Abstract
The p62/SQSTM1 (sequestosome 1) protein, which acts as a cargo receptor for autophagic degradation of ubiquitinated targets, is up-regulated by various stressors. Induction of the p62 gene by oxidative stress is mediated by NF-E2-related factor 2 (NRF2) and, at the same time, p62 protein contributes to the activation of NRF2, but hitherto the mechanisms involved were not known. Herein, we have mapped an antioxidant response element (ARE) in the p62 promoter that is responsible for its induction by oxidative stress via NRF2. Chromatin immunoprecipitation and gel mobility-shift assays verified that NRF2 binds to this cis-element in vivo and in vitro. Also, p62 docks directly onto the Kelch-repeat domain of Kelch-like ECH-associated protein 1 (KEAP1), via a motif designated the KEAP1 interacting region (KIR), thereby blocking binding between KEAP1 and NRF2 that leads to ubiquitylation and degradation of the transcription factor. The KIR motif in p62 is located immediately C-terminal to the LC3-interacting region (LIR) and resembles the ETGE motif utilized by NRF2 for its interaction with KEAP1. KIR is required for p62 to stabilize NRF2, and inhibition of KEAP1 by p62 occurs from a cytoplasmic location within the cell. The LIR and KIR motifs cannot be engaged simultaneously by LC3 and KEAP1, but because p62 is polymeric the interaction between KEAP1 and p62 leads to accumulation of KEAP1 in p62 bodies, which is followed by autophagic degradation of KEAP1. Our data explain how p62 contributes to activation of NRF2 target genes in response to oxidative stress through creating a positive feedback loop.
Keywords: Autophagy, Gene Regulation, Oxidative Stress, Protein Degradation, Transcription Regulation, ARE, KEAP1, NRF2, p62
Introduction
Oxidative stress represents an overproduction of reactive oxygen species in a cell relative to its ability to detoxify such species, and this can lead to cellular damage (1). The Kelch-like ECH-associated protein 1 (KEAP1)3-NF-E2-related factor 2 (NRF2) signaling pathway can be activated by pro-oxidants and electrophiles as an adaptive response to agents that cause oxidative stress. The target genes that transcription factor NRF2 up-regulates all contain a DNA regulatory sequence called the antioxidant response element (ARE) in their promoters (2–5). Such genes include those encoding phase-II drug-metabolizing enzymes, antioxidant proteins, and anti-inflammatory proteins. In addition, NRF2 regulates turnover of oxidized protein via the proteasome (6).
Under non-stressed conditions, NRF2 protein is rapidly degraded within the cell by the 26 S proteasome following its redox-sensitive ubiquitylation (7). The short half-life of NRF2 is principally controlled by KEAP1, which is a substrate adaptor for the Cul3 ubiquitin ligase complex (8, 9). KEAP1 uses its C-terminal β-propeller Kelch-repeat domain to interact directly with the Neh2 domain of NRF2, and its BTB domain to recruit Cul3 to the complex (2). The binding between NRF2 and KEAP1 is believed to occur primarily in the cytoplasm (10), although several groups have also proposed that the two proteins interact in the nucleus (11–14).
Upon exposure to pro-oxidants or electrophiles, such as sulforaphane, KEAP1 is modified through oxidation or adduction of one or more of its cysteine residues, resulting in a putative conformational change that affects its ability to serve as an ubiquitin ligase substrate adaptor, thereby increasing the abundance of the transcription factor NRF2. Among the most functionally important cysteine residues in mammalian KEAP1 are Cys-151, Cys-273, and Cys-288 (15). Whether the effect of Cys modification results in a complete dissociation of NRF2 from KEAP1 is somewhat controversial (2). Within its N-terminal Neh2 domain, NRF2 possesses two binding sites for KEAP1 (DLG and ETGE motifs) and is, therefore, able to interact with both subunits of a KEAP1 dimer (16, 17). However, the interaction mediated by the DLG motif is weaker than that mediated by the ETGE motif, and it has been suggested that only the former interaction is lost as a consequence of modification of KEAP1 by inducing agents (2). The net effect is that NRF2 is no longer degraded, and the protein accumulates in the nucleus where it induces transcription of target genes.
The p62/SQSTM1 (sequestosome 1) protein, also known as A170 (mouse) and ZIP (rat), was originally identified because it bound to the tyrosine kinase Lck (18). Subsequently, p62 was found to bind atypical protein kinase C (19, 20) and shown to act as a scaffold or adaptor protein in NFκB signaling pathways following activation of tumor necrosis factor-α (21), interleukin-1 (22), and nerve growth factor (23) receptors. Also, p62 is involved in activation of caspase-8 upon stimulation of cell death receptors (24). p62 is required for Ras-induced tumorigenesis in vitro and in vivo and is up-regulated in different human tumors (25). Increased p62 protein levels correlate with aggressive progression of breast and prostate cancers (26–28).
During the evolutionary conserved process of autophagy (here understood as macroautophagy), isolation membranes embrace and envelop part of the cytoplasm, sequestering the content into a double-membrane vesicle called an autophagosome. In turn, autophagosomes fuse with lysosomes, and their contents are degraded by lysosomal hydrolases (29). Because p62 binds to ubiquitin and to LC3, it is both a selective autophagy substrate and a cargo receptor for autophagic degradation of ubiquitinated targets (30–34). p62 forms cytosolic inclusion bodies distinct from aggresomes, which contain ubiquitinated protein aggregates that subsequently can be degraded by autophagy (30, 34). Using conditional Atg7 knock-out mice, it has been reported that, when autophagy is abolished in the liver, p62 accumulates in aggregates, phase II drug-metabolizing enzymes and antioxidant proteins are strongly induced, and the liver becomes grossly enlarged and suffers loss of function. Hepatic dysfunction in such mice is relieved when p62 is also knocked out (32). If p62 is not turned over by autophagy, pathogenic conditions arise that are characterized by the accumulation of p62 in ubiquitin-containing inclusions. A recent study showed that the intracellular increase in p62 protein caused by inhibition of autophagy is highly tumorigenic in apoptosis-deficient cells (35). Evidence suggests p62 is a stress response protein that is strongly induced at the mRNA and protein levels by exposure to oxidants, sodium arsenite, cadmium, ionophores, proteasomal inhibitors, or overexpression of polyQ-expanded proteins (36, 37). p62 is a member of the protein battery induced by Nrf2 in response to oxidative stress, and induction of p62 is severely inhibited in cells from Nrf2 knock-out mice (38). Recent studies have suggested that p62 may contribute to the induction of NRF2, but the mechanism has not been elucidated (39).
In the present report, we have mapped an ARE in the promoter/enhancer region of the p62 gene that is responsible for its induction in response to oxidative stress. ChIP analyses verified that endogenous NRF2 is bound to this region of the p62 promoter/enhancer in vivo. Recombinant NRF2 and MAFG bind, presumably as heterodimers, to the p62 ARE in vitro. We show p62 interacts directly with KEAP1 via a DPSTGE motif that resembles the DEETGE sequence (i.e. the high affinity ETGE motif) employed by NRF2 to bind KEAP1. A model in which p62 competes with NRF2 for interaction with KEAP1 is envisaged. Hence, p62 is able to set up a positive feedback loop to activate NRF2, which in turn stimulates increased transcription of the p62 gene. In this manner, p62 protein contributes to a sustained activation of NRF2 in response to oxidative and electrophile stress.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were used: rabbit anti-Nrf2 antibody (Santa Cruz Biotechnology, SC-13032), rabbit anti-mouse Keap1 antibody (16), monoclonal anti-p62 antibody (BD Transduction Laboratories), anti-acetylated histone H3 antibody (Upstate), anti-actin antibody (Sigma, A 2066), anti-FLAG antibody (Stratagene, 200471), DsRED monoclonal antibody (Clontech), anti-Myc antibody (Santa Cruz Biotechnology, 9E10), and horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies (BD Pharmingen). Bafilomycin A1 (B 1793), sulforaphane (S 4441), and N-acetyl cysteine (A 9165) were purchased from Sigma. l-[35S]Methionine was obtained from PerkinElmer.
Plasmid Constructs
Plasmids used in this study are listed in Tables 1 and 2. They were made by conventional restriction enzyme-based cloning or by use of the Gateway recombination system (Invitrogen). Point mutants were made using the QuikChange site-directed mutagenesis kit (Stratagene). Gateway LR reactions were performed as described in the Gateway cloning technology instruction manual (Invitrogen). Oligonucleotides for mutagenesis, PCR, and DNA sequencing reactions were obtained from Invitrogen. All plasmid constructs were verified by restriction digestion and/or DNA sequencing (BigDye, Applied Biosystems). Details of their construction are available upon request.
TABLE 1.
Plasmids used in this study
| Plasmids | Description | Source |
|---|---|---|
| Gateway cloning vectors | ||
| pENTR1A, -2B and -3C | Gateway entry vectors | Invitrogen |
| pDONR221 | Gateway donor vector | Invitrogen |
| pENTR-EGFP | EGFP without stop codon in Gateway entry vector | (34) |
| pDest15 | Bacterial GST fusion expression vector; T7 promoter | Invitrogen |
| pDEST-TH1 | Bacterial MBP fusion expression vector; tac promoter | (60) |
| pDestEGFP-C1 | Mammalian EGFP fusion expression vector; CMV promoter | (40) |
| pDest-mCherry-C1 | Mammalian mCherry fusion expression vector, backbone as pDestEGFP-C1 | (34) |
| pDest-3xFLAG | Mammalian triple flag tag fusion expression vector; CMV promoter, backbone as p3xFLAG-CMV (Sigma, E4026) | This study |
| pDest-myc | Mammalian myc tag fusion expression vector; backbone as pcDNA3.1 | (40) |
| Other vectors | ||
| pGEX-2T | Bacterial GST fusion expression vector; tac promoter | Amersham |
| pCMV-βgal | Mammalian expression vector for bacterial β-galactosidase; CMV promoter | Open Biosystems |
| pcDNA3.1 | Mammalian expression vector; CMV and T7 promoters | Invitrogen |
| pmCherry-C1 | Mammalian mCherry fusion expression vector, backbone as pEGFP-C1 | (34) |
| Promoter constructs used in reporter gene assays | ||
| pGL3-Basic | Mammalian Luciferase reporter plasmid without promoter or enhancer | Promega |
| Nqo1-ARE-Luc | Murine NAD(P)H:quinine oxidoreductase 1 (Nqo1) promoter (−1016/) with functional ARE subcloned into pGL3-Basic | (61) |
| Nqo1-ΔARE-Luc | Same plasmid as above, but with ARE deleted | (61) |
| pGL3-Pp62(−1781/+46) | Human p62 promoter (−1781/+46) subcloned into pGL3-Basic | (28) |
| pGL3-Pp62(−1475/+46) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−1158/+46) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−355/+46) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−107/+46) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−1781/+46) Δ(−1464 to −1273) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−1781/+46) Δ(−1315 to −1273) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−1781/+46) Δ(−1464 to −1426) | Made by deletion of pGL3-Pp62(−1781/+46) | This study |
| pGL3-Pp62(−1781/+46)-3xMut | Made by site-directed mutagenesis of pGL3-Pp62(−1781/+46); the region from −1302 to −1298 mutated from TGAGT to AGGGA | This study |
| cDNA constructs made by traditional subcloning or site-directed mutagenesis | ||
| pcDNA3-V5-mNrf2 | Mammalian expression vector of murine Nrf2 N-terminally tagged with V5; backbone as pcDNA3.1 | (7) |
| pDONR221-NRF2 | Human NRF2 (from ImaGenes clone IRAUp969G0565D) inserted by gateway BP reaction into pDONR221 | This study |
| pDONR221-MAFG | Human MAFG in Gateway donor vector (HsCD00042726) | DF/HCC DNA Resource Core, Harvard |
| pENTR-KEAP1 | Human KEAP1 (from image clone 3955118) subcloned into pENTR3C | This study |
| pENTR-KEAP1(1–307) | Made by deletion of pENTR-KEAP1 | This study |
| pENTR-KEAP1(308–624) | Made by deletion of pENTR-KEAP1 | This study |
| pENTR-KEAP1 R380A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 N382A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 R415A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 H436A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 R483A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 Y525A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-KEAP1 Y572A | Made by site-directed mutagenesis of pENTR-KEAP1 | This study |
| pENTR-EGFP-KEAP1 | Made by subcloning of KEAP1 into pENTR-EGFP | This study |
| pENTR-p62 | Human p62 in entry vector | (40) |
| pENTR-p62 Δ371–385 | Made by deletion of pENTR-p62 | This study |
| pENTR-p62 D347A | Made by site-directed mutagenesis of pENTR-p62 | This study |
| pENTR-p62 T350A | Made by site-directed mutagenesis of pENTR-p62 | This study |
| pENTR-p62 G351A | Made by site-directed mutagenesis of pENTR-p62 | This study |
| pENTR-p62 E352A | Made by site-directed mutagenesis of pENTR-p62 | This study |
| pENTR-p62 R21A/D69A | p62 R21A/D69A (monomeric) in entry vector | (62) |
| pENTR-p62(321–342) | p62(321–342) in entry vector | (34) |
| pENTR-p62(321–370) | p62(321–370) in entry vector | (34) |
| pENTR-p62(321–358) | Made by deletion of pENTR-p62(321–370) | This study |
| pENTR-p62(339–358) | Made by deletion of pENTR-p62(321–370) | This study |
| pENTR-p62(339–370) | Made by deletion of pENTR-p62(321–370) | This study |
| pENTR-p62 MutNLS | p62 lacking both nuclear location signals made by site-directed mutagenesis of pENTR-p62. Mutations: R186A/K187A/K264A/R265A | This study |
| pENTR-p62 G351A/MutNLS | Made by site-directed mutagenesis of pENTR-p62 MutNLS | This study |
| pENTR-p62(124–440) | p62 ΔPB1 (monomeric) in entry vector | (30) |
| pENTR-p62 W338A/L341A | p62 LIR mutant made by site-directed mutagenesis of pENTR-p62 | This study |
| pENTR-p62 Δ123–319 | Made by deletion of pENTR-p62 | This study |
| cDNA constructs made by Gateway LR reactions (previously published) | ||
| pDest-TH1-p62 | Human p62 in pDest-TH1 | (34) |
| pDest-TH1-p62 Δ123–169 | p62 Δ123–169 in pDest-TH1 | (63) |
| pDest-TH1-p62 Δ170–256 | p62 Δ170–256 in pDest-TH1 | (63) |
| pDest-TH1-p62 Δ256–319 | p62 Δ256–319 in pDest-TH1 | (63) |
| pDest-TH1-p62 Δ256–370 | p62 Δ256–370 in pDest-TH1 | (63) |
| pDest-TH1-p62 Δ386–440 | p62 ΔUBA in pDest-TH1 | (63) |
| pDest15-LC3B | Human LC3B in pDest15 | (34) |
| pDest-myc-p62 | Human p62 in pDest-myc | (40) |
| pDest-myc-p62 R21A/D69A | p62 R21A/D69A in pDest-myc | (62) |
| pDest-myc-p62(1–385) | p62 ΔUBA in pDest-myc | (30) |
| pDest-mCherry-p62 | Human p62 in pDest-mCherry-C1 | (34) |
| pDest-mCherry-EGFP | mCherry-EGFP double tag expression vector | (34) |
| pDest-mCherry-EGFP-LC3B | Human LC3B fused to mCherry-EGFP double tag | (34) |
TABLE 2.
cDNA constructs made by Gateway LR reactions in this study
| Destination plasmids | Destination plasmids | Destination plasmids |
|---|---|---|
| pDest-TH1-p62 Δ371–385 | pDest-TH1-p62 D347A | pDest-TH1-p62 T350A |
| pDest-TH1-p62 G351A | pDest-TH1-p62 E352A | pDest-TH1-p62 R21A/D69A |
| pDest15-MAFG | pDest15-NRF2 | pDest15-p62(321–342) |
| pDest15-p62(321–370) | pDest15-p62(321–358) | pDest15-p62(339–358) |
| pDest15-p62(339–370) | pDest-myc-KEAP1 | pDest-myc-KEAP1(1–307) |
| pDest-myc-KEAP1(308–624) | pDest-myc-KEAP1 R380A | pDest-myc-KEAP1 N382A |
| pDest-myc-KEAP1 R415A | pDest-myc-KEAP1 H436A | pDest-myc-KEAP1 R483A |
| pDest-myc-KEAP1 Y525A | pDest-myc-KEAP1 Y572A | pDestmyc-p62 G351A |
| pDest-myc-p62 MutNLS | pDest-myc-p62 G351A/MutNLS | pDest-myc-p62 W338A/L341A |
| pDest-myc-p62(124–440) | pDest-myc-p62 Δ123–319 | pDest-3xFLAG-p62 |
| pDestEGFP-KEAP1 | pDestEGFP-KEAP1 Y572A | pDest-mCherry-KEAP1 |
| pDest-mCherry-KEAP1 Y572A | pDest-mCherry-EGFP-KEAP1 | pDest-mCherry-p62 G351A |
Cell Culture and Transfections
HeLa cells were grown in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (Biochrom AG, S0615), non-essential amino acids, 2 mm l-glutamine, and 1% streptomycin-penicillin (Sigma, P4333). The p62−/− MEFs and HEK293 cells were grown in Dulbecco's modified Eagle's medium (Sigma, D6046) supplemented with 10% fetal bovine serum and antibiotics described above. FlpIn T-Rex 293 cells (Invitrogen, R780-07) with GFP-p62 or -NBR1 integrated at the FRT site4 were grown in the same medium to which had been added 100 μg/ml hygromycin B (Calbiochem, 400051) and 7.5 μg/ml blasticidin (Invitrogen, R210-01). Subconfluent cells were transfected with plasmids using Metafectene Pro (Biontex) or TransIT-LT1 (Mirus, MIR2300) following the supplier's instructions and analyzed 24 or 48 h after transfection. Transfection with siRNAs was carried out with RNAiMAX (Invitrogen, 13778-075), as recommended by the supplier. The siRNAs used were p62 siRNAs (Dharmacon, d-010230-02), NRF2 (NFE2L2) siRNAs (Dharmacon, M-003755-02), and KEAP1 siRNAs (Dharmacon, M-012453-00). Non-targeting Silencer siRNA (Ambion, AM4635) was used as a negative control.
Reporter Gene Assays
HEK293 and p62−/− MEF cells were seeded at a density of 1.5 × 104 cells/well in 24-well plates and transfected either 48 or 24 h later, respectively, using Metafectene Pro. Transfections were performed with 100 ng of the various NRF2 and p62 expression plasmids together with 60 ng of the luciferase reporter plasmids. The β-galactosidase expressing pCMV-βgal vector (5 ng) (Stratagene) was included to correct for variations in transfection efficiency. Cells were harvested 24 h post transfection, and luciferase activities measured using the Dual Light luciferase and β-galactosidase kit (TROPIX) on a Luminoskan RT dual injection luminometer (Labsystems). Unless otherwise indicated, all reporter gene assays were carried out in three parallel experiments and repeated at least three times. The luciferase values varied 1–12% between the parallels.
Light Microscopy Analyses
Cultured cells were fixed in 4% paraformaldehyde and examined using a Zeiss Axiovert 200 microscope with a 40×, 1.2W C-Apochroma objective, equipped with an LSM510-META confocal module. Staining with antibodies was performed as described previously (40). Images were processed using Canvas version 9 (ACD systems).
Flow Cytometry
Expression of GFP-p62 or -NBR1 in stably transfected FlpIn T-Rex 293 cells was induced by adding 1 μg/ml tetracycline (Sigma, T7660) to the medium for 24 h. After removal of tetracycline, degradation of GFP-p62 or -NBR1 was measured by flow cytometry using a FACSAria cell sorter running FACSDiva software version 5.0 (BD Biosciences). GFP fluorescence was collected through a 616/23 bandpass filter in the C detector. Data were collected from a minimum of 5000 singlet events per tube, and the median GFP-A value was used for quantification. To obtain single-cell suspensions, cells were trypsinized and passed through cell strainer caps (BD Biosciences, 352235).
Immunoprecipitations and Western Blot Experiments
For immunoprecipitation experiments, cells were lysed 24 h after transfection in HA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 1 mm EGTA, 1% Triton X-100) with phosphatase inhibitor mixture set II (Calbiochem) and Complete Mini, EDTA-free protease inhibitor mixture (Roche Applied Science). For Western blot experiments with total cell extracts, cells were lysed directly in 2× SDS-PAGE loading buffer (125 mm Tris-HCl, pH 7.4, 4% SDS, 0.04% bromphenol blue, 8% sucrose, 30 mg/ml dithiothreitol) or in a boiling solution of 1% SDS and 10 mm Tris-HCl, pH 7.4. The latter buffer was used for the analysis of levels of NRF2, KEAP1, and p62 in HEK293, HeLa, HepG2, DU145 (prostate carcinoma), HT29 (colon carcinoma), HT1080 (colon actabulum fibrosarcoma), hTertBJ1 (foreskin fibroblast), Kelly (neuroblastoma), and PC3 (prostate) cells. Immunoprecipitation and Western blotting were performed as described previously (40).
GST and MBP Pulldown Experiments
GST and GST-tagged proteins were expressed in Escherichia coli BL21(DE3), and MBP and MBP-tagged proteins in E. coli DH5α. Purification of GST- and MBP-tagged proteins, as well as GST- and MBP-pulldown assays with in vitro translated 35S-labeled proteins, was done as described previously (34).
Gel Mobility-shift Assays
Gel mobility-shift assays were performed essentially as described elsewhere (41), using the following binding buffer 20 mm HEPES, pH 7.9, 220 mm KCl, 5 mm dithiothreitol, 4 mm MgCl2, 1 mm EDTA, 100 μg/ml bovine serum albumin, 24 ng/μl poly(dIC). Double-stranded oligonucleotides containing 20 nucleotides (position −1303/−1388) of the ARE wild-type p62 promoter or the ARE mut p62 promoter were end-labeled by using [32P]ATP and used as probes. For competition experiments, 1 μg of the same unlabeled double-stranded oligonucleotides was used.
ChIP Assays
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (42). Around 1.5 × 107 HeLa cells were used for each tested condition, cross-linked for 10 min at room temperature and sonicated for 20 min. PCR primers used to amplify the p62 promoter were 5′-CTCTCAGGCGCCTGGGCTGCTGAG-3′ and 5′-CGGCGGTGGAGAGTGGAAAATGCC-3′.
RESULTS
Mapping of an NRF2 Binding Site in the p62 Gene Promoter
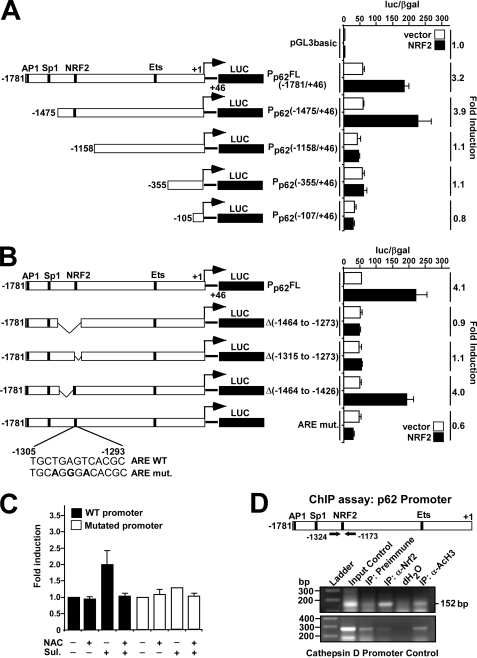
It has been reported previously that NRF2 contributes to the induction of p62 upon oxidative stress (38), and NRF2 overexpression increases p62 mRNA levels (43). To examine whether NRF2 can transactivate the p62 gene, we used a reporter construct containing −1781 to +46 base pairs of the human p62 promoter fused to Luciferase (called pGL3-Pp62(1781/+46)-Luc) (28). Previously mapped binding sites for AP1, Sp1 and Ets transcription factors in the p62 promoter/enhancer are indicated in Fig. 1A (25, 28, 44). As shown in Fig. 1A, co-transfection of HEK293 cells with an NRF2 expression construct and the pGL3-Pp62(1781/+46)-Luc plasmid resulted in >3-fold induction of reporter gene activity; HEK293 cells were chosen for these studies, because they contain little or no detectable endogenous NRF2 (see below). A series of 5′ truncations of the p62 promoter showed that the region between nucleotides −1158 and −1475, which contains several putative ARE enhancers, is required for NRF2-mediated induction. Transient transfections using various constructs containing internal deletions within this region identified a functional ARE between nucleotides −1273 and −1315 (Fig. 1B). This enhancer was further mapped by introduction of three single nucleotide point mutations, at −1298, −1300, and −1302, into the predicted ARE core. These mutations completely abolished transactivation of p62 by NRF2 suggesting that the sequence 5′-TGCTGAGTCAC-3′ between nucleotides −1305 and −1295 represents a minimal functional ARE (in reverse orientation) of the type that contains an embedded AP1 site (Fig. 1B). Because p62 is induced upon oxidative stress in an NRF2-dependent manner, we next tested the effect of sulforaphane on the wild-type and mutant p62 promoter in reporter gene assays, because the isothiocyanate can induce ARE-driven gene expression (45). Treatment with sulforaphane stimulated a 2-fold induction of luciferase driven by the wild-type p62 promoter, whereas the isothiocyanate had only a minor effect on reporter gene activity driven by the mutant p62 promoter (Fig. 1C). Addition of the antioxidant N-acetylcysteine counteracted the effect of sulforaphane, supporting the conclusion that the ARE identified at positions −1305/−1293 is responsible for NRF2-mediated induction of p62 during oxidative stress.
FIGURE 1.
Mapping of an NRF2 binding site in the p62 promoter/enhancer. A and B, reporter gene assays were performed using wild-type (−1781/+46) or the indicated deleted or mutated p62 promoter/enhancer constructs. HEK293 cells were co-transfected with an empty vector (pcDNA3.1) (100 ng) or a plasmid encoding murine Nrf2 (pcDNA3.1-V5-mNrf2) (100 ng) together with the indicated p62 promoter constructs (60 ng). Cells were harvested 24 h after transfection. The relative promoter activities are expressed as the ratio between measured luciferase and β-galactosidase activities. The data shown are the mean activities obtained in one experiment performed in triplicate, and are representative of three or more independent experiments. For each p62 promoter construct, NRF2-mediated fold-activation is shown to the right. C, HEK293 cells co-transfected with wild-type or mutated p62 promoter constructs were analyzed as in A. The cells were treated for 20 h with sulforaphane (Sul, 15 μm) or N-acetylcysteine (NAC, 5 mm) as indicated. The relative activity of the wild-type promoter was set to one. The mean -fold activation obtained in three independent experiments performed in triplicate is presented. D, ChIP assays show that NRF2 can associate with the p62 promoter. Extracts from HeLa cells (1.5 × 107 cells per antibody) were immunoprecipitated with preimmune serum, polyclonal anti-NRF2 antibody, or anti-acetylated histone H3 antibody as a positive control. Input control (1:50) was also included. PCR analyses of the immunoprecipitated chromatin were carried out using primers flanking the ARE (position −1324 and −1173, respectively) (upper panel). PCR analyses of the precipitated chromatin using primers aligning to position −3351 and −3069 of the cathepsin D promoter were used as a control.
To show that NRF2 interacts with the p62 promoter in vivo, ChIP analyses of HeLa cell extracts were performed using NRF2 antibodies and PCR primers targeting a 152-bp DNA fragment containing the ARE. In comparison with HEK293 cells, HeLa cells have higher levels of NRF2 (see below), making them better suited to this type of analysis. Upon ChIP analysis, the p62 promoter fragment co-precipitated with NRF2 antibodies (Fig. 1D, upper panel), indicating that this transcription factor is associated with the upstream regulatory region of p62 in HeLa cells. ChIP of acetylated histone H3 was used as a positive control confirming that the p62 promoter was transcriptionally active. PCR of a region 3 kbp upstream of the cathepsin D gene promoter was included as a negative control in the ChIP experiments (Fig. 1D, lower panel).
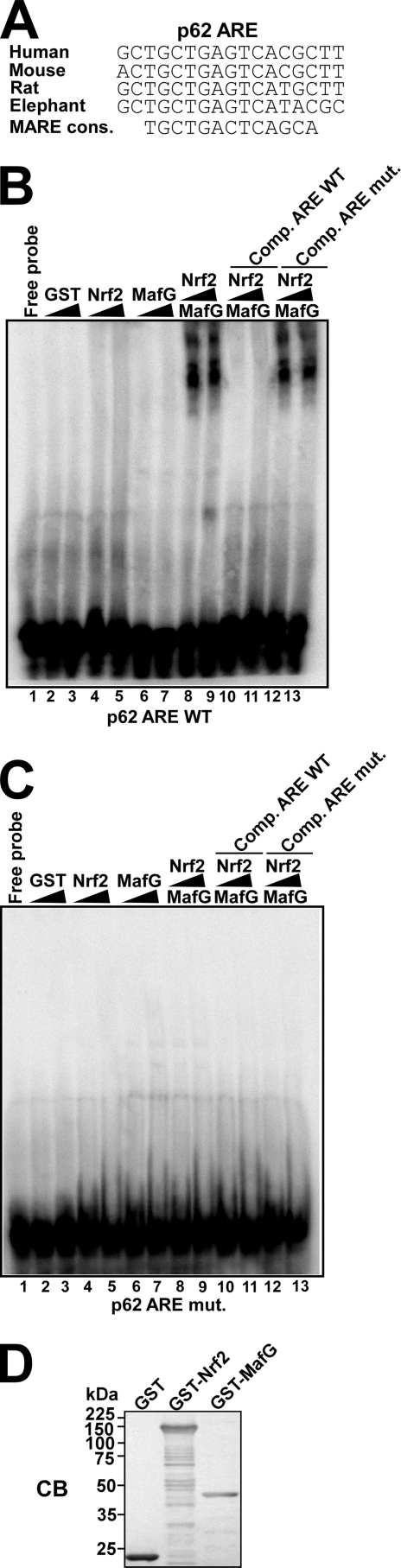
NRF2 Binds to the ARE of the p62 Promoter in Vitro
The ARE sequence in the promoter of p62 is conserved among mammalian species. Its homology with the MARE consensus sequence (43) suggested that MAF proteins may be recruited to the p62-ARE as a heterodimer with NRF2 (Fig. 2A). To test this hypothesis, we produced full-length GST-NRF2 and GST-MAFG fusion proteins in E. coli and examined their ability to interact with a DNA fragment containing the ARE in gel mobility-shift assays (GMSA) in vitro. Neither GST-NRF2 nor GST-MAFG bound the ARE-containing sequence when incubated alone, but when added together they exhibited DNA-binding activity (Fig. 2B). We conclude that NRF2 binds to the p62 promoter in vitro as a heterodimer with MAFG or another small MAF protein. The binding was efficiently inhibited by addition of an excess of unlabeled p62 promoter fragment, but not by addition of a fragment with three point mutations in the ARE (Fig. 2B). We also performed the whole experiment using the mutated p62 promoter fragment as a probe and found the mobility of this fragment was not shifted during electrophoresis (Fig. 2C). In conclusion, the GMSA experiment verifies that NRF2 can associate with the p62 ARE in vitro.
FIGURE 2.
NRF2-MAFG binds to the ARE in the p62 promoter in vitro. A, the ARE in the p62 promoter is conserved in human, mouse, rat, and elephant species, and constitutes a subclass of cis-elements that can be classified as MAREs (Maf recognition elements). B and C, gel mobility-shift assays demonstrating binding of NRF2-MAFG to the wild-type NRF2-responsive ARE element in the p62 promoter (B) but not to the mutated ARE site (C). GST, GST-NRF2, and GST-MAFG proteins, expressed and purified from E. coli, were incubated with [γ-32P]ATP-labeled oligonucleotides (position −1303/−1288) containing the wild-type ARE motif in the p62 promoter (B) or the mutated ARE motif (C). The amounts of recombinant proteins incubated with DNA were 5 μg or 10 μg of GST (lanes 1 and 2), 0.75 μg or 3.75 μg of GST-NRF2 (lanes 3 and 4), 0.5 or 2.5 μg of GST-MAFG (lanes 5 and 6), 0.5 μg of GST-MAFG or 1.5 μg (lanes 7, 9, and 11), or 3.75 μg (lanes 8, 10, and 12) of GST-NRF2. Competition experiments with cold oligonucleotides (1 μg) containing the wild-type ARE (lanes 11 and 12) or mutated ARE (lanes 13 and 14) designed from the p62 promoter show that binding is specific for the wild-type element. D, Coomassie staining of an SDS-PAGE gel showing the GST fusion proteins used in B and C.
p62 Interacts with KEAP1 via a Conserved Sequence Motif
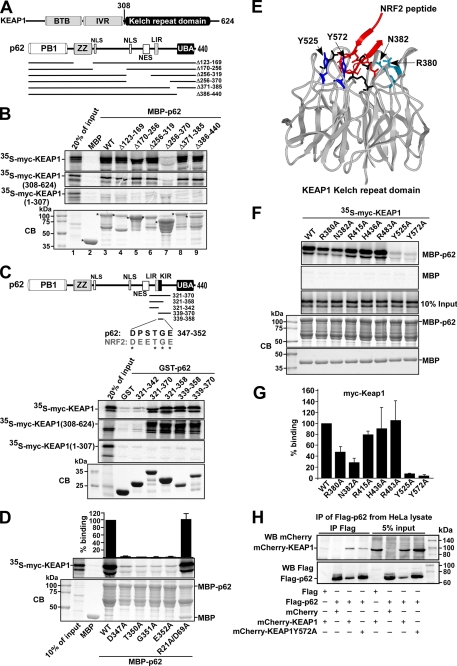
Although p62 is subject to NRF2-mediated induction during oxidative stress, p62 protein has itself been reported to be capable of increasing NRF2 activity (39). To test if p62 binds directly to any of the proteins involved in NRF2-mediated gene expression, we expressed a MBP-p62 fusion protein in E. coli and tested in a MBP pulldown assay for its interaction with either in vitro translated NRF2 or its negative regulator KEAP1. These experiments indicated that a strong interaction exists between p62 and KEAP1 (Fig. 3B), but we detected no interaction between p62 and NRF2 (data not shown). Thereafter a series of MBP-p62 deletion constructs were tested in similar MBP pulldown experiments. Because MBP-p62Δ256–370 did not interact while MBP-p62Δ256–319 and MBP-p62Δ371–385 bound to KEAP1, the interaction with KEAP1 was found to require amino acids 319–370 in p62 (lanes 6–8 in Fig. 3B). Moreover, p62 interacted with the isolated Kelch-repeat domain of KEAP1, but not with a construct lacking this domain (Fig. 3B).
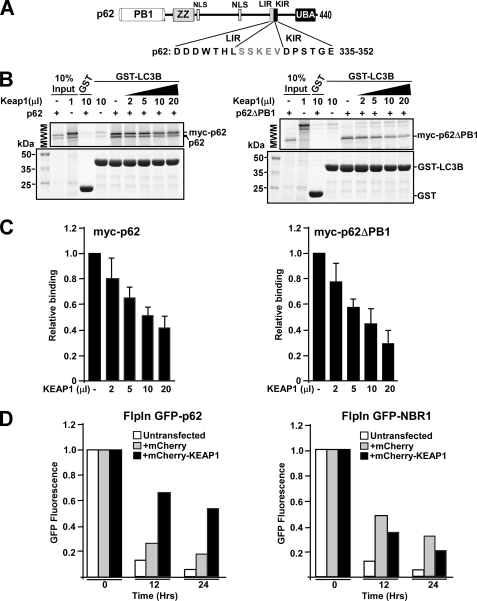
FIGURE 3.
The p62 protein interacts with KEAP1 via a short, conserved sequence motif. A, maps of KEAP1 and p62 indicating positions of domains and the deletion constructs employed in B to map the interaction between the two proteins. B, MBP pulldown assays showing that amino acids 321–370 in p62 are essential for binding to KEAP1, and that p62 interacts with the Kelch-repeat domain of KEAP1. The KEAP1 constructs that are depicted were in vitro translated in the presence of [35S]methionine, and tested in MBP pulldown assays for interaction with the indicated MBP-p62 constructs. The bottom gel panel shows a Coomassie Blue (CB)-stained SDS-polyacrylamide gel with the various MBP-p62 constructs used as input in the pulldown assays. Full-length proteins are indicated with asterisks. C, GST pulldown assays showing that amino acids 339–358 of p62 are sufficient for the interaction with KEAP1. The GST-p62 constructs, which contained the indicated peptides from p62, were tested in GST pulldown assays for interaction with the indicated Myc-tagged portions of KEAP1 produced by in vitro translation; full-length KEAP1 (1–624 amino acids), a C-terminal Kelch-repeat-containing fragment (308–624 amino acids), and an N-terminal BTB domain- and IVR-containing fragment (1–307 amino acids) were tested. The figure at the top shows the p62 constructs used and an alignment between p62 and NRF2 of the conserved DXXTGE motif (amino acids 347–352 in p62). D, MBP pulldown assays showing the effect of single point mutations in the DXXTGE motif of p62. Quantifications of the mean % binding with standard deviations from three independent experiments are shown above the autoradiograph. E, schematic based on the crystal structure of the Neh2 peptide from NRF2 bound to the Kelch-repeat domain of KEAP1. Seven amino acid residues located in loops of the Kelch-repeat were selected for mutational analyses to determine their effect on binding to p62. F and G, mapping of amino acid residues in the Kelch-repeat domain important for interaction with p62. The indicated single-point mutation constructs of KEAP1 were tested in MBP pulldown assays with MBP-p62. Quantification of the mean % binding with standard deviations from three independent experiments are shown. H, mCherry-KEAP1 co-immunoprecipitated with FLAG-p62 from HeLa cell extracts. Cells were co-transfected with the indicated constructs and FLAG-p62 immunoprecipitated with FLAG antibodies 24 h after transfection. Precipitated FLAG-p62 and co-precipitated mCherry-KEAP1 were detected by Western blotting using the indicated antibodies. For B, C, and H, data representative of three independent experiments with similar results are shown.
To verify that amino acids 319–370 in p62 are able to interact with the Kelch-repeat domain of KEAP1, a series of GST-p62 fusion constructs were expressed in E. coli that contained different fragments of the region of interest in the cargo receptor protein. These were tested in GST pulldown assays for their ability to bind to KEAP1 generated by in vitro translation (Fig. 3C). As anticipated, GST-p62(321–370) interacted strongly, with both full-length KEAP1 and the isolated Kelch-repeat domain. The shortest p62 construct found to interact with KEAP1 was p62(339–358) (Fig. 3C). Analysis of amino acids 339–358 in p62 revealed the presence of a linear sequence (DPSTGE, amino acids 347–352) that resembles the DEETGE sequence utilized by NRF2 for its interaction with the Kelch-repeat domain of KEAP1 (Fig. 3C). Point mutants of full-length p62 protein containing substitutions within this motif (D347A, T350A, G351A, and E352A) all displayed a marked loss of binding activity toward KEAP1 in MBP pulldown assays (Fig. 3D). This suggested that the KEAP1-NRF2 and KEAP1-p62 interactions share similar features. Hence, seven point mutants in KEAP1, previously shown to affect its ability to bind NRF2 (46), were tested for their effects on the KEAP1-p62 interaction. The residues that were mutated are present on loops protruding from the bottom of the Kelch-repeat domain and do not affect folding of the β-propeller structure (Fig. 3E). We found that two of these mutations abolished the interaction of KEAP1 with p62 (Y525A and Y572A), two had an intermediary inhibitory effect (R380A and N382A), whereas three showed little or no effect (R415A, H436A, and R483A) (Fig. 3, F and G). This suggests that p62 binds to KEAP1 in a similar, although not identical, manner as NRF2.
The strong in vitro interaction between KEAP1 and p62 suggested that KEAP1 is present in p62-containing structures in vivo. We therefore performed Western blot experiments to test if KEAP1 is present in the same complex as p62 in HeLa cells co-transfected with FLAG-p62 and mCherry-KEAP1. FLAG antibodies were used to immunoprecipitate p62, and mCherry antibodies were employed to detect co-precipitated mCherry-KEAP1. As expected, mCherry-KEAP1 co-precipitated with FLAG-p62, indicating that the interaction between these proteins is very likely to occur in vivo. A point mutant of KEAP1 (Y572A) with reduced affinity for p62 was also tested, and despite a higher level of the mutant in the extract, much less of it was co-precipitated with FLAG-p62 (Fig. 3H).
p62 Induces Its Own Gene by Creating a Positive Feedback Loop in the KEAP1-NRF2 Pathway
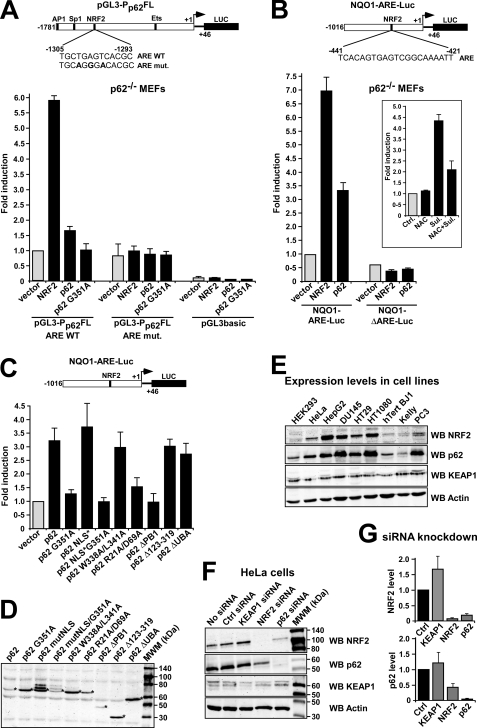
By binding to KEAP1, p62 may inhibit the interaction between KEAP1 and NRF2, thereby increasing ARE-driven gene expression. Because NRF2 transactivates the p62 gene, we hypothesized that the p62 protein contributes to its own up-regulation. To test the effect of p62 on its own promoter, the pGL3-Pp62(−1781/+46)-Luc reporter plasmid (28) was transiently transfected into p62−/− MEFs; the effect of p62 could then be tested by co-expression of Myc-p62. Co-transfection of an expression vector for Myc-p62 and pGL3-Pp62(−1781/+46)-Luc resulted in a 1.7-fold increase in luciferase activity that was relatively low when compared with the effect of NRF2 overexpression (5.8-fold) on reporter gene activity. However, the effect of p62 was consistently observed in several independent experiments, indicating that it is indeed capable of contributing to the induction of its own promoter. We also found that overexpression of p62 had no significant effect on a p62 promoter construct carrying the TGAGT to AGGGA ARE mutation shown above to make the promoter unresponsive to NRF2 (Fig. 4A). Interestingly, forced expression of p62 had a more pronounced effect on expression of other members of the ARE-gene battery as assessed by the mouse Nqo1-ARE-Luc reporter construct (Fig. 4B). This gene was strongly induced by both p62 and NRF2, whereas a negative control for Nqo1-ARE-Luc, which lacked its ARE, was neither induced by p62 nor by NRF2 (Fig. 4B). To address the question of whether it is necessary for p62 to interact with KEAP1 for ARE-driven gene induction to occur, we tested the G351A mutant of p62 that was shown above to be impaired in its ability to interact with KEAP1. Co-expression of this point mutant had no effect on luciferase reporter activity tested, indicating that the ability to interact with KEAP1 is essential for the ability of p62 to induce ARE-driven gene expression (Fig. 4, A and C). Together, our data indicate that p62 protein may contribute to the expression of p62 and to the induction of other NRF2-regulated genes in response to oxidative stress. We therefore considered whether p62 is needed for the induction of the Nqo1-ARE-Luc reporter construct in response to the addition of electrophiles. However, sulforaphane had a clear effect on luciferase activity also when tested in p62−/− MEFs (Fig. 2B). Thus we concluded that NRF2 activation by electrophiles is not dependent on p62, and presumably p62 and electrophiles act via independent mechanisms.
FIGURE 4.
Reciprocal regulation of p62 and NFR2. A and B, ARE-element containing promoters are induced by p62. Reporter gene assays testing activation of the indicated p62 promoter (A) and Nqo1 promoter (B) reporter constructs by co-expression of Nrf2 or p62 in p62−/− MEFs. Cells were co-transfected with 100 ng of empty vector, V5-Nrf2, Myc-p62, or Myc-p62 G351A, together with 60 ng of the indicated Luciferase reporter constructs. The inset in B shows the effect on the activation of the Nqo1-ARE-Luc reporter of treating p62−/− MEFs with N-acetylcysteine (NAC) and sulforaphane (Sul) as indicated. C, reporter gene assays testing activation of the Nqo1-ARE-Luc reporter by co-expression of wild type and various mutants of p62 in p62−/− MEFs. Cells were co-transfected with the indicated Myc-p62 constructs together with 60 ng of Nqo1-ARE-Luc reporter. Cells were analyzed 24 h after transfection. The data in A, B, and C show the mean -fold induction with standard deviations based on three independent experiments performed in triplicate. D, Western blot experiment demonstrating expression of the various Myc-tagged p62 constructs in transfected p62−/− MEFs. The cell extracts were harvested 24 h post transfection using anti-Myc antibodies. E, the endogenous expression level of p62 correlates with the expression level of endogenous NRF2 in various human cell lines. Extracts of the indicated human cell lines were analyzed by Western blotting using the shown antibodies. The anti-actin blot shows equal amounts of proteins in the cell lysates loaded on the gel. F and G, knockdown of p62 in HeLa cells reduce the level of NRF2 and vice versa. HeLa cells transfected with KEAP1, NRF2, or p62 siRNAs, respectively, were analyzed by Western blotting using the indicated antibodies. Scrambled siRNA and mock transfection were used as controls. The quantifications are based on three independent experiments.
We next asked if p62 needs to enter the nucleus in order to activate NRF2. In particular, p62 continuously shuttles between the cytoplasm and the nucleus and we have recently mapped two nuclear import signals (NLSs) and a nuclear export signal that are essential for its nuclear shuttling (47). It is generally believed that the interaction between KEAP1 and NRF2 occurs principally in the cytoplasm (reviewed in Ref. 2), but KEAP1 can also be found in the nucleus, and it has been suggested that KEAP1 can down-regulate NRF2 in this organelle (11–14). We found that a construct for p62 lacking both NLS sequences (MutNLS; R186A/K187A/K264A/R265A) activated the Nqo1-ARE-Luc reporter even more efficiently than did the wild-type construct (Fig. 4C). This indicates that the positive effect p62 exerts on NRF2 activity probably occurs in the cytoplasm, and that nuclear shuttling of p62 is not essential for it to inhibit KEAP1 and thereby activate NRF2.
We then examined whether other regions of p62 are essential for its ability to induce the Nqo1-ARE-Luc reporter. The 440-amino acid-long p62 protein contains an N-terminal PB1 domain, followed by a ZZ type zinc finger domain, nuclear localization, and nuclear export signals, an LIR motif that binds to LC3, and a C-terminal ubiquitin-binding UBA domain (31, 34, 47, 48) (Fig. 3A). The PB1 domain is required for homo-polymerization and for heterodimeric interactions with atypical protein kinases Cs, as well as with MEK5 and NBR1 (40). In these experiments, the reporter plasmid was co-transfected with expression constructs for deletion or point mutants of p62 (Fig. 4C). Expression of the various p62 constructs co-expressed with the reporter plasmid was verified by Western blot experiments (Fig. 4D). An interesting observation was that p62 lacking its PB1 domain did not activate the Nqo1-ARE-Luc reporter (Fig. 4C). Also, an expression construct for a monomeric full-length p62 with two point mutations in its PB1 domain (R21A/D69A) was unable to up-regulate the reporter gene, indicating that polymerization of the scaffold protein is important for NRF2 activation. It is noteworthy that the R21A/D69A mutant can recruit atypical protein kinase C, and therefore phosphorylation of NRF2 probably does not account for the observed activation of ARE-driven gene expression upon forced expression of wild-type p62. Another interesting observation was that a p62 construct mutated in its LIR motif (W338A/L341A) increased the Nqo1-ARE-Luc reporter to the same extent as did wild-type p62 (Fig. 4C); the LIR mutant construct is not efficiently degraded by autophagy, and we therefore conclude that autophagic degradation of KEAP1 is probably not an essential step in the p62-mediated activation of NRF2. Expression constructs for p62 lacking either a large internal region (Δ123–319), which encompassed the ZZ domain, two NLSs, and a nuclear export signal, or the C-terminal ubiquitin-binding UBA domain (ΔUBA) were also equally capable of increasing reporter gene activity as the wild-type protein (Fig. 4C), indicating that these regions of p62 do not contain domains or motifs important for the activation of NRF2.
The Level of p62 Correlates with the Level of NRF2
Because NRF2 induces expression of p62, an accumulation of NRF2 should correlate with an increase in the amount of p62. At the same time, the interaction between p62 and KEAP1 may inhibit proteasomal degradation of NRF2, and p62 may increase the level of NRF2 protein through this mechanism. To investigate the relationship between the levels of p62 and NRF2 protein, extracts from nine different human cell lines were analyzed by Western blotting. The data revealed that a strong correlation exists between the endogenous levels of these two proteins (Fig. 4E). All cell lines with a high or intermediate level of NRF2 had similarly high levels of p62. In contrast, the three cell lines with no or a very low level of NRF2 protein (HEK293, hTertBA1, and Kelly) contained the lowest level of p62 protein among the cell lines tested. To further investigate the interdependency between p62 and NRF2, HeLa cells were transfected with different siRNAs (Fig. 4F). As expected, knockdown of NRF2 reduced the cellular level of p62 (Fig. 4F). In addition, knockdown of p62 reduced the cellular level of NRF2 (Fig. 4F). The level of KEAP1 protein seemed to be rather similar in the different human cell lines examined (Fig. 4E), and it was not significantly affected by knockdown of either p62 or NRF2 (Fig. 4F). Collectively, our data suggest that the levels of p62 and NRF2 are mutually interdependent and that during stress the two proteins cooperate to maintain high levels of each other.
KEAP1 Competes with LC3 for Binding to p62
In the p62 protein its KEAP1 interacting motif (amino acids 347–352) is located immediately adjacent to the C-terminal end of the LC3/GABARAP interacting LIR motif (amino acids 332–342) (Fig. 5A). To examine if p62 can simultaneously interact with KEAP1 and LC3B, we carried out in vitro GST pulldown competition experiments that were designed to test whether increasing the amount of KEAP1 influenced the ability of p62 to interact with GST-LC3B. Because a polymer of p62 may bind to several proteins simultaneously, we tested both wild-type p62 and a monomeric mutant lacking the PB1 domain. Interestingly, KEAP1 appeared to inhibit the interaction of both wild-type p62 and p62ΔPB1 with LC3B (Fig. 5, B and C), indicating that a single p62 molecule cannot interact with both LC3B and KEAP1 simultaneously.
FIGURE 5.
KEAP1 competes with LC3B for the interaction with p62. A, map of p62 illustrating the proximal location of the LIR (LC3-interacting region) and KIR (KEAP1 interacting region) motifs. B and C, GST pulldown assays demonstrating competition between LC3B and KEAP1 for binding to p62. GST-LC3B was incubated with in vitro translated wild-type p62 (polymeric) or a PB1 p62 deletion mutant (monomeric), in the presence or absence of increasing amounts of in vitro translated KEAP1. The data show that with increasing concentrations of KEAP1 there is a reduction in the amount of p62 bound to LC3B. The amount of p62 and LC3B is constant in all reactions. D, KEAP1 inhibits autophagic degradation of GFP-p62, but not GFP-NBR1, in HEK293 Flp-In T-Rex cells stably expressing the GFP-tagged proteins from a tetracycline-inducible promoter. Cells were grown overnight in rich medium (10% serum) in the presence of tetracycline, resulting in the accumulation of the GFP-tagged protein. Then tetracycline was removed (promoter shut-off), and degradation of the GFP-tagged protein followed for 24 h. Degradation of the GFP-tagged protein was measured at indicated time points by flow cytometry, and the readout was a loss of green fluorescence. Cells that were analyzed were either untransfected or transiently transfected with mCherry or mCherry-KEAP1. Expression of mCherry or mCherry-KEAP1 was verified by flow cytometry.
In view of the fact that KEAP1 competes with LC3B for binding to p62, we next examined whether overexpression of KEAP1 might inhibit autophagic degradation of a GFP-p62 fusion protein. To this end, we used a HEK293 Flip-In cell line stably expressing GFP-p62 from an inducible promoter4; autophagic degradation could then be measured by flow cytometry as a loss of green fluorescence after the inducible expression of GFP-p62 had been turned off. Intriguingly, the rate of degradation of the fusion protein was greatly diminished in cells that co-expressed mCherry-KEAP1, indicating that KEAP1 inhibited the autophagic degradation of p62 (Fig. 5D). We have previously observed that transfection of DNA by itself has a small negative effect on the degradation of GFP-p62 in this autophagy reporter cell line.4 Hence, we expected co-expression of mCherry would exert a minor inhibitory effect on p62 degradation. However, the rate of decrease in fluorescence from GFP-p62 observed in cells that co-expressed mCherry-KEAP1 was much slower than could be attributed to mCherry alone (Fig. 5D). Similar experiments were also carried out in HEK293 Flip-In cells stably expressing GFP-NBR1, and degradation of GFP-NBR1 was even less affected by co-expression of mCherry-KEAP1 than by co-expression of mCherry (Fig. 5D). This indicates that the effect of KEAP1 overexpression on the levels of p62 is not via inhibition of autophagy per se, but through a specific effect on p62 degradation caused by the binding of KEAP1 to the KIR motif in p62.
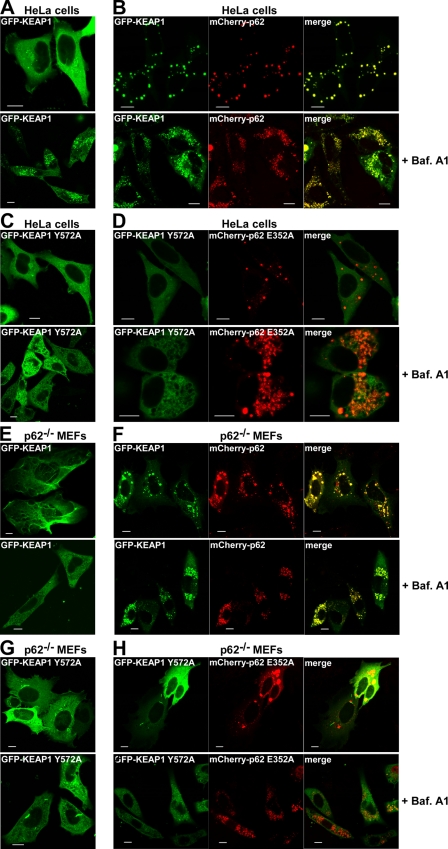
Overexpressed KEAP1 Is Recruited to p62 Bodies and Degraded by Autophagy
We have previously shown that p62 is itself a substrate for selective autophagy and can also act as a cargo receptor for autophagy of ubiquitinated proteins aggregated into so-called p62 bodies (30, 34). Therefore, we next explored whether KEAP1 is recruited to p62 bodies. As mono-specific antibodies that could be used to stain cells for endogenous KEAP1 were not available, we studied an ectopic fusion protein. The localization of ectopic GFP-KEAP1 in HeLa cells was mainly diffuse (Fig. 6A). However, the fusion protein was also localized in p62 bodies (data not shown); p62 bodies are present constitutively in a fraction of HeLa cells due to the presence of the endogenous protein, but their number and size can be dramatically increased when p62 is transiently overexpressed. We observed that GFP-KEAP1 was efficiently redistributed into p62 bodies formed by co-expressed mCherry-p62 (Fig. 6B). Twenty-four hours after transfection, these two proteins co-localized in cytoplasmic aggregates in >95% of the cells that expressed the fusion proteins. No such redistribution of GFP-KEAP1 into p62 bodies was observed if mutants with reduced affinity for each other were co-expressed in HeLa cells (Fig. 6, C and D). Hence, a direct physical interaction between the two proteins is required for KEAP1 to accumulate in p62 bodies. To examine the localization of GFP-KEAP1 in cells lacking endogenous p62, the same set of transfection experiments were performed using p62−/− MEFs. The results were closely similar to the data obtained with HeLa cells and supported the conclusion that KEAP1 is recruited to p62 bodies (Fig. 6, E–H). One interesting difference was that overexpression of KEAP1 in p62−/− MEFs, in particular when expressed alone, appeared to affect the morphology of the cells (Fig. 6E). A similar actin-like bundling of GFP-KEAP1 was not observed in transfected HeLa cells.
FIGURE 6.
Overexpressed KEAP1 is recruited to p62 bodies and degraded by autophagy. A–D, HeLa cells were transfected as indicated with wild-type or mutated GFP-KEAP1 (Y572A), either alone (A and C) or together with mCherry-p62 (B and D). Bafilomycin A1 (16 h) was added as indicated (lower panels). E–H, p62−/− MEFs were transfected as indicated with wild-type or mutated GFP-KEAP1 (Y572A), either alone (E and G) or together with mCherry-p62 (F and H). Bafilomycin A1 (16 h) was added as indicated. A–H, cells were analyzed by confocal microscopy 24 h after transfection. Bars, 10 μm.
Because the fraction of KEAP1 that accumulates in p62 bodies is likely to be degraded by autophagy, we wondered if overexpressed KEAP1 accumulates in acidic vesicles. To test this possibility, the autophagy inhibitor bafilomycin A1 was added, because it causes accumulation of autophagic vesicles whose contents can no longer be acidified and degraded. As a consequence, p62 and other autophagy substrates accumulate in these structures, giving a characteristic localization pattern as seen for mCherry-p62 in Fig. 6B. Treatment with bafilomycin A1 resulted in GFP-KEAP1 localizing in these structures indicating that it is indeed degraded by autophagy (Fig. 6B). Also, we noted that GFP-KEAP1 expressed alone was recruited to these vesicles (Fig. 6A), but this was expected because HeLa cells possess high levels of endogenous p62. However, the KEAP1 Y572A mutant construct was not recruited into autophagic vesicles following treatment of cells with bafilomycin A1 (Fig. 6, C and D). We therefore conclude that overexpressed KEAP1 is degraded by autophagy and that this depends on a direct physical interaction between KEAP1 and p62. Again, the same experiments were performed with p62−/− MEFs, and the results obtained resembled the results obtained with HeLa cells (Fig. 6, E–H). However, there was one important difference: when expressed alone, wild-type GFP-KEAP1 did not accumulate in autophagic vesicles of fibroblasts that had been treated with bafilomycin A1 (Fig. 6E). This result was of course anticipated, because the mutant MEFs lack p62. However, in cells co-expressing mCherry-p62, we observed a strong co-accumulation of GFP-KEAP1 and mCherry-p62 into these structures (Fig. 6F).
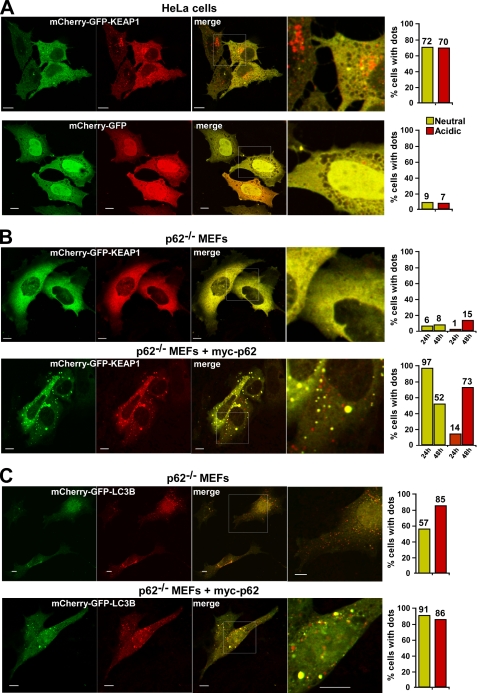
To verify that KEAP1 can be recruited into acidic vesicles, we next expressed KEAP1 fused to the mCherry-GFP double tag in HeLa cells and in p62−/− MEFs. The red fluorescence is stable in an acidic environment, and the accumulation of KEAP1 in red only structures indicates an accumulation in acidic vesicles. When expressed in HeLa cells, a large fraction (70%) of the transfected cells was found to accumulate mCherry-GFP-KEAP1 in acidic vesicles 24 h after transfection. As a control, only 7% of the cells expressing mCherry-GFP alone contained the double tag in acidic vesicles (Fig. 7A). This is consistent with the data obtained using bafilomycin A1 and reflects the presence of endogenous p62 in this cell line. When expressed alone in p62−/− MEFs, mCherry-EGFP-KEAP1 displayed a diffuse staining pattern, and there was little accumulation of the protein in acidic vesicles (Fig. 7B). However, the co-expression of Myc-p62 strongly increased the fraction of cells with mCherry-GFP-KEAP1 in acidic vesicles (Fig. 7B). What was particularly noteworthy is that almost all cells co-transfected with mCherry-GFP-KEAP1 and Myc-p62 contained large aggregates 24 h after transfection. Using this double tag strategy, we have previously observed that the accumulation of p62 aggregates in response to its transient transfection often delays its autophagic degradation. We therefore routinely also investigate p62-transfected cells 48 h after transfection. Indeed, 48 h after transfection the fraction of cells with mCherry-GFP-KEAP1 present in aggregates was reduced and a major fraction of the cells had accumulated the protein in acidic vesicles (Fig. 7B). This showed that KEAP1 is efficiently recruited into acidic vesicles in cells expressing p62, but much less so in cells lacking p62. In comparison, LC3B fused to the mCherry-GFP double tag was efficiently recruited to acidic vesicles both in the absence and presence of p62 (Fig. 7C). To summarize, it is therefore highly likely that NRF2-mediated induction of p62 in response to oxidative stress first results in the accumulation of KEAP1 into p62 bodies, and this is followed by a subsequent autophagic degradation of KEAP1.
FIGURE 7.
Overexpressed KEAP1 accumulates in acidic vesicles. A, HeLa cells transfected with mCherry-GFP-KEAP1 or mCherry-GFP were analyzed by confocal microscopy 24 h after transfection. The fraction of cells with mCherry-GFP-KEAP1 in neutral dots (green and red) and acidic dots (only red) was counted, and the result of a representative experiment based on counting of >300 transfected cells is shown to the right. B, p62−/− MEFs transfected with mCherry-GFP-KEAP1, either alone or together with Myc-p62, were analyzed by confocal microscopy 24 and 48 h after transfection. C, p62−/− MEFs transfected with mCherry-GFP-LC3B, either alone or together with Myc-p62, were analyzed by confocal microscopy 24 h after transfection. To the right is shown quantifications of the percentage of cells with neutral or acidic dots for representative experiments, each based on counting of more than 150 transfected cells. In A–C, bars represent 10 μm.
DISCUSSION
Expression of the p62 gene is induced by NRF2 upon exposure to electrophiles, reactive oxygen species, and nitric oxide (38, 43, 49). Furthermore, p62 protein has been reported to stimulate expression of genes containing an ARE in their promoter regions (39). From these previous studies it appeared that a positive interrelationship of some sort exists between NRF2 and p62, which influences the ARE-gene battery, but the exact mechanism was unresolved. Herein we have shown that NRF2 can induce p62 expression by binding directly to a conserved ARE in its promoter/enhancer and that the p62 protein is able to augment its own expression via this element. Specifically, we found p62 stimulated NRF2 activity by binding to KEAP1, sequestering it, and directing its degradation by autophagy. This association between p62 and KEAP1 leads to stabilization of NRF2, enabling the transcription factor to induce gene expression from ARE-containing promoters.
The p62 ARE identified in the present study is conserved in mammals (Fig. 2A), and it contains the 5′ flanking region TGC-, and the core sequence -TGAGTCA- classifying it as a MAF-like recognition element (MARE). The NRF2 responsive p62 MARE contains a GC to CG substitution in the 3′ flanking region compared with the consensus MARE (43, 50). It has been reported that MARE-like sequences with a G to C substitution in the 3′ flanking region preferentially bind to small MAF:NRF2 heterodimers (51). This is consistent with our GMSA results in which we found MAFG homodimers did not bind the p62 MARE, but could bind the cis-element when incubated together with NRF2. The small MAF proteins (MAFG, -K, and -F) are important components of cellular stress responses and are also involved in neuronal differentiation (52, 53). Furthermore, homodimers and heterodimers composed exclusively of small MAF proteins can act as repressive competitors to MAF:NRF2 heterodimers (reviewed in Ref. 50). The human NQO1 promoter, which contains a MARE element with a G to C substitution in the 3′ end that is similar to the p62 MARE, has been shown to be activated or repressed by MAFG overexpression, depending on the balance between MAFG:MAFG homodimers and MAFG:NRF2 heterodimers (51). Whether p62 expression is down-regulated by homodimers of small MAF proteins, or small MAF protein as a heterodimer with repressors such as BACH1 and BACH2, is an interesting question to address, especially because the small MAF proteins themselves are up-regulated by stressors or signaling induced by nerve growth factors (52, 53). However, our GMSA results showed that MAFG homodimers do not bind to the p62 MARE. Furthermore, the Nrf2 A502Y mutant, which has a binding specificity similar to MAFG, does not induce p62 mRNA (43). Taken together, these observations suggest that the p62 MARE has low affinity for MAFG homodimers.
Prothymosin alpha has previously been reported to compete with Nrf2 for binding to the Kelch-repeat domain of Keap1 (11), and recently, p21 was shown to compete with Keap1 for binding to the DLG and ETGE motifs of Nrf2 (54). In both of these cases, as we have shown here for p62, the net result is activation of Nrf2. Our data suggest the following three mechanisms are involved in the p62-mediated induction of NRF2: (i) p62 protein interacts directly with KEAP1 to stabilize NRF2; (ii) p62 protein sequesters KEAP1 into inclusion bodies; and (iii) p62 protein mediates degradation of KEAP1 by autophagy. In the first of these we show a direct interaction between a KIR motif in p62 and the Kelch-repeat domain in KEAP1. The finding that p62 interacts with KEAP1 in a similar manner as NRF2, suggests that p62 can compete with NRF2 for binding to KEAP1. This conclusion is confirmed by the recent work of Komatsu and his colleagues, published immediately before submission of this report (55), in which they showed a direct interaction between murine p62 and Keap1, and describe the structure of the KIR peptide motif bound to the Kelch-repeat domain of Keap1. In the structure presented by Komatsu et al., a 19-amino acid KIR peptide from murine p62 formed hydrogen bonds with 8 amino acids of Keap1. We have mutated three of these residues and found inhibitory effects for two of them (R380A and N832A). In our hands, mutation of the third, R415A, had a relatively minor effect on the interaction between the two proteins. In common with the findings of Komatsu et al., we have discovered that R483, which was important for the Nrf2-Keap1 interaction, had no effect on the p62-KEAP1 interaction when it was mutated to alanine. This apparent anomaly is explained by the structural data that revealed R483 is not involved in binding the KIR peptide. The two mutants we found to have the most profound inhibitory effect as single point mutations, Y525A and Y572A, constitute part of the binding pocket anchoring the hairpin formed by the E/STGE core motif shared by Nrf2 and p62.
According to the data presented by Komatsu and his colleagues, the affinity of monomeric p62 for Keap1 is not sufficiently strong to compete with the high affinity ETGE motif of Nrf2 (55). They suggest that p62 specifically inhibits the interaction between Keap1 and the low affinity DLG site on Nrf2. Our data along with those of Komatsu et al., indicate that a single binding surface in p62 is involved in the KEAP1 interaction. There are, however, a number of other mechanisms by which p62 might activate NRF2. Firstly, p62 is a polymeric protein, and as a consequence of its quaternary structure, multimerized forms of p62 may be capable of interacting with both of the subunits within a KEAP1 dimer, thereby completely blocking the interaction between KEAP1 and NRF2. Secondly, we have found that KEAP1 is recruited to p62 inclusion bodies upon its overexpression. This sequestering of KEAP1 into p62 bodies may result in the displacement of KEAP1 from locations where it would otherwise interact with Cul3 and promote the ubiquitylation and turnover of NRF2. Thirdly, p62 targets KEAP1 for degradation by autophagy. We observed that when KEAP1 is overexpressed in mammalian cell lines it is efficiently recruited into autophagic vesicles, strongly suggesting that endogenous KEAP1 is similarly degraded by autophagy.
The lack of antibodies suitable for efficient immunostaining of endogenous KEAP1 made it impossible for us to test the mechanisms by which p62 antagonized the endogenous BTB-Kelch protein. However, our data showed that autophagic degradation of overexpressed KEAP1 strongly depended on the presence of p62 and on its ability to interact with KEAP1. One intriguing question was whether the close proximity of the LIR and KIR motifs within p62 implies that it is unable to associate with KEAP1 and LC3B simultaneously. Our in vitro data showed that increasing amounts of KEAP1 protein inhibited the interaction between p62 and LC3B. We speculate that the competitive nature of the binding of these two proteins for p62 may be necessary to avoid complete degradation of endogenous KEAP1 under normal growth conditions. It is clear that ectopically expressed KEAP1 may not reflect physiologically relevant situations accurately. The in vivo situation is also complicated by the fact that p62 is a polymeric protein. It is very likely that a p62 chain may interact with both KEAP1 and LC3B simultaneously. We therefore favor the idea that the fraction of KEAP1 that associates with p62 polymers will be degraded by autophagy along with p62.
We have no evidence that p62 under normal conditions is needed for the initial activation of NRF2 in response to oxidative stress. Reporter gene assays performed using p62−/− MEFs revealed that ARE-driven gene expression is efficiently induced by sulforaphane in cells that lack p62 protein. Hence, the presence of p62 is not essential for the initial activation of NRF2 in response to inducing agents. We hypothesize that the role of p62 under conditions of prolonged cellular stress is to mediate a sustained activation of the oxidative stress response after an initial p62-independent activation of NRF2. This is consistent with previous findings showing that Nrf2 accumulates a few hours before p62 protein levels increase in response to treatment with the anti-Parkinsonian drug deprenyl (56).
Our results show that p62 creates a positive feedback loop in the KEAP1-NRF2 pathway. It is therefore reasonable to ask the question: how is this loop broken when sustained activation of ARE response genes is no longer needed? As p62 is rapidly degraded by autophagy under normal conditions, we conclude that autophagic degradation of p62 will break the loop. Alternatively, as discussed above, the accumulation of small MAF proteins, which occurs upon activation of NRF2, may lead to repression of p62 through small MAF proteins binding to the ARE element in its promoter as homodimers, or possibly as heterodimers with BACH1 and BACH2 repressors.
Interestingly, we found a positive correlation between p62 and NRF2 expression in all human cell lines tested, indicating constitutive activation of NRF2 in cell lines with high p62 protein levels. This is consistent with the results presented recently suggesting p62 as an endogenous protein inducer of NRF2 (55). Using autophagy-deficient mouse models, Komatsu et al. showed accumulation of several NRF2 target proteins leading to pathological conditions; presumably one evolutionary reason why autophagy might be linked with NRF2 is that many of the genes for proteasomal subunits are regulated through the KEAP1-NRF2 pathway (2). This suggests that autophagy is an important process for down-regulation of NRF2-mediated cellular responses induced by stressors. Thus, when autophagy is impaired, p62 accumulates and will by itself activate NRF2 constitutively by sequestering KEAP1 into inclusion bodies. If the hypothesis is correct that p62 is an endogenous protein inducer of NRF2, then other processes that result in an increase in p62 protein levels ought to influence NRF2 activity. Thus, the p62/KEAP1 axis may integrate many stress pathways.
The p62 protein acts both as an adapter or scaffold protein in cellular signaling pathways and as a cargo receptor for degradation of ubiquitinated targets by autophagy (30, 31, 34, 57, 58). Because p62 is itself degraded by selective autophagy it will act as a receptor for autophagic degradation of proteins that bind firmly to it. Hence, ubiquitination is not required per se for p62 to shuttle proteins to autophagosomes. It has recently been shown that a mutant of superoxide dismutase 1 that causes amyotrophic lateral sclerosis binds directly to p62 and is degraded by autophagy (59). Now we show that this is also the case for KEAP1. Consequently, it is very likely that the roles of p62 in cellular signaling may in many cases be intimately linked to its ability to act as a cargo receptor for selective autophagy.
Acknowledgments
We are grateful to J. P. Brody for the generous gift of the pGL3-Pp62(−1781/+46)-Luc reporter plasmid. We thank the BioImaging FUGE core facility at the Institute of Medical Biology, University of Tromsø, for use of instrumentation and expert assistance.
Addendum
Very recently, Copple et al. (64) showed that siRNA depletion of p62 in mouse hepatoma cells results in an increase in the half-life of Keap1, further indicating that p62 regulates the degradation of Keap1.
This work was supported by grants from the FUGE programme of the Norwegian Research Council, the Norwegian Cancer Society, and the Blix Foundation (to T. J.) and by Cancer Research-UK (project grant C4909/A9990 to J. D. H.).
Larsen, K. B., Lamark, T., Øvervatn, A., Harneshaug, I., Johansen, T., and Bjørkøy, G., submitted for publication.
- KEAP1
- Kelch-like ECH-associated protein 1
- ARE
- antioxidant response element
- ChIP
- chromatin immunoprecipitation
- EGFP
- enhanced green fluorescent protein
- GFP
- green fluorescent protein
- GMSA
- gel mobility shift assay
- MARE
- Maf recognition element
- MEF
- mouse embryonic fibroblast
- NRF2
- NF-E2 related factor 2
- NQO1
- NADP(H):quinone oxidoreductase 1
- PB1
- Phox and Bem1p
- wt
- wild type
- siRNA
- small interfering RNA
- CMV
- cytomegalovirus
- MBP
- myelin basic protein
- NLS
- nuclear localization signal
- GST
- glutathione S-transferase.
REFERENCES
- 1.Halliwell B., Gutteridge J. M. (eds) (2007) Free Radicals in Biology and Medicine, 4th Ed., pp. 187–267, Oxford University Press, Oxford, UK [Google Scholar]
- 2.Hayes J. D., McMahon M. (2009) Trends Biochem. Sci. 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 3.Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 4.Motohashi H., Yamamoto M. (2004) Trends Mol. Med. 10, 549–557 [DOI] [PubMed] [Google Scholar]
- 5.Nguyen T., Nioi P., Pickett C. B. (2009) J. Biol. Chem. 284, 13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak M. K., Wakabayashi N., Greenlaw J. L., Yamamoto M., Kensler T. W. (2003) Mol. Cell Biol. 23, 8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) J. Biol. Chem. 278, 21592–21600 [DOI] [PubMed] [Google Scholar]
- 8.Cullinan S. B., Gordan J. D., Jin J., Harper J. W., Diehl J. A. (2004) Mol. Cell Biol. 24, 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004) Mol. Cell Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watai Y., Kobayashi A., Nagase H., Mizukami M., McEvoy J., Singer J. D., Itoh K., Yamamoto M. (2007) Genes Cells 12, 1163–1178 [DOI] [PubMed] [Google Scholar]
- 11.Karapetian R. N., Evstafieva A. G., Abaeva I. S., Chichkova N. V., Filonov G. S., Rubtsov Y. P., Sukhacheva E. A., Melnikov S. V., Schneider U., Wanker E. E., Vartapetian A. B. (2005) Mol. Cell Biol. 25, 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T., Sherratt P. J., Nioi P., Yang C. S., Pickett C. B. (2005) J. Biol. Chem. 280, 32485–32492 [DOI] [PubMed] [Google Scholar]
- 13.Sun Z., Zhang S., Chan J. Y., Zhang D. D. (2007) Mol. Cell Biol. 27, 6334–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velichkova M., Hasson T. (2005) Mol. Cell Biol. 25, 4501–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D. D., Hannink M. (2003) Mol. Cell Biol. 23, 8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J. D. (2006) J. Biol. Chem. 281, 24756–24768 [DOI] [PubMed] [Google Scholar]
- 17.Tong K. I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. (2006) Mol. Cell Biol. 26, 2887–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joung I., Strominger J. L., Shin J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5991–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puls A., Schmidt S., Grawe F., Stabel S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6191–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez P., De Carcer G., Sandoval I. V., Moscat J., Diaz-Meco M. T. (1998) Mol. Cell Biol. 18, 3069–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz L., Sanchez P., Lallena M. J., Diaz-Meco M. T., Moscat J. (1999) EMBO J. 18, 3044–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wooten M. W., Seibenhener M. L., Neidigh K. B., Vandenplas M. L. (2000) Mol. Cell Biol. 20, 4494–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 25.Duran A., Linares J. F., Galvez A. S., Wikenheiser K., Flores J. M., Diaz-Meco M. T., Moscat J. (2008) Cancer Cell 13, 343–354 [DOI] [PubMed] [Google Scholar]
- 26.Kitamura H., Torigoe T., Asanuma H., Hisasue S. I., Suzuki K., Tsukamoto T., Satoh M., Sato N. (2006) Histopathology 48, 157–161 [DOI] [PubMed] [Google Scholar]
- 27.Rolland P., Madjd Z., Durrant L., Ellis I. O., Layfield R., Spendlove I. (2007) Endocr. Relat. Cancer 14, 73–80 [DOI] [PubMed] [Google Scholar]
- 28.Thompson H. G., Harris J. W., Wold B. J., Lin F., Brody J. P. (2003) Oncogene 22, 2322–2333 [DOI] [PubMed] [Google Scholar]
- 29.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 33.Lamark T., Kirkin V., Dikic I., Johansen T. (2009) Cell Cycle 8, 1986–1990 [DOI] [PubMed] [Google Scholar]
- 34.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 35.Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii T., Yanagawa T., Yuki K., Kawane T., Yoshida H., Bannai S. (1997) Biochem. Biophys. Res. Commun. 232, 33–37 [DOI] [PubMed] [Google Scholar]
- 37.Nagaoka U., Kim K., Jana N. R., Doi H., Maruyama M., Mitsui K., Oyama F., Nukina N. (2004) J. Neurochem. 91, 57–68 [DOI] [PubMed] [Google Scholar]
- 38.Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. (2000) J. Biol. Chem. 275, 16023–16029 [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., Kern J. T., Walker J. R., Johnson J. A., Schultz P. G., Luesch H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5205–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. (2003) J. Biol. Chem. 278, 34568–34581 [DOI] [PubMed] [Google Scholar]
- 41.Rekdal C., Sjøttem E., Johansen T. (2000) J. Biol. Chem. 275, 40288–40300 [DOI] [PubMed] [Google Scholar]
- 42.Sjøttem E., Rekdal C., Svineng G., Johnsen S. S., Klenow H., Uglehus R. D., Johansen T. (2007) Nucleic Acids Res. 35, 6648–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura M., Yamamoto T., Zhang J., Itoh K., Kyo M., Kamiya T., Aburatani H., Katsuoka F., Kurokawa H., Tanaka T., Motohashi H., Yamamoto M. (2007) J. Biol. Chem. 282, 33681–33690 [DOI] [PubMed] [Google Scholar]
- 44.Du Y., Wooten M. C., Wooten M. W. (2009) Neurobiol. Dis. 35, 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prestera T., Holtzclaw W. D., Zhang Y., Talalay P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2965–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M. (2006) EMBO J. 25, 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S., Lamark T., Bruun J. A., Øvervatn A., Bjørkøy G., Johansen T. (2010) J. Biol. Chem. 285, 5941–5953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vadlamudi R. K., Joung I., Strominger J. L., Shin J. (1996) J. Biol. Chem. 271, 20235–20237 [DOI] [PubMed] [Google Scholar]
- 49.Kosaka K., Mimura J., Itoh K., Satoh T., Shimojo Y., Kitajima C., Maruyama A., Yamamoto M., Shirasawa T. (2010) J. Biochem. 147, 73–81 [DOI] [PubMed] [Google Scholar]
- 50.Motohashi H., O'Connor T., Katsuoka F., Engel J. D., Yamamoto M. (2002) Gene. 294, 1–12 [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto T., Kyo M., Kamiya T., Tanaka T., Engel J. D., Motohashi H., Yamamoto M. (2006) Genes Cells 11, 575–591 [DOI] [PubMed] [Google Scholar]
- 52.Katsuoka F., Motohashi H., Ishii T., Aburatani H., Engel J. D., Yamamoto M. (2005) Mol. Cell Biol. 25, 8044–8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Töröcsik B., Angelastro J. M., Greene L. A. (2002) J. Neurosci. 22, 8971–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen W., Sun Z., Wang X. J., Jiang T., Huang Z., Fang D., Zhang D. D. (2009) Mol. Cell 34, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 56.Nakaso K., Nakamura C., Sato H., Imamura K., Takeshima T., Nakashima K. (2006) Biochem. Biophys. Res. Commun. 339, 915–922 [DOI] [PubMed] [Google Scholar]
- 57.Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 59.Gal J., Ström A. L., Kwinter D. M., Kilty R., Zhang J., Shi P., Fu W., Wooten M. W., Zhu H. (2009) J. Neurochem. 111, 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammarström M., Hellgren N., van Den Berg S., Berglund H., Härd T. (2002) Protein Sci. 11, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) Biochem. J. 374, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirkin V., Lamark T., Sou Y. S., Björköy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 63.Clausen T. H., Lamark T., Isakson P., Finley K., Larsen K. B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., Johansen T. (2010) Autophagy. 6, 330–344 [DOI] [PubMed] [Google Scholar]
- 64.Copple I. M., Lister A., Obeng A. D., Kitteringham N. R., Jenkins R. E., Layfield R., Foster B. J., Goldring C. E., Park B. K. (2010) J. Biol. Chem. 285, 16782–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]