Abstract
Apicomplexan parasites (including Plasmodium spp. and Toxoplasma gondii) employ a four-carbon pathway for de novo heme biosynthesis, but this pathway is distinct from the animal/fungal C4 pathway in that it is distributed between three compartments: the mitochondrion, cytosol, and apicoplast, a plastid acquired by secondary endosymbiosis of an alga. Parasite porphobilinogen synthase (PBGS) resides within the apicoplast, and phylogenetic analysis indicates a plant origin. The PBGS family exhibits a complex use of metal ions (Zn2+ and Mg2+) and oligomeric states (dimers, hexamers, and octamers). Recombinant T. gondii PBGS (TgPBGS) was purified as a stable ∼320-kDa octamer, and low levels of dimers but no hexamers were also observed. The enzyme displays a broad activity peak (pH 7–8.5), with a Km for aminolevulinic acid of ∼150 μm and specific activity of ∼24 μmol of porphobilinogen/mg of protein/h. Like the plant enzyme, TgPBGS responds to Mg2+ but not Zn2+ and shows two Mg2+ affinities, interpreted as tight binding at both the active and allosteric sites. Unlike other Mg2+-binding PBGS, however, metal ions are not required for TgPBGS octamer stability. A mutant enzyme lacking the C-terminal 13 amino acids distinguishing parasite PBGS from plant and animal enzymes purified as a dimer, suggesting that the C terminus is required for octamer stability. Parasite heme biosynthesis is inhibited (and parasites are killed) by succinylacetone, an active site-directed suicide substrate. The distinct phylogenetic, enzymatic, and structural features of apicomplexan PBGS offer scope for developing selective inhibitors of the parasite enzyme based on its quaternary structure characteristics.
Keywords: Allosteric Regulation, Enzymes, Heme, Parasite Metabolism, Parasitology, Plasmodium falciparum, Toxoplasma gondii, Apicomplexan Parasites, Apicoplast Metabolism, Morpheeins
Introduction
Heme is essential for cellular metabolism, and most organisms synthesize this tetrapyrrole cofactor de novo. Formation of aminolevulinic acid (ALA),3 the first committed precursor in tetrapyrrole biosynthesis, is an important regulatory step in the pathway and is achieved differently in photosynthetic versus non-photosynthetic eukaryotes. The former use the “C5” pathway to adapt glutamyl tRNA using the enzymes glutamate tRNA reductase and glutamate semialdehyde aminomutase, whereas the latter use aminolevulinic acid synthase to condense glycine and succinyl coenzyme A via the “C4” pathway (1, 2). ALA is then converted into tetrapyrroles (heme, chlorophyll, B12, etc.) using a multistep pathway, the first three enzymes of which are common to all organisms. In photosynthetic eukaryotes, the complete pathway for tetrapyrrole biosynthesis is located in the chloroplast, whereas in non-photosynthetic eukaryotes, the enzymes are typically distributed between the mitochondria and cytosol (1).
The first step in ALA utilization is catalyzed by the enzyme porphobilinogen synthase (PBGS; also known as δ-aminolevulinate dehydratase; EC 4.2.1.24), which converts two molecules of ALA into porphobilinogen (3). PBGS enzymes are highly conserved metalloproteins, with well documented enzymatic and structural properties (3, 4). Unusual characteristics include extensive variation in metal ion usage at the active and allosteric sites (5) and an unusually dynamic quaternary structure (6, 7). Cytosolic (animal/fungal) PBGS enzymes are Zn2+-dependent, whereas the plastidic (plant/algal) PBGSs respond to Mg2+. Sequence conservation and activity assays suggest that the latter contain two distinct Mg2+ binding sites: one presumed to be analogous to the catalytic Zn2+ in animal/fungal PBGS and an allosteric site present at an interdimer interface (5). Both the human (HsPBGS) (7) and Pisum sativum (PsPBGS) (6) enzymes exhibit allosteric regulation based on quaternary structure; homooctamers are highly active, whereas homohexamers are inactive; dissociation and conformational change at the dimer level is required for hexamer-octamer interconversion. Misregulation of the human PBGS quaternary structure equilibrium is associated with porphyric disease (8). Proteins exhibiting such alternative quaternary structure assemblies are called “morpheeins” (9).
The protozoan parasites Plasmodium falciparum and Toxoplasma gondii (both human pathogens in the eukaryotic phylum Apicomplexa) are capable of de novo heme biosynthesis, and inhibition of this pathway results in parasite death (10, 11). Phylogenetic studies reveal that apicomplexan heme biosynthesis enzymes exhibit a mosaic of plant and animal origins and are distributed between the parasite mitochondrion, plastid (apicoplast), and cytosol (12, 43). The plastid-associated enzymes are of plant origin, and plastid PBGS is sufficiently distinct from human PBGS to offer potential as a therapeutic target. P. falciparum PBGS forms active octamers and exhibits an unusual metal ion-independent activity (13).
In this report, we describe the cloning of multiple cDNAs encoding T. gondii PBGS (TgPBGS), identification and expression of the physiologically relevant isoform, and kinetic characterization of this enzyme. Unlike PsPBGS and Pseudomonas aeruginosa PBGS (PaPBGS), Mg2+ is not required for TgPBGS octamer stability, which appears to be conferred by an unusual C-terminal amino acid extension found in all apicomplexan PBGSs.
MATERIALS AND METHODS
Molecular Procedures
A Lambda Zap II T. gondii cDNA library (11) was used to PCR-amplify various fragments of the TgPBGS gene (see supplemental Table S1 for a list of primers), and full-length cDNA sequences were obtained by a combination of library screening and rapid amplification of cDNA ends (RACE). Quantitative “real-time” PCR of TgPBGS and T. gondii stromal processing peptidase (TgSPP) transcripts was carried out using poly(A)+ mRNA and the Power SYBR Green kit (Applied Biosystems); absolute levels were determined based on amplification of standards over a range of concentrations from 1 fg to 100 pg. Bradyzoite and sporozoite RNA was kindly provided by Drs. Florence Dzierszinski (McGill University) and Michael White (University of South Florida), respectively. A recombinant fragment of TgPBGS (amino acids 363–658) fused to a C-terminal His tag was expressed in Escherichia coli, purified on Ni2+-agarose beads (Qiagen), and used to prepare mouse polyclonal antiserum (Cocalico Inc.) for Western blotting and immunofluorescence according to standard protocols. See below for production and purification of recombinant enzyme.
Parasite Cultures
RH strain T. gondii tachyzoites were used throughout this study, following standard methods to propagate parasites in primary human foreskin fibroblast cell monolayers (14). Freshly isolated parasites were used to obtain genomic DNA (Roche Applied Science High Pure PCR template preparation kit) and total RNA (Qiagen RNeasy+ minikit) according to the manufacturers' protocols. Parasites, expressing firefly luciferase under control of the β-tubulin promoter (15), were employed for growth and inhibition assays.
TgPBGS Localization and Transit Peptide Cleavage
Transgenic parasites were engineered to express an N-terminal fragment of TgPBGS (amino acids 1–394; Fig. 1E) driven by an upstream β-tubulin promoter and fused to either enhanced green fluorescent protein (eGFP) (for subcellular localization) or hemagglutinin-tagged glutathione S-transferase (for mapping the transit peptide cleavage site). Tachyzoites transiently transfected with the TgPBGSN-eGFP plasmid (14) were incubated 18 h prior to fixation in 3.7% paraformaldehyde, permeabilization with 0.25% Triton X-100, and staining with polyclonal rabbit anti-T. gondii acyl carrier protein (43) followed by Alexa fluor-594 goat anti-rabbit IgG (Invitrogen). DNA was stained by a 5-min incubation in 2.8 μm 4′,6-diamidino-2-phenylindole dye, and imaging was carried out on an Olympus IX70 inverted microscope equipped with a mercury vapor lamp, appropriate barrier/emission filters (DeltaVision), and a Photometrics CoolSNAP high resolution digital CCD camera.
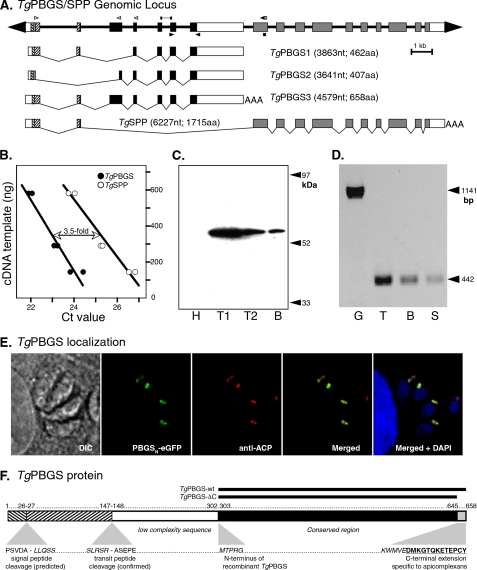
FIGURE 1.
TgPBGS organization, expression, and localization. A, 23 kb spanning the T. gondii PBGS/SPP locus on chromosome III. Exons 1 and 2 encode a bipartite apicoplast-targeting signal (signal sequence + plastid-targeting domain; hatched boxes), fused to mature coding sequences for TgPBGS (exons 3–7; black boxes) or TgSPP (exons 8–17; gray boxes). The white boxes indicate 5′- and 3′-untranslated regions (precise polyadenylation sites have not been mapped for TgPBGS1 and -2). The open arrowheads indicate primers used to define the 5′-end of TgPBGS and TgSPP transcripts by reverse transcription-PCR or RACE, bars indicate fragments amplified for quantitative PCR analysis of mRNA abundance, and closed arrowheads indicate primers used for assessing TgPBGS expression in different parasite stages. Three distinct transcripts including PBGS sequences were identified. B, quantitation of TgPBGS and TgSPP transcripts by quantitative real-time PCR using gene-specific primer pairs (supplemental Table S1); TgPBGS was amplified above the Ct threshold ∼2 cycles earlier than TgSPP, corresponding to a 3.5-fold difference in mRNA abundance (replicate samples at three different dilutions). C, Western blotting of parasite cell lysates detects the 56-kDa PBGS3 protein. H, primary human foreskin fibrobast host cell lysate; T1, 5 × 106 T. gondii tachyzoites; T2, 106 tachyzoites; B, 400 T. gondii bradyzoite cysts (purified from a chronically infected mouse brain). D, a 442-bp PCR product specific for TgPBGS3 mRNA was detected in tachyzoites (T), bradyzoites (B), and sporozoites (S). G, amplification of genomic DNA using the same primers yields a 1.1-kb product. E, subcellular localization of TgPBGS in transgenic parasites expressing the N-terminal leader (394 amino acids) fused to eGFP (green). The apicoplast (red) was detected by immunostaining for T. gondii acyl carrier protein (ACP). 4′,6-diamidino-2-phenylindole (DAPI) (blue) stains the apicoplast and both parasite and host cell nuclei. DIC, differential interference contrast. F, schematic diagram of TgPBGS protein features showing the bipartite plastid-targeting region (hatched), predicted signal peptide, and experimentally determined transit peptide cleavage sites and the portions expressed as recombinant proteins. aa, amino acids; nt, nucleotides.
A stable transgenic parasite line expressing TgPBGSN-glutathione S-transferase-hemagglutinin protein was obtained using standard methods for chloramphenicol selection and clonal isolation (14). Protein was purified from a lysate of 1010 transgenic parasites using glutathione-coupled agarose beads (Amersham Biosciences), separated by SDS-PAGE, and stained with SimplyBlue SafeStain (Invitrogen), and the mature protein band was excised for N-terminal protein sequencing (Cambridge Peptides Inc.).
Purification of Recombinant TgPBGS
cDNA clones of wild type TgPBGS (amino acids 303–658, including all conserved PBGS sequences and the C terminus) or TgPBGS-ΔC (amino acids 303–645; lacking the 13-amino acid apicomplexan-specific C-terminal extension; see Fig. 1E) were cloned into expression vector pET3a (Stratagene) and used to transform E. coli BLR(DE3) cells. Single colonies were inoculated into 1 liter of LB media containing 100 μg/ml ampicillin, 12.5 μg/ml tetracycline, and 0.4% glucose; incubated 12 h at 37°C in a shaking incubator; pelleted and resuspended in 1 liter fresh LB without glucose; and equilibrated to 15 °C. Protein expression was induced by the addition of 100 μm isopropyl 1-thio-β-d-galactopyranoside and allowed to proceed for 15 h, followed by centrifugation at 4 °C to harvest. A 10-g pellet was resuspended in 20 ml of buffer containing 0.4 mg/ml lysozyme (Sigma), 50 mm KH2PO4, pH 8.0, 170 mm KCl, 5 mm EDTA, 10 mm β-ME, and 0.1 mm phenylmethylsulfonyl fluoride. After stirring for 20 min, this mixture was diluted 1:1 with 20 ml of 100 mm KH2PO4, pH 7.0, 12 mm MgCl2, 40 μm ZnCl2, 10 mm β-ME, and 0.1 mm phenylmethylsulfonyl fluoride, followed by the addition of DNase I (Sigma) to a final concentration of 25 μg/ml and a 20-min incubation at 25 °C with stirring.
Bacteria were lysed by several cycles of freezing, thawing, and sonication and sequentially precipitated using a cut between 20 and 45% ammonium sulfate. TgPBGS protein precipitated in the 45% pellet and was resuspended in 30 mm KH2PO4, pH 7.5, 5% ammonium sulfate, and 0.1 mm MgCl2. The protein was applied to a 75-ml phenyl-Sepharose CL-4B column (GE Healthcare) and eluted at 4 °C using a 750-ml gradient from 30 to 2 mm KH2PO4 (pH 7.5) and from 15 to 0% ammonium sulfate, in the presence of 0.1 mm MgCl2. 10-ml fractions were collected at a flow rate of 1 ml/min, and those containing enzyme activity were pooled and applied to a 75-ml Q-Sepharose column (GE Healthcare) equilibrated with 10 mm Tris, pH 7.0, and 1 mm MgCl2. The column was eluted at room temperature using a 1-liter Tris gradient from 10 mm to 1 m at pH 7 in the presence of 1 mm MgCl2. 10-ml aliquots were collected at 3 ml/min, and those spanning the enzyme activity peak were pooled based on purity (assessed by SDS and native gel electrophoresis) and concentrated to ∼6 mg/ml. A 24-ml Superdex 200 analytical gel filtration column (GE Healthcare) equilibrated in 100 mm Tris, pH 7.0, 1 mm MgCl2, 10 mm β-ME was used to determine protein size after calibration with standard markers. All chromatography columns were run using the AKTA system (GE Healthcare). A portion of the resulting TgPBGS protein was dialyzed against several changes of 10 mm Tris, pH 7, containing 10 mm β-ME to remove Mg2+ ions.
Monitoring Parasite Heme Synthesis by 14C-ALA Incorporation
Freshly isolated, extracellular tachyzoite stage parasites were washed and resuspended in minimal essential medium (Invitrogen) containing 1% dialyzed fetal calf serum. 108 parasites were preincubated for 2 h at 37 °C under 5% CO2 in 1 ml of minimal essential medium containing 0, 0.1, 0.25, 0.5, 1, or 2 mm succinylacetone (pH 7.3), followed by the addition of 1 μCi of 14C-ALA (HCl salt) and incubation for a further 6–8 h. Radiolabeling was stopped by centrifugation and washing with fresh minimal essential medium. Heme was extracted as described previously (16), and radioactivity was estimated by scintillation counting.
Enzyme Activity Assays
PBGS assays were run in 100 mm Bistris propane-HCl buffer containing 10 mm β-ME, as previously reported (17), using 10 μg/reaction protein except when testing protein concentration dependence. Reaction mixtures were preincubated at 37 °C for 10 min before starting the assay with the addition of ALA (10 mm unless otherwise stated). Assays were typically run for 20 min, followed by the addition of 0.1 m HgCl2 in 20% trichloroacetic acid, development for 8 min in Ehrlich's reagent, and measurement of absorbance at 555 nm using the quartz dip probe on a Cary50 spectrometer. Enzyme activity was measured in parasite lysates using 200 μg of total protein (∼108 parasites) per assay and a 1-h reaction time.
Homology-based Structural Model of TgPBGS
The PaPBGS crystal structure (Protein Data Bank entry 1GZG) was used as a template for constructing a structural homology model of TgPBGS amino acids 324–643. Alignments were constructed in MolIDE (18) using PSI-BLAST (≥47% identity). Gaps in the alignment map to loops and were modeled independently for each monomer using Loopy (19). Side chain coordinates were retained from the template structure for identical residues; non-identical side chains were modeled using SCWRL 3.0 (20). Side chains for each subunit of the octamer were modeled in the presence of the other subunits to impart appropriate conformational constraints. PaPBGS forms an octamer in solution, and the crystal structure contains two monomers in its asymmetric dimeric unit. The TgPBGS octamer was therefore modeled using one template monomer for subunits A, C, E, and G and the other for subunits B, D, F, and H.
Because no template crystal structure is available for the C-terminal extension of apicomplexan PBGS, TgPBGS(644–658) (VEDMKGTQKFTEPCY) was modeled based on similar sequences in other proteins. The Protein Data Bank provides at least nine sequences (32 structures) containing ≥5 conserved and ≥4 identical amino acids. Five proteins include the sequence VEDMK, which is always helical. The sequence GTQKFT forms a loop and helix in several structures. In three different protein families containing the sequence FTEPCY, PCY was always a non-helical, non-strand loop region, and FTE was variable. Secondary structures for each subsequence were combined using ϕ-ψ angles from available crystal structures, and examination of the resulting possibilities produced only two sterically feasible combinations, one of which was clearly superior based on its interface with the rest of the TgPBGS model. This structure was added to each subunit of the dimer model, with manual adjustment to remove steric hindrance and improve the interaction surface with the two monomeric units. The resulting model was then overlaid with each of the dimers of the octameric 1GZG to form an octamer model of TgPBGS. The protein preparation workflow in Maestro (Schrödinger, LLC) was used to add hydrogen atoms and identify overlapping atoms in the octameric structure, and the entire model was manually refined to accommodate all interfacing residues. The quality of the resulting model was estimated using the Protein Interfaces, Surfaces, and Assemblies Service (21). See supplemental Text T1 for PDB coordinates of the TgPBGS structure model.
RESULTS
Genomic Organization and Expression of T. gondii PBGS Gene
Analysis of the T. gondii ME49 reference genome (see the ToxoDB Web site) identifies a single candidate PBGS gene, on chromosome III (TGME49_053900). Few expressed sequence tags map to this locus, and microarray evidence suggests low transcript abundance, but several independent proteomic studies have identified TgPBGS peptides (see the ToxoDB Web site). A combination of library screening and reverse transcription-PCR revealed four alternatively spliced transcripts defining 17 exons (Fig. 1A). TgPBGS1 (DQ029337, 3863 nucleotides) fuses exons 1-2-4-5-6-7; TgPBGS2 (DQ029338, 3641 nucleotides) fuses exons 1-3-4-5-6-7, using an alternative splice donor and acceptor that truncate exons 1 and 3; TgPBGS3 (DQ029339, 4579 bp) transcripts appear to initiate 129 nucleotides upstream of TgPBGS1 and -2 and fuse exons 1-2-3-4-5-6-7. All three of these transcripts include conserved PBGS sequences. The fourth cDNA includes no mature PBGS sequence, fusing exons 1 and 2 to exons 8–17 (confirmed by reverse transcription-PCR using primer pairs, as indicated in Fig. 1A, and by RACE; not shown) and is predicted to encode a plastid stromal processing peptidase (TgSPP). Production of these two unrelated proteins from the same genetic locus is also observed in P. falciparum (22). Quantitative real-time PCR was used to calculate steady-state abundance of TgPBGS in RH strain tachyzoites as 155.2 ± 31.3 fg/μg of poly(A)+ mRNA, versus 44.7 ± 2.2 fg/μg for TgSPP (i.e. TgPBGS is 3.45 ± 0.53-fold more abundant than TgSPP) (Fig. 1B).
Sequences upstream of the first four ATGs differ from the consensus translational start site for T. gondii (caaaATG) 23), are not conserved in the related parasite Neospora caninum (see the ToxoDB Web site), and fail to initiate a long open reading frame, but the fifth ATG permits synthesis of proteins of 462, 658, or 1715 amino acids (for TgPBGS1, TgPBGS3, and TgSPP, respectively). Use of the fourth ATG would correct for the frameshift introduced by TgPBGS2 intron 1, yielding an open reading frame of 407 amino acids. Only TgPBGS3 is predicted to encode all conserved PBGS sequences (exons 3–7), and Northern blot hybridization reveals a 4.6-kb TgPBGS3 mRNA (not shown). Similarly, Western blotting using polyclonal mouse antiserum raised against TgPBGS (see “Materials and Methods”) detected a ∼56-kDa protein (Fig. 1C), larger than the predicted size of TgPBGS1 or -2 but consistent with the predicted size of TgPBGS3 after cleavage of the apicoplast-targeting signal. TgPBGS transcripts were detected in cDNA derived from three major parasite life cycle stages: tachyzoites, bradyzoites, and sprozoites (Fig. 1D).
Subcellular Localization and Mapping the Correct N-terminal Region of Mature TgPBGS
Exons 1 and 2 are predicted to encode a bipartite apicoplast-targeting signal (24), consisting of a secretory signal sequence fused to a plastid-targeting domain, and are presumed to target both PBGS and SPP to the apicoplast (22). We confirmed apicoplast targeting of TgPBGS3 by expressing the N-terminal 394 amino (derived from exons 1–4) fused to eGFP in a transgenic parasite line (see “Materials and Methods” and supplemental Table S1 for primers). Co-localization with a validated marker (acyl carrier protein) (43) confirmed apicoplast localization (Fig. 1E). In order to precisely map the transit peptide cleavage site, thereby defining the correct N terminus of mature TgPBGS, a cDNA encoding the same N-terminal leader sequence was expressed in T. gondii tachyzoites as a C-terminal glutathione S-transferase-hemagglutinin fusion. This protein was isolated from parasite lysates using glutathione-Sepharose beads, and its N-terminal sequence was determined by mass spectrometry. The mature N terminus begins with alanine 184, after cleavage between positions 183 and 184 (SSLRSR183↓A184SEP), as shown in Fig. 1F. Starting from Ala184, only TgPBGS3 mRNA would be capable of encoding the ∼56-kDa (510 amino acids) mature protein detected by Western blotting (Fig. 1C).
Cloning and Purification of Recombinant TgPBGS
In order to study its kinetic properties, a recombinant version of the enzyme encoded by TgPBGS3 was overexpressed and purified from E. coli (see “Materials and Methods”). TgPBGS-wt (Fig. 1F) excludes the N-terminal targeting signals (amino acids 1–147) and low complexity sequence (amino acids 148–301), but includes amino acids 303–658 spanning all conserved PBGS sequences and a C-terminal extension that distinguishes apicomplexan PBGS from most other species (see supplemental Fig. S1). A mutant TgPBGS lacking the C-terminal 13 amino acids was also engineered (TgPBGS-ΔC; Fig. 1F) in order to compare its native structure and kinetic properties with the wild type enzyme.
TgPBGS-wt activity eluted as a single peak from a phenyl-Sepharose column (supplemental Fig. S2A, top left), yielding 90 ml of pooled fractions at ∼1 mg/ml protein. Subsequent fractionation of the phenyl-Sepharose peak on Q-Sepharose (supplemental Fig. S2A, top right) provided a 40-ml pooled fraction volume of enzyme activity peak at ∼1.2 mg/ml. All column buffers contained 1 mm MgCl2 (see “Materials and Methods” and supplemental Table S2 for further purification details). TgPBGS-ΔC activity eluted from the phenyl-Sepharose column with a slightly extended shoulder before the peak, and the partially purified protein displayed a broad elution profile from Q-Sepharose (supplemental Fig. S2A, bottom). The size and purity of recombinant proteins were confirmed by SDS-PAGE (supplemental Fig. S2B); because TgPBGS-ΔC exhibits very low activity, positive fractions were pooled and concentrated. Purified TgPBGS-ΔC protein formed a fluffy precipitate upon extended storage. No contaminating E. coli PBGS was detected (25), based on assays of TgPBGS-ΔC phenyl-Sepharose peak without metals, with Zn2+ alone, Mg2+ alone, or Zn2+ plus Mg2+ (data not shown).
Kinetic Properties and Metal Ion Dependence of Recombinant TgPBGS
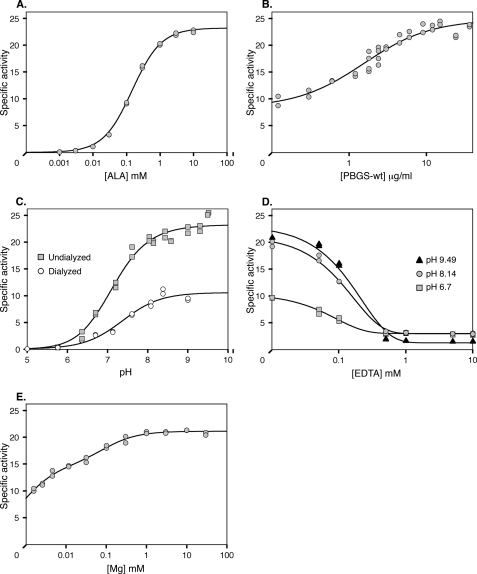
Under optimal conditions (see “Materials and Methods”), the wild type TgPBGS enzyme exhibits specific activity of 23–25 μmol of porphobilinogen/mg of protein/h and a Km of 0.149 mm (Fig. 2A), comparable with previous reports for PBGS from other species (7, 17, 26, 27). TgPBGS activity is also dependent on protein concentration, reaching maximal activity at ∼10 μg/ml (∼0.25 μm) and an apparent Kd of ∼0.04 μm (Fig. 2B), comparable with values reported for PsPBGS (17) and Bradyrhizobium japonicum PBGS (26). In other systems, concentration dependence is associated with regulation based on quaternary structure because high protein concentration favors the formation of high activity octamers over low activity hexamers. The specific activity of TgPBGS does not approach zero at low protein concentrations, however, suggesting a different quaternary structure equilibrium. The excellent fit to standard Michaelis-Menten kinetics over substrate concentrations from 1 μm to 10 mm (Fig. 2A) also argues against alternative quaternary structure assemblies with differing Km values (6, 29).
FIGURE 2.
Kinetic properties of wild type TgPBGS. A, specific activity as a function of ALA concentration. Fitting to the Michaelis-Menten equation yields a Km value of 0.149 mm and Vmax of 23.2 μmol porphobilinogen/mg of protein/h. B, TgPBGS specific activity is dependent on enzyme concentration, reaching maximal activity at ∼10 μg/ml (∼0.25 μm). The data are fitted to a simple hyperbolic equation with a y (offset). C, specific activity of TgPBGS as a function of pH and Mg2+. Undialyzed protein (squares) contains ∼2 μm Mg2+, and dialyzed protein (circles) contains <0.01 μm Mg2+; ALA contributed an additional ∼0.18 μm Mg2+ to the assay. D, effect of EDTA on TgPBGS enzyme activity (using undialyzed TgPBGS without added Mg2+). E, MgCl2 titration of TgPBGS specific activity. Two Kd values of ∼0.7 and 84 μm were determined, based on fitting a double hyperbolic equation. Unless otherwise specified, standard assay conditions included 10 μg/ml TgPBGS (except in B), 10 mm ALA (except in A), pH 8.14 (except in C and D), 1 mm MgCl2 (except in C–E), and 10 mm β-ME; assays were initiated by the addition of ALA and incubated for 20 min at 37 °C.
Many PBGS enzymes are completely dependent on metal ions, but some Mg2+-binding enzymes exhibit partial metal ion independence (13, 30). Because protein sequence alignments suggested that TgPBGS can bind Mg2+ at both the active site and an allosteric site, the enzyme was purified in 1 mm Mg2+ (see “Materials and Methods”). Close to maximal activity was observed using undialyzed TgPBGS-wt in assays containing ∼2 μm Mg2+ (Fig. 2C, squares). Thus, if Mg2+ is required for activity, it has a very high affinity for the protein because the binding site seems to be significantly populated even at low Mg2+ concentrations. ∼40% maximal activity was detected even after dialysis depleted Mg2+ to <0.3 ions/octamer, as determined by atomic absorption spectroscopy (Fig. 2C, circles). EDTA treatment eliminated the activity of undialyzed protein (Fig. 2D), consistent with the hypothesis of a tight binding catalytic Mg2+. Varying the concentration of Mg2+ added to the reaction suggests two affinities, with Kd values of ∼0.7 and 84 μm (Fig. 2E), which we attribute to the active and allosteric sites, respectively. These affinities are significantly tighter than the Kd values of 35 μm and 2 mm reported for PsPBGS (17).
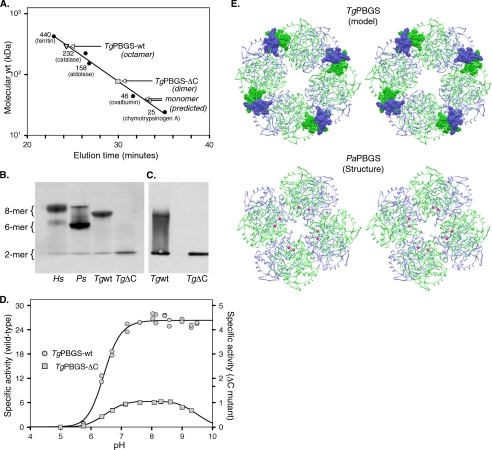
Evaluation of the TgPBGS Quaternary Structure
The molecular mass of recombinant TgPBGS was determined by gel filtration under optimal conditions (see “Materials and Methods”). Comparison with standard marker proteins indicates a molecular mass of ∼323-kDa TgPBGS, consistent with an octameric structure (Fig. 3A), as observed in other PBGS enzymes (7, 31). Unlike HsPBGS or PsPBGS, however, hexamers were not evident in TgPBGS, even as a shoulder of the main peak. The allosteric Mg2+ ions of octameric PBGS reside at a subunit-subunit interface, and their removal from PsPBGS destabilizes the octamer, resulting in hexamer accumulation (6). Purification of PaPBGS without Mg2+ yields a dimeric complex, and dialysis against millimolar levels of Mg2+ produces an octameric assembly. Substrate has also been shown to stabilize octameric assembly in PBGS proteins prone to oligomer dissociation (6, 25, 31, 32, 33). Native gel electrophoresis of TgPBGS reveals that this protein is primarily present as an octamer (Fig. 3B), and even dialysis to reduce Mg2+ to <0.3 Mg2+/octamer does not affect the quaternary structure. The absence of substrate or the presence of EDTA also did not affect the stability of TgPBGS octamer (data not shown).
FIGURE 3.
Quaternary structure analysis of TgPBGS-wt and TgPBGS-ΔC. A, determination of molecular mass by gel filtration chromatography on Superdex 200 (0.1 m Tris, pH 8.5, 10 mm MgCl2, 10 mm β-ME, 0.2 m NaCl). Enzyme elution was assessed at A280, and the column was calibrated using size markers as shown (Amersham Biosciences). For comparison, the expected elution times for monomeric TgPBGS-wt and TgPBGS-ΔC are also indicated. B, native PAGE analysis of HsPBGS (octamer > hexamer ≫ dimer), PsPBGS (hexamer > octamer ≫ dimer), TgPBGS-wt (octamer ≫ dimer), and TgPBGS-ΔC (dimer only). C, Western blotting (of native gels) confirms that the TgPBGS-ΔC mutant fails to form octamers. Comparison with B suggests that this antibody displays higher affinity for the dimer than the octamer form of TgPBGS. D, activity profile demonstrates ∼25-fold lower specific activity for TgPBGS-ΔC relative to TgPBGS-wt (0.1 m Bistris propane, 1 mm MgCl2, 10 mm β-ME, 10 mm ALA; pH range as indicated). E, homology model of TgPBGS octamer (top) and crystal structure of PaPBGS octamer (Protein Data Bank entry 1GZG; bottom), displayed as stereo images. Green, subunits A, C, E, and G; blue, subunits B, D, F, and H. Magenta, allosteric magnesium ions stabilizing the octamer form of PaPBGS. TgPBGS octamers are stable even in the absence of magnesium, probably because of dimer-dimer interactions mediated by the C-terminal extension unique to apicomplexan parasites (amino acids 646–658; shown in CPK).
As noted above, apicomplexan PBGS enzymes are distinguished by their unusual termini, including the N-terminal extension required for targeting to the apicoplast (which is cleaved post-translationally) and C-terminal extension unique to these parasites (Vibrio cholera also displays a shorter, unrelated C-terminal extension; supplemental Fig. S1). TgPBGS-ΔC, a mutant form of the enzyme lacking the C-terminal 13 amino acids, purified as dimers according to gel filtration analysis (Fig. 3A). The absence of higher order TgPBGS-ΔC oligomers was also confirmed by native gel electrophoresis (Fig. 3B) and Western blotting (Fig. 3C). This striking difference in the native structure of TgPBGS-wt versus TgPBGS-ΔC suggests that the C-terminal 13-amino acid extension may be required for octamer stability. Specific activity of TgPBGS-ΔC was ∼25-fold lower than for TgPBGS-wt (∼1 μmol/mg protein/h; Fig. 3D), but it is not clear if this activity is intrinsic to the dimer or due to transient octamer formation in the presence of substrate, which is known to stabilize the octamer form of PBGS in other species.
In order to further explore the role of the TgPBGS C-terminal extension, we constructed a homology model based on the known structure of PaPBGS (see “Materials and Methods”). Individual PBGS subunits form a ∼300-amino acid α8β8-barrel, with an extended N-terminal arm of variable length that interacts with other subunits in oligomeric forms of the enzyme (4). Available crystal structures also show that the C terminus, which is often disordered in the crystal structure, points toward the N terminus and might therefore influence higher order oligomer formation as well. The structural model of TgPBGS (Fig. 3E) predicts that the C-terminal extension increases the interaction surface area from 1604 to 2023 Å2. In addition, the solvation free energy of the protein interface (ΔiG; calculated by the Protein Interfaces, Surfaces, and Assemblies Service, EBI) was −14.6 kcal/mol for TgPBGS-wt versus −4.4 kcal/mol in the TgPBGS-ΔC mutant. For comparison, the average ΔiG for the equivalent interface in crystal structures of human, mouse, yeast, Chlorobium, and E. coli PBGS are −15.4 ± 5.4 kcal/mol, strongly arguing that the C-terminal extension of TgPBGS is required for the formation of an energetically favorable interface. This contrasts with PsPBGS and PaPBGS but is consistent with the observation that TgPBGS octamers remain stable in the absence of Mg2+ (see above), presumably due to stability provided by the C-terminal extension.
Inhibition of TgPBGS and Heme Biosynthesis in T. gondii
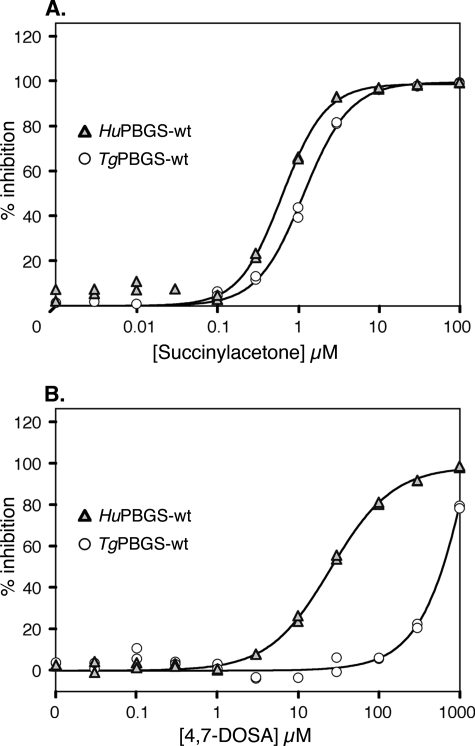
Previous studies have described various substrate mimics that specifically inhibit PBGS (28, 34–37, 41), including succinylacetone, which mimics ALA and is a potent irreversible inhibitor (38). As shown in Fig. 4A, the dose-response curve for succinylacetone inhibition of TgPBGS is comparable with that of HsPBGS, with IC50 values of ∼1 μm. This is to be expected because the inhibitor binds covalently to a conserved active site lysine, causing suicide inhibition (39). In contrast, the active site-directed, species-specific irreversible inhibitor 4,7-dioxosebacic acid, which is most effective against Zn2+-requiring PBGS (28, 37, 40), shows ∼40-fold selectivity for HsPBGS relative to TgPBGS (Fig. 4B). The specificity of 4-oxosebacic acid for E. coli PBGS is also understood at a mechanistic level (37, 40, 41); this compound failed to inhibit either TgPBGS or HsPBGS (not shown), even at concentrations as high as 3 mm and a preincubation time up to 24 h.
FIGURE 4.
Inhibition of PBGS activity by active site-directed suicide inhibitors. Inhibition of TgPBGS and HsPBGS enzyme activity (under optimal conditions) by succinylacetone (A, preincubated 30 min) or 4,7-dioxosebacic acid (4,7-DOSA) (B, preincubated 100 min).
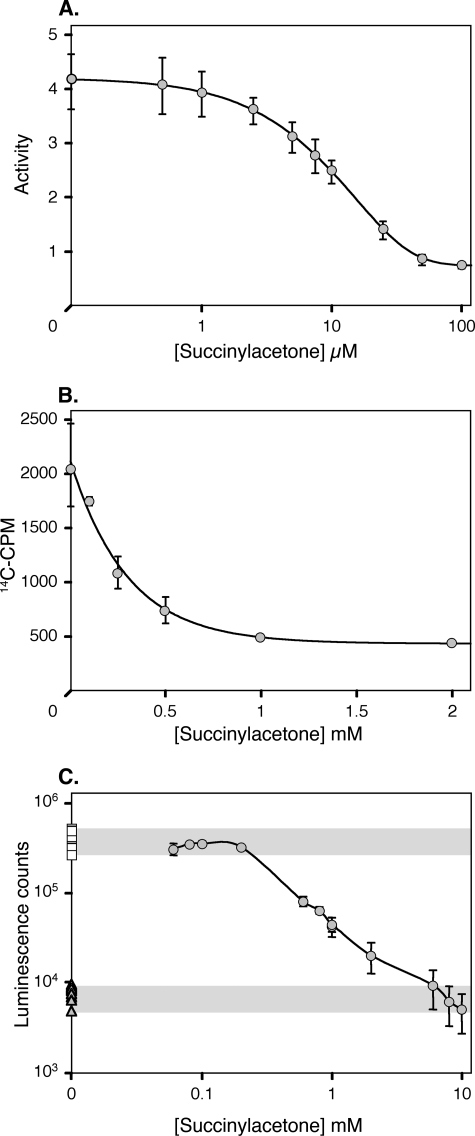
Robust heme synthesis is observed during active stages of the T. gondii life cycle and can be monitored by following the incorporation of 14C-ALA. PBGS enzyme activity in parasite cell lysates was inhibited by succinylacetone in a dose-dependent manner (Fig. 5A). Succinylacetone treatment also markedly decreased parasite heme biosynthesis (Fig. 5B). The effect of succinylacetone on the survival of T. gondii parasites was monitored using transgenic lines expressing a luciferase reporter gene (15) (Fig. 5C). Once again, succinylacetone decreased T. gondii growth in a dose-dependent manner, ultimately killing the parasites. Similar dose-response curves were obtained for heme incorporation and parasite survival. Comparable results have been reported previously for Plasmodium species (10), suggesting that PBGS activity and heme biosynthesis may be essential for apicomplexan parasite survival.
FIGURE 5.
Effect of succinyl acetone on T. gondii. A, inhibition of PBGS activity, assayed as porphobilinogen production by T. gondii tachyzoite lysates (preincubated 20 min with inhibitor in 100 mm Bistris propane buffer, pH 9.0, at 37 °C prior to 1-h incubation with substrate). B, inhibition of heme biosynthesis, assayed as 14C-ALA incorporation into heme by extracellular tachyzoite stage parasites. C, inhibition of T. gondii growth by succinylacetone. Transgenic parasite tachyzoites expressing luciferase (circles) were used to monitor growth in primary human foreskin fibrobast cells. Shading indicates positive and negative controls. Squares, untreated parasites; triangles, parasites lacking the luciferase transgene. Error bars indicate standard deviation.
DISCUSSION
The Unusual PBGS Gene Structure of Apicomplexan Parasites
Heme biosynthesis in T. gondii and P. falciparum is distinct from their animal hosts in that three enzymes of the common pathway, including PBGS, are located in the apicoplast (12). Motifs required for plastid targeting (42) can be identified in the sequence of TgPBGS and P. falciparum PBGS, and apicoplast localization has been confirmed in both species (12, 13, 43). In both species, PBGS sequences (TGME49_053900 and PF14_0381) lie immediately upstream of a stromal processing peptidase gene (pitrilysin; SPP; TGME49_053890 and PF14_0382). These two genes form a single genetic locus, exploiting the same upstream exons to encode an apicoplast-targeting signal (Fig. 1E), which is post-translationally removed by signal peptidase and pitrilysin to yield two distinct mature proteins (Fig. 1A). At the transcript level, TgPBGS is ∼3–5-fold more abundant than TgSPP (Fig. 1B); the analogous transcripts in Plasmodium are equally abundant (22). Mapping the mature N terminus of TgPBGS confirms that processing occurs near the 3′-end of exon 2, removing most of the sequences shared by TgPBGS and TgSPP (Fig. 1F). Three alternatively spliced cDNAs encoding PBGS sequence were identified in T. gondii cDNA libraries (Fig. 1A), but bioinformatic, RNA, protein, and biochemical analyses indicate that TgPBGS3 is by far the most abundant (Fig. 1C) and likely to be the only functional species.
TgPBGS Structure and Function
The activity of recombinant TgPBGS (synthesized without the N-terminal plastid-targeting signals and low complexity sequence; Fig. 1F), is comparable with HsPBGS but lower than that reported for PsPBGS. Efforts to express the full-length mature wild type TgPBGS protein (amino acids 148–658) were unsuccessful, probably because of low complexity sequence in the N-terminal region (amino acids 148–301). This domain is not well conserved between Plasmodium and Toxoplasma or even between T. gondii and its close relative N. caninum. Several insertions and deletions in the low complexity region are observed in different T. gondii isolates. We therefore expect that the TgPBGS-wt protein used for these studies (amino acids 302–658) accurately reflects the enzyme in vivo.
Dialysis, EDTA treatment, and Mg2+ titration experiments lead us to conclude that TgPBGS requires Mg2+, which is bound very tightly, but that the importance of Mg2+ to enzyme quarternary structure differs from that observed in plant/algal PBGS. In PsPBGS, Mg2+ is required for activity, binding to both the active site and an allosteric site located at a subunit interface. The latter is required for the assembly of active octamers rather than inactive hexamers (6, 31). Similarly, the removal of Mg2+ from PaPBGS promotes the formation of inactive dimers. Although titration demonstrates the importance of Mg2+ for optimal TgPBGS activity (Fig. 2E), which was abolished by complete Mg2+ removal (Fig. 2D), TgPBGS purified in 1 mm MgCl2 retained its octameric structure and ∼40% maximal enzyme activity even after removal of Mg2+ by dialysis (Fig. 2C). Atomic absorption spectroscopy confirmed very low levels of Mg2+ ions in dialyzed TgPBGS, but the addition of 10 mm ALA to the assay introduces trace Mg2+ that may be sufficient for activity. PsPBGS shows no activity under such conditions because its affinity for Mg2+ is much lower (see the legend to Fig. 2C). Mg2+ concentration in the chloroplast varies between ∼1 and 10 mm to support chlorophyll synthesis; although Mg2+ concentration in the apicoplast is unknown, it is likely to be far lower because parasites do not synthesize chlorophyll. Tight binding of Mg2+ may therefore be necessary for TgPBGS function.
Quaternary structure has been shown to dramatically affect the activity of PBGS from many species (29). The plant and human enzymes are now known to exist as a mixture of low activity hexamers and high activity octamers whose interconversion occurs through a conformational change in the dissociated dimeric assembly. These non-additive quaternary isoforms of the enzyme are called “morpheein forms,” and proteins that behave in this fashion are called “morpheeins” (9). Interconversion of morpheein forms requires oligomer disassembly, conformational change in the dissociated state, and reassembly as a structurally and functionally distinct oligomer. The equilibrium of quaternary structure assemblies of PBGS is affected by the presence of metal ions and substrate molecules, both of which favor formation of high activity octamers (7, 25). For PsPBGS, the equilibrium between octamers and hexamers results in a protein concentration dependence of the specific activity. Under optimal conditions, the specific activity of TgPBGS also exhibits protein concentration dependence (Fig. 2B), although there is no evidence for hexamers (Fig. 3, A–C). Thus, the quaternary structure equilibrium for TgPBGS is unlike that of the prototype plant PBGS.
A key structural difference between TgPBGS and PsPBGS is the C-terminal extension present in the former. Protein sequence alignments show that all apicomplexan PBGS enzymes contain such a C-terminal extension, although the precise sequence is not conserved (supplemental Fig. S1). Removal of this sequence in the mutant enzyme TgPBGS-ΔC completely eliminated its ability to assemble beyond the dimeric state (Fig. 3, A–C), pointing to the C terminus as likely to be responsible for observed differences in quaternary structure equilibria between TgPBGS and other plant-type PBGS enzymes. Fig. 3E provides a model suggesting how the C-terminal extension may hold the octamer together. Note that the C-terminal extension is predicted to lie at the edge of PBGS dimers (blue-green pairs), interacting with the C-terminal extension of adjacent dimers in an energetically favored conformation. Why TgPBGS might have evolved to use an octamer-stabilizing C-terminal extension whereas allosteric Mg2+ is sufficient to stabilize the octameric structure in PsPBGS is unclear but provides scope for further studies on morpheein structure and function and structural differences that might be exploited for therapeutic development. Crystallographic data for TgPBGS would certainly help to expedite these goals.
Capitalizing on Apicomplexan PBGS for Drug Discovery
PBGS is likely to be essential for parasite survival, both by analogy to other systems and based on the observations that treatment with succinylacetone inhibits heme biosynthesis and kills T. gondii (albeit only at micromolar concentrations, as also observed in other eukaryotic systems). Most known inhibitors of PBGS are substrate mimics that bind the highly conserved active site, making it challenging to design small molecules that can discriminate between the human and parasite enzymes (although note that 4,7-dioxosebacic acid is far more active against HsPBGS than TgPBGS). Allosteric inhibitors may be more readily developed to exploit phylogenetic differences in protein sequence and structure. Recent studies have demonstrated the potential of species-specific allosteric structure-based inhibitors of PBGS that function by trapping the protein in its inactive hexameric assembly (33, 44). Such inhibitors, termed “morphlocks,” have been selected through in silico screening for molecules that bind to a phylogenetically variable surface cavity found only in the low activity PBGS hexamer. A similar structure-based approach may be envisioned for discriminating human versus parasite PBGS, focusing on the parasite-specific C-terminal domain required for active octamer formation.
Supplementary Material
Acknowledgments
We acknowledge Linda Stith for technical assistance, Dr. Manami Nishi for assistance with microscopic imaging, Prof. Roland Dunbrack and the Fox Chase Cancer Center Molecular Modeling Facility for assistance with construction of the TgPBGS model, and Prof. Reinhard Neier for 4,7-dioxosebacic acid.
This work was supported, in whole or in part, by National Institutes of Health Grants AI28724 (to D. S. R.), ES003654 (to E. K. J.), T32CA00935 (to the Institute for Cancer Research), AI077577 (to E. K. J.), and CA006927 (to Fox Chase Cancer Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, Text S1, and Figs. S1 and S2.
- ALA
- aminolevulinic acid
- eGFP
- enhanced green fluorescent protein
- PBGS
- porphobilinogen synthase
- HsPBGS
- H. sapiens PBGS
- PsPBGS
- P. sativum PBGS
- PaPBGS
- P. aeruginosa PBGS
- TgPBGS
- T. gondii PBGS
- RACE
- rapid amplification of cDNA ends
- SPP
- stromal processing protease (pitrilysin)
- TgSPP
- T. gondii SPP
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- GFP
- green fluorescent protein
- β-ME
- β-mercaptoethanol.
REFERENCES
- 1.Papenbrock J., Grimm B. (2001) Planta 213, 667–681 [DOI] [PubMed] [Google Scholar]
- 2.Heinemann I. U., Jahn M., Jahn D. (2008) Arch. Biochem. Biophys. 474, 238–251 [DOI] [PubMed] [Google Scholar]
- 3.Jaffe E. K. (2004) Bioorg. Chem. 32, 316–325 [DOI] [PubMed] [Google Scholar]
- 4.Jaffe E. K. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 115–128 [DOI] [PubMed] [Google Scholar]
- 5.Jaffe E. K. (2003) Chem. Biol. 10, 25–34 [DOI] [PubMed] [Google Scholar]
- 6.Breinig S., Kervinen J., Stith L., Wasson A. S., Fairman R., Wlodawer A., Zdanov A., Jaffe E. K. (2003) Nat. Struct. Biol. 10, 757–763 [DOI] [PubMed] [Google Scholar]
- 7.Tang L., Stith L., Jaffe E. K. (2005) J. Biol. Chem. 280, 15786–15793 [DOI] [PubMed] [Google Scholar]
- 8.Jaffe E. K., Stith L. (2007) Am. J. Hum. Genet. 80, 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe E. K. (2005) Trends Biochem. Sci. 30, 490–497 [DOI] [PubMed] [Google Scholar]
- 10.Surolia N., Padmanaban G. (1992) Biochem. Biophys. Res. Commun. 187, 744–750 [DOI] [PubMed] [Google Scholar]
- 11.Srivastava P., Pandey V. C. (1998) Exp. Parasitol. 88, 60–63 [DOI] [PubMed] [Google Scholar]
- 12.Wu B. (2006) Heme Biosynthetic Pathway in Apicomplexan Parasites, Ph.D. thesis, University of Pennsylvania, Philadelphia, PA [Google Scholar]
- 13.Dhanasekaran S., Chandra N. R., Chandrasekhar Sagar B. K., Rangarajan P. N., Padmanaban G. (2004) J. Biol. Chem. 279, 6934–6942 [DOI] [PubMed] [Google Scholar]
- 14.Roos D. S., Donald R. G., Morrissette N. S., Moulton A. L. (1994) Methods Cell Biol. 45, 27–63 [DOI] [PubMed] [Google Scholar]
- 15.Matrajt M., Nishi M., Fraunholz M. J., Peter O., Roos D. S. (2002) Mol. Biochem. Parasitol. 120, 285–289 [DOI] [PubMed] [Google Scholar]
- 16.Bonday Z. Q., Taketani S., Gupta P. D., Padmanaban G. (1997) J. Biol. Chem. 272, 21839–21846 [DOI] [PubMed] [Google Scholar]
- 17.Kervinen J., Dunbrack R. L., Jr., Litwin S., Martins J., Scarrow R. C., Volin M., Yeung A. T., Yoon E., Jaffe E. K. (2000) Biochemistry 39, 9018–9029 [DOI] [PubMed] [Google Scholar]
- 18.Canutescu A. A., Dunbrack R. L., Jr. (2005) Bioinformatics 21, 2914–2916 [DOI] [PubMed] [Google Scholar]
- 19.Xiang Z., Soto C. S., Honig B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canutescu A. A., Shelenkov A. A., Dunbrack R. L., Jr. (2003) Protein Sci. 12, 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 22.van Dooren G. G., Su V., D'Ombrain M. C., McFadden G. I. (2002) J. Biol. Chem. 277, 23612–23619 [DOI] [PubMed] [Google Scholar]
- 23.Seeber F. (1997) Parasitol. Res. 83, 309–311 [DOI] [PubMed] [Google Scholar]
- 24.Roos D. S., Crawford M. J., Donald R. G., Kissinger J. C., Klimczak L. J., Striepen B. (1999) Curr. Opin. Microbiol. 2, 426–432 [DOI] [PubMed] [Google Scholar]
- 25.Jaffe E. K., Ali S., Mitchell L. W., Taylor K. M., Volin M., Markham G. D. (1995) Biochemistry 34, 244–251 [DOI] [PubMed] [Google Scholar]
- 26.Petrovich R. M., Litwin S., Jaffe E. K. (1996) J. Biol. Chem. 271, 8692–8699 [DOI] [PubMed] [Google Scholar]
- 27.Frankenberg N., Jahn D., Jaffe E. K. (1999) Biochemistry 38, 13976–13982 [DOI] [PubMed] [Google Scholar]
- 28.Kervinen J., Jaffe E. K., Stauffer F., Neier R., Wlodawer A., Zdanov A. (2001) Biochemistry 40, 8227–8236 [DOI] [PubMed] [Google Scholar]
- 29.Selwood T., Tang L., Lawrence S. H., Anokhina Y., Jaffe E. K. (2008) Biochemistry 47, 3245–3257 [DOI] [PubMed] [Google Scholar]
- 30.Bollivar D. W., Clauson C., Lighthall R., Forbes S., Kokona B., Fairman R., Kundrat L., Jaffe E. K. (2004) BMC Biochem. 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokona B., Rigotti D. J., Wasson A. S., Lawrence S. H., Jaffe E. K., Fairman R. (2008) Biochemistry 47, 10649–10656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffe E. K. (1995) J. Bioenerg. Biomembr. 27, 169–179 [DOI] [PubMed] [Google Scholar]
- 33.Lawrence S. H., Ramirez U. D., Tang L., Fazliyez F., Kundrat L., Markham G. D., Jaffe E. K. (2008) Chem. Biol. 15, 586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senior N. M., Brocklehurst K., Cooper J. B., Wood S. P., Erskine P., Shoolingin-Jordan P. M., Thomas P. G., Warren M. J. (1996) Biochem. J. 320, 401–412 [PMC free article] [PubMed] [Google Scholar]
- 35.Gacond S., Frère F., Nentwich M., Faurite J. P., Frankenberg-Dinkel N., Neier R. (2007) Chem. Biodivers. 4, 189–202 [DOI] [PubMed] [Google Scholar]
- 36.Heinemann I. U., Schulz C., Schubert W. D., Heinz D. W., Wang Y. G., Kobayashi Y., Awa Y., Wachi M., Jahn D., Jahn M. (2010) Antimicrob. Agents Chemother. 54, 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarret C., Stauffer F., Henz M. E., Marty M., Lüönd R. M., Bobálová J., Schürmann P., Neier R. (2000) Chem. Biol. 7, 185–196 [DOI] [PubMed] [Google Scholar]
- 38.Tschudy D. P., Hess R. A., Frykholm B. C. (1981) J. Biol. Chem. 256, 9915–9923 [PubMed] [Google Scholar]
- 39.Erskine P. T., Newbold R., Brindley A. A., Wood S. P., Shoolingin-Jordan P. M., Warren M. J., Cooper J. B. (2001) J. Mol. Biol. 312, 133–141 [DOI] [PubMed] [Google Scholar]
- 40.Erskine P. T., Coates L., Newbold R., Brindley A. A., Stauffer F., Wood S. P., Warren M. J., Cooper J. B., Shoolingin-Jordan P. M., Neier R. (2001) FEBS Lett. 503, 196–200 [DOI] [PubMed] [Google Scholar]
- 41.Jaffe E. K., Kervinen J., Martins J., Stauffer F., Neier R., Wlodawer A., Zdanov A. (2002) J. Biol. Chem. 277, 19792–19799 [DOI] [PubMed] [Google Scholar]
- 42.Waller R. F., Keeling P. J., Donald R. G., Striepen B., Handman E., Lang-Unnasch N., Cowman A. F., Besra G. S., Roos D. S., McFadden G. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12352–12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato S., Clough B., Coates L., Wilson R. J. (2004) Protist 155, 117–125 [DOI] [PubMed] [Google Scholar]
- 44.Lawrence S. H., Ramirez U. D., Selwood T., Stith L., Jaffe E. K. (2009) J. Biol. Chem. 284, 35807–35817 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.