Abstract
Human microsomal cytochrome P450 (CYP) 2E1 is widely known for its ability to oxidize >70 different, mostly compact, low molecular weight drugs and other xenobiotic compounds. In addition CYP2E1 oxidizes much larger C9–C20 fatty acids that can serve as endogenous signaling molecules. Previously structures of CYP2E1 with small molecules revealed a small, compact CYP2E1 active site, which would be insufficient to accommodate medium and long chain fatty acids without conformational changes in the protein. In the current work we have determined how CYP2E1 can accommodate a series of fatty acid analogs by cocrystallizing CYP2E1 with ω-imidazolyl-octanoic fatty acid, ω-imidazolyl-decanoic fatty acid, and ω-imidazolyl-dodecanoic fatty acid. In each structure direct coordination of the imidazole nitrogen to the heme iron mimics the position required for native fatty acid substrates to yield the ω-1 hydroxylated metabolites that predominate experimentally. In each case rotation of a single Phe298 side chain merges the active site with an adjacent void, significantly altering the active site size and topology to accommodate fatty acids. The binding of these fatty acid ligands is directly opposite the channel to the protein surface and the binding observed for fatty acids in the bacterial cytochrome P450 BM3 (CYP102A1) from Bacillus megaterium. Instead of the BM3-like binding mode in the CYP2E1 channel, these structures reveal interactions between the fatty acid carboxylates and several residues in the F, G, and B′ helices at successive distances from the active site.
Keywords: Crystal Structure, Cytochrome P450, Fatty Acid Metabolism, Protein Conformation, Protein Structure
Introduction
Enzymes in the cytochrome P450 (CYP)2 superfamily are involved in the monooxygenation of a diverse repertoire of foreign and endogenous compounds. Of the 57 human enzymes in this superfamily, perhaps a third to a fourth are responsible for the metabolism of xenobiotics, including pharmaceuticals. However, a number of these enzymes also have putative roles in the metabolism of endogenous physiologically important compounds. Cytochrome P450 2E1 (CYP2E1) falls into this class.
CYP2E1 is probably best known for its ability to oxidize xenobiotic compounds such as ethanol, the alcoholism treatment disulfiram (Antabuse), and the analgesic acetaminophen. In fact, the action of CYP2E1 on acetaminophen creates a metabolite responsible for hepatotoxicity and death in acetaminophen overdose cases. However, CYP2E1 is also known to oxidize endogenous fatty acids, including lipids associated with signaling mechanisms such as arachidonic acid (1) and epoxyeicosatrienoic acids (2). Specifically, biochemical investigations have revealed that CYP2E1 can bind a range of mid-length fatty acids and catalyze the (primarily ω-1) hydroxylation of saturated fatty acids and the epoxidation of the alkenes of unsaturated fatty acids. The metabolism of fatty acids by CYP may also be important in maintaining homeostasis of fatty acid levels. The role of CYP2E1 in the metabolism of these molecules may be related to its regulation by disease states such as obesity (3) and diabetes (3, 4) in rat models.
There is a wealth of biochemical information about the metabolism of fatty acids by mammalian CYP enzymes but no structural information to date. The closest structural information currently in hand is for the P450 domain of the fatty acid hydroxylase of Bacillus megaterium (BM3) bound to either palmitoleic acid (5), N-palmitoylglycine (6), or N-(10-imidazolyl-dodecanoyl)-l-leucine (7). In all three cases, the hydrocarbon chains extend out from the active site in a largely hydrophobic channel toward the loop between the F and G helices. This channel is open to bulk solvent in the substrate-free BM3 structure.
We recently reported the first structures of CYP2E1, cocrystallized with indazole or with 4-methylpyrazole (8). These structures revealed a small active site insufficient to bind fatty acid compounds. However, two adjacent voids were observed, a small enclosed void and a sinuous channel that extends to the surface, similar to that observed for BM3. Based on analysis of this structure and the binding modes observed for fatty acid compounds in the soluble cytochrome P450 BM3, it was proposed that rotation of Phe478 would allow the active site to merge with the adjacent channel to the surface and accommodate fatty acid binding in an orientation similar to that observed in BM3. This orientation would be consistent with the observed ω-1 metabolites and place the carboxylate end so that it could interact with conserved CYP2E1 amino acid residues Gln216, Asn219, and/or Asn220 in the channel (8).
To test this hypothesis, studies were initiated to determine how fatty acids bind to CYP2E1. Fatty acid analogs have been previously designed to probe lauric acid ω-hydroxylation by rat CYP4A1 (9). Many heme-coordinating Lewis base-type functional groups were screened with the most effective heme-binding analogs being ω-imidazolyl fatty acid analogs (10). These compounds had high affinity and gave a characteristic spectral shifts indicative of direct nitrogen binding to the heme iron of CYP4A1. Although CYP2E1 oxidizes fatty acids with chain lengths from C9 (11) to C20 (1), the fatty acid oxidized at the highest rate for both CYP4A1 and CYP2E1 is lauric acid. This suggests that ligands designed to functionally probe the CYP4A1 active site might also bind to CYP2E1, thus providing a spectral reporter for the binding mode of fatty acid compounds in this human enzyme, stabilizing CYP2E1/fatty acid interactions, and increasing the solubility of the fatty acid for cocrystallization.
To test our hypothesis about fatty acid binding to CYP2E1, a series of these ω-imidazolyl-fatty acid analogs were used. Fatty acid analogs with successively elongated hydrocarbon chains were synthesized, their binding affinities were determined, and structures were solved with three of the compounds in the CYP2E1 active site. The resulting structures identify a fatty acid carboxylate binding mode distinct from that suggested by the BM3 structures. Comparison of these new CYP2E1 structures with those with small molecules in the active site reveal the structural accommodation required to facilitate CYP2E1 binding of these two disparate classes of compounds.
MATERIALS AND METHODS
Protein Engineering, Expression, and Purification
The CYP2E1 protein used in these studies is modified at the N terminus and His-tagged at the C terminus to yield MAKKTSSKGKLPPGP…PRSHHHH (non-native sequence underlined). The protein was also expressed in Escherichia coli and purified using immobilized metal affinity, ion exchange, and gel filtration chromatography as reported previously (8).
Synthesis
The synthesis of ω-imidazolyl-hexanoic acid, ω-imidazolyl-octanoic acid, ω-imidazolyl-decanoic acid, and ω-imidazolyl-dodecanoic acid was performed as previously described (9, 10) with the following modifications. First, methylation of the bromo-fatty acids was performed using thionyl chloride and methanol rather than diazomethane. Second, purification of ω-imidazolyl-hexanoic acid, ω-imidazolyl-octanoic acid, and ω-imidazolyl-decanoic acid was performed using anion exchange (Dowex 1X8 200, formate counter ion) (12) rather than cation exchange. Finally, purification of ω-imidazolyl-dodecanoic acid was performed by crystallization as the lithium salt.
Protein Crystallization
All of the crystals were grown by hanging drop vapor diffusion at 25 °C.
ω-Imidazolyl-decanoic Acid
The CYP2E1·ω-imidazolyl-decanoic acid complex (0.91 mm CYP2E1, 25 mm ω-imidazolyl-decanoic acid in buffer of 120 mm potassium phosphate, pH 7.4, 0.5 m sucrose, and 1 mm EDTA) was equilibrated against 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, and 20% PEG 2000 MME. Crystallization drops were initially seeded with microcrystals of the CYP2E1·indazole complex to initiate nucleation of CYP2E1·ω-imidazolyl-decanoic acid crystals. To ensure crystal homogeneity and the absence of indazole, portions of the resulting crystals were then used to seed fresh crystallization drops of the CYP2E1·ω-imidazolyl-decanoic acid complex. The resulting crystals were immersed in 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, 30% PEG 2000 MME as a cryoprotectant before being flash cooled in liquid nitrogen for data collection. A single native data set was collected on Beamline 17-BM at the Advanced Photon Source.
ω-Imidazolyl-octanoic Acid
The CYP2E1·ω-imidazolyl-octanoic acid complex (0.60 mm CYP2E1, 33 mm ω-imidazolyl-octanoic acid in buffer of 120 mm potassium phosphate pH 7.4, 1.0 m sucrose, and 1 mm EDTA) was equilibrated against 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, and 22% PEG 2000 MME. In this case crystallization drops were initially seeded with microcrystals of the CYP2E1·ω-imidazolyl-decanoic complex to initiate nucleation. As described above, portions of the resulting crystals were then used to seed fresh crystallization drops of the CYP2E1·ω-imidazolyl-octanoic acid complex. The crystals were immersed in 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, 1.4 m sucrose and flash cooled prior to data collection. A single native data set was collected on Beamline 9-2 at the Stanford Synchrotron Radiation Lightsource.
ω-Imidazolyl-dodecanoic Acid
The CYP2E1·ω-imidazolyl-dodecanoic acid complex (0.91 mm CYP2E1, 5 mm ω-imidazolyl-dodecanoic acid in the same buffer used above) was equilibrated against 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, and 22% PEG 2000 MME. Crystallization drops were initially seeded with microcrystals of the CYP2E1·indazole complex. Portions of the resulting crystals were used to serially seed fresh CYP2E1·imidazolyl-dodecanoic acid crystallization drops. The crystals were immersed in 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, 1.4 m sucrose, or 0.1 m NaHEPES, pH 7.5, 5% iso-propanol, 30% PEG 2000 MME as a cryoprotectant before being flash cooled in liquid nitrogen for collection of a native data set on Beamline 9-2 at the Stanford Synchrotron Radiation Lightsource.
Data Collection, Structure Determination, and Structure Analysis
The data were processed using Mosflm (13) and Scala (14) (the octanoic and dodecanoic complexes) or HKL2000 (15) (the decanoic complex). All three structures were solved by molecular replacement using the program PHASER (16) and a search model consisting of CYP2E1 from the complex with indazole (Protein Data Bank code 3E6I). Iterative model building and refinement were performed using COOT (17) and REFMAC (18). Structure validation was performed using WHATCHECK (19) and PROCHECK (20). For the octanoic complex, 99% of the residues are in the core/allowed areas of the Ramachandran plot, whereas 0.8 and 0.3% are in the generously allowed and disallowed regions, respectively. In the decanoic complex, 98.6% are in the core or allowed regions of the Ramachandran plot, 0.9% in the generously allowed, and 0.5% in the disallowed regions. For the dodecanoic complex, 98.6% of amino acids fall into the core/allowed Ramachandran plot regions, whereas 0.9 and 0.5% fall into the generously allowed and disallowed regions, respectively. Probe-occupied voids were calculated using VOIDOO (21) with a probe radius of 1.4 Å and a grid mesh of 0.3 Å. CAVER (22) calculations were performed using a grid mesh of 0.8 Å.
RESULTS
CYP2E1 Binding of Fatty Acid Analogs
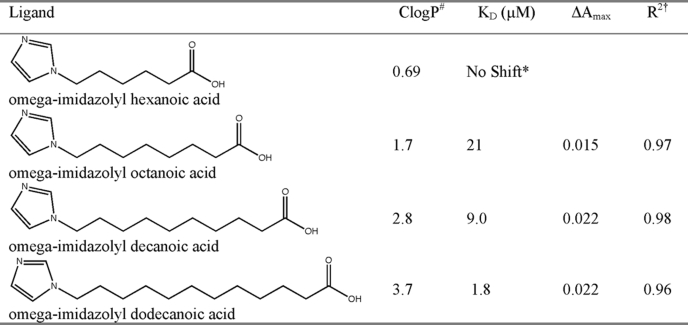
The first structures of CYP2E1 recently revealed the smallest known active site for a human xenobiotic-metabolizing P450, a binding site consistent with its many small molecular weight substrates but insufficient to accommodate the medium and long chain fatty acids that are also substrates for CYP2E1. To understand this two-pronged substrate selectivity of CYP2E1, we have investigated the binding of a series of fatty acid analogs of increasing hydrocarbon chain lengths. Fatty acid analogs with imidazole appended to the ω terminus were used instead of native fatty acids for three reasons. First, addition of the imidazole significantly increased the solubility of the compounds to be cocrystallized (Table 1) to improve saturation of CYP2E1, which is at millimolar concentrations under crystallization conditions. Second, interaction of the imidazole with the heme iron was expected to stabilize binding of the hydrocarbon terminus in an orientation similar to that required for the ω-1 hydroxylation that is predominantly observed for most fatty acid substrates. Third, the expected interaction of the imidazole nitrogen lone pair with the heme iron provides a spectral shift by which binding of the analogs can be monitored.
TABLE 1.
Ligand binding data
* No shift was observed at concentrations up to 230 μm ligand.
† R2, Residual for fit to ΔA = ΔAmax × [ligand]/KD + [ligand].
# ClogP, calculated log of the octanol/water partitioning coefficient.
ω-Imidazolyl-fatty acid analogs were prepared essentially as described (9) with even chain lengths from C6 (ω-imidazolyl-hexanoic acid) to C12 (ω-imidazolyl-dodecanoic acid). Simple modifications were made to the synthesis as described under “Materials and Methods,” largely to avoid the use of diazomethane.
Although the purified CYP2E1 protein has a split Soret peak (390 and 414 nm), the addition of most of the fatty acid analogs resulted in a shift to a single peak at 424 nm as expected for a Type II ligand (nitrogen bound directly to the heme iron). Titration experiments revealed that the ligand with the highest affinity for CYP2E1 is ω-imidazolyl-dodecanoic acid with a KD of 1.8 μm (Table 1). Spectral binding affinities decrease with shorter chain lengths for the decanoic and octanoic compounds. Titration with ω-imidazolyl-hexanoic acid resulted in no observable binding up to 230 μm ligand (230 times the protein concentration).
Overall Structures
X-ray diffraction data from three single crystals allowed the determination of the following structures: to 2.9 Å for CYP2E1 crystallized with ω-imidazolyl octanoic fatty acid, to 2.7 Å with ω-imidazolyl decanoic fatty acid, and to 3.1 Å with the ω-imidazolyl dodecanoic fatty acid. The data collection and refinement statistics are presented in Table 2. Coordinates for the octanoic, decanoic, and dodecanoic analog structures have been deposited in the Protein Data Bank (codes 3KOH, 3GPH, and 3LC4, respectively).
TABLE 2.
Data collection and refinement statistics
| CYP2E1/ω-imidazolyl octanoic fatty acid | CYP2E1/ω-imidazolyl decanoic fatty acid | CYP2E1/ω-imidazolyl dodecanoic fatty acid | |
|---|---|---|---|
| Data collection | |||
| Space group | P43 | P43 | P43 |
| Cell dimensions (Å) | 71.2, 71.2, 224.9 | 70.7, 70.7, 222.8 | 70.7, 70.7, 224.0 |
| Resolution (Å)a | 38.01-2.90 (2.98-2.90) | 50-2.70 (2.80-2.70) | 38.01-3.10 (3.18-3.10) |
| Total/unique reflections | 102,605/24,282 | 192,248/28,548 | 245,170/19,925 |
| Rsyma | 0.123 (0.436) | 0.095 (0.438) | 0.189 (0.405) |
| I/σIa | 9.4 (2.5) | 12.1 (2.4) | 11.3 (4.9) |
| Completenessa (%) | 98.3 (99.1) | 95.4 (93.4) | 100 (100) |
| Redundancya | 4.2 (4.3) | 6.7 (6.3) | 12.3 (7.4) |
| Refinement | |||
| Resolution (Å) | 35.59-2.90 | 31.31-2.70 | 37.93-3.10 |
| No. reflections | 23,000 | 27,040 | 18,847 |
| Rwork/Rfree (%) | 20.6/29.1 | 21.1/28.3 | 19.8/27.0 |
| No. atoms | |||
| Protein | 7554 | 7536 | 7502 |
| Ligand | 30 | 34 | 38 |
| Heme | 86 | 86 | 86 |
| Water | 11 | 25 | 5 |
| B-factors | |||
| Protein | 50.4 | 28.9 | 47.12 |
| Ligand | 60.6 | 29.9 | 53.1 |
| Heme | 36.9 | 24.8 | 29.6 |
| Water | 32.5 | 14.8 | 18.1 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.013 | 0.019 | 0.012 |
| Bond angles (°) | 1.527 | 1.909 | 1.504 |
a The values in parentheses are for the highest resolution shell.
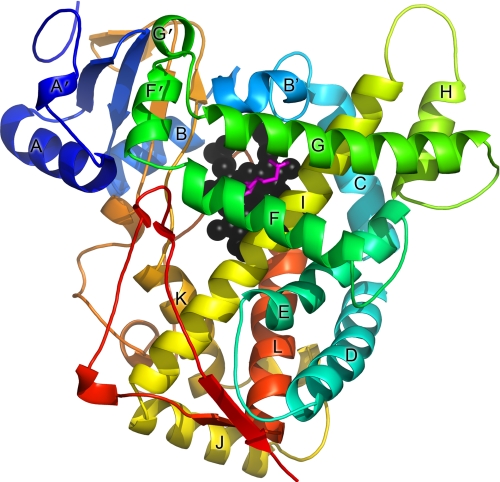
All three of the new CYP2E1 structures reported herein have the global P450 fold (Fig. 1) and the same overall conformation as each other and as the previously described CYP2E1·indazole and CYP2E1·4-methylpyrazole complexes. Pairwise comparisons of these structures reveal Cα root mean square deviation values in the range of 0.3–0.5 Å, similar to the root mean square deviation values between the two monomers of the asymmetric units.
FIGURE 1.
Overall structure of CYP2E1 with ω-imidazolyl decanoic fatty acid. The polypeptide is colored from blue at the N terminus to red at the C terminus with the major helices labeled. Heme is shown as black spheres, whereas the ligand is shown as magenta sticks. All of the figures were generated with PyMOL.
Binding Mode of ω-Imidazolyl Fatty Acids
All three of the fatty acid analogs generally occupy the same space in the CYP2E1 protein (Fig. 2A). The imidazole interacts with the heme iron, whereas the hydrocarbon chain extends above the I helix to terminate with the carboxylate terminus generally located in a space between the I, G, and B′ helices that was a void in the CYP2E1 structures with small molecules (Fig. 2B).
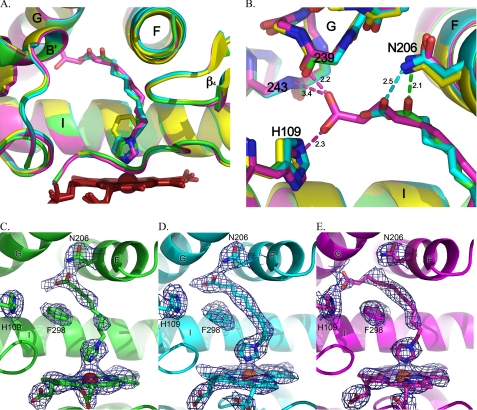
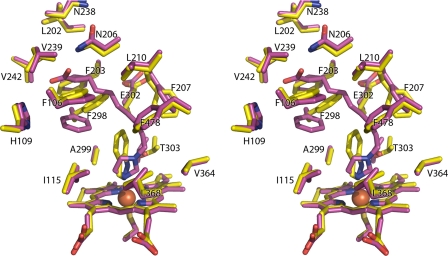
FIGURE 2.
Comparisons of CYP2E1 complexes with indazole (yellow), ω-imidazolyl octanoic fatty acid (green), ω-imidazolyl decanoic fatty acid (cyan), and ω-imidazolyl dodecanoic fatty acid (magenta). A, superposition of all four complexes. B, CYP2E1 interactions with the carboxylate termini of imidazolyl-fatty acids via Asn206 (octanoic, green; decanoic, cyan) or via contacts with both the G helix backbone and His109 (dodecanoic, magenta). C, total omit map of ω-imidazolyl octanoic fatty acid contoured at 0.9 σ. D, total omit map of ω-imidazolyl decanoic fatty acid contoured at 1.0 σ. E, total omit map of ω-imidazolyl dodecanoic fatty acid contoured at 0.9 σ.
As expected from the type II spectra, in each structure the nonalkylated imidazole nitrogen interacts with the heme iron with distances of 2.3–2.6 Å. This is consistent with nitrogen-iron distances observed for other CYP enzymes in complex with Type II ligands. In contrast to the previous indazole and 4-methylpyrazole structures, in this case the second imidazole nitrogen is alkylated and lies outside of hydrogen bonding distance from the CYP2E1 hydrogen bond donor/acceptor, T303. The lack of H-bonding by this imidazole nitrogen is logical because the lone pair electrons of the alkylated imidazole nitrogen would be delocalized in the aromatic ring and thus decrease its affinity for accepting hydrogen bonds.
The various length hydrocarbon chains extend from the imidazole ring to the carboxylate anion located 9.4–11.8 Å away. In the decanoic ligand, positioning of this chain is strongly supported by continuous electron density at 1 σ for both molecules of the asymmetric unit (Fig. 2D). For the lower resolution octanoic and dodecanoic structures, however, the electron density for the hydrocarbon chains is slightly less substantial. In molecule A of the octanoic ligand, there is a short discontinuity in the middle of the chain at 0.9 σ (Fig. 2C), whereas molecule B has less density. For the dodecanoic ligand, the density is weaker where the hydrocarbon connects with the imidazole ring (Fig. 2E) and is similar in the two molecules. The poorer electron density may be due to the lower resolution of these structures, the increase in flexibility for the dodecanoic chain, or, in the case of the octanoic ligand, the lower affinity and partial occupancy. Regardless, the hydrocarbon chains are reasonably well defined and essentially overlie each other as they extend toward the G helix. This extended lipophilic portion of the ligands is surrounded by a “necklace” of phenylalanine side chains: Phe298, Phe116, Phe106, Phe478, and Phe207. The only charged amino acid side chain in this region, Glu302 on the I helix, is directed away from the hydrocarbon.
The carboxylate ends of the fatty acid analogs, although each occupying space between the G, I, and B′ helices, have distinct interactions with the protein (Fig. 2B). In the case of the octanoic fatty acid, the carboxylate is only 2 Å from the side chain nitrogen of Asn206, whereas the second ligand carboxylate oxygen is ∼3.4 Å distant. The decanoic carboxylate extends two carbons farther but also interacts with Asn206, although the distances are longer (2.5 and 3.3 Å in the two molecules of the asymmetric unit). The dodecanoic carboxylate extends even further into the void and past Asn206. Instead one oxygen interacts both with the backbone carbonyl of Val239 (2.2 Å) and the amide nitrogen of Lys243 (3.4 Å) in successive turns of the G helix, whereas the other carboxylate oxygen interacts with the imidazole of His109 (2.3 Å). These interactions position the carboxylate head group of the dodecanoic ligand midway between Val239 and His109 and nearly at the surface of the enzyme. Thus the fatty acids extend from the active site toward the surface of the protein between the G and B′ helices. Mutations of either residue (N206L and H109F) produced inactive, unstable forms of the 2E1 enzyme.
CYP2E1 Conformational Adaptations to Ligand Binding
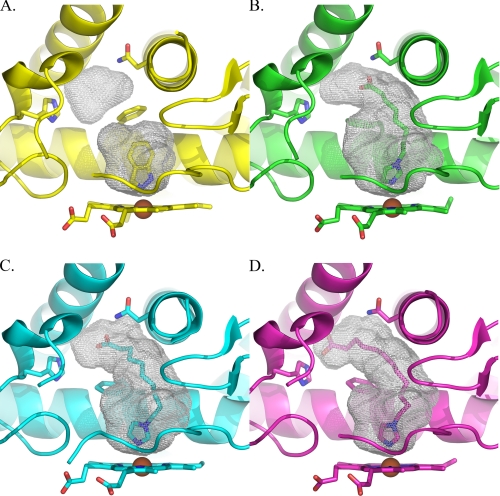
The size and topology of the active site cavity is significantly altered in the presence of these fatty acid ligands. Instead of the ∼190 Å3 active site cavities observed when the small molecules 4-methylpyrazole or indazole are bound (Fig. 3A), the cavity volumes are more than doubled for even the shortest of the fatty acid ligands. The volumes are 420, 440, and 473 Å3, respectively, for the octanoic, decanoic, and dodecanoic ligands (Fig. 3, B–D). In addition to the increasing cavity size, the ligands fill progressively more of the available cavities as their sizes increase. In other words the octanoic acid cavity has additional volume available at the carboxylate end that is not occupied, but the dodecanoic ligand does not.
FIGURE 3.
Comparison of CYP2E1 voids with indazole (A), ω-imidazolyl octanoic acid (B), ω-imidazolyl decanoic acid (C), and ω-imidazolyl dodecanoic acid (D).
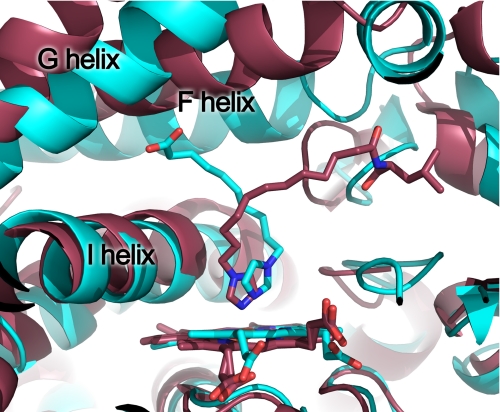
These large changes in the active site geography result not from large scale displacements or alterations in the helical elements surrounding the active site, as observed in other mammalian cytochrome P450 enzymes, but instead originate primarily from the reorientation of a single amino acid side chain, that of Phe298 (Fig. 4). In the indazole-bound structure this side chain adopts a favored Phe rotamer position (33%, χ1 = 177) and forms a barrier between the active site void and a small additional void located between the G, I, and B′ helices. In the ω-imidazolyl-decanoic acid-bound structure, the Phe298 side chain adopts a position very similar to the next most favorable Phe rotamer (13%, χ1 = 59), rotating it significantly toward the B′ helix. As a result, the two voids are connected, allowing the fatty acid to extend into the space between the B′ and F helices and beneath the G helix. This repositioning of Phe298 does not require changes in either the backbone of the I helix in which it is found or the B′ helix. The same rotation of Phe298 is also clearly observed in both molecules of the ω-imidazolyl-dodecanoic acid-bound structure and in one of the two molecules of the ω-imidazolyl-octanoic acid-bound structure. The second protein molecule of the ω-imidazolyl-octanoic acid structure has density supporting this same rotation of Phe298 but also has some density that bridges to the ligand and overlies the position that Phe298 adopts in the indazole structure. In combination with the discontinuous ligand density, this may suggest that in this molecule the Phe can adopt either position, depending on the occupancy of the ligand. However, the resolution of the structure does not support modeling of alternate conformations.
FIGURE 4.
Stereo view of CYP2E1 with indazole (yellow sticks) and with ω-imidazolyl decanoic acid (magenta sticks) demonstrates that the primary difference between the two structures consists of the rotation of Phe298, which allows merging of the two spaces observed in the indazole structure.
Several other residues make minor shifts to acclimate to the fatty acids, including Phe106, Phe207, and Phe478. For example, the plane of the aromatic ring of Phe207 is altered by ∼60° to pack flat against the hydrocarbon backbones of all the fatty acid ligands instead of adopting the more edge-on positioning seen in the small molecule structures. Overall, these shifts appear to be either in adjustment to the rotation of Phe298 or the presence of the hydrophobic methylene chain of the ligand. In contrast, neither Asn206 nor His109, both of which interact with the carboxylates, has significant alterations in their positions in the small molecular weight molecule versus fatty acid structures.
DISCUSSION
Comparison with Fatty Acid Binding Modes in Other Cytochrome P450 Enzymes
Previous structural work examining fatty acid binding in cytochrome P450 enzymes is limited to the soluble bacterial enzymes BM3 (CYP102A1), P450 BioI (CYP107H), and P450BSβ (CYP152A1). Fatty acids and analogs have been cocrystallized with BM3 and occupy a channel extending from the active site to the protein surface adjacent to the B′ helix and the A helix. Fatty acids have been hypothesized to bind to CYP2E1 in the same binding mode (23), and the first structures of CYP2E1 revealed such a channel separated from the small CYP2E1 active site by essentially a single Phe side chain that was likely to be flexible. Located in the channel at an appropriate distance from the iron was the conserved patch of residues 216QXXNN220 that we hypothesized might act as hydrogen bond donors to the carboxylate of fatty acid substrates to orient them with the (ω-1)-carbon in the active site for metabolism. However, in the current structures there is no electron density supporting fatty acid analog binding in this channel. Instead these analogs are oriented in a very different manner.
A comparison of the BM3 structure and the present CYP2E1 dodecanoic structure (Fig. 5) illustrates the differences in binding mode. The two proteins bind fatty acid analogs in very different orientations. These ligands are oriented in opposite directions with respect to each other in the enzyme active sites. Both enzymes catalyze the subterminal hydroxylation of fatty acids (CYP2E1 mainly ω-1 and BM3 mainly ω-2 but also ω-1 and ω-3) (24). Thus, whereas the functionality of these proteins is very similar, the structural solutions to fatty acid binding and metabolism are divergent. CYP4A enzymes are important fatty acid ω and ω-1 hydroxylases, and it is unknown which binding mode might be applicable for these enzymes.
FIGURE 5.
Comparison of fatty acid analog binding in CYP2E1 (cyan) and BM3 (rose). The ligand in CYP2E1 is ω-imidazolyl decanoic fatty acid (cyan sticks). The ligand in BM3 is ω-imidazoyl-dodecanoic-l-leucine (Protein Data Bank code 3BEN).
Structures of fatty acid complexes are also known for P450BioI, a protein involved in biotin synthesis that sequentially hydroxylates midchain C7 and C8 atoms to perform oxidative cleavage. Structures reveal that various length fatty acid ligands linked to an acyl carrier protein adopt a U-shape to position the C7 and C8 atoms above the heme (25). Comparison of this structure with the current 2E1 structures reveals that the head group of the P450BioI ligands extends from the active site in the general direction of what is the original channel in 2E1 and similar to that of fatty acid ligands in BM3 (supplemental Fig. S1). In P450BioI the alkyl chain terminus extends somewhat along the direction of the fatty acid analogs in 2E1. Finally, structures are also available for P450BSβ, a long chain fatty acid peroxygenase that hydroxylates the α and β atoms at the carboxylate end of fatty acids. Consistent with these products, a structure with palmitic acid shows the ligand Cα and Cβ atoms located 5–6 Å from the heme iron with the alkyl chain extending from the active site in what would be approximately the channel in CYP2E1 (supplemental Fig. S1) (26). Thus the binding mode observed for CYP2E1, with the carboxylate located between the G, I, and B′ helices, is a different binding mode than that observed thus far for fatty acid-metabolizing cytochrome P450 enzymes.
The Basis for Differential Fatty Acid Binding Affinity
The binding data show that the highest affinity ligand of the ω-imidazolyl-fatty acid series examined is the dodecanoic acid. The structures reveal that although the interactions of the imidazole moieties are similar among the three ligands, the length of the dodecanoic ligand allows its carboxylate to be positioned at the right position to have the most substantial interactions with the protein, with good hydrogen bonding between both oxygens and the protein (His109 and backbone). The shorter decanoic and octanoic ligands cannot extend to this position. In addition, the amount of hydrophobic interaction between the ligand hydrocarbon chains and the hydrophobic surfaces of the protein is maximal in the case of the dodecanoic ligand. Instead the decanoic and octanoic ligands interact with Asn206, which is closer to the active site, as well as having less hydrophobic interaction with CYP2E1 residues lining the active site. In mammals, Asn206 is conserved. His109 is also fairly well conserved in a variety of mammalian species but is Phe in rat, mouse, and rabbit CYP2E1 enzymes. Of the range of medium chain saturated fatty acids that CYP2E1 hydroxylates at the ω-1 position, the highest turnover is observed for lauric acid (12:0) (27), which might be expected to take advantage of the interactions shown for the decanoic ligand. The alkylated nitrogen of the imidazole in the decanoic acid analog may approximate the position of the subterminal carbon of lauric acid and is positioned 4.5 Å from the heme iron. Regardless, the overall topography of the CYP2E1 active site alters to form a binding site that seems to be complementary to a fatty acid, essentially a hydrophobic tubular space capped by sequentially located residues capable of interacting with the carboxylate. This is compatible with the ability of CYP2E1 to bind and oxidize a range of fatty acid chain lengths.
Access Channel
The program CAVER has been used to find the most accessible path from the active site to the surface of the protein. In previous CYP2E1 structures, the most accessible exit path left the active site between Leu103 and Phe478 and then followed the access channel to the surface of the protein, exiting the protein between the B-B′ loop, the β1 sheet system, and beneath the F′ and G′ helices. In fact, the top four such paths all described the same basic route, transitioning from the active site to the access channel on one side or the other of Phe478.
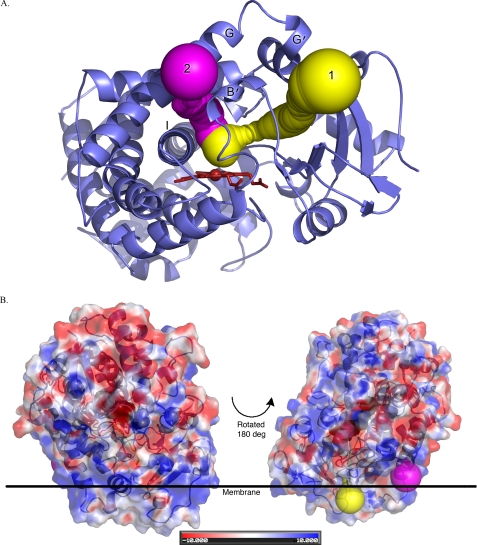
The same analysis of the most likely route for ligand entry and egress in the CYP2E1·ω-imidazolyl-decanoic acid structure identified a path that still exited the protein by traversing the channel as described above (Fig. 6A). However, the second highest scoring result followed the general position of the fatty acid ligand binding cavity to exit the protein between the B′ and G helices above the I helix (Fig. 6A).
FIGURE 6.
Exit routes from CYP2E1·imidazolyl-decanoic fatty acid structure as computed by CAVER. A, the top ranked route from the active site is shown in yellow and follows the access channel, whereas the next most likely route is shown in magenta and follows the fatty acid-binding site. B, relationship of the CAVER exit routes to the putative membrane surface.
Mapping the exit points of these two pathways may suggest different roles of the two paths. Path 1 exits the protein close to the putative membrane-bound portion of the protein, whereas path 2 exits on a face of the protein that is expected to be more exposed (Fig. 6B). This could provide different access/egress routes for substrates that are either more or less hydrophobic or for substrates versus more polar metabolites.
Extrapolation of this Binding Mode to Arachidonic Acid
As previously mentioned, in addition to the metabolism of medium length fatty acids, CYP2E1 is known for the oxidation of arachidonic acid. Two important arachidonic acid metabolites generated by CYP2E1 (along with other CYPs) are 20-hydroxyeicosatraenoic acid and 19-hydroxyeicosatraenoic acid. These metabolites are potent signaling molecules. 20-Hydroxyeicosatraenoic acid has been shown to be a renal and cerebral vasculature autacoid and potent vasoconstrictor (28, 29), whereas 19-hydroxyeicosatraenoic acid has been shown to be a potent vasodilator of renal preglomerular vessels (30). In previous work, a binding mode for arachidonic acid was proposed along the large channel (path 1), but the terminus of arachidonic acid that is oxidized (C19 and C20) might also be able to penetrate to the active site via the new route indicated as part of the fatty acid analog binding site (supplemental Fig. S2A). In addition, CYP2E1 forms epoxides from arachidonic acid (14,15-, 11,12-, and 8,9-epoxyeicosatrienoic acids) (1) and linoleic acid (9,10- and 12,13-epoxyeicosatrienoic acids) (31). It was previously hypothesized that arachidonic acid might be able to bind with the carboxylate anion in the access channel, penetrate past Phe478 to position midchain atoms over the heme, and then penetrate further past Phe298 so that the ω terminus was in the enclosed void of the CYP2E1·indazole structure. With these new structures it seems more likely that arachidonic acid binding might be opposite to this. That is, the carboxylate head group of arachidonic acid might have similar interactions to those observed for the fatty acid analogs examined herein, extension of the hydrocarbon chain to place the unsaturated bond over the heme for epoxidation, and further penetration of the ω terminus past Phe478 into the observed channel (supplemental Fig. S2B). This would require repositioning of Phe478, which appears possible from the structures elucidated thus far but has not actually been observed.
In summary, the structures of CYP2E1 bound to fatty acid ω-imidazolyl analogs of various chain lengths have revealed that these compounds do not bind in the CYP2E1 access channel, as proposed based on fatty acid binding to BM3. Instead these ligands occupy the merged volumes of the original compact active site and a void located between the I, G, and B′ helices. The fatty acid-binding site more than doubles in volume, primarily with the rotation of a single side chain, Phe298. In addition, these structures appear to present another route for potential access/egress to/from the active site. Future work is needed to determine whether this binding mode is conserved in other mammalian P450 enzymes that metabolize fatty acids and to define the CYP2E1 active site topography in the absence of ligand or with other ligands, particularly even larger fatty acids such as arachidonic acid, whose metabolism has a significant physiological impact.
Supplementary Material
Acknowledgments
Thanks are due to Jennifer Laurence for critical suggestions regarding protein stabilization, to Robert Hanzlik who provided initial samples of the imidazole ligands, and to group members Melanie Blevins and Andria Skinner who attempted to make the unstable CYP2E1 mutant proteins. Crystals were grown using the facilities of the Protein Structure Laboratory core facility at the University of Kansas (supported by National Institutes of Health Grant RR017708). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the United States Department of Energy Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, the National Institutes of Health National Center for Research Resources Biomedical Technology Program, and the National Institute of General Medical Sciences. Use of the Industrial Macromolecular Crystallography Association Collaborative Access Team Beamline 17-BM at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago. Use of the Advanced Photon Source was supported by the Office of Basic Energy Sciences of the United States Department of Energy Office of Science under Contract W-31-109-Eng-38.
This work was supported, in whole or in part, by National Institutes of Health Grant GM076343 (to E. E. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3KOH, 3GPH, and 3LC4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CYP
- cytochrome P450
- BM3
- cytochrome P450 BM3 (CYP102A1) from B. megaterium
- PEG
- polyethylene glycol
- MME
- monomethyl ether.
REFERENCES
- 1.Laethem R. M., Balazy M., Falck J. R., Laethem C. L., Koop D. R. (1993) J. Biol. Chem. 268, 12912–12918 [PubMed] [Google Scholar]
- 2.Roy U., Joshua R., Stark R. L., Balazy M. (2005) Biochem. J. 390, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas D., Farez C., Bardou L. G., Vaisse J., Attali J. R., Valensi P. (1998) Fundam. Clin. Pharmacol. 12, 553–558 [DOI] [PubMed] [Google Scholar]
- 4.McCarver D. G., Byun R., Hines R. N., Hichme M., Wegenek W. (1998) Toxicol. Appl. Pharmacol. 152, 276–281 [DOI] [PubMed] [Google Scholar]
- 5.Li H., Poulos T. L. (1997) Nat. Struct. Biol. 4, 140–146 [DOI] [PubMed] [Google Scholar]
- 6.Haines D. C., Tomchick D. R., Machius M., Peterson J. A. (2001) Biochemistry 40, 13456–13465 [DOI] [PubMed] [Google Scholar]
- 7.Haines D. C., Chen B., Tomchick D. R., Bondlela M., Hegde A., Machius M., Peterson J. A. (2008) Biochemistry 47, 3662–3670 [DOI] [PubMed] [Google Scholar]
- 8.Porubsky P. R., Meneely K. M., Scott E. E. (2008) J. Biol. Chem. 283, 33698–33707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alterman M. A., Chaurasia C. S., Lu P., Hardwick J. P., Hanzlik R. P. (1995) Arch. Biochem. Biophys. 320, 289–296 [DOI] [PubMed] [Google Scholar]
- 10.Lu P., Alterman M. A., Chaurasia C. S., Bambal R. B., Hanzlik R. P. (1997) Arch. Biochem. Biophys. 337, 1–7 [DOI] [PubMed] [Google Scholar]
- 11.Fukuda T., Imai Y., Komori M., Nakamura M., Kusunose E., Satouchi K., Kusunose M. (1994) J. Biochem. 115, 338–344 [DOI] [PubMed] [Google Scholar]
- 12.Bookser B. C., Zhu S. (2001) J. Comb. Chem. 3, 205–215 [DOI] [PubMed] [Google Scholar]
- 13.Leslie A. G. (1998) MOSFLM 6.0, Cambridge, UK [Google Scholar]
- 14.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 15.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 16.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 21.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 22.Petrek M., Otyepka M., Banás P., Kosinová P., Koca J., Damborský J. (2006) BMC Bioinformatics 7, 316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S. V., Koley A. P., Dai R., Robinson R. C., Leong H., Markowitz A., Friedman F. K. (2000) Biochemistry 39, 5731–5737 [DOI] [PubMed] [Google Scholar]
- 24.Celik A., Sperandio D., Speight R. E., Turner N. J. (2005) Org. Biomol. Chem. 3, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 25.Cryle M. J., Schlichting I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15696–15701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D. S., Yamada A., Sugimoto H., Matsunaga I., Ogura H., Ichihara K., Adachi S., Park S. Y., Shiro Y. (2003) J. Biol. Chem. 278, 9761–9767 [DOI] [PubMed] [Google Scholar]
- 27.Adas F., Salaün J. P., Berthou F., Picart D., Simon B., Amet Y. (1999) J. Lipid Res. 40, 1990–1997 [PubMed] [Google Scholar]
- 28.McGiff J. C., Quilley J. (1999) Am. J. Physiol. 277, R607–R623 [DOI] [PubMed] [Google Scholar]
- 29.Gebremedhin D., Lange A. R., Lowry T. F., Taheri M. R., Birks E. K., Hudetz A. G., Narayanan J., Falck J. R., Okamoto H., Roman R. J., Nithipatikom K., Campbell W. B., Harder D. R. (2000) Circ. Res. 87, 60–65 [DOI] [PubMed] [Google Scholar]
- 30.Carroll M. A., Balazy M., Margiotta P., Huang D. D., Falck J. R., McGiff J. C. (1996) Am. J. Physiol. 271, R863–R869 [DOI] [PubMed] [Google Scholar]
- 31.Moran J. H., Mitchell L. A., Bradbury J. A., Qu W., Zeldin D. C., Schnellmann R. G., Grant D. F. (2000) Toxicol. Appl. Pharmacol. 168, 268–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.