Abstract
Three-dimensional structures of β2-microglobulin (β2m) from chicken and various mammals have been described previously, but aside from genomic sequences, very little is known about the three-dimensional structures of β2m in species other than warm-blooded vertebrates. Here, we present the first three-dimensional structure of β2m from bony fish grass carp (Ctid-β2m), resolved at 2.1 Å. The key structural differences between this new structure and previously published structures are two new hydrogen bonds at positions Ile37 and Glu38 in strand C and Lys66 in strand E, and a hydrophobic pocket around the center of the protein found in Ctid-β2m. Importantly, Ctid-β2m has a short D strand and a long loop between stands C and D, rather than the flexible region found in other β2m structures that serves as a putative binding region for the major histocompatibility complex heavy chain. Comparing the Ctid-β2m structure with those of bovine and human β2ms, the Cα root mean square deviation of the latter are 1.3 Å and 1.8 Å, respectively. Compared with the constant domains of Lamprey T cell receptor-like receptor (Lamp-TCRLC) and Amphioxus V and C domain-bearing protein (Amphi-VCPC), Ctid-β2m exhibits very different topology. The three-dimensional structures of domains predicted from Amphi-VCPC/Lamp-TCRLC are distinctly lacking in strand A of β2ms. There are 18 amino acids at the N terminus of Amphi-VCPC that may have evolved into strand A of β2ms. A mutation in the BC loops of Amphi-VCPC may have led to the novel topology found in β2m. Based on these results, Ctid-β2m may well reflect evolutionary characteristics of ancestral C set molecules.
Keywords: Evolution, Immunology, MHC, Protein Conformation, Protein Structure, B2m, Bony Fish, Crystal Structure, Evolution, IgSF
Introduction
β2-microglobulin (β2m)3 is classified as part of the set of immunoglobulin superfamily (IgSF) constant (C) proteins based on its amino acid sequence, number of strands, and folding topology (1, 2). Many protein molecules possessing the IgSF C domain exist in numerous organisms, but only two forms have been discovered in the immune system. One form is composed of IgSF domains, comprising the B cell receptor, the major histocompatibility complex (MHC) and the T cell receptor (TCR), in which IgSF C is combined with the IgSF variable (V) domain (3). The other form consists of independent molecules, such as the β2m molecules (4, 5). Both forms, including dependent and independent molecules, play important roles in the adaptive immune system. β2m is a functional molecule that noncovalently associates with MHC class I molecules to stabilize their heavy chain three-dimensional structure, which is required to bind foreign antigen peptides and to facilitate T helper lymphocyte, cytotoxic T lymphocyte, and natural killer cells during the immune response (6–8). β2m genes are conserved across species, including humans, mammals, and birds. The genomic structure of these genes contains four exons and three introns. The β2m genes are genetically unlinked and are located outside of the MHC region (9).
The three-dimensional structure of bovine β2m was first solved by Becker et al. (5). To date, the structures of β2m in humans, mice, and chickens have been determined either in their monomer form or as part of MHC I complexes (4, 5, 10, 11). The known β2m structures are composed of 99 mature residues with a seven-stranded β-sandwich fold and thus belong to the typical IgSF C1 set of molecules. In the reported three-dimensional structures of β2m, strands A, B, and E comprise one β-sheet; strands C, F, and G form the second β-sheet; and the D strand runs between the layers. A central disulfide bond, bridging Cys25 and Cys80 on the B and F stands, respectively, contributes to protein stability (14). The D strand is divided by a two-residue β-bulge into two short two-residue β-strands (D1 and D2). This β-bulge has been found in all solved β2m structures and is considered a common feature of β-sheet proteins. The loss of the β-bulge, together with changes in the positions of three residues in the CD loop linking the C and D strands, results in the formation of a new continuous six-residue β-sheet. Structural rearrangements of the D strand by a possible edge-to-edge mechanism have been implicated in human β2m amyloidosis (12). Interestingly, the IgSF C domains are not only highly conserved in B cell receptor, TCR, and MHC class I and II but also appear in FcγR I, CD2, CD4, and CD8; the C domain has even been found in lower animals, such as in the lamprey TCR-like receptor (Lamp-TCRLC) and in the Amphioxus V and C domain-bearing protein (Amphi-VCPC) (13, 14). However, it generally has been believed that the β2ms emerged suddenly in the antigen presentation system of the adaptive immune system and are present only in jawed vertebrates. Very little is known about the three-dimensional structure of β2m in the early vertebrate bony fishes, and, therefore, its evolutionary origin cannot be explained adequately.
Fish β2m genes were first reported by Ono and co-workers (15) and Dixon et al. in 1993 (16). To date, β2m genes have been reported in 16 fish species, including grass carp (17), zebrafish (15), carp and tilapia (16), trout (9), channel catfish (18), Atlantic salmon (19), and sturgeon (20). Research has demonstrated that the 116 amino acids encoded by the fish β2m genes (except for in sturgeon) are three residues shorter than those of humans, mammals, and chickens. Fish β2m genes have a number of unique characteristics that are not shared by their mammalian homologues, such as a deletion of two amino acids in the mature protein and a single N-linked glycosylation site in grass carp (17) and catfish (18). In addition, as first discovered in rainbow trout, the fish β2m locus consists of three linked genes: two similar expressed genes and one gene that is incomplete and not expressed (9).
We have reported previously a representative bony fish grass carp β2m (Ctid-β2m) gene (17). Here, we resolved the first three-dimensional structure of Ctid-β2m at 2.1 Å. Ctid-β2m closely resembles the previously described human and bovine monomeric β2m structures, with some significant variations. Two new hydrogen bonds and a hydrophobic pocket were found in Ctid-β2m. In particular, the structure of Ctid-β2m revealed an unusually flexible conformation in the region of the CD loop. This β2m structure highlights the evolutionary propensity toward stability with the existence of an unusually flexible CD loop in bony fish. Because fish β2m is evidently the evolutionary turning point of the IgSF C set of molecules, the three-dimensional structures of Lamp-TCRLC and Amphi-VCPC were predicted, and their topologies were compared with that of Ctid-β2m. Based on the structures presented here, Ctid-β2m may well reflect evolutionary characteristics tracing back to ancestral C-set molecules.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
The Ctid-β2m gene was amplified from the plasmid p2X-Ctid-β2m, which was constructed previously by our research group using PCR (21). This plasmid contains a unique NdeI restriction site, a stop codon and a unique XhoI restriction site (21, 22). The products were ligated into a pET21a vector (Novagen) and transformed into Escherichia coli strain BL21 (DE3) for protein expression. The recombinant proteins were expressed as inclusion bodies, which were then lysed using a sonicator and centrifuged at 2,000 × g. The pellet was washed three times with a solution containing Triton X-100 (20 mm Tris-HCl, 100 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol) and then once with the same solution without Triton X-100. The inclusion body was dissolved overnight in urea buffer (8 m urea, 50 mm Tris-HCl pH 8.0, 100 mm NaCl, 10 mm EDTA, 10% (v/v) glycerine, 10 mm dithiothreitol) using ∼1 ml of urea buffer per 30 mg of protein. The Ctid-β2m was refolded by the gradual dilution method using the following refolding buffer: 100 mm Tris-HCl, 2 mm EDTA, 400 mm l-arginine-HCl, 0.5 mm oxidized glutathione, 5 mm reduced glutathione, 0.1 mm phenylmethylsulfonyl fluoride, and 0.1 mm NaN3, pH 8.0. After stirring for 48 h at 4 °C, the remaining soluble proteins were concentrated and purified using a Superdex G-200 (Amersham Biosciences) size-exclusion column followed by Resource-Q (Amersham Biosciences) ion-exchange chromatography (21, 23).
Crystallization, Data Collection, and Processing
The purified Ctid-β2m was adjusted to a concentration of 10 mg/ml with crystallization buffer (10 mm Tris-HCl, 10 mm NaCl). An initial crystallization trial was set up with Crystal Screens I and II (Hampton Research) at 18 °C using the hanging drop method. The drop, containing equal volumes (1 μl each) of protein solution and reservoir crystallization buffer, was placed over a well containing 200 μl of reservoir solution using VDX plates. Crystals suitable for data collection were grown in 3–5 days under optimized conditions using 0.1 m MES pH 6.5, 12% polyethylene glycol 20000, 3% (v/v) ethanol, and a protein concentration of 5 mg/ml. For data collection, the crystals were soaked for several minutes in reservoir solution supplemented with 20% glycerol as a cryoprotectant and then flash cooled directly in liquid nitrogen. X-ray diffraction data were collected to 2.1 Å resolution on a Rigaku MicroMax007 rotating-anode x-ray generator operated at 40 kV and 20 mA (CuKα; λ = 1.5418 Å) equipped with an R-AXIS VII2+ image plate detector. The data were processed and scaled using DENZO and SCALEPACK as implemented in HKL-2000 (24). The data collection statistics of the Ctid-β2m crystals are shown in Table 1.
TABLE 1.
Statistics for data and refinement of Ctid-β2m
Values in parentheses are given for the highest resolution shell. Rfree is calculated over reflections in a test set (5%) not included in atomic refinement. r.m.s., root mean square.
| Data statistics | |
| No. of reflections (unique/total) | 6,433 (82,112) |
| Resolution (Å) | 2.1 (2.10-2.18) |
| Completeness (%) | 92.2 (55.9) |
| Rmerge (%) | 10.7 (33.4) |
| I/σ | 25.3 (5.4) |
| Unit cell parameters (Å) | 38.74, 40.60, 71.09 |
| Space group | P212121 |
| Wavelength (Å) | 1.5418 |
| Refinement statistics | |
| Resolution range (Å) | 35.56–2.1 |
| No. of protein atoms | 781 |
| No. of water molecules | 196 |
| Average B-factor (Å2) | 30.93 |
| r.m.s. deviation | |
| Bonds (Å) | 0.006 |
| Angle | 1.082° |
| Rwork (%) | 19.30 |
| Rfree (%) | 22.34 |
Structure Solution, Refinement, and Analysis
The crystal structure of Ctid-β2m was solved by molecular replacement using human β2m (PDB code 1LDS) as a search model for the CNS program (24, 25). Residues that differed between Ctid-β2m and the search model were manually rebuilt in the O program (26) under the guidance of Fo − Fc and 2 Fo − Fc electron density maps. After refinement of the model with the CNS program using stimulated annealing, energy minimization, restrained individual B factors, and the addition of 196 water molecules, the Rwork and Rfree dropped to 19.3 and 22.34%, respectively for all data between 35 and 2.1 Å. The course of refinement was monitored by calculating Rfree based on a subset containing 3% of the total number of unique reflections. The coordinate error estimated by the Luzzati plot in CNS (21, 22) for the Ctid-β2m structure is 0.41 Å. The average real-space fit value for Ctid-β2m, as calculated by O (26), is 0.95. Model geometries were verified using the PROCHECK program (27).
Homology Modeling of the IgSF C Domains in Jawless Vertebrates and Protochordates
Although fusion molecules, including the IgSF C domain, exist widely in the adaptive immune system, only two proteins, Amphi-VCPC and Lamp-TCRLC, were predicted to possess the IgSF C domain in jawless vertebrates and protochordates (13, 14). Because the three-dimensional structures of Amphi-VCPC and Lamp-TCRLC are not yet available, but are necessary to determine the evolutionary origin of the IgSF C set, the three-dimensional structures of Amphi-VCPC and Lamp-TCRLC were predicted by amino acid homology modeling using the SWISS-MODEL server based on the existing three-dimensional structures of ligand binding protein (Lingo-1) (PDB code 2ID5) and T cell surface glycoprotein CD4 (PDB code 2NY4) in the Protein Data Bank. DNAMAN was used to analyze the differences among these molecules and the PyMOL Molecular Graphics System (DeLano Scientific) was used for figure preparation.
RESULTS
Overall Structure of Ctid-β2m
The mature Ctid-β2m contains 97 amino acids (compared with 99 for human and mice). As expected, the Cα root mean square deviation of Ctid-β2m (PDB code 3GBL) is 1.3 and 1.8 Å, similar to values for the structures of monomer bovine β2m (PDB code 1BMG) and human β2m (PDB code 1LDS). The chains are folded into a typical “β-barrel” configuration dominated by two antiparallel pleated sheets; one sheet is composed of four strands, and the other sheet is composed of three strands (Fig. 1A). The Ctid-β2m structure is composed of two face-to-face β-sheets of different sizes: the “large β-sheet” is composed of strands A (6–11 aa), B (21–28 aa), D (55–57 aa), and E (60–70 aa), and the “small β-sheet” is formed by strands C (36–41 aa), F (78–84 aa), and G (87–92 aa). Six loop regions (AB, BC, CD, CE, EF, and FG) connect these strands. The two β-sheets are linked by a Cys25-Cys80 disulfide bridge, which is highly conserved in all known β2ms and provides strong geometric constraints on the surrounding residues. A cluster of hydrophobic residues (Ile7, Ile9, and Tyr10 on strand A; Leu23, Ile24, Ytr26, and Val27 on strand B; Ile37, Leu39, and Leu40 on strand C; Phe54 on strand D; Trp69, Leu64, Thr65, Val68, and Ple70 on strand E; Typ78 and Val82 on strand F; and Thr88 and Val92 on strand G) that are likely to form a strong hydrophobic pocket is found on both the large and small β-sheets (Fig. 1B).
FIGURE 1.
Overall structure of Ctid-β2m. A, Ctid-β2m is shown in a ribbon representation and colored orange. β-strands are labeled with letters A to G. The CD loop and the intrachain disulfide bridge are labeled in red and purple, respectively. B, the hydrophobic pocket in Ctid-β2m. The hydrophobic residues (Ile7, Ile9, and Tyr10 on strand A; Leu23, Ile24, Ytr26, and Val27 on strand B; Ile37, Leu39, and Leu40 on strand C; Phe54 on strand D; Trp69, Leu64, Thr65, Val68, and Ple70 on strand E; Typ78 and Val82 on strand F; and Thr88 and Val92 on strand G) are labeled in blue.
Comparison of the Three-dimensional Structures of β2ms in Vertebrates
An alignment of Ctid-β2m with zebrafish, trout, catfish, salmon, chicken, bovine, mouse, and human β2ms revealed 37 identical amino acids (38% homology). Without Ctid-β2m, 48 amino acids are identical among an alignment of the other β2ms (48% homology) (Fig. 2A). In Ctid-β2m, there are 20 amino acid residues that are distinct from the other homologues. These residues are randomly distributed throughout the β-sheets and loops. Previous studies have shown that most β2ms in teleosts are mature proteins composed of 97 amino acids. They have two amino acid deletions at positions 91 and 92, in which they differ from mammalian β2ms (9, 17). Compared with human β2m, it has been indicated that bovine monomeric β2m has an additional deletion at position 48 (28). The function of the amino acid deletion is as yet unknown. The strands of Ctid-β2m are slightly different compared with previously described β2ms. Strand B spans residues 21–28, which is the same as in mouse β2m, whereas this strand in human β2m is composed of residues 21–30. Strand G is longer than in other described β2m structures. Because of the two amino acids deleted in Ctid-β2m at positions 91 and 92, loop FG is shorter than its human β2m counterpart (Fig. 2, B and C). Strand E is two amino acids longer than in human β2m and corresponds to residues 60–70 as opposed to 62–70 in human β2m. Compared with bovine and human β2ms, the total Cα root mean square deviation values of Ctid-β2m are 1.3 and 1.8Å, respectively, using the program DALI (Table 1). The main difference between Ctid-β2m and human β2m is found around the loops from His11 to Asn20 and from His83 to Lys86, which correspond to Arg13 to Phe23 and Asn84 to Lys92 in human β2m (PDB code 1LDS). Compared with bovine β2m, the loops in Ctid-β2m are different in sequence, from residue Glu56 to Trp59 and from His83 to Lys86, which correlate to Ser56 to Ser60 and Lys82 to Arg90 in bovine β2m (Fig. 2). Altogether, these data indicate small changes in β-sheet composition among different species.
FIGURE 2.
Analysis of Ctid-β2m with β2ms from fish, chicken, and mammals. A, sequence alignment of Ctid-β2m with β2ms from fish and mammals. The sources of the sequences are as follows: Ctid-β2m (GenBankTM accession no. AB190815, PDB code 3GBL), zebrafish (GenBankTM accession no. L05383), trout (GenBankTM accession no. L49056), catfish (GenBankTM accession no. AF016042), salmon (GenBankTM accession no. AF180488), chicken (GenBankTM accession no. M84767), bovine (GenBankTM accession no. NM_173893), mouse (GenBankTM accession no. NM_009734), and human (GenBankTM accession no. NM_004048). Numbers over the alignment denote residues that form Ctid-β2m. Black arrows above the alignment indicate β-strand. T, toil. Residues highlighted in red are absolutely conserved, whereas those with blue squares are highly conserved (80%). Green numbers denote residues that form disulfide bonds. The alignment was generated using the program Clustal X and drawn with ESPript. B, structural superpositions of Ctid-β2m with the β2m monomer from chickens. Ctid-β2m and chicken β2m are colored in orange and green, respectively. C, structural superpositions of Ctid-β2m with β2m monomers from humans and bovines. Ctid-β2m, human, and bovine β2ms are colored in orange, green, and red, respectively. The superposition was created by PyMOL, using the C atoms of the globular segment. Ctid-β2m (PDB code 3GBL), chicken (PDB code 3BEW), HLA β2m (PDB code 1LDS), and bovine (PDB code 1BMG).
Two Additional Hydrogen Bonds Found in Ctid-β2m
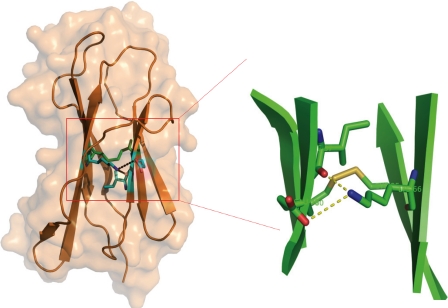
A total of ∼60 intramolecular hydrogen bonds (mostly main chain to main chain) stabilize the folded human β2m (28). The hydrogen bonds of Ctid-β2m between strands D and E are conserved, as compared with human β2m. Both human β2m and Ctid-β2m form hydrogen bonds. Ctid-β2m has hydrogen bonds between Gln50–Ser67, Thr52–Thr65, and Ala55–His63, whereas the corresponding bonds of human β2m are Glu50–Tyr67, Ser52–Leu65, and Ser55–Tyr63. Asp53 is highly conserved in all β2ms and forms significant hydrogen bonds to the heavy chain in the human HLA-β2m complex structure. In human β2m, Asp53 is located in the β-bulge and forms hydrogen bonds to Gln32, Arg35, and Arg48 in the α1 domain of the HLA heavy chain, whereas the counterpart residues in the grass carp heavy chain are Gln31, Tyr34, and Lys45. These three residues (Gln32, Arg35, and Arg48) are also highly conserved in Ctid-β2m. However, two new hydrogen bonds in Ctid-β2m appear between the directly adjacent C and E strands (Fig. 3A). Two hydrogen bonds are formed at positions Ile37 and Glu38 in strand C to integrate with Lys66 in strand E. A hydrogen atom from the ϵ-amino group of Lys and an oxygen atom provided by the carboxyl group of Ile formed one hydrogen bond. Another hydrogen bond was formed by the above mentioned hydrogen atom and an oxygen atom from the γ-carboxyl group of Glu. The lengths of these hydrogen bonds are 2.75 and 3.54 Å, respectively, whereas the distance between the two strands is 7.65 Å. Lys66 is a fish species-specific residue not present in mammalian and chicken β2ms, whereas Ile37 is highly conserved (Fig. 3B). The disulfide bond formed by residues Cys25 and Cys80 occurs between strands B and F and is highly conserved among all β2ms, including Ctid-β2m. The two new hydrogen bonds between strands C and D, which have not been found in any other resolved β2m structures, and the newly described hydrophobic pocket indicate a stable interaction between strands C and D in Ctid-β2m.
FIGURE 3.
Details of the Ctid-β2m contacts of the two extra hydrogen bonds and the disulfide bond. Hydrogen bonds are illustrated as dotted lines. The disulfide bond is shown as a yellow stick. Residues forming hydrogen bonds are shown in the stick model and colored by atom types: blue, N; red, O. All of the residues are labeled.
The Unusual Flexible CD Loop in Ctid-β2m
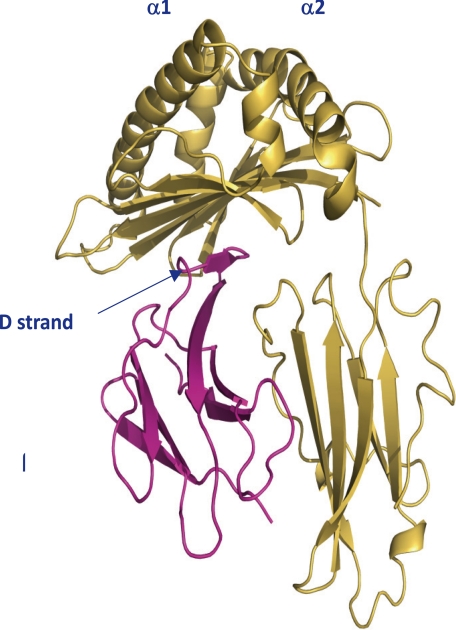
The most structurally important part of the β2ms is strand D, which binds to the surface of the MHC heavy chain (6, 23). Residues 50–56 (D1–D2 strands) interact with the heavy chain and are part of a region formed by two small strands. Residues Glu50–His51 comprise D1 and Ser55–Phe56 comprise D2. Between the D1–D2 strands, a noticeable β-bulge is formed by Asp53–Leu54 in the human structure (6). The β-bulge is found in all β2m structures described to date, as well as in monomeric bovine β2m (5). However, strand D in Ctid-β2m starts at positions 55–57, and only a short β-sheet was observed. Ctid-β2m is different from human monomeric β2m in that a longer β-sheet begins at positions 51–56. Much attention has been paid to the short D strand region, which can be involved in contacts with the heavy chain. A rather flexible domain from the CD loop to the D strand was found in Ctid-β2m (Fig. 4). Thus, Ctid-β2m appears to form a relatively unusual flexible region, which contributes greatly to the instability of its MHC class I complex.
FIGURE 4.
Structural superposition of Ctid-β2m with Human HLA-A*0201. Ctid-β2m is shown in a ribbon representation and colored orange, and the CD loop is indicated by a box. The PyMOL Molecular Graphics System was used to prepare the figure, and the geometry of the refined structure was validated according to Ramachandran plot criteria.
Comparison of Ctid-β2m with Lamp-TCRLC and Amphi-VCPC
In jawless vertebrates, Lamp-TCRLC possessing the IgSF C domain was found to have 20.7% identity with Ctid-β2m at the amino acid level (13). To determine the evolutionary origin of the IgSF C set, the three-dimensional structure of Lamp-TCRLC was predicted using amino acid homology modeling. Using the first approach mode in SWISS-MODEL, the three-dimensional structure of the Lingo-1 molecule (PDB code 2ID5; 29) was found to have 24.68% identity with the Lamp-TCRLC at the amino acid level. The Expect-value of the constructed Lamp-TCRLC three-dimensional structure is 2.20e−8. The three-dimensional structure of Lamp-TCRLC is composed of 76 residues (amino acids 19–94) that form a six-stranded β-sandwich fold (Fig. 5A), but it is not a typical IgSF C molecule. In the three-dimensional structure of Lamp-TCRLC, strands A (20–24 aa) and C (56–59 aa) comprise one β-sheet, whereas strands B (34–38 aa), E (73–81 aa), and F (84–92 aa) form the second β-sheet, and strand D (66–69 aa) runs between the layers (Fig. 5B). A central disulfide bond between Cys25 and Cys76 on loop AB and strand E, respectively, may stabilize the protein. Strand D lies on top of the two β-sheets. Altogether, the topology of Lamp-TCRLC is very different from that of Ctid-β2m (Fig. 5C).
FIGURE 5.
The predicted three-dimensional structures of Lamp-TCRLC, Amphi-VCPC, and a structural overlay with Ctid-β2m. A, sequence alignment of Ctid-β2m with Lamp-TCRLC. Three-dimensional structure of Lamp-TCRLC alone (B) and overlaid with Ctid-β2m (C). Using the first approach mode in SWISS-MODEL, the three-dimensional structure of the Lingo-1 molecule (2ID5A) is found to share 24.68% identity with the IgSF C domain of Lamp-TCRLR at the amino acid level. The best E-value of the constructed Lamp-TCRLC three-dimensional structure is 2.20e−8. The three-dimensional structure of Lamp-TCRLC is composed of 76 residues (amino acids in the 19–94 region) with a six-stranded β-sandwich fold. D, sequence alignment of Ctid-β2m with Amphi-VCPC. Structure of Amphi-VCPC alone (E) and overlaid with Ctid-β2m (F). The three-dimensional structure of Amphi-VCPC was predicted by amino acid homology modeling, based on part of a CD4 complex (PDB code 2NY4B). Amphi-VCPC shares 16.88% identity with human T cell surface glycoprotein CD4. The E-value of the constructed three-dimensional structure is 1.00e−8. The predicted three-dimensional structure of the Amphi-VCPC domain is composed of 75 residues (amino acids in the 8–82 region) with a six-stranded β-sandwich fold.
In protochordates, only a single gene, which is referred to as Amphi-VCP, has been predicted to have the IgSF C domain (14). Amphi-VCP was found to have 17.6% identity with Ctid-β2m at the amino acid level. To further determine the evolutionary origin of the IgSF C set, the three-dimensional structure of Amphi-VCPC was predicted using amino acid homology modeling based on part of the CD4 complex (PDB code 2NY4B; 30). Amphi-VCPC was found to have 16.88% identity with the human T cell surface glycoprotein CD4 molecule (PDB code 2NY4B). The E-value of the constructed three-dimensional structure is 1.00e−8. The predicted three-dimensional structure of the Amphi-VCPC domain is composed of 75 residues (in the 8–82 region) that form a six-stranded β-sandwich fold (Fig. 5D), which is not typical for IgSF C set molecules. In the three-dimensional structure of Amphi-VCPC, strands A (17–20 aa) and C (48–51 aa) comprise one β-sheet, whereas B (31–35 aa), E (62–69 aa), and F (72–79 aa) form the second β-sheet. Strand D (56–58 aa) is positioned on top of the two β-sheets (Fig. 5E). However, the predicted central disulfide bond connecting Cys21 and Cys65 on loop AB and strand E is not found in the predicted three-dimensional structure. Compared with Ctid-β2m, the topology of Amphi-VCPC is quite different (Fig. 5F), whereas the three-dimensional structures of Amphi-VCPC and Lamp-TCRLC are remarkably similar.
The Evolutionary Origin of the IgSF C Set
By examining the topology of the bony fish β2m, Amphi-VCPC and Lamp-TCRLC molecules, features indicating the evolutionary origin of the IgSF C set were found. The β2m molecule might be traceable as a descendent of the ancestor of Amphi-VCPC/Lamp-TCRLC-like molecules. In support of this possibility, 1) 18–19 aa in the N terminus of Amphi-VCPC/Lamp-TCRLC could have evolved to become the A strand of β2m, and 2) a mutation in the BC loops of Amphi-VCPC/Lamp-TCRLC molecules may have led to the novel topology found in β2m.
DISCUSSION
In the study, we determined to 2.1 Å the three-dimensional structure of the β2m molecule in bony fish, which is the first β2m described in non-warm-blooded animals. The results show that Ctid-β2m is composed of A, B, D, and E strands and C, F, and G strands like a standard IgSF C set molecule. Two extra hydrogen bonds at positions Ile37 and Glu38 and a hydrophobic pocket around the center of Ctid-β2m were also found. Importantly, a short D strand and a longer CD loop might compose a flexible region for binding to the MHC heavy chain in bony fish. These features are different from the structures of mammalian and chicken β2ms, which have two strands, D1 and D2. To explore the evolutionary origin of the unattached IgSF C set, we homology modeled Amphi-VCPC and Lamp-TCRLC at the three-dimensional level. The proteins in the fusion form as well as the IgSF C might be precursors of β2ms, although β2m emerged suddenly in fish species. These results highlight the evolutionary propensity toward stability with the presence of an unusual flexible CD loop that co-evolved with the MHC class I molecules 400 million years ago.
IgSF domains can be classified as variable, constant, strand-switched, or hybrid based on their β-strand topology (2). Although the IgSF domains exist widely in the adaptive immune system of vertebrates, only two genes, Amphi-VCP and Lamp-TCRL, have highlighted the significance of the C domains in the jawless vertebrates and protochordates (13, 14). To approach the evolutionary origin of the IgSF C, the three-dimensional structures of Amphi-VCPC and Lamp-TCRLC were predicted, although homology modeling is not precise. The three-dimensional structure of Lamp-TCRLC is composed of 76 residues that form a six-stranded β-sandwich fold but is not typical for IgSF C set molecules. The three-dimensional structure of Amphi-VCPC is composed of 75 residues and also forms a six-stranded β-sandwich fold. Surprisingly, the topologies of Lamp-TCRLC and Amphi-VCPC are identical, although Amphi-VCPC and Lamp-TCRLC molecules split more than 500 million years ago. We hypothesized that the precursors of β2ms are the IgSF C set-related molecules as well as Amphi-VCP/Lamp-TCR-like molecules. In support of this hypothesis, the C domains of Amphi-VCPC and Lamp-TCRLC lack the A strand, which is similar to Ctid-β2m. That might imply that the B strand of β2m is the hodiernal A strand in Amphi-VCPC/Lamp-TCRLC. There are 18–19 aa at the N terminus of the Amphi-VCPC/Lamp-TCRLC molecules that could have evolved to become an A strand, as in β2m. In addition, a mutation event might have occurred in the BC loops of Amphi-VCPC/Lamp-TCRLC that led to the origination of a new strand. If so, the topology of Amphi-VCPC/Lamp-TCRLC might have either evolved to be the precursor of β2m or led to the creation of new fusion molecules, such as B cell receptor, TCR, Ig, MHC, CD4, and CD8.
In conclusion, the crystal structure of Ctid-β2m was solved by molecular replacement. In the three-dimensional structure of Ctid-β2m, two new hydrogen bonds and a strong hydrophobic pocket were discovered, resulting in two more stable β-sheets. On the other hand, a single D strand and long CD loop were found, which are indicative of an unusual flexible D strand in Ctid-β2m. The three-dimensional structure of Ctid-β2m highlights the evolutionary propensity toward stability, with the presence of an unusual flexible CD loop for binding to the MHC class I heavy chain. We hypothesize that β2ms evolved from C set molecules, such as Amphi-VCPC and Lamp-TCRLC molecules, in evolutionarily earlier animals. The predicted three-dimensional structures of both Amphi-VCPC and Lamp-TCRLC distinctly lacked the A and D strands of β2m. Furthermore, a region of 18–19 aa in the N terminus of Amphi-VCPC and Lamp-TCRLC could have evolved to be the A strand of β2ms. Altogether, a mutation occurring in the BC loops of Amphi-VCPC and Lamp-TCRLC-like molecules may have led to a new topology, forming the basis for what is now a standard C set molecule.
This work was supported by the National Basic Research Program (Project 973) of China (2007CB815805) and the National Natural Science Foundation of China (Grant 30371098).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) AF016042, AF180488, M84767, NM_173893, NM_009734, and NM_004048.
The atomic coordinates and structure factors (code 3GBL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- β2m
- β2-microglobulin
- TCRLC
- T cell receptor-like receptor
- VCPC
- V and C domain-bearing protein
- IgSF
- immunoglobulin superfamily
- MHC
- major histocompatibility complex
- C
- constant
- V
- variable
- PDB
- Protein Data Bank
- aa
- amino acids.
REFERENCES
- 1.Cooper M. D., Alder M. N. (2006) Cell 124, 815–822 [DOI] [PubMed] [Google Scholar]
- 2.Bork P., Holm L., Sander C. (1994) J. Mol. Biol. 242, 309–320 [DOI] [PubMed] [Google Scholar]
- 3.Hernández Prada J. A., Haire R. N., Allaire M., Jakoncic J., Stojanoff V., Cannon J. P., Litman G. W., Ostrov D. A. (2006) Nat. Immunol. 7, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trinh C. H., Smith D. P., Kalverda A. P., Phillips S. E., Radford S. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9771–9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker J. W., Reeke G. N., Jr. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4225–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. (1987) Nature 329, 506–512 [DOI] [PubMed] [Google Scholar]
- 7.Lancet D., Parham P., Strominger J. L. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 3844–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaëlsson J., Achour A., Rölle A., Kärre K. (2001) J. Immunol. 166, 7327–7334 [DOI] [PubMed] [Google Scholar]
- 9.Magor K. E., Shum B. P., Parham P. (2004) J. Immunol. 172, 3635–3643 [DOI] [PubMed] [Google Scholar]
- 10.Achour A., Persson K., Harris R. A., Sundbäck J., Sentman C. L., Lindqvist Y., Schneider G., Kärre K. (1998) Immunity 9, 199–208 [DOI] [PubMed] [Google Scholar]
- 11.Koch M., Camp S., Collen T., Avila D., Salomonsen J., Wallny H. J., van Hateren A., Hunt L., Jacob J. P., Johnston F., Marston D. A., Shaw I., Dunbar P. R., Cerundolo V., Jones E. Y., Kaufman J. (2007) Immunity 27, 885–899 [DOI] [PubMed] [Google Scholar]
- 12.Richardson J. S., Richardson D. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancer Z., Mayer W. E., Klein J., Cooper M. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13273–13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu C., Dong M., Wu X., Li S., Huang S., Su J., Wei J., Shen Y., Mou C., Xie X., Lin J., Yuan S., Yu X., Yu Y., Du J., Zhang S., Peng X., Xiang M., Xu A. (2005) J. Immunol. 174, 3493–3500 [DOI] [PubMed] [Google Scholar]
- 15.Ono H., Figueroa F., O'hUigin C., Klein J. (1993) Immunogenetics 38, 1–10 [DOI] [PubMed] [Google Scholar]
- 16.Dixon B., Stet R. J., van Erp S. H., Pohajdak B. (1993) Immunogenetics 38, 27–34 [DOI] [PubMed] [Google Scholar]
- 17.Hao H. F., Yang T. Y., Yan R. Q., Gao F. S., Xia C. (2006) Fish Shellfish Immunol 20, 118–123 [DOI] [PubMed] [Google Scholar]
- 18.Criscitiello M. F., Benedetto R., Antao A., Wilson M. R., Chinchar V. G., Miller N. W., Clem L. W., McConnell T. J. (1998) Immunogenetics 48, 339–343 [DOI] [PubMed] [Google Scholar]
- 19.Persson A. C., Stet R. J., Pilström L. (1999) Immunogenetics 50, 49–59 [DOI] [PubMed] [Google Scholar]
- 20.Lundqvist M. L., Appelkvist P., Hermsen T., Pilström L., Stet R. J. (1999) Immunogenetics 50, 79–83 [DOI] [PubMed] [Google Scholar]
- 21.Chen W., Chu F., Peng H., Zhang J., Qi J., Jiang F., Xia C., Gao F. (2008) Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 64, 200–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao H. F., Li X. S., Gao F. S., Wu W. X., Xia C. (2007) Protein Expr. Purif. 51, 120–125 [DOI] [PubMed] [Google Scholar]
- 23.Chu F., Lou Z., Chen Y. W., Liu Y., Gao B., Zong L., Khan A. H., Bell J. I., Rao Z., Gao G. F. (2007) J. Immunol. 178, 944–952 [DOI] [PubMed] [Google Scholar]
- 24.Smith K. J., Reid S. W., Harlos K., McMichael A. J., Stuart D. I., Bell J. I., Jones E. Y. (1996) Immunity 4, 215–228 [DOI] [PubMed] [Google Scholar]
- 25.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 28.Rosano C., Zuccotti S., Bolognesi M. (2005) Biochim. Biophys. Acta 1753, 85–91 [DOI] [PubMed] [Google Scholar]
- 29.Mosyak L., Wood A., Dwyer B., Buddha M., Johnson M., Aulabaugh A., Zhong X., Presman E., Benard S., Kelleher K., Wilhelm J., Stahl M. L., Kriz R., Gao Y., Cao Z., Ling H. P., Pangalos M. N., Walsh F. S., Somers W. S. (2006) J. Biol. Chem. 281, 36378–36390 [DOI] [PubMed] [Google Scholar]
- 30.Zhou T., Xu L., Dey B., Hessell A. J., Van Ryk D., Xiang S. H., Yang X., Zhang M. Y., Zwick M. B., Arthos J., Burton D. R., Dimitrov D. S., Sodroski J., Wyatt R., Nabel G. J., Kwong P. D. (2007) Nature 445, 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]