Abstract
The cyclooxygenases (COX-1 and COX-2) are membrane-associated heme-containing homodimers that generate prostaglandin H2 from arachidonic acid (AA). Although AA is the preferred substrate, other fatty acids are oxygenated by these enzymes with varying efficiencies. We determined the crystal structures of AA, eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA) bound to Co3+-protoporphyrin IX-reconstituted murine COX-2 to 2.1, 2.4, and 2.65 Å, respectively. AA, EPA, and docosahexaenoic acid bind in different conformations in each monomer constituting the homodimer in their respective structures such that one monomer exhibits nonproductive binding and the other productive binding of the substrate in the cyclooxygenase channel. The interactions identified between protein and substrate when bound to COX-1 are conserved in our COX-2 structures, with the only notable difference being the lack of interaction of the carboxylate of AA and EPA with the side chain of Arg-120. Leu-531 exhibits a different side chain conformation when the nonproductive and productive binding modes of AA are compared. Unlike COX-1, mutating this residue to Ala, Phe, Pro, or Thr did not result in a significant loss of activity or substrate binding affinity. Determination of the L531F:AA crystal structure resulted in AA binding in the same global conformation in each monomer. We speculate that the mobility of the Leu-531 side chain increases the volume available at the opening of the cyclooxygenase channel and contributes to the observed ability of COX-2 to oxygenate a broad spectrum of fatty acid and fatty ester substrates.
Keywords: Arachidonic Acid, Cyclooxygenase (COX) Pathway, Eicosapentaenoic Acid (EPA), Protein Structure, X-ray Crystallography
Introduction
The cyclooxygenase enzymes (COX-1 and COX-2) are membrane-associated heme-containing bifunctional enzymes that catalyze the first committed step in prostaglandin (PG)2 biosynthesis (reviewed in Refs. 1, 2). The product, PGH2, is produced as a result of two sequential reactions that are performed in separate but functionally linked active sites. In the first reaction, arachidonic acid (AA; 20:4 ω-6) bound in the cyclooxygenase channel undergoes a bis-oxygenation to form the intermediate PGG2. The released intermediate is then bound in the peroxidase active site, where the 15-hydroperoxide group of PGG2 is reduced to form PGH2 in the second reaction. Both COX-1 and COX-2 require a preliminary catalytic turnover at the peroxidase active site to generate an oxy-ferryl porphyrin radical that is subsequently transferred to Tyr-385 for the initiation of cyclooxygenase catalysis.
Understanding the similarities and differences associated with the structure and function of COX-1 and COX-2 and the rationale for the existence of two isoforms has been the focus of much recent research (3–5). COX-1 and COX-2, which are encoded by separate genes (6), display ∼60% sequence identity within the same species and greater than 85% sequence identity among orthologs from different species (7). Accordingly, the crystal structures of COX-1 and COX-2 are virtually superimposable, and the catalytic mechanism is conserved between isoforms (1, 7). The major difference between COX-1 and COX-2 is in their observed expression patterns (8). In most cases, COX-1 is expressed constitutively, producing PGs that mediate “housekeeping” functions, whereas COX-2 is expressed in response to growth factors, tumor promoters, or cytokines (8). PGs produced via COX-2 are intimately involved in a number of pathologies, including inflammation, pain, and colon cancer (9). Other differences between isoforms include the following: (a) the lower concentration of reducing cosubstrate required by COX-2 for activation compared with COX-1, allowing for the utilization of AA when it is a small constituent in a fatty acid pool in vivo (4, 5, 10); and (b) the ability of aspirin-acetylated COX-2 to oxygenate AA, which shifts the reaction specificity such that (15R)-hydroxyeicosatetraenoic acid ((15R)-HETE) is formed instead of PGH2 (11, 12).
Although AA is the preferred substrate for COX-1 and COX-2, other fatty acids can be oxygenated by both isoforms with varying efficiencies (13–15). A prime example is the 30% efficiency (compared with AA) by which COX-2 can oxygenate the fatty acid eicosapentaenoic acid (EPA) (20:5 ω-3) as opposed to the less than 5% efficiency by which COX-1 oxygenates this substrate (14, 16). COX-2 can also oxygenate a more extensive array of substrates compared with COX-1, presumably because of an ∼20% larger active site created by a highly conserved I523V substitution between isoforms (17). Marnett and co-workers (18–20) have established that COX-2, but not COX-1, oxygenates the endocannabinoids 2-arachidonylglycerol, arachidonylethanolamide, and N-arachidonylglycine, utilizing the same general catalytic mechanism derived for AA. These substrates are larger than AA due to the additional chemical constituent attached to the δ-end of the fatty acid.
Crystal structures of ovine (ov) COX-1 have been elucidated in the presence of AA and other fatty acid substrates to moderate resolutions (∼3.0 Å) (16, 21–23). The fatty acids in these structures are all oriented in the cyclooxygenase channel such that the carboxylate of the substrate interacts with the guanidinium group of Arg-120 and the ω-end of the substrate binds in a hydrophobic groove above Ser-530, resulting in the placement of C-13 below Tyr-385 (7). The ovCOX-1:fatty acid crystal structures provided experimental verification at the molecular level of fundamental components of the cyclooxygenase reaction previously identified via biochemical studies, including the following: the required interaction between Arg-120 and the carboxylate of the fatty acid substrate (22, 24); the positioning of the 13-pro-S hydrogen below Tyr-385 for subsequent abstraction (2, 22); and the placement of the ω-end of the fatty acid substrate within the hydrophobic groove, where the terminal carbon lies adjacent to Gly-533 (22, 25). Furthermore, analysis of the spatial arrangement of AA within the cyclooxygenase channel of the ovCOX-1:AA crystal structure allowed for the proposal of a valid sequence of events for the generation of PGG2 and recycling of the tyrosyl radical during cyclooxygenase catalysis (22). AA bound in its productive conformation in ovCOX-1 has been utilized as a model to wed structure with functional studies carried out on both COX-1 and COX-2 that have investigated the catalytic process and molecular determinants governing substrate binding, specificity, and product formation (16, 19–21, 23, 26–30).
Attempts to study the structure of AA bound in the cyclooxygenase channel of COX-2 have yielded interesting but somewhat ambiguous results. Kiefer et al. (31) reported the structure of AA bound to the apoenzyme form of murine (mu) COX-2 in an attempt to prevent oxygenation of the substrate during crystallization. The electron density observed in the cyclooxygenase channel of the resulting structure suggested a mixture of AA and PGG2. The authors concluded that contamination of the protein preparation with heme-containing COX-2 led to the oxygenation of AA during the crystallization process. In another attempt, a peroxidase-inactive mutant (H207A) of muCOX-2 was used to form a complex with AA. Analysis of the electron density within the cyclooxygenase channel of this structure showed that AA had indeed bound, but in a nonproductive conformation in which the carboxylate of AA hydrogen-bonded to Tyr-385 and Ser-530 at the apex of the channel instead of near the opening. We report here studies designed to further investigate the structural basis of fatty acid substrate binding to COX-2. We engineered a His6-tagged N580A muCOX-2 construct to produce milligram quantities of apoenzyme for crystallization trials. The apoenzyme was reconstituted with Co3+-protoporphyrin IX and fatty acid substrate to generate the appropriate enzyme-substrate complexes for x-ray crystallographic analyses. Crystal structures of muCOX-2 in complex with AA, EPA, and docosahexaenoic acid (DHA) (22:6 ω-3) were elucidated to resolutions of 2.1, 2.4, and 2.65 Å, respectively. The structures were utilized to detail and functionally characterize the nuances involved in the binding of fatty acid substrates in the cyclooxygenase channel of COX-2.
EXPERIMENTAL PROCEDURES
Materials
AA ((5Z,8Z,11Z,14Z)-eicosatetraenoic acid), EPA ((5Z,8Z,11Z,14Z,17Z)-eicosapentaenoic acid), and DHA ((4Z,7Z,10Z,13Z,16Z,19Z)-docosahexaenoic acid) were purchased from Cayman Chemical (Ann Arbor, MI). Decyl maltoside and β-octylglucoside (βOG) were purchased from Anatrace (Maumee, OH). Co3+-protoporphyrin IX was purchased from Porphyrin Products (Logan, UT). The QuikChange mutagenesis kit was purchased from Stratagene (La Jolla, CA), and the Bac-to-Bac baculovirus expression kit and associated reagents, including Spodoptera frugiperda 21 (Sf21) insect cells, fetal bovine serum, fungizone, penicillin/streptomycin, and sf-900 II serum-free media were purchased from Invitrogen. Hi-Trap HP chelating and HiPrep Sephacryl S200-HR chromatography columns were purchased from GE Healthcare. Oligonucleotides used for site-directed mutagenesis were purchased from Integrated DNA Technologies (Coralville, IA).
Preparation of His6 N580A Murine COX-2 and Leu-531 Mutants
Native muCOX-2 was cloned into the NotI and BamHI sites of the vector pFastbac1. The pFastbac1 construct was then used as a template to insert six histidine residues behind the signal sequence and between Asn-19 and Pro-20 of muCOX-2 using the QuikChange mutagenesis kit as described in Ref. 32 and the following primer (note: forward primer listed only; inserted amino acids are boldface and underlined): 5′- CTCAGCCAGGCAGCAATTCATCACCATCACCATCACCCTTGCTGTTCCAATCCA-3′. As muCOX-2 is variably glycosylated at Asn-580, we further generated an N580A mutant construct to remove this N-glycosylation site utilizing the QuikChange mutagenesis kit and the His6 muCOX-2 construct in pFastbac1 generated above as the template, with the following primer (forward primer only; site of mutation boldface and underlined): 5′-ACAGCCACCATCGCTGCAAGTGCCTCC-3′. Finally, Leu-531 mutants were generated using the QuikChange mutagenesis kit and the His6 N580A muCOX-2 construct as a template, utilizing the following primers (forward primer only; site of mutation bolded and underlined): L531A, 5′-GGAGCACCATTCTCCGCGAAAGGACTTATGGG-3′; L531P, 5′-GGAGCACCATTCTCCCCGAAAGGACTTATGGG-3′; L531T, 5′-GGAGCACCATTCTCCACGAAAGGACTTATGGG-3′; L531F, 5′-GGAGCACCATTCTCCTTCAAAGGACTTATGGG-3′. Proper insertion of the His6 tag, N580A, and Leu-531 mutations were confirmed by DNA sequencing, which was performed at the Roswell Park Cancer Institute DNA Sequencing Laboratory.
Generation of Recombinant Baculovirus and Expression of Murine COX-2
Positive baculovirus was generated for His6 N580A muCOX-2 and mutant constructs via the transformation of max efficiency DH10Bac cells with the appropriate construct in pFastbac1, followed by transfection of Sf21 insect cells with the isolated bacmid DNA. The initial p1 virus stock was amplified in 50 ml of Sf21 insect cells using a multiplicity of infection (MOI) of 0.1 plaque-forming unit/ml. The resulting p2 virus was harvested 48 h post-infection, plaque-assayed, and used to generate large quantities of p3 virus for subsequent large scale expressions. For expression, Sf21 insect cells were grown at 27 °C in sf-900 II serum-free media supplemented with 1.5% (v/v) fetal bovine serum and the appropriate antibiotics and additives and maintained in suspension in an 8-liter spinner flask equipped with an air-sparging line for proper aeration. The cells were infected with p3 virus stock at a multiplicity of infection of 1.0 once the cell density reached ∼2.0 × 106 cells/ml in the spinner flask and harvested via centrifugation 72–96 h post-infection in conjunction with a drop in cell viability to below 80%. Cell pellet was harvested via centrifugation, pooled, and stored at −80 °C.
Purification of Murine COX-2
The cell pellet from 2 liters of culture was resuspended in 50 mm Tris, pH 8.0, 300 mm NaCl, 10 mm imidazole, and 1 mm 2-mercaptoethanol (2 ml/g cell pellet) and subsequently ruptured using a microfluidizer (Microfluidics, Newton, MA). Decyl maltoside was added to the cell lysate to a final concentration of 0.7% (w/v), and the protein was solubilized for 1 h with stirring at 4 °C. The mixture was then centrifuged at 40,000 rpm at 4 °C for 1 h to remove insoluble debris. The clarified supernatant was loaded on a 5-ml HiTrap chelating HP column and washed with buffer A (50 mm Tris, pH 8.0, 300 mm NaCl, 20 mm imidazole, 1 mm 2-mercaptoethanol, and 0.5% (w/v) decyl maltoside). An intermediate wash step consisting of 10 column volumes was carried out using buffer B (buffer A containing a final concentration of 60 mm imidazole), followed by elution with buffer C (buffer A containing 200 mm imidazole) over 5 column volumes. Functionally active fractions were subsequently pooled and dialyzed overnight at 4 °C against 50 mm Tris, pH 8.0, 300 mm NaCl, and 0.53% (w/v) βOG to initiate detergent exchange for crystallization. The sample was concentrated to ∼2 ml using a Millipore Ultrafree-15 spin concentrator with a 50-kDa nominal molecular mass cutoff. The concentrated protein sample was then trypsin-digested for 20–90 min at 25 °C with a 30:1 ratio COX-2:trypsin (33). The reaction was terminated via the addition of 2 mm phenylmethylsulfonyl fluoride. Trypsin digestion has no deleterious affect on COX-2 function, as digested protein contained cyclooxygenase and peroxidase activity comparable with the undigested sample. The protein was then subjected to size-exclusion chromatography utilizing a HiPrep 16/60 Sephacryl S-300 HR column equilibrated in 25 mm Tris, pH 8.0, 150 mm NaCl, and 0.53% (w/v) βOG. The peak fractions of the purified His6 N580A muCOX-2 constructs were pooled and concentrated to 3 mg/ml for crystallization trials.
Cyclooxygenase and Peroxidase Assays
For measurement of cyclooxygenase activity, the initial rate of O2 uptake was measured at 37 °C using a model 5300 biological oxygen monitor (YSI Inc., Yellow Springs, OH), equipped with an oxygen electrode. Each standard assay mixture contained 3 ml of 100 mm Tris, pH 8.0, 1 mm phenol, 5 μm hematin, and 1–200 μm substrate. Reactions were initiated by the addition of up to 20 μg of protein in a volume of 20 μl. Km and Vmax values were determined by measuring oxygen uptake using 1–200 μm fatty acid substrate and fitting the data to the Michaelis-Menten equation using GraphPad Prism 5.0 for Windows (GraphPad Software, San Diego). Peroxidase activity was measured spectrophotometrically at 25 °C. The reaction mixture in the cuvette contained 100 mm Tris, pH 8.0, 100 μm N,N,N′,N′-tetramethylphenylenediamine, 1.7 μm hematin, and up to 100 μg of protein. Reactions were initiated by adding 100 μl of 300 μm H2O2, and the oxidation of N,N,N′,N′-tetramethylphenylenediamine was monitored at 610 nm over time.
Crystallization and Data Collection
For crystallization, purified apo-His6 N580A muCOX-2 (or the L531F mutant) was reconstituted with a 2-fold molar excess of Co3+-protoporphyrin IX, followed by dialysis overnight at 4 °C against 20 mm Tris, pH 8.0, 100 mm NaCl, 0.6% (w/v) βOG. Just prior to setup, a 10-fold molar excess of substrate was added to the reconstituted protein. Initial crystallization screening was carried out on the muCOX-2:AA complex using the 1536 microbatch-under-oil screen offered by the High Throughput Crystallization Laboratory, at The Hauptman-Woodward Medical Research Institute (34). One subsequent crystallization lead was further optimized to generate crystals of the COX-2:fatty acid complexes utilized in this study. Specifically, crystals were grown in sitting-drops by combining 3 μl of protein solution with 3 μl of 23–34% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, 20 mm MgCl2, and 0.6% (w/v) βOG and equilibrating over reservoir solutions containing 23–34% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, and 20 mm MgCl2. Trays were incubated at 23 °C, and plate-like crystals were formed in 3 days up until approximately 4 weeks. Crystals were cryopreserved via transfer to a solution consisting of 30% polyacrylic acid 5100, 100 mm HEPES, pH 7.5, 20 mm MgCl2, 0.6% (w/v) βOG, and 10% ethylene glycol. Following incubation in cryoprotectant for 10–120 min, the crystals were looped and flash-cooled directly into the nitrogen gas stream prior to diffraction analysis. Data were collected on beamline A1 at the Cornell High Energy Synchrotron Source (Ithaca, NY) at a wavelength of 0.9789 Å using an Area Detector Systems CCD Quantum-210 Detector. Datasets were integrated and scaled using MOSFLM and SCALA, respectively, in the CCP4 suite of programs (35). For the L531F mutant COX-2:AA structure, the dataset was assembled from diffraction measurements from two crystals. The data from each crystal was processed individually using MOSFLM and then scaled together using SORTMTZ and SCALA. Details of the data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Crystallographic parameter | COX-2:AA | L531F:AA | COX-2:EPA | COX-2:DHA |
|---|---|---|---|---|
| Space group | I222 | I222 | I222 | I222 |
| No. in asymmetric unit | 2 | 2 | 2 | 2 |
| Unit cell length | ||||
| a | 119.98 Å | 120.79 Å | 121.85 Å | 119.29 Å |
| b | 132.55 Å | 133.04 Å | 132.34 Å | 131.71 Å |
| c | 180.52 Å | 180.65 Å | 180.29 Å | 179.24 Å |
| α = β = γ | 90° | 90° | 90° | 90° |
| Wavelength | 0.9789 Å | 0.9777 Å | 0.9789 Å | 0.9789 Å |
| Resolution | 20.0-2.10 Å | 20.0-2.40 Å | 20.0-2.40 Å | 20.0-2.65 Å |
| Highest resolution shella | 2.21-2.10 Å | 2.53-2.40 Å | 2.53-2.40 Å | 2.79-2.65 Å |
| Rmergeb | 8.4 (58.2) | 17.4 (32.2) | 9.6 (55.9) | 12.4 (48.9) |
| RPIMb | 4.1 (29.5) | 7.2 (23.6) | 5.6 (33.7) | 5.7 (22.6) |
| Total observations | 423,435 (56,778) | 320,496 (18,069) | 223,307 (30,395) | 244,651 (34,902) |
| Total uniquec | 83,526 (12,044) | 55,133 (7387) | 56,982 (8181) | 41,205 (5946) |
| I/σ(I) | 12.9 (2.4) | 8.0 (2.1) | 11.0 (2.4) | 10.9 (3.4) |
| Completeness | 99.6% (99.3%) | 96.8% (90.2%) | 99.8% (99.6%) | 99.8% (100%) |
| Multiplicity | 5.1 (4.7) | 5.8 (2.4) | 3.9 (3.7) | 5.9 (5.9) |
| Wilson B factor | 28.5 Å2 | 38.5 Å2 | 40.7 Å2 | 50.4 Å2 |
| No. of atoms in refinement | 10,230 | 9920 | 9766 | 9565 |
| Rwork | 0.169 (0.245) | 0.179 (0.226) | 0.176 (0.248) | 0.181 (0.263) |
| Rfreed | 0.210 (0.293) | 0.228 (0.298) | 0.223 (0.325) | 0.234 (0.347) |
| Average B factor, protein | 33.6 Å2 | 29.1 Å2 | 34.8 Å2 | 38.2 Å2 |
| Average B factor, solvent | 26.5 Å2 | 22.0 Å2 | 23.5 Å2 | 14.0 Å2 |
| Average B factor, fatty acid | ||||
| Monomer A | 43.7 Å2 | 49.2 Å2 | 44.5 Å2 | 64.7 Å2 |
| Monomer B | 52.6 Å2 | 51.8 Å2 | 45.6 Å2 | 64.3 Å2 |
| Mean positional errore | 0.284 Å | 0.292 Å | 0.296 Å | 0.348 Å |
| r.m.s.d. in bond length | 0.008 Å | 0.008 Å | 0.008 Å | 0.006 Å |
| r.m.s.d. in bond angle | 1.121° | 1.490° | 1.104° | 1.021° |
a The values in parentheses represent the values in the outermost resolution shell.
b Rmerge and RPIM are as defined in Ref. 57.
c Data represent reflections with F > 0 σF, which were used in the refinement.
d 5.0% of the total reflections were used to generate the test set.
e Coordinate error was calculated by Luzatti plot.
Structure Solution and Refinement
Although crystal structures of both COX-1 and COX-2 have been elucidated, extensive care was taken to remove model bias during the structure solution and refinement process given the crystallographic observations reported previously for AA in complex with apo- and H207A muCOX-2 (31). As such, difference Fourier maps were not utilized to generate initial phases for the enzyme-substrate complexes. Instead, a modified version of monomer A of Protein Data Bank entry 1CVU (31) was created in which all ligands and waters as well as residues 33–144, 320–325, 344–391, and 500–553 were deleted. The regions deleted encompass the N and C termini, as well as the membrane binding domain and walls of the cyclooxygenase channel. The remaining structure (∼60% of the total residues) was then utilized as the search model for molecular replacement calculations on each dataset using the program PHASER (36). The subsequent phases were then used as input to ARP/wARP (37) utilizing the “automated model building starting from experimental phases” option, which further minimizes the introduction of model bias (38). ARP/wARP was able to build the majority of missing portions of the protein model. Iterative cycles of manual model building and refinement, using the programs COOT (39) and REFMAC5 (40), were then carried out to fit the remaining residues and to add waters, substrate, sugar moieties, and other ligand molecules. The coordinates and stereochemical dictionaries for AA, EPA, and DHA were downloaded from the PRODRG server (41). TLS refinement (42), utilizing the TLSMD web server (43, 44), was carried out on each structure during the final rounds of REFMAC5 refinement. Final refinement statistics are summarized in Table 1.
To verify that the observed differences between the conformations of AA and EPA in each monomer were significant, we performed the following exercise in both the muCOX-2:AA and muCOX-2:EPA crystal structures. The substrate model built in the nonproductive conformation in monomer A was replaced with the substrate model built in the productive conformation from monomer B and vice versa. Cycles of REFMAC5 refinement were carried out, followed by the calculation of 2Fo − Fc and Fo − Fc electron density maps. In the areas where the carbon positions differ between the substrates, we would expect to see the appropriate “positive” and “negative” electron density peaks in the calculated Fo − Fc electron density maps. A similar method has been used to verify the changes observed between fatty acid substrates bound in the cyclooxygenase channel of ovCOX-1 (16, 23). Inspection of the resulting electron density maps clearly shows positive difference density in the hydrophobic groove above Ser-530 in monomer B, when AA and EPA are modeled in their nonproductive conformations. Similarly, negative difference density is observed for the misplacement of the ω-end of each substrate in the hydrophobic groove above Ser-530 in monomer A when AA and EPA are modeled in their productive conformations. Moreover, negative difference density is localized around the side chain of Leu-531 in each monomer of the muCOX-2:AA crystal structure as this side chain is forced to reposition to accommodate the different modeled conformations of AA.
Structural Analysis
van der Waals and hydrogen bond interactions were calculated using the program COOT. The upper limit on distance for consideration as a van der Waals contact is 4.0 Å. Superposition of coordinates between structures was done using the program LSQAB within the CCP4 suite of programs and the coordinates for all Cα atoms unless otherwise stated. The root mean square deviation reported for the side chain of Arg-120 was derived from the measured distances between the positions of the Cα and all side chain atoms of this residue (eight total atoms) in the structures compared. Simulated annealing omit maps were calculated using CNS (45). Model validation was carried out using MOLPROBITY (46). Figures were created in CCP4MG (47). The coordinates and structure factors for Co3+-protoporphyrin IX-reconstituted muCOX-2 in complex with AA, EPA, DHA, as well as for the L531F muCOX-2:AA complex have been deposited in the Protein Data Bank (codes 3HS5, 3HS6, 3HS7, and 3KRK, respectively).
RESULTS
To characterize the binding of fatty acid substrates within the cyclooxygenase channel of COX-2, we generated a construct of muCOX-2 that was optimized for structural biology studies. A His6 tag was engineered into muCOX-2 two residues beyond the signal sequence at the N terminus of the enzyme to facilitate purification of the enzyme (32). In addition, we mutated Asn-580, one of four known N-glycosylation sites in COX-2 (48) to alanine. Roughly 50% of the COX-2 molecules are N-glycosylated at Asn-580 (48). Subsequent replacement of this residue with alanine results in the removal of N-glycosylation heterogeneity. The resulting His6 N580A muCOX-2 construct was utilized in an insect cell-based expression system to overproduce recombinant enzyme, and a two-step purification protocol, consisting of immobilized metal affinity and size-exclusion chromatography steps, was implemented to generate milligram quantities of the protein. The purified His6 N580A muCOX-2 enzyme was functionally characterized by spectrophotometric measurement of the peroxidase activity and determination of the oxygen consumption kinetics using an oxygen electrode, with AA, EPA, and DHA utilized as fatty acid substrates. The enzyme retains complete peroxidase activity with respect to untagged, natively glycosylated wild-type muCOX-2 (data not shown). Moreover, calculated kcat and Km values are nearly identical to those reported previously for huCOX-2 and muCOX-2 when AA and EPA are utilized as substrates (Table 2) (14, 15). DHA is a poor substrate for huCOX-2, exhibiting a specific activity of ∼10% relative to the oxygenation of AA (15). When His6 N580A muCOX-2 was tested with DHA, we observed a similar relative activity to that reported for huCOX-2, with a Km value ∼7-fold higher than that of AA (Table 2). Taken together, the His6 and N580A modifications that were introduced into the native muCOX-2 construct to facilitate the steps associated with generating the required quantities of protein for x-ray crystallographic studies do not influence or alter the ability of the enzyme to oxygenate fatty acid substrates.
TABLE 2.
Fatty acid substrate oxygenation by His6 N580A muCOX-2
Oxygen consumption was measured using an oxygen electrode with the fatty acids AA, EPA, and DHA as described under “Experimental Procedures.” kcat and Km values calculated for the utilization of AA and EPA were from duplicate measurements, and those for DHA were from triplicate measurements. We define the relative rate as the rate at which His6 N580A muCOX-2 utilizes EPA and DHA as substrates normalized to the utilization of AA, which is set to 100%.
| Substrate | kcat | Km | kcat/Km | Relative rate |
|---|---|---|---|---|
| s−1 | μm | % | ||
| AA (20:4 ω-6) | 27.0 ± 0.40 | 5.14 ± 0.29 | 5.24 | 100 |
| EPA (20:5 ω-3) | 8.70 ± 0.21 | 9.45 ± 0.73 | 0.92 | 32.3 |
| DHA (22:6 ω-3) | 3.45 ± 0.15 | 36.8 ± 4.25 | 0.09 | 12.8 |
Crystallographic Structure Determination of Native and Mutant COX-2:Fatty Acid Complexes
To generate stable complexes of the fatty acid substrates within the cyclooxygenase channel, we reconstituted purified apo-His6 N580A muCOX-2 with Co3+-protoporphyrin IX to create a native-like enzyme that lacks both peroxidase and cyclooxygenase activity and hence does not form PG product when incubated with substrates (49). This method has been used successfully to trap AA and other fatty acid substrates within the cyclooxygenase channel of ovCOX-1 for structural studies (16, 21–23). We subsequently determined the crystal structures of His6 N580A muCOX-2 in complex with the fatty acid substrates AA, EPA, and DHA using synchrotron radiation. (Accordingly, we use the naming conventions of muCOX-2:AA, muCOX-2:EPA, and muCOX-2:DHA, when describing the co-crystal structures.)
Each of the muCOX-2:fatty acid structures contains two monomers in the crystallographic asymmetric unit (termed “monomer A” and “monomer B” for comparison purposes) that form the canonical biological dimer. Interpretable electron density was observed for Co3+-protoporphyrin IX and the carbohydrate moieties linked to Asn-68, Asn-144, and Asn-410 in each monomer (residue numbering based on ovCOX-1 numbering scheme), and a single βOG molecule was observed at a crystallographic special position for each dimer (data not shown). No electron density was observed for C-terminal residues beyond Gln-584 in monomer A and Val-583 in monomer B in any of the muCOX-2:fatty acid complexes. The domain architecture, which includes the N-terminal epidermal growth factor domain, the membrane-binding domain, and the catalytic domain, is well resolved for each monomer in all three structures. There are no significant structural differences between monomers within each muCOX-2:fatty acid structure (A versus B) or between monomers of the different muCOX-2:fatty acid structures (A versus A, B versus B, and A versus B) when compared, as calculated root mean square deviations (r.m.s.d.) between monomers are within the mean positional error of these structures (supplemental Table S1).
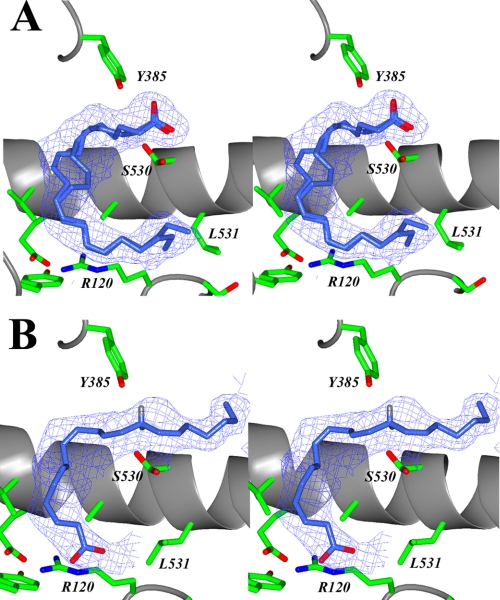
Interpretable electron density was observed for fatty acid substrate in the cyclooxygenase channel of each monomer of the muCOX-2 dimer for the structures determined. However, there are significant conformational differences observed for AA within the cyclooxygenase channels in the muCOX-2:AA crystal structure and EPA in the muCOX-2:EPA crystal structure when their corresponding monomers (monomer A and monomer B) are examined. Specifically, AA is bound in monomer A such that the carboxylate of the fatty acid substrate interacts with Tyr-385 and Ser-530 at the apex of the channel, similar to that observed by Kiefer et al. (31) and generally referred to as a nonproductive binding mode for AA (Fig. 1A). In stark contrast, AA is bound in monomer B such that the carboxylate lies near Arg-120 and Tyr-355 at the opening of the cyclooxygenase channel, similar to the productive binding mode of AA observed in the ovCOX-1:AA crystal structure (Fig. 1B) (22). Similarly, EPA binds in a nonproductive conformation in monomer A (supplemental Fig. S1) and a productive conformation in monomer B (Fig. 2A) in the muCOX-2:EPA crystal structure. Interestingly, DHA adopts similar global substrate conformations in both monomers in the muCOX-2:DHA crystal structure, with the carboxylate group located near Arg-120 and Tyr-355 (Fig. 2, B and C). The observed binding mode of substrate within these structures is described in detail below.
FIGURE 1.
AA bound in the cyclooxygenase channel of the muCOX-2:AA complex. A, stereo view of AA bound in the “nonproductive” conformation in the cyclooxygenase channel of monomer A. Alternate positions are observed for the C8–C9 double bond of AA bound in the nonproductive conformation. B, stereo view of AA bound in the “productive” conformation in the cyclooxygenase channel of monomer B. The positions of the hydrogen atoms have been modeled onto C-13 of AA. The 13-pro-S hydrogen lies 2.9 5Å below the phenolic oxygen of Tyr-385. The Fo − Fc-simulated annealing omit map density, contoured at 4σ, is shown with the final refined models of AA (dark blue).
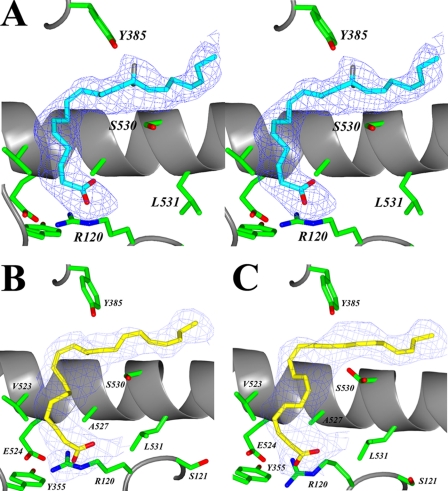
FIGURE 2.
EPA and DHA bound in the cyclooxygenase channel of the muCOX-2:EPA and muCOX-2:DHA complexes. A, stereo view of EPA bound in the productive conformation in the cyclooxygenase channel of monomer B of the muCOX-2:EPA crystal structure. The positions of the hydrogen atoms have been modeled onto C-13 of EPA. The 13-pro-S hydrogen lies 2.96 Å below the phenolic oxygen of Tyr-385. The observed conformation of DHA in monomer A is shown in B, and monomer B in the muCOX-2:DHA crystal structure is shown in C. DHA binds in each cyclooxygenase channel such that the carboxylate interacts with the side chains of Arg-120 and Tyr-355 at the opening of the channel. The Fo − Fc-simulated annealing omit map density, contoured at 4σ, is shown with the final refined models of EPA (cyan) and DHA (yellow).
AA and EPA Bound in a Productive Conformation within the Cyclooxygenase Channel of COX-2
The conformations of AA and EPA observed in monomer B of the muCOX-2:AA and muCOX2:EPA crystal structures exhibit the “L-shaped” configuration observed for AA and other fatty acid substrates bound in the cyclooxygenase channel of ovCOX-1 (Fig. 3, A and C) (16, 22, 23). In both cases, the carboxylate of the substrate is positioned at the channel opening near the side chains of Arg-120 and Tyr-355, and the ω-end is encased by residues Phe-205, Phe-209, Val-228, Val-344, Phe-381, and Leu-534, which form a hydrophobic groove above Ser-530. The central substrate carbons of AA and EPA weave around the side chain of Ser-530, properly placing C-13 of each substrate 2.95 and 2.96 Å, respectively, below the phenolic oxygen of Tyr-385 for abstraction of the 13-pro-S hydrogen. As such, the observed conformations of AA and EPA in monomer B of these structures can be considered as productive substrate conformations given that they exhibit the proper positioning and stereochemical alignment necessary for initiation of the cyclooxygenase reaction (22).
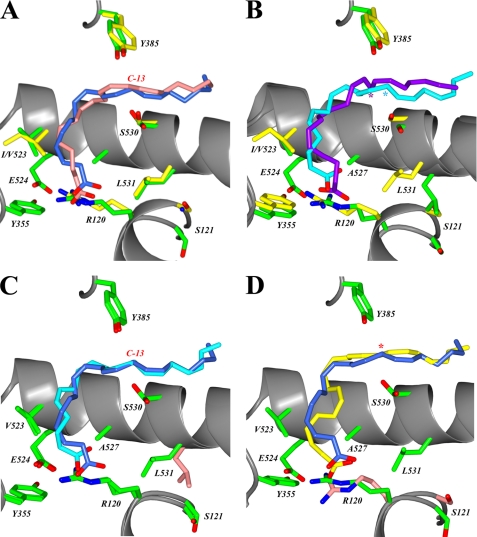
FIGURE 3.
Comparison of the binding modes of fatty acid substrates in the cyclooxygenase channel. The superposition of fatty acid substrates within the cyclooxygenase channel are shown. A, superposition of the productive conformation of AA in monomer B of muCOX-2:AA (dark blue) and ovCOX-1:AA (Protein Data Bank code 1DIY; pink). Carbon-13 of AA is labeled accordingly. B, superposition of the productive conformation of EPA in monomer B of muCOX-2:EPA (cyan) with the nonoptimally aligned conformation observed in the crystal structure of ovCOX-1:EPA (Protein Data Bank code 1IGX; purple). The position of C-13 for EPA in each structure is labeled with a cyan and purple asterisk. The positions of cyclooxygenase active site residues in A and B are shown in green for COX-2 and yellow for COX-1. C, superposition of the productive conformation of AA in monomer B of muCOX-2:AA (dark blue) with the productive conformation of EPA in monomer B of muCOX-2:EPA (cyan). The location of C-13 is labeled accordingly, and the position of the side chain of Leu-531 in the muCOX-2:EPA structure is shown in pink. D, superposition of the productive conformation of AA in monomer B of muCOX-2:AA (dark blue) with the conformation of DHA observed in monomer B of muCOX-2:DHA (yellow). The locations of C-13 of AA and carbon-15 of DHA are denoted by a red asterisk. The positions of the side chains of Arg-120 and Ser-121 in monomer B of muCOX-2:DHA are shown in pink.
AA and EPA make a total of 54 and 56 contacts, respectively, with 19 residues that line the cyclooxygenase active site in the muCOX-2:AA and muCOX-2:EPA crystal structures (supplemental Table S2 and S3). The 19 residues participating in these interactions are essentially conserved with respect to those observed to form contacts with fatty acid substrates bound in the cyclooxygenase channel of ovCOX-1 (16, 22, 23). The major difference between the active site residue contacts made by AA and EPA in the muCOX-2:AA and muCOX-2:EPA crystal structures compared with those made between substrate and active site residues in ovCOX-1 is the lack of an interaction between the carboxylate of these two substrates and the side chain of Arg-120. This interaction is a required molecular determinant for the binding of fatty acid substrates to COX-1 (24), but not COX-2, and our structural data confirm previous mutagenesis studies to this effect (50). Indeed, only one hydrophilic contact is made by AA and EPA with cyclooxygenase active site residues in these structures, a hydrogen bond between the carboxylate oxygen O1 of each substrate and the phenolic hydroxyl of Tyr-355, compared with three in structures of fatty acids bound to ovCOX-1 (16, 22, 23). The lack of a required interaction between the carboxylate of AA and EPA and side chain of Arg-120 coupled with the larger volume of the COX-2 cyclooxygenase channel also provides these substrates with greater conformational freedom within the active site of COX-2. This is borne out in r.m.s.d. values of 1.04 and 0.92 Å calculated for the productive conformations of AA and EPA in the muCOX-2:AA and muCOX-2:EPA structures, respectively, compared with the conformation exhibited by AA in the ovCOX-1:AA structure (supplemental Table S4 and Table S5).
COX-1 is essentially inactivated when EPA is utilized as a substrate due to EPA binding in a strained conformation within the cyclooxygenase channel, which results in the misalignment of C-13 below Tyr-385 (14, 16). The conformation of EPA within the cyclooxygenase channel of monomer B of the muCOX-2:EPA crystal structure is significantly different from that observed in the ovCOX-1:EPA crystal structure, with a calculated r.m.s.d. of 2.28 Å between the 22 atoms of the substrate (supplemental Table S6). The observed conformation of EPA in monomer B is more closely related to that observed for AA in both the ovCOX-1:AA and muCOX-2:AA (monomer B) crystal structures (Fig. 3, B and C). In this orientation, C-13 of EPA is properly aligned below Tyr-385, consistent with kinetic data showing that COX-2 can utilize EPA as a substrate (Table 2) (14). The decreased efficiency observed for the oxygenation of EPA by COX-2 compared with AA may be explained by the presence of an additional double bond at the ω-end of the substrate. The increased rigidity likely constrains the conformational flexibility at the ω-end, which in turn limits its maneuverability within the cyclooxygenase channel during catalysis. Indeed, there is an ∼2-fold increase in the Km value for AA and EPA binding to muCOX-2 (Table 2).
The spatial positions of the side chains of the cyclooxygenase active site residues in monomer B of the muCOX-2:AA and muCOX-2:EPA crystal structures remain mostly unchanged compared with their positions in the ovCOX-1:AA and ovCOX-1:EPA crystal structures. The only notable exceptions are differences exhibited by the side chains of Arg-120, Ser-530, and Leu-531 (Fig. 3, A–C). The side chain of Ser-530 exhibits a dual conformation in the muCOX-2:AA crystal structure. The mobility and flexibility of this side chain likely facilitates access of the ω-end of the substrate to the hydrophobic groove and, although not essential for catalysis, may contribute to the proper alignment of C-13 below Tyr-385 (7). Shifts in the position of the side chain of Ser-530 have been observed previously in the ovCOX-1:DHLA crystal structure (23). A reduction in the mobility of the side chain of Ser-530 upon aspirin treatment could be a factor in the generation of (15R)-HETE by aspirin-acetylated COX-2 (11, 27). The conformation of the side chain of Arg-120 is different in the muCOX-2:AA and muCOX-2:EPA crystal structures, compared with its positioning in the ovCOX-1:AA and ovCOX-1:EPA crystal structures (r.m.s.d. between side chain atoms of 1.27 and 1.61 Å, respectively; Fig. 3, A and B). The Nϵ, Nη1, and Nη2 side chain atoms of Arg-120 forms a bifurcated interaction with both the carboxylate of the substrate and the Oϵ2 atom of Glu-524 in the cyclooxygenase channel of ovCOX-1 (16, 22, 23). In contrast, the lack of interaction between the carboxylate of the substrate and the side chain of Arg-120 in COX-2 results in both the Nη1 and Nη2 atoms of Arg-120 engaging the Oϵ1 and Oϵ2 atoms of Glu-524 in a dual ionic interaction that is reinforced by a hydrogen bond, which contributes to the stabilization of both side chains and the closing of the cyclooxygenase channel entrance upon substrate binding. Finally, we observe an alternate rotamer conformation for the side chain of Leu-531 in monomer B of the muCOX-2:EPA crystal structure. The implications of the mobility of this side chain are discussed below.
DHA Bound in the Cyclooxygenase Channel of COX-2
Despite being two carbons longer and possessing two additional unsaturated double bonds with respect to AA, DHA binds in an L-shaped conformation in both monomers of the muCOX-2:DHA crystal structure, similar to that observed for 18 and 20 carbon fatty acid substrates bound to ovCOX-1 (16, 22, 23) and muCOX-2 (Fig. 2, B and C). The substrate is constrained within the known boundaries constituting the cyclooxygenase channel, such that the carboxylate of DHA interacts with Arg-120 and Tyr-355 at the base of the channel and the ω-end abuts the side chain of Ile-377 near Gly-533 in the hydrophobic groove above Ser-530 (7, 16, 22, 23). To facilitate binding, carbons C1–C9 of DHA take on a compacted and coiled orientation, similar to that observed for EPA binding to the cyclooxygenase channel of ovCOX-1 (Fig. 3D) (16). The distinct orientation of these carbons is necessary to accommodate the rigidity associated with the C4–C5 and C7–C8 unsaturated bonds. The constrained conformation of DHA places carbon-15 ∼3.0 Å below the phenolic oxygen of Tyr-385. The oxygenation of DHA by COX-2 is inefficient (Table 2) and results in the generation of monohydroxy fatty acid products (15, 51). Hence, it is clear from the observed conformation of DHA that the inherent rigidity and compacted nature of binding of this substrate in the cyclooxygenase channel would favor the production of monohydroxy fatty acids and preclude the formation of a cyclic endoperoxide product. The residues lining the cyclooxygenase channel that contact DHA are also conserved with respect to those making contacts with AA and EPA when bound to muCOX-2 (supplemental Tables S7 and S8). Not surprisingly, the constrained orientation of DHA results in this fatty acid making ∼23% more contacts with residues lining the cyclooxygenase channel, including two salt links between the carboxylate oxygen and Arg-120 and a hydrogen bond between the carboxylate and phenolic oxygen of Tyr-355. The conformations observed for DHA are virtually identical within each monomer of the muCOX-2:DHA crystal structure, with an r.m.s.d. of 0.71 Å between the 24 atoms of the substrate (supplemental Table S9), which would be expected for a rigid substrate bound in the cyclooxygenase channel.
Nonproductive Conformations of AA and EPA
AA and EPA exhibit the nonproductive binding conformation in monomer A of the muCOX-2:AA and muCOX-2:EPA crystal structures (Fig. 1A and supplemental Fig. S1). In this conformation, the carboxylate of the fatty acid is stabilized by two hydrogen bonds, one each to the phenolic oxygen of Tyr-385 and the hydroxyl group of the side chain of Ser-530 and two water molecules occupy the empty hydrophobic groove above Ser-530. The carbon atoms of both AA and EPA are directed toward the opening of the cyclooxygenase channel. The ω-end of each substrate occupies the space above Arg-120 and abuts the side chain of Leu-531, whose side chain is now forced into a different rotameric conformation compared with its location in monomer B of the muCOX-2:AA and muCOX-2:DHA crystal structures and the crystal structures of ovCOX-1 complexed with fatty acid substrates (16, 22, 23). An alternate conformation of the nonproductive binding mode is also observed for AA, as the C8–C9 unsaturated bond can be modeled in two different positions (Fig. 1A). The observed nonproductive conformations of both AA and EPA are very similar to that seen for AA in the H207A apo muCOX-2 crystal structure (supplemental Fig. S2) (31).
Mobility of Leu-531 Influences Substrate Binding to COX-2
To investigate the importance of the difference in the observed side chain position of Leu-531 in our muCOX-2:fatty acid crystal structures, we constructed site-directed mutants of Leu-531 and determined the ability of these mutants to oxygenate AA. Specifically, Leu-531 was substituted with Ala, Phe, Pro, and Thr utilizing the optimized His6 N580A muCOX-2 construct as a template, followed by expression and purification as described above. All four mutant constructs were purified to levels equivalent to wild-type His6 N580A muCOX-2 utilized in crystallization trials and also exhibited wild-type levels of peroxidase activity when measured spectrophotometrically (data not shown). Interestingly, none of the four Leu-531 substitutions resulted in significant loss of cyclooxygenase activity (Table 3). There is less than a 2-fold reduction in relative Vmax compared with wild-type muCOX-2 when AA is utilized as the substrate. Moreover, the L531A and L531P constructs exhibit lower Km values for AA than wild-type enzyme (Table 3). In stark contrast, substitution of Leu-531 to Ala in ovCOX-1 results in a mutant construct with greater than 21-fold reduction and 27-fold increase in relative specific activity and Km values, respectively, compared with wild-type ovCOX-1 (26). Substitutions of Leu-531 with Asn, Asp, Ile, and Val in ovCOX-1 also result in 5–14-fold reductions in relative specific activity (26, 52). However, as is the case for the COX-2 substitutions, Km values for these ovCOX-1 substitutions remain unchanged or are slightly lower compared with wild-type enzyme (26, 52). Hence, Leu-531 substitutions are more tolerated in muCOX-2, whereas a reduction in size or conservative replacement of this side chain in ovCOX-1 significantly affects the proper alignment of substrate for oxygenation (26).
TABLE 3.
Oxygenation of AA by Leu-531 mutants of His6 N580A muCOX-2
Oxygen consumption was measured using an oxygen electrode and AA as the fatty acid substrate as described under “Experimental Procedures.” The oxygenation of AA by Leu-531 mutant constructs is normalized to the oxygenation of AA by wild-type His6 N580A muCOX-2 and reported as a relative Vmax. Relative Vmax values for L531T and L531F were measured at an AA concentration of 25 μm in duplicate. The relative Vmax and Km values reported for L531A and L531P represent results from triplicate measurements. ND, not determined.
| Construct | Relative Vmax | Km |
|---|---|---|
| % | μm | |
| Wild-type | 100 | 5.14 ± 0.29 |
| L531A | 58 | 3.65 ± 0.32 |
| L531P | 64 | 2.95 ± 0.27 |
| L531T | 56 | ND |
| L531F | 89 | ND |
To investigate the structural consequences of removing the flexibility associated with the Leu-531 side chain on AA binding in the cyclooxygenase channel of COX-2, we determined the crystal structure of Co3+-protoporphyrin IX-reconstituted L531F muCOX-2 in complex with AA (denoted L531F:AA). As with the other muCOX-2:fatty acid complexes, the domain architecture, Co3+-protoporphyrin IX, and carbohydrate moieties are well resolved in the crystal structure, and there are no significant structural differences between monomers (data not shown). In contrast to what we observe in the muCOX-2:AA and muCOX-2:EPA crystal structures, the L531F mutation causes AA to bind in the same global conformation in both monomers of the L531F:AA crystal structure, with the carboxylate of AA located near the entrance of the cyclooxygenase channel in each monomer (Fig. 4, A and B). The ω-end of the substrate is bound in the hydrophobic groove above Ser-530, and the contacts made between AA and the residues lining the cyclooxygenase channel are conserved with respect to those identified when fatty acid substrates bind productively in the cyclooxygenase channel (supplemental Tables S10 and S11) (16, 22, 23).
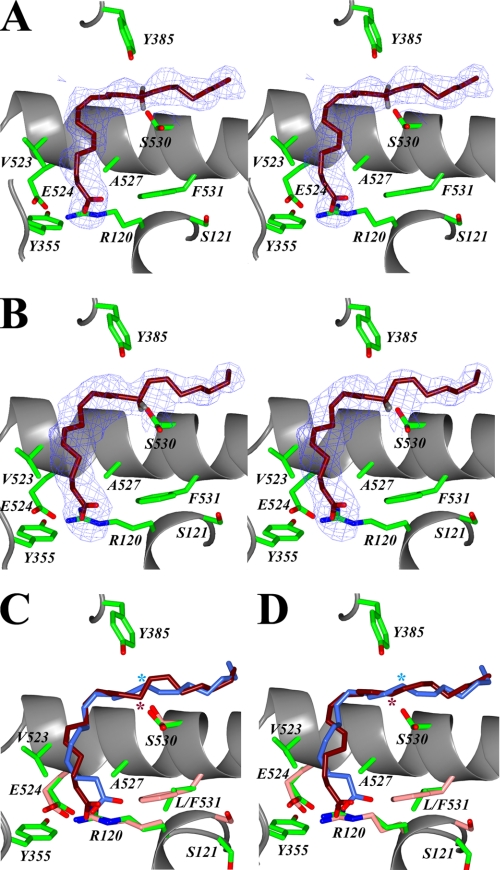
FIGURE 4.
AA bound in the cyclooxygenase channel of the L531F:AA complex. Stereo view of AA bound in the cyclooxygenase channel of monomer B (A) and monomer A (B) of the L531F:AA crystal structure. The L531F mutation causes AA to bind with its carboxylate near Arg-120 and Tyr-355 at the opening of the channel in each monomer of the biological dimer. The Fo − Fc-simulated annealing omit map density, contoured at 4σ, is shown, and the final refined models of AA (magenta) and hydrogen atoms have been modeled onto C-13 of AA. The superposition of the productive conformation of AA in monomer B of muCOX-2:AA (dark blue) with the observed conformation of AA in monomer A (C) and monomer B (D) of L531F:AA (magenta) is shown. The location of C-13 of AA in each structure is denoted with a dark blue and magenta asterisk accordingly. The positions of the side chains of Arg-120, Glu-524, Ser-530, and Phe-531 in the L531F:AA structure are colored in pink.
The side chain of Leu-531 does not interact with fatty acid substrate when it is bound in a productive conformation to muCOX-2 or when it is bound to ovCOX-1 (16, 22, 23). Instead, it forms hydrophobic interactions with the side chain of Arg-120, suggesting a stabilizing role for this side chain in the maintenance of high affinity binding of substrate to ovCOX-1 (26). In the L531F:AA crystal structure, the side chain of Phe-531 lies above helix D of the membrane-binding domain in the vicinity of Arg-120, where it exhibits a single rotamer conformation (Fig. 4, A and B). Substitution of Leu for Phe makes the side chain long enough to make an additional hydrophobic contact with C-1 of AA. This in turn influences the binding of the carboxylate of AA at the opening of the cyclooxygenase channel. The carboxylate is perturbed with respect to its position in monomer B of the muCOX-2:AA crystal structure, forming two interactions with the Nϵ and Nη1 atoms of Arg-120 in addition to its interaction with the side chain atoms of Glu-524 and analogous to that observed for AA and other fatty acid substrates bound to ovCOX-1 (16, 22, 23).
The similar L-shaped conformations of AA observed in monomer A and monomer B of the L531F:AA crystal structure shows that the L531F substitution results in a decrease in the volume of the cyclooxygenase channel available for AA binding (Fig. 4, C and D). This loss of volume combined with the rigidity of the Phe-531 side chain impedes AA from binding in a nonproductive conformation in monomer A. Moreover, further inspection of AA binding in each monomer of the L531F:AA crystal structure indicates that AA does in fact bind differently in each monomer despite their similar global orientations in the cyclooxygenase channel. As observed for AA and EPA in the muCOX-2:AA and muCOX-2:EPA crystal structures, AA is bound productively in monomer B, with C-13 located ∼3.2 Å below Tyr-385 for hydrogen abstraction (Fig. 4, A and D). Accordingly, the observed longer distance between C-13 and the phenolic oxygen of Tyr-385 compared with that observed in the muCOX-2:AA crystal structure likely explains the slight decrease in relative specific activity for this mutant (Table 3). However, AA binds in a nonoptimal conformation in monomer A, as C-13 is displaced with respect to its position in monomer B such that the modeled 13-pro-S hydrogen points away from Tyr-385 in a manner similar to that observed for EPA binding to ovCOX-1 (Fig. 4, B and C) (16). Hence, our functional analysis of Leu-531 mutants in COX-2 coupled with the L531F:AA crystal structure suggests that the side chain of Leu-531 does not play a significant role in high affinity binding of substrate to COX-2 and only a minor role in aligning C-13 optimally below Tyr-385 for hydrogen abstraction. Rather, the mobility observed for the side chain of Leu-531 results in an increase in active site volume at the opening of the cyclooxygenase channel that influences the binding of certain fatty acid substrates.
DISCUSSION
In light of the difficulties and nuances associated with earlier attempts to elucidate the structure of AA bound to muCOX-2, we applied techniques developed to produce ovCOX-1:fatty acid substrate complexes for crystallographic studies in an attempt to determine the crystal structures of fatty acid substrates bound to muCOX-2. All of the structures reported here crystallized with I222 space group symmetry, with a biological dimer in the asymmetric unit of the crystal. Thus, there is no crystallographic symmetry relating one monomer to the other (as is the case with ovCOX-1:fatty acid crystal structures), and the biological dimer observed in these muCOX-2:fatty acid structures can be considered to contain two independent monomers and hence two independent cyclooxygenase active sites. As such, significant efforts were taken to eliminate the introduction of model bias during the building and refinement of fatty acid substrates within the cyclooxygenase channels so that potential differences between substrate conformations or active site residues in each monomer could be identified.
The conformation of AA, EPA, and DHA observed in monomer B of their respective crystal structures provides the first insights at the molecular level for the productive binding of these substrates in the cyclooxygenase channel of COX-2. The observed productive conformations of AA and EPA are in accord with early models put forth for AA binding in the cyclooxygenase channel of COX-2 that placed the carboxylate near the opening of the channel near Arg-120 and the ω-end in a hydrophobic groove above Ser-530 (25, 50). The sequence of events put forth for the cyclooxygenase reaction based on the conformation of AA bound to ovCOX-1 (22) is also valid for AA and EPA, given their similar overall L-shaped conformations and the optimal positioning of C-13 beneath the phenolic oxygen of Tyr-385 for hydrogen abstraction. Moreover, the observed constrained conformation of DHA provides for an explanation at the molecular level as to why monohydroxy acids are formed upon oxygenation of this substrate by COX-2, rather than a cyclic endoperoxide product.
When the binding of AA and EPA in COX-1 and COX-2 are compared, the first notable difference in cyclooxygenase active site residues is related to the flexibility of Ser-530, which exhibits alternate conformations for its side chain in the muCOX-2:AA crystal structure. The observed movement of the Ser-530 side chain may provide for easier access of the ω-end of the substrate into the hydrophobic groove and also aid in the proper positioning of C-13 for catalysis (7). Furthermore, it is possible that a reduction of the flexibility of the Ser-530 side chain plays a role in the generation of (15R)-HETE upon aspirin acetylation of COX-2.
The second notable difference is the lack of interaction between the carboxylate of AA and EPA and the side chain of Arg-120 in the muCOX-2 crystal structures. An ionic interaction between the carboxylate and Arg-120 is required for fatty acid binding to COX-1 but not COX-2, and the muCOX-2:AA and muCOX-2:EPA crystal structures confirm previous mutagenesis results to this effect. Rieke et al. (50) postulated that the lack of a required interaction between the carboxylate of AA and the side chain of Arg-120 in COX-2 suggested that the hydrophobic residues lining the cyclooxygenase channel contributed more significantly to fatty acid substrate binding to COX-2. Indeed, carbons C13–C20 of AA and EPA make 40 and 38 of the 54 and 56 observed contacts (74 and 68%), respectively, with residues lining the hydrophobic groove above Ser-530 within the cyclooxygenase channel in monomer B of the muCOX-2:AA and muCOX-2:EPA crystal structures, compared with 26 of the 52 contacts (50%) made by the equivalent carbons of AA in the ovCOX-1:AA crystal structure. Hence, the lack of interaction between the carboxylate and Arg-120 is now compensated for by an increased number of hydrophobic contacts at the ω-end of the substrate, ultimately providing AA and EPA with the ability to achieve greater conformational flexibility for the carboxylate end of the substrate in the cyclooxygenase channel of COX-2. This is supported by the greater than 0.9 Å r.m.s.d. calculated for these substrates compared with the position of AA bound in the cyclooxygenase channel of ovCOX-1 and in agreement with molecular dynamics simulations carried out to explore the equilibrium behavior of AA bound to COX-1 and COX-2 (29).
The side chain of Leu-531 must move to accommodate the ω-end of the substrate when AA is bound nonproductively in COX-2. Movement of this side chain is the only notable difference when the positions of equivalent residues are compared with the active sites of ovCOX-1:AA and monomer B of muCOX-2:AA. In contrast to ovCOX-1, Leu-531 substitutions in muCOX-2 did not result in a significant loss of cyclooxygenase activity or binding affinity when AA was utilized as the substrate. The side chain of Leu-531 does not interact with fatty acid substrates bound productively to ovCOX-1 (16, 22, 23) or muCOX-2. In both ovCOX-1 and monomer B of the muCOX-2:AA structure, the side chain of Leu-531 forms interactions with the side chain of Arg-120. This interaction has been postulated to provide stabilization of Arg-120 for high affinity binding of substrate to ovCOX-1 (26). When AA is bound in a nonproductive conformation, the ω-end of the substrate interacts with the side chains of both Arg-120 and Leu-531, effectively replacing the direct interaction between the two side chains, which occurs when substrate is bound productively. It is unclear from a structural perspective whether or not the observed flexibility of Leu-531 in the muCOX-2:AA crystal structure is also a feature of the COX-1 homodimer, given that the known crystal structures of ovCOX-1 in complex with fatty acid substrates all crystallized with a single monomer in the asymmetric unit. However, the significant decrease in specific activity observed for conservative substitutions of Leu-531 in ovCOX-1 coupled with the strict requirement for the carboxylate of the fatty acid substrate to interact with the side chain of Arg-120 for high affinity binding suggests that Leu-531 would not exhibit the same side chain flexibility in the cyclooxygenase channel of COX-1.
Structural elucidation of the L531F:AA crystal structure revealed AA bound in the same global conformation in each monomer, with the side chain of Phe-531 exhibiting a single rotamer conformation. The L531F substitution introduces rigidity at this site that decreases the volume of the cyclooxygenase channel available for substrate binding, which in turn impedes the binding of AA in the nonproductive conformation. The substitution also introduces a hydrophobic contact between this side chain and carbon-1 of AA that influences the carboxylate of AA to form the requisite ionic and hydrogen bond interactions with the side chain of Arg-120, analogous to that observed for substrate binding to ovCOX-1. Thus, our results indicate that the flexibility of the side chain of Leu-531 is important for increasing the volume available for substrate binding at the opening of the cyclooxygenase channel of COX-2. As such, we postulate that the observed increase in active site volume achieved by this movement is a significant contributor to the observed ability of COX-2 to bind a broad spectrum of fatty acid substrates and esters compared with COX-1 (18, 20, 53).
Why do we observe significantly different conformations of AA and EPA in each monomer of the biological dimer in the muCOX-2:AA and muCOX-2:EPA crystal structures? It has recently been demonstrated that the cyclooxygenase enzymes oxygenate fatty acid substrates via a half-of-sites reactivity mechanism (54). Accordingly, it has been postulated that at any given time only one monomer of the COX-2 homodimer is functional, and it alone accounts for the cyclooxygenase activity of the enzyme. Yuan et al. (54) initially speculated that only one substrate molecule bound in the cyclooxygenase channel of one monomer, which then could cause the partner monomer to undergo a conformational change that inhibits substrate from binding to this monomer. Subsequent analyses from the same laboratory have since demonstrated that substrate indeed binds concurrently to each monomer of the dimer. As such, the current model of half-of-sites activity for cyclooxygenase involves one monomer of the biological dimer binding substrate with a higher affinity, which can then act to allosterically modulate cyclooxygenase catalysis in the partner monomer, although the residues and mechanism involved in mediating the cross-talk between monomers have not been identified (15).
The crystal structures presented here agree with aspects of the model proposed for half-of-sites activity for cyclooxygenase. Specifically, we see fatty acid substrate bound in the cyclooxygenase channel of both monomers of the canonical cyclooxygenase homodimer. Moreover, only one substrate is bound in an optimal conformation for subsequent cyclooxygenase catalysis. In all of our COX-2:fatty acid crystal structures, the substrate in monomer A is bound in either a nonproductive conformation (muCOX-2:AA and muCOX-2:EPA) or in a nonoptimal or perturbed conformation that results in the misalignment of C-13 (C-15 of DHA) for catalysis (muCOX-2:DHA and L531F:AA), whereas substrate is always bound productively in monomer B. It remains unclear as to whether or not the nonproductive binding mode observed for AA and EPA in monomer A of their respective crystal structures represents an “allosteric” conformation that contributes to the signaling at the dimer interface (15). It is interesting to note that the global conformational differences observed for AA and EPA in monomer A of the muCOX-2:AA and the muCOX-2:EPA crystal structures versus the binding of DHA and AA in monomer A of the COX-2:DHA and L531F:AA crystal structures correlate with the ability of these substrates to attenuate Cu2+/o-phenanthroline-induced cross-linking studies carried out using P127C/S541C ΔC huPGHS-2 (15). Based on these results, the authors suggest that there are differences in the way that select substrates interact within the cyclooxygenase channel of COX-2 (15).
The different Leu-531 side chain conformations are the most significant changes observed when the areas in and around the cyclooxygenase channel of monomer A and monomer B of these crystal structures are compared. It is too early to speculate as to whether or not the movement of this side chain is directly or indirectly involved in mediating the cross-talk between monomers. We also do not observe any significant changes in residues at the dimer interface between monomers, which suggests that a large backbone movement is not the mechanism by which the monomers relay the allosteric signal. It should be noted, however, that large conformational backbone changes are not the exclusive means by which proteins transmit allosteric signals. Indeed, there are emerging ideas and new ways of thinking about allostery and cross-talk that do not involve a significant movement or change in shape (55, 56), and it is clear that additional structural, functional, and biophysical analyses will be required to shed light on the issue with respect to COX-1 and COX-2.
In summary, the crystal structures of Co3+-protoporphyrin IX-reconstituted muCOX-2 in complex with AA, EPA, and DHA presented here provide the first molecular insight into the productive binding of these substrates in the cyclooxygenase channel of COX-2. Moreover, our structure-function analyses contribute to the verification of previous biochemical studies undertaken to characterize the role that Arg-120 and Leu-531 play in fatty acid substrate binding to COX-1 and COX-2. Finally, we show that the mobility of the side chain of Leu-531 influences the binding of AA and EPA within the cyclooxygenase channel of COX-2 and suggest that this inherent flexibility contributes to increase the volume available at the opening of the cyclooxygenase channel so that COX-2 can oxygenate a wide ranging array of fatty acid substrates and esters compared with COX-1. The structural elucidation of COX-2 complexed with these fatty acid-based COX-2-specific substrates will be required to validate our hypothesis.
Supplementary Material
Acknowledgments
We thank Drs. Alan Brash and Claus Schneider in the Department of Pharmacology, Vanderbilt University, for supplying the cDNA for native murine COX-2.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM077176 (to M. G. M.) from the NIGMS. This work was also supported by an Arthritis Investigator award (to M. G. M.) from the Arthritis Foundation as part of the Segal Osteoarthritis Initiative. This work was based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation Award DMR-0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by National Institutes of Health Award RR-01646 from NCRR.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S11, Figs. S1 and S2, and additional references.
The atomic coordinates and structure factors (codes 3HS5, 3HS6, 3HS7, and 3KRK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PG
- prostaglandin
- AA
- arachidonic acid
- (15R)-HETE
- (15R)-hydroxyeicosatetraenoic acid
- EPA
- eicosapentaenoic acid
- DHA
- docosahexaenoic acid
- ov
- ovine
- mu
- murine
- βOG
- β-octylglucoside
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Rouzer C. A., Marnett L. J. (2003) Chem. Rev. 103, 2239–2304 [DOI] [PubMed] [Google Scholar]
- 2.Smith W. L., DeWitt D. L., Garavito R. M. (2000) Annu. Rev. Biochem. 69, 145–182 [DOI] [PubMed] [Google Scholar]
- 3.Rouzer C. A., Marnett L. J. (2009) J. Lipid Res. 50, S29–S34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith W. L. (2008) Trends Biochem. Sci. 33, 27–37 [DOI] [PubMed] [Google Scholar]
- 5.Smith W. L., Langenbach R. (2001) J. Clin. Invest. 107, 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y. J., Mbonye U. R., DeLong C. J., Wada M., Smith W. L. (2007) Prog. Lipid Res. 46, 108–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garavito R. M., Malkowski M. G., DeWitt D. L. (2002) Prostaglandins Other Lipid Mediat. 68, 129–152 [DOI] [PubMed] [Google Scholar]
- 8.Tanabe T., Tohnai N. (2002) Prostaglandins Other Lipid Mediat. 68, 95–114 [DOI] [PubMed] [Google Scholar]
- 9.Smyth E. M., Grosser T., Wang M., Yu Y., FitzGerald G. A. (2009) J. Lipid Res. 50, S423–S428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulmacz R. J. (2005) Biochem. Biophys. Res. Commun. 338, 25–33 [DOI] [PubMed] [Google Scholar]
- 11.Lecomte M., Laneuville O., Ji C., DeWitt D. L., Smith W. L. (1994) J. Biol. Chem. 269, 13207–13215 [PubMed] [Google Scholar]
- 12.Rowlinson S. W., Crews B. C., Goodwin D. C., Schneider C., Gierse J. K., Marnett L. J. (2000) J. Biol. Chem. 275, 6586–6591 [DOI] [PubMed] [Google Scholar]
- 13.Laneuville O., Breuer D. K., Xu N., Huang Z. H., Gage D. A., Watson J. T., Lagarde M., DeWitt D. L., Smith W. L. (1995) J. Biol. Chem. 270, 19330–19336 [DOI] [PubMed] [Google Scholar]
- 14.Wada M., DeLong C. J., Hong Y. H., Rieke C. J., Song I., Sidhu R. S., Yuan C., Warnock M., Schmaier A. H., Yokoyama C., Smyth E. M., Wilson S. J., FitzGerald G. A., Garavito R. M., Sui de X., Regan J. W., Smith W. L. (2007) J. Biol. Chem. 282, 22254–22266 [DOI] [PubMed] [Google Scholar]
- 15.Yuan C., Sidhu R. S., Kuklev D. V., Kado Y., Wada M., Song I., Smith W. L. (2009) J. Biol. Chem. 284, 10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malkowski M. G., Thuresson E. D., Lakkides K. M., Rieke C. J., Micielli R., Smith W. L., Garavito R. M. (2001) J. Biol. Chem. 276, 37547–37555 [DOI] [PubMed] [Google Scholar]
- 17.Kurumbail R. G., Stevens A. M., Gierse J. K., McDonald J. J., Stegeman R. A., Pak J. Y., Gildehaus D., Miyashiro J. M., Penning T. D., Seibert K., Isakson P. C., Stallings W. C. (1996) Nature 384, 644–648 [DOI] [PubMed] [Google Scholar]
- 18.Kozak K. R., Rowlinson S. W., Marnett L. J. (2000) J. Biol. Chem. 275, 33744–33749 [DOI] [PubMed] [Google Scholar]
- 19.Kozak K. R., Prusakiewicz J. J., Rowlinson S. W., Schneider C., Marnett L. J. (2001) J. Biol. Chem. 276, 30072–30077 [DOI] [PubMed] [Google Scholar]
- 20.Prusakiewicz J. J., Kingsley P. J., Kozak K. R., Marnett L. J. (2002) Biochem. Biophys. Res. Commun. 296, 612–617 [DOI] [PubMed] [Google Scholar]
- 21.Harman C. A., Rieke C. J., Garavito R. M., Smith W. L. (2004) J. Biol. Chem. 279, 42929–42935 [DOI] [PubMed] [Google Scholar]
- 22.Malkowski M. G., Ginell S. L., Smith W. L., Garavito R. M. (2000) Science 289, 1933–1937 [DOI] [PubMed] [Google Scholar]
- 23.Thuresson E. D., Malkowski M. G., Lakkides K. M., Rieke C. J., Mulichak A. M., Ginell S. L., Garavito R. M., Smith W. L. (2001) J. Biol. Chem. 276, 10358–10365 [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharyya D. K., Lecomte M., Rieke C. J., Garavito M., Smith W. L. (1996) J. Biol. Chem. 271, 2179–2184 [DOI] [PubMed] [Google Scholar]
- 25.Rowlinson S. W., Crews B. C., Lanzo C. A., Marnett L. J. (1999) J. Biol. Chem. 274, 23305–23310 [DOI] [PubMed] [Google Scholar]
- 26.Thuresson E. D., Lakkides K. M., Rieke C. J., Sun Y., Wingerd B. A., Micielli R., Mulichak A. M., Malkowski M. G., Garavito R. M., Smith W. L. (2001) J. Biol. Chem. 276, 10347–10357 [DOI] [PubMed] [Google Scholar]
- 27.Schneider C., Boeglin W. E., Prusakiewicz J. J., Rowlinson S. W., Marnett L. J., Samel N., Brash A. R. (2002) J. Biol. Chem. 277, 478–485 [DOI] [PubMed] [Google Scholar]
- 28.Schneider C., Boeglin W. E., Brash A. R. (2004) J. Biol. Chem. 279, 4404–4414 [DOI] [PubMed] [Google Scholar]
- 29.Furse K. E., Pratt D. A., Porter N. A., Lybrand T. P. (2006) Biochemistry 45, 3189–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furse K. E., Pratt D. A., Schneider C., Brash A. R., Porter N. A., Lybrand T. P. (2006) Biochemistry 45, 3206–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiefer J. R., Pawlitz J. L., Moreland K. T., Stegeman R. A., Hood W. F., Gierse J. K., Stevens A. M., Goodwin D. C., Rowlinson S. W., Marnett L. J., Stallings W. C., Kurumbail R. G. (2000) Nature 405, 97–101 [DOI] [PubMed] [Google Scholar]
- 32.Smith T., Leipprandt J., DeWitt D. (2000) Arch. Biochem. Biophys. 375, 195–200 [DOI] [PubMed] [Google Scholar]
- 33.Rowlinson S. W., Kiefer J. R., Prusakiewicz J. J., Pawlitz J. L., Kozak K. R., Kalgutkar A. S., Stallings W. C., Kurumbail R. G., Marnett L. J. (2003) J. Biol. Chem. 278, 45763–45769 [DOI] [PubMed] [Google Scholar]
- 34.Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., DeTitta G. T. (2003) J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 35.Dodson E. J., Winn M., Ralph A. (1997) Methods Enzymol. 277, 620–633 [DOI] [PubMed] [Google Scholar]
- 36.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen S. X., Ben Jelloul M., Long F., Vagin A., Knipscheer P., Lebbink J., Sixma T. K., Lamzin V. S., Murshudov G. N., Perrakis A. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 42.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 43.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 44.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 45.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 46.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 48.Otto J. C., DeWitt D. L., Smith W. L. (1993) J. Biol. Chem. 268, 18234–18242 [PubMed] [Google Scholar]
- 49.Malkowski M. G., Theisen M. J., Scharmen A., Garavito R. M. (2000) Arch. Biochem. Biophys. 380, 39–45 [DOI] [PubMed] [Google Scholar]
- 50.Rieke C. J., Mulichak A. M., Garavito R. M., Smith W. L. (1999) J. Biol. Chem. 274, 17109–17114 [DOI] [PubMed] [Google Scholar]
- 51.Liu W., Cao D., Oh S. F., Serhan C. N., Kulmacz R. J. (2006) FASEB J. 20, 1097–1108 [DOI] [PubMed] [Google Scholar]
- 52.Shimokawa T., Smith W. L. (1992) J. Biol. Chem. 267, 12387–12392 [PubMed] [Google Scholar]
- 53.Turman M. V., Kingsley P. J., Marnett L. J. (2009) Biochemistry 48, 12233–12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan C., Rieke C. J., Rimon G., Wingerd B. A., Smith W. L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6142–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai C. J., del Sol A., Nussinov R. (2008) J. Mol. Biol. 378, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.del Sol A., Tsai C. J., Ma B., Nussinov R. (2009) Structure 17, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.