocytes (33). The dynamic nature of tyrosine phosphorylation of focal adhesion proteins, the requirement for focal adhesions in IL-1 signaling (4), and our previous demonstration of a requirement for the protein-tyrosine phosphatase SHP-2 in IL-1 signaling to ERK (12) led us to examine the role of protein-tyrosine phosphatases in IL-1-induced MMP3 release. Human gingival fibroblasts were treated with IL-1 or vehicle in the presence or absence of the PTP inhibitor IV (10 μm; 4 h). This time period was the minimum duration required for detection of MMPs released into the culture medium; longer treatments (>8 h) were associated with apoptosis (data not shown). In two separate experiments using cells from different passages, MMP3 secretion after IL-1 treatment was blocked by inhibition of PTPs (Fig. 2, A–D), indicating that the catalytic activities of PTPs are important for focal adhesion-dependent, IL-1-induced signaling and MMP3 release. We pursued this idea by examining another PTP as a test of the specificity of IL-1-induced ERK activation and MMP3 release. PTPα was studied because of its central role in IL-1 signaling through focal adhesions (28). Human gingival fibroblasts were transfected with control siRNA (green fluorescent protein) or siRNA specific for PTPα. Following knockdown, cells were incubated with or without IL-1. Control cells exhibited marked phosphorylation of ERK and MMP3 release in response to IL-1, whereas cells with PTPα knockdown exhibited no ERK activation (Fig. 2E) or MMP3 release (Fig. 2F).
FIGURE 2.
MMP3 release depends on protein-tyrosine phosphatase activity. A–D, human MMP arrays were used to screen-conditioned media collected from human gingival fibroblasts grown on fibronectin, treated with vehicle control (A and B), or PTP inhibitor IV (10 μm; 4 h; C and D), and stimulated with vehicle (A and C) or IL-1 (40 ng/ml; 4 h; B and D). Negative (Neg) and positive (Pos) controls were also screened. E, human gingival fibroblasts (HGF) were transfected with GFP-siRNA (control-siRNA) or with PTPα-siRNA, plated on fibronectin-coated dishes and treated with or without IL-1 (40 ng/ml overnight). Cell-conditioned media were concentrated 10 times, separated on SDS-PAGE, and immunoblotted for MMP3. Right panel shows effectiveness of PTPα knockdown in human gingival fibroblasts using GFP-siRNA as control or PTPα-siRNA. F, whole cell lysates prepared from similarly transfected human gingival fibroblasts, which were plated on fibronectin and stimulated with vehicle or IL-1 (20 ng/ml; 30 min) were immunoblotted for phospho-ERK and ERK.
Accumulation of SHP-2 in Focal Adhesions Requires PTPα
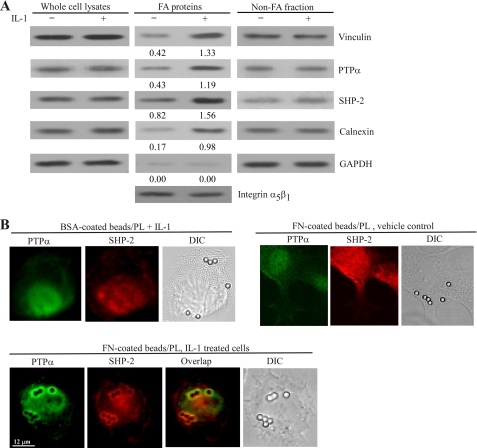
Among the numerous PTPs in focal adhesions, PTPα and SHP-2 have been shown, independent of one another, to be crucial for the assembly of, and the signaling through, focal adhesions (12, 18, 21, 22, 26, 37). First, we determined whether IL-1 affected the abundance of PTPα and SHP-2 in focal adhesions. Cells were incubated with collagen-coated magnetite beads and magnetically separated bead-associated proteins (31) were immunoblotted for PTPα, SHP-2, and vinculin (a focal adhesion protein) (Fig. 3A) (29). IL-1 enhanced PTPα, SHP-2, and vinculin abundance in the bead preparations. Consistent with these data, by immunofluorescence, we found that in cells plated on fibronectin-coated beads (but not bovine serum albumin-coated beads), SHP-2 colocalized with PTPα in focal adhesions after IL-1 treatment of 30 min (Fig. 3B).
FIGURE 3.
IL-1 induces recruitment of SHP-2 to focal adhesion and endoplasmic reticulum and colocalization of SHP-2 with PTPα in focal adhesions. A, cells were incubated with collagen-coated magnetite beads for 30 min at 37 °C to induce focal adhesion formation and treated with vehicle or IL-1 (20 ng/ml at 37 °C for 30 min). Focal adhesion (FA) preparations and nonfocal adhesion fractions were isolated as described under “Experimental Procedures.” Equivalent amounts of whole cell lysates, focal adhesion-associated proteins, and nonfocal adhesion proteins were separated on SDS-polyacrylamide gels and immunoblotted for vinculin, PTPα, SHP-2, and GAPDH. Focal adhesion proteins were probed for α5β1 integrin as a loading control. Numeric values below focal adhesion proteins indicate the density ratios relative to the corresponding integrin α5β1 levels in the presence or absence of IL-1. B, Rat2 cells were grown on poly-l-lysine (PL)-coated coverslips previously incubated with fibronectin (FN) or BSA-coated microbeads for 3–4 h. After stimulation with IL-1 or vehicle, cells were coimmunostained for PTPα and SHP-2 and viewed by fluorescence microscopy. DIC, differential interference contrast.
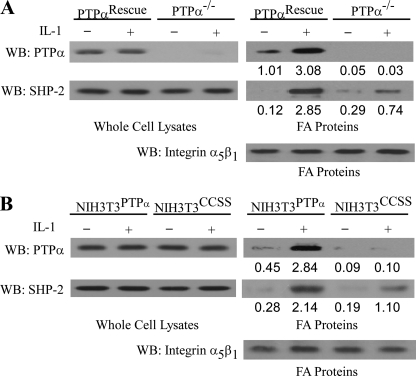
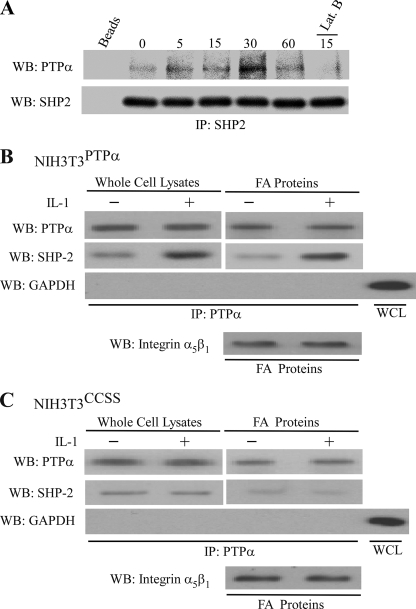
We determined whether the recruitment of SHP-2 and PTPα to focal adhesions in response to IL-1 was dependent on PTPα. IL-1 treatment enhanced the accumulation of SHP-2 and PTPα in focal adhesion fractions isolated from PTPα-null cells reconstituted with wild-type PTPα (designated as PTPαRescue; Fig. 4A) or in modified NIH 3T3 cells that overexpressed wild-type PTPα (designated NIH3T3PTPα; Fig. 4B).
FIGURE 4.
Functional PTPα is required for IL-1 induced SHP-2 accumulation in focal adhesions. A, PTPα-null cells transfected with or without wild-type PTPα (PTPαRescue or PTPα−/−, respectively) treated with IL-1 (20 ng/ml) or vehicle control for 30 min at 37 °C. Focal adhesion (FA)-associated proteins and whole cell lysates were prepared, separated by SDS-polyacrylamide gels, and immunoblotted (WB) for PTPα, SHP-2, and α5β1 integrin. B, same experiment as in A, repeated using NIH3T3 cells overexpressing either wild-type PTPα (NIH3T3PTPα) or catalytically inert mutant, CCSS (NIH3T3CCSS).
By contrast, in PTPα-null cells (PTPα−/−), IL-1-induced SHP-2 recruitment to focal adhesion fractions was ∼3-fold less. SHP-2 recruitment was dependent on the catalytic activity of PTPα because this process was also reduced by ∼2-fold in NIH3T3 cells overexpressing a catalytically inert PTPα mutant (C433S/C723S; referred to as NIH3T3CCSS; Fig. 4B) functioning here as a dominant-negative.
By contrast, in the absence of PTPα (in PTPα-null cells, PTPα−/−) or when the catalytic activity of PTPα was absent (in dominant-negative NIH3T3 cells overexpressing a catalytically inert PTPα mutant C433S/C723S, NIH3T3CCSS), IL-1-induced SHP-2 recruitment to focal adhesion fractions was reduced by ∼2- or 3-fold, respectively (Fig. 4B).
Associations between PTPα and SHP-2
We considered whether PTPα and SHP-2 may associate with each other in focal adhesions in human gingival fibroblasts. Accordingly, cells were incubated with collagen-coated beads and treated with IL-1, and the bead-associated proteins were solubilized and immunoprecipitated with SHP-2 antibody. When the SHP-2 immunoprecipitates were blotted for PTPα, there was a time-dependent enhancement of the association between PTPα and SHP-2 (Fig. 5A), which was blocked in cells pretreated with latrunculin B (1 μm; 15 min) to prevent the formation of focal adhesions (38).
FIGURE 5.
IL-1 induced association between PTPα and SHP-2 requires intact phosphatase domains of PTPα. A, SHP-2 immunoprecipitates were prepared from focal adhesion (FA)-associated proteins derived from human gingival fibroblasts that had been treated with IL-1 for indicated times. Immunoprecipitates (IP) were immunoblotted (WB) for SHP-2 and PTPα. For Lat B sample, cells were pretreated with latrunculin B (1 μm) for 15 min prior to IL-1 treatment to dissipate focal adhesions. B and C, PTPα was purified by immunoprecipitation (IP) from whole cell lysates (WCL) and focal adhesion fractions, prepared from genetically modified NIH3T3 cells lines expressing either wild-type PTPα (NIH3T3PTPα; B) or catalytically inert PTPα (NIH3T3CCSS; C) that were stimulated with IL-1 (20 ng/ml) or vehicle for 30 min at 37 °C. PTPα immunoprecipitates were immunoblotted for PTPα, SHP-2, and GAPDH. GAPDH was used to assess the purity of the immunoprecipitates.
We examined the requirement for catalytically active PTPα for the association between PTPα and SHP-2. NIH3T3PTPα (Fig. 5B) or NIH3T3CCSS (Fig. 5C) cells were treated with vehicle or IL-1. PTPα was immunoprecipitated from whole cell lysates and focal adhesion preparations, and the immunoprecipitates were immunoblotted for SHP-2 and PTPα. The association between SHP-2 and PTPα was dependent on the catalytic activity of PTPα because in cells expressing the catalytically defective PTPα (C433S/C723S mutant), SHP-2 did not coprecipitate with PTPα, even after IL-1 treatment (Fig. 5C).
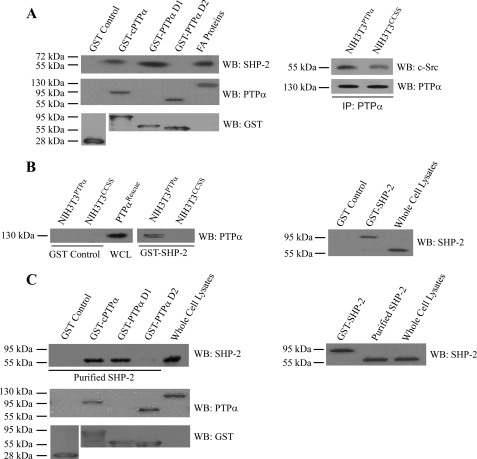
Potential associations between SHP-2 and PTPα were examined in pulldown assays using purified, GST-tagged PTPα proteins. These bacterially expressed mutant proteins included either the entire cytosolic domain (GST-cPTPα) or the catalytic domain D1 alone (GST-PTPαD1) or D2 alone (GST-PTPαD2). The bead-bound proteins were incubated with focal adhesion preparations that had been prepared from cells expressing wild-type SHP-2. The PTPα-associated proteins were isolated and immunoblotted for SHP-2 (Fig. 6A; left panel) after loading equal amounts of proteins. Equal loading was assessed by BCA protein assay, as indicated by the immunoblots for PTPα and GST. We note that the PTPα antibody used here was directed against the membrane distal catalytic (D2) domain; thus, it failed to recognize the GST-PTPαD1 in the immunoblots of the GST pulldown assay (Fig. 6, A, left panel, and C). The data from these experiments demonstrated that SHP-2 in focal adhesion preparations bound to the cytosolic domain of PTPα and in particular with the D1 catalytic domain of PTPα.
FIGURE 6.
Pulldown assays of PTPα and SHP-2. A, left panel, association between PTPα and SHP-2 was assessed by pulldown assays using glutathione-Sepharose bead-bound bacterial fusion proteins: GST-cPTPα, GST-PTPα D1, and GST-PTPα D2 or GST control. GST proteins were incubated with focal adhesion (FA) fractions prepared from PTPαRescue cells, PTPα-null cells transfected with wild-type PTPα. Proteins bound to beads were immunoblotted (WB) for SHP-2, PTPα, and GST. Right panel, PTPα was immunoprecipated (IP) from NIH3T3 cell lines that have been genetically modified to express wild-type PTPα (NIH3T3PTPα) or catalytically inert PTPα (NIH3T3CCSS) and immunoblotted for SHP-2 and PTPα. B, glutathione-Sepharose bead-bound bacterial GST-SHP-2 wild-type fusion protein or bead-bound GST alone were incubated with lysates of genetically modified NIH3T3 cells lines that express wild-type PTPα (NIH3T3PTPα) or double mutant PTPα (NIH3T3CCSS). The bead-bound materials were analyzed by immunoblotting for PTPα. C, glutathione-Sepharose bead-bound bacterial GST-cPTPα, GST-PTPα D1, and GST-PTPα D2 fusion proteins or GST control were incubated with purified SHP-2 by removal of GST tag by enzymatic cleavage from GST-SHP-2 bacterial fusion protein. The bead-associated proteins were analyzed by immunoblotting for SHP-2, PTPα, and GST. As controls, GST-SHP-2, purified SHP-2, and whole cell lysates were probed with antibodies to SHP-2.
Previously, we have reported that interactions between PTPα and the Ca2+ release channel IP3R1, required the membrane proximal domain (D1) of PTPα (28). In contrast, the membrane distal domain has been implicated in interactions between PTPα and other receptor phosphatases (39). However, the associations between Fyn (a Src family kinase) and PTPα is apparently independent of the catalytic activity of PTPα (40). Hence, we examined the interactions between PTPα and c-Src in genetically modified NIH3T3 cell lines expressing either wild-type PTPα (NIH3T3PTPα) or phosphatase-dead mutant PTPα (NIH3T3CCSS) to determine whether or not catalytic activity was required for their interaction (Fig. 6A, right panel).
We performed the converse experiments using purified GST-SHP-2 (wild-type) or GST beads that were incubated with cell lysates from NIH3T3 cells overexpressing either the PTPα wild-type (NIH3T3PTPα) or catalytically inert CCSS mutant (NIH3T3CCSS). When the GST-SHP-2-associated proteins were immunoblotted for PTPα, we noted that SHP-2 associated with PTPα only in cells expressing wild-type PTPα (Fig. 6B).
The experiments described above used cell lysates to examine potential associations between SHP-2 and PTPα. These experiments preclude the ability to assess direct interactions between these proteins and complicate the interpretation of data because of the possibility of multiple interacting proteins. Accordingly, the various PTPα domain constructs (cPTPα, cytosolic; PTPαD1, catalytic domain D1 only; PTPαD2, catalytic domain D2 only) conjugated to GST were incubated in a cell-free system with bacterially expressed SHP-2 and GST-associated proteins were immunoblotted for SHP-2. These data showed that purified SHP-2 interacted with the whole cytosolic domain of PTPα as well as the D1 catalytic domain but barely with catalytic domain D2 (Fig. 6C).
Functional Regulation of SHP-2 by PTPα
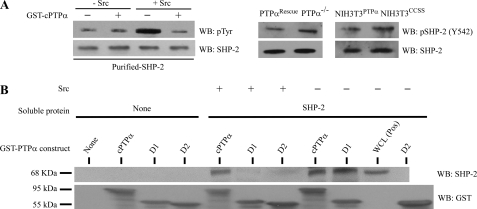
The requirement for the D1 catalytic domain of PTPα to mediate interactions with SHP-2 suggested that SHP-2 may be dephosphorylated by PTPα and that this process in turn may affect their interaction. We examined this possibility using c-Src kinase to phosphorylate purified SHP-2 in vitro. GST-bound, cytosolic PTPα was incubated with phosphorylated SHP-2 to assess PTPα–mediated dephosphorylation of SHP-2. Immunoblotting of phosphotyrosine residues of SHP-2 bound to beads showed that the presence of cPTPα in the in vitro reactions resulted in loss of SHP-2 phosphorylation mediated by c-Src (Fig. 7A, left panel). In addition, we found that in the absence of PTPα or its phosphatase activity, there was a slight increase in SHP-2 phosphorylation at tyrosine 542 (the residue phosphorylated with IL-1 treatment; (12); Fig. 7A, middle and right panels). Next, we examined how the phosphorylation status of SHP-2 (catalyzed by Src) influenced its interaction with PTPα, by incubating purified SHP-2 with the previously described GST-PTPα mutant proteins bound to beads. Prior to incubation SHP-2 was phosphorylated in vitro by c-Src kinase or with kinase buffer alone. In the absence of c-Src-induced phosphorylation of SHP-2, cPTPα, and the catalytic domain D1 of PTPα bound to SHP-2, but there was no binding of SHP-2 to the catalytic domain D2 of PTPα. In contrast, when SHP-2 was phosphorylated by c-Src kinase, cPTPα interactions were reduced, and binding to PTPαD1 was undetectable (Fig. 7B). These data indicated that the catalytic domain 1 of PTPα is important for interactions with SHP-2 and that this interaction may be regulated by the phosphorylation status of SHP-2, which, in turn, is dependent on the catalytic activity of PTPα.
FIGURE 7.
Functional interaction between PTPα and SHP-2. A, left panel, in vitro tyrosine phosphorylation was analyzed by first incubating bacterially expressed and purified SHP-2 in the presence or absence of active Src and then with GST-cPTPα. Materials bound by the GST pulldown assay were immunoblotted (WB) for SHP-2 and phosphotyrosine (pTyr) residues. Middle and right panels, SHP-2 phosphorylation was examined by immunoblotting for phosphotyrosine 542 of SHP-2 in the absence of PTPα in wild-type and null cells (middle panel) as well as in the absence of phosphatase activity of PTPα in cells that overexpress wild-type or catalytically inert mutant (right panel). B, bacterially expressed SHP-2 was purified and either prephosphorylated by c-Src or not and incubated with GST-PTPα domain constructs (cPTPα, cytoplasmic; D1, catalytic domain D1 only; D2, catalytic domain D2 only) in buffer (50 mm Tris, 1 mm EDTA, pH 6.8) for 30 min at 37 °C. Proteins eluted by GST pulldown assay were immunoblotted for SHP-2. Equal loading was assessed by GST immunoblots.
Phosphatase Activity of PTPα Is Important for IL-1-induced ERK Activation and MMP3 Release
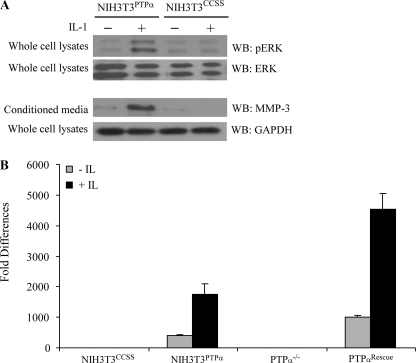
We examined IL-1-induced ERK activation and MMP3 release in NIH3T3 cells expressing wild-type PTPα (NIH3T3PTPα) or cells expressing the catalytically inert PTPα mutant (NIH3T3CCSS), which does not permit interactions of PTPα with SHP-2. In contrast to wild-type cells, NIH3T3CCSS cells did not exhibit ERK activation or MMP3 release in response to IL-1 (Fig. 8A). We ascertained whether PTPα regulated transcription of MMP3 in response to IL-1. NIH3T3 cells and PTPα-null and rescue cells were treated with IL-1 and mRNA levels were measured by quantitative real-time-PCR following reverse transcription. These data showed that in the presence of catalytically active PTPα, there were 4–5-fold increases of MMP3 mRNA after IL-1 treatment compared with vehicle controls (p < 0.01). MMP3 mRNA was detected at very low levels in those cells expressing the catalytically inert PTPα mutant (NIH3T3CCSS) or in cells that were null for PTPα (Fig. 8B).
FIGURE 8.
MMP3 expression requires intact catalytic domains of PTPα. A, genetically modified NIH3T3 cells lines expressing wild-type PTPα (NIH3T3PTPα) or C433S/C723S double mutant (NIH3T3CCSS) were plated on fibronectin and treated with or without IL-1 (40 ng/ml; overnight). Conditioned media were collected, concentrated 10 times, separated by SDS-PAGE, and immunoblotted (WB) for MMP3. Whole cell lysates prepared from same experiment were immunoblotted for GAPDH (upper panels). Whole cell lysates derived from cells stimulated with vehicle or IL-1 (20 ng/ml; 30 min) were immunoblotted for phospho-ERK and ERK (lower panels). B, same cells in A, as well as PTPα-null cells and PTPα-null cells transfected with wild-type PTPα vector were treated with or without IL-1 (40 ng/ml; 2 h) and analyzed by quantitative RT-PCR as described under “Experimental Procedures.” Data are mean ± S.E. of fold differences relative to the nontreated controls of PTPα−/− or NIH3T3CCSS in MMP3 mRNA levels normalized to GAPDH mRNA levels.
DISCUSSION
IL-1-induced expression and release of MMPs is a crucial factor in the pathogenesis of rheumatoid arthritis (41), periodontal diseases (42), postmyocardial infarction remodeling (43), and in the remodeling of normal human endometrium (44). Furthermore, PTPα is important in cellular transformation (45). In view of the data described here showing that PTPα regulates MMP3 expression, there is a potential role of PTPα and its interactions with SHP-2 in oncogenesis as well as in the regulation of inflammatory lesions.
Here, we screened a limited number of MMPs and TIMPs and found that IL-1 stimulation of human gingival fibroblasts appears to selectively promote expression of MMP3. As MMP3 expression in response to IL-1 is mediated in part by the ERK mitogen-activated protein kinases (46), and as IL-1-induced ERK signaling is critical for expression of AP-1 transcription factors (32), which are important for MMP3 expression (47), we examined how PTPs may be involved in this signaling system. Previously, we described a role for PTPα (28) and for SHP-2 (12, 29) in IL-1 signaling and demonstrated the significance of PTPα in focal adhesion maturation (21). Our major findings here are that interactions between SHP-2 and PTPα and IL-1-induced MMP3 release are dependent on the integrity of the catalytic domains of PTPα. The interactions between PTPα and SHP-2 required the membrane proximal domain of PTPα (D1) and may be important for targeting of SHP-2 and PTPα to focal adhesions, which is an important requirement for IL-1-induced signaling in fibroblasts (28, 29).
SHP-2 and PTPα are both classical phosphotyrosine-specific phosphatases, but the former is a cytosolic PTP, and the latter is a receptor-like PTP (48–51). In general, the receptor-like PTPs exhibit a variable extracellular domain, a transmembrane domain, and a cytosolic domain containing one or two catalytic domains, only one of which is usually catalytically active (20). In contrast, the soluble intracellular PTPs, including SHP-2, often exhibit a multidomain structure containing a conserved catalytic domain and additional regulatory or targeting/binding modules such as SH2, PDZ, FERM (F ezrin-radixin-moesin homology), or proline-rich domains. We found that the membrane proximal phosphatase domain of PTPα, in addition to being essential for catalysis, provides a potential interaction site for SHP-2. As indicated by our data, dephosphorylated SHP-2 may interact more avidly with PTPα than phosphorylated SHP-2, suggesting that the catalytic site of PTPα may not be limited to mediating catalytic activity but may also influence protein-protein interactions. Because SHP-2 does not contain focal adhesion or membrane-targeting domains, the localization of PTPα to cell membranes, its enrichment in focal adhesions (25) and its binding to SHP-2 may enable the targeting of SHP-2 to focal adhesions, which is enhanced by IL-1 stimulation (12). Previous data have shown that in substrate-dependent cells, focal adhesions are essential for IL-1-induced ERK activation and calcium release (4, 32) and that SHP-2 in focal adhesions is required for the generation of these downstream signals (12, 29). Therefore, PTPα may provide essential focal adhesion-dependent anchorage motifs for PTPs such as SHP-2 that are essential for IL-1 signaling through focal adhesions.
Our data on the importance of the integrity of the catalytic domain of PTPα to permit interaction with SHP-2 indicate that the membrane proximal catalytic domain of PTPα, in addition to mediating phosphatase activity, may provide opportunities for protein-protein interactions. These interactions are evidently regulated by the phosphorylation status of SHP-2 as phosphorylated (and purified) SHP-2 has less affinity for PTPα compared with the nonphosphorylated SHP-2. Conceivably, IL-1-induced dephosphorylation of SHP-2 by PTPα augments interactions between these two proteins, which, in turn, enhance the localization of SHP-2 to focal adhesions (12), where it is required for signal transduction leading to ERK activation. Previously, interactions between phosphatase domains within the same PTPα molecule have been considered to be one system for regulation of phosphatase activity (3). In this interaction, the inhibitory helix-turn-helix wedge motif from one domain occludes the active site of the partner domain (52). Conceivably, reciprocal occlusion of the active sites might dampen catalytic activity. The novelty of our report is that the catalytic domains of PTPα may also provide docking sites for other PTPs, like SHP-2, which then provide appropriate intracellular localization to focal adhesions and evidently affect the phosphorylation status of these proteins.
This work was supported by grants from the Canadian Institutes of Health Research (MOP 84254; to C. A. M. and G. P. D.). This work was also supported in part by National Institutes of Health Grant HL090669 (to G. P. D.) and funds from the Harold and Mary Zirin Chair in Pulmonary Biology at National Jewish Health (to G. P. D.).
- PTP
- protein-tyrosine phosphatase
- IL
- interleukin
- TIMP
- tissue inhibitor of MMP
- ERK
- extracellular signal-regulated kinase
- MMP
- matrix metalloproteinase
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- GST
- glutathione S-transferase
- siRNA
- short interfering RNA
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- PIPES
- 1,4-piperazinediethanesulfonic acid
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- cPTP
- cytoplasmic protein-tyrosine phosphatase.
REFERENCES
- 1.Ostman A., Böhmer F. D. (2001) Trends Cell Biol. 11, 258–266 [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. (2000) Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 3.Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 4.Arora P. D., Ma J., Min W., Cruz T., McCulloch C. A. (1995) J. Biol. Chem. 270, 6042–6049 [DOI] [PubMed] [Google Scholar]
- 5.Zaidel-Bar R., Milo R., Kam Z., Geiger B. (2007) J. Cell Sci. 120, 137–148 [DOI] [PubMed] [Google Scholar]
- 6.Zaidel-Bar R., Cohen M., Addadi L., Geiger B. (2004) Biochem. Soc. Trans. 32, 416–420 [DOI] [PubMed] [Google Scholar]
- 7.Kirchner J., Kam Z., Tzur G., Bershadsky A. D., Geiger B. (2003) J. Cell Sci. 116, 975–986 [DOI] [PubMed] [Google Scholar]
- 8.Ilić D., Furuta Y., Kanazawa S., Takeda N., Sobue K., Nakatsuji N., Nomura S., Fujimoto J., Okada M., Yamamoto T. (1995) Nature 377, 539–544 [DOI] [PubMed] [Google Scholar]
- 9.Klinghoffer R. A., Sachsenmaier C., Cooper J. A., Soriano P. (1999) EMBO J. 18, 2459–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carragher N. O., Frame M. C. (2004) Trends Cell Biol. 14, 241–249 [DOI] [PubMed] [Google Scholar]
- 11.Panetti T. S. (2002) Front Biosci. 7, d143–150 [DOI] [PubMed] [Google Scholar]
- 12.MacGillivray M., Herrera-Abreu M. T., Chow C. W., Shek C., Wang Q., Vachon E., Feng G. S., Siminovitch K. A., McCulloch C. A., Downey G. P. (2003) J. Biol. Chem. 278, 27190–27198 [DOI] [PubMed] [Google Scholar]
- 13.Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., Kasuga M. (1996) Mol. Cell Biol. 16, 6887–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuda M., Matozaki T., Fukunaga K., Fujioka Y., Imamoto A., Noguchi T., Takada T., Yamao T., Takeda H., Ochi F., Yamamoto T., Kasuga M. (1998) J. Biol. Chem. 273, 13223–13229 [DOI] [PubMed] [Google Scholar]
- 15.Yu D. H., Qu C. K., Henegariu O., Lu X., Feng G. S. (1998) J. Biol. Chem. 273, 21125–21131 [DOI] [PubMed] [Google Scholar]
- 16.Oh E. S., Gu H., Saxton T. M., Timms J. F., Hausdorff S., Frevert E. U., Kahn B. B., Pawson T., Neel B. G., Thomas S. M. (1999) Mol. Cell Biol. 19, 3205–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kodama A., Matozaki T., Fukuhara A., Kikyo M., Ichihashi M., Takai Y. (2000) Mol. Biol. Cell 11, 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera Abreu M. T., Wang Q., Vachon E., Suzuki T., Chow C. W., Wang Y., Hong O., Villar J., McCulloch C. A., Downey G. P. (2006) J. Cell Physiol. 207, 132–143 [DOI] [PubMed] [Google Scholar]
- 19.Wang Q., Downey G. P., Herrera-Abreu M. T., Kapus A., McCulloch C. A. (2005) J. Biol. Chem. 280, 8397–8406 [DOI] [PubMed] [Google Scholar]
- 20.Poole A. W., Jones M. L. (2005) Cell Signal 17, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 21.Herrera Abreu M. T., Penton P. C., Kwok V., Vachon E., Shalloway D., Vidali L., Lee W., McCulloch C. A., Downey G. P. (2008) Am. J. Physiol. Cell Physiol. 294, C931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harder K. W., Moller N. P., Peacock J. W., Jirik F. R. (1998) J. Biol. Chem. 273, 31890–31900 [DOI] [PubMed] [Google Scholar]
- 23.Ponniah S., Wang D. Z., Lim K. L., Pallen C. J. (1999) Curr. Biol. 9, 535–538 [DOI] [PubMed] [Google Scholar]
- 24.Su J., Muranjan M., Sap J. (1999) Curr. Biol. 9, 505–511 [DOI] [PubMed] [Google Scholar]
- 25.Lammers R., Lerch M. M., Ullrich A. (2000) J. Biol. Chem. 275, 3391–3396 [DOI] [PubMed] [Google Scholar]
- 26.von Wichert G., Jiang G., Kostic A., De Vos K., Sap J., Sheetz M. P. (2003) J. Cell Biol. 161, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., Chen S. C., Pallen C. J. (2006) J. Biol. Chem. 281, 11972–11980 [DOI] [PubMed] [Google Scholar]
- 28.Wang Q., Rajshankar D., Branch D. R., Siminovitch K. A., Herrera Abreu M. T., Downey G. P., McCulloch C. A. (2009) J. Biol. Chem. 284, 20763–20772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Herrera Abreu M. T., Siminovitch K., Downey G. P., McCulloch C. A. (2006) J. Biol. Chem. 281, 31093–31105 [DOI] [PubMed] [Google Scholar]
- 30.Zheng X. M., Resnick R. J., Shalloway D. (2000) EMBO J. 19, 964–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plopper G., Ingber D. E. (1993) Biochem. Biophys. Res. Commun. 193, 571–578 [DOI] [PubMed] [Google Scholar]
- 32.Lo Y. Y., Luo L., McCulloch C. A., Cruz T. F. (1998) J. Biol. Chem. 273, 7059–7065 [DOI] [PubMed] [Google Scholar]
- 33.Luo L., Cruz T., McCulloch C. (1997) Biochem. J. 324, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qwarnstrom E. E., Page R. C., Gillis S., Dower S. K. (1988) J. Biol. Chem. 263, 8261–8269 [PubMed] [Google Scholar]
- 35.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. (2003) J. Cell Sci. 116, 4605–4613 [DOI] [PubMed] [Google Scholar]
- 37.von Wichert G., Haimovich B., Feng G. S., Sheetz M. P. (2003) EMBO J. 22, 5023–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal G., Lee W., Arora P. D., McKee M., Downey G., McCulloch C. A. (2001) J. Cell Sci. 114, 119–129 [DOI] [PubMed] [Google Scholar]
- 39.Blanchetot C., den Hertog J. (2000) J. Biol. Chem. 275, 12446–12452 [DOI] [PubMed] [Google Scholar]
- 40.Bhandari V., Lim K. L., Pallen C. J. (1998) J. Biol. Chem. 273, 8691–8698 [DOI] [PubMed] [Google Scholar]
- 41.Vincenti M. P., Brinckerhoff C. E. (2002) Arthritis research 4, 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Havemose-Poulsen A., Holmstrup P. (1997) Crit. Rev. Oral Biol. Med. 8, 217–236 [DOI] [PubMed] [Google Scholar]
- 43.Nian M., Lee P., Khaper N., Liu P. (2004) Circulation research 94, 1543–1553 [DOI] [PubMed] [Google Scholar]
- 44.Osteen K. G., Keller N. R., Feltus F. A., Melner M. H. (1999) Gynecol. Obstet. Invest. 48, 2–13 [DOI] [PubMed] [Google Scholar]
- 45.Zheng X. M., Wang Y., Pallen C. J. (1992) Nature 359, 336–339 [DOI] [PubMed] [Google Scholar]
- 46.DiBattista J. A., Pelletier J. P., Zafarullah M., Fujimoto N., Obata K., Martel-Pelletier J. (1995) J. Rheumatol. Suppl. 43, 123–128 [PubMed] [Google Scholar]
- 47.Mauviel A., Kähäri V. M., Kurkinen M., Evans C. H., Uitto J. (1992) Journal of cellular biochemistry 50, 53–61 [DOI] [PubMed] [Google Scholar]
- 48.Tonks N. K. (2005) Cell 121, 667–670 [DOI] [PubMed] [Google Scholar]
- 49.Alonso A., Sasin J., Bottini N., Friedberg I., Friedberg I., Osterman A., Godzik A., Hunter T., Dixon J., Mustelin T. (2004) Cell 117, 699–711 [DOI] [PubMed] [Google Scholar]
- 50.Jones M. L., Poole A. W. (2004) Methods Mol. Biol. 273, 169–178 [DOI] [PubMed] [Google Scholar]
- 51.Tsui H. W., Siminovitch K. A., de Souza L., Tsui F. W. (1993) Nat. Genet. 4, 124–129 [DOI] [PubMed] [Google Scholar]
- 52.Bilwes A. M., den Hertog J., Hunter T., Noel J. P. (1996) Nature 382, 555–559 [DOI] [PubMed] [Google Scholar]