Abstract
Survivin and Plk1 kinase are important mediators of cell survival that are required for chromosome alignment, cytokinesis, and protection from apoptosis. Interference with either survivin or Plk1 activity manifests many similar outcomes: prometaphase delay/arrest, multinucleation, and increased apoptosis. Moreover, the expression of both survivin and Plk1 is deregulated in cancer. Given these similarities, we speculated that these two proteins may cooperate during mitosis and/or in cell death pathways. Here we report that survivin and Plk1 interact during mitosis and that Plk1 phosphorylates survivin at serine 20. Importantly, we find that overexpression of a non-phosphorylatable version, S20A, is unable to correct chromosomes connected to the spindle in a syntelic manner during prometaphase and allows cells harboring these maloriented chromosomes to enter anaphase, evading the spindle tension checkpoint. By contrast, the constitutive phosphomimic, S20D, completes congression and division ahead of schedule and, unlike S20A, is able to support proliferation in the absence of the endogenous protein. Despite the importance of this residue in mitosis, its mutation does not appear to affect the anti-apoptotic activity of survivin in response to TRAIL. Together, these data suggest that phosphorylation of survivin at Ser20 by Plk1 kinase is essential for accurate chromosome alignment and cell proliferation but is dispensable for its anti-apoptotic activity in cancer cells.
Keywords: Apoptosis, Cell Division, Mitosis, Protein Phosphorylation, RNA Interference (RNAi), Polo-like Kinase 1, Survivin
Introduction
Survivin is a protein with multiple functions, whose expression is deregulated in cancer. It is best known for its participation in the chromosomal passenger protein (CPP)2 complex during mitosis, and its ability to inhibit apoptosis (reviewed in Refs. 1–3). During mitosis, survivin is regulated by the kinases Aurora-B and Cdk1. Mutation of the Aurora-B phosphorylation site at Thr117 (4) or treatment with an Aurora-B inhibitor alters the affinity of survivin for centromeres and interferes with the error correction process facilitated by CPPs that ensures proper alignment of chromosomes at the metaphase plate (5–7). Furthermore, data from a constitutive phosphomimic suggest that phosphorylation of survivin by Aurora-B prevents the completion of cytokinesis, implying a critical requirement for dephosphorylation of this site (7). Cdk1 phosphorylates survivin at Thr34 in its BIR domain (8). Mutational analysis has shown that expression of a Thr34 phosphomimic, T34E, greatly reduces the rate of cell proliferation and cannot support cell division in the absence of the endogenous protein, whereas expression of the non-phosphorylatable counterpart, T34A, supports cell growth (9, 10). Intriguingly, T34A sensitizes cells to apoptotic stimuli and is being explored as a potential therapeutic tool (2, 11), whereas T34E potently inhibits cell death (9, 12, 13). Thus, phosphorylation by Cdk1 is one means of separating the mitotic and anti-apoptotic roles of survivin.
Plk1 (Polo-like kinase 1) is also an essential, multitasking protein, whose expression is deregulated in cancer. First identified in Drosophila (14), Plk1 regulates mitotic entry, centrosome separation, spindle assembly, chromosome alignment, APC/C activation, and cytokinesis and has been implicated as a mediator of apoptosis (15). In cultured mammalian cells, Polo disruption has been achieved using a number of different techniques, including chemical genetics (16, 17), small molecule inhibition (18–21), and RNAi (22, 23). As expected for a protein with many roles, its loss has pleiotropic effects, including the generation of monopolar spindles, polyploidy, and increased apoptosis. Although the majority of Plk1 is centrosomal in early mitosis, a subpopulation associates with the kinetochores (24) and has been implicated in mediating the spindle checkpoint (22, 23). Mad2 and BubR1 are checkpoint proteins that are recruited to the kinetochores of chromosomes that are not properly attached to the spindle. Mad2 is recruited due to the absence of microtubule attachments, whereas BubR1 is recruited when paired kinetochores are not under tension. Interestingly, treatment of Plk1 or survivin-depleted cells with microtubule poisons has suggested that Plk1 stabilizes Mad2 recruitment at kinetochores (22), whereas survivin stabilizes BubR1 at these sites (25, 26). Supporting this notion, simultaneous depletion of survivin and Plk1 eliminates both spindle checkpoint signals, and consequently cells exit mitosis inappropriately and undergo mitotic catastrophe (22). However, Matsumura et al. (23), recently reported that Plk1 interacts directly with BubR1 and that phosphorylation of BubR1 by Plk1 is required for correct chromosome orientation during prometaphase but not for its recruitment to kinetochores or for spindle checkpoint activation. Thus, although Plk1 and survivin may have complementary roles in the maintenance of the spindle checkpoint, direct links between Plk1 and BubR1 also exist that facilitate chromosome biorientation. In cells that enter anaphase normally, Plk1 is found at the central spindle and midbody, where it colocalizes with the CPPs and is required to facilitate cytokinesis through communication with the microtubule organizers, MKLP1, MKLP2, and PRC1, and the RhoA signaling cascade (27–29).
In the present study, we report that survivin and Plk1 kinase interact during mitosis and that survivin is a Plk1 substrate. We identify Ser20 as a principle target of Plk1 within the survivin protein and discover that inhibiting phosphorylation at this site interferes with the correction of syntelically attached chromosomes. Inhibiting phosphorylation at this site also prevents cell proliferation in the absence of the endogenous protein but does not affect cellular response to an apoptotic stimulus. We conclude that phosphorylation of survivin by Plk1 is essential to prevent aneuploidy caused by maloriented chromosomes. Further, these data demonstrate a second phosphorylation event, distinct from that of Cdk1, capable of divorcing the mitotic and anti-apoptotic roles of survivin.
EXPERIMENTAL PROCEDURES
Unless otherwise stated, all cell culture reagents were from Invitrogen, and general chemicals were from Sigma.
Molecular Biology
Site-directed mutagenesis was carried out by QuikChange site-directed mutagenesis (Stratagene) using wild type survivin cDNA with a silent mutation in its RNAi targeting region, cloned in pBluescript, as template (see Ref. 30). Once sequences were verified, the constructs were cut and pasted into pcDNA3.1 with a C-terminal GFP tag for expression in mammalian cells or into pGEX4T1 for NH2-terminal GST tagging and recombinant expression.
Wild type full-length plk1 cDNA was amplified from IMAGE clone 2822226 (Au2-e5; Geneservice) using the 5′-primer GCTTGAATTCATGAGTGCTGCAGT and the 3′-primer GCTTCTCGAGTTAGGAGGCCTTCGA, containing an EcoRI and XhoI site, respectively, for subsequent cloning procedures. The region encoding the Polo binding domain (PBD) and the Polo kinase domains were extracted by PCR from the full-length template using similar flanking enzymes and appropriately designed primers.
Cell Culture and Drug Treatments
U2OS cells were maintained at 37 °C with 5% CO2 in DMEM, 10% FCS (PAA), with penicillin/streptomycin, and fungizome. U2OS cells stably expressing the various forms of survivin were established by FuGene 6-mediated transfection with 1–2 μg of the relevant pcDNA3.1 constructs. Twenty-four hours post-transfection, cells were exposed to 500 μg/ml G418, and 7–10 days later, single GFP-expressing colonies were selected. Pools consisting of a minimum of four separate clones were used for analysis. Cells were arrested in prometaphase by overnight incubation in 0.2 μg/ml colcemid or 2 μm dimethylastron (Axxora) and released by extensive washing in PBS (room temperature) and reincubation in complete DMEM at 37 °C. To inhibit Plk1 activity, cells were incubated overnight with 50 nm BI 2536 (Tocris). When imaging cells live, regular DMEM was substituted for CO2-independent medium without phenol red.
Recombinant Protein Expression
For bacterial expression of recombinant survivin, Plk1, and their variants, pGEX4T1 vectors encoding the relevant cDNAs were transformed into BL21 cells. Protein expression was induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 30 °C, and lysates were prepared as in Ref. 30. Recombinant GST-tagged proteins were then bound to glutathione-Sepharose 4B beads (GE Healthcare) and eluted in 50 mm Tris-HCl (pH 8) containing 10 mm glutathione. When stated, GST was cleaved off the recombinant protein/beads using thrombin, which was then removed with benzamidine-agarose.
For GST pull-down experiments, the GST-tagged proteins bound to beads were incubated with an in vitro translated “partner” protein expressed from pcDNA or pBluescript using a TNT-T7 transcription-translation kit (Promega) and [35S]methionine (PerkinElmer Life Sciences) as tracer (see Ref. 30 for detailed methods).
Fluorescence Microscopy
To localize Plk1 kinase, cells were fixed with 4% formaldehyde in PBS for 5 min, washed with PBS, permeabilized with 0.15% Triton for 2 min, and blocked with 1% bovine serum albumin in PBS for 15 min and immunoprobed with anti-Plk1 (AbCam, Ab14209, 1:100), anti-CENPC (AbCam, Ab50974, 1:250), anti-Aurora-B (AbCam, Ab2254, 1:1000), anti-borealin (polyclonal, in house), or anti-BubR1 (1:250, gift from S. S. Taylor (Manchester, UK)) antibodies, followed by Texas Red anti-rabbit, anti-sheep, or anti-mouse antibodies as appropriate (1:200, Vector Laboratories). For tubulin localization, cells were probed with 1:2000 anti-α-tubulin (B512) and Texas Red anti-mouse (1:200, Vector Laboratories). To visualize F-actin, formaldehyde-Triton-fixed cells were incubated for 30 min at room temperature with 200 nm rhodamine-phalloidin prepared in PBS. All cell preparations were counterstained with the DNA stain DAPI upon mounting in Vectashield (Vector Laboratories). Fixed cells were viewed using an inverted fluorescence Olympus microscope fitted with a ×60 (numerical aperture 1.4), or ×100 (numerical aperture 1.3) oil immersion objective, and images were captured using DeltaVision software (Applied Precision). Two-dimensional projections were generated from deconvolved Z-stacks, and images were prepared using Adobe Photoshop.
High resolution live cell imaging was performed using an Olympus-based personal Delta Vision work station at ×100 (numerical aperture 1.4, oil). A Z-sweep of 40 0.3-μm sections was acquired at each time point every 2 min, using both differential interference contrast and GFP optics. Subsequent off-line image preparation was carried out using Volocity software (Improvision) and finalized with Adobe Photoshop. Images in Fig. 5H were acquired using a Leica DMIRB microscope, fitted with a ×40 oil immersion lens, using Open Lab software (Improvision).
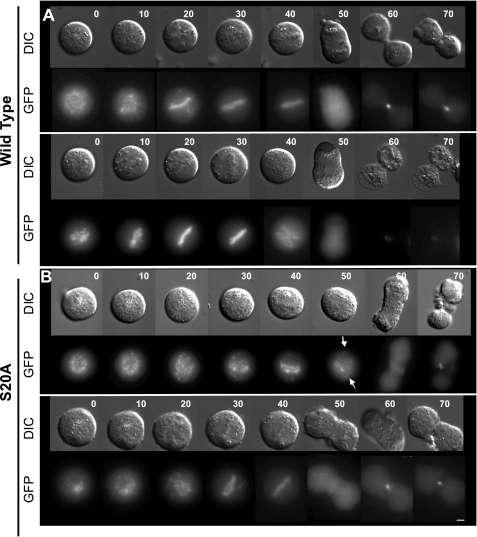
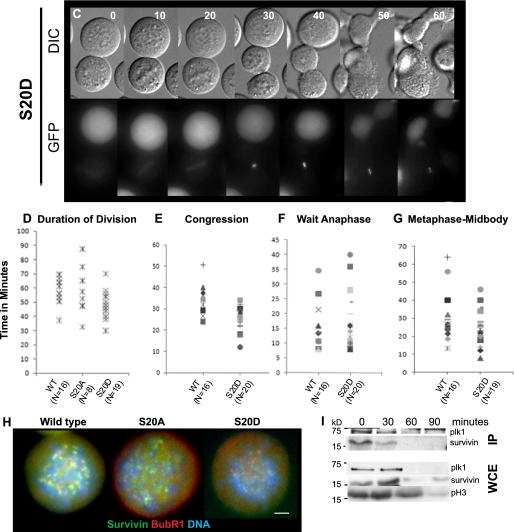
FIGURE 5.
S20A cannot correct syntelically oriented chromosomes. U2OS lines stably expressing survivin-GFP (wt) (A), S20A-GFP (S20A) (B), or S20D-GFP (S20D) (C), as indicated, were treated overnight with 2 μm dimethylastron and harvested by mitotic shake off before extensive washing in PBS and release into prewarmed drug-free CO2-independent medium. A Z-sweep of 40 images (0.3 μm) was recorded for each sample at multiple positions in differential interference contrast (DIC; upper images in each set) and GFP (lower images) at 2-min intervals, using a ×100 (numerical aperture 1.4) oil immersion lens. Gallery images are two-dimensional projections of Z-stacks of every fifth time point in each sequence (i.e. 10 min apart, as indicated numerically at the top right). S20A cells entered anaphase despite the persistence of misaligned chromosomes (arrows), whereas S20D cells divided more rapidly. D–G, timing of specific mitotic events. Because S20A cells rarely achieved chromosome alignment, only the total duration of division could be assessed (D), whereas the time taken to congress chromosomes to the metaphase plate (E), the duration of time spent at the metaphase plate (F), and time taken from achieving alignment to midbody formation are given for wild type and S20D lines. G, wide field imaging of cells treated for 16 h with dimethylastron and immunoprobed with anti-BubR1 antibodies (red) revealed diminished BubR1 signal at kinetochores of S20A cells. I (top), immunoprecipitation of endogenous Plk1 using anti-Plk1 antibodies revealed detectable co-immunoprecipitation of endogenous survivin at 0 and 30 min after release from dimethylastron. Whole cell extracts (WCE) were probed with anti-phospho-H3 antibodies to confirm transition through mitosis (bottom). Bars, 5 μm.
Immunoprocedures
For immunoprecipitation, whole cell lysates were prepared from 3 × 106 cells by a 1 h incubation at 4 °C in Nonidet P-40 buffer (50 mm Tris-HCl (pH 8), 150 mm NaCl, 10 mm EDTA, 1% Nonidet P-40). After clearing by centrifugation, the supernatant was then incubated with rotation for 2 h with 2–4 μg of antibody, and then 25 μl of protein G beads were added for a further 2 h (or overnight) at 4 °C. To immunoprecipitate Plk1, a mixed population of anti-Plk1 monoclonal antibodies was used (AbCam catalog no. 14210; 2 μg/3 × 106 cells), and to immunoprecipitate GFP, an in-house polyclonal rabbit antibody was used at 4 μg/3 × 106 cells). Beads were washed with Nonidet P-40 buffer, and proteins were boiled off the beads with 5× Laemmli sample buffer.
Standard procedures were used for SDS-PAGE and immunoblotting with a 0.22-μm nitrocellulose membrane. ECL-plus (GE Healthcare), x-ray film (GE Healthcare), and a Storm PhosphorImager were used to detect signals. After SDS-PAGE and transfer to nitrocellulose membrane, immunoblots were probed with polyclonal anti-survivin antibodies (1:2000, in-house), anti-Plk1 kinase (rabbit, AbCam 14209, 1:100), anti-GFP (mouse monoclonal 3E1, 1:500, CR-UK), anti-cyclin B1 (1:500, BD Biosciences), anti-actin (1:5000), anti-GFP (1:5,000), anti-phosphorylated histone H3 (Upstate, 1:1000), anti-tubulin (B512, 1:2000), or anti-GST (AbCam, Ab9085; 1:500). Horseradish peroxidase-conjugated secondary antibodies (DAKO) were diluted 1:2000 in 3% milk.
For Far Western analysis, 5 μg of untagged recombinant survivin was phosphorylated, subjected to SDS-PAGE, and then transferred to Hybond-C nitrocellulose membrane. The membrane was then blocked in AC buffer (20 mm Tris-HCl (pH 7.5), 100 mm NaCl, 0.5 mm EDTA, 0.1% Tween 20), and 2% milk for 1 h. Next the membrane was incubated overnight at 4 °C with 5 μg/ml recombinant GST-tagged Polo binding domain in AC buffer with 2% milk and 1 mm dithiothreitol. To assess GST-PBD binding, the membrane was probed with polyclonal goat anti-GST antibodies (AbCam, Ab6613, 1:5000) for 2 h and detected using standard horseradish peroxidase/ECL methods, as above.
In Vitro Kinase Assays
Recombinant Plk1 kinase (Cell Signaling) or Plk1 immunoprecipitated with protein-G beads (CR-UK) and anti-Plk1 antibodies (mixed monoclonal mouse antibodies, AbCam, Ab14210, 2 μg/mg cell extract) were used for in vitro kinase assays.
2 μl of recombinant Plk1 kinase or 10 μl of Plk1-protein G beads were incubated with 2 μg of recombinant substrate in Plk1 kinase buffer (25 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm NaCl, 1 mm dithiothreitol, 0.5 mm EDTA), and a reaction was started by the addition of 1 μl of 2 mm ATP and 0.5 μl of [γ-32P]ATP (5 μCi/reaction; PerkinElmer Life Sciences). The reaction (final volume 20 μl) was incubated for 10–40 min at 37 °C and stopped with sample buffer. To inhibit Plk1 activity in vitro, 500 nm GW843682X (Tocris) was added to the reaction.
In vitro phosphorylation with other recombinant kinases (for Fig. 2E) was performed similarly using purified Cdk1 (Cell Signaling), Aurora-B-INCENP (IN-box; a gift from Dr. P. Eyers), or CK2 (a gift from Prof. E. Pinna).
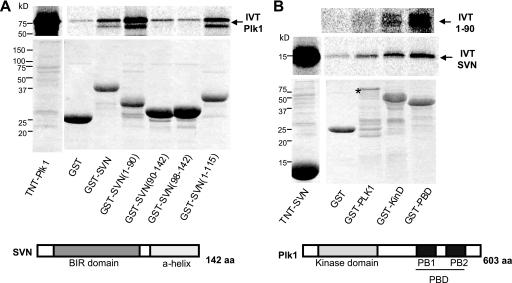
FIGURE 2.
Survivin and Plk1 interact directly in vitro. In vitro pull-down assays. GST or GST-tagged versions of survivin (A) or GST-tagged versions of Plk1 (B) were incubated with in vitro translated (IVT) [35S]methionine-labeled Plk1 (A), survivin (SVN), or the C-terminally truncated form of survivin, 1–90 (B). Plk1 interacted directly with GST-survivin and survivin truncations lacking the C-terminal α-helix. Conversely, survivin interacted with full-length Plk1, its kinase domain (KinD), and its PBD, which encompasses two Polo boxes (PB1 and PB2), and survivin 1–90 showed strongest interaction with GST-PBD. Note the reduced expression of full-length Plk1, probably due to decreased stability as indicated by the asterisk. Stick models of survivin and Plk1 are shown to illustrate the relevant domains (not drawn to scale). aa, amino acids.
siRNA
Endogenous survivin was eliminated from U2OS cells using a double pulse procedure with siRNA oligonucleotides directed against nucleotides 54–65 (26). 5 × 104 cells were reverse transfected with 3 pmol of survivin siRNA using Hyperfect (Qiagen) in antibiotic-free DMEM in 24-well plates. 24 h later, cells were exposed to a further pulse of 3 pmol of siRNA. Thereafter, cell proliferation was monitored at 24-h intervals using a hemocytometer and trypan blue exclusion. Other analyses were carried out 48–96 h after the first pulse.
FACS Profiling
Cells were harvested in ice-cold PBS and fixed with 70% ethanol (−20 °C) for a minimum of 2 h. They were then washed with PBS and treated with 100 μg/ml RNase and stained with 100 μg/ml propidium iodide for at least 15 min at 25 °C. Samples were analyzed using a FACS Canto (BD Biosciences).
Apoptosis Assay
To assess the ability of cells to inhibit apoptosis, 105 cells were seeded into 24-well plates on day 0, and the following day, apoptosis was induced by the addition of 250 ng/ml recombinant human TRAIL (Peprotech) for 30, 60, 90, or 120 min, as indicated. Cells were then lysed in 150 μl of mammalian protein extraction reagent (MPER; Perbio) with 1 mm EDTA in the presence of 1 μg/ml each of the protease inhibitors pepstatin A and 4-(2-aminoethyl)-benzenesulfonyl fluoride. To assess apoptotic activity, 40 μl of each lysate (in triplicate) was incubated per well of a 96-well plate with 200 μl of caspase assay buffer (20 mm Tris (pH 7.5), 10% glycerol, 2 mm dithiothreitol) and 4 ng of the caspase-3/7-specific substrate, Ac-DEVD-7-amino-4-methylcoumarin (Biomol). After incubation at 37 °C for a minimum of 1 h, caspase activity was assessed fluorogenically using a SpectraMax Gemini Spectrofluorometer set at 380 nm (excitation) and 440 nm (emission).
RESULTS
Survivin and Plk1 Associate in Vivo during Mitosis
To begin our investigation, we first used immunolocalization to determine whether Plk1 colocalized with survivin-GFP in our system (Fig. 1A). U2OS cells stably expressing survivin-GFP, probed with antibodies to Plk1, revealed that during early mitosis (prometaphase and metaphase), although the majority of Plk1 kinase was present on the centrosomes, a subpopulation localized to the kinetochores adjacent to the survivin-GFP at the centromeres. Thereafter, all Plk1 kinase colocalized with survivin-GFP, decorating the midzone microtubules during anaphase and the midbody during cytokinesis.
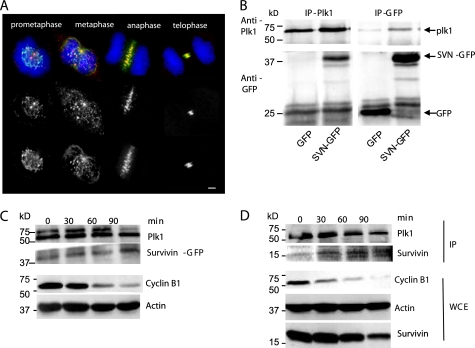
FIGURE 1.
Colocalization and interaction between survivin and Plk1 during mitosis. A, immunolocalization of Plk1 kinase (Ab14209; red) in formaldehyde-fixed U2OS cells expressing survivin-GFP (green), counterstained with DAPI to visualize the chromosomes (blue). A subpopulation of Plk1 colocalizes (as indicated in yellow) with survivin at the centromeres during prometaphase and metaphase. At anaphase and telophase, all Plk1 transfers to the central spindle and midbody, where it colocalizes with survivin. Bar, 5 μm. B, reciprocal immunoprecipitation (IP) of GFP or survivin-GFP using anti-GFP antibodies and of Plk1 kinase using anti-Plk1 antibodies in U2OS cells transiently transfected with plasmids encoding GFP and Plk1 or encoding survivin-GFP and Plk1, as indicated. Survivin-GFP co-immunoprecipitated with Plk1, and conversely, Plk1 co-immunoprecipitated with survivin-GFP but not with GFP alone. IB, immunoblot. C and D, immunoprecipitation using anti-Plk1 kinase antibodies was repeated on synchronized cell extracts prepared from survivin-GFP-expressing U2OS cells (C) or U2OS cells with no ectopic survivin (D). Cells were arrested in mitosis using a sequential thymidine-nocodazole regime (time 0) and released for 30, 60, or 90 min as indicated. Accompanying whole cell extracts (WCE) were probed with anti-cyclin B1 antibodies to indicate release from mitosis, and anti-actin was included as a loading control for the whole cell extracts. Both survivin-GFP (C) and endogenous survivin (D) showed the greatest association with ectopic Plk1 kinase when cyclin B1 levels were at their lowest, indicating increased affinity between the proteins as cells exit mitosis.
Next we asked whether the two proteins associate in vivo. Reciprocal co-immunoprecipitations were performed using Plk1 or GFP antibodies in U2OS cells cotransfected with pcDNA vectors expressing GFP and Plk1 or survivin-GFP and Plk1 (Fig. 1B). When Plk1 was immunoprecipitated, an abundant band was visible in the survivin-GFP lane but not in the GFP control lane (Fig. 1B, left). Conversely, when antibodies to GFP were used, a strong Plk1 band co-immunoprecipitated with survivin-GFP-expressing cells but not with the GFP control (Fig. 1B, right). Thus, survivin and Plk1 colocalize and co-immunoprecipitate in U2OS cells, which both confirms the recent findings of Feng et al. (31) and validates our system.
Because survivin-GFP and Plk1 kinase show greater colocalization during anaphase, telophase, and cytokinesis than in prometaphase or metaphase, we next asked whether the ability of survivin-GFP to associate with Plk1 kinase increased as cells progressed through mitosis. The Plk1 immunoprecipitation experiment was repeated using extracts prepared from survivin-GFP-expressing U2OS cells, after transient transfection with cDNA to Plk1 kinase at 30-min intervals after release from a nocodazole-induced prometaphase arrest. As shown in Fig. 1C, survivin-GFP coimmunoprecipitated with ectopic Plk1 kinase at all time points, and its association grew stronger as cells exited mitosis, as indicated by the reduction in cyclin B1 expression in the accompanying whole cell extracts. When the same experiment was carried out in U2OS cells expressing no exogenous survivin, the overexpressed Plk1 kinase immunoprecipitated endogenous survivin and displayed a similar profile of association during mitosis (Fig. 1D). We also noted that this increased association occurred despite an overall decline in endogenous survivin levels, which occurs as cells exit mitosis. Further immunoprecipitations revealed that the endogenous forms also co-associate (see Fig. 5H).
Survivin and Plk1 Kinase Interact Directly in Vitro
To assess whether the interaction between survivin and Plk1 was direct, we turned to in vitro analysis. GST, GST-survivin, or various NH2- and C-terminally truncated forms were incubated with in vitro 35S-labeled full-length Plk1 kinase. As shown in Fig. 2A, Plk1 bound to full-length survivin and more strongly to the C-terminally truncated forms of survivin, 1–90, and 1–115. By contrast, binding to the NH2-terminal truncations, 90–142 and 98–142, was comparable with the GST control. Thus, we conclude that the interaction with Plk1 is mediated by the NH2 end of survivin. The reciprocal experiment was then performed to map the part(s) of Plk1 that interact with survivin or the NH2 90 amino acids of survivin (1–90). GST, GST-Plk1, the NH2 half containing the kinase domain, GST-KD, or the C-terminal half with the Polo binding domain, GST-PBD, was incubated with in vitro translated 35S-labeled full-length survivin or survivin 1–90 (Fig. 2B). Here survivin bound to GST-Plk1 but not to GST alone, confirming that the two proteins interact directly in vitro. Survivin interacted with both the kinase and the Polo binding domains, suggesting that it can bind Plk1 via two distinct sites, whereas the NH2-terminal 1–90 bound most tightly to the GST-PBD region.
Plk1 Phosphorylates Survivin at Ser20
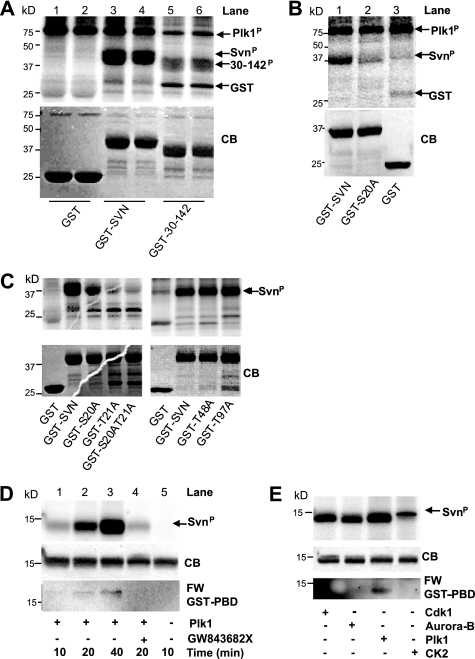
Having established that survivin can bind Plk1 both in vivo and in vitro, we then asked whether survivin is a Plk1 substrate. In vitro kinase assays were performed with either recombinant or immunoprecipitated Plk1 (Fig. 3 and data not shown). Plk1 phosphorylated itself (Fig. 3A, plk1P) and full-length GST-survivin (Fig. 3A, lanes 3 and 4, SvnP) but not the GST control (Fig. 3A, lanes 1 and 2). Of all of the truncations tested (see Table 1), the longest version to show significant reduction in phosphorylation was 30–142 (Fig. 3A, lanes 5 and 6), suggesting that the primary Plk1 phosphosite is within the NH2-terminal 29 amino acids. Plk1 frequently targets serines or threonines in close proximity (32), and within the first 29 amino acids of survivin, serine 20 and threonine 21 are adjacent. In silico modeling studies have suggested that Ser20 is an externally exposed residue, whereas Thr21 forms a strong hydrogen bond with a neighboring residue and is deeply embedded in the BIR domain. Consistent with its structural importance, we found that mutating Thr21 (T21A or T21V) greatly altered the solubility of the protein, presumably due to problems in its folding in vitro. Given the insolubility of Thr21 mutants, we have interpreted in vitro experiments using these mutants cautiously and have focused our attention primarily on Ser20.
FIGURE 3.
Survivin is a Plk1 substrate. A–C, in vitro kinase assays showing autoradiograms in the upper panels and corresponding Coomassie Blue (CB) staining of each gel in the lower panels. A, recombinant GST, GST-survivin, or GST-survivin lacking the first 29 amino acids (fragment 30–142), was incubated in vitro with recombinant Plk1 in the presence of [γ-32P]ATP. Plk1 phosphorylated GST-survivin (SvnP; lanes 3 and 4) but not the GST control (lanes 1 and 2). 32P incorporation was greatly reduced in the N-terminal truncation, GST-30–142 (lanes 5 and 6), suggesting that Plk1 phosphorylation occurs within the first 30 amino acids. We also noted that Plk1 phosphorylated itself (Plk1P). B, in vitro Plk1 kinase assay to investigate Ser20. Plk1 phosphorylated wild type survivin (GST-SVN; lane 1) but not GST-S20A (lane 2) or GST (lane 3). C, the Plk1 kinase assay was repeated using the point mutants indicated. Note that although Thr21 and S20A/T21A mutants showed reduced phosphorylation, they were only partially soluble, and these preparations required considerably more extract/beads than other samples. T48A and T97A mutants were phosphorylated as wild type. D, Far Western. Untagged recombinant survivin (GST tag removed by thrombin cleavage) was phosphorylated in vitro with Plk1, as described above. The reaction was denatured, and survivin and Plk1 were separated by electrophoresis and transferred to nitrocellulose. The membrane was then incubated with GST-PBD and probed with anti-GST antibodies. Top, autoradiogram showing increased phosphorylation (32P incorporation) from 10 to 40 min (lanes 1–3) and the ability of GW843682X to inhibit phosphorylation (lane 4). Lane 5 is included as a negative control. Middle, Coomassie-stained gel (CB) to demonstrate equality in loading. Bottom, Far Western (FW) showing increasing association between denatured survivin and GST-PBD with increasing phosphorylation (lanes 1–3). This interaction is abolished in the presence of the Plk1 inhibitor GW843682X (lane 4). E, Far Western using survivin prephosphorylated with Cdk1, Aurora-B, Plk1, or CK2, as indicated. Only Plk1 phosphorylation promoted GST-PBD binding under these conditions.
TABLE 1.
Mapping Plk1 phosphorylation site(s) of survivin
| Survivin construct | Phosphorylation | Expression | Solubility |
|---|---|---|---|
| 1–142 (full-length) | ++ | Good | Soluble |
| 1–115 | ++ | Good | Soluble |
| 1–90 | ++ | Good | Soluble |
| 90–142 | − | Good | Soluble |
| 98–142 | − | Good | Soluble |
| 30–142 | − | Good | Soluble |
| S20A | − | Good | Soluble |
| T21A | − | Good | Mostly insoluble |
| S20A/T21A | − | Good | Mostly insoluble |
| T48A | ++ | Good | Soluble |
| T97A | ++ | Good | Soluble |
To determine whether serine 20 was a Plk1 target site, we mutated it to a non-phosphorylatable alanine (S20A) and repeated the Plk1 kinase assay. As shown in Fig. 3B, although GST-survivin shows abundant incorporation of 32P (lane 1), this was reduced to background levels in the GST-S20A mutant (compare lane 2 with GST control, lane 3), indicating that Ser20 is phosphorylated by Plk1.
As mentioned above, Thr21 is also a potential Plk1 site, and a further site, Thr97, which, although outside the region we identified by GST mapping, lies within an appropriate consensus sequence for Plk1 (see “Discussion”). Thus, we next tested whether substituting these threonines for alanine could also inhibit Plk1 phosphorylation of survivin in vitro. As shown in Fig. 3C (left), the substitution T21A, also reduced 32P incorporation in the presence of Plk1, as did the S20A/T21A double mutant; thus, Thr21 too may be targeted by Plk1. By contrast, mutating Thr97 had no effect on the ability of Plk1 to phosphorylate survivin; nor did mutation of a putative CK2 site, T48A (Fig. 3C, right). Thus, we conclude that Plk1 can target survivin at Ser20 and potentially Thr21 but not Thr97 in vitro.
To ascertain whether Plk1 phosphorylation of survivin could influence binding of the PBD of Plk1, we performed a Far Western experiment (Fig. 3D). Untagged recombinant survivin was in vitro phosphorylated with recombinant Plk1, and the two proteins were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was then incubated with recombinantly expressed GST-PBD and probed with anti-GST antibodies. Under these conditions, GST-PBD binding to survivin was initially very low, but it increased as phosphorylation of survivin increased. Thus, binding of the PBD of Plk1 can be regulated by phosphorylation, and Plk1 itself can prime this association. We next tested a panel of other kinases known to phosphorylate survivin, Cdk1, Aurora-B, and CK2.3 Despite efficient phosphorylation by each kinase as determined by 32P incorporation (Fig. 3E, top), GST-PBD only bound to the Plk1-phosphorylated form, confirming that Plk1 can regulate its own binding to survivin.
Characterization of Survivin Phosphorylation Mutants
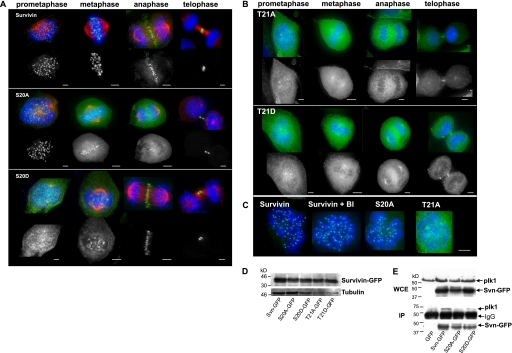
To begin to understand the functional significance of Plk1-mediated regulation of survivin, U2OS cell lines stably expressing GFP, survivin-GFP, or C-terminally GFP-tagged Ser20 and Thr21 mutants were established, and their localization was recorded. As shown in Fig. 4A, the non-phosphorylatable S20A concentrated at centromeres from early prometaphase through to metaphase, whereas the corresponding (putative) phosphomimic, S20D, was diffusely localized during early stages of mitosis but nevertheless gained access to the centromeres as the chromosomes congressed. After metaphase, both S20A and S20D localized at the midzone, like wild type, although a tighter association with these structures was often apparent for S20D, compared with S20A. During interphase, when survivin is overexpressed, it is predominantly cytoplasmic, and this localization remained unchanged for Ser20 mutants (data not shown).
FIGURE 4.
Analysis of Ser20 and Thr21 survivin mutants. U2OS lines stably expressing survivin-GFP or survivin-GFP bearing the mutations S20A, S20D, T21A, and T21D, as indicated, were examined during mitosis using fluorescence microscopy. In A, anti-tubulin was used to immunoprobe cells for microtubules (red). Tubulin localization was omitted in B to aid clarity of GFP signal, which was more diffuse. In all panels, chromosomes are stained with DAPI (blue). S20A was clearly localized at the centromeres from prophase through to metaphase, whereas this localization was weaker and more variable for S20D. Interestingly, the converse was the case during anaphase and telophase, with S20D showing more complete localization to the midzone and midbody than S20A. B, neither T21A nor T21D displayed localization typical of survivin-GFP; instead, these forms were diffusely distributed throughout the cell, with some concentration at the centrosomes. C, cells expressing wild type survivin were incubated in the absence (survivin) or presence of BI 2356 (survivin + BI) and survivin-GFP localization compared with the distribution of survivin bearing alanine substitutions at Ser20 or Thr21, as indicated. BI 2356 treatment induced a prometaphase arrest and did not affect survivin-GFP accumulation at the centromeres. S20A phenocopied BI treatment, whereas T21A localization was distinct. Bars, 5 μm. D, immunoblot with anti-GFP antibodies and anti-tubulin as a loading control to compare levels of expression in each cell line. E, immunoprecipitation (IP) using anti-GFP antibodies in the cell lines indicated after transient expression of Plk1 kinase. WCE, whole cell extracts probed with anti-Plk1 and anti-survivin antibodies revealed that transient Plk1 expression was similar in each line, and survivin variants were also present at similar levels. Anti-GFP antibodies were used to immunoprecipitate GFP or the survivin-GFP variant of interest, and immunoblots were probed with anti-Plk1 antibodies. S20A exhibited reduced binding to Plk1 compared with wild type survivin or S20D under these conditions.
In contrast to the Ser20 mutants and despite similar expression levels (Fig. 4D), neither T21A nor T21D localized to any of the structures that typically recruit survivin (Fig. 4B). Instead they were distributed diffusely throughout the cells with some accumulation at the centrosomes. The similarity in localization between T21A and T21D suggests that T21D probably represents another non-phosphorylatable form, as opposed to a phosphomimic. Despite this aberrant localization, but consistent with the viability of these stably expressing cell lines, aurora-B and borealin localization remained unperturbed in the presence of all forms of ectopically expressed survivin, and no gross abnormalities in chromosome arrangements were observed at any specific stage (supplemental Fig. 1) (data not shown).
To ascertain whether the localization of alanine-substituted Ser20 or Thr21 was representative of a non-Plk1-phosphorylated survivin, we compared their localization to survivin-GFP in cells treated with BI 2356, a small molecule inhibitor of Plk1 (Fig. 4C). BI 2356-treated cells arrested in prometaphase and displayed abundant centromeric survivin-GFP, phenocopying the distribution of S20A, further indicating that Ser20 is the principal Plk1 target of survivin. We therefore focused the remainder of this study on the Ser20-mutated forms.
Next, we asked whether mutation of Ser20 altered Plk1-survivin liaisons in vivo. Immunoprecipitation of the ectopic proteins using anti-GFP antibodies from asynchronous cell lines expressing GFP, survivin-GFP, S20A, or S20D (Fig. 4E), which had been transiently transfected with Plk1 kinase, revealed that S20A reduced the Plk1-survivin interaction. These data are consistent with the lack of S20A phosphorylation in vitro (Fig. 1B) and the ability of Plk1 phosphorylation to facilitate binding of the PBD in the Far Western assay (Fig. 1E).
S20A Is a Dominant Negative Mutant That Cannot Correct Syntelically Attached Chromosomes
To determine whether Ser20 mutants influenced the accuracy and timing of mitosis, we turned to live imaging analyses. When released from mitotic arrest using colcemid and observed at low magnification, no major changes were observed between the different cell lines, although S20D-expressing cells appeared to complete division and readhere to the substrate slightly more readily (supplemental Fig. 2) (data not shown). Similarly, no clear differences in cell division were visible during initial observations of asynchronous cells, even at high resolution. However, one of the principle roles of the CPPs is to correct erroneously attached chromosomes during their congression to the metaphase plate, and such defects are often difficult to detect. Thus, to discover whether Ser20 mutants were competent in this role, we exacerbated the presence of misaligned chromosomes by invoking a mitotic arrest using the Eg5 inhibitor dimethylastron, which inhibits centrosome separation, causing formation of a monopolar spindle to which all chromosomes attach in a syntelic fashion. Because this treatment is reversible, we assayed the ability of the different cell populations to correct positioning of maloriented chromosomes generated by this treatment and their subsequent ability to divide. Gratifyingly, this experiment revealed that the non-phosphorylatable version of survivin, S20A, was unable to correct all maloriented chromosomes within the cell, and strikingly, cells expressing S20A were able to proceed into anaphase and cytokinesis regardless of the misaligned chromosomes and without a prolonged delay (Fig. 5, B (top) and D), although alignment was achieved in some cases (see Fig. 5B, bottom). In contrast to the malfunctioning S20A-expressing cells, both the control survivin- and S20D-expressing cells successfully corrected all erroneously connected chromosomes (Fig. 5, compare A and C), with S20D completing the tasks of chromosome alignment and cytokinesis slightly faster than those expressing wild type survivin (Fig. 5, A and C–G). Note that the time spent waiting for anaphase was similar between these two populations (Fig. 5F). From these data, we conclude that S20A is a dominant negative mutant that cannot correct maloriented chromosomes, whereas S20D operates more efficiently during prometaphase and cytokinesis than its wild type counterpart. Immunostaining of each line after overnight treatment with dimethylastron demonstrated that the spindle checkpoint protein, BubR1, was highly abundant on the kinetochores of suntelic chromosomes in populations expressing wild type and S20D survivin but reduced, although not completely absent, in cells expressing S20A (Fig. 5H).
Finally, to demonstrate interaction between survivin and Plk1 during correction of syntelic chromosomes, we immunoprecipitated endogenous Plk1 from extracts prepared at 30-min intervals after release from dimethylastron treatment. Immunoblotting of whole cell extracts indicated that endogenous Plk1 expression declines as cells exit mitosis, concomitant with the reduction in phosphorylated H3 levels (Fig. 5I, bottom). Accordingly, when immunoprecipitated, endogenous Plk1 was detectable at 0 and 30 min post-release, at which times endogenous survivin co-immunoprecipitated with it (Fig. 5I, top). Thus, although our overexpression data in Fig. 1 demonstrated that we can detect interactions between Plk1 and survivin when they are in sufficient quantity, when monitoring interaction of the endogenous forms, the greater abundance of Plk1 during early mitotic events makes their liaisons more readily detectable during chromosome congression.
SurvivinS20A Cannot Support Cell Proliferation
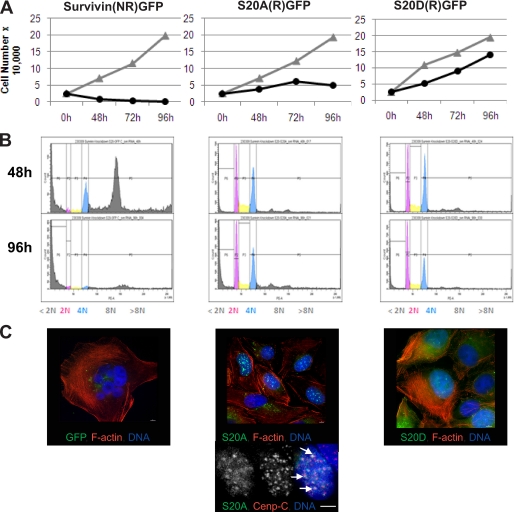
Next, we used siRNA to deplete endogenous survivin from cells expressing survivin-GFP or siRNA-resistant Ser20 mutants and assessed their ability to support cell division. Initially upon removal of the endogenous protein (48 h), both Ser20 mutants localized normally during mitosis, with S20D gaining access to the centromeres more efficiently in its absence (data not shown). Depletion of survivin from the control culture expressing siRNA-sensitive survivin-GFP inhibited cell proliferation with no increase in cell number apparent even at the earliest time point (Fig. 6A, left). FACS profiling of this population at 48 h indicated a loss of G1 cells with 2 n content, an accumulation of cells with >4 n, and a concomitant increase in cells with sub-2 n DNA, indicative of cells undergoing apoptosis (Fig. 6B, left). By 96 h, few cells remained in the control population, and those that did were highly multinucleated, as judged by fluorescence imaging of F-actin and DNA (Fig. 6C, left). By contrast, although their growth rates were reduced, populations expressing S20A and S20D continued to proliferate up to 72 h post-RNAi (Fig. 6A, center and right). FACS profiling at 48 h indicated an accumulation of cells with 4 n content in both populations but negligible sub-2 n or >4 n cells (Fig. 6B, center and right). Interestingly, although S20A sustained growth and viability for 72 h post-RNAi, the persistent absence of the endogenous form eventually inhibited these cells from proliferating, whereas S20D cells continued to grow, as indicated at the 96 h time point. To confirm the differences in their proliferative capacity, we assessed the clonogenic potential of cells from each population 96 h post-RNAi by replating them at low density and counting colony formation 7 days later. In accordance with the proliferation data, cells expressing S20A were unable to form viable colonies, but those expressing S20D formed colonies of >50 cells (data not shown). Despite cessation of proliferation, DNA-FACS profiling indicated that the cell cycle distribution of the S20A population at 48 and 96 h was comparable with the distribution of the S20D population, which was still growing. Finally, we used fluorescence imaging to assess the phenotype of these cells at 96 h. As expected, any remaining cells in the control population were polyploid (Fig. 6C, left), whereas the S20A and S20D populations had nuclei of normal size (Fig. 6C, right). Strikingly, however, the nuclei in the S20A population showed a strong accumulation of S20A in foci (Fig. 6C, middle). Immunoprobing with anti-CENPC antibodies at high resolution revealed that some of these foci were sites of centromere clustering (Fig. 6C, bottom).
FIGURE 6.
S20D, but not S20A, supports cell proliferation. U2OS cells expressing survivin-GFP, sensitive (i.e. not resistant (NR)) to siRNA or siRNA-resistant (R) S20A-GFP, or S20D-GFP were subjected to siRNA. Data presented are from a representative experiment, performed three independent times. A, cell proliferation was monitored for 96 h in control or survivin-specific siRNA-exposed cultures. In the control population expressing siRNA-sensitive survivin (NR)-GFP, survivin-specific siRNA prevented cell proliferation from 48 h. B, FACS profiling of DNA content 48 h (top) and 96 h (bottom) after depletion of endogenous survivin. In contrast, neither S20A nor S20D cells became polyploid at 48 or 96 h. C, cells were examined microscopically at 96 h after staining with rhodamine-phalloidin and DAPI to visualize F-actin (red) and DNA (blue), respectively. All GFP signal (green) was eliminated in cells expressing siRNA-sensitive survivin-GFP, and any remaining cells were multinucleated. Cells expressing S20A-GFP were mononucleated, and S20A-GFP was present in bright nuclear foci. S20D-GFP cells were also mononucleated, but S20D-GFP was mostly cytoplasmic. High resolution imaging (×100) of S20A cells immunoprobed with anti-CENPC antibodies 96 h post-survivin siRNA revealed that some of these nuclear foci were sites of centromere clustering (arrows). Bars, 5 μm.
Ser20 Survivin Mutants Can Inhibit Apoptosis
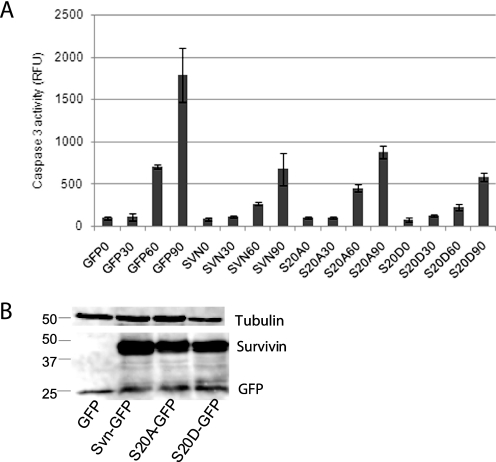
In addition to their essential functions in mitosis, survivin and Plk1 kinase are also implicated in cell death pathways. To test whether mutation of the Plk1-relevant phosphosite of survivin affected its ability to inhibit apoptosis, U2OS cells expressing GFP, wild type survivin, S20A, or S20D were treated with TRAIL at 30-min intervals and analyzed using a tetrapeptide cleavage assay for caspase-3 activity. As shown in Fig. 7, wild type survivin, S20A, and S20D expression all protected cells from apoptosis. These data suggest that the phosphorylation status of Ser20 is not important for the anti-apoptotic function of survivin within the extrinsic apoptosis pathway.
FIGURE 7.
Survivin IAP activity is retained in both Ser20 mutants. A, a fluorogenic (Ac-DEVD-7-amino-4-methylcoumarin) caspase activity assay was performed on extracts prepared from U2OS cells expressing GFP, survivin-GFP, S20A-GFP, or S20D-GFP, which had been exposed to TRAIL for 0, 30, 60, or 90 min. Immunoblotting with anti-GFP antibodies and anti-tubulin as a loading control revealed that all versions were expressed at similar levels (B) and were able to suppress caspase-3 activity. RFU, relative fluorescence units. Error bars, standard deviation.
DISCUSSION
Survivin and Plk1 kinase are both cancer-relevant proteins that are involved in cell division and cell death. Given the similarity in their expression, localization, and the response of cells to their ablation, we hypothesized that they may act in concert during these events to fulfill their duties. Here we show that survivin and Plk1 do indeed interact in vivo, as recently reported by Feng et al. (31), and extend these observations to demonstrate that their interaction can occur directly. Importantly, we show for the first time that survivin is a Plk1 substrate and identify Ser20 as the principle residue targeted by Plk1 and that phosphorylation at this site is required for cell division and to correct malorientated chromosomes during congression. Finally, we demonstrate that although Ser20 phosphorylation is essential for cell proliferation, it does not affect the ability of survivin to inhibit apoptosis.
Plk1 Phosphorylates Survivin Principally on Ser20
At the outset of this study, we were excited to discover that within the highly conserved central region of survivin, residues 95–100 (ELT97LGE), follow precisely the canonical Plk1 phosphorylation consensus: (D/E)X(S/T)ΨX(D/E) (where X is any residue, S/T is the phosphotarget, and Ψ is hydrophobic) (33). This region is engaged in a multitude of survivin activities, including its homodimerization (34, 35) and its interaction with its mitotic partner, borealin (36, 37), and is also an integral part of its nuclear exportation signal (38, 39). However, despite the apparently perfect consensus, its ideal positioning to act as a phosphor switch, and the precedence that nuclear-cytoplasmic shuttling of cyclin B1 (40) and MKLP1 are regulated by Plk1 (27, 41), in vitro phosphorylation of survivin by Plk1 was unaffected when threonine 97 was substituted for a non-phosphorylatable alanine (T97A). Instead, GST phosphomapping and site-directed mutagenesis revealed Ser20 as the principle Plk1 target site, a site previously shown to be regulated by cAMP protein kinase A (42). We also noted that the residue neighboring Ser20, Thr21, may be targeted by Plk1, but due to its structural importance within the molecule, we have interpreted these data with caution. Moreover, despite considerable effort, phosphopeptide analysis of this region has so far evaded detection by mass spectrometry. Treatment with BI 2356, however, revealed that cells arrested in prometaphase in response to the Plk1 inhibition have abundant survivin-GFP at their centromeres, a pattern closely phenocopying S20A localization and distinct from the distribution of T21A (this study) and T97A (38), lending further credence to our conclusion that Ser20 is the principle Plk1 target of survivin.
Survivin Is a Late Docking Partner of Plk1
In addition to its phosphorylation consensus, binding of the PBD of Plk1 can be modulated both positively and negatively by prior phosphorylation of its substrate (32). For example, Cdk1 does both; it primes Plk1 binding and phosphorylation of early mitotic partners, such as cyclin B1 and cdc25, thereby promoting mitotic entry (40, 43) while simultaneously inhibiting binding and phosphorylation of anaphase relevant Plk1 substrates, including the microtubule-binding proteins MKLP2 and PRC1 (28, 29). Docking and phosphorylation of these late mitotic Plk1 partners is mediated by kinases distinct from Cdk1 and can be regulated by Plk1 itself (28, 29). Our data show that Plk1 phosphorylates survivin at Ser20 and that although binding can occur independently of phosphorylation, binding of the PBD can be enhanced by Plk1 phosphorylation but not Cdk1, Aurora-B, or CK2, suggesting that it is a docking partner of Plk1 during, but not exclusively in, late mitosis. Consistent we this, our expression studies indicate that Plk1 and survivin can associate throughout mitosis. Survivin is not the only CPP regulated by Plk1; indeed, Goto et al. (44) reported that Cdk1 phosphorylates INCENP at Thr388 and that phosphorylation at this site is required for Plk1 recruitment to the kinetochores and anaphase onset (44), suggesting that it is also a Plk1 docking partner. It will be interesting to discover whether there is a sequential pattern of CPP phosphorylation during mitosis and whether distinct subcomplexes are regulated by these events. Moreover, because protein kinase A also targets Ser20 (42), presumably it too can facilitate PBD binding, raising questions as to whether there is cooperation between Plk1 and protein kinase A in the regulation of survivin during the cell cycle.
Phosphorylation of Survivin by Plk1 Is Required for Spindle Checkpoint Response and Cell Division
The most striking observation from our overexpression studies is that S20A, which cannot be phosphorylated by Plk1, proceeds through anaphase and cytokinesis despite the presence of maloriented chromosomes. Moreover, in addition to being unable to correct these erroneously attached chromosomes, cells expressing S20A do not delay entrance into anaphase and divide within the normal schedule experienced by cells expressing wild type survivin. This dominant negative phenotype is reminiscent of the response of survivin-depleted cells to monastrol and taxol, conditions that satisfy the checkpoint protein Mad2, forcing the cells to rely completely on the tension sensor BubR1 to detect any errors (25, 26). Using RNAi, in these studies, we and others demonstrated that the persistence of BubR1 at centromeres lacking tension is dependent upon the presence of survivin. Further work revealed that although survivin is phosphorylated by Aurora-B during prometaphase, to achieve chromosome alignment, the Aurora-B target of survivin (Thr117), must be dephosphorylated (7). The current data extend these observations confirming that the presence of survivin at the centromere alone is insufficient to sustain the spindle tension checkpoint, as indicated by BubR1 immunolocalization, and demonstrate for the first time that survivin must be phosphorylated at Ser20 by Plk1 kinase for accurate congression and safe passage into anaphase. Furthermore, data obtained using our (putative) phosphomimic, S20D, suggest that Plk1-phosphorylated survivin corrects chromosome alignment and executes cytokinesis more efficiently than the wild type version.
Using RNAi complementation, we observed that expression of S20A alone inhibited cell proliferation, whereas S20D supported cell growth, further demonstrating that phosphorylation of survivin at Ser20 is critical for cell division. Cell cycle analysis by FACS profiling and phenotypic inspection of the S20A mutant showed that there was an accumulation of cells with 4 n content, from 48 h, and that a slight increase in cell death was occurring at 96 h. At this late time point, S20A was abnormally present and abundant in nuclear foci. These observations suggest that phosphorylation of survivin by Plk1 at the metaphase-anaphase transition may facilitate its transfer from the centromeres to the anaphase spindle, which could result from either an increased affinity for microtubules or alternatively decreased affinity for the centromere. However, at least one report has suggested that the transfer of the CPPs from the centromere to the cleavage furrow can occur in the absence of Plk1 (22).
Ser20 Phosphorylation Is Dispensable for Survivin IAP Activity
Ser20 was previously identified as a protein kinase A target (42). These authors further reported that the mutation S20A augmented the IAP activity of survivin and attributed the enhanced cytoprotection conferred by its expression to an increased affinity for XIAP, an IAP family member with which survivin has been shown to cooperate to inhibit apoptosis (42, 45). In agreement with this report, we find that S20A is able to inhibit TRAIL-mediated apoptosis but find that the phosphomimic is also protective. Thus, we conclude that phosphorylation at this site, which can be achieved by Plk1 or protein kinase A, does not play a primary role in the regulation of survivin IAP activity.
In summary, our data presented herein are the first to demonstrate that survivin is a Plk1 substrate and have identified Ser20 as the major Plk1 phosphorylation site. Using paired Ser20 mutants, we have shown that phosphorylation at Ser20 is required for survivin chromosome alignment and appropriate response to the spindle tension checkpoint. We also provided data indicating that phosphoregulation of Ser20 by Plk1 provides a new route to separate the mitotic and anti-apoptotic functions of survivin. Intriguingly, we recently reported that the “dual” roles of survivin could be bifurcated by altering the status of its Cdk1 site, Thr34 (9), and it is of note that many mitotic proteins are regulated by both Cdk1 and Plk1. Clearly, phosphoregulation of survivin alone and as an integral member of the chromosomal passenger complex is not one-dimensional but part of a highly complex phosphorylation network, posing considerable challenge to unraveling the molecular consequences of these modifications both in mitosis and apoptosis.
Supplementary Material
Acknowledgments
We thank Dr. C. Connell for preparing and analyzing samples for FACS profiling; Dr. P. Eyers and Prof. E. Pinna for purified Aurora-B and CK2 kinases, respectively; Dr. S. S. Taylor for BubR1 antibodies; Dr. N. Brissett for discussions on survivin structure; Dr. H. Hochegger for assistance with live imaging, and Drs. P. Jones and S. Morley for critically reading the manuscript.
The majority of this work was supported by Cancer Research-UK via a senior fellowship (to S. P. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
R. M. Barrett, R. Colnaghi, and S. P. Wheatley, unpublished observations.
- CPP
- chromosomal passenger protein
- BIR
- baculovirus inhibitor of apoptosis repeat
- IAP
- inhibitor of apoptosis protein
- INCENP
- inner centromeric protein
- PBD
- Polo binding domain
- TRAIL
- tumor necrosis factor receptor apoptosis-inducing ligand
- RNAi
- RNA interference
- PBS
- phosphate-buffered saline
- DMEM
- Dulbecco's modified Eagle's medium
- GST
- glutathione S-transferase
- DAPI
- 4′,6-diamidino-2-phenylindole
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- FACS
- fluorescence-activated cell sorting.
REFERENCES
- 1.Altieri D. C. (2003) Oncogene 22, 8581–8589 [DOI] [PubMed] [Google Scholar]
- 2.Altieri D. C. (2008) Nat. Rev. Cancer 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 3.Wheatley S. P., McNeish I. A. (2005) Int. Rev. Cytol. 247, 35–88 [DOI] [PubMed] [Google Scholar]
- 4.Wheatley S. P., Henzing A. J., Dodson H., Khaled W., Earnshaw W. C. (2004) J. Biol. Chem. 279, 5655–5660 [DOI] [PubMed] [Google Scholar]
- 5.Beardmore V. A., Ahonen L. J., Gorbsky G. J., Kallio M. J. (2004) J. Cell Sci. 117, 4033–4042 [DOI] [PubMed] [Google Scholar]
- 6.Delacour-Larose M., Thi M. N., Dimitrov S., Molla A. (2007) Cell Cycle 6, 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheatley S. P., Barrett R. M., Andrews P. D., Medema R. H., Morley S. J., Swedlow J. R., Lens S. M. (2007) Cell Cycle 6, 1220–1230 [DOI] [PubMed] [Google Scholar]
- 8.O'Connor D. S., Grossman D., Plescia J., Li F., Zhang H., Villa A., Tognin S., Marchisio P. C., Altieri D. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13103–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett R. M., Osborne T. P., Wheatley S. P. (2009) Cell Cycle 8, 278–283 [DOI] [PubMed] [Google Scholar]
- 10.Yue Z., Carvalho A., Xu Z., Yuan X., Cardinale S., Ribeiro S., Lai F., Ogawa H., Gudmundsdottir E., Gassmann R., Morrison C. G., Ruchaud S., Earnshaw W. C. (2008) J. Cell Biol. 183, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKay T. R., Bell S., Tenev T., Stoll V., Lopes R., Lemoine N. R., McNeish I. A. (2003) Oncogene 22, 3539–3547 [DOI] [PubMed] [Google Scholar]
- 12.Hoffman W. H., Biade S., Zilfou J. T., Chen J., Murphy M. (2002) J. Biol. Chem. 277, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 13.Jones M. K., Padilla O. R., Webb N. A., Norng M. (2008) J. Cell Physiol. 215, 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunkel C. E., Glover D. M. (1988) J. Cell Sci. 89, 25–38 [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Erikson R. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkard M. E., Randall C. L., Larochelle S., Zhang C., Shokat K. M., Fisher R. P., Jallepalli P. V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkard M. E., Maciejowski J., Rodriguez-Bravo V., Repka M., Lowery D. M., Clauser K. R., Zhang C., Shokat K. M., Carr S. A., Yaffe M. B., Jallepalli P. V. (2009) PLoS Biol. 7, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007) Curr. Biol. 17, 304–315 [DOI] [PubMed] [Google Scholar]
- 19.Steegmaier M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., Grauert M., Adolf G. R., Kraut N., Peters J. M., Rettig W. J. (2007) Curr. Biol. 17, 316–322 [DOI] [PubMed] [Google Scholar]
- 20.Santamaria A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., Barr F. A., Nigg E. A. (2007) Mol. Biol. Cell 18, 4024–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scutt P. J., Chu M. L., Sloane D. A., Cherry M., Bignell C. R., Williams D. H., Eyers P. A. (2009) J. Biol. Chem. 284, 15880–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Vugt M. A., van de Weerdt B. C., Vader G., Janssen H., Calafat J., Klompmaker R., Wolthuis R. M., Medema R. H. (2004) J. Biol. Chem. 279, 36841–36854 [DOI] [PubMed] [Google Scholar]
- 23.Matsumura S., Toyoshima F., Nishida E. (2007) J. Biol. Chem. 282, 15217–15227 [DOI] [PubMed] [Google Scholar]
- 24.Arnaud L., Pines J., Nigg E. A. (1998) Chromosoma 107, 424–429 [DOI] [PubMed] [Google Scholar]
- 25.Lens S. M., Wolthuis R. M., Klompmaker R., Kauw J., Agami R., Brummelkamp T., Kops G., Medema R. H. (2003) EMBO J. 22, 2934–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho A., Carmena M., Sambade C., Earnshaw W. C., Wheatley S. P. (2003) J. Cell Sci. 116, 2987–2998 [DOI] [PubMed] [Google Scholar]
- 27.Liu X., Zhou T., Kuriyama R., Erikson R. L. (2004) J. Cell Sci. 117, 3233–3246 [DOI] [PubMed] [Google Scholar]
- 28.Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E. A., Sillje H., Barr F. A. (2007) Nat. Cell Biol. 9, 436–444 [DOI] [PubMed] [Google Scholar]
- 29.Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E. A., Mayer T. U., Barr F. A. (2003) J. Cell Biol. 162, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noton E. A., Colnaghi R., Tate S., Starck C., Carvalho A., Ko Ferrigno P., Wheatley S. P. (2006) J. Biol. Chem. 281, 1286–1295 [DOI] [PubMed] [Google Scholar]
- 31.Feng Y. B., Lin D. C., Shi Z. Z., Wang X. C., Shen X. M., Zhang Y., Du X. L., Luo M. L., Xu X., Han Y. L., Cai Y., Zhang Z. Q., Zhan Q. M., Wang M. R. (2009) Int. J. Cancer 124, 578–588 [DOI] [PubMed] [Google Scholar]
- 32.Elia A. E., Cantley L. C., Yaffe M. B. (2003) Science 299, 1228–1231 [DOI] [PubMed] [Google Scholar]
- 33.Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. (2003) J. Biol. Chem. 278, 25277–25280 [DOI] [PubMed] [Google Scholar]
- 34.Chantalat L., Skoufias D. A., Kleman J. P., Jung B., Dideberg O., Margolis R. L. (2000) Mol. Cell 6, 183–189 [PubMed] [Google Scholar]
- 35.Verdecia M. A., Huang H., Dutil E., Kaiser D. A., Hunter T., Noel J. P. (2000) Nat. Struct. Biol. 7, 602–608 [DOI] [PubMed] [Google Scholar]
- 36.Bourhis E., Hymowitz S. G., Cochran A. G. (2007) J. Biol. Chem. 282, 35018–35023 [DOI] [PubMed] [Google Scholar]
- 37.Jeyaprakash A. A., Klein U. R., Lindner D., Ebert J., Nigg E. A., Conti E. (2007) Cell 131, 271–285 [DOI] [PubMed] [Google Scholar]
- 38.Colnaghi R., Connell C. M., Barrett R. M., Wheatley S. P. (2006) J. Biol. Chem. 281, 33450–33456 [DOI] [PubMed] [Google Scholar]
- 39.Knauer S. K., Mann W., Stauber R. H. (2007) Cell Cycle 6, 518–521 [DOI] [PubMed] [Google Scholar]
- 40.Toyoshima-Morimoto F., Taniguchi E., Shinya N., Iwamatsu A., Nishida E. (2001) Nature 410, 215–220 [DOI] [PubMed] [Google Scholar]
- 41.Liu X., Erikson R. L. (2007) Biochem. Biophys. Res. Commun. 353, 960–964 [DOI] [PubMed] [Google Scholar]
- 42.Dohi T., Xia F., Altieri D. C. (2007) Mol. Cell 27, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian Y. W., Erikson E., Taieb F. E., Maller J. L. (2001) Mol. Biol. Cell 12, 1791–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., Nigg E. A., Inagaki M. (2006) Nat. Cell Biol. 8, 180–187 [DOI] [PubMed] [Google Scholar]
- 45.Arora V., Cheung H. H., Plenchette S., Micali O. C., Liston P., Korneluk R. G. (2007) J. Biol. Chem. 282, 26202–26209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.