Abstract
Background
Increased ethanol intake, a major predictor for the development of alcohol use disorders, is facilitated by the development of tolerance to both the aversive and pleasurable effects of the drug. The molecular mechanisms underlying ethanol tolerance development are complex and are not yet well understood.
Methods
To identify genetic mechanisms that contribute to ethanol tolerance, we examined the time course of gene expression changes elicited by a single sedating dose of ethanol in Drosophila, and completed a behavioral survey of strains harboring mutations in ethanol-regulated genes.
Results
Enrichment for genes in metabolism, nucleic acid binding, olfaction, regulation of signal transduction, and stress suggests that these biological processes are coordinately affected by ethanol exposure. We also detected a coordinate up-regulation of genes in the Toll and Imd innate immunity signal transduction pathways. A multi-study comparison revealed a small set of genes showing similar regulation, including increased expression of 3 genes for serine biosynthesis. A survey of Drosophila strains harboring mutations in ethanol-regulated genes for ethanol sensitivity and tolerance phenotypes revealed roles for serine biosynthesis, olfaction, transcriptional regulation, immunity, and metabolism. Flies harboring deletions of the genes encoding the olfactory co-receptor Or83b or the sirtuin Sir2 showed marked changes in the development of ethanol tolerance.
Conclusions
Our findings implicate novel roles for these genes in regulating ethanol behavioral responses.
Keywords: Alcohol, Ethanol Tolerance, Rapid Tolerance, Drosophila, Behavior
Alcohol use disorders are biologically complex and are strongly influenced by both genetic predisposition and environmental factors. While much is known about the anatomical sites of action and the intercellular signaling mechanisms engaged by ethanol, less is known about how short-term ethanol exposure leads to changes in neural activity and behavioral responses. Alcohol intoxication causes both pleasurable (euphoria and social disinhibition) and aversive (disorientation and sedation) effects. Increased ethanol intake is facilitated by the development of tolerance, most simply defined as the acquired resistance to the aversive and to a lesser extent the pleasurable effects of the drug (Fadda and Rossetti, 1998). Three distinct forms of tolerance have been defined in animal models (Kalant et al., 1971): acute tolerance develops during the course of a single ethanol exposure; rapid tolerance is induced by a single acute ethanol exposure and is measured after ethanol from the first exposure is metabolized; and chronic tolerance develops following prolonged or repeated ethanol exposures. Each form of tolerance engages distinct and overlapping biological processes; however, the molecular mechanisms underlying tolerance development remain largely enigmatic (Fadda and Rossetti, 1998). Selective breeding studies in rodents support genetic contributions to the development of each form of tolerance (Erwin and Deitrich, 1996; Rustay and Crabbe, 2004; Tabakoff et al., 1980). Moreover, the ability to develop tolerance is a predictor for the later development of alcohol use disorders, and therefore it is important to gain a greater understanding of how ethanol exposure leads to its development.
In recent years, the fruit fly Drosophila has been developed as a model to study drugs of abuse, including ethanol, cocaine, and nicotine (Bainton et al., 2000; McClung and Hirsh, 1998; Moore et al., 1998; Wolf and Heberlein, 2003). The behaviors induced by single or repeated ethanol exposures in Drosophila are remarkably similar to those observed in mammals, and many of the neurotransmitter and second messenger systems that mediate these behaviors appear to be conserved. Naive flies exhibit locomotor activation at low doses and locomotor incoordination and sedation at higher doses of ethanol. Furthermore, flies develop tolerance to the locomotor incoordinating and sedating effects of ethanol within hours after a single exposure to a moderate dose of ethanol (Berger et al., 2004; Scholz et al., 2000). This tolerance is defined as functional, as it reflects changes in the behaviors elicited without underlying changes in ethanol intake or metabolism. Since tolerance persists after the ethanol absorbed from the first exposure is completely metabolized, this paradigm most closely resembles rapid tolerance paradigms defined in rodents (Kalant et al., 1971). Several parameters of tolerance, such as its maximal extent and kinetics of dissipation, are also similar in flies and rodents. Genetic and pharmacologic studies indicate that rapid tolerance in Drosophila requires the integrity of octopaminergic and γ-aminobutyric acid B (GABAB) receptor pathways, a large conductance calcium-sensitive potassium (BK) channel, the Homer neuronal adaptor protein, and a novel stress pathway defined by the gene hangover (Cowmeadow et al., 2005; Dzitoyeva et al., 2003; Scholz et al., 2005; Urizar et al., 2007). Thus, at least some of the neurotransmitter and signaling systems that mediate these behaviors are evolutionarily conserved (e.g., the GABAB and BK-type potassium channels; Pietrzykowski et al., 2004; Zaleski et al., 2001), and studies in flies can provide novel candidate genes for mammalian and human studies (Riley et al., 2006).
Here, we describe a molecular and genetic survey designed to identify genes whose expression is regulated by ethanol exposure and that contribute to ethanol behavioral responses, including acute sensitivity and the development of rapid tolerance. The results implicate both novel and previously identified genes in ethanol behavioral responses. Analysis of gene expression following a single sedating ethanol dose revealed coordinated regulation of genes involved in stress, immunity, and olfaction, and a multi-study comparison of gene expression changes following ethanol exposure in flies identified metabolic genes, including 3 for serine biosynthesis, as being commonly regulated (Morozova et al., 2006; Urizar et al., 2007). A genetic survey of flies carrying lesions associated with a select set of ethanol-responsive genes uncovered roles for genes involved in olfaction, chromatin structure, immunity, and metabolism in regulating ethanol sensitivity and tolerance. Evidence from mammalian studies suggests that each of these biological processes may play roles in the actions of drugs of abuse, and suggests that the effects of ethanol at the molecular level are more broadly conserved than previously appreciated.
MATERIALS AND METHODS
Strains and Husbandry
All Drosophila strains were outcrossed for 5 to 10 generations to the Berlin genetic background containing the eye color marker mutation w1118 (Berlin). Flies were maintained on standard cornmeal/molasses/yeast media at 25°C and 70% relative humidity with an approximately 16/8 h light/dark schedule. Strains used in this study were obtained from the Bloomington Stock Center, Harvard University, the Drosophila Genetics Resource Center (Japan), the National Institute of Genetics (Japan), the Szeged Stock Center (Hungary), Petros Ligoxygakis (d03636), and Leslie Vosshall (Or83b1, Or83b2). Insertion lines (4.43, 5.10, 10.219, 11.247, 11.83) of the transposon P{GawB} were generated in the Heberlein lab.
Behavioral Assays
For behavioral testing, 25 virgins and males were mated for 2 days in bottles with 50 ml standard media with a few added grains of Baker’s yeast, and groups of 20 to 25 young adult male progeny were collected into standard food vials without yeast 11 days later. Flies were allowed to recover from CO2 anesthesia for 2 days and were tested behaviorally between 11 am and 6 pm. Behavioral tests were carried out at 25°C with constant illumination. Each experiment consisted of a group of 20 to 25 genetically identical flies that were progeny from 1 parental cross. Replicate experiments were carried out with groups collected from distinct parental crosses and were carried out on different days. This design was used to take day-to-day and potential genetic variation into account. Ethanol was delivered as a continuous vapor stream that was mixed with a humidified air stream to achieve a flow rate of 5.5 l/min. Ethanol concentration is expressed as the percentage of ethanol vapor: [(ethanol vapor)/(ethanol vapor + humidified air)] × 100. Behavioral experiments were performed in the 8 chambered booz-o-mat, and locomotor tracking analysis was carried out as described previously (Wolf et al., 2002). Briefly, flies were allowed to acclimate to the testing chamber for 9 minutes in a stream of humidified air and were then switched to a stream of ethanol vapor. Fly locomotion was filmed starting 2 minutes before ethanol vapor application. Locomotor activity was quantified as the population average distance traveled over time over 20-second intervals at specific time-points using an automated locomotor tracking device. To induce rapid ethanol tolerance, flies were exposed to ethanol vapor for 26 minutes, and were allowed to rest for 3 to 3.5 hours before receiving a second ethanol exposure of the same length and concentration. Distance traveled (Dist) is defined as the area under the curve from 2 to 25 minutes as determined by trapezoidal summation, and ΔDist is Distexposure2 – Distexposure1. Peak speed (PS) is the greatest speed achieved from 2 to 25 minutes. Sedation sensitivity (Sed) was quantified as the fraction of flies that lost the ability to right themselves following 26 minutes of ethanol exposure. Sedation tolerance (Sed Tol) is Sedexposure1 – Sedexposure2; n is the number of groups of flies tested, where groups are 20 to 25 genotypically identical male flies. Ethanol absorption was measured by exposing groups of 25 flies in the booz-o-mat locomotor tracking device to either ethanol vapor (47%) or humidified air for 15 minutes. Flies were frozen on dry ice immediately and the ethanol concentration in whole fly homogenates was measured with an alcohol dehydrogenase-based spectrophotometric assay (Diagnostic Chemicals, Ltd., Charlottetown, PE, Canada). Statistical analyses were carried out with Minitab 15.1.30.0 (State College, PA), and various derivative measures were calculated in Microsoft (Redmond, WA) Excel 2003. All error bars are standard error of the mean, except standard deviation for quantitative polymerase chain reaction (PCR).
Microarray Experiment and Analysis
Microarray analysis of gene expression was carried out on the Berlin genetic background strain. For each treatment condition, 3 groups of 200 male flies each were treated identically and combined prior to RNA extraction; this design helped to minimize variations in ethanol dose and time of individual experiments. Treatment conditions were exposure to either humidified air (flow rate: 3.7 l/min) or ethanol vapor (saturated ethanol vapor diluted to 60% with humidified air to 3.7 l/min) for 30 minutes at 25°C in perforated 50 ml conical tubes. Treated flies were allowed to recover in standard culturing conditions (food containing vials kept at 25°C and 70% humidity) for 0, 30, 60, 90, 120, 180, and 210 minutes. Additionally, 2 independent groups of flies were given no treatment. Flies were quickly frozen and heads were isolated. RNA from the resulting 16 treatment conditions was hybridized to Affymetrix Drosophila 2.0 oligonucleotide microarray chips at the Partners HealthCare Center for Personalized Genetic Medicine Microarray Facility (Harvard University).
The microarray data were preprocessed utilizing gcrma (robust multi-array averaging with sequence composition correction; http://www.bioconductor.org) in 4 steps: background correction, normalization between chips, signal estimation, and adjustments for false hybridization with GC content as a surrogate. Genes with low expression values were excluded from further analysis. Differential expression for the log base 2 normalized ethanol and air time courses was assessed by using linear models and empirical Bayes methods with the limma package in the R statistical program (Smyth, 2004). For each probe set, a quadratic regression model was fit to the expression values for time and treatment. We used the p value from the moderated F statistic (limma interaction p value) to test for time-dependent differential expression between the ethanol- and air-treated samples. The false discovery rate (fdr) was also determined from the p value adjusted for multiple comparisons utilizing the Benjamini and Hochberg method in limma. Because specific genes were later validated by quantitative PCR (qPCR) and mutant analysis (see below), we did not use the fdr for further analysis (fdr values are included in Table S1). Genes with p < 0.05 difference in ethanol versus air exposure (1,807 genes) were clustered with the HOPACH algorithm (a bidirectional clustering method that maximizes cluster homogeneity; Salomonis et al., 2005). Gene ontology (GO) terms showing significant enrichment within each cluster were identified using the MAPPFinder algorithm in the GenMAPP package (http://www.genmapp.org). The microarray data have been deposited in NCBIs Gene Expression Omnibus and are accessible through GEO Series accession number GSE18208 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18208).
The quality of the microarray data was validated using qPCR carried out according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA) on an ABI PRISM 7900 Sequence Detection System, using expression of RpL32 as a standard to normalize sample concentrations. All qPCR validation experiments utilized a time-course set of RNA samples from air- and ethanol-exposed flies that were derived independently from those used for microarray analysis. The relative transcript expression levels for genes in ethanol- and air-treated flies were determined by comparing to untreated controls. Taqman probesets (Applied Biosystems) used in this study were aay: Dm01822491_s1, AcCoAS: Dm01798031_g1, Akap200: Dm01803695_g1, aru: Dm01814168_g1, DnaJ-H: Dm01790940_g1, elk: Dm01842361_m1, for: Dm01808235_g1, jhamt: Dm01791790_g1, Idgf1: Dm01842859_g1, lush: Dm01823120_ g1, puc: Dm02135504_m1, RpL32: Dm02151827_g1, Sir2: Dm01844783_g1, Tdc1: Dm01815580_m1, Tsp42Eg: Dm01817973_ g1, Tps1: Dm01801466_g1, wdb: Dm02145441_g1. Gene data are from Flybase (http://www.flybase.org) release FB2009_05, as of May 29, 2009.
RESULTS
Measures of Behavioral Change in an Ethanol Rapid Tolerance Paradigm
Repeated ethanol exposures result in alterations in the effects of ethanol on the maintenance of postural control, locomotor activity levels, sedation sensitivity, and sedation recovery kinetics in flies (Berger et al., 2004; Scholz et al., 2000). The locomotor response of flies to ethanol vapor exposure is multi-phasic and dose-dependent (Fig. 1B to D; Wolf et al., 2002). Prior to ethanol exposure, flies typically exhibit low levels of locomotor activity (“A” in Fig. 1B). When a stream of ethanol vapor commences, flies show an immediate and transient olfactory startle response that achieves peak magnitude in 15 to 20 seconds, and dissipates by 45 to 60 seconds. Following a brief quiescence, flies enter into a more prolonged period of locomotor activity (2 to 25 minutes in Fig. 1B), the hyperactive phase, which correlates with accumulating internal ethanol concentrations of 15 to 40 mM (Wolf et al., 2002). As ethanol continues to accumulate, the hyperactive phase is gradually terminated as locomotor incoordination and sedation increase (10 to 25 minutes in Fig. 1C, 6 to 25 minutes in Fig. 1D). Previous studies utilizing a rapid tolerance paradigm showed that 3.5 hours after being exposed to a moderately high ethanol vapor concentration (ethanol vapor mixed with humidified air to 60%), flies given a second exposure of the same concentration exhibited increased locomotor activity levels throughout the hyperactive phase (Scholz et al., 2000). This effect is likely due in part to the development of tolerance to the sedative and locomotor-incoordinating effects of ethanol (Berger et al., 2004).
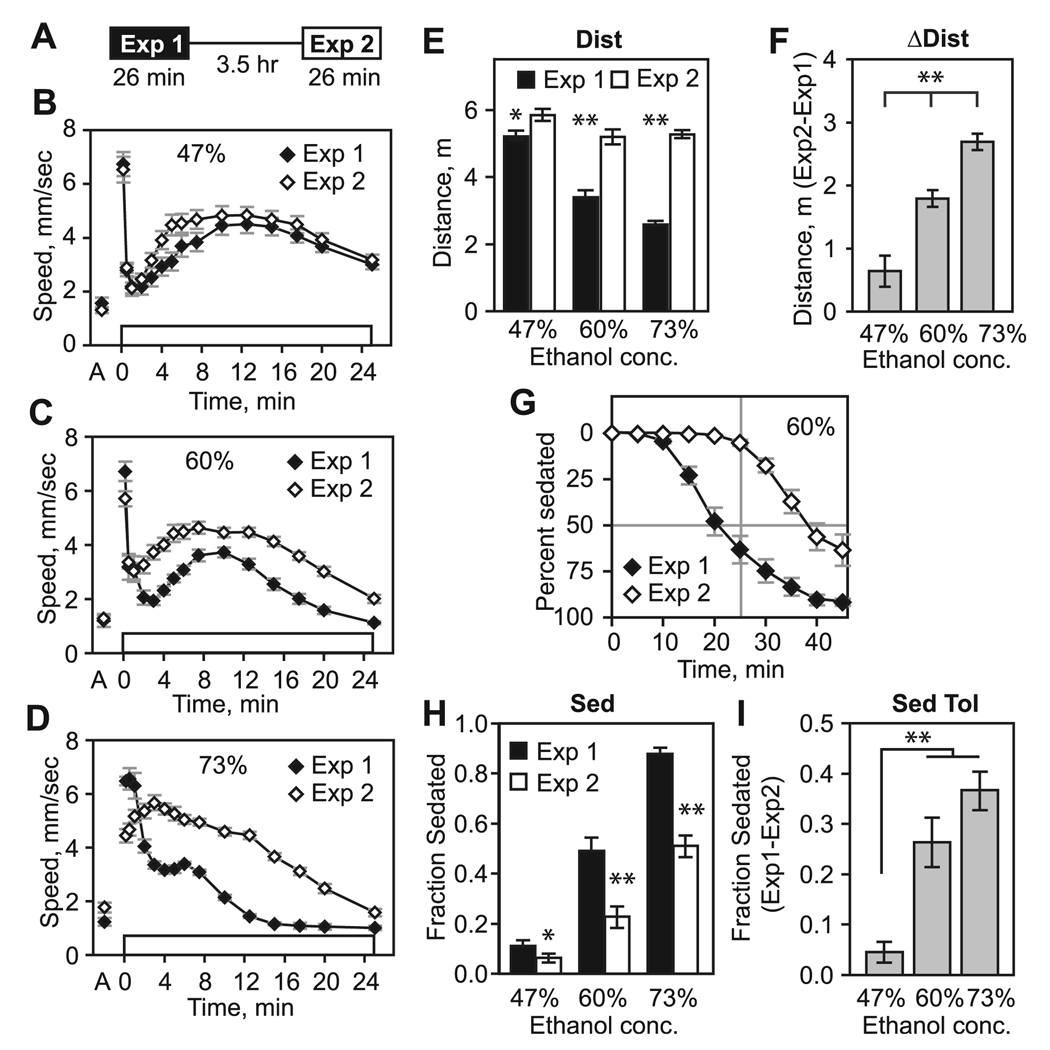
Fig. 1.
Dose-dependent alterations in ethanol-induced hyperactivity and sedation sensitivity in a rapid ethanol tolerance paradigm. (A) Exposure scheme for induction and detection of ethanol rapid tolerance. Groups of genetically identical flies are given two 26-minute exposures to ethanol vapor, separated by a 3.5-hour rest under standard culturing conditions. (B) Prior to ethanol exposure (“A” on horizontal axis) flies exhibit low levels of locomotor activity. Exposure of naive flies to a continuous stream of ethanol vapor (47% concentration, open box) results in an olfactory startle response that subsides by 1 minute, followed by a hyperactive phase from approximately 2 to 25 minutes (Exp 1). Hyperactivity declines with increasing locomotor incoordination and sedation. Following a 3.5-hour rest, flies exposed a second time to the same ethanol vapor concentration exhibit a small increase in hyperactivity during its onset (Exp 2). (C and D) Exposure to higher concentration ethanol vapor results in greater differences in the hyperactive phase between Exp 1 and Exp 2. (E) Distance traveled (Dist) from 2 to 25 minutes ethanol vapor exposure. Distance traveled was significantly greater during Exp 2 versus Exp 1 (47%: *p = 0.037, 60%: **p < 0.0005, 73%: **p < 0.0005, paired t-test, n = 15). (F) Change in distance traveled (ΔDist) between Exp 2 and Exp 1 (**p < 0.0005, ANOVA, all samples different by Tukey’s multiple comparison test). (G) Time course of ethanol sedation at 60% ethanol vapor. (H) The fraction of flies sedated (Sed) following 26-minute ethanol exposure. Fewer flies were sedated following the second ethanol exposure at all concentrations tested (47%: *p = 0.038, 60%: **p < 0.0005, 73%: **p < 0.0005, paired t-test, n = 15). (I) Sedation tolerance (Sed Tol) is the difference in the fraction of flies sedated between Exp 1 and Exp 2 (**p < 0.0005, ANOVA, 47% sample different by Tukey’s multiple comparison test).
To determine how locomotor behaviors are modified by previous ethanol exposure, we developed 2 measures that allow quantitative assessment of locomotor activity levels, and we performed an ethanol dose–response in the rapid tolerance paradigm (Fig. 1B to D). For the hyperactive phase, we determined the area under the curve from 2 to 25 minutes of ethanol exposure, termed the distance traveled (Dist) for simplicity. The difference in Dist between exposures (ΔDist) provided an index of the locomotor alterations elicited by repeated ethanol exposures. Naive flies showed a dose-dependent decrease in hyperactivity as ethanol concentrations increased (Wolf et al., 2002), which was reflected in a decrease in Dist (Fig. 1E). We also observed dose-dependent effects on ΔDist with increased ΔDist at higher ethanol concentrations (Fig. 1F). At 47% ethanol vapor, we observed a small increase in Dist during exposure 2 relative to exposure 1 (Fig. 1B and E). Because the increase in locomotor activity appeared to be limited to hyperactivity onset at 47% ethanol vapor (but not at higher concentrations; Fig. 1B), we determined the time of PS during the first exposure to 47% ethanol vapor (12.5 minutes) and split Dist into Dist onset (2 to 12.5 minutes) and Dist offset (12.5 to 25 minutes). This revealed that ΔDist increased during hyperactivity onset (2.32 m Exp 1 vs. 2.76 m Exp 2, p = 0.013, paired t-test) but not offset (2.89 m Exp 1 vs. 3.10 m Exp 2, p = 0.156) at 47% ethanol vapor concentrations. Earlier onset of locomotor activation during the second ethanol exposure at this low ethanol concentration may reflect sensitization to the locomotor activating effects of ethanol (Lister, 1987). Higher ethanol vapor concentrations that lead to overall increased hyperactivity in the rapid tolerance paradigm may be due to a mix of sensitization and tolerance.
We next determined the dose-dependent effects of ethanol vapor on sedation sensitivity and tolerance using a loss-of-righting assay (Fig. 1G to I). Flies showed sigmoidal sedation kinetics when exposed to a continuous stream of the moderate 60% ethanol vapor, with a time to 50% sedation of 25.6 ± 2.2 minutes (Fig. 1G). Because measuring the time to 50% sedation is labor intensive, we counted the number of flies sedated at 26 minutes (Sed) for subsequent analyses. Consistent with previous findings, naive flies showed increased sedation with increased ethanol concentration (Fig. 1H; Urizar et al., 2007). Flies developed robust tolerance to the sedating effects of ethanol (Sed Tol) that increased with increasing dose (Fig. 1I). In sum, these dose–response experiments revealed a correlation of 2 measures of behavioral plasticity in the rapid tolerance paradigm, ΔDist and Sed Tol, with both measures increasing with increasing ethanol concentration. Additionally, a low concentration of ethanol vapor (47%) revealed a potentiation of locomotor activity during hyperactivity onset of exposure 2 that may reflect sensitization to the locomotor activating effects of ethanol. These dose–response experiments defined the dynamic range of the ethanol-induced behaviors measured here and provided a valuable framework for interpreting altered behavioral responses in mutant strains. We used 60% ethanol vapor for most of the remaining experiments, as this concentration allowed us to detect bidirectional alterations for each behavioral measure.
Regulation of Gene Expression by Ethanol Exposure
To identify genes that may regulate the development of ethanol tolerance, we determined gene expression levels in the heads of flies exposed for 30 minutes to either 60% ethanol vapor or humidified air as a control (Fig. 2). We captured the dynamic effects of ethanol exposure on gene expression levels by performing a time-course microarray analysis on samples collected before, immediately after, and at 6 additional time-points extending to 3.5 hours following the termination of ethanol exposure. Gene expression changes were quantified using a linear models approach and ranked by the magnitude of the difference in expression change for ethanol versus air exposure (Table S1; Smyth, 2004). Genes with significant expression changes (p < 0.05, 1,807 genes) were then clustered into groups with similar patterns of expression. This identified 9 distinct patterns of gene expression that included changes in expression induced by exposure to ethanol, air, or both (Fig. 2B). Gene expression changes following ethanol exposure were generally of greater magnitude than following air exposure. However, some genes (e.g., Cp1 and Cyp4p2) showed greater air regulation, and others (CG32602 and CG13422) showed regulation by both ethanol and air. In general, ethanol-regulated genes showed either transient changes (clusters 1, 2, 8, and 9) or only later changes (clusters 3, 6, and 7). A search for over-represented GO terms associated with these clusters identified, among others, terms for odorant binding, metabolism, regulation of signal transduction, nucleic acid binding, Toll signaling, Ras signal transduction, and cytoskeletal organization and biogenesis (Table S2). Remarkably, similar categories were identified in 2 mammalian studies that modeled specific aspects of addiction: an analysis of gene expression differences between alcohol preferring and nonpreferring mice, and an analysis of the effects of chronic cocaine exposure on gene expression (Mulligan et al., 2006; Renthal et al., 2009). The quality of the microarray analysis was confirmed by qPCR, with 11 of 14 genes tested showing similar regulation of expression by ethanol exposure (Fig. 2C to F and data not shown).
Fig. 2.
Time course microarray analysis of gene expression levels following a single exposure to a moderate dose of ethanol. (A) Treatment scheme for microarray analysis. Fourteen groups of 200 flies were exposed to either 60% ethanol vapor or humidified air for 30 minutes, allowed to recover for 0 to 210 minutes and then frozen at the time-point indicated by the triangles. Two additional groups were frozen as no exposure controls (time 0). RNA extracted from the heads of the resulting 16 samples was hybridized to Drosophila 2.0 microarrays (Affymetrix). (B) Clustered gene expression patterns (rows) for exposure to ethanol vapor (left) and humidified air (right) at the given time-points (columns), represented as the medoid values for each cluster. Green and magenta indicate reduced and increased expression relative to the averaged unexposed controls, respectively. (C to F) Example expression level time courses from the microarray analysis (C and E) and qPCR confirmations using RNA isolated from a separate exposure series (D and F) for the genes tyramine decarboxylase 1 (Tdc1; C and D) and Sir2 (E and F). Data are expressed as fold change relative to unexposed controls. Time of the 30-minute ethanol or air exposure is indicated as a box on the horizontal axis. E (red) and A (blue) indicate gene expression levels in ethanol- and air-exposed samples, respectively. Error bars indicate standard deviation of 3 sample replicates for qPCR. (G) All genes (rows) in cluster 1. Genes in area bracketed by red bars are predominantly olfactory specific, showing a large, coordinate decrease in expression of many, but not all, olfactory specific genes. (H) Overlap of genes identified in this and 2 previous microarray studies (Morozova et al., 2006 and Urizar et al., 2007) of gene regulation by ethanol exposure in Drosophila.
Patterns of expression regulation were apparent for genes that function in olfaction, heat-shock responses, and immunity. In Drosophila, genes specific to olfaction include the odorant receptor (OR) family of 60 genes and the odorant-binding protein (OBP) family of 51 genes. OBPs are found in the fluid of the sensillum surrounding the dendrites of the OR neurons (Hallem et al., 2006). We observed a large, graded, and coordinated down-regulation of many but not all olfactory-specific genes following ethanol exposure (Fig. 2G). Examples of down-regulated olfactory genes include those coding for the olfactory co-receptor Or83b, the OBPs Lush and OS-E, and the pheromone-binding proteins Pbprp1, Pbprp3, Pbprp4, and Pbprp5 (Table S1). Exposure of flies to high concentration ethanol vapor results in activity-dependent olfactory neuron apoptosis and olfactory organ damage that is always accompanied by a blackening of the main olfactory organ, the third segment of the antennae (R.L. French, personal communication). Exposing flies to ethanol vapor under conditions identical to those used for our microarray study resulted in blackening of only 0.5% of antennae (n = 216). These data suggest that the down-regulation of olfactory genes by ethanol exposure occurs at lower ethanol concentrations than those that cause visible tissue damage. Moreover, expression of the OR gene Or43b, and the antennal-specific genes OS9 and pinocchio were not decreased following ethanol exposure (Table S1; Morozova et al., 2006; Raha and Carlson, 1994; Rollmann et al., 2005). Down-regulation of olfactory genes could serve as a general protective mechanism against ethanol-induced excitotoxicity. Consistent with decreased olfactory function in ethanol-exposed flies, we found that the magnitude of the olfactory startle response was reduced during the second ethanol exposure (PS decreased from 8.8 mm/s for exposure 1 to 7.4 mm/s for exposure 2, p = 0.007, paired t-test).
Of the 20 most significantly ethanol-regulated genes, 5 encoded heat-shock proteins (HSPs; Table S1). Additionally, nearly half of all Drosophila Hsp genes (9 of 21) showed significantly increased expression (p < 0.05) following ethanol exposure (Fig. S1). Three of the 4 most strongly regulated Hsp genes belonged to the Hsp70 family that function as molecular chaperones, and regulate diverse biological and cellular processes including apoptosis, immunity, and thermotolerance (Gong and Golic, 2006). As is the case with stressors such as heat, ethanol induced expression of Hsp70 family members to remarkably high expression levels, including a 50-fold induction for Hsp70Ab. Interestingly it was previously reported that heat shock can induce ethanol tolerance, and genetic analyses uncovered both stress-dependent and stress-independent ethanol tolerance pathways (Scholz et al., 2005). HSPs may therefore play an important role in one of these tolerance pathways.
In Drosophila, bacterial and fungal infection results in an innate immune response that engages the Toll, Imd, and melanization pathways (Brennan and Anderson, 2004). Toll and Imd signal transduction pathways are similar to the Toll-like and tumor necrosis factor pathways in vertebrates, respectively, and both converge on members of the nuclear factor-κB (NFκB) family of transcriptional regulators. In our microarray experiment, ethanol exposure resulted in increased expression of immunity genes of the Toll (cact, Myd88, Tl), Imd (imd, Rel), and melanization (Spn27A) pathways (Fig. S2). Additionally, a small subset of genes of unknown function that are induced by bacterial infection were also induced by ethanol exposure (IM2, IM10, IM23; Uttenweiler-Joseph et al., 1998). The temporal patterns of gene expression induction segregated into 2 classes: an early and transient induction and a slowly increasing induction. Interestingly, genes in the Toll and Imd pathway all showed the early and transient pattern of induction. These data suggest that expression of the Toll and Imd pathways may be coordinately regulated by ethanol exposure.
Multi-Study Comparison of Ethanol-Regulated Genes
Comparison of the most significantly regulated genes in our study (p < 0.01, 737 genes; Table S1) to those identified in 2 previous studies of gene expression regulation by ethanol exposure in Drosophila revealed a small set of genes detected by all studies (Fig. 2H; Morozova et al., 2006; Urizar et al., 2007). Of the 29 commonly identified genes, 25 showed the same direction of expression regulation (increase or decrease) in all 3 studies (Table S3). Twelve of the commonly regulated genes are annotated with GO terms for metabolic and biosynthetic processes, and these included 3 up-regulated genesencoding proteins involved in serine biosynthesis (aay, CG3011, CG8129; Table 1). l-Serine is important in protein and phospholipid synthesis, and d-serine can act as a co-agonist with glutamate at N-methyl-d-aspartate (NMDA) receptors (Oliet and Mothet, 2009). Serine is mainly synthesized from the glycolysis intermediate 3-phosphoglycerate in a 3-step enzymatic process; the Drosophila gene astray encodes the terminal enzyme 3-phosphoserine phosphatase. Serine is also synthesized from glycine by glycine hydroxylmethyltransferase, which is encoded by the Drosophila gene CG3011.Serine racemase converts l-serine to d-serine (Oliet and Mothet, 2009) and is encoded by the Drosophila gene CG8129. While d-serine has not yet been described as a neurotransmitter in flies, coregulation of the biosynthetic enzymes by ethanol raises the possibility that d-serine contributes to ethanol responding. Finally, we note the ethanol-induced down-regulation of the high mobility group protein gene HmgZ that is predicted to regulate chromatin structure, and of the lipophorin protein gene Rfabg that has been implicated in long range intercellular signaling (Panakova et al., 2005; Ragab et al., 2006). Ethanol-regulated genes in common with our study and one other study include those encoding DNA-binding proteins (CTCF, fkh, odd, Side), chromatin structure (mbf1, Rpd3), signal transduction (Akap200, Cks85A, mats, Pp1alpha-96A, Ptr, tsl), innate immunity (dnr1, dro5, Spn27A), and many other genes whose products are known or predicted to function in lipid and other metabolic pathways (Table S4). Finally, we note that the genes identified in the 2 previous studies show greater overlap with our set of regulated genes than with each other, suggesting that surveying the time course of gene expression changes was effective in capturing a broad range of ethanol-responsive genetic programs.
Table 1.
Strains Showing Greatest Change in Dist or ΔDist
| Symbol | Name or function | Lesion | Cluster | Dist, m | ΔDist, m | Sed | Sed Tol | EtOH, mM |
|---|---|---|---|---|---|---|---|---|
| aay | 3-Phosphoserine phosphatase | NP6536 | 7 | 4.9 (0.43) | 0.6 (0.26) | 0.2 (0.05) | 0.0 (0.02) | 26.4 (4.50) |
| AcCoAS | Acetyl-CoA synthase | f03474 | 7 | 2.1 (0.05) | 0.4 (0.09) | 0.7 (0.09) | 0.1 (0.06) | 19.0 (2.81) |
| Akap200 | A kinase anchoring protein | NP6271 | 9 | 4.5 (0.38) | 0.2 (0.37) | 0.4 (0.05) | 0.2 (0.03) | 14.1 (1.49) |
| Arc2 | Activity-regulated, cytoskeletal-associated protein | EY14313 | 8 | 6.7 (0.36) | 0.2 (0.26) | 0.2 (0.07) | 0.1 (0.04) | 27.2 (2.80) |
| CG5508 | Glycerol-3-phosphate-O-acyltransferase | NP0597 | 8 | 7.1 (0.82) | 1.8 (0.38) | 0.3 (0.06) | 0.4 (0.03) | 27.1 (2.13) |
| CG9238 | Protein phosphatase 1 regulatory subunit | EP3518 | 7 | 3.3 (0.22) | 3.4 (0.44) | 0.7 (0.06) | 0.5 (0.08) | 21.8 (1.54) |
| CG17734 | HIG1 domain family | KG06609 | 9 | 6.4 (0.62) | −0.6 (0.33) | 0.3 (0.09) | 0.2 (0.04) | 19.6 (1.09) |
| CG32512 | Similar to mouse Tmem205 | f01175 | 7 | 4.6 (0.37) | 0.1 (0.26) | 0.5 (0.09) | 0.1 (0.10) | 23.5 (1.40) |
| CG42270 | Disabled interacting protein | NP3418 | 7 | 4.8 (0.49) | 1.8 (0.19) | 0.5 (0.09) | 0.5 (0.08) | 21.9 (2.10) |
| dpr9 | Ig-domain transmembrane receptor | c06869 | 8 | 2.3 (0.19) | 1.0 (0.19) | 0.6 (0.04) | 0.3 (0.06) | 25.1 (3.67) |
| fu12 | 1-Acylglycerol-3-phosphate-O-acyltransferase | EP1138 | 7 | 5.9 (0.43) | 0.7 (0.19) | 0.2 (0.05) | 0.1 (0.06) | 29.6 (4.71) |
| Jhedup | Juvenile hormone esterase | e01859 | 1 | 4.5 (0.45) | −0.2 (0.31) | 0.2 (0.06) | 0.1 (0.04) | 18.3 (0.73) |
| Or83b | Olfactory coreceptor | 1 | 1 | 2.2 (0.21) | 3.0 (0.64) | 0.6 (0.08) | 0.3 (0.11) | 26.4 (2.62) |
| Ptr | Patched related receptor | NP2732 | 1 | 6.8 (0.86) | −0.2 (0.37) | 0.2 (0.03) | 0.0 (0.03) | 16.5 (0.85) |
| rho-4 | Rhomboid 4 | NP3452 | 7 | 5.1 (0.33) | 2.3 (0.50) | 0.8 (0.03) | 0.3 (0.06) | 27.6 (2.07) |
| scb | Integrin | NP7060 | 7 | 6.2 (0.47) | −1.3 (0.34) | 0.2 (0.09) | 0.1 (0.07) | 28.0 (3.40) |
| Sir2 | Sirtuin | 2A–7–11 | 2 | 2.9 (0.21) | 0.2 (0.17) | 0.3 (0.04) | 0.1 (0.03) | 26.9 (2.51) |
| Spn27A | Serpin 27A | e02539 | 7 | 2.1 (0.17) | 1.5 (0.49) | 0.8 (0.07) | 0.2 (0.07) | 20.7 (1.54) |
| Tdc1 | Tyramine decarboxylase 1 | f03311 | 8 | 2.3 (0.15) | 1.8 (0.09) | 0.7 (0.03) | 0.4 (0.08) | nd |
| Vrp1 | Verprolin | EY02177 | 7 | 2.4 (0.13) | 0.6 (0.43) | 0.4 (0.08) | 0.2 (0.15) | 16.5 (1.58) |
| Genetic background | 3.1 (0.08) | 1.2 (0.08) | 0.6 (0.02) | 0.4 (0.02) | 22.1 (0.88) | |||
Cluster indicates grouping in the cluster analysis as depicted in Fig. 2B. Standard error of the mean for each ethanol response measure is indicated in parentheses. Shaded numbers indicate measures that differed from matched controls (p < 0.05, paired t-test). For ethanol absorption, there was an effect of genotype (p < 0.01, ANOVA) but no difference from the genetic background control was detected by Dunnett’s comparisons test.
Survey for Ethanol Behavioral Mutants
We wished to determine if genes that show alterations in transcript levels following ethanol exposure also contribute to ethanol behavioral responses. To accomplish this, we assayed 107 strains that harbored transposon insertion lesions in 61 ethanol-regulated genes for locomotor activity and sedation sensitivity in response to ethanol vapor in the rapid tolerance paradigm (Table S5). We chose genes for mutant analysis based on one or more of the following criteria: (i) highly significant gene expression regulation by exposure to ethanol vapor but little to no regulation by exposure to humidified air, (ii) onset of gene expression regulation within 90 minutes of termination of ethanol vapor exposure to bias toward genes that may contribute to tolerance, (iii) sequence similarity to mammalian gene products, (iv) previous data implicating the gene or related genes in cellular processes in the central nervous system, and (v) availability of transposon insertional lesions or other genetic tools that allowed for rapid behavioral testing. To minimize the influence of genetic background, all strains were backcrossed to the Berlin parental strain prior to behavioral testing. The Berlin strain was tested behaviorally alongside the surveyed strains to provide a standard metric for daily variations in behavioral responses, and its behavior was essentially identical to the averaged response of all 107 strains that we tested (Fig. S3). All strains were tested at the moderate 60% ethanol vapor concentration.
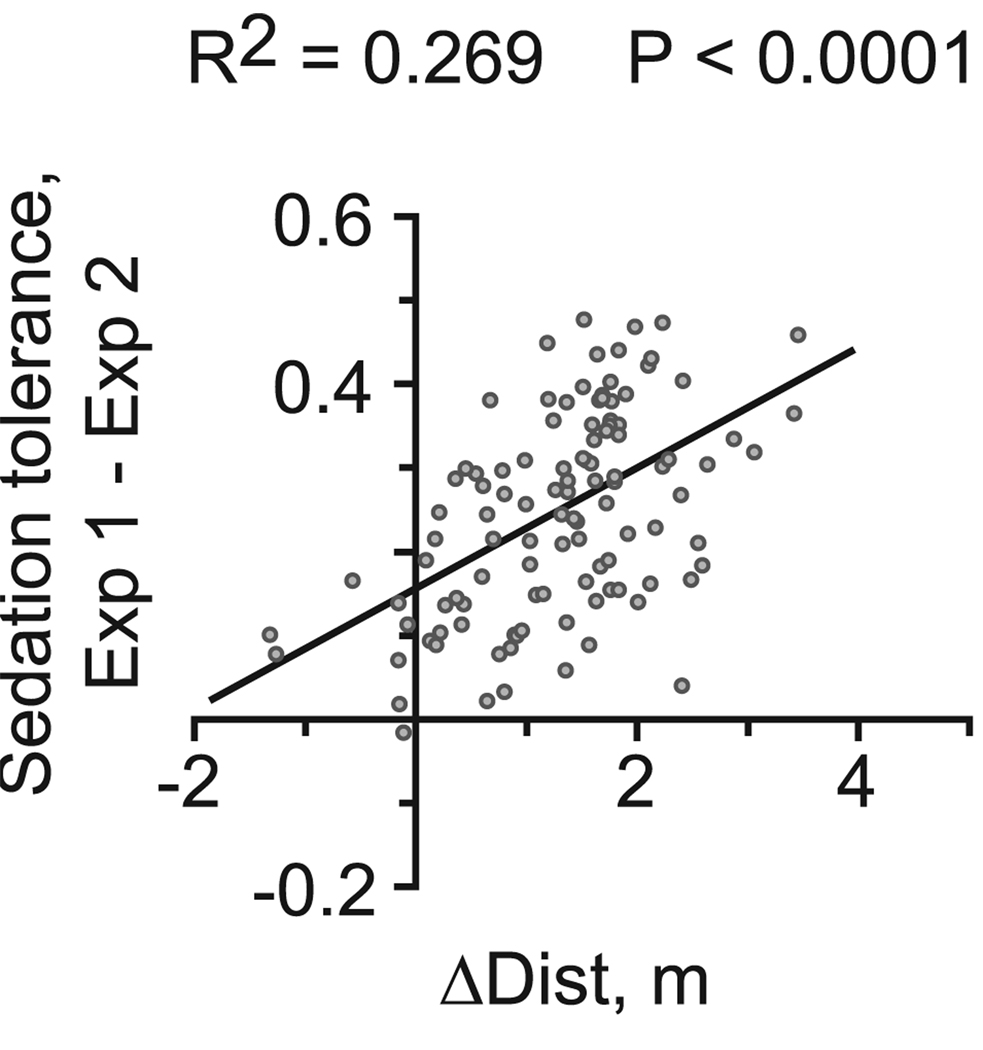
We observed a broad distribution of locomotor activity and sedation sensitivity responses to ethanol among the surveyed strains (Fig. S4). To ascertain the relationship between the locomotor and sedative effects of ethanol exposure, we performed regression analyses. We found a moderate correlation between our 2 measures of behavioral plasticity, ΔDist and sedation tolerance (R2 = 0.269, p < 0.001; Fig. 3), and only mild correlations between Dist and both ΔDist (R2 = 0.109, p < 0.001) and sedation sensitivity (R2 = 0.149, p < 0.001). Additionally, a stronger correlation was observed between ethanol sedation sensitivity and sedation tolerance (R2 = 0.484, p < 0.001). Correlation of these measures is also dose dependent (Fig. 1), suggesting that some of the observed strain differences may be due to shifts in either the actual or perceived ethanol dose. However, all measures of ethanol responses could be separated genetically. For example, animals carrying the NP6006 lesion at the X11L locus exhibited higher Dist and near normal sedation sensitivity, and animals carrying the EP2336 lesion at the Tsp42El locus exhibited an increase in ΔDist and essentially no sedation tolerance (Table S5). Furthermore, when tested at different ethanol doses, phenotypic covariation for individual strains did not always hold (e.g., see Fig. 7). Therefore, the locomotor and sedative effects of ethanol exposure have both common and distinct substrates that can be distinguished by genetic and pharmacologic means.
Fig. 3.
Correlation of behavioral measures for ethanol tolerance in strains surveyed for ethanol behavioral alterations. Graph depicts values for all strains surveyed (gray dots) for ΔDist on the horizontal axis, and Sed Tol on the vertical axis. Linear regression analysis (line) indicated a moderate correlation between ΔDist and Sed Tol.
Fig. 7.
Behavioral effects of deletion of Sir2. (A) The Sir2 locus consists of a single transcriptional unit that is predicted to overlap with the DnaJ-H gene at the 5′-end of both genes. The 2A–7–11 targeted deletion, depicted below the Sir2 locus, removes the Sir2 translation start site and the majority of the open reading frame (shaded). Probes for qPCR are indicated as bars above the genes. The NP1145 transposon insertion is located just upstream of the Sir2 translation start site. (B) Transcript levels of DnaJ-H are unaffected in 2A–7–11 homozygotes, when compared with genetic background controls. (C) Loco-motor activity profile for Sir22A–7–11, compared with matched Berlin controls, tested in the rapid tolerance paradigm and exposed to 60% ethanol vapor; n = 8. (D to G) Measures of ethanol responses derived from the experimental data depicted in C (D: p = 0.562; E: *p = 0.027; F: **p = 0.006; G: *p = 0.027; 2-sample t-test with pooled variance). (H) Locomotor activity profile for Sir22A–7–11, compared with matched Berlin controls, tested in the rapid tolerance paradigm and exposed to 73% ethanol vapor; n = 3. (I to L) Measures of ethanol responses derived from the experimental data depicted in H (I: p = 0.729; J: **p = 0.003; K: p = 0.225; L: p = 0.697; paired t-test).
We chose the 10 highest and lowest scoring strains for the Dist and ΔDist metrics as the strongest candidate ethanol behavioral mutants (Table 1). Where more than 1 lesion in or near a gene resulted in similar behavioral scores, we chose 1 strain as representative for that gene. Additionally, some strains showed strong behavioral scores for both Dist and ΔDist. Ethanol absorption was unaffected in these strains, indicating that the observed behavioral alterations were not due to altered ethanol pharmacokinetics (Table 1). While we did not explicitly screen for ethanol sedation sensitivity and sedation tolerance mutants, 11 additional strains showed strong effects for either one or both measures (Table S6). Here we briefly discuss a subset of the strongest Dist and ΔDist mutants, followed by a more in-depth analysis of 4 mutant strains in which the genetic lesion completely disrupts the associated gene.
We tested 4 strains carrying distinct transposon insertions in or near the aay locus. All 4 strains exhibited increased ethanol-induced hyperactivity, and 3 exhibited reduced ethanol sedation sensitivity and sedation tolerance (Table 1 and Table S5). One transposon insertion, S042314, was previously shown to affect the aay locus, and when homozygous resulted in axon guidance defects in the embryonic peripheral nervous system (Prokopenko et al., 2000). Additionally, flies homozygous for a transposon insertion in the CG3011 locus, encoding the enzyme that converts glycine to serine, also exhibited increased ethanol-induced hyperactivity (Table S5). Expression of aay and CG3011 were strongly up-regulated by ethanol exposure in all studies carried out to date in flies (Morozova et al., 2006; Urizar et al., 2007). These data suggest that genes in the serine synthesis pathway negatively regulate ethanol-induced hyperactivity and may also promote ethanol sedation sensitivity.
CG9238 encodes a protein phosphatase 1 (PP1) regulatory subunit, sharing 41% amino acid identity with mammalian protein targeting to glycogen (PTG). Mammalian PTG is an activator of PP1, promoting dephosphorylation of glycogen synthase and the production of glycogen as an energy store. Three independent transposon insertions in the CG9238 locus, 2 in the upstream regulatory region and one in the 5′-untranslated region of the gene, resulted in increased ΔDist with little apparent effect on Dist, sedation sensitivity, or sedation tolerance (Table 1 and Table S5). By microarray analysis CG9238 expression was transiently increased twofold, peaking 30 minutes after the termination of ethanol vapor exposure (Table S1). Transcript levels for CG9238 in the transposon insertion strain BG02516 were decreased to 53% ± 2% of controls. These data are consistent with CG9238 having an inhibitory effect on increased locomotor activity during ethanol exposure 2, suggesting that glycogen synthesis impacts ethanol tolerance in flies. Indeed, insulin signaling promotes glycogen synthesis in a PP1-dependent manner, and the insulin signaling pathway regulates ethanol sensitivity in flies (Brady and Saltiel, 2001; Corl et al., 2005).
Acetyl-Coenzyme A Synthase
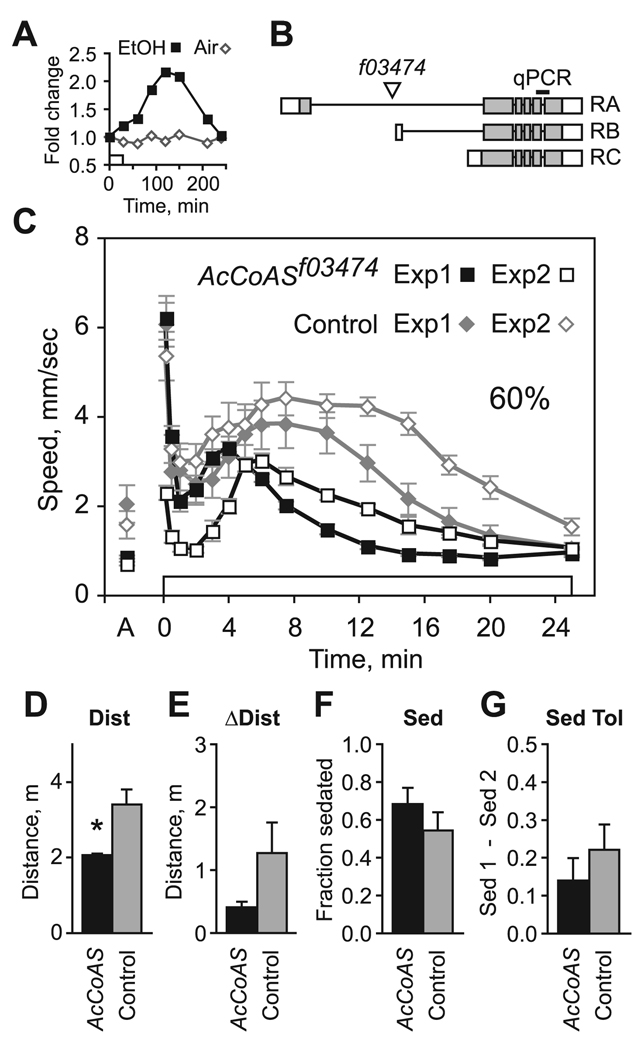
Acetyl-coenzyme A (acetyl-CoA) is an intermediate in many biochemical pathways, including those for ethanol metabolism, the citric acid cycle, the mevalonate pathway, and acetylcholine synthesis. One means of acetyl-CoA synthesis is by ligation of acetate and CoA by acetyl-CoA Synthase (AcCoAS). Microarray analysis showed that ethanol exposure transiently and robustly induced expression of AcCoAS by over twofold (Fig. 4A). Flies homozygous for the transposon f03474, inserted in AcCoAS (Fig. 4B), were viable, fertile, and showed no morphological or overt behavioral defects. AcCoAS transcripts were barely detectable in f03474 homozygotes (0.4% ± 0.08% of controls), suggesting that AcCoASf03474 animals may completely lack AcCoAS activity.AcCoASf03474 flies exhibited a marked reduction in Dist (Fig. 4C,D). Reduced hyperactivity was not accompanied by altered ethanol sedation sensitivity (Fig. 4F), and sedated flies recovered fully prior to the second ethanol exposure (not shown). Additionally, AcCoASf03474 flies were able to respond normally to external stimuli as evidenced by the normal magnitude ethanol olfactory startle response in naive animals (mean PS Berlin: 6.1 ± 0.49 mm/s, AcCoASf03474: 6.2 ± 0.49 mm/s, p = 0.823, paired t-test, n = 6). Upon second exposure, AcCoASf03474 showed a markedly reduced startle response (mean PS Berlin: 5.4 ± 0.54 mm/s, AcCoASf03474: 2.3 ± 0.16 mm/s, p = 0.003, paired t-test, n = 6) and a delayed onset of hyperactivity, but showed a normal increase in ΔDist during the descending phase of the hyperactivity response (Fig. 4E). Additionally, AcCoASf03474 flies showed normal ethanol sedation tolerance (Fig. 4G). These data suggest that biochemical pathways that depend on acetyl-CoA production via AcCoAS contribute to ethanol-induced hyper-activity in naive animals.
Fig. 4.
Behavioral effects of mutation of AcCoAS. (A) Expression levels by microarray analysis of AcCoAS following exposure to ethanol (squares) or air (diamonds) relative to untreated controls (time 0). (B) The AcCoAS locus consists of 3 transcriptional units (RA, RB, RC; shaded region indicates extent of open reading frame). The transposon f03474 is inserted just upstream of the RB transcription start site. The probeset used to detect AcCoAS transcripts by qPCR spanned the last intron. (C) Locomotor activity profile for AcCoASf03474 compared with matched controls (Berlin) during two 26-minute exposures to 60% ethanol vapor separated by a 3.5-hour rest. Locomotor activity in humidified air (A) is measured just prior to ethanol exposure (0 to 26 minutes); n = 6. (D to G) Ethanol behavioral measures derived for experimental data depicted in C (D: *p = 0.014; E: p = 0.152; F: p = 0.134; G: p = 0.371, paired t-test).
Olfactory Co-Receptor Or83b
The microarray and ethanol startle data presented above suggested that olfactory sensory acuity may be reduced during the development of rapid ethanol tolerance. Bilateral surgical removal of the third antennal segment that houses the majority of the olfactory sensory neurons was previously shown to block the ethanol olfactory startle response (Wolf et al., 2002), but we found that it also reduced ethanol absorption by an unknown mechanism (not shown), confounding this direct analysis of olfactory input to rapid tolerance. Or83b is an OR present in approximately 70% of all olfactory neurons that is required for both the localization to dendrites and function of co-expressed ORs as heteromeric ion channels (Benton et al., 2006; Sato et al., 2008). Or83b expression levels were markedly reduced by ethanol exposure (Fig. 5A). Surprisingly, flies harboring a targeted deletion of Or83b (Or83b1) exhibited a normal ethanol-induced olfactory startle response (PS for Berlin: 5.9 ± 0.46 mm/s, Or83b1: 5.1 ± 0.90 mm/s, p = 0.471, paired t-test, n = 6), suggesting that ethanol odor is detected by Or83b-independent means, presumably by the 30% of OR neurons that retain functionality (Fig. 5A). Or83b mutant flies showed 2 specific alterations in ethanol-induced behaviors, reduced Dist during the first ethanol exposure, and an increased ΔDist (Fig. 5B,C). Neither behavioral change in Or83b mutants could be attributed to altered ethanol sedation sensitivity, and there was also no effect of the mutation on ethanol sedation tolerance (Fig. 5D,E). These data suggest that an Or83b-mediated olfactory mechanism promotes acute locomotor responding to ethanol, and that this mechanism is independent of the olfactory-mediated startle response. Down-regulation of Or83b gene expression following acute ethanol exposure may therefore contribute to increased locomotor activity that accompanies the development of rapid tolerance.
Fig. 5.
Behavioral effects of deletion of Or83b. (A) Locomotor activity profile of Or83b1 compared with matched Berlin controls. n = 6. Inset: expression levels by microarray analysis of Or83b following exposure to ethanol or humidified air relative to untreated controls (time 0). (B to E) Measures of ethanol responses derived from experimental data depicted in A (B: p = 0.051; C: *p = 0.032; D: p = 0.085; E: p = 0.581, paired t-test).
Immune Response Gene Spn27A
Upon infection in Drosophila, a melanization pathway is activated that is involved in sequestering and eliminating invading bacteria (Brennan and Anderson, 2004). The melanization pathway consists of a cascade of serine proteases that are inactivated by serpins, including Spn27A, that irreversibly bind serine protease catalytic sites. Transcript levels for the gene-encoding Spn27A were induced nearly 5-fold 3.5-hour following ethanol exposure termination (Fig. 6A). Bacterial infection causes a similar temporal pattern of Spn27A induction, and this up-regulation is thought to help limit the melanization reaction in time and space (De Gregorio et al., 2002). To begin to test the role of immune genes in ethanol responses, we characterized the effects of disrupting Spn27A. The transposon e02539 is inserted into the coding region of the Spn27A locus and is predicted to result in truncation of the Spn27A protein (Fig. 6B). Animals homozygous for e02539 exhibited maternal-effect lethality and increased spontaneous melanization, detected as discrete dark spots through the cuticle. These phenotypes are consistent with e02539 causing a strong loss-of-function for Spn27A (De Gregorio et al., 2002; Ligoxygakis et al., 2002). Spn27Ae02539 homozygotes were viable and had no gross morphological or behavioral deficits. Acute ethanol exposure resulted in reduced Dist (Fig. 6C,D,F). However, the ability to develop rapid tolerance was unaffected (Fig. 6E,G). These data indicate that Spn27A regulates acute responding to ethanol exposure.
Fig. 6.
Behavioral effects of disruption of Spn27A. (A) Expression levels by microarray analysis of Spn27A following exposure to ethanol or humidified air relative to untreated controls (time 0). (B) The Spn27A locus consists of a single transcriptional unit with no introns and a single open reading frame (shaded). The transposon e02539 is inserted into the open reading frame. (C) Locomotor activity profile for Spn27Ae02539, compared with matched Berlin controls; n = 6. (D to G) Measures of ethanol responses derived from the experimental data depicted in C (D: **p = 0.001; E: p = 0.994; F: p = 0.092; G: p = 0.431; paired t-test).
Histone/Protein Deacetylase Sir2
Sir2 encodes the Drosophila homolog of the class III histone/protein deacetylase (HDAC) Sir2/Sirt1 that is evolutionarily conserved from yeast to man, and it has been implicated in the regulation of gene expression, protein activity, lifespan, and, more recently, in circadian and cocaine behaviors (Furuyama et al., 2004; Nakahata et al., 2009; Ramsey et al., 2009; Renthal et al., 2009). Expression of Sir2 in fly heads was transiently reduced to about half of normal levels by ethanol exposure (Fig. 2E,F). Expression of Sir2 was also found to be decreased following 2 brief exposures to high concentration ethanol vapor (Morozova et al., 2006). Of the 4 other class III HDAC-encoding genes in flies, Sirt2 was also down-regulated by ethanol exposure in the microarray analysis. Additionally, the HDAC class I-encoding gene Rpd3 was up-regulated, but expression of other HDAC-encoding genes were unaffected by ethanol exposure (Table S1). A targeted deletion of Sir2, 2A-7–11, removes most of the Sir2 coding region without affecting the expression levels of the neighboring DnaJ-H gene (Fig. 7A,B). These Sir2 null mutant flies were fully viable and fertile, and showed no obvious morphological or behavioral defects. Exposure of Sir22A-7–11 flies to ethanol vapor in the rapid tolerance paradigm revealed 3 phenotypes: reduced sedation sensitivity, reduced sedation tolerance, and a near complete lack of ΔDist (Fig. 7C to G). Because sedation tolerance and ΔDist covary by ethanol dose (Fig. 1F,I), we determined whether this was also the case in Sir2 nulls by testing for ethanol responses at a higher ethanol dose (73%). Strikingly, at this higher ethanol concentration Sir22A-7–11 mutants retained the decreased ΔDist phenotype but exhibited normal sedation sensitivity and sedation tolerance (Fig. 7H to L). A second transposon insertion, NP1145, did not complement Sir22A-7–11 for ΔDist and as a homozygote exhibited a similar dose-independent reduction of ΔDist and dose-dependent effect on sedation sensitivity and sedation tolerance (not shown). Sir2 was expressed in the nervous system in both glia and neurons (Fig. S5). Thus, Sir2 regulates locomotor behavioral plasticity in the ethanol rapid tolerance paradigm and ethanol sedation sensitivity when ethanol is presented at moderate concentrations.
DISCUSSION
By using a combination of global gene expression analysis and a mutant survey, we demonstrate that genes important for immunity, metabolism, olfaction, and protein acetylation regulate ethanol behavioral responses in Drosophila. Evidence for genes involved in each of these biological processes in regulating the effects of drugs of abuse in higher organisms has been documented, but the mechanisms by which they contribute to ethanol behavioral responses and the development of alcohol use disorders are not well understood. Our studies greatly extend the genetic overlap of how ethanol affects behavior from flies to mammals and provide a basis for dissecting the roles of specific biological processes and their inter-relationships in a relatively simple genetic system.
To find genes regulated by exposure to ethanol, we determined the time course of gene expression changes from immediately following a just sedating ethanol exposure to 3.5 hours later, when accumulated ethanol has been completely metabolized and expression of ethanol rapid tolerance is high (Scholz et al., 2000). This approach has 2 main benefits over single time-point sampling used in previous studies of ethanol regulation of gene expression: assumptions about sampling time are minimized, and small but temporally consistent changes in gene expression can be detected. For example, we detected the coordinate up-regulation of immunity genes that was largely missed previously (Fig. S2). Moreover, we sampled gene expression in whole fly heads that included the entire brain (excluding the thoracic ganglion), trachea, fat bodies, and musculature. Regional effects of ethanol exposure on gene expression could result in relatively small alterations in overall expression levels that are nonetheless critical for the behavioral actions of ethanol. We detected gene expression changes that overlapped with those identified in 2 previous studies of the effects of ethanol exposure in Drosophila (Morozova et al., 2006; Urizar et al., 2007). The overlap of gene expression regulation detected was small, but the direction of the regulation was remarkably consistent, indicating both that the quality of the expression data from all studies was high, and that some biological processes are engaged by ethanol presented at varied concentrations, lengths of time, and frequency. Increased expression of 3 genes-encoding serine synthesis enzymes was a prominent effect of ethanol exposure that was detected across studies, and our preliminary mutational analysis of 2 of these genes (the 3-phosphoserine phosphatase-encoding aay and the glycine hydroxymethyl-transferase encoding CG3011) supports a role for serine in behavioral responses to acute and repeated ethanol exposures (Table S5). Serine is involved in protein and phospholipid synthesis, but also acts as a co-agonist for NMDA signaling (Oliet and Mothet, 2009). Any or all of these roles for serine may be affected by increased serine synthesis following ethanol exposure. Alterations in membrane lipid physiology correlate with resistance to lethal ethanol concentrations in flies (Montooth et al., 2006), and phospholipid synthesis and signaling pathway changes are induced by ethanol consumption in the mammalian liver (Baraona and Lieber, 1979). In flies, the fat bodies perform many functions of the mammalian liver and thus may be the site of serine regulation by ethanol. The role of the fat bodies in ethanol behavioral responses is not known. While a role for d-serine has yet to be uncovered in flies, the presence of the enzyme serine racemase indicates that it can be synthesized from l-serine, and up-regulation of serine racemase expression by ethanol suggests that ethanol exposure results in increased d-serine availability. Moreover, glutamatergic NMDA receptor signaling is a major target of ethanol in mammals at concentrations similar to those used for our microarray study (Lovinger et al., 1989). It will be important to determine the mechanisms by which the regulation of serine synthesis contributes to the effects of ethanol.
Olfaction is a potent source of information about the reward and valuational properties of a variety of environmental stimuli, and there is increasing evidence that olfaction may play a direct role in the effects of ethanol on brain functions that are relevant to addiction. Ethanol vapor evokes a strong olfactory response in many species, and in humans it elicits activity in specific areas of the brain, including the nucleus accumbens and ventral tegmental area, that are critical for the rewarding properties of alcohol (Kareken et al., 2004). Moreover, the magnitude of activity observed in the nucleus accumbens positively correlates with high alcohol use. Studies in rodents have shown that prenatal exposure to ethanol can increase the behavioral response to ethanol odors and increased ethanol intake (Youngentob and Glendinning, 2009). Acute ethanol ingestion can also interfere with olfactory discrimination, suggesting that there exists a complex interplay between the sensory and central effects of ethanol (Patel et al., 2004). A relationship between the smell of ethanol and its behavioral effects may also exist in insects. In the honeybee, ingestion of ethanol can impair olfactory learning and discrimination (Mustard et al., 2008). In Drosophila, ethanol is attractive at low concentrations, influences neural activity in olfactory neurons, and evokes locomotor activity (Hallem and Carlson, 2006; Kim et al., 1998; Wolf et al., 2002). Moreover, Drosophila lines selected for ethanol sedation resistance exhibit reduced attraction to ethanol (Hoffmann and Cohan, 1987). Here, we showed that ethanol exposure causes a strong down-regulation of specific olfactory genes, suggesting that olfactory responses to this ethologically relevant odor are modified at the level of gene expression. Whether ethanol elicits this response directly in the olfactory receptor neurons, or indirectly through olfactory transduction circuits or at other sites remains to be explored. Additionally, we found that, when 1 of the down-regulated genes, the olfactory co-receptor encoding Or83b, was ablated, the increase in ethanol-induced hyperactivity upon a second exposure was magnified. Most simply, these data indicate that olfaction in Drosophila regulates plasticity in the behavioral response to ethanol.
Inflammatory immune responses associated with chronic and heavy drinking in mammals have been documented in the liver and the brain, and have been studied largely in the context of alcohol-related disease and tissue damage (Goral et al., 2008). Under these conditions, innate immune signaling, mediated by the Toll-like receptors, can be either suppressed or enhanced, depending on ethanol dose, length of exposure, and tissue type. Recent studies indicate that expression of genes that encode components of the innate immune signaling pathways are also regulated in brain tissue following exposure to moderate levels of ethanol or cocaine. Most consistently, altered expression of transcriptional regulator NFκB was identified in the midbrain following acute and chronic ethanol exposures (Rulten et al., 2006), in whole brains in comparisons of high and low alcohol preferring mouse strains (Mulligan et al., 2006), and in the nucleus accumbens following chronic cocaine exposure (Ang et al., 2001). These studies demonstrate that NFκB signaling pathways are regulated in the brain by drugs of abuse. To date, only 1 functional study for immune pathways in the brain in drug responses has been done, implicating NFκB in cocaine reward in the accumbens (Russo et al., 2009). Our microarray study detected a marked up-regulation of genes in the innate immune signaling pathways Tl and Imd following acute ethanol exposure. Both Tl and Imd pathways converge on members of the NFκB family, indicating that regulation of NFκB signaling by ethanol is an evolutionarily conserved phenomenon. Preliminary behavioral analysis of flies carrying a transposon insertion in the NFκB homolog Rel locus has suggested that Rel may regulate ethanol sedation sensitivity (Table S6). This suggests that Rel, and by extension the Imd pathway, contributes to ethanol responses in flies. As these signaling pathways are used in many contexts during development and in postembryonic physiology, it will be important to map their actions in space and time to pinpoint their function in ethanol behavioral responses.
Our findings also implicate the serine protease inhibitor Spn27A in promoting ethanol-induced hyperactivity. In early development, Spn27A targets the Easter serine protease that cleaves and activates the Toll ligand Spätzle. In adults, Spn27A acts on 2 different serine proteases, MP1 and MP2, to regulate the activation of phenoloxidase in the production of melanin during bacterial infection (Tang et al., 2006). The target for Spn27A in ethanol responses is not known. While Easter is dispensable for immune responses, it is expressed in adult heads, and Spn27A has been shown to interact with the Toll pathway by an as yet incompletely understood mechanism during bacterial infection (Ligoxygakis et al., 2002). Moreover, we were unable to detect any gross increase in melanization in adult flies lacking Spn27A following ethanol exposure, as occurs in infection (De Gregorio et al., 2002), despite escalating the dose and number of exposures (not shown). While not conclusive, these data suggest that Spn27A expression regulation by ethanol exposure may utilize this immunity pathway in a manner distinct from bacterial infection. In addition, infection-induced immune responses are limited to the tissues that contact the invading organism, including the tracheal epithelia and the fat bodies that directly interface with the hemolymph. Ethanol, by contrast, diffuses throughout all tissues, and may be able to activate immune response pathways in tissues protected from invaders, such as the brain. Finally, we note that the kinetics of Spn27A transcript accumulation following ethanol exposure are similar to that elicited by infection, suggesting that a similar mechanism of regulating Spn27A levels is engaged by these distinct signals.
Our data demonstrate that the HDAC Sir2 promotes changes in locomotor behavior elicited by repeated ethanol exposure, and also promotes sensitivity to the sedative actions of ethanol. Recent findings have shown that the sirtuins, through their actions as HDACs, influence animal behavior, including circadian rhythms and the rewarding properties of cocaine (Nakahata et al., 2009; Renthal et al., 2009). It is likely that histone acetylation also regulates the physiological effects of ethanol exposure: HDAC activity is decreased in the amygdala following acute ethanol exposure in rats, and hepatocyte cell culture studies identified increased histone acetylation following ethanol exposure (Pandey et al., 2008; Park et al., 2003). Additionally, repeated exposure of Drosophila to benzyl alcohol, which may affect flies in a manner similar to ethanol, revealed tolerance to recovery from sedation that is accompanied by increased histone acetylation, and these effects were mimicked by an HDAC inhibitor (Wang et al., 2007). Sir2 is likely expressed in all tissues, and ethanol-dependent changes in protein acetylation levels may be equally widespread.
What genes might be regulated by ethanol in a Sir2-dependent manner? Drosophila Sir2 targets include, among others, those bound by the Hairy transcriptional repressor and the E(Z) histone methyltransferase complex that is involved in chromatin silencing (Bianchi-Frias et al., 2004; Furuyama et al., 2004). Association of Hairy with specific genomic loci is cell-type or developmental stage specific, suggesting that additional factors direct Hairy and by extension Sir2 to DNA. Studies of Hairy- and Sir2-dependent histone acetylation patterns in ethanol-exposed flies could provide candidate genes for the regulation of behavioral plasticity. Nuclear non-histone protein targets of mammalian Sir2 homolog SIRT1 include p53, FOXO proteins, and NFκB, and these evolutionarily conserved proteins may also play a role in regulating ethanol behavioral responses (Denu, 2005).
Acetyl-CoA synthase and Sir2 may provide a link between the metabolic consequences of ethanol exposure and behavioral response patterns. Unique among the histone deacetylases, the sirtuins require nicotinamide adenine dinucleotide (NAD+) for activity, and therefore are tied to the metabolic state of the cell. Ethanol metabolism leads to decreases in NAD+ levels through the activities of alcohol dehydrogenase and acetaldehyde dehydrogenase (Zakhari, 2006). Lowered NAD+ levels may lead to decreased Sir2 activity, and decreased Sir2 expression levels following ethanol exposure may reflect a means of tuning Sir2 availability to the metabolic state of cells. Additionally, acetyl-CoA, a major product of ethanol metabolism, is the source of acetyl groups for the histone acetyltransferases. The net effect of ethanol metabolism may favor histone acetylation over deacetylation, resulting in increased gene expression following ethanol exposure. Additionally, as noted above, acetyl-CoA is an intermediate for many cellular pathways, including acetylcholine synthesis and the mevalonate pathway. The mevalonate pathway in insects leads to the synthesis of many cellular products, including prenylation that anchors proteins such as the Ras small GTPase family to lipid membranes, and juvenile hormone (Belles et al., 2005). If ethanol metabolism plays a role in the behavioral effects of ethanol exposure, it is clearly only one of many mechanisms for the the actions of ethanol, and it is likely that drugs of abuse engage specific molecular pathways through multiple means. For example, cocaine is metabolized by means that are completely distinct from ethanol, and our data and that of others show that regulation of histone deacetylases is important for the behavioral effects of both of these addictive drugs (Kumar et al., 2005).
Supplementary Material
ACKNOWLEDGMENTS
We thank Rachael French for information ahead of publication, Mark Eddison and Roland Bainton alleles of foraging and Tre1, respectively, and members of the Heberlein lab for helpful discussions. This work was supported by NIAAA grants AA014594-03 and AA017072-01.
Footnotes
The authors declare that they had no competing interests.
REFERENCES
- Ang E, Chen J, Zagouras P, Magna H, Holland J, Schaeffer E, Nestler EJ. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. J Neurochem. 2001;79:221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT-Y, Singh CM, Moore MS, Neckameyer WS, Heberlein U. Dopamine modulates acute responses to cocaine, nicotine, and ethanol in Drosophila. Curr Biol. 2000;10:187–194. doi: 10.1016/s0960-9822(00)00336-5. [DOI] [PubMed] [Google Scholar]
- Baraona E, Lieber CS. Effects of ethanol on lipid metabolism. J Lipid Res. 1979;20:289–315. [PubMed] [Google Scholar]
- Belles X, Martin D, Piulachs MD. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu Rev Entomol. 2005;50:181–199. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Heberlein U, Moore MS. Rapid and chronic: two distinct forms of ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2004;28:1469–1480. doi: 10.1097/01.alc.0000141817.15993.98. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady MJ, Saltiel AR. The role of protein phosphatase-1 in insulin action. Recent Prog Horm Res. 2001;56:157–173. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- Corl AB, Rodan AR, Heberlein U. Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster. Nat Neurosci. 2005;8:18–19. doi: 10.1038/nn1363. [DOI] [PubMed] [Google Scholar]
- Cowmeadow RB, Krishnan HR, Atkinson NS. The slowpoke gene is necessary for rapid ethanol tolerance in Drosophila. Alcohol Clin Exp Res. 2005;29:1777–1786. doi: 10.1097/01.alc.0000183232.56788.62. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H. Gamma-aminobutyric acid B receptor 1 mediates behavior-impairing actions of alcohol in Drosophila: adult RNA interference and pharmacological evidence. Proc Natl Acad Sci USA. 2003;100:5485–5490. doi: 10.1073/pnas.0830111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin VG, Deitrich RA. Genetic selection and characterization of mouse lines for acute functional tolerance to ethanol. J Pharmacol Exp Ther. 1996;279:1310–1317. [PubMed] [Google Scholar]
- Fadda F, Rossetti ZL. Chronic ethanol consumption: from neuroadaptation to neurodegeneration. Prog Neurobiol. 1998;56:385–431. doi: 10.1016/s0301-0082(98)00032-x. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Banerjee R, Breen TR, Harte PJ. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol. 2004;14:1812–1821. doi: 10.1016/j.cub.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Loss of Hsp70 in Drosophila is pleiotropic, with effects on thermotolerance, recovery from heat shock and neurodegeneration. Genetics. 2006;172:275–286. doi: 10.1534/genetics.105.048793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goral J, Karavitis J, Kovacs EJ. Exposure-dependent effects of ethanol on the innate immune system. Alcohol. 2008;42:237–247. doi: 10.1016/j.alcohol.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Cohan FM. Olfactory responses of Drosophila melanogaster selected for knockdown resistance to ethanol. Behav Genet. 1987;17:307–312. doi: 10.1007/BF01065509. [DOI] [PubMed] [Google Scholar]
- Kalant H, LeBlanc AE, Gibbins RJ. Tolerance to, and dependence on, some non-opiate psychotropic drugs. Pharmacol Rev. 1971;23:135–191. [PubMed] [Google Scholar]
- Kareken DA, Claus ED, Sabri M, Dzemidzic M, Kosobud AE, Radnovich AJ, Hector D, Ramchandani VA, O’Connor SJ, Lowe M, Li TK. Alcohol-related olfactory cues activate the nucleus accumbens and ventral tegmental area in high-risk drinkers: preliminary findings. Alcohol Clin Exp Res. 2004;28:550–557. doi: 10.1097/01.alc.0000122764.60626.af. [DOI] [PubMed] [Google Scholar]
- Kim MS, Repp A, Smith DP. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics. 1998;150:711–721. doi: 10.1093/genetics/150.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister RG. The effects of repeated doses of ethanol on exploration and its habituation. Psychopharmacology (Berl) 1987;92:78–83. doi: 10.1007/BF00215483. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, White G, Weight FF. Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science. 1989;243:1721–1724. doi: 10.1126/science.2467382. [DOI] [PubMed] [Google Scholar]
- McClung C, Hirsh J. Stereotypic behavioral responses to free-base cocaine and the development of behavioral sensitization in Drosophila. Curr Biol. 1998;8:109–112. doi: 10.1016/s0960-9822(98)70041-7. [DOI] [PubMed] [Google Scholar]
- Montooth KL, Siebenthall KT, Clark AG. Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J Exp Biol. 2006;209:3837–3850. doi: 10.1242/jeb.02448. [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh SM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustard JA, Edgar EA, Mazade RE, Wu C, Lillvis JL, Wright GA. Acute ethanol ingestion impairs appetitive olfactory learning and odor discrimination in the honey bee. Neurobiol Learn Mem. 2008;90:633–643. doi: 10.1016/j.nlm.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliet SH, Mothet JP. Regulation of N-methyl-d-aspartate receptors by astrocytic d-serine. Neuroscience. 2009;158:275–283. doi: 10.1016/j.neuroscience.2008.01.071. [DOI] [PubMed] [Google Scholar]
- Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PH, Miller R, Shukla SD. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem Biophys Res Commun. 2003;306:501–504. doi: 10.1016/s0006-291x(03)01040-4. [DOI] [PubMed] [Google Scholar]
- Patel SJ, Bollhoefer AD, Doty RL. Influences of ethanol ingestion on olfactory function in humans. Psychopharmacology (Berl) 2004;171:429–434. doi: 10.1007/s00213-003-1612-x. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, et al. Alcohol tolerance in large-conductance, calcium-activated potassium channels of CNS terminals is intrinsic and includes two components: decreased ethanol potentiation and decreased channel density. J Neurosci. 2004;24:8322–8332. doi: 10.1523/JNEUROSCI.1536-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, He Y, Lu Y, Bellen HJ. Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics. 2000;156:1691–1715. doi: 10.1093/genetics/156.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab A, Thompson EC, Travers AA. High mobility group proteins HMGD and HMGZ interact genetically with the Brahma chromatin remodeling complex in Drosophila. Genetics. 2006;172:1069–1078. doi: 10.1534/genetics.105.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Carlson J. OS9: a novel olfactory gene of Drosophila expressed in two olfactory organs. J Neurobiol. 1994;25:169–184. doi: 10.1002/neu.480250208. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HEr, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BP, et al. Alcohol dependence is associated with the ZNF699 gene, a human locus related to Drosophila hangover, in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) sample. Mol Psychiatry. 2006;11:1025–1031. doi: 10.1038/sj.mp.4001891. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Mackay TF, Anholt RR. Pinocchio, a novel protein expressed in the antenna, contributes to olfactory behavior in Drosophila melanogaster. J Neurobiol. 2005;63:146–158. doi: 10.1002/neu.20123. [DOI] [PubMed] [Google Scholar]
- Rulten SL, Ripley TL, Hunt CL, Stephens DN, Mayne LV. Sp1 and NFkappaB pathways are regulated in brain in response to acute and chronic ethanol. Genes Brain Behav. 2006;5:257–273. doi: 10.1111/j.1601-183X.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V, Renthal W, Graham A, Birnbaum SG, Green TA, Robison B, Lesselyong A, Perrotti LI, Bolanos CA, Kumar A, Clark MS, Neumaier JF, Neve RL, Bhakar AL, Barker PA, Nestler EJ. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustay N, Crabbe J. Genetic analysis of rapid tolerance to ethanol’s incoordinating effects in mice: inbred strains and artificial selection. Behav Genet. 2004;34:441–451. doi: 10.1023/B:BEGE.0000023649.60539.dd. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Cotte N, Zambon AC, Pollard KS, Vranizan K, Doniger SW, Dolganov G, Conklin BR. Identifying genetic networks underlying myometrial transition to labor. Genome Biol. 2005;6:R12. doi: 10.1186/gb-2005-6-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, et al. Characterization of acute and chronic tolerance in mice selected for inherent differences in sensitivity to ethanol. Alcohol Clin Exp Res. 1980;4:70–73. doi: 10.1111/j.1530-0277.1980.tb04794.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila. J Biol Chem. 2006;281:28097–28104. doi: 10.1074/jbc.M601642200. [DOI] [PubMed] [Google Scholar]
- Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27:4541–4551. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci USA. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. Drug-induced epigenetic changes produce drug tolerance. PLoS Biol. 2007;5:e265. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FW, Heberlein U. Invertebrate models of drug abuse. J Neurobiol. 2003;54:161–178. doi: 10.1002/neu.10166. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngentob SL, Glendinning JI. Fetal ethanol exposure increases ethanol intake by making it smell and taste better. Proc Natl Acad Sci USA. 2009;106:5359–5364. doi: 10.1073/pnas.0809804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhari S. Overview: how is alcohol metabolized by the body? Alcohol Res Health. 2006;29:245–254. [PMC free article] [PubMed] [Google Scholar]
- Zaleski MJ, et al. GABA(B) receptors play a role in the development of tolerance to ethanol in mice. Psychopharmacology (Berl) 2001;153:415–424. doi: 10.1007/s002130000581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.