Abstract
Lymphocyte egress from lymph nodes requires the G-protein-coupled sphingosine 1-phosphate receptor-1 (S1P1). The activation antigen CD69 associates with and inhibits the function of S1P1, inhibiting egress. Here we undertook biochemical characterization of the requirements for S1P1-CD69 complex formation. Domain swapping experiments between CD69 and the related type II transmembrane protein, NKRp1A, identified a requirement for the transmembrane and membrane proximal domains for specific interaction. Mutagenesis of S1P1 showed a lack of requirement for N-linked glycosylation, tyrosine sulfation, or desensitization motifs but identified a requirement for transmembrane helix 4. Expression of CD69 led to a reduction of S1P1 in cell lysates, likely reflecting degradation. Unexpectedly, the S1P1-CD69 complex exhibited a much longer half-life for binding of S1P than S1P1 alone. In contrast to wild-type CD69, a non-S1P1 binding mutant of CD69 failed to inhibit T cell egress from lymph nodes. These findings identify an integral membrane interaction between CD69 and S1P1 and suggest that CD69 induces an S1P1 conformation that shares some properties of the ligand-bound state, thereby facilitating S1P1 internalization and degradation.
Keywords: Cellular Immune Response, Chemotaxis, G Protein-coupled Receptors (GPCR), Immunology, Immunosupressor, Lymphocyte, Membrane Proteins, Protein-Protein Interactions, Lymph Node, Sphingosine-1-phosphate
Introduction
Sphingosine 1-phosphate receptor-1 (S1P1)3 and S1P are required for T and B cell egress from lymphoid organs (1) Following exposure to inflammatory mediators, lymphocyte egress from lymphoid organs can be transiently shutdown, a process that is thought to improve the ability to mount a local immune response (2, 3). Shutdown mediated by IFNα/β is strongly dependent on induction of CD69 expression in the lymphocyte (4). Lymphocyte egress is also inhibited by small molecule modulators of S1P1 function (5). Prolonged systemic inhibition of egress has an immunosuppressive effect and one small molecule that targets S1P receptors, FTY720, has recently completed phase III clinical trials for treatment of multiple sclerosis (6).
CD69 is a type II transmembrane protein of the C-type lectin family and the Cd69 gene is encoded within the NK C-type lectin cluster (7–9). CD69 is a disulfide-linked dimer, and crystal structure analysis established the ectodomain has a C-type lectin fold, though whether it retained a sugar-binding site was unclear (10, 11). In contrast to other members of the NK C-type lectin cluster, CD69 has not been established to have a role in NK cell recognition of target cells. In transgenic overexpression studies, CD69 inhibited T cell egress from the thymus (12, 13). We found that endogenous CD69 inhibits the function of S1P1 in T and B cells (4). When CD69 was overexpressed in cells, it caused down-modulation of S1P1 (4). Reciprocally, when cells lacked expression of S1P1 because of targeted gene deletion, CD69 was detected on the cell surface (1). These data suggested that the presence of S1P1 keeps the low amounts of CD69 produced in naïve T cells from reaching the cell surface. Further evidence that S1P1 can antagonize CD69 expression came from the identification of S1P1 in a genetic screen for molecules that suppress surface CD69 expression in Jurkat T cells (14). These combined observations have suggested that CD69 and S1P1 interact in a variety of lymphocyte cell types and that an overabundance of either molecule can suppress the expression of the other. Evidence for a biochemical interaction between these molecules came from co-immunoprecipitation experiments of epitope-tagged receptors, and from a reporter assay showing that cell surface cross-linking of S1P1 led to co-crosslinking of CD69 (4). However, the properties of this interaction have not been defined.
Here we perform mutagenesis and domain swapping experiments to map regions of CD69 and S1P1 required for complex formation and receptor down-modulation. We use binding studies to show that the complex has an increased binding strength for S1P, and we show that S1P1 protein amounts are reduced in the presence of abundant CD69. Finally, we demonstrate that an S1P1 non-binding mutant of CD69 is ineffective in blocking T cell egress from lymph nodes.
EXPERIMENTAL PROCEDURES
Cell Culture
WEHI-231 cells maintained in RPMI complete (10% fetal bovine serum, supplemented with penicillin/streptomycin, 10 mm HEPES, l-glutamine, and 50 μm β-mercaptoethanol. Cells were split before reaching confluence, but were used for co-IP experiments when the concentration of cells was over 106/ml and >95% viable.
Constructs and Retroviral Transduction
Construction of the MSCV2.2 retroviral vector expressing a Flag-tagged full-length mouse S1P1, upstream of a IRES and a cytoplasmic domain-truncated human CD4, has been described (15). Full-length mouse S1P3 was also cloned into this vector. Mouse CD23, CD69, and human NKRp1A were cloned from splenic cDNA into MSCV2.2 upstream of an IRES and GFP reporter element. Chimeric constructs were produced by PCR with primers overlapping the junctions. All constructs were sequenced. The protein sequences of each mutant or chimeric construct are described in supplemental data. Cultures of Phoenix-E packaging cell line were transfected with these transfer vectors, and supernatants containing retrovirus were collected, and WEHI-231 cells were transduced as described.
Immunoprecipitation and Western Blotting
Immunoprecipitation was done as previously described (4). Briefly, cell pellets were lysed in 0.875% Brij97, 0.125% Nonidet P-40, 150 mm NaCl, 10 mm Tris-HCl pH 7.4, 0.02% NaN3 buffer containing protease inhibitors (Sigma). Samples were resolved by 10% SDS-PAGE (NuPAGE, Invitrogen) and transferred to Immobilon-FL membranes (Millipore). Membranes were blocked with LI-COR buffer and stained with rabbit anti-actin (Sigma), anti-Flag M1 (Sigma), anti-HA biotin 3F10 (Roche). Products were detected with goat-anti-mouse IRDye 680, IRDye 800CW (LI-COR Biosciences), or donkey-anti-rabbit IRDye 700DX (Rockland) and imaged on an Odyssey Infrared Imaging System (LI-COR Biosciences).
Flow Cytometry
Data were acquired on a FACSCalibur or LSRII (Becton Dickinson) and analyzed with FlowJo software (Treestar). Fluorochrome- or biotin-conjugated antibodies were from BD Pharmingen or eBioscience. Flag M2 bio (Sigma) was used for staining the S1P receptor-tagged cells. All constructs, except N6N-DS, N6N-stalk, and 69ISNKE, which were not recognized by any available antibodies, were tested for surface expression (supplemental Fig. S1).
S1P Binding Assay
Labeled sphingosine d-erythro-1-phosphate [33P] (S1P, American radiolabeled chemicals) was resuspended in binding reaction buffer (20 mm Tris pH 7.4, 0.5% fatty acid free bovine serum albumin, 100 mm NaCl, 15 mm NaF, 2 mm 4-dexypyridoxine, 200 μm phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture). WEHI-231 cells were washed 2× with phosphate-buffered saline, and 2 × 105 cells were added to each binding reaction tube. The 33P-labeled S1P was added at a final concentration of 200 pm and allowed to bind on ice for 30′. Unlabeled S1P was then added at 2 μm, and the cells collected on 1.3 cm GF/F filters (Whatman) by vacuum filtration at the indicated times and immediately washed two times with 0.5 ml of binding buffer. The filters were collected into scintillation vials and 1 ml of Ecolume (MP Biomedicals) was added to each tube and mixed before reading on a LS6500 scintillation counter (Beckman).
Transwell Migration Assay
Chemotaxis was across bare 5-μm transwell filters (Corning Costar Corp.) over 3 h in response to S1P (Sigma) or SDF1α (Peprotech) as described (1).
In Vivo Cell Transfer
Spleens from donor mice were mechanically dissociated on 70-μm nylon cell strainers. Cells were washed and resuspended at 5 × 106/ml, and 1 ml was added per well of a 24-well plate coated with anti-CD3 (3 μg/ml) and anti-CD28 (0.5 μg/ml) antibody in phosphate-buffered saline at 37 °C for 3 h. Cells were activated for 24 h and then transduced by spinfection with fresh Phoenix E supernatant as described (16). Cells were then rested for 48 h, collected, and checked for transduction efficiency. Transduced cells were labeled with 5 μm CMTMR (Invitrogen) for 20′ at a 37 °C and washed 2× before combining. 5 × 107 cells were transferred per mouse and cells were analyzed at 24-h post-transfer. At that time, half of the recipients were treated i.v. with 100 μg of both anti-α4 and anti-αL antibody to block LN entry and analyzed 18 h post-injection.
Structure Modeling
Structural models were generated by modeler (17) utilizing the β1-adrenergic receptor (pdb ID 2VT4) as a model (18). Figures were generated with Pymol (19).
RESULTS
Mapping CD69 Interaction Sites
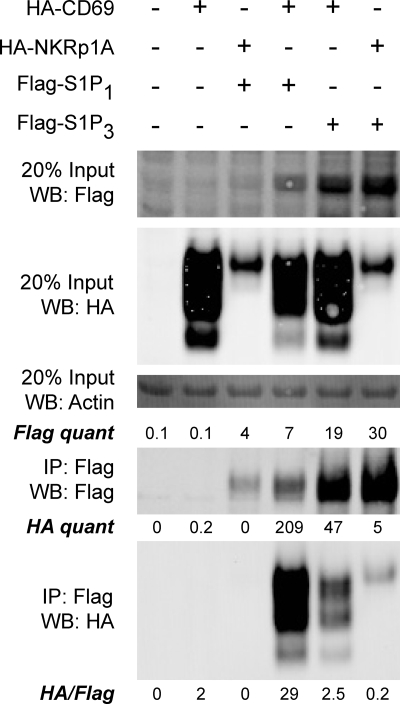
To test whether immune-specific molecules were required for the CD69-S1P1 interaction we transduced 3T3 fibroblasts with retroviruses expressing HA-tagged CD69 or the related type II transmembrane protein, human NKRp1A, and Flag-tagged S1P1 or S1P3. Immunoprecipitation experiments with lysates from cotransduced cells revealed that S1P1 co-immunoprecipitated CD69 but not NKRp1A, whereas S1P3 co-immunoprecipitated little of either lectin molecule (Fig. 1). These results suggest a lack of immune-specific molecule requirement for the CD69-S1P1 interaction.
FIGURE 1.
CD69 and S1P1 interact in mouse 3T3 fibroblast cells. Cells expressing or co-expressing the indicated constructs were lysed and subjected to immunoprecipitation with anti-Flag M2 beads. The material in the IP was then run on SDS-PAGE and Western blotted for Flag to detect the S1P receptor, and for HA to detect the C-type lectin construct and actin as a loading control. The amount of each protein in the cell lysate was compared with the amount immunoprecipitated by Flag-S1P1 with anti-Flag antibody. When equivalent amounts of Flag were immunoprecipitated, there was a 10-fold greater interaction between CD69 and S1P1 when compared with the interaction with control proteins NKRp1A and S1P3. This result is representative of two experiments.
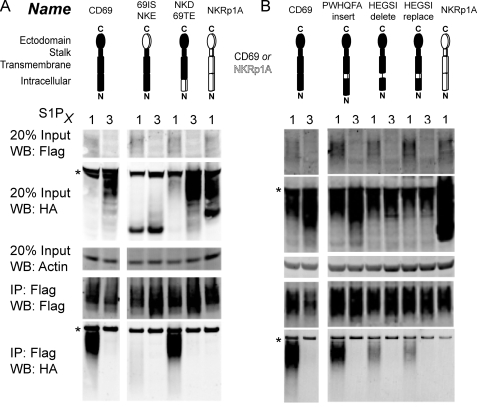
To map the sites on CD69 involved in the interaction, we used a domain swapping approach in WEHI-231 B lymphoma cells with NKRp1A. A construct containing the transmembrane and intracellular domains of CD69 (69ITNKE) retained the ability to interact with S1P1, though with reduced efficiency compared with wild type (Fig. 2A) whereas the converse construct where CD69 regions were replaced with the NKRp1A sequence (NKIT69E) disrupted the interaction (Fig. 2A). All these constructs were expressed at similar levels on the surface of cells lacking FLAG-S1P1 as assessed by flow cytometry (supplemental Fig. S1). In the Western blot analysis for these HA-tagged constructs, the amount of CD69 and 69ITNKE was lower in the lysates from S1P1 than S1P3 expressing cells, perhaps because interaction with S1P1 promotes degradation of some of the complex (Fig. 2A). Consistent with the loss being due to complexing, NKIT69E was present in similar amounts in the S1P1- and S1P3-expressing cells (Fig. 2A). This correlation between interaction with S1P1 and reduction in the total HA-tagged construct detected in cell lysates was noted for many of the subsequently tested constructs.
FIGURE 2.
Interaction between mutant CD69, containing targeted sequence swaps with hNKRp1A, and S1P1 or S1P3 in WEHI-231 cells. A, cells co-expressing the indicated constructs were lysed and subject to immunoprecipitation with anti-Flag M2 beads and then Western blotted to detect Flag-S1P1 or Flag-S1P3 and HA-CD69, HA-NKRp1A, or HA-CD69/NKRp1A chimeric constructs. Diagrams indicate regions of CD69 (filled) or NKRp1A (open) used in each construct. B, co-IP and Western blotting of further chimeric molecules derived by expanding the minimal predicted transmembrane domain to include the stalk or to include an additional six intracellular amino acids. These results are representative of two experiments. Nonspecific bands are indicated with an asterisk.
Selective transfer to NKRp1A of the CD69 transmembrane region alone (N6N) was insufficient to transfer the interaction (Fig. 2A), but inclusion of the membrane proximal intracellular sequence, HEGSI, (N6N-Δ31; Δ31 designating a six amino acid expansion of the minimal transmembrane domain achieved by beginning the CD69 sequence 31 amino acids from the N terminus) conferred an ability to interact with S1P1 (Fig. 2B). This may partly have reflected enhanced expression as the N6N construct showed poor surface expression compared with N6N-Δ31 (supplemental Fig. S1). However, analysis of constructs selectively lacking the HEGSI motif (described below) further suggested a role for this region in the interaction with S1P1. Unexpectedly, the N6N-Δ31 construct also interacted with S1P3 (Fig. 2B). On the extracellular side, inclusion of the membrane proximal stalk region (N6N-stalk) was not sufficient to confer an ability to interact (Fig. 2B). However, the stalk conferred interaction specificity when it was added to the construct containing the CD69 transmembrane and HEGSI motif (converting N6N-Δ31 to N6N-DS; Fig. 2B). The altered molecular weight of some constructs likely reflects altered glycosylation states of the chimeric proteins. Although constructs containing the CD69 ectodomain (NKIT69E) or the entire CD69 intracellular region (69INKTE) did not show interaction (Fig. 2A), when these domains were transferred into NKRp1A together (6N6 and 6N6-stalk) they conferred some ability to interact (Fig. 2, A and B). This might reflect a weak interaction conferred by both the ectodomain and intracellular domain that only achieves sufficient strength to be detectable in co-immunoprecipitation when both domains are present together.
Further mapping of the CD69 stalk segment showed that the membrane proximal half was needed for conferring interaction specificity; when this region of CD69 was replaced with the corresponding region from hNKRp1A (69NKS1 and 69NKS12), the chimeric molecule now interacted with S1P3 as well as with S1P1 (Fig. 3A). By contrast, the membrane distal half did not affect interaction specificity (69NKS2, Fig. 3A). Further subdividing the membrane proximal half into two segments identified the most membrane proximal QKSSIEK sequence as conferring S1P receptor interaction specificity (69NKS1N, Fig. 3B).
FIGURE 3.
Interaction between mutant CD69, containing stalk sequence swaps with hNKRp1A, and S1P1 or S1P3. A, co-IP and Western blotting of Flag-S1P receptors and HA-CD69 constructs containing swaps of small sequences of the CD69 stalk to the analogous sequence in NKRp1A, and HA-NKRp1A construct with the entire CD69 stalk. B, co-IP of CD69 and NKRpA1 stalk mutant 69NKS1N, with NKRp1A sequence QKSSIEK, and stalk mutant 69NKS1C, with the N-terminal stalk sequence SVDIQQS. These data are representative of two experiments with similar results. Nonspecific bands are indicated with an asterisk.
On the intracellular side, the conserved membrane proximal HEGSI motif was necessary for the wild-type interaction strength of construct NKD69TE (Fig. 4A). When starting from the wild-type CD69, deletion of this motif (HEGSI delete) or replacement by the corresponding PWHQFA motif of NKRp1A (HEGSI replace) led to reduced binding, while simply shifting the motif away from the membrane by inserting the NKRp1A sequence (PWHQFA insert) had little effect (Fig. 4B). Moreover, deletion of the HEGSI motif from the swap construct containing the CD69 ecto- and intracellular domains (6N6-stalk) in construct 6N6-DS led to a loss of binding (Fig. 2B). All of these constructs showed strong surface expression by flow cytometry (supplemental Fig. S1). Alanine scanning of this sequence did not identify a particular residue that was responsible for a significant portion of the binding (supplemental Fig. S2). These findings suggest CD69 has a multipartite interaction surface that includes the transmembrane, membrane proximal, and ectodomains and mediates a specific interaction with S1P1.
FIGURE 4.
Interaction between chimeric CD69 molecules, including constructs with modifications of the HEGSI motif, and S1P1 or S1P3. A, two 3-domain constructs analyzed compared with wild-type CD69 as in Fig. 2. B, co-IP and Western blotting of further HEGSI mutants to narrow down the contribution to binding. These data are representative of two experiments with similar results. Nonspecific bands are indicated with an asterisk.
Mapping S1P1 Interaction Sites
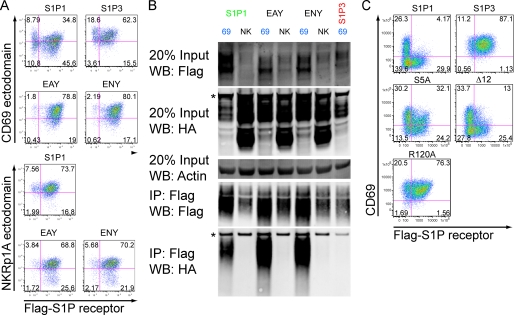
Because CD69 is a member of the C-type lectin family, we initially tested the impact on complex formation of mutating the single surface-exposed consensus N-glycosylation (N30D) site (20) as well as the tyrosine sulfation sites (Y-sulfo) of S1P1 (21). Mutation of these sites or replacement of the entire N terminus with the corresponding region of S1P3 (S1P3 + 1) had little or no effect on S1P1 interaction with CD69 and the S1P1 N terminus alone was not sufficient to confer binding (S1P1 + 3) (Fig. 5A). We also determined that the human sequence of S1P1 was able to interact with CD69 normally (huS1P1). We next generated a series of swap mutants between S1P1 and S1P3. Exchanging all the intracellular loops of S1P3 with those in S1P1 (S1P31313) was not sufficient to confer binding (Fig. 5A). Previously described receptor desensitization mutants, S5A and Δ12 (22, 23) had no effect on interaction (Fig. 5B). In addition, S1P binding was not required for the formation of a S1P1-CD69 complex because the S1P non-binding construct R120A (24) interacted with CD69 at least as well as wild type (Fig. 5B).
FIGURE 5.
Co-IP of CD69 with ectodomain S1P1 mutants. A, tyrosine sulfation (Y-Sulfo), N-glycosylation (N30D), human S1P1 (huS1P1), reciprocal N-terminal swap mutants (S1P1+3 and S1P3+1), and intracellular loop mutant (S1P31313) were compared with S1P1 and S1P3 in their ability to co-IP mouse HA-CD69 as in Fig. 2. B, co-IP of S1P1 desensitization mutants, S5A, Δ12 are shown along with the S1P non-binding mutant R120A. These data are representative of two experiments with similar results.
Replacement of transmembrane helix 4 and associated membrane flanking loop regions of S1P1 by the corresponding region in S1P3 caused a marked reduction in CD69 binding in construct S1P13D (Fig. 6), an effect that was not seen following individual swapping of the other membrane helices and flanking regions (Fig. 6). All of the swap mutant constructs, with the exception of S1P13B, showed robust surface expression by flow cytometry (supplemental Fig. S1). Further mutagenesis revealed that transmembrane helix 4 (S1P13D2) is necessary for most of the interaction with CD69 when compared with the flanking intracellular and extracellular regions (S1P13D1 and S1P13D3, respectively) (Fig. 7A). Replacement of helix 4 in S1P3 with the corresponding region of S1P1 (S1P31D) was sufficient to confer an ability to interact with CD69 but not NKRp1A (Fig. 7B). Analogous with the CD69 HEGSI motif, no single position was responsible for a majority of the binding when each of the eight differences between the S1P1 transmembrane domains were mutated to the corresponding S1P3 residue (supplemental Fig. S3).
FIGURE 6.
Co-IP of CD69 with S1P1 mutants containing swapped transmembrane regions. The schematics above the labels designate the different helices of S1P1 (solid lines and filled rectangles) that were swapped with the analogous sequence for S1P3 (dashed lines and unfilled rectangles). These constructs are labeled S1P13A-G where A refers to transmembrane domain 1 and G to transmembrane domain 7. Co-IP and Western blotting was performed as in Fig. 2. The densitometry readings show the ratio of HA signal to Flag signal indicating the relative ability of CD69 to co-IP with the mutant S1P1 constructs. These data are representative of two experiments with similar results.
FIGURE 7.
The S1P1 helix 4 is necessary and sufficient for most of the interaction with CD69. A, co-IP of S1P13D1–3 constructs where the intracellular loop (D1), minimal predicted transmembrane domain (D2), or extracellular loop (D3) of the S1P1D (membrane helix 4) construct have been swapped with the corresponding region of S1P3. B, co-IP of the S1P31D is the converse construct from the S1P13D with all regions from S1P3 except helix 4. Schematics showing helix 4 of the construct used designate the S1P1 regions of the construct with solid lines and filled rectangles, whereas the S1P3 regions are designated by dashed lines and unfilled rectangles. These data are representative of two experiments with similar results. Nonspecific bands are indicated with an asterisk.
Differing Requirements for CD69- and S1P1-mediated Modulation of the Complex
Under conditions of co-expression, cells with high expression of CD69 down-modulate S1P1 and reciprocally cells with high expression of S1P1 down-modulate CD69 (4). To test the fate of S1P1 that has complexed with CD69 we compared Flag-S1P1-expressing cells that had been transduced with intermediate (+) or high (++) amounts of CD69. In the CD69 intermediate samples, the cells fall mostly into two mutually exclusive populations: cells with surface Flag-S1P1 staining and little CD69, and cells with CD69 surface expression and no Flag-S1P1 (Fig. 8A, HA-CD69+). Examination of reporter gene expression in these exclusive populations shows that the Flag-S1P1 surface-negative cells have higher amounts of the CD69 reporter (GFP) than the Flag-S1P1 surface-positive cells, whereas both have similar amounts of the Flag-S1P1 reporter (hCD4) (Fig. 8A, histogram plots) in agreement with previous findings (4). In cells transduced to express very high amounts of CD69 (Fig. 8A, HA-CD692+) the surface exposure of Flag-S1P1 was almost completely suppressed. By contrast, in Flag-S1P3-expressing cells, the intensity of Flag and CD69 staining was minimally affected by the presence of even very high amounts of CD69. High expression of CD69 not only reduces surface S1P1 abundance but leads to a reduction in total S1P1 protein within the cell (Fig. 8B), suggesting the protein is targeted for degradation. By contrast, although S1P3 was less abundantly expressed, the total amount was unaffected by high CD69 expression (Fig. 8B).
FIGURE 8.
CD69-mediated down-modulation of S1P1 is associated with protein degradation. A, flow cytometric analysis of Flag-S1P1 or Flag-S1P3 when co-transduced and sorted for low or high levels of CD69 in WEHI-231 cells. The relative amount of CD69 expression (+ or ++) was determined by expression of an IRES GFP reporter. Histogram overlays on the right show GFP reporter and hCD4 reporter expression for cells in quadrant 1 and 2 of the top left plot, indicating the relative expression of the CD69-IRES-GFP construct and the Flag-S1P1-IRES-hCD4 construct, respectively. B, co-IP of S1P1 or S1P3 with CD69 from the cells shown in the flow cytometric analysis. Densitometry readings are indicated showing the intensity of Flag and HA signal and the calculation of the ratio of HA signal to Flag signal is shown beneath the lower panel. These data are representative of two experiments using standard lysis buffer (see “Experimental Procedures”) and one experiment using 1% Triton X in place of Brij97 and Nonidet P-40 with similar results. Nonspecific bands are indicated with an asterisk.
Previously we found that pertussis toxin treatment led to recovery of CD69 expression in S1P1 dominated cells, while not allowing recovery of S1P1 expression in CD69-dominated cells (4), suggesting different requirements for down-modulation of the complex depending on which partner is more abundantly expressed. By mutating the ERY motif of S1P1 (to EAY or ENY), we found that an intact G-protein-coupling motif (25) is needed for down-modulation of the complex, irrespective of whether CD69 or S1P1 is dominant (Fig. 9A). That is, rather than observing evidence for mutually exclusive surface expression of Flag-S1P1 and CD69 (as seen with wild-type S1P1 in Fig. 9A) the molecules were coexpressed on the cell surface (Fig. 9A, EAY and ENY). As expected, cells expressing NKRp1A as a control showed no evidence of Flag-S1P1 modulation (Fig. 9A, NKRp1A-stained plots). Despite the effect of the EAY and ENY mutations on modulation, these mutations did not disrupt the ability of CD69 and S1P1 to interact in the co-immunoprecipitation assay (Fig. 9B). Disruption of a desensitization motif in the C terminus of S1P1 by mutating 5 serines to alanines (S5A) (22) prevented S1P1 from mediating efficient down-modulation of CD69 without blocking CD69 down-modulation of S1P1 (Fig. 9C). That is, the cells expressing high amounts of mutant Flag-S1P1 were mostly co-expressing surface CD69 (Fig. 9C, S5A) in contrast to the suppression of CD69 expression on the cells expressing high amounts of wild-type Flag-S1P1 (Fig. 9C). This suggests a requirement for this serine-rich motif in S1P1-mediated modulation of CD69, but not in CD69-mediated modulation of S1P1. Deletion of the last 12 amino acids from the S1P1 C terminus, a region that contains further residues involved in ligand-mediated desensitization (23) did not affect down-modulation of the CD69-S1P1 complex (Fig. 9C, Δ12). Notably, mutation of S1P1 residue Arg-120 to Ala, a mutation that disrupts S1P binding (24), prevented efficient down-modulation of the CD69-S1P1 complex (Fig. 9C, R120A). This suggests that acquisition of a ligand-bound conformation might be necessary for efficient modulation of the CD69-S1P1 complex, both in cells dominated by CD69 and in cells dominated by S1P1.
FIGURE 9.
Interaction between CD69 and S1P1-containing mutations in the G-protein-interacting ERY motif or the cytoplasmic tail. A, flow cytometric analysis of S1P1 ERY motif mutants, EAY and ENY, or S1P3 as a control, co-transduced with CD69 and hNKRp1A in WEHI-231 cells. Cells are costained for the CD69 ectodomain and the Flag-S1P receptors and mutants as indicated. B, co-IP experiment for the S1P1 mutants or S1P3 as indicated. These data are representative of two experiments with similar results. C, flow cytometric analysis of desensitization mutants (S5A, Δ12) and S1P non-binding mutant (R120A), co-transduced with CD69 and hNKRp1A in WEHI-231 cells. Cells are co-stained for the CD69 ectodomain and the Flag-S1P receptors and mutants as indicated. Nonspecific bands are indicated with an asterisk.
CD69 Enhances S1P1 Binding of S1P
To determine whether CD69 altered the ligand binding properties of S1P1, we used a radiolabeled ligand binding assay to test the half-life of S1P binding to S1P1 or the S1P1-CD69 complex in transduced WEHI-231 cells. Compared with WEHI-231 cells expressing S1P1 alone, the cells expressing both S1P1 and CD69 had about 10-fold lower amounts of surface S1P1 as detected by flow cytometry (supplemental Fig. S1). Unexpectedly, the co-transduced WEHI-231 cells bound 2-fold greater amounts of [33P]S1P than cells transduced with S1P1 alone, and the decay of cell-associated labeled-S1P when competed with an excess of unlabeled S1P was ∼10-fold slower (Fig. 10). Because the increased binding cannot be explained by an increased number of surface S1P1 receptors, these findings fit best with the interpretation that CD69 stabilizes a high affinity ligand binding conformation of S1P1.
FIGURE 10.

Dissociation data to determine the half-life for S1P binding to the S1P1-CD69 complex. WEHI-231 cells (2 × 105) transduced with S1P1 or co-transduced with CD69 and S1P1 were treated with 200 pm [33P]S1P on ice for 30′. Cold S1P (2 mm) was added, and the amount of labeled S1P that was cell associated following the incubation times shown was measured. S1P1 half-life was measured to be 1.33 ± 0.6 min (n = 2) and S1P1-CD69 half-life was measured to be 13 ± 5 min (n = 4).
S1P1 Non-binding CD69 Mutant Fails to Inhibit Lymphocyte Egress
Previous work established that CD69 is necessary for inhibition of lymphocyte egress following IFNα/β exposure and correlated this requirement with the ability of CD69 to inhibit S1P1 function (4). To further test whether CD69 inhibition of egress depends on its ability to interact with and inhibit S1P1, we tested the activity of wild type and an S1P1 non-binding mutant of CD69 (6N6-Δ31, Fig. 2B) at inhibiting T cell migration to S1P and egress from lymph nodes (Fig. 11). Consistent with the inability to interact with S1P1, 6N6-Δ31 had no inhibitory activity on S1P1-mediated cell migration to S1P (Fig. 11A). The reciprocal construct, N6N-Δ31 that has some ability to interact with S1P1 (Fig. 2B) showed partial inhibition of migration (Fig. 11A). Interestingly, construct 6N6-stalk that contains the CD69 ectodomain and scores positive in the interaction assay (Fig. 2B), failed to inhibit migration (Fig. 11A). Wild-type CD69 strongly inhibited migration to S1P as expected, while not affecting the response to the chemokine SDF1α (CXCL12) (Fig. 11A). Together, these findings highlight the need for the CD69 transmembrane and HEGSI motif for the migration inhibitory effect but also suggest that additional interaction surfaces cooperate to achieve the full migration block.
FIGURE 11.
Requirement for CD69 interaction with S1P1 to inhibit lymphocyte chemotaxis and egress from lymph nodes. A, transwell migration assay testing CD69 and relevant CD69/NKRp1A chimeric molecules for their ability to inhibit S1P1-dependent S1P migration. Migration to SDF-1α, a CXCR4 ligand, is shown as a negative control. B, schematic of cell transfer experiment. Donor cells were activated for 24 h with anti-CD3/CD28 antibodies before transduction of the indicated recombinant retroviruses. These three CMTMR and congenically marked cell populations were then mixed and injected into recipient mice. Half of these mice were analyzed 24 h post-transfer and half where treated at 24 h with α4 and αL integrin-neutralizing antibodies to block LN entry and LN cells were analyzed 18 h later. C, flow cytometric analysis showing transferred CD4 cells, distinguishing 6N6-Δ31-transduced cells (CMTMR−) from CD69-transduced cells (CMTMR+) in the same LN preparation. Transduced cells with high GFP reporter expression were gated and the percent of these cells among total LN cells is shown. D, ratio of transduced cells remaining in LNs following 18 h of α4 plus αL antibody treatment compared with the starting number of cells. This result is representative of two independent experiments.
Activated T cells were transduced with retroviruses encoding wild type or 6N6-Δ31 CD69, or NKRp1A as a negative control, and then transferred to recipient mice (Fig. 11B). After allowing 24 h for cells to enter LNs, further entry was blocked by injection of α4 and αL integrin-neutralizing antibodies (15, 26) and the amount of decay in cell numbers was measured 18 h later (Fig. 11B). Cells transduced with CD69 increased their representation in the LN over 2-fold during the α4 and αL blockage time period, whereas the cells transduced with the interaction negative construct, 6N6-Δ31, showed a decline in their relative LN representation, despite both transduced populations showing similar CD69 surface staining (Fig. 11C). When these cells were quantitated and compared between the two time points of α4 and αL blockade, the wild-type CD69 almost fully blocked any decay in cell number over the 18 h, while cells expressing the S1P1 non-binding CD69 mutant decayed in number identically to cells transduced with the NKRp1A control construct or to non-transduced recipient T cells (Fig. 11D). These findings indicate that the egress inhibiting function of CD69 depends on its ability to interact with S1P1.
DISCUSSION
The above findings identify an integral membrane interaction between the type II transmembrane protein CD69 and the G-protein-coupled receptor S1P1. This interaction involves the transmembrane and membrane proximal regions of CD69 and helix 4 of S1P1. The C-type lectin domain is insufficient to promote interaction but augments interactions mediated by the transmembrane and membrane proximal domains. CD69 promotes a high affinity ligand-binding state in S1P1. We propose a model where the interaction of CD69 with helix 4 of S1P1 stabilizes a conformation of S1P1 that mimics some properties of the ligand-bound state, promoting internalization and subsequent degradation of the receptor. In vivo experiments establish that the egress inhibiting function of CD69 depends on its ability to couple with S1P1 via this membrane-domain interaction.
The recently determined β-adrenergic receptor structure allows for a reasonable structure model of S1P receptors to be produced (18). In this structural model of S1P1, helix 4 displays 6 out of 8 residues that differ with S1P3 on the surface, but a calculation of electrostatic surface potential yields a surface that looks similar between S1P1 and S1P3 (supplemental Fig. S4). This model along with the data presented above showing that single residue changes from the S1P1 sequence to S1P3 have little effect on interaction suggest that hydrophobic interactions would dominate the interaction surface in the membrane as opposed to hydrogen bonds.
The region of CD69 that we have identified with regards to S1P1 specificity when compared with that of S1P3 has been localized to five extracellular amino acids of CD69 that directly flank the predicted transmembrane helix. Because the extracellular regions vary extensively between members of the S1P receptor family, there is a possibility that specific interactions occur between S1P1 and CD69 in this region. Though the interaction between CD69 and S1P1 and S1P3 seemed to occur similarly for some constructs, we could not detect a function of this binding effect on S1P3 under either surface co-modulation or in S1P migration assays (data not shown).
In vivo studies demonstrated the ability of CD69 to act downstream of IFNα/β signaling and inhibit lymphocyte egress from lymph nodes (4). Our studies provide strong evidence that this inhibition depends on CD69 interaction with S1P1 via its transmembrane domain and HEGSI motif. We show that one mode of CD69-mediated inhibition of S1P1 is likely to be through down-regulation and degradation of the receptor. However, an additional mode of regulation may be through stabilization of a high affinity conformation of the receptor, possibly disrupting the ability of the receptor to sense a ligand gradient. Recent work with the stable high affinity S1P1 agonist, FTY720, has shown it can disrupt receptor function by causing constitutive receptor signaling (27). The mechanism by which CD69 promotes down-regulation and possible degradation of S1P1 is not yet clear. CD69-mediated surface down-regulation is PTX resistant (4) yet does require the S1P1 ERY signaling motif and an intact S1P ligand-binding site. This may indicate a requirement for G-protein interaction with the CD69-S1P1 complex even if Gi activation is not needed. We have so far not been able to detect coimmunoprecipitation of Gαi2 or Gαi3 with the CD69-S1P1 complex. Future studies with more sensitive reagents and Gαi-deficient control cells will be needed to address this issue.
Previous PTX experiments suggested that the requirements for CD69-S1P1 complex modulation differ depending on which molecular partner is in excess (4), and this is reinforced here by the finding that the S5A mutant of S1P1 is partially defective in down-modulation when S1P1 is in excess but not when CD69 is the more abundant receptor. These five serine residues have previously been implicated in ligand-mediated modulation of S1P1 and may be targets of G-protein-coupled receptor kinases (23). Thus, in S1P1 dominant conditions, surface down-modulation may occur by a similar pathway to ligand-mediated modulation.
The present experiments have been performed with transduced, epitope-tagged forms of S1P1 and CD69 because we have found that available reagents against endogenous mouse S1P1 and CD69 have limited sensitivity for Western blot analysis. In the studies performed so far, there is close agreement between conclusions from overexpression experiments and findings with cells from S1P1- or CD69-deficient mice (4), suggesting that our biochemical findings will extend to the requirements for endogenous interactions of S1P1 and CD69. To more fully characterize the endogenous complex, it will be important in future work to generate antibodies for mouse S1P1 and CD69 that permit sensitive detection of the molecules in immunoprecipitation studies.
The finding that CD69 could inhibit lymphocyte egress raised the possibility that a soluble form of CD69 might be a novel small molecule regulator of S1P1. Our studies suggest that the CD69 ectodomain is not essential for the CD69-S1P1 interaction and when expressed in cells in association with a non-interacting transmembrane domain, is unable to regulate S1P1, arguing against utility of this domain as an egress regulator. However, our experiments raise the possibility that a membrane-inserting peptide modeled on the CD69 transmembrane and membrane-flanking domains might have inhibitory effects on S1P1.
Despite GPCRs representing the largest class of membrane proteins (∼950 genes), there are few well-defined examples of GPCR regulation by associating transmembrane proteins. Some odorant receptors require receptor transporting proteins (RTP-1 or -2) as chaperones during protein folding and transport through the ER (28). Receptor activity modifying proteins (RAMPs) are three related single transmembrane proteins that influence both trafficking and ligand binding of the calcitonin-like receptor (29, 30). Melanocortin 2 receptor accessory protein (MRAP) is essential for surface expression of the MC2R and thus for receptor function. It is notable that, like CD69, the RTPs and MRAP are thought to be type II transmembrane proteins (31, 32). However, each of these GPCR accessory proteins exhibits a positive influence on receptor function. CD69 may represent the first physiological example of a transmembrane accessory protein acting as a negative regulator of GPCR function. Further characterization of how the integral membrane interaction we define here mediates inhibition of GPCR function may lead to a more general understanding of how GPCRs can be regulated by interacting molecules.
Supplementary Material
Acknowledgments
We thank Ying Xu and Jinping An for help with molecular cloning and Jesse Green and Marcus Zachariah for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI074847.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and data.
- S1P1
- sphingosine 1-phosphate receptor-1
- S1P3
- sphingosine 1-phosphate receptor-3
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- GPCR
- G-protein-coupled receptor
- IP
- immunoprecipitate
- LN
- lymph node.
REFERENCES
- 1.Matloubian M., Lo C. G., Cinamon G., Lesneski M. J., Xu Y., Brinkmann V., Allende M. L., Proia R. L., Cyster J. G. (2004) Nature 427, 355–360 [DOI] [PubMed] [Google Scholar]
- 2.Hall J. G., Morris B. (1965) Br. J. Exp. Pathol. 46, 450–454 [PMC free article] [PubMed] [Google Scholar]
- 3.Sprent J., Miller J. F., Mitchell G. F. (1971) Cell Immunol. 2, 171–181 [DOI] [PubMed] [Google Scholar]
- 4.Shiow L. R., Rosen D. B., Brdicková N., Xu Y., An J., Lanier L. L., Cyster J. G., Matloubian M. (2006) Nature 440, 540–544 [DOI] [PubMed] [Google Scholar]
- 5.Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G. J., Card D., Keohane C., Rosenbach M., Hale J., Lynch C. L., Rupprecht K., Parsons W., Rosen H. (2002) Science 296, 346–349 [DOI] [PubMed] [Google Scholar]
- 6.Kappos L., Radue E. W., O'Connor P., Polman C., Hohlfeld R., Calabresi P., Selmaj K., Agoropoulou C., Leyk M., Zhang-Auberson L., Burtin P. (2010) N. Engl. J. Med. 362, 387–401 [DOI] [PubMed] [Google Scholar]
- 7.Hamann J., Fiebig H., Strauss M. (1993) J. Immunol. 150, 4920–4927 [PubMed] [Google Scholar]
- 8.Ziegler S. F., Ramsdell F., Hjerrild K. A., Armitage R. J., Grabstein K. H., Hennen K. B., Farrah T., Fanslow W. C., Shevach E. M., Alderson M. R. (1993) Eur. J. Immunol. 23, 1643–1648 [DOI] [PubMed] [Google Scholar]
- 9.Ziegler S. F., Levin S. D., Johnson L., Copeland N. G., Gilbert D. J., Jenkins N. A., Baker E., Sutherland G. R., Feldhaus A. L., Ramsdell F. (1994) J. Immunol. 152, 1228–1236 [PubMed] [Google Scholar]
- 10.Natarajan K., Sawicki M. W., Margulies D. H., Mariuzza R. A. (2000) Biochemistry 39, 14779–14786 [DOI] [PubMed] [Google Scholar]
- 11.Llera A. S., Viedma F., Sánchez-Madrid F., Tormo J. (2001) J. Biol. Chem. 276, 7312–7319 [DOI] [PubMed] [Google Scholar]
- 12.Feng C., Woodside K. J., Vance B. A., El-Khoury D., Canelles M., Lee J., Gress R., Fowlkes B. J., Shores E. W., Love P. E. (2002) Int. Immunol. 14, 535–544 [DOI] [PubMed] [Google Scholar]
- 13.Nakayama T., Kasprowicz D. J., Yamashita M., Schubert L. A., Gillard G., Kimura M., Didierlaurent A., Koseki H., Ziegler S. F. (2002) J. Immunol. 168, 87–94 [DOI] [PubMed] [Google Scholar]
- 14.Chu P., Pardo J., Zhao H., Li C. C., Pali E., Shen M. M., Qu K., Yu S. X., Huang B. C., Yu P., Masuda E. S., Molineaux S. M., Kolbinger F., Aversa G., de Vries J., Payan D. G., Liao X. C. (2003) J. Biol. 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo C. G., Xu Y., Proia R. L., Cyster J. G. (2005) J. Exp. Med. 201, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swift S., Lorens J., Achacoso P., Nolan G. P. (1999) Curr. Prot. Imm 10.17.14–10.17.29 [DOI] [PubMed] [Google Scholar]
- 17.Eswar N., Marti-Renom M. A., Webb B., Madhusudhan M. S., Eramian D., Shen M., Pieper U., Sali A. (2006) Curr. Prot. Bioinformatics 5.6.1–5.6.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLano W. (2006) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 20.Kohno T., Wada A., Igarashi Y. (2002) FASEB J. 16, 983–992 [DOI] [PubMed] [Google Scholar]
- 21.Fieger C. B., Huang M. C., Van Brocklyn J. R., Goetzl E. J. (2005) FASEB J. 19, 1926–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oo M. L., Thangada S., Wu M. T., Liu C. H., Macdonald T. L., Lynch K. R., Lin C. Y., Hla T. (2007) J. Biol. Chem. 282, 9082–9089 [DOI] [PubMed] [Google Scholar]
- 23.Watterson K. R., Johnston E., Chalmers C., Pronin A., Cook S. J., Benovic J. L., Palmer T. M. (2002) J. Biol. Chem. 277, 5767–5777 [DOI] [PubMed] [Google Scholar]
- 24.Parrill A. L., Wang D., Bautista D. L., Van Brocklyn J. R., Lorincz Z., Fischer D. J., Baker D. L., Liliom K., Spiegel S., Tigyi G. (2000) J. Biol. Chem. 275, 39379–39384 [DOI] [PubMed] [Google Scholar]
- 25.Rovati G. E., Capra V., Neubig R. R. (2007) Mol. Pharmacol. 71, 959–964 [DOI] [PubMed] [Google Scholar]
- 26.Pham T. H., Baluk P., Xu Y., Grigorova I., Bankovich A. J., Pappu R., Coughlin S. R., McDonald D. M., Schwab S. R., Cyster J. G. (2010) J. Exp. Med. 207, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullershausen F., Zecri F., Cetin C., Billich A., Guerini D., Seuwen K. (2009) Nat. Chem. Biol. 5, 428–434 [DOI] [PubMed] [Google Scholar]
- 28.Cooray S. N., Chan L., Webb T. R., Metherell L., Clark A. J. (2009) Mol. Cell Endocrinol 300, 17–24 [DOI] [PubMed] [Google Scholar]
- 29.Christopoulos G., Perry K. J., Morfis M., Tilakaratne N., Gao Y., Fraser N. J., Main M. J., Foord S. M., Sexton P. M. (1999) Mol. Pharmacol. 56, 235–242 [DOI] [PubMed] [Google Scholar]
- 30.Armour S. L., Foord S., Kenakin T., Chen W. J. (1999) J. Pharmacol. Toxicol. Methods 42, 217–224 [DOI] [PubMed] [Google Scholar]
- 31.Metherell L. A., Chapple J. P., Cooray S., David A., Becker C., Rüschendorf F., Naville D., Begeot M., Khoo B., Nürnberg P., Huebner A., Cheetham M. E., Clark A. J. (2005) Nat. Genet. 37, 166–170 [DOI] [PubMed] [Google Scholar]
- 32.Sebag J. A., Hinkle P. M. (2009) J. Biol. Chem. 284, 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.