Abstract
Fermenting microbial communities generate hydrogen; its removal through the production of acetate, methane, or hydrogen sulfide modulates the efficiency of energy extraction from available nutrients in many ecosystems. We noted that pathway components for acetogenesis are more abundantly and consistently represented in the gut microbiomes of monozygotic twins and their mothers than components for methanogenesis or sulfate reduction and subsequently analyzed the metabolic potential of two sequenced human gut acetogens, Blautia hydrogenotrophica and Marvinbryantia formatexigens in vitro and in the intestines of gnotobiotic mice harboring a prominent saccharolytic bacterium. To do so, we developed a generally applicable method for multiplex sequencing of expressed microbial mRNAs (microbial RNA-Seq) and, together with mass spectrometry of metabolites, showed that these organisms have distinct patterns of substrate utilization. B. hydrogenotrophica targets aliphatic and aromatic amino acids. It increases the efficiency of fermentation by consuming reducing equivalents, thereby maintaining a high NAD+/NADH ratio and boosting acetate production. In contrast, M. formatexigens consumes oligosaccharides, does not impact the redox state of the gut, and boosts the yield of succinate. These findings have strategic implications for those who wish to manipulate the hydrogen economy of gut microbial communities in ways that modulate energy harvest.
Keywords: Bacterial Metabolism, Bacterial Transcription, Gene Expression, Intestine, Metabolic Regulation, Acetogenesis, Gnotobiotic Mice, Human Gut Hydrogen Economy, Human Gut Microbiota/Microbiome, Microbial RNA-Seq
Introduction
Mammals harbor large and diverse communities of gut microbes (1). One important function of the gut microbiota is to break down otherwise indigestible components of the diet, many of which are derived from plants (2). Fermentation of dietary polysaccharides and proteins is accomplished by syntrophic interactions between microbes linked in a metabolic food web that yields short chain fatty acids (e.g. acetate, propionate, and butyrate), other organic acids (e.g. formate), and gases (H2 and CO2) (3). In a primary fermenter, the accumulation of H2 inhibits reoxidation of NADH, thereby reducing the yield of ATP and short chain fatty acids, including acetate (4). Short chain fatty acids account for up to 10% of human energy requirements (5). Thus, changes in the microbiota that affect production of short chain fatty acids (i.e. fermentation efficiency) may also affect host energy balance.
Three groups of gut microbes, methanogens, acetogens, and sulfate-reducing bacteria (SRB),3 have the capacity to consume H2. The end products of H2 oxidation generated by each of these groups (methane, acetate, and H2S, respectively) have diverse and important effects on host physiology and/or the environment (6–9). Understanding the metabolic roles that methanogens, acetogens, and SRB play in the H2 economy of the gut has implications for understanding and potentially manipulating our energy balance.
In considering the different groups of H2-consuming organisms as potential therapeutic targets for manipulating the efficiency of gut microbial fermentation and host energy balance, it is important to understand the range of niches encompassed by members of each group, the degree of niche overlap that exists within and between groups, and the various factors that govern the levels of colonization of these groups. For example, there is evidence that SRB may gain an advantage depending upon the levels of mucin (10) or sulfate (11) available and that acetogens may compete more effectively in the absence of methanogens (12). Attempts to replace methanogens with acetogens in ruminants, motivated by a desire to reduce global greenhouse gas emissions and improve the efficiency of fuel partitioning between diet and host (13) have been generally unsuccessful (14). Nonetheless, experiments performed in gnotobiotic lambs have shown that hydrogen-dependent acetogenesis can sustain a functional rumen and replace methanogens as a sink for H2 (15).
To successfully manipulate the human gut bioreactor, we need to understand its operations at a mechanistic level. Although deep draft and finished genome sequences are now available for over a hundred gut microbial species, including several H2 consumers, there is a lack of broadly applicable tools that enable microbiologists to go beyond descriptions of “what gene is present” in a genome or a community to understanding the mechanisms by which microbes gain access to and metabolize nutrients within their habitats and influence one another and their hosts (16).
To begin to unravel the niches of these organisms, the substrates they use, and the factors that affect their metabolism, we have developed a method for massively parallel sequencing of expressed microbial mRNAs (microbial RNA-Seq) that utilizes gut microbe-directed rRNA depletion oligonucleotides and size selection to increase the fraction of mRNA in the total microbial RNA population by more than 10-fold. We validated the quantitative precision of this method and used simulations to determine the sequencing depth necessary to detect expression changes in bacterial communities where members are present at different abundances. To focus the application of this technique to understanding the niches (professions) of H2 consumers in the gut, we first analyzed human gut microbiome data sets, obtained by shotgun sequencing of fecal microbial community DNA samples obtained from six families of monozygotic twins and their mothers, and showed that pathway components for acetogenesis are more abundantly and consistently represented than components for methanogenesis or sulfate reduction. We then focused on characterizing the interactions between two recently sequenced human gut-derived acetogens and a prominent sequenced human gut saccharolytic bacterium, Bacteroides thetaiotoamicron, in the distal intestines of gnotobiotic mice. We employed microbial RNA-Seq to profile their transcriptomes in defined locations of the mouse intestine and subsequently verified our RNA-Seq-based predictions of the metabolic activities of these organisms in vivo by using mass spectrometric and biochemical analyses. In principle, this multi-faceted approach can be broadly applied to explore the niches and metabolic activities of members of myriad microbial communities.
MATERIALS AND METHODS
Bacterial Strains
Genome assemblies for Blautia hydrogenotrophica strain DSM 10507 and Marvinbryantia formatexigens DSM 14469 were downloaded from GenBankTM (accession numbers NZ_ACBZ00000000 and NZ_ACCL00000000).
RNA-Seq
Cecal and fecal samples obtained from mice at the time that they were sacrificed and bacteria cultured using the conditions described in the supplemental text were immediately frozen at −80 °C and maintained at this temperature prior to processing. All of the samples were treated with RNAProtect (Qiagen). An aliquot (∼100 mg) of each frozen cecal sample was suspended in a solution containing 500 μl of acid-washed glass beads (Sigma-Aldrich), 500 μl of extraction buffer A (200 mm NaCl, 20 mm EDTA), 210 μl of 20% SDS, and 500 μl of a mixture of phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5; Ambion) and lysed by using a bead beater (BioSpec Products). Cellular debris was removed by centrifugation (8,000 × g; 3 min). The extraction was repeated, and the nucleic acids were precipitated with isopropanol and sodium acetate (pH 5.5). A similar protocol was used for fecal samples and for cell pellets obtained from in vitro cultures.
Details about protocols used for removing residual DNA from RNA preparations, rRNA depletion, double-stranded cDNA synthesis, and multiplex sequencing with the Illumina GA-II instrument as well as our data analysis pipeline are provided in the supplemental text. The methods for analyzing fecal microbiome data sets for the presence of H2-consuming metabolic pathways, in vitro culture conditions, and quantitative biochemical and GC/MS assays of metabolites are also provided in the supplemental text.
RESULTS AND DISCUSSION
The Prominence of Acetogens in the Human Gut Microbiota
Known acetogens comprise a phylogenetically diverse group of bacteria, distributed among 22 different genera belonging to three phyla (17). Most are members of the Firmicutes, which together with the Bacteroidetes constitute the dominant bacterial phyla in the distal human gut (18, 19). The reductive acetyl-CoA pathway (also known as the Wood-Ljungdahl pathway) enables some bacterial species to convert glucose stoichiometrically to acetate (homoacetogenesis) and/or to grow under conditions where CO2 and H2 are the sole sources of carbon and energy (20, 21). In this situation, acetogens use four molecules of H2 to reduce two molecules of CO2, generating ATP and acetate as the main end products of their metabolism (17). Acetogenesis from CO2 via the acetyl-CoA pathway accounts for ∼35% of the acetate produced from glucose in fecal suspensions from subjects that do not carry methanogens (22); acetate in the distal gut is readily absorbed and used in the liver as a substrate for de novo lipogenesis.
Previous attempts at determining the abundance of acetogens in the human gut microbiota were based on growth on H2 and CO2, a trait that it is not conserved among all acetogens (12, 23). In addition, the polyphyletic nature of acetogens precludes the use of a culture-independent 16S rRNA gene-based approach to study them as a functional group (17). Co-occurrence of genes coding for acetyl-CoA synthase (acsB), carbon monoxide dehydrogenase (acsA), and both subunits of the corrinoid iron-sulfur protein (acsC-acsD) is a good marker for the presence of the Wood-Ljungdahl pathway in bacteria (supplemental Fig. S1A) (24). Therefore, we searched the fecal microbiomes of 18 adult females living in the United States (19) for sequences bearing significant homology to each one of these four genes (six sets of adult monozygotic twins and their mothers; average of 100 Mb generated per fecal sample by 454 FLX shotgun pyrosequencing; see supplemental text for details). Sequences matching enzymes of the acetyl-CoA pathway were present in all samples (note that this method targets all acetogens, not just H2-consuming species).
To compare the abundance of the acetyl-CoA pathway relative to other hydrogen consumers, we searched for sequences matching genes specific for methanogens (mcrA; encodes methyl coenzyme M reductase subunit A, a conserved enzyme in the methanogenesis pathway) and sulfate reducers (dsrB; encodes dissimilatory sulfite reductase β-subunit). We detected mcrA in only 22% of the samples and dsrB in 66%. Even in samples where these two genes were present, sequences matching the acetyl-CoA pathway were ∼6–7-fold more abundant (p < 0.005, analysis of variance; supplemental Fig. S1B). These results indicate that (i) acetogenesis is a dominant and consistently represented metabolic process in the distal guts of the sampled individuals and (ii) the three different types of H2 consumers co-exist in at least a subset of these people.
Acetogenesis Pathways Represented in Two Human Gut Acetogens
Despite their abundance in myriad ecosystems, only three known acetogens have been sequenced to date: Moorella thermoacetica, a soil-associated acetogen (24), and two human gut isolates (both Firmicutes), B. hydrogenotrophica (formerly known as Ruminococcus hydrogenotrophicus; strain S5a36; isolated from feces of a non-methane-excreting human subject (25)) and M. formatexigens (formerly known as Bryantella formatexigens; strain I-52T; does not grow with H2 and CO2 and requires formate for acetogenesis (26)). Deep draft assemblies of the genomes of these two organisms were generated by the Washington University Genome Center under the auspices of the Human Gut Microbiome Initiative. In silico analysis of the B. hydrogenotrophica genome disclosed that it possesses all of the genes involved in the Wood-Ljungdahl pathway (supplemental Table S1A and Fig. S1A), including eight genes predicted to encode subunits of iron-only hydrogenases (supplemental Table S1B). However, it has a limited capacity to access polysaccharides (30 glycoside hydrolases were identified using the classification scheme in the CAZy database (27) (supplemental Table S1C)). In contrast, the deep draft assembly of the M. formatexigens genome lacks a complete Wood-Ljungdahl pathway, i.e. it does not possess the selenium-containing subunit of formate dehydrogenase, thereby explaining the requirement of the organism for formate for acetogenesis, at least in vitro (26). Unlike B. hydrogenotrophica, M. formatexigens is richly endowed with glycoside hydrolases (103, which is more than most gut Firmicutes sequenced to date (supplemental Table S1D)).
Developing an RNA-Seq Platform for Microbial Gut Communities
The task of identifying the metabolic niches of these newly sequenced gut acetogens posed a challenge that is pervasive throughout microbiology; the surge in genome sequencing has led to a massive increase in the number of species for which there are no available tools to move beyond descriptions of gene content to assessments of gene function. DNA microarrays currently represent one of the most commonly used tools for this purpose. However, their flexibility is limited: custom arrays may have to be fabricated for each species or strain being characterized. Moreover, problems of cross-hybridization potentially limit the number of community members that can be assayed simultaneously. RNA-Seq provides a more general approach to RNA quantitation that obviates some of the limitations of DNA microarrays. Although RNA-Seq has been successfully applied to several organisms (28, 29), additional problems hinder its applicability to microbial communities, including the need to enrich the mRNA portion of total RNA relative to the much larger pool of rRNA and the need to determine the depth of sequencing necessary to detect transcriptional changes in species present at different relative abundances.
Therefore, we developed a method for microbial mRNA profiling that uses the massively parallel Illumina GA-II instrument and is broadly applicable to sequenced gut microbes. Bacterial mRNA lacks a poly(A) tail, which prevents specific targeting of the mRNA versus the much larger rRNA pool when generating cDNA for sequencing. Our approach combined multiple column washes to deplete 5S rRNA and tRNAs by size fractionation, with a hybridization-based pulldown of 16S and 23S rRNAs using custom-designed biotinylated oligonucleotides that contain short rDNA sequences conserved across a set of 37 human gut-derived sequenced microbial genomes (see supplemental text, “Optimization of RNA Depletion” for details). We achieved up to 13-fold enrichment of mRNA using this method (mRNA represented on average 51% of sequences present in 89 rRNA-depleted microbial RNA samples described below). The rRNA-depleted RNA preparations were then used for random hexanucleotide-primed production of double-stranded cDNA and subsequent sequencing. Read lengths of 36 nucleotides were more than sufficient to accurately assign a transcript (cDNA) to a given locus in the genomes of the sequenced members of a model two-member human gut microbiota, assembled in gnotobiotic mice, composed of a saccharolytic bacterium and one or the other sequenced acetogen (see below and supplemental text). The degree of rRNA depletion achieved allowed us to pool up to 10 double-stranded cDNA samples, each labeled with a unique 4-bp barcode, per lane of the eight-lane Illumina GA-II flow cell for multiplex sequencing.
A total of 89 RNA samples obtained from in vitro and in vivo experiments were analyzed in this fashion. An average of 6.1 ± 2.5 million reads were generated per lane (n = 33 lanes), yielding a total of 7.3 gigabases of sequence data. Subsampling of the microbial RNA-Seq data sets indicated that the correlation between in vitro biological replicates of cultures either of the two human gut acetogens or of the saccharolytic human gut bacterium, Bacteroides thetaiotaomicron, reached an asymptote above 0.9 when the number of mRNA reads/sample was >300,000 (see supplemental text, “Sufficient Sequencing,” and Fig. 1A). The reliability of this method for profiling mRNA was further established by directly comparing RNA-Seq data sets generated from rRNA-depleted samples containing B. thetaiotaomicron RNA with 14 data sets generated from the starting nondepleted total RNA sample with custom Affymetrix GeneChips (these microarrays contain probesets to >98% of predicted genes in the organism's genome). We found a high correlation between both methods (r = 0.86) (Fig. 1B; also see supplemental text, “Comparisons with Affymetrix GeneChips”).
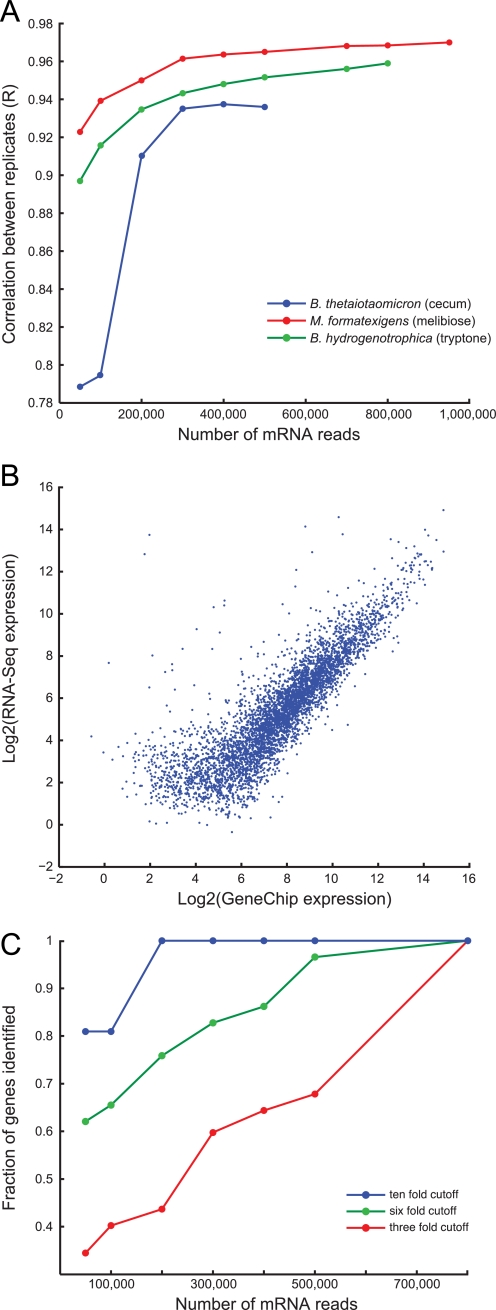
FIGURE 1.
Validation of microbial RNA-Seq. A, the correlation between biological replicates at different sequencing depths reached an asymptote above 0.9 for in vivo samples (blue circles, B. thetaiotaomicron in cecum) and in vitro samples (red circles, M. formatexigens grown on MBf medium with melibiose as a carbon source; green circles, B. hydrogenotrophica grown on MA4 medium with tryptone as a carbon source). B, mRNA abundances for RNA samples prepared from the ceca of B. thetaiotaomicron mono-associated mice (n = 4 samples from 4 animals) were highly correlated (r = 0.86) between Affymetrix GeneChip and RNA-Seq platforms. Comparable results were also found for bi-associations of B. thetaiotaomicron with B. hydrogenotrophica and M. formatexigens (n = 5 replicates each). Most of the discrepancies between the two methods involved transcripts detected at higher levels by RNA-Seq compared with GeneChip. C, the sequencing depth required to detect differential gene expression for microbial community members at different relative abundances was estimated by calculating the overlap between the fraction of genes differentially expressed at a given fold change threshold for a complete sample (M. formatexigens RNA; each of two replicate cultures grown in MBf medium plus melibiose or xylose) versus subsamples of the complete sample. With 200,000 reads, all 10-fold changes were found with zero false positives (blue circles). 6- and 3-fold changes (green and red circles, respectively) required greater sequencing depths to identify >80% of the differentially expressed genes. See Figs. S4 and S5 for additional analyses.
To estimate the necessary sequencing depth for RNA profiling community members at variable abundances in the microbiota, we calculated the overlap between the fraction of genes differentially expressed during growth in vitro at a given fold change threshold for a complete sample (two replicates of 800,000 reads) and subsamples of the complete sample (either 50,000, 100,000, 300,000, 400,000, or 500,000 double-stranded cDNA reads; see supplemental text, “Sufficient Sequencing” for details). More than 80% of the 10-fold differentially expressed genes from the full sample were detectable with only 50,000 mRNA reads (Fig. 1C). Given the sequencing capacity of the Illumina GA-IIx instrument at the time these experiments were performed (∼18 million reads/lane) and our current mRNA enrichment ability, this suggests that the majority of 10-fold expression changes can be detected in all of the species present at 1.7% or more of a complex microbial community.
Characterizing the Metabolic Potential of Acetogens in the Distal Gut Using Gnotobiotic Mice
Groups of 8-week-old germ-free mice were colonized with B. thetaiotaomicron alone or with B. thetaiotaomicron for 2–3 days followed by B. hydrogenotrophica or M. formatexigens for 14 days (n = 4–5 mice/experiment; n = 3 independent experiments). In the presence of B. thetaiotaomicron, both acetogens robustly colonized the distal gut where they represented 13 ± 3% (B. hydrogenotrophica) and 22 ± 2% (M. formatexigens) of the two-member model communities (see Fig. 2 legend for absolute numbers). These levels of colonization are sufficient to detect expression changes in all community members with RNA-Seq.
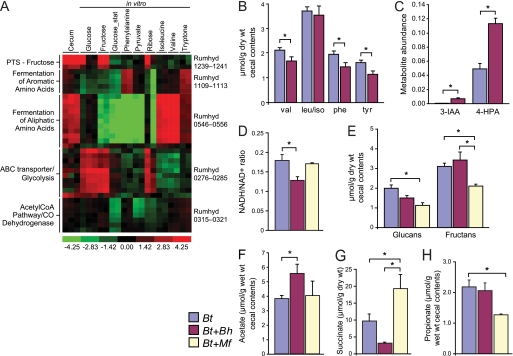
FIGURE 2.
The impact of two human gut acetogens on microbial metabolism in the ceca of gnotobiotic mice. A, multiplex RNA-Seq analysis of the B. hydrogenotrophica transcriptome. The heat map shows selected B. hydrogenotrophica genes, expressed in the ceca of B. thetaiotaomicron (Bt)/B. hydrogenotrophica (Bh) bi-associated mice and in vitro during growth of B. hydrogenotrophica in MA4 medium containing nine different substrates. PPDE > 0.99. Carbon sources were supplied at 1% w/v. The maximal relative expression across a row is red; the minimum is green. stat, stationary phase (note that all other cultures were harvested during mid-log phase). Rumhyd is the locus tag for B. hydrogenotrophica (previously known as R. hydrogenotrophicus). PTS, phosphotransferase system. B, targeted tandem mass spectrometry analysis of amino acid levels. C, nontargeted GC/MS analysis of cecal contents. The data are expressed as unit-less peak areas, normalized to the total ion chromatogram for the cecal sample. 3-IAA, 3-indoleacetic acid; 4-HPA, 4-hydroxyphenylacetic acid. D–H, cecal contents from mice, colonized with either acetogen plus B. thetaiotaomicron or with B. thetaiotaomicron alone, were assayed for NAD+, NADH (D; n = 4–5/group), polyglucose- and polyfructose-containing polysaccharides (E; n = 14–15/group), acetate (F; n = 14–15 animals/group), succinate (G; n = 14–15/group), and propionate (H; n = 14–15/group). The mean values ± S.E. are plotted. *, p < 0.05 based on analysis of variance, except B and C (Student's t test). Quantitative PCR assays revealed the following levels of colonization in the different groups of mice studied: B. thetaiotaomicron mono-association, 7.5 × 1010 ± 1.3 × 1010 genome equivalents/g of wet cecal content, B. thetaiotaomicron/B. hydrogenotrophica bi-association, 6.5 × 1010 ± 9.3 × 109 (B. thetaiotaomicron), 9.4 × 109 ± 2.1 × 109 (B. hydrogenotrophica), and B. thetaiotaomicron/M. formatexigens (Mf) bi-association, 3.1 × 1010 ± 6.3 × 109 (B. thetaiotaomicron), 8.4 ± 109 ± 6.3 × 108 (M. formatexigens); n = 4–5 animals group (three independent experiments).
Analysis of B. hydrogenotrophica in Vivo and in Vitro
We applied microbial RNA-Seq to (i) frozen samples of cecal luminal contents harvested from B. thetaiotaomicron/B. hydrogenotrophica bi-associated mice consuming a standard chow diet at the time of their sacrifice (n = 3) and (ii) samples of in vitro cultures representing nine different conditions (mid-log phase in media containing glucose, fructose, ribose, pyruvate, phenylalanine, valine, isoleucine, or tryptone as carbon sources plus stationary phase in cells grown with glucose) (n = 2–3 cultures/condition; Fig. 2A and supplemental Table S2, A–I). The choice of these carbon sources was guided by a list of 556 B. hydrogenotrophica genes whose expression was defined as significantly increased in the ceca of bi-associated mice compared with growth in vitro with glucose (fold difference>3; PPDE > 0.99; supplemental Table S2A). This list included 16 genes up-regulated in vivo and predicted to be involved in fermentation of aliphatic and aromatic amino acids (Fig. 2A and supplemental Table S2A). By identifying genes that B. hydrogenotrophica uses for growth in medium containing each of these carbon sources, RNA-Seq allowed us to define and interpret the metabolic pathways that it deploys in the ceca of gnotobiotic mice. For example, for in vitro growth on aliphatic amino acids, B. hydrogenotrophica up-regulates genes encoding an ABC transporter and enzymes required for the transformation of valine, leucine, and isoleucine to oxidized branched chain fatty acids (isobutyrate, isovalerate, and 2-methylbutyrate, respectively); these enzymes include an amino acid aminotransferase (Rumhyd0546), a 2-keto acid oxidoreductase (Rumhyd0549–Rumhyd0551), a phosphate acyltransferase (Rumhyd0555), and a fatty acid kinase (Rumhyd0554) (Fig. 2A and supplemental Table S2, C and H). B. hydrogenotrophica also induces a pathway for fermentation of aromatic amino acids in vitro and in vivo that has been described in Archaea, where the transaminated forms of these amino acids are subjected to oxidative decarboxylation by an indolepyruvate ferredoxin oxidoreductase (30) (Fig. 2A and supplemental Table S2D).
Tandem mass spectrometry confirmed that bi-associated mice have significantly lower cecal levels of aliphatic and aromatic amino acids compared with B. thetaiotaomicron mono-associated animals (Fig. 2B and supplemental Table S3A). Furthermore, end products of aromatic amino acid fermentation were present at significantly higher levels in the ceca of the bi-associated mice (Fig. 2C and supplemental Table S3B). Fermentation of aliphatic and aromatic amino acids yields reducing equivalents and CO2 that can be captured via Wood-Ljungdahl pathway for production of acetyl-CoA. Microbial RNA-Seq verified that genes in the pathway are highly expressed in vitro in the presence of phenylalanine, valine, and isoleucine and established that, in the cecum, pathway components account for ∼5% of all expressed B. hydrogenotrophica mRNAs.
A locus encoding an ABC sugar transporter system (Rumhyd0280–Rumhyd0282) plus several genes involved in glycolysis (glucokinase (Rumhyd0283), fructose-bisphosphate aldolase (Rumhyd0284), and phosphofructokinase (Rumhyd0285)) that is highly expressed in the presence of glucose and other simple sugars (i.e. fructose and ribose) during log phase growth in vitro exhibited significantly lower levels of expression in the ceca of bi-associated mice (Fig. 2A). Follow-up GC/MS revealed that levels of simple sugars including glucose, tagatose/fructose/sorbose, and ribose were low and in some cases below the limit of detection in the ceca of bi-associated mice consuming a complex plant polysaccharide-rich chow diet (supplemental Table S3B).
In a primary fermenter, such as B. thetaiotaomicron, accumulation of H2 inhibits reoxidation of NADH. We found that the presence of B. hydrogenotrophica caused a significant decrease in the cecal [NADH]/[NAD+] ratio (Fig. 2D), despite increased consumption of cecal glucans, in bi-compared with mono-associated animals, i.e. more bacterial fermentation occurred with co-colonization (Fig. 2E). In addition, GC/MS revealed that compared with B. thetaiotaomicron mono-associated controls, co-colonization (bi-association) with B. thetaiotaomicron and B. hydrogenotrophica resulted in significantly higher levels of acetate in the cecum (Fig. 2F) and a 10% increase in serum acetate levels (p < 0.05; Student's t test). Integrating our RNA-Seq and metabolic profiling results, we concluded that the increased level of acetate observed in bi-associated compared with B. thetaiotaomicron mono-associated mice is likely a consequence of H2 removal by B. hydrogenotrophica, which promotes bacterial fermentation and acetogenesis from amino acids, fructose, H2, and CO2. Acetogens typically require a higher partial pressure of H2 for efficient consumption compared with methanogens (20), so their ability to use organic carbon sources as well as H2 and CO2 likely conveys an advantage while competing for nutrients in the distal gut.
B. hydrogenotrophica Benefits from the Presence of B. thetaiotaomicron
In the presence of B. thetaiotaomicron, levels of B. hydrogenotrophica colonization are ∼1000-fold greater as judged by quantitative PCR (9.4 × 109 ± 2.1 × 109 genome equivalents/g of wet cecal contents (n = 15 bi-associated mice) versus 8.1 × 106 ± 3.1 × 106 (n = 5 mono-associated animals; p < 0.01, t test)). Removal of H2 by B. hydrogenotrophica allows B. thetaiotaomicron to regenerate NAD+, which can then be used for glycolysis (Fig. 2D). However, cecal levels of B. thetaiotaomicron colonization (Fig. 2 legend) and its transcriptional profile are not significantly impacted by the presence or absence of B. hydrogenotrophica (criteria, fold change > 3; PPDE > 0.99). Note that the cecal levels of B. hydrogenotrophica in mono-associated mice were too low to obtain sufficient microbial biomass from individual animals for RNA-Seq profiling of the acetogen's transcriptome.
Analysis of M. formatexigens in Vivo and in Vitro
The biochemical changes elicited by M. formatexigens in the ceca of B. thetaiotaomicron-containing gnotobiotic mice contrast sharply with the changes caused by the H2-consuming B. hydrogenotrophica. Despite relatively high levels of M. formatexigens colonization (27 ± 2% of the levels achieved by B. thetaiotaomicron; n = 15 bi-associated animals), the concentration of acetate in the ceca and sera of bi-associated animals was not significantly different compared with B. thetaiotaomicron mono-associated controls (Fig. 2F).
GC/MS analysis disclosed that the concentrations of formate in the ceca of B. thetaiotaomicron mono-associated and B. thetaiotaomicron/M. formatexigens bi-associated mice were equivalent (2.7 ± 0.5 mm versus 2.5 ± 0.3, respectively; n = 5 animals assayed/group; p > 0.5 analysis of variance) but 20-fold lower than the concentration shown to result in homoacetogenesis in vitro (i.e. where acetate is the sole end product of fermentation by M. formatexigens) (31). In vitro studies had indicated that at this low level of formate, M. formatexigens ferments carbohydrates to a mixture of succinate, acetate, and lactate in vitro (31). We found that a similar effect occurs in vivo; succinate levels were significantly higher in the ceca of B. thetaiotaomicron/M. formatexigen bi-associated animals compared with B. thetaiotaomicron mono-associated (or B. thetaiotaomicron/B. hydrogenotrophica co-colonized) mice (Fig. 2G). Furthermore, RNA-Seq studies of M. formatexigens growing in vitro in defined minimal medium with glucose revealed that varying formate levels from 0 to 45 mm did not affect the levels of expression of acetyl-CoA pathway components (supplemental text). This observation suggests that the acetyl-CoA pathway is not highly regulated by the addition of its substrates, at least in the presence of these organic carbon sources. A similar conclusion was made based on RNA-Seq assays of the response of B. hydrogenotrophica to the addition of H2 or CO2 to the headspace of in vitro cultures. The molecular details of how acetogens regulate the acetyl-CoA pathway remain obscure (17). A study with M. thermoacetica, a soil acetogen that can use nitrate as an electron acceptor, showed that in some cases the presence of nitrate resulted in a small decrease in the level of mRNA encoding carbon monoxide dehydrogenase and lower enzymatic activity (32, 33).
Compared with B. thetaiotaomicron mono-associated controls, the presence of M. formatexigens produced a modest but statistically significant 2.5-fold decrease (p < 0.05) in the cecal levels of B. thetaiotaomicron (Fig. 2 legend). There was also a concomitant and statistically significant decrease in the cecal concentration of propionate (Fig. 2H), a principal end product of B. thetaiotaomicron fermentation that M. formatexigens does not generate (39) (the M. formatexigens genome lacks genes coding for key enzymes in propionate-generating pathways, such as propionate CoA transferase/lactate CoA transferase, lactoyl-CoA dehydratase (acrylate pathway), and methylmalonyl-CoA decarboxylase (conversion of succinate to propionate)).
To assess whether B. thetaiotaomicron and M. formatexigens compete for similar substrates, we examined the transcriptional responses of M. formatexigens and B. thetaiotaomicron when they co-colonized the ceca of mice, when B. thetaiotaomicron was the sole inhabitant, and when M. formatexigens was in the log phase of anaerobic growth in vitro with glucose as its sole source of carbon. The B. thetaiotaomicron genome contains 261 genes annotated in the CAZy database as encoding known or predicted glycoside hydrolases and polysaccharide lyases; most of these genes occur in genomic clusters, termed polysaccharide utilization loci (PULs); 88 of these PULs have been identified to date, contain 866 genes, and comprise almost 18% of its genome (34, 35). In the presence of M. formatexigens, B. thetaiotaomicron increased the expression of only two PULs relative to mono-association (>3-fold, PPDE > 0.99; supplemental Table S4). One of these PULs is involved in degrading host mucin O-glycans (BT0865–0867), a prominent host substrate that B. thetaiotaomicron forages in vivo (34). There were no down-regulated PULs. Thus, it appears that M. formatexigens, despite its arsenal of glycoside hydrolases (supplemental Table S1D), has a small impact on the carbohydrate metabolism of B. thetaiotaomicron.
Microbial RNA-Seq identified more than 500 M. formatexigens genes whose expression is significantly greater in the ceca of bi-associated mice compared with log phase cultures in MBf medium (defined in the supplemental text) (criteria; >3-fold difference; PPDE > 0.99). Eighteen loci, composed of genes encoding ABC-type sugar transporters and in many cases adjacent co-expressed glycoside hydrolases, were among the most highly up-regulated in vivo (supplemental Table S5A) (note that BLAST analysis against 123 sequenced bacterial genomes, representing various cultured human gut-derived phylotypes, revealed that these modules of transcriptionally co-regulated genes involved in carbohydrate catabolism are commonly found in other Firmicutes but not in Bacteroidetes).
To identify the substrates that activate expression of these loci, we applied RNA-Seq to M. formatexigens during log phase growth in MBf medium containing one of 15 different carbon sources. These substrates were selected based on the predicted functions of genes that exhibited significantly higher expression in vivo versus in vitro with glucose as the sole carbon source, as well as from a panel of 30 substrates that we tested for their ability to support growth of this acetogen in vitro (supplemental Table S6). The results (supplemental Table S5, B–Q) revealed that M. formatexigens has the capacity to degrade naturally occurring plant-derived oligosaccharides, in particular substrates containing α-galactosidic linkages that are not accessible to the host (e.g. stachyose and raffinose). RNA-Seq also confirmed that the metabolism of these oligosaccharides is associated with the induction of a subset of the loci referred to above that encode a predicted ABC transporter, an extracellular substrate binding protein, and one or more membrane-spanning or intracellular glycoside hydrolases.
RNA-Seq indicated that M. formatexigens consumes microbial and host-derived glycans such as N-acetylglucosamine in vivo; three ABC transporters plus enzymes involved in N-acetylglucosamine metabolism (e.g. NAG-deacetylase) that are induced in vitro by N-acetylglucosamine are up-regulated in the cecum (supplemental Fig. S2). An analogous set of transcriptional responses was observed with several mono- and disaccharides including galactose, fructose, xylose, arabinose, cellobiose, and trehalose (supplemental Fig. S2).
Together, these results suggest that B. thetaiotaomicron and M. formatexigens occupy different niches in the distal gut; the former feasts on complex host and plant-derived polysaccharides (e.g. pectins, starch, and mannans) (36), whereas the acetogen consumes mono- and oligosaccharides. GC/MS provided support for this conclusion; oligosaccharides such as cellobiose, lactulose, and lactobionic acid were present in the ceca of B. thetaiotaomicron mono-associated mice but were depleted in bi-associated animals (supplemental Table S3B).
Comparing the Transcriptomes of B. hydrogenotrophica, M. formatexigens, and B. thetaiotaomicron in Feces versus the Cecum
Using feces instead of cecal contents to assess the transcriptomes of members of model human gut microbial communities provides the great advantage of being able to evaluate the niches of component organisms over time and in response to defined perturbations, such as changes in diet or additions of other organisms, in a single animal so that it can serve as its own control. As a prelude to such future studies, we examined how the different microbes included in the present study modified their transcriptomes as they exited the cecum and were freshly expelled in feces. RNA-Seq of B. thetaiotaomicron disclosed a set of genes that are differentially expressed in feces relative to the cecum in mono-associated mice as well as in both groups of bi-associated animals (threshold cut-offs for fold differences in expression, >10 and <−10; PPDE ≥ 0.99; n = 8 fecal RNA samples from 2–3 mice profiled per group; supplemental Table S7, A–C). A B. thetaiotaomicron locus (BT2420–2425) encoding an osmosensitive sensing and K+ transporting system was among the most highly up-regulated in feces compared with cecum (supplemental Fig. S3A and Table S7A). Because potassium serves as an osmoprotectant and accumulates in the microbe in response to elevated external osmolarity (37), this locus likely helps to prevent cellular dehydration in the fecal environment. A similar system was also up-regulated by M. formatexigens in feces relative to cecum (supplemental Table S7D and Fig. S3B). The genome of B. hydrogenotrophica encodes homologs of these genes (Rumhyd3698–3705) that are also expressed at higher levels in feces relative to the ceca of bi-associated animals. However, these changes do not pass our stringent cut-off for significance; in fact, B. hydrogenotrophica did not exhibit any statistically significant differences in gene expression between feces and cecum in bi-associated mice, including genes involved in the acetyl-CoA pathway. Thus, transcriptional changes observed in feces relative to the cecum are mostly stress- and osmotic balance-related, suggesting that microbial gene expression profiles obtained from feces may serve as useful proxies for exploring organismal niches in more proximal regions of the gut.
Prospectus
In this report, we describe a generally applicable approach for characterizing the niches of sequenced members of the human gut microbiota that (i) uses microbial RNA-Seq to profile their transcriptomes in defined locations of the intestines of gnotobiotic mice, (ii) further interprets the resulting data sets through multiplex RNA-Seq-based profiling of the organisms during in vitro growth on substrates whose utilization is suggested by in silico metabolic reconstructions of in vivo mRNA profiling data, and (iii) takes advantage of the RNA-Seq data to guide quantitative mass spectrometric assays of metabolites present in the gut habitat.
Applying this paradigm to B. hydrogenotrophica and M. formatexigens has provided insights about the role of acetogenic bacteria in the mammalian gut. Pioneering work in wood-feeding termites had shown that acetogenesis constitutes a major H2 sink and that production of acetate from H2 and CO2 via the acetyl-CoA pathway makes an important contribution to host nutrition (38–40). In addition, previous studies have indicated that acetogens can thrive in a wide variety of other environments including the intestines of rodents, ruminants, and kangaroos (23, 41, 42). Our findings in gnotobiotic mice are consistent with the notion that a human gut-derived strain of B. hydrogenotrophica can function as a component of a H2 sink in its distal gut habitat. In contrast, the effects documented in vivo with M. formatexigens suggest that at least under the conditions tested (i.e. a simplified community), this bacterium does not have an impact on community redox status (as evidenced by the [NADH]/[NAD+] ratios) and does not generate significant amounts of acetate despite its ability to consume carbohydrates. It is not clear whether the requirement of formate for acetogenesis seen in M. formatexigens is a common trait among human gut acetogens. However, whereas H2 is generally considered as the principal currency for interspecies electron transfer, formate can serve the same role (43). Thus, it seems likely that in the context of complex communities, where more formate may be produced, M. formatexigens may help improve fermentation and produce acetate as the main product of its metabolism (26).
Several reports have suggested that acetogens, methanogens, and sulfate reducers compete with one another for H2 in the human distal colon (10, 11, 44), whereas other studies indicate that they may co-exist at high levels (45). A greater change in Gibbs free energy is associated with the reduction of sulfate by H2 than with the reduction of CO2 by H2 to methane or acetate (46). In addition, under pure culture conditions, the average H2 threshold (i.e. lowest concentration of H2 that can be utilized) of acetogens is significantly higher than the average H2 threshold of SRB and methanogens (20, 47). Our metagenomic analysis disclosed that components of the Wood-Ljundahl pathway for acetogenesis were more broadly and more highly represented in the human families surveyed than components of pathways for methanogenesis or sulfate reduction. In addition, our transcriptional and metabolic analyses in gnotobiotic mice demonstrated that acetogens can access a broad range of substrates for growth, including mono-, di-, tri-, and tetrasaccharides as well as aromatic and aliphatic amino acids. Given these findings, we hypothesize that this broad substrate range confers a fitness advantage to acetogens while competing for H2 and formate, even when methanogens and SRB are present. This hypothesis is supported by studies of termite gut acetogens indicating that their ability to grow mixotrophically (i.e. to simultaneously utilize organic substrates and H2 for energy) confers a growth advantage over H2-consuming methanogens (48, 49). Nonetheless, the dynamic nature of the gut ecosystem (e.g. fluctuations in its chemical environment including pH, dietary substrates, mucus glycans, peristaltic movement of its luminal contents, and morphologic variations along its cephalocaudal axis) may provide a variety of ecological niches that favor the co-existence of different types of acetogens and other classes of H2 consumers.
Nutritional status is among the most important, modifiable determinants of global human health. The nutritional value of food is not an absolute entity but rather is influenced in part by the collection of microbes present in the guts of consumers. Understanding how the gut microbiota affects the nutritional status of a host requires that we determine the metabolic niches of key microbial community members, such as H2-consuming bacteria and archaeons. Characterizing interactions among acetogens, methanogens, and SRB, the niches they occupy in the presence of various other gut bacterial phylotypes as a function of diet, and their metabolic impact on the host may provide new therapeutic avenues for enhancing our nutritional status and modifying our energy balance. Creating a defined human gut microbiota in gnotobiotic mice composed of sequenced members with or without various groups of H2 consumers, coupled with RNA-Seq and GC/MS analyses, should be useful for conducting proof-of-principle and proof-of-mechanism tests of this idea.
Supplementary Material
Acknowledgments
We thank David O'Donnell, Maria Karlsson, and Sabrina Wagoner for help with various aspects of gnotobiotic husbandry; Jill Manchester for assistance with biochemical assays; Jessica Hoisington-López for participation in cDNA sequencing; Bernard Henrissat for CAZy annotations; and Eric Martens, Henning Seedorf, Liz Hansen, and Andrew Goodman for helpful comments during the course of these studies. Stephan Baumann (Agilent Technologies, Santa Clara, CA) provided substantial assistance with nontargeted metabolite profiling by GC/MS.
This work was supported, in whole or in part, by National Institutes of Health Grants DK70977, DK078669, and DK30292. This work was also supported by the Crohn's and Colitis Foundation of America.
RNA-Seq data have been deposited in Gene Expression Omnibus (GEO) under accession number GSE 21906.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S8, and Figs. S1–S5.
- SRB
- sulfate-reducing bacteria
- GC/MS
- gas chromatography-mass spectrometry
- microbial RNA-Seq
- massively parallel sequencing of expressed microbial mRNAs
- PUL
- polysaccharide utilization locus
- PPDE
- posterior probability of differential expression.
REFERENCES
- 1.Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. (2008) Nat. Rev. Microbiol. 6, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. (2008) Nat. Rev. Microbiol. 6, 121–131 [DOI] [PubMed] [Google Scholar]
- 3.Macfarlane S., Macfarlane G. T. (2003) Proc. Nutr. Soc. 62, 67–72 [DOI] [PubMed] [Google Scholar]
- 4.Wolin M. J., Miller T. L. (1983) Fed. Proc. 42, 109–113 [PubMed] [Google Scholar]
- 5.McNeil N. I. (1984) Am. J. Clin. Nutr. 39, 338–342 [DOI] [PubMed] [Google Scholar]
- 6.Attene-Ramos M. S., Wagner E. D., Plewa M. J., Gaskins H. R. (2006) Mol Cancer Res. 4, 9–14 [DOI] [PubMed] [Google Scholar]
- 7.Ferchaud-Roucher V., Pouteau E., Piloquet H., Zaïr Y., Krempf M. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E716–E720 [DOI] [PubMed] [Google Scholar]
- 8.Havlikova M., Kroeze C., Huijbregts M. A. (2008) Sci. Total Environ. 396, 121–131 [DOI] [PubMed] [Google Scholar]
- 9.Nakamura N., Lin H., McSweeney C. S., Mackie R. I., Gaskins H. R. (2010) Annu. Rev. Food Sci. Technol. 1, 363–395 [DOI] [PubMed] [Google Scholar]
- 10.Gibson G. R., Cummings J. H., Macfarlane G. T. (1988) J. Appl. Bacteriol. 65, 241–247 [DOI] [PubMed] [Google Scholar]
- 11.Christl S. U., Gibson G. R., Cummings J. H. (1992) Gut 33, 1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dore J., Pochart P., Bernalier A., Goderel I., Morvan B., Rambaud J. (1995) FEMS Microbiol. Ecol. 17, 279–284 [Google Scholar]
- 13.Blaxter K. L., Clapperton J. L. (1965) Br. J. Nutr. 19, 511–522 [DOI] [PubMed] [Google Scholar]
- 14.Williams Y. J., Popovski S., Rea S. M., Skillman L. C., Toovey A. F., Northwood K. S., Wright A. D. (2009) Appl. Environ. Microbiol. 75, 1860–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonty G., Joblin K., Chavarot M., Roux R., Naylor G., Michallon F. (2007) Appl. Environ. Microbiol. 73, 6391–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., Lozupone C. A., Knight R., Gordon J. I. (2009) Cell Host Microbe 6, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake Harold K. K., Matthies Carola (2006) in The Prokaryotes (Falkow S., Rosenberg E., Schleifer K. H., Stackenbrandt E., Dworkin M. eds) pp. 354–420 [Google Scholar]
- 18.Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. (2005) Science 308, 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. (2009) Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclerc M., Bernalier A., Donadille G., Lelait M. (1997) Anaerobe 3, 307–315 [DOI] [PubMed] [Google Scholar]
- 21.Kerby R., Zeikus J. G. (1983) Curr. Microbiol. 8, 27–30 [Google Scholar]
- 22.Miller T. L., Wolin M. J. (1996) Appl. Environ. Microbiol. 62, 1589–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doré J., Morvan B., Rieu-Lesme F., Goderel I., Gouet P., Pochart P. (1995) FEMS Microbiol. Lett. 130, 7–12 [DOI] [PubMed] [Google Scholar]
- 24.Pierce E., Xie G., Barabote R. D., Saunders E., Han C. S., Detter J. C., Richardson P., Brettin T. S., Das A., Ljungdahl L. G., Ragsdale S. W. (2008) Environ. Microbiol. 10, 2550–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernalier A., Willems A., Leclerc M., Rochet V., Collins M. D. (1996) Arch. Microbiol. 166, 176–183 [DOI] [PubMed] [Google Scholar]
- 26.Wolin M. J., Miller T. L., Collins M. D., Lawson P. A. (2003) Appl. Environ. Microbiol. 69, 6321–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoder-Himes D. R., Chain P. S., Zhu Y., Wurtzel O., Rubin E. M., Tiedje J. M., Sorek R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3976–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passalacqua K. D., Varadarajan A., Ondov B. D., Okou D. T., Zwick M. E., Bergman N. H. (2009) J. Bacteriol. 191, 3203–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mai X., Adams M. W. (1994) J. Biol. Chem. 269, 16726–16732 [PubMed] [Google Scholar]
- 31.Wolin M. J., Miller T. L. (1993) Appl. Environ. Microbiol. 59, 3551–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arendsen A. F., Soliman M. Q., Ragsdale S. W. (1999) J. Bacteriol. 181, 1489–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fröstl J. M., Seifritz C., Drake H. L. (1996) J. Bacteriol. 178, 4597–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens E. C., Chiang H. C., Gordon J. I. (2008) Cell Host Microbe 4, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens E. C., Koropatkin N. M., Smith T. J., Gordon J. I. (2009) J. Biol. Chem. 284, 24673–24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. (2005) Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 37.Laimins L. A., Rhoads D. B., Epstein W. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 464–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breznak J. A. (1994) in Acetogenesis (Drake H. L. ed) pp. 303–330, Chapman & Hall, New York [Google Scholar]
- 39.Breznak J. A., Switzer J. M. (1986) Appl. Environ. Microbiol. 52, 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odelson D. A., Breznak J. A. (1983) Appl. Environ. Microbiol. 45, 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prins R. A., Lankhorst A. (1977) FEMS Microbiol. Lett. 1, 255–258 [Google Scholar]
- 42.Ouwerkerk D., Maguire A., McMillen L., Klieve A. (2007) Recent Adv. Anim. Nutr. Austr. 16, 101–104 [Google Scholar]
- 43.Boone D. R., Johnson R. L., Liu Y. (1989) Appl. Environ. Microbiol. 55, 1735–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibson G. R., Macfarlane G. T., Cummings J. H. (1988) J. Appl. Bacteriol. 65, 103–111 [DOI] [PubMed] [Google Scholar]
- 45.Stewart J. A., Chadwick V. S., Murray A. (2006) Lett. Appl. Microbiol. 43, 58–63 [DOI] [PubMed] [Google Scholar]
- 46.Thauer R. K., Jungermann K., Decker K. (1977) Bacteriol. Rev. 41, 100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cord-Ruwisch R., Seitz H. J., Conrad R. (1988) Arch. Microbiol. 149, 350–357 [Google Scholar]
- 48.Breznak J. A., Switzer-Blum J. M. (1991) Arch. Microbiol. 156, 105–110 [Google Scholar]
- 49.Graber J. R., Breznak J. A. (2004) Appl. Environ. Microbiol. 70, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.