Abstract
Steroids exert powerful effects on the brains and behavior of many species, but measures and manipulations of endocrine physiology in songbirds often reveal unexplained connections between steroids and the brain. The zebra finch song system, a sensorimotor neural circuit sensitive to steroids throughout life, organizes and functions largely in apparent independence from gonadally derived steroids. We tested the hypothesis that the zebra finch brain has the capacity for de novo steroidogenesis and that neurally synthesized steroids, neurosteroids, may impact the song system. Using multiple techniques, we demonstrate that the steroidogenic acute regulatory protein (StAR), cytochrome P450 side-chain cleavage (CYP11A1), and 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase, the first three factors in the steroidogenic pathway, are expressed in both developing and adult zebra finch brain. Detailed expression mapping at posthatch d 20 (P20) and adult reveals widespread area-specific expression and coexpression patterns for steroidogenic acute regulatory protein, CYP11A1, and 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase, which suggest neurosteroids may modulate multiple brain functions, including sensory and motor systems. Notably, whereas expression of other steroidogenic genes such as aromatase has been essentially absent from the song system, each of the major song nuclei express at least a subset of steroidogenic genes described here, establishing the song system as a potential steroidogenic circuit.
The neural song system has long been used to investigate hormonal effects on the central nervous system, including actions on the developing brain, adult neural plasticity, the control of complex motor output, and cellular mechanisms underlying learning. Manipulations of circulating steroid levels in several species of songbirds, however, reveal discrepancies between the global steroid environment and steroid sensitive behaviors (cf. Ref. 1). The disconnection between gonadally derived steroids and steroid sensitive neural systems is particularly striking in the zebra finch song circuit. Only male zebra finches sing, and males have a larger and more complex song system than females (2). Estrogens and androgens contribute to male song learning and crystallization (3), and the adult male song system is sensitive to the activational effects of steroids (4). Unlike most sexually dimorphic brain regions, however, the song system is not dependent on gonadally derived steroids for many of its organizational and activational events (5, 6). For example, estrogen administration to females early in development functionally masculinizes the song system (7–9), but genetic females induced to develop testes retain a feminine song system (10, 11). Furthermore, whereas estrogens seem to drive early song system masculinization, it is for the most part androgen receptors (ARs), not estrogen receptors, that are present within song nuclei (12, 13), and ARs are necessary for circuit masculinization (14). Later in posthatch development, estrogen can be synthesized by cultured brain slices and can masculinize some song system components (15). Thus, the zebra finch song system is steroid sensitive at all ages, but its incomplete dependence on gonadally derived steroids suggests that the brain may provide steroids essential to its organization and function.
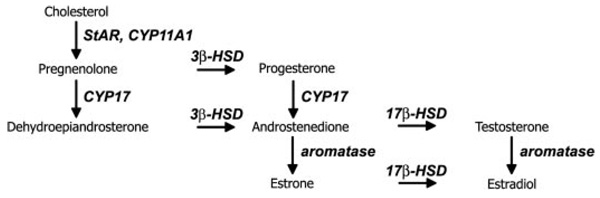
We tested the hypothesis that developing and adult zebra finch brains express all of the genes necessary for de novo neurosteroidogenesis throughout posthatch life and that the expression pattern of steroidogenic genes would suggest a role for neurosteroids in song system function at posthatch d 20 (P20) and adult. The idea that the brain can synthesize steroids is not unique to the zebra finch; it has been postulated and demonstrated in various other animals including humans, rodents, nonsongbirds, and amphibians (16–22). The zebra finch song system, however, may be a particularly useful model in which to study neurosteroidogenesis because the song system may be a biologically relevant functional end point for neurosteroid action. Our investigation focused on the androgen-synthetic pathway because ARs are expressed in most song nuclei and are necessary for song system organization, and the estrogen-synthetic enzyme aromatase has already been extensively investigated (4, 23). Three enzymes, cytochrome P450 side chain cleavage (CYP11A1), 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase (3β-HSD), and cytochrome P450 17α-hydroxylase/17, 20 lyase (CYP17), and a cholesterol transport protein, the steroidogenic acute regulatory protein (StAR), are necessary for androgen synthesis (Fig. 1). Here we focus on StAR, CYP11A1, and 3β-HSD; CYP17 was cloned and generally described (24), but because comprehensive analysis of multiple steroidogenic genes has not been performed in songbirds, it will also be considered in the broader conclusions. We cloned zebra finch StAR, CYP11A1, and 3β-HSD cDNAs, used multiple molecular techniques to confirm their neural expression, and comprehensively mapped their expression patterns in male and female brains at P20 and adult. All techniques demonstrate that steroidogenic genes are expressed in the zebra finch brain throughout posthatch life, and in situ hybridization at P20 and adult indicates widespread and specific neural expression of each gene. Furthermore, if the mRNA distribution described reflects the protein distribution, locally synthesized steroids could modulate multiple brain functions including general sensory and motor integration and the sensorimotor functions of major song nuclei.
FIG. 1.
The estrogen-synthetic portion of the steroidogenic pathway. Aromatase, Cytochrome P450 aromatase (CYP19).
Materials and Methods
Animals were bred at the University of California, Los Angeles and Michigan State University vivariums. Tissue used for RNA-based techniques were rapidly dissected, frozen on dry ice or collected into RNA-later (Ambion, Austin, TX), and stored at −80 C until use. Protocols were approved by the University of California, Los Angeles, Chancellor’s Animal Research Care Committee and the Michigan State University Institutional Animal Care and Use Committee.
RT-PCR and subcloning
Total RNA extraction and reverse transcription were performed as described elsewhere (24). RT-PCR was first performed with either primers based on the chicken cDNA sequences (GenBank) or degenerate primers based on several species sequences. PCR products were sub-cloned, sequenced (Big Dye, version 3; Applied Biosystems, Foster City, CA), and used to design zebra finch specific primers. RT-PCR was performed with cDNA prepared from telencephalon, diencephalon, optic tectum (TeO), and cerebellum from P1–7 and adult, males and females, using zebra finch-specific primers as previously described (24). Subcloned PCR products were ligated into PCRScript Amp (Stratagene, La Jolla, CA) and sequenced (BigDye version 3; Applied Biosystems). Subclone identity was confirmed by BLAST analysis (http://www.ncbi.nlm.nih.gov/blast/).
cDNA cloning and 5′rapid amplification of cDNA ends (RACE)
Two zebra finch cDNA libraries (λZAP; Stratagene) were constructed from testes or male whole brain. The testicular library was screened for 3β-HSD, CYP11A1, and StAR with zebra finch subclones randomly primed and labeled with α32P-dCTP. The brain library was screened for 3β-HSD. Hybridization was performed as previously described (24). After three rounds of screening, positive pBS (SK−) clones were rescued, sequenced in the antisense and sense directions (BigDye version 3; Applied Biosystems), and subjected to BLAST homology comparisons to confirm identities. Based on sequence comparison with the chicken cDNA sequences, the zebra finch CYP11A1 and StAR clones were truncated at the 5′ end and lacked the putative ATG translational start codon.
To complete the CYP11A1 and StAR cDNA sequences, 5′RACE was performed on testicular cDNA according to the manufacturer’s manual (GeneRacer; Invitrogen, Carlsbad, CA). RACE products were subcloned into PCRScript Amp (Stratagene) and sequenced. The sequences of both the CYP11A1 and StAR 5′RACE subclones overlapped with the respective clone sequences. Therefore, for CYP11A1 and StAR, composite clone and subclone cDNA sequences were constructed.
Northern analysis
Total RNA was extracted (RNeasy; QIAGEN, Valencia, CA) from pooled tissue samples. For P1–5 male and female whole brains, approximately 30 brains were pooled for each sex. For adult tissues, four males and four females were used to extract RNA from telencephalon and the rest of the brain. Total RNA was also extracted from four adult pairs of ovaries and testes. Poly (A+) RNA was isolated from each of these samples (Oligotex; QIAGEN) and electrophoresed (5 µg for brain samples, 2 µg for gonad samples) as described elsewhere (24). Blots were hybridized with α32P-dCTP labeled cDNA probes (DECAprime; Ambion) derived from zebra finch subclones for each gene essentially as outlined in Ref. 24. The Northern blot for StAR analysis was constructed with total RNA from adult male and female telencephalon, hindbrain, ovary, and testis (10 µg for all samples) in a comparable technique. All blots were exposed to autoradiographic film (X-OMAT AR; Eastman Kodak, Rochester, NY) with intensifying screen at −80 C.
Relative quantitative RT-PCR
SYBR Green relative quantitative (rq) RT-PCR was performed to determine the relative abundance of 3β-HSD, CYP11A1, and StAR mRNA in male and female brain during early development. For 3β-HSD and CYP11A1 rqRT-PCR, whole brains from P5 males and females (n = 3 for each sex) were assayed. RNA was extracted (Trizol; Invitrogen) and treated with DNase I to remove contaminating genomic DNA. Standard PCR experiments confirmed the absence of genomic DNA. Primers for P5 PCRs were designed to cross predicted exon-intron boundaries based on theoretical comparison of rat gene structures and zebra finch cDNA sequences (Spidey; http://www.ncbi.nlm.nih.gov/spidey). An initial RNA concentration gradient determined that 280 ng of each RNA sample was appropriate for the rqRT-PCR experiment. Tubes were provided with primers for 3β-HSD, CYP11A1, or β-actin, the control gene. Detailed experimental design and procedures were as explained elsewhere (24). Fluorescence was measured after each at each cycle at 83 C to remove confounds from primer dimer fluorescence. Threshold cycles (Ct) were analyzed on Applied Biosystems software (SDS).
To determine relative StAR mRNA levels in males and females during development, rqRT-PCR was performed on telencephalon from P5 (n = 6 of each sex) and P1 (n = 7 of each sex) males and females. These assays were performed with a few modifications from that described for the P5 3β-HSD and CYP11A1 experiment above. DNase I-treated total RNA extracted from each individual whole brain was reverse transcribed in a reaction with oligo-T primers and SuperScript II. Instead of β-actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was the control amplification gene for the StAR rqRT-PCR experiments. The PCR cycling conditions were: 50 C for 2 min, a 10-min hot start at 95 C, and then 40 cycles of 95 C for 15 min and 60 C for 1 min. Data were analyzed with Applied Biosystems software (SDS).
To obtain relative abundance measures, the average Ct of each triplicate sample was calculated. The average Ct for each sample was converted to the ratio of steroidogenic gene Ct (3β-HSD, CYP11A1, StAR) to control gene Ct [β-actin (3β-HSD and CYP11A1), GAPDH (StAR)]. Ratios for each gene and age were analyzed for quantitative sex differences with one-way ANOVA (P < 0.05) (SPSS Inc., Chicago, IL).
3β-HSD activity measurement
Previous studies detected 3β-HSD activity in zebra finch tissues, including brain, with multiple stringent controls to validate that the biochemical procedure specifically measured 3β-HSD activity (25–27). Here several control experiments ensured that our biochemical analyses measured specific 3β-HSD activity converting 3H-dehydroepiandrosterone (DHEA) to 3H-androstenedione (3H-AE) in P5 brains. All control reactions were done in triplicate. First, radioinert 3H-AE (1.5 µg; Steraloids, Newport, RI) was included in the tissue incubations. This cold trap reduces conversion of the 3H-AE product into downstream steroids, increasing the levels of detectable 3H-AE by minimizing production of 3H-estrogens. Second, because activity of type I 3β-HSD is dependent on the cofactor nicotinamide adenine dinucleotide (NAD+), 1.1 mmNAD+ were substituted with 1.1 mm nicotinamide adenine dinucleotide phosphate in control reactions. This should prevent conversion of 3H-DHEA to 3H-AE. Third, specific pharmacological blockers of both 3β-HSD (Trilostane, a gift from Micron Technologies, Exton, PA) and aromatase (Fadrozole, a gift from Novartis Pharma, East Hanover, NJ) were singly included in tissue incubations. Tissue was incubated with either 4 mm Trilostane or 200 nm Fadrozole, concentrations previously confirmed to effectively block enzyme activity (26–28). In addition, background control tubes containing all reaction components minus tissue were run in parallel, and tubes containing known amounts of 3H-AE and 3H-estrone (E1) were included to measure steroid recovery efficiency.
We measured 3β-HSD activity in the telencephalon, diencephalon, TeO, and cerebellum of male and female P5 birds (n = 6 for each sex). Tissue was homogenized in cold 0.05 m sucrose phosphate buffer (pH 7.4) and incubated for 3 h at 41 C with 200 nm 1,2,6,7-3H-DHEA (specific activity = 74 Ci/mmol; PerkinElmer, Wellesley, MA) in the presence of NAD+ and ATP cofactors in conditions previously determined by dose-and time-course experiments to produce quantifiable amounts of 3HDHEA within a linear range of enzyme activity (27). Steroid isolation, purification, and thin-layer chromatography extraction techniques were performed as previously described (25–27). Briefly, steroids were isolated via ether extraction and then purified by phenolic partition. Estrogens were collected in ethyl acetate and androgens in carbon tetrachloride. Estrogen and androgen partitions were chromatographed on thin-layer chromatography plates in 3:1 ether to hexane. Bands of individual steroids were scraped from the plates, suspended in methanol, and quantified in a scintillation counter. To normalize steroid conversion by tissue sample size, the quantity of proteins contained in each sample was obtained by the Bradford method with BSA protein standards. Values are reported as femtomoles of 3H-steroid produced per milligram of protein. Statistical analysis was performed with two-way ANOVA (SPSS).
In situ hybridization
Whole brains from P20 and adult males and females (n = 3 for each sex at each age) were sectioned to 20 µm on a cryostat. Sections from one male and one female were always processed together to facilitate between-sex comparisons. Zebra finch specific subclones constructed as described above were linearized and served as templates for in vitro transcription of 33P-UTP-labeled cRNA riboprobes. Sense configured probes were one negative control. In addition, for both StAR and CYP11A1, two nonoverlapping antisense and sense probe sets were used to confirm hybridization specificity. Only one 3β-HSD riboprobe set was used because enzymatic activity measures and successful brain library screens validated neural expression. Slides were processed essentially as described elsewhere (24). Hybridization was performed at 59 C, and the final high stringency was conducted in 0.1× SSC at 63 C for 30 min. After in situ hybridization, slides were exposed to autoradiographic film (X-OMAT MR, Eastman Kodak) for 48–100 h, dipped in liquid emulsion (NTB-2, Eastman Kodak), and exposed at 4 C until developed and fixed (Kodak D-19, Kodak Fixer; Eastman Kodak).
Each brain area listed as a site of expression met several criteria: 1) containing labeling both on film and emulsion dipped slides; 2) bilateral labeling that extended across several sections; 3) labeling in multiple birds; 4) lack of hybridization with sense probes, compared with that seen with antisense probes; and 5) for StAR and CYP11A1, the two nonoverlapping antisense probes for each gene needed to hybridize to the same areas.
Neuroanatomical hybridization distribution was determined by comparing film images, emulsion-dipped slides viewed under dark-field illumination, and adjacent Nissl-stained sections. Published bird brain atlases (29,30) guided anatomical determination, and nomenclature was derived from Reiner et al. (31). Mapping was performed on sections spanning the entire brain, but the investigation of three genes in both males and females at two different ages precluded photomicrographic presentation of the entire brain of all of these groups. Shown are selected regions demonstrating either hybridization over specific nuclei, including song nuclei, or general patterns of hybridization at several anterior-posterior levels of the brain.
A complete description of hybridization is presented in Table 1. Relative abundance of StAR, CYP11A1, and 3β-HSD hybridization was subjectively determined and divided into four categories: high (+++), medium (++), low (+), and absent (−). When no silver grain accumulation was detected in both film images and emulsion-dipped slides, compared with adjacent areas of background and sense control sections, the area was designated as absent. The high, medium, and low designations were assigned by first identifying a brain area that had the darkest label (high) and the lowest label (low) for one gene at one age. Then a brain region having an intermediate hybridization level was designated medium. These nuclei served as the standards for hybridization to which the mean hybridization intensity of all identifiable brain regions were compared. The same nuclei were used as standards for all brains of a particular gene and age, and the process was performed for each gene and age independently. Thus, for example, medium in one gene may not be exactly the same hybridization intensity as medium in another gene. By virtue of this nucleus by nucleus subjective categorization process, qualitative determination of sex differences in hybridization distribution and intensity was also conducted.
TABLE 1.
Areas labeled by in situ hybridization in adult (Ad) and P20 brains
| StAR | CYP11A1 | 3β-HSD | StAR | CYP11A1 | 3β-HSD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain area | Ad | P20 | Ad | P20 | Ad | P20 | Brain area | Ad | P20 | Ad | P20 | Ad | P20 |
| Telencephalon | Midbrain/hindbrain | ||||||||||||
| HA | + | + | + | + | + | + | HM | − | − | + | + | − | − |
| M | + | + | + | + | + | + | HL | − | − | + | + | − | − |
| HD | + | + | + | + | + | ++ | Tu | − | − | ++ | ++ | − | − |
| MSt | + | + | + | + | + | + | SNc | ++ | ++ | ++ | ++ | − | − |
| N | ++ | ++ | ++ | ++ | + | + | TeO | − | − | ++ | ++ | − | − |
| S | − | − | ++ | ++ | ++ | ++ | ICo/MLd | ++ | ++ | ++ | ++ | ++ | ++ |
| TFM | − | − | − | ++ | − | − | VTA | ++ | ++ | ++ | ++ | − | − |
| CoS | + | + | +++ | +++ | − | − | MPv | − | − | +++ | +++ | − | − |
| nCPa | + | + | +++ | +++ | − | − | IM/IPC | +++ | +++ | ++ | ++ | ++ | ++ |
| HP | + | + | ++ | ++ | +++ | +++ | Pt | − | − | +++ | +++ | ++ | ++ |
| GP | − | − | + | + | − | − | SRt | − | − | ++ | ++ | − | − |
| LV | − | − | + | + | − | − | SGC | +++ | +++ | ++ | ++ | +++ | +++ |
| A | + | + | ++ | ++ | ++ | ++ | GCt | ++ | ++ | ++ | ++ | + | + |
| Tn | − | − | ++ | ++ | ++ | ++ | III V | − | − | − | − | − | − |
| Hypothalamus | EM | ++ | ++ | + | + | ++ | ++ | ||||||
| POA | ++ | ++ | ++ | ++ | ++ | ++ | SLu | ++ | ++ | ++ | ++ | ++ | ++ |
| PVN | ++ | ++ | ++ | ++ | ++ | ++ | EW | +++ | +++ | ++ | ++ | ++ | ++ |
| VMN | ++ | ++ | ++ | ++ | ++ | ++ | Ru | ++ | ++ | +++ | +++ | ++ | ++ |
| LHy | + | + | + | + | ++ | ++ | nOMd/v | +++ | ++ | +++ | +++ | ++ | ++ |
| Thalamus | mnV | − | − | ++ | ++ | − | − | ||||||
| GLv | ++ | ++ | ++ | ++ | ++ | ++ | PL/PM | +++ | +++ | +++ | +++ | ++ | ++ |
| LA | ++ | ++ | ++ | ++ | ++ | ++ | VLV | +++ | +++ | +++ | +++ | − | − |
| rot | ++ | ++ | ++ | +++ | ++ | ++ | RPgc | +++ | +++ | +++ | +++ | ++ | ++ |
| DLA | ++ | ++ | ++ | ++ | ++ | ++ | IO | +++ | +++ | ++ | ++ | ++ | ++ |
| DLL | ++ | ++ | ++ | ++ | ++ | ++ | LoC | ++ | ++ | ++ | ++ | ++ | ++ |
| ICT | ++ | ++ | ++ | ++ | ++ | ++ | OS | +++ | +++ | +++ | +++ | − | − |
| VLT | ++ | ++ | ++ | ++ | ++ | ++ | nPrV | +++ | +++ | +++ | +++ | ++ | ++ |
| ov | +++ | +++ | ++ | +++ | ++ | ++ | MV | +++ | +++ | +++ | +++ | ++ | ++ |
| RSd/v | − | − | +++ | +++ | − | − | Dcn | +++ | +++ | +++ | +++ | ++ | ++ |
| SPC | ++ | ++ | ++ | ++ | − | − | Vest | ++ | ++ | ++ | ++ | ++ | ++ |
| DLP | ++ | ++ | ++ | ++ | ++ | ++ | Song system | ||||||
| DMP | ++ | ++ | ++ | ++ | − | − | HVC | + | ++ | + | ++ | + | + |
| DLM | ++ | ++ | ++ | ++ | ++ | ++ | RA | + | + | + | ++ | + | + |
| PMI | ++ | ++ | ++ | ++ | ++ | ++ | LMAN | ++ | + | ++ | + | + | + |
| DIP | ++ | ++ | ++ | ++ | ++ | ++ | Area X | − | − | + | + | + | + |
| DP | ++ | ++ | − | − | − | − | |||||||
| SpM | +++ | +++ | +++ | +++ | ++ | ++ | |||||||
| PPC | − | − | ++ | ++ | − | − | |||||||
| SP | +++ | +++ | +++ | +++ | − | − | |||||||
| SpL | +++ | +++ | +++ | +++ | ++ | ++ | |||||||
| IS | − | − | ++ | ++ | − | − | |||||||
| AP | ++ | ++ | ++ | ++ | ++ | ++ | |||||||
The hybridization level for each area was subjectively classified as relatively high (+++), medium (++), low (+), or absent (−) based on label intensity observed after film exposure and is intended only as a comparative measure of hybridization levels within an age and gene. Subjective analysis identified brain areas with the highest (+++) and lowest (+) hybridization levels for one gene at one age, then a nucleus of intermediate level (++), to serve as standards for each hybridization level for each gene and age. Brain areas not showing specific label were designated absent (−). The mean hybridization intensity level of each identifiable brain area was subjectively compared with these standard regions on the same film exposure to derive the hybridization level for each gene. No distinction between male and female hybridization is indicated here, and levels are a consensus of all brains at an age hybridized for each gene. MSt, Medial striatum; S, septal nuclei; CoS, commissuralis septi; nCPa, cells of the nucleus commissural pallii; GP, globus pallidus; LV, lateral ventricle; Tn, nucleus taeniae; PVN, paraventricular nucleus; VMN, ventromedial nucleus; LHy, lateral hypothalamus; GLv, lateral geniculate; LA, lateral anterior nucleus; rot, nucleus rotundus; DLA, anterior dorsolateral; DLL, lateral dorsolateral; ICT, nucleus intercalatus; VLT, ventrolateral nucleus; RSd/v, dorsal and ventral superior reticular nuclei; SPC, nucleus superficialis parvocellularis; DLP, posterior dorsolateral; DMP, posterior dorsomedial nucleus; DLM, medial dorsolateral; PMI, paramedianus internis; DIP, posterior dorsointermedius; nucleus PPC, principalis precommissuralis; SP, nucleus subpretectalis; IS, nucleus interstitialis; AP, area pretectalis; Tu, tuberis; SNc, substantia nigra pars compacta; IM, magnocellular isthmus nuclei; IPC, parvocellular isthmus nuclei; Pt, nucleus pretectalis; SRt, nucleus subrotundus; GCt, central gray; EM, ectomammillaris; SLu, nucleus semilunaris; EW, Edinger-Westphal; Ru, nucleus ruber; nOMd/v, distinct cells of the nucleus of the III cranial nerve; PM, medial pons; PL, lateral pons; VLV, ventral portion of the lateral lemniscus; RPgc, gigantocellularis; IO, isthmoopticus; LoC, locus ceruleus; nPrV, principal nucleus of the cranial nerve; MV, motor nucleus of the cranial nerve; N, nidopallium; M, mesopallium; HD, hyperpallium densocel-lulare; HA, hyperpallium apicale; HP, hippocampus; A, arcopallium; LSt, lateral striatum; POA, preoptic area; ov, ovoidalis; SpL, lateral nucleus of the spiriform complex; SpM, medial nucleus of the spiriform complex; SPC, nucleus superficialis parvocellularis; HM, medial habenula; HL, lateral habenula; TeO, optic tectum; SGC, cellular layer stratum griseum cellulare; ICo, nucleus intercollicularis; MLd, dorsal lateral mesencephalic nucleus; VTA, ventral tegmental area; IP, interpeduncularis nucleus; PPT, pedunculopontine tegmental nucleus; dcn, deep cerebellar nuclei; vest, vestibular nuclei; OS, superior olive; PC, Purkinje cells; RA, robust nucleus of the arcopallium; LMAN, lateral magnocellular nucleus of the nidopallium.
Image analysis
Exposed autoradiographic films of Northern blots were scanned into Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA). Contrast was modified to clarify bands and nonspecific background spots were minimized. Dark-field in situ hybridization images were captured into MR-Grab (Carl Zeiss, Inc., Thornwood, NY) with an AxioCam MRc digital camera (Zeiss) mounted on an inverted light microscope (Axioskop 20; Zeiss). Film in situ hybridization images were scanned and magnified using VueScan (Hamrick Software, http://www.hamrick.com) and an Expression 10000 XL scanner (Epson, Long Beach, CA) to produce images at final publication size and 600 dpi. All in situ images were transferred to Photoshop, where contrast and background levels were optimized and dark-field images were black-white inverted for figure clarity. All modifications to contrast were performed equally to adjacent antisense and sense hybridization pictures prevent distortion of data representations.
Results
RT-PCR and subcloning
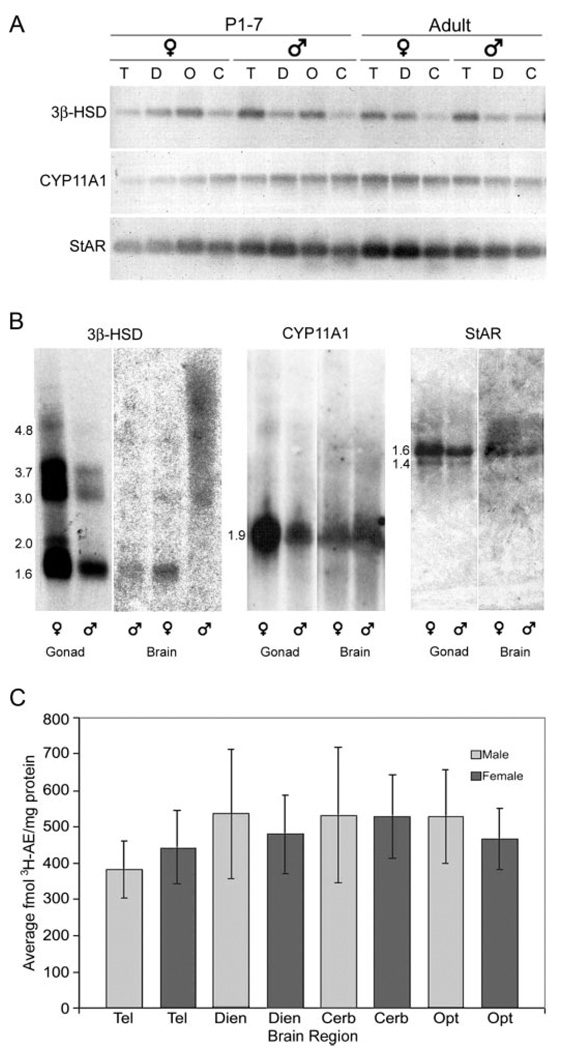
Amplification of StAR, CYP11A1, and 3β-HSD cDNA from developing and adult brain regions showed a band of the expected size in each lane for P1–7 male and female telencephalon, diencephalon, TeO, and cerebellum (Fig. 2A). Reverse transcription minus samples showed no amplification (not shown). Comparison of subclone sequences to known chicken cDNA sequences confirms PCR product identities (StAR: 85% homologous; CYP11A1: 86%; 3β-HSD: 86%). Zebra finch-specific subclones derived from RT-PCR products served as templates for cDNA library, Northern blot, and in situ hybridization probes.
FIG. 2.
3β-HSD, CYP11A1, and StAR are expressed in male and female brain. A, RT-PCR shows specific amplification of 3β-HSD, CYP11A1, and StAR cDNAs from P1–7 telencephalon (T), diencephalon (D), TeO (O), and cerebellum (C) and adult T, D, and C. B, Northern blots hybridized with 3β-HSD, CYP11A1, and StAR probes confirm the presence of mRNAs in brain. Shown are a male and female brain band from the same age with the exception of 3β-HSD. Three bands were shown for 3β-HSD, from left to right: adult male rest of brain, P1–5 female whole brain, and P1–5 male whole brain; the P1–5 male band is smeared. Gonads from adult male and female serve as positive control. Approximate transcript sizes in kilobases for each band are listed to the left of the gonadal blots for each gene. C, Steroid biochemistry detects 3β-HSD activity in telencephalon (Tel), dien-cephalon (Dien), cerebellum (Cerb), and TeO (Opt) of P5 males and females. Error bars (C), ± sem.
cDNA cloning and 5′ RACE
3β-HSD
Comparison of the zebra finch 3β-HSD brain and testis clones to the chicken cDNA sequence showed both clones contained the entire open reading frame (ORF). The testis and whole-brain nucleotide sequences were 96% identical, and only two predicted amino acids were different. These rare differences may have been introduced during the reverse transcription process; it is unknown whether these sequences are functionally distinct. Both zebra finch 3β-HSD clones had ORF of 1134 bp and a predicted protein of 377 amino acids, similar to that in other species. Both included the predicted translational start sites and contained two possible overlapping canonical AATAAA polyadenylation signals in the 3′untranslated region (UTR). The zebra finch clones show 86% nucleotide sequence identity, 87% amino acid identity, and 94% similarity with the chicken 3β-HSD cDNA. Both clones contain functional motifs such as the putative cofactor binding site and membrane spanning regions and most likely code for functional 3β-HSD proteins. The complete brain sequence was submitted to GenBank (accession no. AY445525).
CYP11A1
Comparison of the zebra finch CYP11A1 cDNA clone sequence to the chicken cDNA sequence showed that the zebra finch clone was truncated and did not include the presumptive ATG translational start codon. Therefore, a 5′ RACE subclone was sequenced and identified as CYP11A1 based on homology to the chicken sequence. A composite of the clone and 5′RACE subclone sequences was constructed and contained a 1524-bp ORF, 39-bp 5′UTR, 248-bp 3′UTR, and a predicted protein length of 508 amino acids. The zebra finch cDNA has high homology to the chicken nucleotide (80%) and amino acid (75% identity, 85% similarity) sequences, and shared putative steroid and heme binding functional domains with other clones (32,33), supporting that this cDNA sequence codes for a functional CYP11A1 protein. The complete sequence was submitted to GenBank (accession no. AY633556).
StAR
A 1492-bp StAR cDNA clone that included the 598-bp 3′UTR and 894 bp of ORF but not the putative ATG translational start site was isolated from the testicular library. The 5′RACE subclone obtained to complete the ORF sequence included the predicted translational start codon and 156 bp of 5′UTR and overlapped with the cDNA clone sequence. A composite of these two sequences was submitted to GenBank (accession no. AY505123). Both the nucleotide and predicted amino acid sequence were homologous to the chicken cDNA sequence: 85% identical with the nucleotide sequence and 86% identical and 92% similar to the predicted amino acid sequence. Additionally, the predicted translation of the zebra finch StAR cDNA includes two conserved canonical protein kinase A recognition sites functionally relevant for steroid synthesis (34). Therefore, the zebra finch cDNA likely codes for a functional protein.
Northern blots
3β-HSD
Multiple 3β-HSD transcripts were detected in brain and gonad samples on Poly (A+) Northern blots (Fig. 2B: adult male rest of brain, P1–5 female and male whole brains, adult gonads). Five transcripts, sized 4.8, 3.7, 3.0, 2.0, and 1.6 kb, hybridized to the zebra finch probe in ovary and testis. At least two of these transcripts, sized 1.6 and 3.0 kb, are apparent in adult male rest of brain and female P1–5 whole brain. 3β-HSD mRNA levels were lower in the brain samples than in the gonad samples, and the ovary hybridization was more intense than the testicular, as expected (35). The major band of 1.6 kb is similar in size to transcripts identified via Northern blotting in other species in which a second transcript, approximately 2.0 kb, was also identified (36–39). Thus, the presence of more than one 3β-HSD transcript is not unique to zebra finches and may reflect the multiple protein isoforms found in other species (40).
CYP11A1
One major transcript, approximately 1.9 kb, hybridized to a CYP11A1 probe in male and female P1–5 whole brains (Fig. 2B: P1–5 male and female whole brains, adult gonads) and adult telencephalon, rest of brain, ovary, and testis. The 1.9-kb transcript size is similar to those in other species in which a second, larger transcript has also been reported on Northern blots (33, 41). This second band probably represents alternative untranslated regions or incompletely processed pre-RNA rather than different ORF lengths because only one CYP11A1 protein has been described.
StAR
One major transcript of approximately 1.6 kb hybridized to the StAR probe in adult testis, ovary, and male and female telencephalon and hindbrain (Fig. 2B: adult male hindbrain, adult female diencephalon, adult gonads). Another transcript, approximately 1.4 kb, also hybridized to the probe in testis, ovary, male hindbrain, and female telencephalon. It is possible that the 1.4-kb mRNA is also present in the other samples but remained below detection limits, although GAPDH control hybridization shows approximately equal RNA loading in all lanes (not shown). More than one transcript is described in other species in which StAR has been analyzed, including a transcript approximately 1.6 kb (42–46); thus, these results are consistent with those from other species.
SYBR Green relative quantitative RT-PCR
To measure the relative abundance of 3β-HSD, CYP11A1, and StAR mRNA in male and female brains, individual P5 whole brains and P1 and P5 telencephalon (StAR) were subjected to rqRT-PCR. The Ct value for all minus reverse transcription control tubes was 45, or undetectable. The Ct ratios (steroidogenic gene Ct/control gene Ct) for individual birds and the average ratios ± sem for each sex and gene are listed in Table 2. The results from one-way ANOVA analysis (α < 0.05) are reported below.
TABLE 2.
Summary of SYBR Green rqRT-PCR results
| StAR | CYP11A1 | 3β-HSD | ||||
|---|---|---|---|---|---|---|
| Ratio | Average ± sem | Ratio | Average ± sem | Ratio | Average ± sem | |
| P5 | ||||||
| Female | ||||||
| A | 1.78 | 1.81 ± 0.145 | 1.89 | 1.85 ± 0.231 | 1.54 | 1.58 ± 0.208 |
| B | 1.83 | 1.85 | 1.59 | |||
| C | 1.81 | 1.81 | 1.61 | |||
| Male | ||||||
| A | 1.84 | 1.86 ± 0.145 | 1.90 | 1.89 ± 0.436 | 1.64 | 1.58 ± 0.285 |
| B | 1.89 | 1.96 | 1.55 | |||
| C | 1.86 | 1.81 | 1.56 | |||
| P1 | ||||||
| Female | ||||||
| A | 1.99 | 1.96 ± 0.130 | ||||
| B | 1.92 | |||||
| C | 1.99 | |||||
| D | 1.92 | |||||
| E | 1.94 | |||||
| F | 1.97 | |||||
| G | 2.00 | |||||
| Male | ||||||
| A | 2.00 | 1.95 ± 0.166 | ||||
| B | 1.96 | |||||
| C | 2.00 | |||||
| D | 1.95 | |||||
| E | 1.89 | |||||
| F | 1.97 | |||||
| G | 1.90 | |||||
Ratio of Ct values for StAR, CYP11A1, and 3β-HSD in individual P5 male and female whole brains (steroidogenic enzyme Ct/β-actin Ct) and individual P1 male and female telencephalon (steroidogenic enzyme Ct/GAPDH Ct). The average Ct ratios for each age, sex, and gene are listed ± sem.
3β-HSD
No significant difference in 3β-HSD mRNA levels between P5 male and female whole brain was detected (F1,5 = 0.009, P = 0.930).
CYP11A1
No significant difference was detected in relative CYP11A1 mRNA levels in P5 male and female whole brain (F1,5 = 0.658, P = 0.463).
StAR
No significant sex difference was detected in StAR mRNA levels in P5 telencephalon (F1,11 = 0.084, P = 0.778) or P1 telencephalon (F1,13 = 0.166, P = 0.691).
SYBR Green rqRT-PCR provided highly reproducible results across samples. One-way ANOVA analysis shows that mRNA abundance levels for StAR, CYP11A1, and 3β-HSD are equivalent between the sexes.
3β-HSD activity measurement
Three control tissue incubation manipulations confirm that the steroid biochemistry procedure was specific for 3β-HSD activity in P5 brains. First, the 3H-AE cold trap significantly reduced the metabolism of 3H-AE; synthesis of 3H-E1 was reduced to undetectable levels (14 fmol 3H-E1 per milligram protein without cold trap, 0.0 fmol 3H-E1 per milligram protein with cold trap). Second, inclusion of nicotinamide adenine dinucleotide phosphate instead of NAD+ prevented the conversion of 3H-DHEA to 3H-AE (59.75 fmol 3H-AE per milligram protein reduced to undetectable levels of 3H-AE). Finally, experiments testing the effectiveness of Trilostane and Fadrozole blockage of 3β-HSD and aromatase, respectively, showed reduction in 3H-DHEA metabolism. Trilostane reduced by 63% the femtomoles 3H-AE per milligram protein synthesized, but Fadrozole had no effect on the production of 3H-AE (47.6 fmol/mg protein without and 50.7 fmol/mg protein with Fadrozole) because Fadrozole is specific for aromatase. Both Trilostane and Fadrozole independently reduced the synthesis of 3H-estrogens from the 3H-DHEA to undetectable levels as expected by blocking the production of the androgen substrate for estrogen synthesis (Trilostane) or estrogen synthesis directly (Fadrozole).
Steroid biochemistry detected 3H-AE in P5 male and female brain tissue homogenates incubated with 3H-DHEA and NAD+, indicating type I 3β-HSD activity. 3β-HSD activity was detected in all four major brain regions: telencephalon, diencephalon, TeO, and cerebellum (Fig. 2C). No significant effect of sex (F1,24 = 0.290, P = 0.866), brain region (F3,24 = 0.319, P = 0.811), or interaction (F3,24 = 0.098, P = 0.961) in 3β-HSD activity was detected at P5. These experiments confirm the widespread and relatively even distribution of 3β-HSD protein in P5 males and females shown with RT-PCR techniques and Northern blot analysis.
In situ hybridization
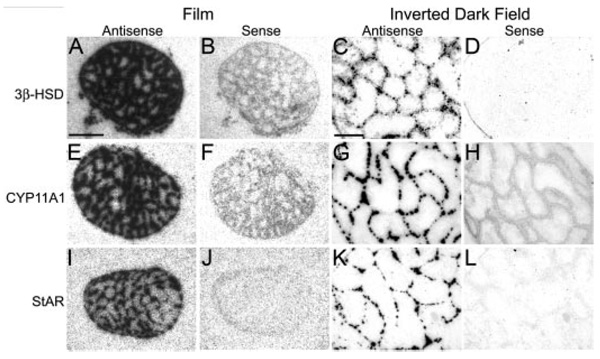
Gonads
Gonads were positive control tissues and hybridized on the same slides as the brain. All three genes hybridized to P20 and adult testes in a pattern consistent with that of interstitial cells, the androgen producing cells (Fig. 3). In females, a single line of follicular cells, most likely the steroid producing granulosa cell layer, was labeled for 3β-HSD, CYP11A1, and StAR (not shown).
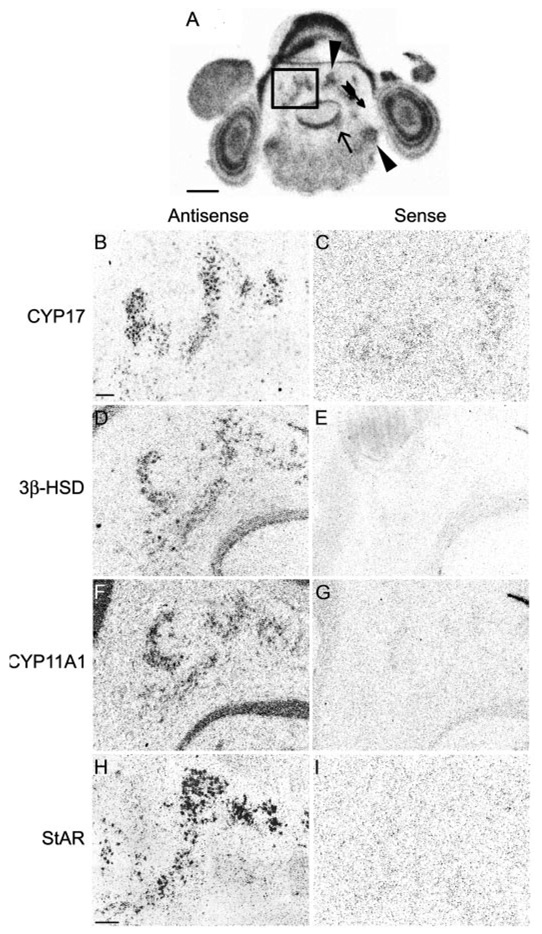
FIG. 3.
Adult testes are positive control for in situ hybridization specificity. Testes hybridized with antisense configured probes for 3β-HSD (A and C), CYP11A1 (E and G), and StAR (I and K) show high levels of hybridization over cells patterned as Leydig cells in film images (A, E, and I) and emulsion dipped slides (C, G, and K). No hybridization is seen with sense configured probes (B, F, and J and D, H, and L). Scale bar (A), 1 mm (A, B, E, F, I, and J); scale bar (C), 100 µm (C, D, G, H, K, and L).
Brain
Generally, 3β-HSD, CYP11A1, and StAR are widely expressed in P20 and adult male and female brains (Table 1). With the exception of HVC, robust nucleus of the arcopallium (RA), lateral nucleus of the anterior nidopallium (LMAN), and area X, sexually dimorphic song nuclei, no systematic sex difference was observed in the qualitative expression distribution of 3β-HSD, CYP11A1, or StAR. Thus, descriptions and relative intensity levels are a consensus summary of both males and females (n = 6 for each gene at each age). Hybridization that met the criteria described in Materials and Methods was either uniformly distributed, i.e. in low levels throughout the telencephalon, or clustered over what appeared to be cells of varying sizes with some small cells as illustrated in dark-field images in Fig. 4 and larger cells presented in Figs. 5 and 6. We were conservative and did not define labeled cell types, but it is possible that these patterns of silver grains represent hybridization over both glia and neurons, as might be expected from previous studies (17, 47–50).
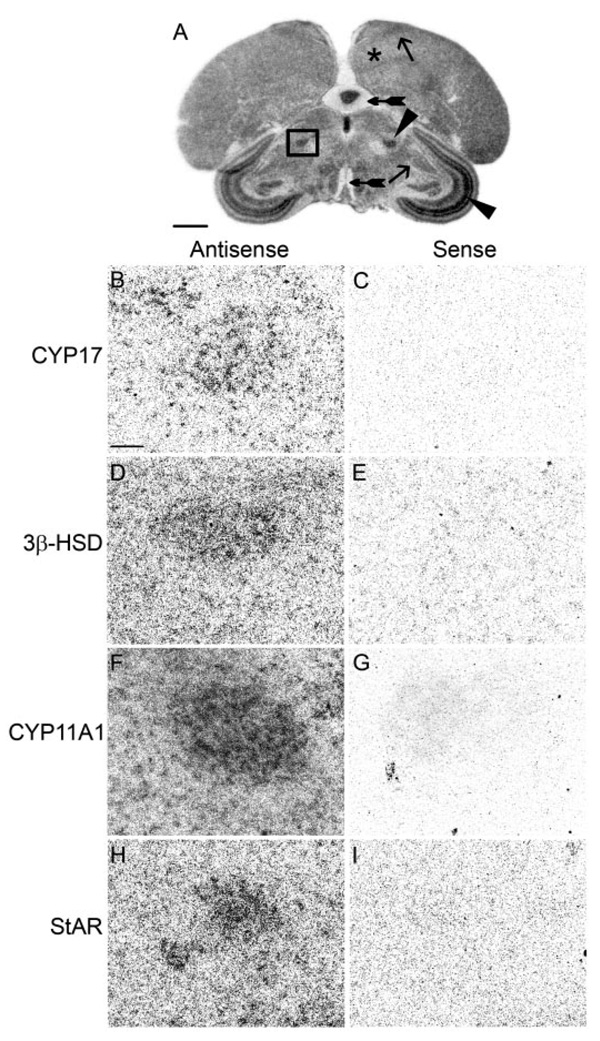
FIG. 4.
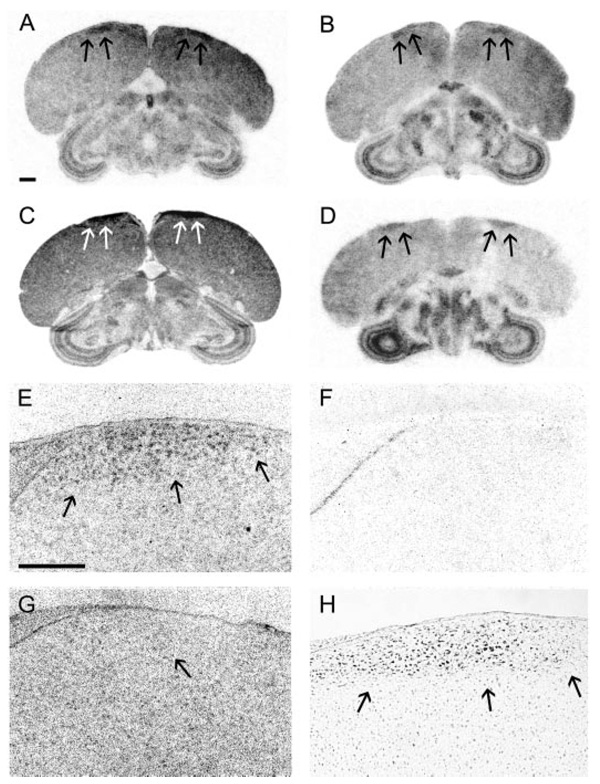
The medial nucleus of the spiriform complex, a sensory integration area, expresses CYP17, 3β-HSD, CYP11A1, and StAR. A, Low-magnification image of a film image showing position of the SpM (darkly stained nucleus in box) in an adult male section hybridized for StAR. B–I, High-magnification dark-field images of area in box. B, D, F, and H, Emulsion-dipped sections hybridized with antisense configured probes for CYP17, 3β-HSD, CYP11A1, and StAR, respectively. Adjacent sections hybridized with sense probes (C, E, G, and I) do not show label. Areas representative of low (+), medium (++), and high (+++) hybridization levels are indicated. The telencephalon with low hybridization is marked with an asterisk; the ICo/MLd and HVC determined to have medium hybridization are marked with an arrow; the SpM and SGC, areas of high levels of hybridization, are marked with an arrowhead; and the nerve tract of the III cranial nerve and the molecular layer of the cerebellum, which did not show hybridization, are indicated with tailed arrows. Scale bar (A), 1 mm; (B), 100 µm (B–I).
FIG. 5.
The multiple dcn, motor output areas, express CYP17, 3β-HSD, CYP11A1, and StAR. A, Low-magnification film image showing the position of the dcn (collection of darkly stained nuclei in box) in an adult male section hybridized for CYP11A1. B–I, Higher magnification of the area in box. B, D, F, and H, Emulsion-dipped sections hybridized with antisense configured probes for CYP17, 3β-HSD, CYP11A1, and StAR, respectively. The adjacent sections hybridized with sense probes (C, E, G, and I) do not show label. Areas representative of absent (−), medium (++), and high (+++) hybridization levels are indicated. No area determined to have low hybridization is apparent on this section. A vest determined have medium hybridization is marked with an arrow; the dcn and OS, areas of high levels of hybridization, are marked with an arrowhead; and a hindbrain region absent of hybridization is indicated with a tailed arrow. Scale bars (A, B, and H), 1 mm (A, B: B–G, H: H, I).
FIG. 6.
StAR, CYP11A1, 3β-HSD, and CYP17 are expressed in male HVC. A–D, Film images show position of HVC hybridized for StAR (A), CYP11A1 (B), 3β-HSD (C), and CYP17 (D) in adult male brain. Arrows indicate position of HVC. E–G, Inverted dark-field images of emulsion-dipped slides hybridized for CYP11A1 show specific cellular hybridization in male HVC hybridized with antisense (E) but not sense-configured probes (F). HVC label is not evident in adult female hybridized with antisense (G) configured probes. Arrow (G) indicates analogous region to HVC in the female. High-magnification image of a nissl-stained section shows the cellular anatomy of male HVC (H). Scale bars (A), 1 mm (A–D) and (E) 300 µm (E-H).
Song system
HVC
HVC was the only major song nucleus to express StAR, CYP11A1, 3β-HSD, and CYP17. This was evident in both film and emulsion-dipped slides viewed under dark-field illumination (Fig. 6). Males, but not females, showed hybridization for all four genes (Fig. 6), although the small HVC of females is extremely difficult to identify under these conditions. In all cases, the cellular hybridization distribution was equivalent across the entire male nucleus.
LMAN
The most distinctive labeling in LMAN was in P20 males but not females hybridized with CYP11A1 riboprobes (Fig. 7). In addition, StAR (Fig. 7) and 3β-HSD were expressed in P20 and adult male LMAN, but CYP17 hybridization was not found in either male or female LMAN (24). In the case of LMAN, hybridization was more obvious in P20 male than adult male brains.
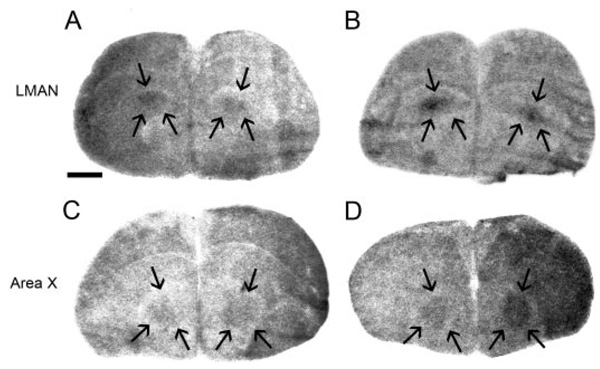
FIG. 7.
StAR and CYP11A1 are expressed in LMAN and area X. Low-magnification film images show StAR (A) and CYP11A1 (B) hybridization in P20 male LMAN and CYP11A1 in P20 (C) and adult (D) area X. Song nuclei are delineated by arrows. Scale bar, 1 mm.
Area X
This study identified CYP11A1 and 3β-HSD, but not StAR, expression in P20 and adult male area X (Fig. 7), and CYP17 mRNA had been previously described in male area X and the analogous area in females at P20 and adult (24). Females did not consistently show label for CYP11A1 or 3β-HSD in the region of area X, suggesting that CYP17 is the only gene specifically expressed in the analogous female area X region.
RA
StAR, CYP11A1 (Fig. 8), and 3β-HSD, but not CYP17 (24), were expressed in P20 and adult male RA. These genes are found in cells scattered throughout the nucleus, although at low levels not higher than those of the surrounding telencephalon (Fig. 8). RA was indistinct in females and no females showed clear signs of RA hybridization for any genes. However, because cells within the entire arcopallium (Fig. 8A) are lightly yet specifically labeled in both sexes, it is possible that the female RA also contains some steroidogenic gene label.
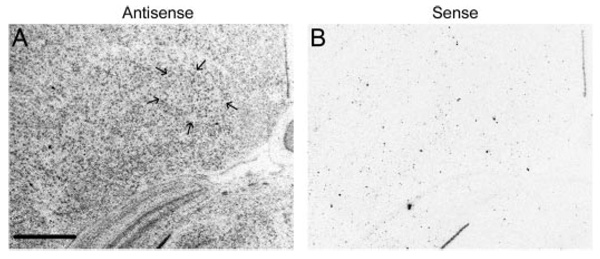
FIG. 8.
CYP11A1 is expressed in adult male RA. High-magnification dark-field images show specific hybridization of adult male RA (delineated by arrows) with CYP11A1 antisense (A) but not sense (B) configured probes. The low level of hybridization within RA is specific but not distinct from hybridization in the surrounding telencephalon. Scale bar, 500 µm.
Telencephalon
In the male and female telencephalon, all brain divisions contained cells with StAR, CYP11A1, and 3β-HSD hybridization, with no discernible sex differences. Labeled telencephalic regions included the medial striatum, nidopallium (N), mesopallium (M), hyperpallium densocellulare (HD), and hyperpallium apicale (HA). The anterior portions of M and HD showed higher levels of StAR and CYP11A1 label than the surrounding tissue, whereas HA tended to have lower hybridization levels. At the level of the anterior commissure (Fig. 9), cells of the nucleus commissural pallii and the commissuralis septi hybridized to StAR and CYP11A1 probes, whereas septal nuclei labeled solely for 3β-HSD, and CYP11A1 was the only gene to label the globus pallidus. 3β-HSD riboprobes also hybridized to a mediodor-sal subregion of the anterior hippocampus (HP). The principal cells of the HP hybridized to riboprobes for all three genes. In the caudal telencephalon, all three genes hybridized to cells in the arcopallium (A), often labeling large, distinct cells. The lateral striatum showed label for StAR and 3β-HSD, and nucleus taeniae showed label for CYP11A1 and 3β-HSD, although not at higher levels than the surrounding telencephalon. In the caudal N, 3β-HSD labeled cells were typically larger and further apart than those hybridized in the A.
FIG. 9.
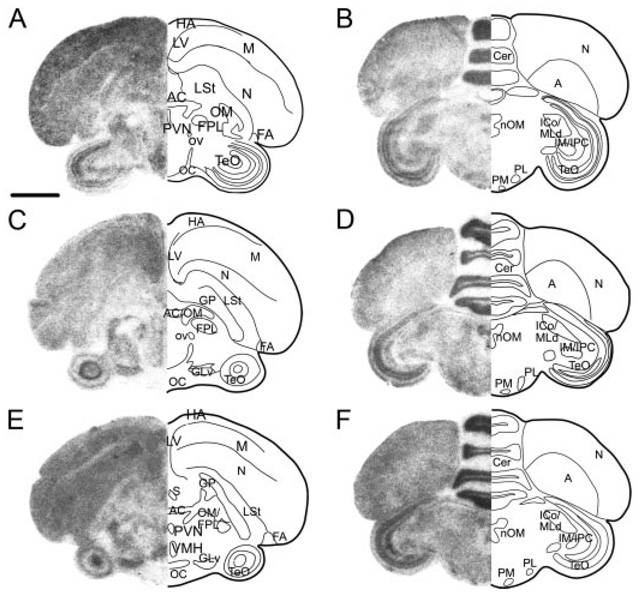
StAR, CYP11A1, and 3β-HSD expression is widely distributed but is specific to brain area and gene. Left side of each panel shows low-magnification film images of StAR (A and B), CYP11A1 (C and D), and 3β-HSD (E and F) at two anterior-posterior levels [A, C, and E at the level of the anterior commissure (AC) and B, D, and F at the level of the nucleus of the III cranial nerve (nOM)]. Right side of each panel is schematic drawing highlighting some major anatomical features of the brain section. Not all nuclei are equally distinct for each gene, and each gene shows a unique specific pattern of hybridization. Anterior sections are illustrated in Fig. 7. LV, Lateral ventricle; OM, tractus occipitomesencephalicus; FPL, lateral forebrain bundle; OC, optic chiasm; FLM, fasciculus longitudinalis medialis; Cer, cerebellum. Scale bar, 1 mm.
Diencephalon
Hypothalamic hybridization for StAR, CYP11A1, and 3β-HSD was equivalent for males and females. Several distinct brain regions showed specific hybridization for all three genes (Fig. 9), including the preoptic area (POA), lateral hypothalamus, paraventricular nucleus, and ventromedial nucleus. Many thalamic nuclei also contained cells that hybridized to riboprobes for all three genes, including the lateral geniculate, lateral anterior nucleus, nucleus intercalatus, and the ventrolateral nucleus. In addition, nucleus rotundus, ovoidalis (ov), paramedianus internis, and the lateral and medial nuclei of the spiriform complex (SpL and SpM, respectively) (Fig. 4), all contained cells that hybridized with all three riboprobes, although CYP11A1 showed the most intense label in SpM and SpL. All components of the dorsal lateral thalamus, the anterior dorsolateral, medial dorsolateral, lateral dorsolateral, posterior dorsolateral, and posterior dorsointermedius, also showed label for StAR, CYP11A1, and 3β-HSD. Three thalamic nuclei contained cells that showed only StAR and CYP11A1 hybridization: nucleus superficialis parvocellularis, posterior dorsomedial nucleus, and the nucleus subpretectalis. CYP11A1 was the only gene to identify specifically labeled cells within the nucleus subrotundus, nucleus interstitialis, nucleus principalis precommissuralis, and the dorsal and ventral superior reticular nuclei. Only riboprobes for 3β-HSD specifically labeled cells in the area pretectalis.
Midbrain and hindbrain
Many cell groups of the midbrain and hindbrain showed high levels of StAR, CYP11A1, and 3β-HSD expression (Fig. 9) in both males and females. Within the TeO, all three genes were found expressed in the cellular layer stratum griseum cellulare (SGC), magnocellular and parvocellular isthmus nuclei, nucleus semilunaris, nucleus intercollicularis (ICo), and dorsal lateral mesencephalic nucleus (MLd). SGC label was particularly striking after hybridization with 3β-HSD riboprobes. Also in the midbrain, the large cells of nucleus ruber, cells in nuclei ectomammillaris and Edinger-Westphal, and the distinct cells of the nucleus of the III cranial nerve accumulated label for StAR, CYP11A1, and 3β-HSD. Nucleus pretectalis, ventral tegmental area (VTA), ventral portion of the lateral lemniscus, and substantia nigra pars compacta contained cells that hybridized only to StAR and CYP11A1 riboprobes, whereas the central gray contained cells that hybridized solely for StAR and 3β-HSD. CYP11A1 was the only gene to show distinct label in the tuberis, interpeduncularis, and pedunculopontine tegmental nuclei.
In the hindbrain, StAR, CYP11A1, and 3β-HSD labeled cells in the medial and lateral pons, isthmoopticus, locus ceruleus, caudal reticular pons, gigantocellularis, and two components of the trigeminal nerve complex: the principal nucleus and the motor nucleus. Some of the most distinctive label for these three genes was found in the deep cerebellar nuclei (dcn) (Fig. 5) and vestibular nuclei (vest). Hybridization for StAR and CYP11A1 was detected in the superior olive (OS), whereas 3β-HSD was the only riboprobe to convincingly show hybridization in Purkinje cells. Several other hindbrain cell groups contained riboprobe label, most likely nuclei for cranial nerves, but were not specifically identifiable.
Discussion
Steroids exert powerful morphological and functional effects on the zebra finch song system throughout life, but with the exception of AR, no sex steroid-related gene has been found widely within song nuclei. Gonadally derived steroids have some activational effects but do not explain the wide range of steroid influences throughout posthatch life. Here we show using standard RT-PCR, real-time rqRT-PCR, Northern blot analyses, in situ hybridization and, for 3β-HSD, biochemical procedures that each of the genes necessary for de novo androgen synthesis, StAR, CYP11A1, 3β-HSD, and CYP17 (24), are expressed in the zebra finch brain and some or all of the major song nuclei in P20 and adult males. The sometimes overlapping, sometimes unique expression distributions throughout the brain suggest that complex local steroid environments might exist. Local steroid concentrations may influence crucial events for the construction and maintenance of the sensorimotor song circuit and other brain areas such as thalamic relay nuclei and cerebellar nuclei integral to general sensory information processing and motor function.
The presence of 3β-HSD, CYP11A1, and StAR mRNAs had not been previously examined in songbirds; therefore, after cloning each cDNA from zebra finch libraries, we identified their expression in major brain areas using several techniques. Standard RT-PCR indicated that all brain regions contained 3β-HSD, CYP11A1, and StAR mRNA in development and adulthood. SYBR Green rqRT-PCR confirmed neural expression during development, but statistical analysis of the rqRT-PCR results did not detect a significant sex difference in mRNA levels. Northern blot analysis demonstrated that 3β-HSD, CYP11A1, and StAR mRNAs could be detected in brains. Although expression levels were low, these transcripts have been very difficult to detect in the brains of other species in which the protein was readily detectable [e.g. CYP11A1 (51)]. This implies that protein levels are higher than mRNA levels. If this mRNA to protein ratio holds for the zebra finch, it may be that protein levels are relatively high in the zebra finch brain, as is true for aromatase and 3β-HSD. Activity measurements of 3β-HSD, the only enzyme studied here with a reliable biochemical assay, indicated that the mRNA detection reflected functional protein in P5 male and female brains. This experiment did not detect a regional or sex difference, although this procedure can detect such differences under select environmental conditions in the adult (27). Attempts are ongoing to develop reliable and sensitive biochemical assays for CYP11A1 and CYP17. The various molecular methodologies establish that StAR, CYP11A1, and 3β-HSD are expressed in the zebra finch brain.
The neural expression of multiple steroidogenic genes that can be regulated independently implies that highly specific steroid environments could be created. Gonads can secrete large quantities of steroids, but these steroids are relatively nonspecific in the context of a complex tissue such as the brain. Global mRNA levels of steroidogenic genes may be low in the brain, but if expression is carefully regulated, then discrete, relevant steroid microenvironments may be created to provide distinct brain regions or cell types with unique combinations of steroids at developmentally or behaviorally appropriate times. For example, two brain areas, the SpM and the dcn, were shown here to express StAR, CYP11A1, 3β-HSD, and CYP17. Hybridization of all four genes appears indistinguishable in the dcn, suggesting equivalent cell type expression and transcript abundance. Hybridization patterns were, however, slightly different for each gene in the SpM, as demonstrated by the varying combinations of large silver grain clusters seen with CYP11A1 hybridization, compared with the smaller, more diffuse silver grain clusters seen with 3β-HSD hybridization. Furthermore, not all brain areas express all four genes; for example, abundant CYP17 expression was described in the habenula (24), but StAR, CYP11A1, and 3β-HSD did not label habenula cells. Finally, in general, the expression distributions for StAR, CYP11A1, and 3β-HSD are more widespread than CYP17 and aromatase. One explanation may be that StAR and CYP11A1 mRNAs are present in many areas to confer widespread capacity for neurosteroidogenesis, 3β-HSD may be widely distributed due to its position as a branch point for synthesis of all classes of steroids, but CYP17 (24) and aromatase (23) mRNAs are more limited in distribution to restrict production of androgens and estrogens. The sometimes unique, sometimes overlapping expression of steroidogenic enzymes in various brain regions, across development and in particular cell types, has been described in other animals (48, 52–56), indicating that regulated neurosteroidogenesis may be important to modulate individual neural functions and minimizing adverse effects on other brain areas.
Neurosteroidogenesis may also broaden the role of steroids in the brain. By uncoupling the synthesis of sex steroids from sex organs, the role of sex steroids may be expanded in the brain beyond those primarily useful for reproductive behaviors. Neurosteroids may act in concert with gonadally derived steroids to modify reproductive function, as evidenced by steroidogenic gene expression in nuclei such as the POA, but neurosteroids could also influence sensory and motor systems beyond the reproductive circuits. For example, expression of StAR, CYP11A1, 3β-HSD, and CYP17 in the spiriform complex, a collection of nuclei that perform sensorimotor processing, suggests neurosteroid regulation in global sensory integration. Furthermore, steroidogenic gene expression in auditory areas such as nucleus ov may be relevant to song processing because ov makes projections to telencephalic areas (57) that are integral to individual song processing (58). Expression of steroidogenic genes in the dcn, efferent targets of the Purkinje cells, suggest that neurosteroids could modulate global cerebellar activity, potentially including the postural and beak movement components of song production. Aromatase does not share this expression pattern (23), furthering the idea that the steroids synthesized by CYP11A1, 3β-HSD, and CYP17 could modulate sensory and motor systems independently from estrogens.
Neurosteroids synthesized by the enzymes studied here have been shown in other systems to exert a myriad of effects on multiple cell attributes, including cell morphology, chemical and electrical signaling, survival, and migration (59–61). Thus, neurosteroids and their nonclassical actions may at least partially explain the often contradictory results of global estrogen and androgen manipulations on the song system (5). This study, along with a previous study reporting CYP17 in area X (24), shows that the major song nuclei have the capacity to synthesize neurosteroids locally, although not all nuclei have the same expression profiles. Steroidogenic gene expression within major song nuclei is not always highly distinct from surrounding brain regions, but with the exception of androgen receptors, expression studies of other steroid-related genes show very limited or no expression within song nuclei, compared with surrounding brain. Thus, the expression of these genes, and the sex differences observed in their expression within the major song nuclei, suggest that neurosteroids are a fundamental component of song nucleus function. Major song nuclei are already identifiable by P20; it may be that the role of the specific combination of locally synthesized neurosteroids is to fine-tune cellular events in the song system that optimize the complex processes of male developmental song learning that start around P20 and the adult production of a highly stereotyped song.
For example, StAR and CYP11A1 are expressed at P20 in LMAN, a nucleus positioned to convey sensory information to motor pathways during song learning (62, 63). Pregnenolone or its sulfated version can modulate N-methyl-d-aspartate receptor (NMDAR) properties (59). During developmental song learning, NMDAR activity is necessary in LMAN (64, 65), and the volume of projections to LMAN is altered (66) in a time course similar to alterations in LMAN NMDAR number and structure (67). Exquisite neurosteroid control of NMDAR activation, mediated by local StAR and CYP11A1 activity, could facilitate anatomical changes necessary for normal song system organization and song learning.
StAR and CYP11A1 are also expressed in RA, the premotor nucleus of the song system. RA activity is synchronized, precise, and predictive of motor output (68). Pregnenolone or Pregnenolone or its sulfated version inhibit γ-aminobutyric acid type A receptors (GABAAR) (61) and pharmacological blockade of GABAAR in RA results in electrical bursts that coincide with involuntary vocalizations (69). Thus, exact control over RA neurons is essential for proper song production. HVC axons synapse directly on GABAergic interneurons and GABAAR-expressing premotor neurons in RA and generally direct RA activity (70). It is possible that interactions between HVC input and neurosteroid regulation of GABAAR could allow RA to send out the accurate and reproducible vocalization signals necessary for production of stereotyped song.
HVC was the only major song nucleus to express all four steroidogenic genes. HVC projects to two nuclei, area X of the sensory pathway and RA of the motor pathway, and integrates both sensory and motor components of the song system (71). It is possible that the most complex mix of steroids is necessary in HVC to facilitate its intricate integrative processing. Local steroid synthesis may be especially potent in HVC; intracerebral steroid implants more profoundly alter cellular HVC characteristics the closer they are to HVC (72). Furthermore, unilateral HVC lesions minimize steroid sensitive masculine traits of area X and RA (73), and the masculine projection from HVC to RA is steroid sensitive (15). Thus, neurosteroidogenesis within HVC may be important not solely for HVC functions but may also have wider relevance to song system masculinization due to its connections with other major song nuclei.
Previous studies revealed that the zebra finch brain is capable of steroid metabolism and synthesis in vivo and in vitro (15, 74, 75) and measured levels of individual steroidogenic enzymes in large tissue regions (Refs. 27, 76 and this paper). This study, by describing the detailed spatiotemporal expression distributions of multiple genes essential for de novo steroid synthesis, meaningfully increases the complexity with which neurosteroidogenesis can be conceived functionally. Comprehensive expression mapping suggests that neurosteroidogenesis may be a crucial mechanism by which the brain controls sensory and motor functions. The implications for local steroid synthesis are especially intriguing for precise regulation of the steroid sensitive song system. It is possible that neurosteroids may reconcile discrepancies between circulating steroid levels and steroid sensitive behaviors seen in many species of songbirds (1), and further investigation of neurosteroidogenesis in the zebra finch song system may be invaluable to understand the role of neurosteroids in natural, behaviorally relevant neural systems.
Acknowledgments
The authors thank Dr. Jim Boulter and Dr. Arthur Arnold for generously sharing resources and knowledge and Noel Alday, Petra Wise, Camilla Peabody, and Yu Ping Tang for technical support.
This work was supported by National Institute of Mental Health Predoctoral NRSA MH65114 (to S.E.L.), Canadian Institutes of Health Research Postdoctoral Fellowship (to D.A.M.), National Institutes of Health Grants R01 MH55488 and K02 MH065907 (to J.W.) and R01 MH61994 (to B.A.S.).
Abbreviations
- A
Arcopallium
- AR
androgen receptor
- Ct
threshold cycle
- CYP17
cytochrome P45017α-hydroxylase/17,20 lyase
- CYP11A1
cytochrome P450 side chain cleavage
- dcn
deep cerebellar nuclei
- DHEA
3H-dehydroepiandrosterone
- E1
3H-estrone
- GABAAR
γ-aminobutyric acid type A receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
hyperpallium apicale
- 3H-AE
3H-androstenedione
- HD
hyperpallium densocellulare
- HP
hippocampus
- 3β-HSD
3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase
- ICo
nucleus inter-collicularis
- LMAN
lateral nucleus of the anterior nidopallium
- M
mesopallium
- MLd
dorsal lateral mesencephalic nucleus
- N
nidopallium
- NAD+
nicotinamide adenine dinucleotide
- NMDAR
N-methyl-d-aspartate receptor
- ORF
open reading frame
- OS
superior olive
- ov
ovoidalis
- P
posthatch day
- PMI
paramedianus internis
- POA
preoptic area
- RA
robust nucleus of the arcopallium
- RACE
rapid amplification of cDNA ends
- rq
relative quantitative
- SGC
stratum griseum cellulare
- SpL
lateral nucleus of the spiriform complex
- SpM
medial nucleus of the spiriform complex
- StAR
steroidogenic acute regulatory protein
- TeO
optic tectum
- UTR
untranslated region
- vest
vestibular nuclei
- VTA
ventral tegmental area
References
- 1.Wingfield JC, Jacobs J, Hillgarth N. Ecological constraints and the evolution of hormone-behavior interrelationships. Ann NY Acad Sci. 1997;15:22–41. doi: 10.1111/j.1749-6632.1997.tb51911.x. [DOI] [PubMed] [Google Scholar]
- 2.Nottebohm F, Arnold AP. Sexual dimorphism in vocal control areas of the songbird brain. Science. 1976;194:211–213. doi: 10.1126/science.959852. [DOI] [PubMed] [Google Scholar]
- 3.Bottjer SW, Johnson F. Circuits, hormones, and learning: vocal behavior in songbirds. J Neurobiol. 1997;33:602–618. doi: 10.1002/(sici)1097-4695(19971105)33:5<602::aid-neu8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Schlinger BA. Sex steroids and their actions on the birdsong system. J Neurobiol. 1997;33:619–631. [PubMed] [Google Scholar]
- 5.Wade J, Arnold AP. Sexual differentiation of the zebra finch song system. Ann NY Acad Sci. 2004;1016:540–559. doi: 10.1196/annals.1298.015. [DOI] [PubMed] [Google Scholar]
- 6.Schlinger BA. Sexual differentiation of avian brain and behavior: current views on gonadal hormone-dependent and independent mechanisms. Annu Rev Physiol. 1998;60:407–429. doi: 10.1146/annurev.physiol.60.1.407. [DOI] [PubMed] [Google Scholar]
- 7.Gurney ME. Hormonal control of cell form and number in the zebra finch song system. J Neurosci. 1981;1:658–673. doi: 10.1523/JNEUROSCI.01-06-00658.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adkins-Regan E, Mansukhani V, Seiwert C, Thompson R. Sexual differentiation of brain and behavior in the zebra finch: critical periods for effects of early estrogen treatment. J Neurobiol. 1994;25:865–877. doi: 10.1002/neu.480250710. [DOI] [PubMed] [Google Scholar]
- 9.Simpson HB, Vicario DS. Early estrogen treatment alone causes female zebra finches to produce learned, male-like vocalizations. J Neurobiol. 1991;22:755–776. doi: 10.1002/neu.480220710. [DOI] [PubMed] [Google Scholar]
- 10.Wade J, Gong A, Arnold AP. Effects of embryonic estrogen on differentiation of the gonads and secondary sexual characteristics of male zebra finches. J Exp Zool. 1997;278:405–411. doi: 10.1002/(sici)1097-010x(19970815)278:6<405::aid-jez8>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Gong A, Freking FW, Wingfield J, Schlinger BA, Arnold AP. Effects of embryonic treatment with fadrozole on phenotype of gonads, syrinx, and neural song system in zebra finches. Gen Comp Endocrinol. 1999;115:346–353. doi: 10.1006/gcen.1999.7318. [DOI] [PubMed] [Google Scholar]
- 12.Nordeen EJ, Nordeen KW, Arnold AP. Sexual differentiation of androgen accumulation within the zebra finch brain through selective cell loss and addition. J Comp Neurol. 1987;259:393–399. doi: 10.1002/cne.902590307. [DOI] [PubMed] [Google Scholar]
- 13.Gahr M, Metzdorf R. Distribution and dynamics in the expression of androgen and estrogen receptors in vocal control systems of songbirds. Brain Res Bull. 1997;44:509–517. doi: 10.1016/s0361-9230(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 14.Grisham W, Lee J, McCormick ME, Yang-Stayner K, Arnold AP. Antiandrogen blocks estrogen-induced masculinization of the song system in female zebra finches. J Neurobiol. 2002;51:1–8. doi: 10.1002/neu.10028. [DOI] [PubMed] [Google Scholar]
- 15.Holloway CC, Clayton DF. Estrogen synthesis in the male brain triggers development of the avian song control pathway in vitro. Nat Neurosci. 2001;4:170–175. doi: 10.1038/84001. [DOI] [PubMed] [Google Scholar]
- 16.Takase M, Ukena K, Yamazaki T, Kominami S, Tsutsui K. Pregnenolone, pregnenolone sulfate, and cytochrome P450 side-chain cleavage enzyme in the amphibian brain and their seasonal changes. Endocrinology. 1999;140:1936–1944. doi: 10.1210/endo.140.4.6641. [DOI] [PubMed] [Google Scholar]
- 17.Tsutsui K, Ukena K, Takase M, Kohchi C, Lea RW. Neurosteroid biosynthesis in vertebrate brains. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1999;124:121–129. doi: 10.1016/s0742-8413(99)00065-1. [DOI] [PubMed] [Google Scholar]
- 18.Forlano PM, Deitcher DL, Myers DA, Bass AH. Anatomical distribution and cellular basis for high levels of aromatase activity in the brain of teleost fish: aromatase enzyme and mRNA expression identify glia as source. J Neurosci. 2001;21:8943–8955. doi: 10.1523/JNEUROSCI.21-22-08943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieu M, Mensah-Nyagan AG, Vallarino M, Do-Rego JL, Beaujean D, Vaudry D, Luu-The V, Pelletier G, Vaudry H. Immunohistochemical localization of 3β-hydroxysteroid dehydrogenase and 5α-reductase in the brain of the African lungfish Protopterus annectens. J Comp Neurol. 2001;438:123–135. doi: 10.1002/cne.1304. [DOI] [PubMed] [Google Scholar]
- 20.Mellon SH, Vaudry H. Biosynthesis of neurosteroids and regulation of their synthesis. Int Rev Neurobiol. 2001;46:33–78. doi: 10.1016/s0074-7742(01)46058-2. [DOI] [PubMed] [Google Scholar]
- 21.Mensah-Nyagan AG, Beaujean D, Luu-The V, Pelletier G, Vaudry H. Anatomical and biochemical evidence for the synthesis of unconjugated and sulfated neurosteroids in amphibians. Brain Res Brain Res Rev. 2001;37:13–24. doi: 10.1016/s0165-0173(01)00110-2. [DOI] [PubMed] [Google Scholar]
- 22.Stoffel-Wagner B. Neurosteroid biosynthesis in the human brain and its clinical implications. Ann NY Acad Sci. 2003;1007:64–78. doi: 10.1196/annals.1286.007. [DOI] [PubMed] [Google Scholar]
- 23.Shen P, Schlinger BA, Campagnoni AT, Arnold AP. An atlas of aromatase mRNA expression in the zebra finch brain. J Comp Neurol. 1995;360:172–184. doi: 10.1002/cne.903600113. [DOI] [PubMed] [Google Scholar]
- 24.London SE, Boulter J, Schlinger BA. Cloning of the zebra finch androgen synthetic enzyme CYP17: a study of its neural expression throughout post-hatch development. J Comp Neurol. 2003;467:496–508. doi: 10.1002/cne.10936. [DOI] [PubMed] [Google Scholar]
- 25.Vanson A, Arnold AP, Schlinger BA. 3β-Hydroxysteroid dehydrogenase/isomerase and aromatase activity in primary cultures of developing zebra finch telencephalon: dehydroepiandrosterone as substrate for synthesis of androstenedione and estrogens. Gen Comp Endocrinol. 1996;102:342–350. doi: 10.1006/gcen.1996.0077. [DOI] [PubMed] [Google Scholar]
- 26.Cam V, Schlinger BA. Activities of aromatase and 3β-hydroxysteroid dehydrogenase/Δ4-Δ5 isomerase in whole organ cultures of tissues from developing zebra finches. Horm Behav. 1998;33:31–39. doi: 10.1006/hbeh.1998.1434. [DOI] [PubMed] [Google Scholar]
- 27.Soma KK, Alday NA, Hau M, Schlinger BA. Dehydroepiandrosterone metabolism by β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase in adult zebra finch brain: sex difference and rapid effect of stress. Endocrinology. 2004;145:1668–1677. doi: 10.1210/en.2003-0883. [DOI] [PubMed] [Google Scholar]
- 28.Wade J, Schlinger BA, Hodges L, Arnold AP. Fadrozole: a potent and specific inhibitor of aromatase in the zebra finch brain. Gen Comp Endocrinol. 1994;94:53–61. doi: 10.1006/gcen.1994.1059. [DOI] [PubMed] [Google Scholar]
- 29.Stokes TM, Leonard CM, Nottebohm F. The telencephalon, diencephalon, and mesencephalon of the canary, Serinus canaria, in stereotaxic coordinates. J Comp Neurol. 1974:337–374. doi: 10.1002/cne.901560305. [DOI] [PubMed] [Google Scholar]
- 30.Breazile JE, Kuenzel WJ. Systema nervosum centrale. In: Baumel JJ, editor. Handbook of avian anatomy: nomina anatomica avium. Cambridge, MA: The Nuttall Ornithological Club; 1993. pp. 493–554. [Google Scholar]
- 31.Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, Wild M, Ball GF, Durand S, Gunturkun O, Lee DW, Mello CV, Powers A, White SA, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED, Guturkun O. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomura O, Nakabayashi O, Nishimori K, Mizuno S. The cDNA cloning and transient expression of a chicken gene encoding cytochrome P-450scc. Gene. 1997;185:217–222. doi: 10.1016/s0378-1119(96)00645-2. [DOI] [PubMed] [Google Scholar]
- 33.Boerboom D, Sirois J. Equine P450 cholesterol side-chain cleavage and 3β-hydroxysteroid dehydrogenase/Δ(5)-Δ(4) isomerase: molecular cloning and regulation of their messenger ribonucleic acids in equine follicles during the ovulatory process. Biol Reprod. 2001;64:206–215. doi: 10.1095/biolreprod64.1.206. [DOI] [PubMed] [Google Scholar]
- 34.LeHoux JG, Fleury A, Ducharme L, Hales DB. Phosphorylation of the hamster adrenal steroidogenic acute regulatory protein as analyzed by two-dimensional polyacrylamide gel electrophoreses. Mol Cell Endocrinol. 2004;215:127–134. doi: 10.1016/j.mce.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Nakabayashi O, Nomura O, Nishimori K, Mizuno S. The cDNA cloning and transient expression of a chicken gene encoding a 3β-hydroxysteroid dehydrogenase/Δ5–>4 isomerase unique to major steroidogenic tissues. Gene. 1995;162:261–265. doi: 10.1016/0378-1119(95)00278-e. [DOI] [PubMed] [Google Scholar]
- 36.Zhao HF, Rheaume E, Trudel C, Couet J, Labrie F, Simard J. Structure and sexual dimorphic expression of a liver-specific rat 3β-hydroxysteroid dehydrogenase/isomerase. Endocrinology. 1990;127:3237–3239. doi: 10.1210/endo-127-6-3237. [DOI] [PubMed] [Google Scholar]
- 37.Bain PA, Yoo M, Clarke T, Hammond SH, Payne AH. Multiple forms of mouse 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase and differential expression in gonads, adrenal glands, liver, and kidneys of both sexes. Proc Natl Acad Sci USA. 1991;88:8870–8874. doi: 10.1073/pnas.88.20.8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorence MC, Naville D, Graham-Lorence SE, Mack SO, Murry BA, Trant JM, Mason JI. 3β-Hydroxysteroid dehydrogenase/Δ5-Δ4-isomerase expression in rat and characterization of the testis isoform. Mol Cell Endocrinol. 1991;80:21–31. doi: 10.1016/0303-7207(91)90139-j. [DOI] [PubMed] [Google Scholar]
- 39.Guennoun R, Fiddes RJ, Gouezou M, Lombes M, Baulieu EE. A key enzyme in the biosynthesis of neurosteroids, 3β-hydroxysteroid dehydrogenase/Δ5-Δ4-isomerase (3β-HSD), is expressed in rat brain. Brain Res Mol Brain Res. 1995;30:287–300. doi: 10.1016/0169-328x(95)00016-l. [DOI] [PubMed] [Google Scholar]
- 40.Payne AH, Abbaszade IG, Clarke TR, Bain PA, Park CH. The multiple murine 3β-hydroxysteroid dehydrogenase isoforms: structure, function, and tissue- and developmentally specific expression. Steroids. 1997;62:169–175. doi: 10.1016/s0039-128x(96)00177-8. [DOI] [PubMed] [Google Scholar]
- 41.Vilchis F, Chavez B, Larrea F, Timossi C, Montiel F. The cDNA cloning and tissue expression of the cytochrome P450scc from Syrian hamster (Me-socricetus auratus) Gen Comp Endocrinol. 2002;126:279–286. doi: 10.1016/s0016-6480(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 42.Clark BJ, Wells J, King SR, Stocco DM. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. [PubMed] [Google Scholar]
- 43.Sugawara T, Lin D, Holt JA, Martin KO, Javitt NB, Miller WL, Strauss JF., III Structure of the human steroidogenic acute regulatory protein (StAR) gene: StAR stimulates mitochondrial cholesterol 27-hydroxylase activity. Biochemistry. 1995;34:12506–12512. doi: 10.1021/bi00039a004. [DOI] [PubMed] [Google Scholar]
- 44.Bauer MP, Bridgham JT, Langenau DM, Johnson AL, Goetz FW. Conservation of steroidogenic acute regulatory (StAR) protein structure and expression in vertebrates. Mol Cell Endocrinol. 2000;168:119–125. doi: 10.1016/s0303-7207(00)00316-6. [DOI] [PubMed] [Google Scholar]
- 45.Menjivar M, Castillo N, Vilchis F, Medina-Campos ON, Pedraza-Chaverri J, Guadalupe Ortiz-Lopez M. Impaired testicular cytochrome P450 side-chain-cleavage (P450scc) and steroidogenic acute regulatory (StAR) protein expression in rats with nephrotic syndrome. Mol Cell Endocrinol. 2003;209:1–7. doi: 10.1016/j.mce.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Manna PR, Huhtaniemi IT, Stocco DM. Detection of hCG responsive expression of the steroidogenic acute regulatory protein in mouse Leydig cells. Biol Proced Online. 2004;6:83–93. doi: 10.1251/bpo76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mensah-Nyagan AG, Do-Rego JL, Beaujean D, Luu-The V, Pelletier G, Vaudry H. Neurosteroids: expression of steroidogenic enzymes and regulation of steroid biosynthesis in the central nervous system. Pharmacol Rev. 1999;51:63–81. [PubMed] [Google Scholar]
- 48.Zwain IH, Yen SS. Neurosteroidogenesis in astrocytes, oligodendrocytes, and neurons of cerebral cortex of rat brain. Endocrinology. 1999;140:3843–3852. doi: 10.1210/endo.140.8.6907. [DOI] [PubMed] [Google Scholar]
- 49.Compagnone NA, Mellon SH. Neurosteroids: biosynthesis and function of these novel neuromodulators. Front Neuroendocrinol. 2000;21:1–56. doi: 10.1006/frne.1999.0188. [DOI] [PubMed] [Google Scholar]
- 50.King SR, Manna PR, Ishii T, Syapin PJ, Ginsberg SD, Wilson K, Walsh LP, Parker KL, Stocco DM, Smith RG, Lamb DJ. An essential component in steroid synthesis. The steroidogenic acute regulatory protein, is expressed in discrete regions of the brain. J Neurosci. 2002;22:10613–10620. doi: 10.1523/JNEUROSCI.22-24-10613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Compagnone NA, Bulfone A, Rubenstein JL, Mellon SH. Expression of the steroidogenic enzyme P450scc in the central and peripheral nervous systems during rodent embryogenesis. Endocrinology. 1995;136:2689–2696. doi: 10.1210/endo.136.6.7750493. [DOI] [PubMed] [Google Scholar]
- 52.Akwa Y, Young J, Kabbadj K, Sancho MJ, Zucman D, Vourc’h C, Jung-Testas I, Hu ZY, Le Goascogne C, Jo DH, et al. Neurosteroids: biosynthesis, metabolism and function of pregnenolone and dehydroepiandrosterone in the brain. J Steroid Biochem Mol Biol. 1991;40:71–81. doi: 10.1016/0960-0760(91)90169-6. [DOI] [PubMed] [Google Scholar]
- 53.Tsutsui K, Ukena K. Neurosteroids in the cerebellar Purkinje neuron and their actions. Int J Mol Med. 1999;4:49–56. [PubMed] [Google Scholar]
- 54.Plassart-Schiess E, Baulieu EE. Neurosteroids: recent findings. Brain Res Brain Res Rev. 2001;37:133–140. doi: 10.1016/s0165-0173(01)00113-8. [DOI] [PubMed] [Google Scholar]
- 55.Mellon SH, Griffin LD, Compagnone NA. Biosynthesis and action of neurosteroids. Brain Res Brain Res Rev. 2001;37:3–12. doi: 10.1016/s0165-0173(01)00109-6. [DOI] [PubMed] [Google Scholar]
- 56.Sierra A. Neurosteroids: the StAR protein in the brain. J Neuroendocrinol. 2004;16:787–793. doi: 10.1111/j.1365-2826.2004.01226.x. [DOI] [PubMed] [Google Scholar]
- 57.Vates GE, Broome BM, Mello CV, Nottebohm F. Auditory pathways of caudal telencephalon and their relation to the song system of adult male zebra finches. J Comp Neurol. 1996;366:613–642. doi: 10.1002/(SICI)1096-9861(19960318)366:4<613::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 58.Mello CV. Identification and analysis of vocal communication pathways in birds through inducible gene expression. An Acad Bras Cienc. 2004;76:243–246. doi: 10.1590/s0001-37652004000200008. [DOI] [PubMed] [Google Scholar]
- 59.Mellon SH, Griffin LD. Neurosteroids: biochemistry and clinical significance. Trends Endocrinol Metab. 2002;13:35–43. doi: 10.1016/s1043-2760(01)00503-3. [DOI] [PubMed] [Google Scholar]
- 60.Schumacher M, Akwa Y, Guennoun R, Robert F, Labombarda F, Desarnaud F, Robel P, De Nicola AF, Baulieu EE. Steroid synthesis and metabolism in the nervous system: trophic and protective effects. J Neurocytol. 2000;29:307–326. doi: 10.1023/a:1007152904926. [DOI] [PubMed] [Google Scholar]
- 61.Belelli D, Lambert JJ. Neurosteroids: endogenous regulators of the GABA(A) receptor. Nat Rev Neurosci. 2005;6:565–575. doi: 10.1038/nrn1703. [DOI] [PubMed] [Google Scholar]
- 62.Bottjer SW, Halsema KA, Brown SA, Miesner EA. Axonal connections of a forebrain nucleus involved with vocal learning in zebra finches. J Comp Neurol. 1989;279:312–326. doi: 10.1002/cne.902790211. [DOI] [PubMed] [Google Scholar]
- 63.Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basham ME, Nordeen EJ, Nordeen KW. Blockade of NMDA receptors in the anterior forebrain impairs sensory acquisition in the zebra finch (Poephila guttata) Neurobiol Learn Mem. 1996;66:295–304. doi: 10.1006/nlme.1996.0071. [DOI] [PubMed] [Google Scholar]
- 65.Aamodt SM, Nordeen EJ, Nordeen KW. Blockade of NMDA receptors during song model exposure impairs song development in juvenile zebra finches. Neurobiol Learn Mem. 1996;65:91–98. doi: 10.1006/nlme.1996.0010. [DOI] [PubMed] [Google Scholar]
- 66.Johnson F, Bottjer SW. Growth and regression of thalamic efferents in the song-control system of male zebra finches. J Comp Neurol. 1992;326:442–450. doi: 10.1002/cne.903260309. [DOI] [PubMed] [Google Scholar]
- 67.Wallhausser-Franke E, Nixdorf-Bergweiler BE, DeVoogd TJ. Song isolation is associated with maintaining high spine frequencies on zebra finch 1MAN neurons. Neurobiol Learn Mem. 1995;64:25–35. doi: 10.1006/nlme.1995.1041. [DOI] [PubMed] [Google Scholar]
- 68.Yu AC, Margoliash D. Temporal hierarchical control of singing in birds. Science. 1996;273:1871–1875. doi: 10.1126/science.273.5283.1871. [DOI] [PubMed] [Google Scholar]
- 69.Vicario DS, Raksin JN. Possible roles for GABAergic inhibition in the vocal control system of the zebra finch. Neuroreport. 2000;11:3631–3635. doi: 10.1097/00001756-200011090-00046. [DOI] [PubMed] [Google Scholar]
- 70.Spiro JE, Dalva MB, Mooney R. Long-range inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J Neurophysiol. 1999;81:3007–3020. doi: 10.1152/jn.1999.81.6.3007. [DOI] [PubMed] [Google Scholar]
- 71.Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 72.Grisham W, Mathews GA, Arnold AP. Local intracerebral implants of estrogen masculinize some aspects of the zebra finch song system. J Neurobiol. 1994;25:185–196. doi: 10.1002/neu.480250209. [DOI] [PubMed] [Google Scholar]
- 73.Herrmann K, Arnold AP. Lesions of HVc block the developmental masculinizing effects of estradiol in the female zebra finch song system. J Neurobiol. 1991;22:29–39. doi: 10.1002/neu.480220104. [DOI] [PubMed] [Google Scholar]
- 74.Schlinger BA, Arnold AP. Brain is the major site of estrogen synthesis in a male songbird. Proc Natl Acad Sci USA. 1991;88:4191–4194. doi: 10.1073/pnas.88.10.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schlinger BA, Arnold AP. Circulating estrogens in a male songbird originate in the brain. Proc Natl Acad Sci USA. 1992;89:7650–7653. doi: 10.1073/pnas.89.16.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vockel A, Prove E, Balthazart J. Sex- and age-related differences in the activity of testosterone-metabolizing enzymes in microdissected nuclei of the zebra finch brain. Brain Res. 1990;511:291–302. doi: 10.1016/0006-8993(90)90174-a. [DOI] [PubMed] [Google Scholar]