Abstract
Our recent work identified store-operated Ca2+ entry (SOCE) as the critical Ca2+ source required for the induction of human myoblast differentiation (Darbellay, B., Arnaudeau, S., König, S., Jousset, H., Bader, C., Demaurex, N., and Bernheim, L. (2009) J. Biol. Chem. 284, 5370–5380). The present work indicates that STIM2 silencing, similar to STIM1 silencing, reduces myoblast SOCE amplitude and differentiation. Because myoblasts in culture can be induced to differentiate into myotubes, which spontaneously contract in culture, we used the same molecular tools to explore whether the Ca2+ mechanism of excitation-contraction coupling also relies on STIM1 and STIM2. Live cell imaging of early differentiating myoblasts revealed a characteristic clustering of activated STIM1 and STIM2 during the first few hours of differentiation. Thapsigargin-induced depletion of endoplasmic reticulum Ca2+ content caused STIM1 and STIM2 redistribution into clusters, and co-localization of both STIM proteins. Interaction of STIM1 and STIM2 was revealed by a rapid increase in fluorescence resonance energy transfer between CFP-STIM1 and YFP-STIM2 after SOCE activation and confirmed by co-immunoprecipitation of endogenous STIM1 and STIM2. Although both STIM proteins clearly contribute to SOCE and are required during the differentiation process, STIM1 and STIM2 are functionally largely redundant as overexpression of either STIM1 or STIM2 corrected most of the impact of STIM2 or STIM1 silencing on SOCE and differentiation. With respect to excitation-contraction, we observed that human myotubes rely also on STIM1 and STIM2 to refill their endoplasmic reticulum Ca2+-content during repeated KCl-induced Ca2+ releases. This indicates that STIM2 is a necessary partner of STIM1 for excitation-contraction coupling. Thus, both STIM proteins are required and interact to control SOCE during human myoblast differentiation and human myotube excitation-contraction coupling.
Keywords: Calcium, Calcium Imaging, Calcium Intracellular Release, Cell Differentiation, Skeletal Muscle, Contraction, Myogenesis, STIM1, STIM2, Excitation
Introduction
STIM1 and STIM2 are endoplasmic reticulum (ER)2 transmembrane proteins that are activated by a drop in Ca2+ content in the ER (2–5). Once activated, STIM1 and STIM2 trigger a Ca2+ influx (also called store-operated Ca2+ entry (SOCE)) through Ca2+-selective Orai channels located at the plasma membrane (6–13). This Ca2+ influx restores the ER Ca2+ content (2–4, 10, 12, 14–26).
Several studies examined whether STIM2 had a specific role, distinct from STIM1, but no clear answer has emerged yet. A lower Ca2+-activation threshold of STIM2 as compared with STIM1 has been suggested (2), although N-terminal Ca2+-binding affinity seems to be similar for STIM1 and STIM2 (27–29). In STIM2 knock-out mice, and also in several cell types in which STIM2 has been silenced, SOCE amplitude is only slightly reduced. This could reflect either a smaller activation of Orai1 by the N-terminal part of STIM2 or a lower expression of STIM2 as compared with STIM1 (3, 4, 24, 30). In other studies, overexpression of STIM2 has been reported to inhibit SOCE (31) or, on the contrary, to restore SOCE in cell types with STIM1 deficiency (4, 32). Hence, findings on STIM2 are still controversial.
Post-natal myogenesis critically relies on Ca2+ influx through the SOCE pathway (1, 33–37). We recently established that the initiation of myoblast differentiation requires, likely among other Ca2+ sources, STIM1/Orai1-dependent cytosolic Ca2+ signals, which are amplified by a plasma membrane hyperpolarization (1, 33, 38, 39). Although distinct mechanisms control postnatal muscle formation and embryonic myogenesis (40), SOCE may be involved in both processes as mice lacking STIM1 die shortly after birth from myopathy (41). Concerning more mature muscle cells, recent evidences show that refilling of Ca2+ stores during repeated muscle contractions not only relies, as traditionally thought, on Ca2+ re-uptake by sarcoplasmic-endoplasmic reticulum calcium ATPase to the ER, but also on a functional STIM1-dependant SOCE (35, 41–45). Accordingly, STIM1 knock-out mice suffer from a muscular contractile defect, and patients with STIM1 deficiency present a muscular hypotonia (32, 46). Whether STIM2 is also involved is still unknown. The present work explores the involvement of STIM2 in human myoblast Ca2+ homeostasis and differentiation and in human myotube excitation-induced Ca2+ release.
EXPERIMENTAL PROCEDURES
Cell Culture
Muscle samples, cell dissociation, and clonal culture from satellite cells were prepared as previously described (33, 47). Human muscle samples were obtained from eight children (operated for clubfoot and less than 4 years old) without any known neuromuscular disease. Nine different satellite-cell clones were used in this study. Differentiation was assessed by quantification of nuclear expression of the myogenic transcription factor MEF2 and myogenin (39). Plasmids (STIM1-YFP from Addgene 19754 (48), STIM1-CFP from Addgene 19755, STIM2-YFP from Addgene 18862, STIM1-wt from ORIGENE Technologies, and STIM2-wt from Addgene 18868) were transfected by electroporation with Amaxa Nucleofector II device (Lonza). Following the manufacturer's protocol, a suspension of 5 × 105 myoblasts, 2 μg of plasmid DNA and/or 0.1–0.2 nm of small interfering RNA (siRNA) were electroporated. The transfection efficiency, assessed by fluorescence-activated cell sorting for STIM1-YFP and STIM2-YFP, was 83 ± 7% and 70 ± 2%, respectively (six independent experiments).
siRNA Knockdown
Myoblasts were transfected in suspension by incubating 4 × 105 cells in a solution containing 500 μl of Opti-MEM, 3 μl of Lipofectamine RNAiMax (Invitrogen), and 20 pmol of a specific siRNA (Ambion, Qiagen, Invitrogen) according to the manufacturers' protocols (Invitrogen). The transfection efficiency assessed by Block-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) measurements was ∼90%. Optimal quantities of Lipofectamine and siRNA were defined by evaluating the strongest effect of siRNA STIM2 on SOCE (see below) and were constantly used thereafter. This optimal effect was obtained 48–60 h post-transfection. Three different siSTIM2 siRNAs from Invitrogen were used: HSS183972, CACAUGAAGUAGAAGUGCAAUACUA; HSS183973, UCUCUCUGAGUUGACAACUUGUUUA; and HSS183974, CCAGAAUAAGCAGCAUCCCACAUGA.
These different siRNAs gave similar results on SOCE, differentiation inhibition, and protein decrease analyzed by Western blots. To simplify, all results shown in this study were obtained with HSS183974 siSTIM2. STIM1 siRNA was the previously described siSTIM1a (1). The siRNA siAllstar Negative Control from Qiagen was used as a negative control.
Immunostaining
Myoblasts were fixed and stained with the appropriate fluorescence markers as described previously (33). Anti-MEF2 antibody (1:200, sc-313, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-myogenin antibody (1:900, clone F5D, BD Biosciences) were used. Secondary antibodies were Alexa 488-labeled goat anti-mouse IgG (1:500, Molecular Probes, Eugene, OR) and Alexa 546-labeled goat anti-rabbit IgG (1:500, Molecular Probes). DAPI (100 ng/ml, Sigma) was used to localize nuclei. Images were acquired using a Zeiss Axiovert S100 TV microscope equipped with an Optoscan monochromator (Cairn Research Ltd., Faversham, UK) and a motorized stage (Märzhäuser Wetzlar GmbH, Wetzlar, Germany). DAPI, Alexa 488, and Alexa 546 fluorescence values were acquired by changing the excitation wavelengths with our Optoscan monochromator and using a triple band filter set (XF2050 dichroic mirror and XF3063 emission filter from Omega Optical Inc., Brattleboro, VT). A motorized stage was used to automatically acquire 10 random fields in each condition (200–400 cells/condition). Analysis to evaluate labeled nuclei was carried out using the Metamorph 7.5.6 software (Molecular Devices Corp., Visitron Systems GmbH, Puchheim, Germany). A cluster of labeled nuclei inside a myotube that clearly expressed MEF2 or myogenin was used to define the threshold above which nuclei were considered as positive for the expression of MEF2 or myogenin, respectively (10 random fields for each clone).
Western Blots and Immunoprecipitations
Western blots were performed as previously described (39). Briefly, myoblasts were lysed using the Nonidet P-40 Extraction Buffer (Invitrogen). Total proteins were separated on a SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated in T-TBS (0.1% Tween 20, 20 mm Tris-HCl, pH 7.5, 137 mm NaCl) and 5% nonfat milk. Blots were incubated with the primary antibodies diluted in T-TBS and nonfat milk as follows: mouse monoclonal antibody anti-GOK/STIM1 1:500 (BD Biosciences), rabbit anti-STIM1 (N- and C-terminal, Sigma-Aldrich) antibodies, rabbit polyclonal anti-STIM2 antibody (1:500, ProSci Inc.), and mouse monoclonal antibody against α-tubulin (clone DM1A, Sigma) 1:10,000. Blots were incubated with horseradish peroxidase-conjugated goat anti-mouse diluted 1:6,000 (Bio-Rad) or with horseradish peroxidase-conjugated goat anti-rabbit diluted 1:6,000 (Bio-Rad), respectively. Antibodies were revealed using ECL reagents and Hyperfilm ECL (Amersham Biosciences). Optiquant 03.00 (Packard instrument Co.) software was used to quantify the level of protein expression. STIM1 immunoprecipitation was performed by lysing ∼2 million myoblasts in Nonidet P-40 buffer. Samples were pre-cleared with agarose beads. Cells were then incubated overnight with IgG2A control antibody or GOK/STIM1 antibody and agarose beads. Precipitate samples were then washed three times before performing the Western blots as formerly described.
Cytosolic Calcium Measurements and Fluorescence Imaging
Myoblasts grown on coverslips were loaded for 30 min at room temperature with the cell-permeant fluorescent Ca2+-indicator Fura-2-AM (Biotium Inc., Chemie Brunschwig AG, Basel, Switzerland). The Fura-2-AM preparation was done as previously described (33). Ratiometric images of Fura-2 fluorescence were monitored using an Axiovert S100 TV microscope (Zeiss AG, Feldbach, Switzerland) equipped with an Optoscan monochromator (Cairn Research Ltd.), which rapidly changed the excitation wavelengths between 340 and 380 nm. Fluorescence emissions were captured through a 510WB40 filter (Omega Optical Inc.) using a CoolSNAP HQ digital camera (Photometrics-Ropper Scientific, Tucson, AZ). Image acquisition and ratio analysis were carried out using the Metafluor 6.3r7 software (Molecular Devices, Visitron Systems GmbH). Rmax was evaluated in all conditions (using a solution containing 4 μm ionomycin and 5 mm Ca2+), ruling out any possible non-linearity between data obtained in myoblasts transfected with different plasmids or siRNA. No significant difference between the various Rmax was observed. Monitoring of cluster formation was performed in myoblasts transiently transfected with either STIM1-CFP or STIM2-YFP. Fluorescence emissions of the two probes were performed using a triple band beamsplitter designed for CFP/YFP/RFP and an emission filterwheel (Ludl Electronic Products) that change emission filters for CFP (470/24 nm) and YFP (535/30 nm) (Chroma Technology Corp., Rockingham VT). Long term experiments following STIM1-CFP and STIM2-YFP cluster formation were monitored using a 40× oil immersion objective at a rate of 0.2 Hz at 37 °C in a CO2-controlled atmosphere, using a micro-incubator (Incubator S, CTI controller 3700 digital and Tempcontrol 37-2, digital, two channels, PeCon GmbH, Erbach, Germany). Analysis of cluster formation was carried out using morphometric journals with Metamorph 7.5.6 (Molecular Devices).
Confocal FRET Measurements
Live cell confocal image acquisition of FRET between STIM1-CFP and STIM1-YFP or STIM2-YFP was performed using a spinning disc confocal microscope. The ratio of transfected plasmids (CFP/YFP) was 1:2. The incident laser beam (440 nm, CUBE 440 nm, Coherent, Inc.) was injected into a Yokogawa spinning disc confocal scan head (QLC100, Visitech International) mounted on an inverted microscope (Zeiss Axiovert 200M). Fluorescence images of CFP (480 nm) and FRET (535 nm) were simultaneously captured using a split view device (Optical Insight Inc.), which splits the two fluorescence emissions of the two probes on the two halves of the chip of a Cascade II 16-bit cooled EMCCD frame transfer camera (Photometrics-Ropper Scientific). Images were acquired, and ratio analysis was carried out using Metafluor 6.3r7 software (Molecular Devices).
Laser Scanning Confocal Image Acquisition
For better morphometric studies, confocal images acquisitions of fixed specimens were made on an LSM510 Meta Laser scanning confocal microscope (Zeiss AG). STIM1-CFP and STIM2-YFP signals were acquired using the 458 and 514 nm laser lines, respectively.
Statistics
Error bars are ±S.D. except for Fura-2 measurements in which ±S.E. is indicated.
RESULTS
STIM2 Silencing Inhibits Myoblast Differentiation and SOCE
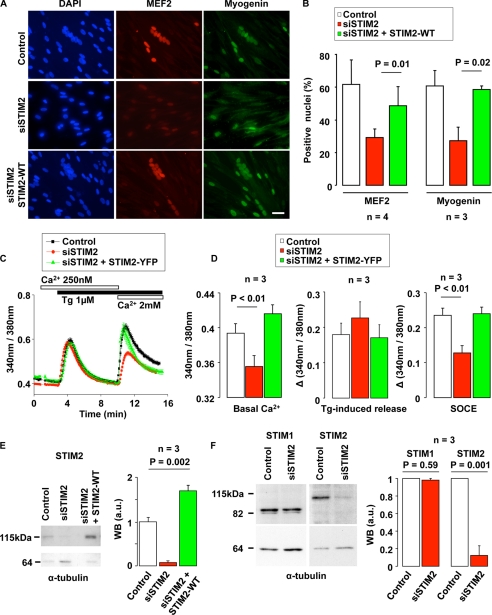
We assessed the role of STIM2 in muscle differentiation and SOCE by inhibiting STIM2 expression in cultured myoblasts derived from single human satellite cells. Myoblasts, grown in proliferation medium, were incubated for 5 h with an siRNA-targeting STIM2 (siSTIM2) and, 42 h after, switched to differentiation medium to induce their differentiation into multinucleated myotubes. As shown by immunofluorescence staining, incubation of myoblasts with siSTIM2 decreased the formation of nuclei clusters, a hallmark of cell differentiation and fusion (Fig. 1A, left images, DAPI staining), as well as the expression of the transcription factors MEF2 and myogenin, early markers of myoblast differentiation (39) (Fig. 1A). MEF2 and myogenin were expressed, respectively, by 61 ± 16% and 60 ± 10% of control myoblasts, and only in 27 ± 5% and 29 ± 8% of siSTIM2-treated cells (Fig. 1B). To exclude the possibility that the inhibition of myogenesis observed in myoblasts transfected with siSTIM2 was due to off-target or nonspecific effects of the siRNA, we attempted to rescue the differentiation process by re-expressing STIM2 in myoblasts treated with siSTIM2. As shown in Fig. 1A (bottom images) and Fig. 1B (right bars), MEF2 and myogenin expression as well as nuclei cluster formation were restored in siSTIM2-treated cells co-transfected with a plasmid encoding for the wild-type STIM2 protein.
FIGURE 1.
STIM2 silencing impedes human myoblast differentiation and SOCE. A, myoblasts were transfected either with siSTIM2 and an empty plasmid (pcDNA3), or with siSTIM2 and a plasmid coding for the wild-type STIM2 protein (STIM2-WT). After transfection, myoblasts were kept for 2 days in proliferation medium and 2 more days in differentiation medium. Nuclei were stained in blue using DAPI. MEF2 and myogenin were stained in red and green, respectively. When siSTIM2 and/or STIM2-WT were not present, myoblasts were transfected with a siRNA negative control (siAllstar) and/or an empty plasmid (pcDNA3). The scale bar is 25 μm. B, fraction of nuclei expressing MEF2 and myogenin assessed in three different myoblast clones (four experiments for MEF2 and three for myogenin). The conditions were the same as in A. C, cytoplasmic Ca2+ was monitored with Fura-2 in proliferating myoblasts, 2 days after co-transfection of either siSTIM2 and a control plasmid (EGFP-N3), or siSTIM2 and a plasmid coding for STIM2-YFP. Intracellular Ca2+ stores were depleted with 1 μm thapsigargin (Tg) in a medium containing 250 nm Ca2+, and thereafter 2 mm Ca2+ were added to reveal SOCE. D, effects of siSTIM2 (with EGFP-N3 as a control plasmid) and of siSTIM2 together with a plasmid coding for STIM2-YFP on basal Ca2+ level, peak amplitude of Tg-induced Ca2+ response, and SOCE response (siAllstar and EGFP-N3 plasmid were used in control conditions). Mean of three experiments in three different clones. E, Western blot illustrating STIM2 disappearance 2 days after siSTIM2 transfection, and STIM2 reappearance when a STIM2-WT plasmid was co-transfected with the siSTIM2. α-Tubulin was used as a loading control. Quantification was obtained out of three different myoblast clones. Mean control STIM2 expression was normalized to 1 (arbitrary units (a.u.)), and, in each experiment, STIM2 expression was normalized to α-tubulin expression. F, Western blot showing the preservation of STIM1 2 days after transfection of siSTIM2. Quantification and normalization were done as in E.
Because the role of STIM2 in SOCE activation is controversial, we evaluated the impact of STIM2 silencing on cell Ca2+ handling in proliferating myoblasts 48 h after siSTIM2 transfection using the fluorescent Ca2+-indicator Fura-2. As shown in Fig. 1C, STIM2 silencing decreased both the resting Ca2+ level and the amplitude of SOCE evoked by re-addition of 2 mm Ca2+ after depletion of the Ca2+ stores with thapsigargin (Tg, 1 μm). The resting Ca2+ level was decreased from a ratio of 0.39 ± 0.01 to 0.35 ± 0.01 by STIM2 silencing, and the peak SOCE response of the Ca2+ influx was reduced by 45 ± 7% (Fig. 1, C and D, n = 3 independent experiments, p < 0.002 for all conditions). Fig. 1 (C and D) also indicate that, as expected, a genetic rescue of STIM2 normalized both the resting Ca2+ level and the amplitude of SOCE. Interestingly, STIM2 silencing did not decrease the peak response of the Tg-induced Ca2+ release (Fig. 1D, middle red bar, n = 3 independent experiments).
The effect of siSTIM2 on protein expression of STIM2 was then assessed by Western blot. Fig. 1E shows that the amount of STIM2, at the onset of differentiation, was reduced by 87 ± 11% in cells treated with siSTIM2, and that STIM2 protein expression was restored by a genetic rescue. Because STIM1 and STIM2 sequences are highly homologous, we also confirmed the specificity of the siSTIM2 by showing that STIM1 expression was unaffected by siSTIM2 (Fig. 1F, n = 3 independent experiments). Finally, two additional siRNAs against STIM2 were tested (see “Experimental Procedures”) and yielded similar results on STIM1 and STIM2 expression, myogenesis inhibition, and Ca2+ homeostasis, confirming the specificity of the siSTIM2 used in this study (data not shown). Altogether, these results indicate that STIM2 is involved in myoblast Ca2+ handling and is required for myoblast differentiation.
STIM1 and STIM2 Form Discrete Clusters during Initiation of Myoblast Differentiation
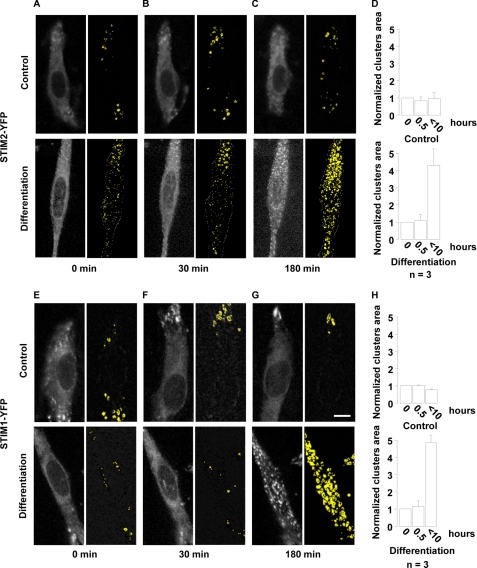
Depletion of ER Ca2+ stores leads to STIM redistribution into clusters (2, 49). To test whether STIM proteins were recruited during myogenesis, we followed the distribution of YFP-conjugated STIM1 and STIM2 using live cell imaging with a wide-field microscope equipped with a micro-incubator.
Fig. 2(A and B) shows that STIM2 was diffusely distributed in proliferation (control) and during the first 30 min of differentiation with only a few clusters detectable (50). During the initiation of differentiation, however, STIM2 formed discrete clusters of progressively increasing size (Fig. 2C, upper images). Initiation of cluster formation never occurred within the first hour of differentiation. Cluster formation could last between 2 and 10 h depending on the clonal cultures, and, in addition, myoblasts within one culture were not synchronized. Nonetheless, lower magnification images showed that clusters appeared eventually in ∼50% of the cultured myoblasts (data not shown). Quantification of cluster formation yielded that the total area of clusters was increased by a factor of 4.3 ± 1 during initiation of differentiation (Fig. 2D, n = 6 independent experiments). Fig. 2 (E–G) shows that STIM1 clusters formed with a similar pattern when differentiation was induced (n = 4 independent experiments). The integrated area of STIM1 clusters increased by a factor 4.8 ± 0.4 with differentiation (Fig. 2H). This characteristic cluster formation suggests that the ER Ca2+ content is recruited during the initial phase of myoblast differentiation. Because we did not observe cyclic cluster formation, we presume that STIM molecules are permanently activated during this early phase of differentiation and probably reflect recurrent store depletions during the initiation of post-natal myogenesis.
FIGURE 2.
STIM1 and STIM2 form characteristics clusters when differentiation is initiated. Myoblasts were transfected with STIM1-YFP or STIM2-YFP and kept for 2 days in proliferation medium. On the day of the experiment, live cell imaging was started in proliferation medium for 2 h and, at time = 0 min and continued in either differentiation medium or a proliferation medium containing 2 mm Ca2+ (control). The scale bar for all images (in G) represents 15 μm. In A–C: Left images: STIM2-YFP fluorescence in differentiation or proliferation (control) conditions. Right images: visualization of clusters using an automatic detection of fluorescence intensity variations called “by edges” (Metamorph 7.5.6, Molecular Devices). Clusters (STIM2-YFP clustering) are shown in yellow. D, quantification of STIM2-YFP clusters are measured in three independent experiments. Results were normalized to the area measured in proliferation conditions. E–H, same experiments as in A–D, except that STIM1-YFP was used instead of STIM2-YFP.
Activated STIM1 and STIM2 Interact inside Clusters
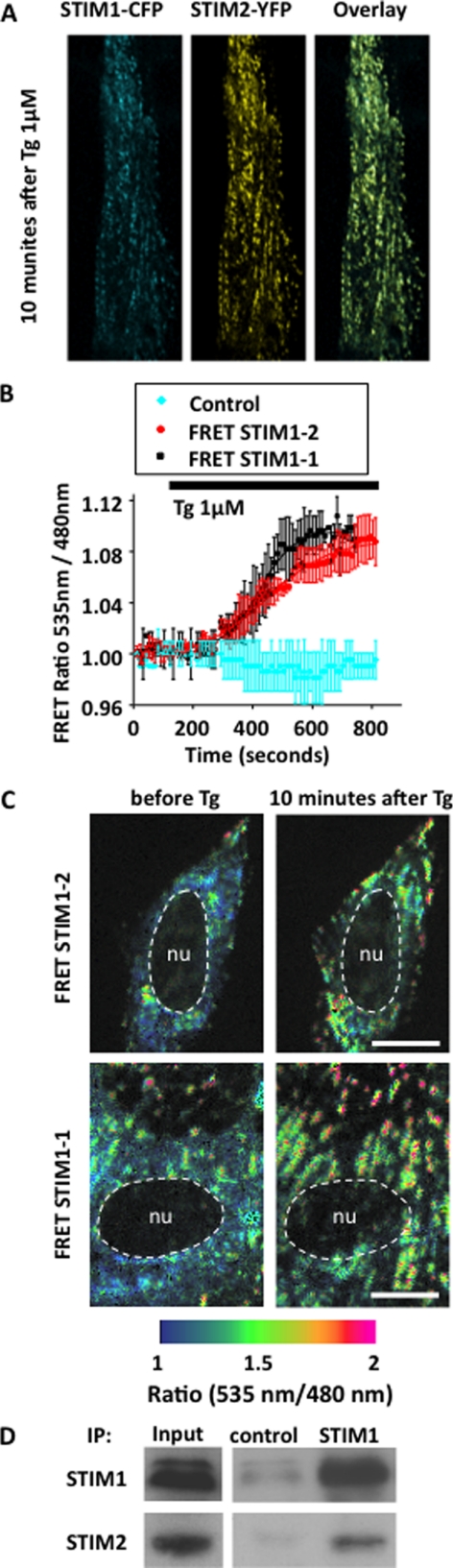
Although STIM1 and STIM2 function independently (4, 32), SOCE involves a synergistic interaction of both proteins as normal SOCE amplitude is greater than the addition of residual SOCE in respective STIM protein silencing (1, 4, 51). Because we observed that both STIM1 and STIM2 form clusters at the beginning of myoblast differentiation, we wondered whether STIM1 and STIM2 molecules could interact together when activated. To follow very accurately and simultaneously STIM1 and STIM2 cluster formation, we fully activated STIM molecules within minutes using 1 μm Tg.
Fig. 3A illustrates confocal images of myoblast co-expressing STIM1-CFP and STIM2-YFP, 10 min after the application of 1 μm Tg. As expected, Tg induced cluster formation of both STIM proteins. Fig. 3A also demonstrates that clusters of STIM1-CFP optically co-localize with clusters of STIM2-YFP (n = 3 independent experiments). To confirm STIM1 and STIM2 proximity, we monitored FRET between the two proteins. Myoblasts co-expressing STIM1-CFP and either STIM1-YFP or STIM2-YFP showed in both cases a rapid Tg-induced increase in FRET (Fig. 3B, n = 3 independent experiments). Fig. 3C shows cluster formation of both STIM proteins, and also that FRET ratio increased mainly inside clusters. Interestingly, the increase in FRET measured on the entire confocal cell slice was identical for STIM1-STIM1 and STIM1-STIM2 interactions, suggesting that a close proximity of both proteins was as essential during SOCE. The proof that SOCE in myoblast relies on a direct interaction between both proteins came from co-immunoprecipitation experiments. As seen in Fig. 3D, by using an antibody against STIM1, endogenous STIM2 co-immunoprecipitated with endogenous STIM1 (n = 3 independent experiments). Co-immunoprecipitations were performed using either myoblasts or myotubes, before and after addition of 1 μm thapsigargin. No obvious difference was observed between proliferation and differentiation conditions, nor before and after thapsigargin addition (data not shown).
FIGURE 3.
Redistribution and interaction between STIM1-CFP and STIM2-YFP induced SOCE activation. A, confocal images of myoblasts co-expressing STIM1-CFP and STIM2-YFP. Cultures were fixed 10 min after the application of 1 μm Tg. The right image shows that STIM1 and STIM2 clusters are nicely co-localized. C, myoblasts were co-transfected 24 h before images acquisition with STIM1-CFP and either STIM1-YFP or STIM2-YFP. The FRET ratio between STIM1-CFP and either STIM2-YFP (upper images) or STIM1-YFP (lower images) before and 10 minutes after 1 μm Tg application are shown. Scale bars represent 10 μm. B, time course of FRET increases between STIM1-CFP and either STIM1-YFP (black) or STIM2-YFP (red) following the application of 1 μm Tg (one representative result out of three independent experiments). Values were normalized to resting FRET before Tg application. The blue curve represents FRET signals in myoblast expressing STIM1-CFP and STIM2-YFP after application of 1 μm DMSO (control). D, Western blot illustrating co-immunoprecipitation of endogenous STIM2 protein using an antibody against STIM1 protein during early differentiation.
Myoblast Differentiation Can Rely on Either STIM1 or STIM2 Alone
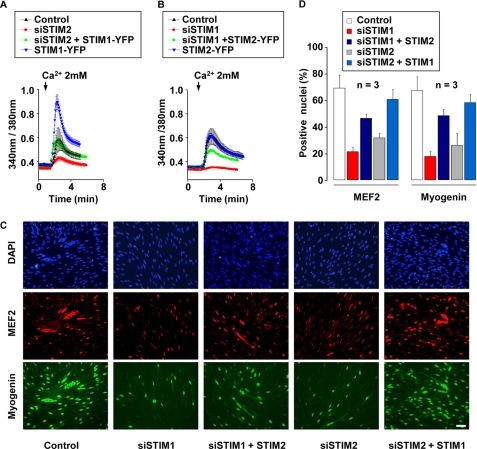
Our findings identified SOCE as a critical Ca2+ source for myoblast differentiation (1). To test whether STIM1 and STIM2 could substitute for each others, STIM2 was overexpressed in STIM1-silenced myoblasts and, conversely, STIM1 in STIM2-silenced myoblasts. We first monitored Tg-induced SOCE in both conditions using Fura-2. It can be seen in Fig. 4(A and B) that overexpression of YFP-conjugated STIM1 in STIM2-silenced cells restored full SOCE amplitude, whereas overexpression of YFP-conjugated STIM2 in STIM1-silenced cells rescued only 62 ± 26% of the Tg-induced Ca2+ influx (n = 3 independent experiments). To exclude that these differences in rescue could simply be attributed to a higher expression of STIM1 compared with STIM2, we compared YFP fluorescence and SOCE amplitude in cells overexpressing either STIM1-YFP (in the presence of siSTIM2 or control siRNA) or STIM2-YFP (in the presence of siSTIM1 or control siRNA). YFP fluorescence is directly linked to STIM expression, because we used vectors encoding for fused STIM-YFP proteins. The mean YFP fluorescence was similar in all tested conditions (p ≥ 0.12, n = 11 cells recorded in each condition). For STIM1-YFP-overexpressing cells, the YFP fluorescence was 907 ± 55 arbitrary units with siSTIM2 versus 843 ± 54 arbitrary units with control siRNA; and for STIM2-YFP overexpressing cells, the YFP fluorescence was 904 ± 70 arbitrary units with siSTIM1 and 835 ± 75 with control siRNA. On the other hand, the mean SOCE amplitude (assessed by Fura-2 fluorescence ratio) was increased by ∼3 times in STIM1-overexpressing cells (0.6 ± 0.2 versus 0.2 ± 0.05, p = 0.01, n = 11 cells recorded). Thus, differentiation can be fully restored in myoblasts expressing high amounts of STIM1 to compensate for STIM2 silencing (p = 0.49), but STIM2 overexpression in STIM1-silenced myoblasts cannot restore complete differentiation (p = 0.015); the proportion of myoblasts expressing MEF2 and myogenin was nevertheless more than doubled (46 ± 3% versus 21 ± 3% and 48 ± 5% versus 17 ± 4%, respectively) when STIM2 was overexpressed in STIM1-silenced myoblasts (Fig. 4, C and D, n = 3 independent experiments). Fig. 4C illustrates also that nuclei clusters, which reflect myotubes formation, are strongly suppressed after STIM1 and STIM2 silencing, whereas some nuclei clusters reappear when STIM1 and STIM2 are overexpressed to rescue the invalidation of STIM2 and STIM1, respectively.
FIGURE 4.
Potential redundancy of STIM1 and STIM2. A and B, cytoplasmic Ca2+ was assessed with Fura-2, 48 h after myoblast transfection. Intracellular Ca2+ stores were depleted with 1 μm Tg in a medium containing 250 nm Ca2+ (not shown), and 2 mm Ca2+ was subsequently added to reveal SOCE (same protocol as in Fig. 1C). In each condition, siRNA and a plasmid were co-transfected. When no specific siRNA and/or plasmid were used, siAllstar siRNA and/or EGFPN3 plasmid were/was added as control. C, transfected myoblasts were kept for 2 days in proliferation medium, and then transferred for 2 more days in differentiation medium. MEF2 and myogenin were stained in red and green, respectively. Nuclei were stained with DAPI. The scale bar represents 50 μm. D, mean myogenin- and MEF2-positive nuclei obtained out of three independent experiments.
STIM2 Is Up-regulated in Late Differentiation and Is Essential for Depolarization-induced Ca2+ Release
STIM1 is supposed to control the store-operated Ca2+ entry required for the development and the contractile function of mouse skeletal muscle (41). We confirmed this result in human myotubes (supplemental Fig. S1), and, because our data suggest that STIM1 and STIM2 interact strongly, we evaluated a potential role for STIM2 in differentiated human myotubes during both store-operated Ca2+ entry and depolarization-induced Ca2+ releases.
Our previous results indicated that, during differentiation, SOCE is greatly increased in myotubes and that STIM1 silencing reduced SOCE by 80 ± 3% in these multinucleated cells (1). Fig. 5A (left part) illustrates that SOCE is also inhibited by 81 ± 14% in STIM2-silenced multinucleated myotubes. STIM2 was silenced without impeding differentiation by triggering myoblast differentiation immediately after siSTIM2 transfection. We should stress that, unlike in Fig. 1, in which STIM2 expression was silenced during the initiation of differentiation, in Fig. 5, STIM2 expression was preserved during the initiation of differentiation allowing differentiation to occur normally (supplemental Fig. S2). This is in agreement with our former report on the critical role of SOCE only during the very first hours of the differentiation process (1). On the other hand, with the protocol used in Fig. 5, STIM2 expression was suppressed by 91 ± 5% in multinucleated differentiated myotubes 60 h after transfection (supplemental Fig. S2C). It can also be seen in Fig. 5A that STIM2 invalidation decreased the resting Ca2+ concentration in myotubes from 0.39 ± 0.01 to 0.34 ± 0.01 (n = 3 independent experiments). To exclude an indirect effect via STIM1, we verified that STIM1 expression was unaffected in STIM2-silenced myotubes (supplemental Fig. S2C).
FIGURE 5.
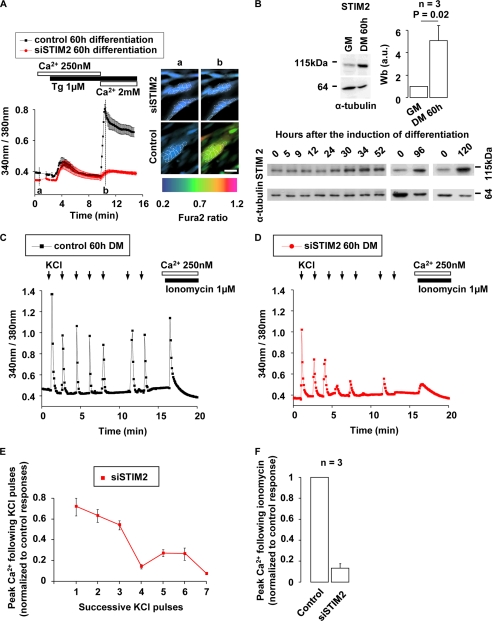
STIM2 is up-regulated in myotubes and participates to the refilling of Ca2+ stores. A, myoblasts were incubated with siSTIM2 for 5 h and immediately triggered to differentiate. 60 h afterward, multinucleated myotubes were formed, and cytoplasmic Ca2+ was assessed using Fura-2. Images illustrate Fura-2 measurements in multinucleated cells (nuclei are circled by discontinuous lines) at time “a” (resting Ca2+) and time “b” (max SOCE) of Fura-2 traces. One representative experiment (n = 3). B, Western blots illustrating the increase of STIM2 expression at various times of differentiation. Quantification was carried out of three Western blots performed on three different clones in proliferation and after 60 h of differentiation. α-Tubulin was used as loading control. C and D, 60 h after siSTIM2 transfection and differentiation induction (large multinucleated myotubes were present), cytoplasmic Ca2+ responses generated by successive KCl (65 mm) applications were assessed using Fura-2. siAllstar siRNA was used as control. E, peak Ca2+ responses to successive KCl pulses in siSTIM2-transfected myotubes. Ca2+ responses were normalized to the first response. F, Fura-2 measurements of remaining Ca2+ store content after seven pulses of 65 mm KCl. Ca2+ store content was assessed by application of 1 μm ionomycin in 250 nm external Ca2+. Results were normalized to those obtained in control conditions (three independent experiments).
The right part of Fig. 5A confirmed that, after the induction of differentiation, clusters of nuclei characteristic of differentiated myotubes appeared in both control and STIM2-silenced myotubes. Because SOCE is larger in myotubes, we evaluated by Western blot the amount of STIM2 protein in myotubes at various times of differentiation and compared it to that in myoblasts. After 60-h differentiation, STIM2 protein expression was found to be up-regulated by a factor 5.1 ± 1.3 in myotubes. This suggests that the stronger effect of STIM2 silencing on SOCE in myotubes (81 ± 14% decrease) as compared with myoblasts (45 ± 7% decrease) may be related to this up-regulation of STIM2 (Fig. 5B, n = 3 independent experiments).
We then evaluated the impact of STIM2 silencing on the Ca2+ signals during depolarization-induced Ca2+ release in myotubes. Cytosolic Ca2+ measurements with Fura-2 showed that control multinucleated myotubes exposed to 7 pulses of 65 mm KCl were able to perform 7 similar cytosolic Ca2+ peaks (Fig. 5C, n = 3 independent experiments). In contrast, STIM2-silenced myotubes were unable to sustain more than three successive Ca2+ peaks (Fig. 5D, n = 3 independent experiments).
Examination of the Ca2+ peaks triggered by KCl depolarizations indicates that the 3 first Ca2+ peaks in STIM2-silenced myotubes were 28 ± 6% smaller than those in the control myotubes and that the following 4–7 Ca2+ transients were at least 75% lower in myotubes in which STIM2 was invalidated (Fig. 5E). The Ca2+ peaks were inhibited even when the delay between successive stimulations was doubled to increase the time for the refilling of the Ca2+ stores (between peaks 5 and 6 in Fig. 5, C and D). Following the seven KCl depolarizations, the Ca2+ content of the intracellular store in STIM2-silenced myotubes was reduced by 87 ± 2% as assessed by total Ca2+ content release using 1 μm ionomycin in 250 nm external Ca2+ medium (Fig. 5F, n = 3 independent experiments) or alternatively 10 μm caffeine and 1.5 μm thapsigargin in 250 nm external Ca2+ (supplemental Fig. S3).
DISCUSSION
In this study, we describe the key roles played by STIM2 in post-natal human myoblast differentiation and in myotube depolarization-induced Ca2+ release. Using siRNA that silenced STIM2 (siSTIM2), we observed (i) a decrease in SOCE amplitude, (ii) an inhibition of myoblast differentiation, assessed by MEF2 and myogenin expression, and (iii) an inability for myotubes to sustain repetitive depolarization-induced Ca2+ releases. STIM2 re-expression in myoblast transfected with siSTIM2 restored SOCE amplitude and differentiation, excluding possible nonspecific effects of siSTIM2. In agreement with former reports, SOCE inhibition was smaller with STIM2 silencing than with STIM1 or Orai1 silencing (3, 4), although SOCE inhibition by siSTIM2 correlated well with the degree of inhibition of myoblast differentiation (1). The smaller effect of STIM2 silencing, as compared with STIM1 silencing, on SOCE amplitude could be explained by differences in the N-terminal part of STIM molecules, which would account for a smaller STIM2-linked (as compared with STIM1) Ca2+ entry (1, 24). We verified by Western blot that STIM2 silencing was as efficient as STIM1 silencing (87 ± 11% versus 80 ± 17% reduction, respectively). Silencing STIM2 in multinucleated myotubes had, however, a much more drastic effect on SOCE than in myoblasts. This difference may be due to the strong up-regulation of STIM2 during late differentiation rather than to subtle differences in Orai1 activation.
STIM1-STIM2 Interactions
We hypothesized that STIM1 and STIM2 interact synergistically during SOCE and myoblast differentiation. As in Feske (51), we defined “synergy” as a greater SOCE observed when STIM1 and STIM2 are acting together than the addition of each SOCE measured when a single STIM molecule is acting and the other silenced. To confirm this hypothesis, we monitored (i) the clustering and co-localization of STIM proteins, (ii) FRET between STIM1-CFP and STIM2-YFP molecules, and (iii) the ability of STIM1 and STIM2 to co-immunoprecipitate after SOCE activation, i.e. after the induction of differentiation or after ER-Ca2+ depletion by Tg.
Our previous work suggests that STIM1-Orai1 activation occurs during the first 10 h of myoblast differentiation (1). In the present study, we find that, in ∼50% of myoblasts, both STIM proteins form stable clusters during the first few hours following the induction of differentiation. Cluster formation being a hallmark of STIM activation (2, 3), the presence of permanent clusters during the initiation of myoblast differentiation suggests that recurrent ER Ca2+ decreases maintain STIM1 and STIM2 molecules activated. Attempts to observe STIM1 and STIM2 clustering in the same myoblast during differentiation were not possible, because myoblasts did not survive more than 1–2 days when STIM1 and STIM2 were expressed together. This might be due to Ca2+ imbalance when both STIM molecules are transfected, although, as reported by Wang et al. (22), cell survival is already drastically reduced by overexpression of STIM2 alone. Consequently, ER Ca2+ depletion by Tg was used to induce STIM1-STIM2 clustering in the same myoblast and, in these conditions, a clear co-localization of STIM1 and STIM2 clusters was observed.
To evaluate further STIM molecules clustering and co-localization, we monitored FRET increase (following ER Ca2+ depletion by Tg) between STIM1-CFP and STIM1-YFP, and between STIM1-CFP and STIM2-YFP. Because FRET only occurs when the distance between two molecules is in the range of 1–10 nm, the increase in FRET observed between STIM1 proteins and between STIM1 and STIM2 proteins suggests further that both STIM molecules relocated and get closer during ER Ca2+ depletion. This experiment shows, in addition, that STIM1 proteins are as intimately associated as STIM1 and STIM2 proteins, because FRET increases between STIM1 proteins and between STIM1 and STIM2 proteins were almost identical.
The third strategy used to assess a possible tight interaction between STIM1 and STIM2 was co-immunoprecipitation (52). As shown in Fig. 3D, we were able to co-immunoprecipitate both STIM molecules using an antibody that binds only to STIM1. We are aware that co-immunoprecipitation does not exclude the presence of a third unknown protein that would link STIM1 and STIM2. This is, however, unlikely because FRET would probably not occur between STIM1 and STIM2 if a third protein was present.
In agreement with previous reports (4, 51), our results suggest that STIM1 and STIM2 are acting synergistically, because the addition of the remaining SOCE under respective STIM silencing is much smaller than the normal myoblast or myotube SOCE (this work and Ref. 1). Indeed, the addition of residual influxes in STIM1- and STIM2-silenced myoblasts is 60 ± 11% (respectively, 29 ± 6% in myotubes) of control SOCE amplitude. Thus, ∼40% (respectively, 71% in myotubes) of max SOCE amplitude may be attributed to STIM1-STIM2 synergy. This synergy can also be deducted from the observation that STIM1 overexpression in myoblasts expressing endogenous STIM2 generated a large increase in SOCE, which was not seen when STIM1 was overexpressed in STIM2-silenced myoblasts (Fig. 4A).
STIM1 and STIM2 Are Able to Function Independently
Our current findings clearly suggest that STIM2 interacts with STIM1 and that this interaction is required to produce adequate SOCE during both myoblast differentiation and depolarization-induced Ca2+ transients. Interestingly, however, our results also show that STIM1 and STIM2 are able to function mostly independently as STIM1 overexpression in STIM2-silenced myoblasts, and, to a certain extent, STIM2 overexpression in STIM1-silenced myoblasts are able to restore both SOCE and myoblast differentiation. This is consistent with a recent work on mice neurons showing that STIM2 regulates SOCE independently of STIM1 (53). A hypothesis would be that STIM2 controls SOCE during small store depletions, whereas STIM1 would be recruited, together with STIM2, during larger store depletions. In this model, during large store depletions, cells would benefit from STIM1-STIM2 interactions that would generate larger SOCE.
Cytosolic Resting Ca2+
Compared with the degree of SOCE inhibition, cytosolic resting Ca2+ was more decreased by STIM2 silencing than by STIM1 silencing (1). These results are in agreement with former reports on resting Ca2+ regulation by STIM2 (2, 22, 24). However, because our former findings clearly identified SOCE rather than resting Ca2+ level as being the crucial parameter involved in myoblast differentiation, we did not investigate further the impact of STIM2 on resting Ca2+ level in myoblasts (1).
Depolarization-induced Calcium Releases
Recent evidences obtained in mice muscle fibers suggest that SOCE and STIM1 are required for Ca2+ store refilling following depolarization-induced Ca2+ transients (41). These observations led us to examine a potential role for STIM2 during recurrent Ca2+ transients elicited in human multinucleated myotubes. We first observed that STIM2 was strongly up-regulated after 60 h of differentiation, and that SOCE reliance on STIM2 was greatly increased after myotube formation. Indeed, SOCE measured in mononucleated proliferating myoblasts was reduced by 45 ± 7% (Fig. 1D) when STIM2 expression was reduced by 87 ± 11% (Fig. 1F), whereas SOCE measured in multinucleated myotubes was reduced by 81 ± 14% when STIM2 expression was reduced by 91 ± 5% (supplemental Fig. S2C). This, on its own, already pointed toward a role for STIM2 in multinucleated myotubes. We then observed that STIM2 silencing impeded recurrent KCl-induced Ca2+ transients. In siSTIM2-silenced myotubes, the ER Ca2+ content was fully depleted following a train of seven KCl pulses. STIM2 is thus crucial for an adequate Ca2+ store refilling in human myotubes. These observations confirm that Ca2+ transients involved in myotube excitation-contraction coupling do not only rely on Ca2+ recycling by sarcoplasmic-endoplasmic reticulum calcium ATPase but also need SOCE. In addition, these experiments also confirm that SOCE depends on the cooperation between STIM1 and STIM2.
Patients with STIM1 Deficiency
In recent reports, three patients with STIM1 deficiency were shown to suffer from muscular hypotonia and immune disorders (32, 54). Our present observations suggest that, although they act synergistically, STIM1 and STIM2 can substitute for each other during post-natal myogenesis. These results could explain the apparent discrepancy between the strong inhibition of post-natal myogenesis observed during acute depletion of STIM1 or STIM2, and the apparent mild clinical manifestation in muscle of patients with STIM1 deficiency.
Together, our new findings show that STIM2 is involved in human myoblast SOCE and differentiation, but that STIM1 is predominant at this stage of muscle formation. On the other hand, we observed that STIM2 is strongly up-regulated in human myotubes, and that, at this later stage of differentiation, STIM2 is required for SOCE and excitation-induced Ca2+ release.
Supplementary Material
Acknowledgments
We thank P. Brawand and C. Pomponio for excellent technical assistance with cell cultures, Dr A. Kaelin for providing muscle biopsies, Dr Anjana Rao for providing STIM1-YFP and STIM1-CFP plasmids, and Dr. Tobias Meyer for providing STIM2-YFP and STIM2-WT plasmids.
This work was supported by the Fonds National Suisse pour la Recherche Scientifique (Grant 310030-124910 to L. B.), the Fondation Suisse pour la Recherche sur les Maladies Musculaires, the Fondation Hans Wilsdorf, and the Fondation Marcel Levaillant.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ER
- endoplasmic reticulum
- SOCE
- store-operated Ca2+ entry
- wt
- wild type
- siRNA
- small interference RNA
- DAPI
- 4′,6-diamidino-2-phenylindole
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- RFP
- red fluorescent protein
- FRET
- fluorescence resonance energy transfer
- siSTIM2
- siRNA-targeting STIM2
- Tg
- thapsigargin.
REFERENCES
- 1.Darbellay B., Arnaudeau S., König S., Jousset H., Bader C., Demaurex N., Bernheim L. (2009) J. Biol. Chem. 284, 5370–5380 [DOI] [PubMed] [Google Scholar]
- 2.Brandman O., Liou J., Park W. S., Meyer T. (2007) Cell 131, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh-Hora M., Yamashita M., Hogan P. G., Sharma S., Lamperti E., Chung W., Prakriya M., Feske S., Rao A. (2008) Nat. Immunol. 9, 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derler I., Fahrner M., Muik M., Lackner B., Schindl R., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 24933–24938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gwack Y., Srikanth S., Oh-Hora M., Hogan P. G., Lamperti E. D., Yamashita M., Gelinas C., Neems D. S., Sasaki Y., Feske S., Prakriya M., Rajewsky K., Rao A. (2008) Mol. Cell. Biol. 28, 5209–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Lu J., Xu P., Xie X., Chen L., Xu T. (2007) J. Biol. Chem. 282, 29448–29456 [DOI] [PubMed] [Google Scholar]
- 9.Lioudyno M. I., Kozak J. A., Penna A., Safrina O., Zhang S. L., Sen D., Roos J., Stauderman K. A., Cahalan M. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2011–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luik R. M., Wu M. M., Buchanan J., Lewis R. S. (2006) J. Cell Biol. 174, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer J. C., Dehaven W. I., Smyth J. T., Wedel B., Boyles R. R., Bird G. S., Putney J. W., Jr. (2006) J. Biol. Chem. 281, 24979–24990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vig M., Peinelt C., Beck A., Koomoa D. L., Rabah D., Koblan-Huberson M., Kraft S., Turner H., Fleig A., Penner R., Kinet J. P. (2006) Science 312, 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziadek M. A., Johnstone L. S. (2007) Cell Calcium 42, 123–132 [DOI] [PubMed] [Google Scholar]
- 15.Hewavitharana T., Deng X., Wang Y., Ritchie M. F., Girish G. V., Soboloff J., Gill D. L. (2008) J. Biol. Chem. 283, 26252–26262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan P. G., Rao A. (2007) Trends Biochem. Sci. 32, 235–245 [DOI] [PubMed] [Google Scholar]
- 17.Liou J., Fivaz M., Inoue T., Meyer T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luik R. M., Wang B., Prakriya M., Wu M. M., Lewis R. S. (2008) Nature 454, 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peinelt C., Vig M., Koomoa D. L., Beck A., Nadler M. J., Koblan-Huberson M., Lis A., Fleig A., Penner R., Kinet J. P. (2006) Nat. Cell Biol. 8, 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soboloff J., Spassova M. A., Tang X. D., Hewavitharana T., Xu W., Gill D. L. (2006) J. Biol. Chem. 281, 20661–20665 [DOI] [PubMed] [Google Scholar]
- 21.Vig M., Beck A., Billingsley J. M., Lis A., Parvez S., Peinelt C., Koomoa D. L., Soboloff J., Gill D. L., Fleig A., Kinet J. P., Penner R. (2006) Curr. Biol. 16, 2073–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y. J., Deng X. X., Zhou Y. D., Hendron E., Mancarella S., Ritchie M. F., Tang X. D., Baba Y., Kurosaki T., Mori Y., Soboloff J., Gill D. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S. L., Yu Y., Roos J., Kozak J. A., Deerinck T. J., Ellisman M. H., Stauderman K. A., Cahalan M. D. (2005) Nature 437, 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y. D., Mancarella S., Wang Y. Y., Yue C. Y., Ritchie M., Gill D. L., Soboloff J. (2009) J. Biol. Chem. 284, 25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vig M., DeHaven W. I., Bird G. S., Billingsley J. M., Wang H., Rao P. E., Hutchings A. B., Jouvin M. H., Putney J. W., Kinet J. P. (2008) Nat. Immunol. 9, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jousset H., Frieden M., Demaurex N. (2007) J. Biol. Chem. 282, 11456–11464 [DOI] [PubMed] [Google Scholar]
- 27.Parvez S., Beck A., Peinelt C., Soboloff J., Lis A., Monteilh-Zoller M., Gill D. L., Fleig A., Penner R. (2008) FASEB J. 22, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stathopulos P. B., Zheng L., Ikura M. (2009) J. Biol. Chem. 284, 728–732 [DOI] [PubMed] [Google Scholar]
- 29.Zheng L., Stathopulos P. B., Li G. Y., Ikura M. (2008) Biochem. Biophys. Res. Commun. 369, 240–246 [DOI] [PubMed] [Google Scholar]
- 30.Schuhmann M. K., Stegner D., Berna-Erro A., Bittner S., Braun A., Kleinschnitz C., Stoll G., Wiendl H., Meuth S. G., Nieswandt B. (2010) J. Immunol. 184, 1536–1542 [DOI] [PubMed] [Google Scholar]
- 31.Soboloff J., Spassova M. A., Hewavitharana T., He L. P., Xu W., Johnstone L. S., Dziadek M. A., Gill D. L. (2006) Curr. Biol. 16, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 32.Picard C., McCarl C. A., Papolos A., Khalil S., Lüthy K., Hivroz C., LeDeist F., Rieux-Laucat F., Rechavi G., Rao A., Fischer A., Feske S. (2009) N. Engl. J. Med. 360, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaudeau S., Holzer N., König S., Bader C. R., Bernheim L. (2006) J. Cell. Physiol. 208, 435–445 [DOI] [PubMed] [Google Scholar]
- 34.Louis M., Zanou N., Van S. M., Gailly P. (2008) J. Cell Sci. 121, 3951–3959 [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg P., Hawkins A., Stiber J., Shelton J. M., Hutcheson K., Bassel-Duby R., Shin D. M., Yan Z., Williams R. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9387–9392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiber J. A., Tabatabaei N., Hawkins A. F., Hawke T., Worley P. F., Williams R. S., Rosenberg P. (2005) Dev. Biol. 287, 213–224 [DOI] [PubMed] [Google Scholar]
- 37.Lorenzon P., Giovannelli A., Ragozzino D., Eusebi F., Ruzzier F. (1997) Eur. J. Neurosci. 9, 800–808 [DOI] [PubMed] [Google Scholar]
- 38.Hinard V., Belin D., Konig S., Bader C. R., Bernheim L. (2008) Development 135, 859–867 [DOI] [PubMed] [Google Scholar]
- 39.Konig S., Hinard V., Arnaudeau S., Holzer N., Potter G., Bader C. R., Bernheim L. (2004) J. Biol. Chem. 279, 28187–28196 [DOI] [PubMed] [Google Scholar]
- 40.Knapp J. R., Davie J. K., Myer A., Meadows E., Olson E. N., Klein W. H. (2006) Development 133, 601–610 [DOI] [PubMed] [Google Scholar]
- 41.Stiber J., Hawkins A., Zhang Z. S., Wang S., Burch J., Graham V., Ward C. C., Seth M., Finch E., Malouf N., Williams R. S., Eu J. P., Rosenberg P. (2008) Nat. Cell Biol. 10, 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurebayashi N., Ogawa Y. (2001) J. Physiol. 533, 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyfenko A. D., Dirksen R. T. (2008) J. Physiol. 586, 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLennan D. H. (2000) Eur. J. Biochem. 267, 5291–5297 [DOI] [PubMed] [Google Scholar]
- 45.Pan Z., Yang D., Nagaraj R. Y., Nosek T. A., Nishi M., Takeshima H., Cheng H., Ma J. (2002) Nat. Cell Biol. 4, 379–383 [DOI] [PubMed] [Google Scholar]
- 46.McCarl C. A., Picard C., Khalil S., Kawasaki T., Röther J., Papolos A., Kutok J., Hivroz C., Ledeist F., Plogmann K., Ehl S., Notheis G., Albert M. H., Belohradsky B. H., Kirschner J., Rao A., Fischer A., Feske S. (2009) J. Allergy Clin. Immunol. 124, 1311–1318.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baroffio A., Aubry J. P., Kaelin A., Krause R. M., Hamann M., Bader C. R. (1993) Muscle Nerve 16, 498–505 [DOI] [PubMed] [Google Scholar]
- 48.Prakriya M., Feske S., Gwack Y., Srikanth S., Rao A., Hogan P. G. (2006) Nature 443, 230–233 [DOI] [PubMed] [Google Scholar]
- 49.Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth J. T., Petranka J. G., Boyles R. R., DeHaven W. I., Fukushima M., Johnson K. L., Williams J. G., Putney J. W., Jr. (2009) Nat. Cell Biol. 11, 1465–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feske S. (2009) Immunol. Rev. 231, 189–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams R. T., Manji S. S., Parker N. J., Hancock M. S., Van S. L., Eid J. P., Senior P. V., Kazenwadel J. S., Shandala T., Saint R., Smith P. J., Dziadek M. A. (2001) Biochem. J. 357, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berna-Erro A., Braun A., Kraft R., Kleinschnitz C., Schuhmann M. K., Stegner D., Wultsch T., Eilers J., Meuth S. G., Stoll G., Nieswandt B. (2009) Sci. Signal. 2, ra67. [DOI] [PubMed] [Google Scholar]
- 54.Feske S., Picard C., Fischer A. (2010) Clin. Immunol. 135, 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.