Abstract
Degradation of DNA during gene delivery is an obstacle for gene transfer and for gene therapy. DNases play a major role in degrading foreign DNA. However, which of the DNases are involved and whether their inactivation can improve gene delivery have not been studied. We have recently identified deoxyribonuclease I (DNase I) and endonuclease G (EndoG) as the major degradative enzymes in the mouse kidney proximal tubule epithelial (TKPTS) cells. In this study, we used immortalized mouse TKPTS cells and primary tubular epithelial cells isolated from DNase I or EndoG knockout (KO) mice and examined the degradation of plasmid DNA during its uptake. DNase I and EndoG KO cells showed a higher rate of transfection by pECFP-N1 plasmid than wild-type cells. In addition, EndoG KO cells prevented the uptake of fluorescent-labeled RNA. Complete inhibition of secreted DNase I by G-actin did not improve plasmid transfection, indicating that only intracellular DNase I affects DNA stability. Data demonstrate the importance of DNase I and EndoG in host cell defense against gene and RNA delivery to renal tubular epithelial cells in vitro.
Introduction
Extracellular DNA uptake occurs during normal and cancer tissue growth (Bergsmedh et al., 2006; Yan et al., 2006) and during viral and bacterial infections (Chu et al., 2006; Metifiot et al., 2007), and is routinely used in genetic manipulations and experimental animals (Tanswell et al., 1998; Glasspool-Malone et al., 2002; Freitas et al., 2007). The entry of foreign DNA (fDNA) is harmful to the host cell (Li et al., 1999), causing DNA-dependent cell death, whereas DNase treatment before transfection prevented cell death (Stacey et al., 1993). The introduction of single-stranded DNA in cells induced DNA damage and apoptotic factors, acting upstream of ATM/p53 in the p53-dependent pathway (Nur et al., 2003). Studies have also demonstrated that the introduction of fDNA induces mutations due to homologous recombination (Thomas and Capecchi, 1986; Torchilin, 2006).
The uptake of DNA is restricted by a number of cell defense enzymes, the core of which consists of DNA endonucleases (Tanswell et al., 1998; Glasspool-Malone et al., 2002). Despite attempts to protect the fDNA by modifications, lipid or viral packaging, increased rate of DNA delivery, or by precise targeting to a tissue, our knowledge about the protection against the cellular defense system remained insufficient (Tanswell et al., 1998; Glasspool-Malone et al., 2002; Freitas et al., 2007). Endocytosis of DNA normally leads to its lysosomal delivery, with DNases playing major role in degrading fDNA (Torchilin, 2006). However, it is still unknown which of these DNases are involved in the degradative process and whether the inactivation of these enzymes would improve gene delivery due to the lack of tools, such as knockout (KO) endonuclease-deficient mice or endonuclease inhibitors.
In our previous studies, we selected two of the nine known cellular cytotoxic endonucleases, which can degrade nonmodified DNA, namely, deoxyribonuclease I (DNase I) and endonuclease G (EndoG). These were the most active endonucleases in kidney tubular epithelial cells (Peitsch et al., 1995; Basnakian et al., 2005; Irvine et al., 2005). DNase I is a 31 kDa cytoplasmic enzyme that digests single- and double-stranded DNA. It can be excreted from the cells. Maximal enzymatic activity of DNase I can be measured when both Ca2+ and Mg2+ ions are present (Basnakian et al., 2002). EndoG is a nuclear-encoded mitochondrial nuclease that translocates to the nucleus during apoptosis (Zhang et al., 2003), is expressed in the cytoplasm as a precursor (33 kDa), and is converted to its mature form (28 kDa) upon entering the mitochondrion (Ikeda and Kawasaki, 2001). EndoG is a preferentially Mn-dependent enzyme (Widlak et al., 2001) that is capable of digesting double- and single-stranded DNA, RNA, and DNA/RNA heteroduplexes (Huang et al., 2006).
As primary cells are known to be resistant to DNA transfection, we have improved DNA delivery to primary tubular epithelial (PTE) cells using Lipofectamine. To determine the role of DNase I and EndoG, PTE cells were isolated from DNase I or EndoG KO mice. Our data provide evidence that in DNase I KO and EndoG KO cells, the efficiency of transfection by plasmid DNA is significantly higher than in wild-type (WT) cells, indicating that these two endonucleases restrict DNA uptake in murine renal cells.
Materials and Methods
Animals
Homozygous DNase I KO mice (CD-1 background) were obtained from Dr. T. Moroy, University of Essen, Germany. EndoG KO mice (129xC57/B6 background) were obtained from Drs. M. Xu and J. Zhang, University of Cincinnati, OH. Because EndoG−/− animals are not viable, the cells were isolated from heterozygous mice (EndoG KO). DNase I KO mice were bred as heterozygotes, and EndoG+/− mice were bred with WT mice (EndoG+/+). All mice were genotyped by PCR as previously described (Zhang et al., 2003; Djurovic et al., 2004). All animal experiments received human care to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences. All animal experiments were approved by the Animal Care and Use Committee of the Central Arkansas Veterans Healthcare System.
Cell cultures
Mouse PTE cells were freshly isolated from DNase I KO, EndoG KO, and WT mice as described by Nowak et al. (2003), and cultured up to 10 days (passages 1 and 2) before experiments. Immortalized mouse kidney proximal tubule epithelial (TKPTS) cells were obtained from Dr. Elsa Bello-Reuss (University of Texas Medical Branch, Galveston, TX) and were cultured as previously described (Ernest and Bello-Reuss, 1995). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 7% fetal bovine serum (Hyclone, Logan, UT). Cells were maintained in a CO2 incubator at 37°C in 5% CO2, fed at 48–72 h intervals, and used within 1 day after reaching confluency, with the exception of RNA interference (RNAi) experiments described below.
Total protein extraction from cultured cells
Cells (2–4 × 106) were grown as described above. Cultured cells were collected by centrifugation (1000 rpm, 3 min, 4°C). After removal of supernatant, the cells were resuspended in phosphate buffered saline and centrifuged again as described above. For protein extraction, cells were diluted in 100 μL Buffer A (50 mM Tris-HCl, pH 7.9; 0.25 M sucrose) and the Complete Mini Proteinase Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) (1 tablet/10 mL), and disrupted with Virsonic 475 (Virtis, Gardiner, NY) (2 × 20 s). Particulate material was precipitated out from the extract by centrifugation (14,000 rpm, 10 min, 4°C), and the supernatant was collected. The extracts were dialyzed against storage buffer (55% Glycerol, 10 mM Tris-HCl pH 7.6, 0.5 mM dithiothreitol) and stored at −20°C for up to 2 weeks without loss of endonuclease activity. Protein was measured using the bicinchoninic acid protein assay (Pierce, Rockford, IL). Bovine serum albumin was used as the standard.
Plasmid incision assay
Endonuclease activity in total protein extracts from kidney cells and in culture medium was determined using the plasmid incision assay (PIA) with pBR322 plasmid (New England Biolabs, Beverly, MA) as the substrate as described previously (Basnakian et al., 2005).
To determine whether Lipofectamine is protecting plasmid DNA from degradation by endonucleases, pBR322 plasmid was pretreated with Lipofectamine before it was exposed to the culture medium. Plasmid and Lipofectamine were diluted separately in serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich), mixed together, and incubated for 20 min at room temperature.
After adding serially diluted samples (1:5) to the reaction mixture (1 μg pBR322 plasmid DNA, 2 mM CaCl2, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.4, and 0.5 mM dithiothreitol) the reaction was incubated for 1 h at 37°C, after which the reaction was terminated by adding Stop-solution (10 mM Tris-HCl, pH 7.4, 1% SDS, 25 mM Na2EDTA, 7.5 mM bromophenol blue). The samples were run on a 1% agarose gel in Tris-acetate-EDTA buffer, pH 8 (7V/cm, 35 min), and the DNA was viewed with ethidium bromide. The EagleEye scanning densitometer (Stratagene, La Jolla, CA) was utilized to quantify the relative amount of endonuclease-treated plasmid DNA present in a covalently closed circular DNA (C), open circular DNA (O), or linear DNA (L), or in digested form (D). One unit was defined as the amount of endonuclease capable of converting 1 μg covalently closed supercoiled plasmid DNA to open circular, linear, or digested DNA in 1 h at 37°C.
This assay was also used for the characterization of endonucleases in primary cells. Endonuclease activity was measured the same way as above in samples containing serially diluted protein (1:5), 1 μg plasmid pBR322 DNA (New England Biolabs), 2 mM CaCl2, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.4, and 0.5 mM dithiothreitol to determine the Ca/Mg-dependent (primarily DNase I) endonuclease activity or 5 mM MnCl2, 10 mM Tris-HCl (pH 7.4), and 0.5 mM dithiothreitol for the Mn-dependent (mainly EndoG) activity.
As opposed to PIA, zymogram gel electrophoresis, previously used by us to assess DNase I activity (Basnakian et al., 2005), was not applicable for EndoG due to low specific activity of the enzyme in the used tubular epithelial cells.
Real-time reverse transcriptase polymerase chain reaction
Our previously described protocol was followed (Basnakian et al., 2006). Briefly, 1 μg of total RNA was reverse-transcribed in a 50-μL reaction mixture followed by real-time reverse transcriptase polymerase chain reaction (RT-PCR) in a 25-μL reaction using SmartCycle (Cepheid, Sunnyvale, CA). Reaction mixture was prepared using Platinum SYBR Green qPCR Supermix-UDG (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. Primers for endonucleases were as follows: 5′-GATGAGACCATCCCTCTGGA-3′ and 5′-ATGTGAGTCAGCCCATCTCC-3′ for EndoG, and 5′-ACTCAATCGGGACAAACCTG-3′ and 5′-ATTTCCACAGGGTTCACAGC-3′ for DNase I. Two-temperature cycles with annealing/extension temperature at 62°C for EndoG and DNase I, and 64°C for 18S mRNA were used. The fluorescence was measured at the end of the annealing step. The melting curve analyses were performed at the end of the reaction after the 45th cycle between 60°C and 95°C to assess the quality of the final PCR products. The threshold cycle C(t) values were calculated by fixing the basal fluorescence at 15 units. cDNA samples for real-time PCR were diluted to 1:5, 1:10, and 1:200 for EndoG, DNase I, and 18S mRNA, respectively. Three replication reactions were performed for each sample, and the average C (t) was calculated. The standard curve of the reaction effectiveness was plotted against serially diluted (five points) mixtures of amplified cDNA samples for EndoG and for 18S mRNA. Calculation of the relative RNA concentration was performed using Cepheid SmartCycle software (Version 2.0d). Data are presented as ratio of EndoG/18S mRNA or DNase I/18S mRNA.
Plasmid transfection
PTE cells were transfected with pECFP-N1 plasmid (Clontech Laboratories, Mountain View, CA) that encodes cyan fluorescent protein (CFP) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were seeded into six-well plates 24 h before transfection. Briefly, 4 μg plasmid DNA and 1:7.5 DNA/liposome ratios were diluted in separate tubes in 250 μL serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich), mixed together, and incubated for 20 min at room temperature. Two milliliters serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich) was added to the cells. Transfection complexes were then added dropwise onto the cells. After 24–48 h incubation at 37°C in 5% CO2 the expression of CFP was detected by fluorescent microscopy using cyan filter.
Small interfering RNA transfection
PTE cells were transfected using TransIT-TKO transfection reagent (Mirus Bio, Madison, WI). Cells were seeded into six-well plates 24 h before transfection. In brief, 18 μL transfection reagent was diluted to 250 μL with serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich) incubated for 15 min, and then 75 μL (1 μM) small interfering RNA (siRNA)/fluorescent siRNA (Label IT RNAi Delivery Control-Fluorescein; Mirus Bio) was added and incubated for further 20 min at room temperature. After incubation 1172 μL serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich) was added to the cells. The transfection complex was then added dropwise to the cells. After 48 h incubation at 37°C in 5% CO2 the expression of fluorescent-labeled siRNA was detected by fluorescent microscopy.
The Label IT RNAi Delivery Control–Fluorescein contains a chemical dye fluorescein, also known as Fluorescein Isothiocyanate. The Fluorescein Isothiocyanate label is attached via a linker molecule and covalently bonded to the nucleotides. The introduction of short RNA duplexes into mammalian cells leads to sequence-specific destruction of target mRNA. These short double-stranded RNAs, referred to as siRNA, which can act catalytically at sub-molar ratios to cleave greater than 95% of the target mRNA in the cell and destruction of the mRNA target, can ultimately lead to decreased expression of the encoded protein. The RNAi effect can be long-lasting and may be detectable after many cell divisions. These properties make siRNA extremely effective at inhibiting target gene expression once introduced into the cell. The sequence of the Label IT RNAi Delivery Control is not homologous to any known mammalian gene and is not known to affect any cellular events. It is designed as a tool to facilitate observation and optimization of double-stranded RNA oligonucleotide delivery during RNAi experiments, both in vitro and in vivo (see manufacturer protocol; Mirus Bio, Lit.# ML039).
TKPTS cells were transfected using TransIT-TKO transfection reagent (Mirus Bio). Cells were seeded into 24-well plates 24 h before transfection. In brief, 4 μL transfection reagent was diluted to 50 μL with serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich) incubated for 15 min, and then 15 μL (1 μM) siRNA was added and incubated for further 20 min at room temperature. After incubation 250 μL serum-free DMEM/Ham's F-12 medium (Sigma-Aldrich) was given to the cells. The transfection complex was then added dropwise to the cells. After 48 h incubation at 37°C in 5% CO2 (medium was replaced-medium with serum-after 2 h) cells were transfected with enhanced CFP plasmid, pECFP-N1 (see Plasmid transfection section in Materials and Methods). After 24–48 h incubation at 37°C in 5% CO2 the expression of CFP was detected by fluorescent microscopy using cyan filter.
EndoG siRNA target sequence was AAAUGCCUGGAACAACCUUGA, DNase I siRNA target sequence was TGACATCGCTGTTATCCAA (Dharmacon, Lafayette, CO), and siCONTROL was Non-Targeting siRNA (Dharmacon).
Statistical analysis
Statistical analysis was performed with a two-way ANOVA and Student's t-test. Results were expressed as mean ±standard error of the mean. p < 0.05 was considered significant.
Results
Endonuclease activity of TKPTS cells to digest plasmid DNA
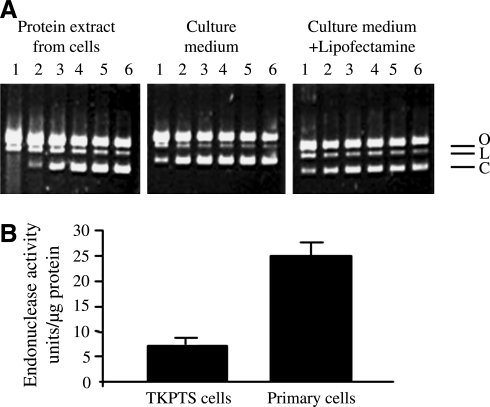
Initial experiments served to determine whether plasmid DNA can be digested by endonucleases present in tubular epithelial cells and excreted to the culture medium. After growing TKPTS cells to confluency, cells and medium were collected separately. Total cellular protein was extracted as described in the Materials and Methods section. Protein concentrations were measured in medium and protein extracts. Endonuclease activity was measured using the PIA. In this experiment, the circular covalently closed and supercoiled pBR322 plasmid DNA was converted by single-stranded breaks and double-stranded breaks to open circular (O) and linear (L) forms, respectively (Fig. 1A). The activity was higher in cell protein extracts than in the external medium, indicating that the destruction of plasmid DNA takes place mainly inside the cells.
FIG. 1.
Endonuclease activity in tubular epithelial cell extract and culture medium. The activity was measured using pBR322 PIA as described in the Materials and Methods section. (A) Endonuclease activity is present both in the cellular protein extracts and in the culture medium (left and middle panels). Pretreatment with Lipofectamine does not protect plasmid DNA against in vitro digestion by endonucleases (right panel). Dilutions (1–6) of cell extract or medium 1:1, 1:5, 1:25, 1:125, 1:625, and 1:3125, respectively. O, open circular DNA (with one or more single-strand DNA breaks but no double-strand breaks); L, linear DNA (with one double-strand DNA break); C, covalently closed circular DNA (without DNA breaks), which is the primary substrate for endonucleases. Endonuclease activity is seen only in the first two dilutions in cell extract, and in nondiluted culture medium. (B) Endonuclease activity in immortalized TKPTS cells and PTE cells. Primary cells have a higher total endonuclease activity (25 ± 2 units/μg protein in primary cells vs. 7 ± 3 units/μg protein in TKPTS cells, n = 3–6, p < 0.001) as measured using the pBR322 PIA in the presence of Ca2+ and Mg2+ ions (2 mM CaCl2 and 5 mM MgCl2), which are the cofactors for most of the cellular endonucleases. PIA, plasmid incision assay; TKPTS, mouse kidney proximal tubule epithelial; PTE, primary tubular epithelial.
Lack of inhibition of extracellular endonuclease activity by lipofectamine
Lipofectamine is preferentially used for plasmid DNA delivery during transfection of cells (Djurovic et al., 2004). To test whether Lipofectamine is capable of protecting plasmid DNA from degradation by endonucleases, pBR322 plasmid was pretreated with Lipofectamine and then exposed to the culture medium. As shown in Figure 1A (right panel), Lipofectamine per se provided no protection against the endonucleases present in culture medium.
Endonuclease activity in immortalized versus primary cells
Primary cells are known to exert some resistance to DNA transfection (Stacey et al., 1993; Welter et al., 2004; Zhong et al., 2005). To determine whether the resistance to fDNA is associated with the high endonuclease activity in primary cells, we compared the total endonuclease activities of protein extracts isolated from immortalized TKPTS cells and from PTE cells.
The PIA shows that endonuclease activity was several times higher in primary cells than in immortalized cells (Fig. 1B), confirming our notion that the resistance of primary cells to transformation is primarily due to their nuclease activity. This conclusion is based on the assumption that the permeability of plasmid to membrane is the same in primary and TPKTS cells.
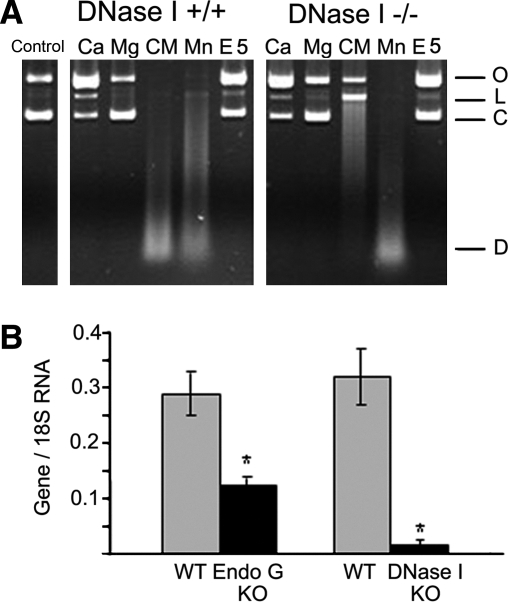
Endonucleases in KO primary cells
As endonucleases have overlapping cation and pH requirements, direct comparison of the activities characteristic to specific endonucleases is usually impossible. Therefore, the use of KO mice provides a unique opportunity to determine individual endonuclease activities belonging to particular endonucleases. DNase I and EndoG endonucleases were chosen since they turned out to be the two most active ones in murine kidney cells (Basnakian et al., 2005; Yin et al., 2007). PTE cells were isolated from WT and KO mice as described, and their protein extracts were tested for endonuclease activities using the PIA in the presence of different cations. Results show that the majority of the endonuclease activity in WT mice is Ca/Mg-dependent DNase I, the major endonuclease in these cells (Fig. 2A). After inactivation of DNase I in DNase I KO cells, the second most active endonuclease could be detected as Mn-dependent endonuclease, corresponding to EndoG. Partial inactivation of EndoG in heterozygous EndoG KO cells was associated with a reduced Mn-dependent activity without any effect on the Ca/Mg-dependent DNase I activity. However, the reduction of Mn-dependent activity in EndoG KO mice did not reach statistical significance, probably because the precision of PIA was not enough to measure incomplete inactivation of the enzyme in heterozygotes (data not shown). Real-time RT-PCR was performed as an alternative approach to determine whether the expression of these two endonucleases is decreased in murine KO cells. Our data indicate a complete fall out (95–100%) of DNase I activity in homozygous DNase I KO mice and a 60–70% loss in heterozygous EndoG in KO mouse kidney cells (Fig. 2B).
FIG. 2.
Activity and expression of endonucleases in PTE cells. (A) In the total protein extracts isolated from DNase I WT mice, the strongest endonuclease activity could be obtained when Ca2+ and Mg2+ ions were added together, resulting in digested DNA. This is characteristic to DNase I, which therefore provides most of the endonuclease activity in the normal kidney. In the kidney tissue extracts obtained from DNase I KO mice, Mn-dependent endonuclease was the most prominent, suggesting that EndoG is the second major endonuclease in the absence of DNase I (Widlak et al., 2001). Vertical row: O, open circular DNA; L, linear DNA; C, covalently closed circular DNA; D, digested DNA. Horizontal row: control nondigested pBR322 DNA; Ca2+, 2 mM CaCl2, pH 7.5; Mg2+, 2 mM MgCl2, pH 7.5; CM [Ca2++Mg2+], 2 mM CaCl2 + 2 mM MgCl2; Mn2+, 2 mM MnCl2, pH 7.5; E5, 2 mM EDTA, no cations, pH 5 (to measure DNase II activity). (B) Expression of endonucleases in WT, EndoG KO, and DNase I KO cells measured using real-time reverse transcriptase polymerase chain reaction. DNase I expression is wiped out almost completely, while EndoG KO is partially inhibited because these cells were isolated from heterozygous animals (n = 4, *p < 0.001). DNase I, deoxyribonuclease I; WT, wild-type; KO, knockout; EndoG, endonuclease G.
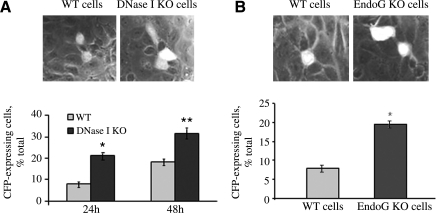
Inactivated DNase I and reduced EndoG contribute to DNA transfection
The inactivation of DNase I and/or reduced activity of EndoG may affect the transfection of DNA into PTE cells. To test this possibility, we compared transfection efficiencies of WT and KO cells by introducing into these cells the pECFP plasmid that encodes the CFP. Figure 3A shows that more CFP entered in DNase I KO cells, indicating that the rate of transfection is significantly higher relative to WT cells. The transfection of EndoG KO cells was also significantly more efficient than that of WT cells (Fig. 3B). These data confirm that the presence of endonucleases reduces the efficiency of DNA transfection.
FIG. 3.
Efficiency of plasmid transfection of PTE cells with active or inactive endonucleases. (A) Expression of CFP after pECFP-N1 plasmid transfection in DNase I KO PTE cells is higher than in WT cells. KO versus WT (21 ± 5% vs. 8 ± 5% transfected cells, n = 3, *p < 0.013; 32 ± 6% vs. 18 ± 5% transfected cells, n = 3, **p < 0.025). Two time-points have been used, 24 h and 48 h incubation after transfection. The second time-point shows higher transfection efficiency and has been chosen for experiment B, where EndoG WT and KO cells have been transfected with the same plasmid. (B) Expression of CFP 48 h after pECFP plasmid transfection in EndoG KO PTE cells is higher than in WT cells (19 ± 5% vs. 8 ± 5% CFP-positive cells, n = 6, *p < 0.001). CFP, cyan fluorescent protein.
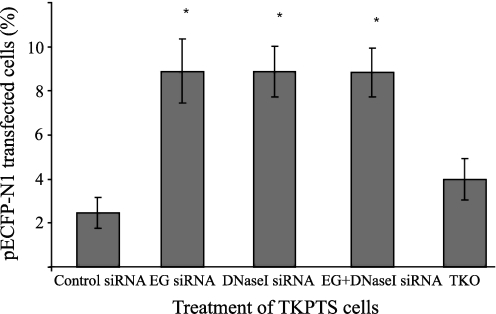
siRNA transfection
KO mice and cells are valuable models, but the results obtained with them are often difficult to interpret, as the long-term effect of an inactive protein may influence the expression of other proteins. To confirm that the inactivation of DNase I or EndoG provides a beneficial effect on plasmid transfection, we have used siRNA to silence these endonucleases. These experiments proved that silencing of either endonuclease strongly increases the transfection efficiency of tubular epithelial cells.
TKPTS cells have been treated with DNase I siRNA, EndoG siRNA, or both and then transfected with pECFP-N1 (cyan) plasmid. The EndoG siRNA target sequence consisted of AAAUGCCUGGAACAACCUUGA. The DNase I siRNA target sequence corresponded to GACATCGCTGTTATCCAA (Dharmacon). Cells treated with DNase I siRNA, with EndoG siRNA or with DNase I + EndoG siRNA, exhibited significantly (*p = 0.001) higher efficiency of transfection (≈9%) with cyan plasmid, than the siRNA control (≈2.5%) or the transfection reagent (TKO) control (≈4%) (Fig. 4). These experiments proved that the silencing of either endonuclease strongly increases transfection efficiency of TKPTS cells.
FIG. 4.
Efficiency of pECFP-N1 plasmid transfection in TKPTS cells after EG siRNA, DNase I siRNA, and EG+DNase I siRNA treatment. TKPTS cells were treated with EndoG, DNase I, and EndoG+DNase I siRNA using TransIT-TKO transfection reagent (TKO), controlled with siCONTROL Non-Targeting siRNA (control siRNA) and transfected with pECFP-N1 cyan plasmid. SiRNA-silenced cells (EG, DNase I and both) show significantly higher plasmid transfection efficiency (8.8% vs. 2.4% of pECFP-N1 transfected cells, *p = 0.001). siRNA, small interfering RNA.
Inactivation of EndoG promotes RNA transfection
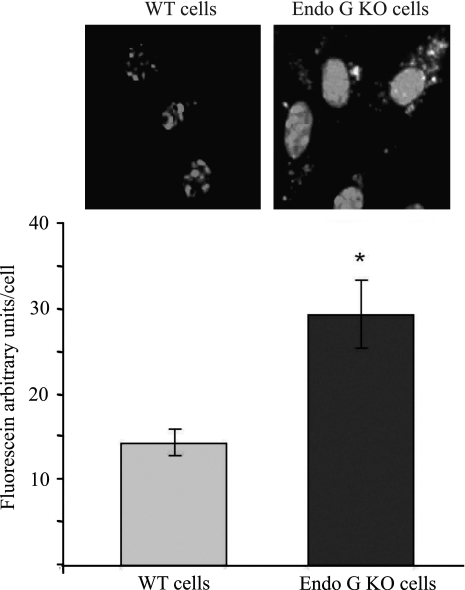
As opposed to DNase I, EndoG degrades both DNA and RNA (Kalinowska et al., 2005). Consequently, besides its anti-DNA activity, EndoG may also be an important factor in RNA transfection of cells. To test the validity of this assumption EndoG KO and WT cells were transfected with a fluorescein-labeled siRNA (Label IT RNAi Delivery Control shRNA; Mirus Bio). The Label IT RNAi Delivery Control consists of fluorescein-labeled double-stranded RNA duplexes that have the same length, charge, and configuration as standard siRNA used in RNAi studies. As suggested by the manufacturer, the sequence of this probe was not homologous to any known mammalian gene and was not known to affect any cellular events. In our experiment, EndoG KO cells had a two times faster rate of siRNA transfection than those of WT cells (Fig. 5). This observation suggests that EndoG is likely to play a dual role in host cell defense; it protects cells from both DNA and RNA invasion.
FIG. 5.
Efficiency of RNA transfection of PTE cells with active or inactive EndoG. Primary EndoG KO or WT mouse tubular epithelial cells were transfected with fluorescent siRNA as described in the Materials and Methods section. About 48 h later RNA transfection was detected using a fluorescent microscope. Blue color of DAPI was used to stain the nuclei. EndoG KO cells show significantly higher rate of siRNA transfection than WT cells (14 ± 2 vs. 29 ± 4 arbitrary fluorescence units per cell, n = 3, *p < 0.01).
Inactivation of extracellular DNase I has no effect on transfection
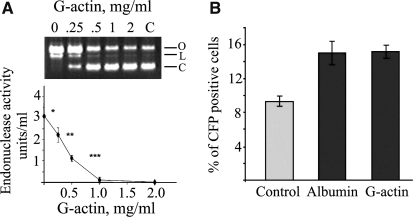
While EndoG is entirely an intracellular enzyme, DNase I is secreted (Lacks, 1981). To determine whether secreted extracellular DNase I influences the efficiency of transfection, it was inhibited by G-actin, a known specific and irreversible inhibitor of DNase I (Lacks, 1981). When choosing G-actin, it was also taken into consideration that G-actin is not toxic to cells and it does not enter cells and thus can be applied specifically to interact with and eliminate the activity of extracellular DNase I. TKPTS cells were exposed to G-actin in the concentration range between 0 and 2 mg/mL in the medium. Endonuclease activity was measured after G-actin treatment using the PIA (Fig. 6A). Complete inhibition of DNase I activity was found at 1 mg/mL G-actin concentration. In another experiment, TKPTS cells were treated either with 1 mg/mL G-actin or albumin as a control and then transfected with pECFP plasmid (Fig. 6B). There was no difference between G-actin and the control albumin treatment, suggesting that extracellular DNase I is unlikely to influence transfection and only intracellular DNase I seems to be involved in the anti-DNA host cell defense. Control cells were not treated, and albumin was used as a protein control. In the experiment there was no difference between the albumin control and G-actin treatment. This explains why extracellular DNase I inactivated by G-actin does not improve transfection.
FIG. 6.
Extracellular DNase I does not influence DNA transfection efficiency. (A) TKPTS cells were treated with different concentration of G-actin in culture medium, and endonuclease activity was measured with PIA. G-actin inhibited DNase I in culture medium (n = 4 per concentration point, *p = 0.018, **p = 0.0052, ***p = 0.006). (B) The efficiency of TKPTS cells transfection with pECFP plasmid in the presence of 1 mg/mL G-actin was not different from the one measured in the presence of 1 mg/mL albumin (15 ± 5 vs. 15 ± 8% of CFP-positive cells, n = 6).
Discussion
We have attempted to protect the fDNA to be introduced in host cells by reducing the DNase I and EndoG activities. This seemingly obvious approach has not been used earlier due to the lack of specific inhibitors and to insufficient data regarding the potential of these enzymes as host cell defense enzymes. There is only one report showing the role of another endonuclease, DNase gamma (DNase 1L3) in host cell defense (Wilber et al., 2002). Our earlier data showed that DNase I is the major endonuclease in tubular epithelial cells and EndoG is the second major degradative enzyme (Yin et al., 2007). Plasmid as fDNA was introduced in host cells, but is subject to endonucleolytic attacks both outside and inside the cells. Lipofectamine increased the efficiency of transfection, but did not protect plasmid DNA from destruction by endonucleases. Cellular extracts turned out to have a much higher endonuclease activity than the nuclease excreted by the cells to the culture medium.
It is to be mentioned that in our experiments, fDNA was attacked by endonucleases, which are compartmentalized and thus are not freely available in the cytoplasm: EndoG is mainly localized in mitochondria, while DNase I is associated with endoplasmic reticulum. Therefore, either there are enough preexisting free cytoplasmic EndoG and DNase I activities to destroy the incoming DNA, or fDNA induces the release of these endonucleases from cellular compartments. Such a release could be part of the host cell apoptosis induced by fDNA. That the introduction of fDNA causes apoptosis has been described by Shimokawa et al. (2000).
We have also observed that the total specific endonuclease activity measured by the PIA was significantly higher in primary cells than in immortalized TKPTS cells. This could explain why primary cells are more resistant to transfection than immortalized cells. The resistance of primary cells to transfection was also reported by others (Stacey et al., 1993; Welter et al., 2004; Zhong et al., 2005). Our strategy to introduce foreign genes in cells is based on the inactivation of the two major endonucleases to contribute to the survival of the internalized plasmid DNA and siRNA. Both DNase I KO and EndoG KO cells had significantly higher rate of transfection compared to WT cells. Moreover, in EndoG KO cells the rate of fluorescent siRNA transfection was also significantly higher than in WT cells. This suggests that EndoG has a dual role in host cell defense: besides degrading fDNA, it protects cells from both DNA and RNA uptake. Inhibition of extracellular DNase I by G-actin did not affect the efficiency of DNA transfection, indicating that only intracellular DNase I is the primary molecular player in the anti-DNA host cell defense. Although there is no doubt that endonucleases play an important if not decisive role in the degradation of foreign and self-DNA (e.g., apoptosis and necrosis), the question arises whether cellular factors other than endo- and exonucleases are involved in the degradation of nucleic acids, the knocking out of which could contribute to the efficiency of transfection. In a recent study, we have confirmed that in the kidney, DNase I is necessary for EndoG induction (Yin et al., 2007). It is thus reasonable to think that there might be some cooperation between these two endonucleases during protection of cells against invasion by nucleic acids. The experiments carried out in the murine renal system (WT and KO) suggested future studies, which will be directed toward (a) selection of specific drugs that inhibit DNase I and EndoG as a new strategy for efficient gene therapy in kidney cells, (b) assessment of the function of these endonucleases in apoptosis in the degradation of fDNA, and (c) determination of whether other factors could modulate DNase I or EndoG activities to increase transfection/transduction efficiency through the partial downregulation of endonucleases. Another viable approach could be the suppression of fDNA-induced apoptosis and, thus, indirect suppression of other anti-fDNA endonuclease activities that may contribute to the DNA destruction. Although fDNA induced apoptosis and DNases in other studies (Stacey et al., 1993; Nur et al., 2003), the inhibition of apoptosis has not been previously used for improving the delivery of fDNA.
The delivery of fDNA into DNase I–deficient cells raises the question of what the physiological consequences of impaired clearance of DNase I could be. According to an earlier view DNase I may protect against development of systemic lupus erythematosus (SLE), suggesting that DNase I treatment may be helpful in preventing onset of the disease. Direct evidence was provided that deficient DNase I function may lead to lupus erythematosus (Napirei et al., 2000). In addition, KO mice lacking DNase I develop antichromatin autoantibodies and glomerulonephritis. These observations indicate that DNase I may protect against SLE by digesting extracellular DNA (Napirei et al., 2006). Although DNase I deficiency was shown to produce a lupus-like syndrome in mice and lupus patients often have low levels of DNase I, there is no evidence that any human SLE is caused by DNase I deficiency alone (Walport, 2000). Consequently, supplementing the enzyme is unlikely to offer SLE patients more than partial symptom relief, as DNase I deficiency is only one of the factors in lupus disorder (Napirei et al., 2000).
In conclusion, our data reflect the importance of DNase I and EndoG in host cell defense against gene delivery in PTE cells. Future studies will shed light on the cooperative nature of these two endonucleases in destroying fDNA. Temporary and targeted inhibition of these endonucleases may provide new modalities to improve DNA stability during gene delivery.
Acknowledgments
This research was supported in part by VA Merit Review grant and a grant from the National Institutes of Health (A.G.B.). We gratefully acknowledge Anna Stewart for excellent technical assistance.
Disclosure Statement
No competing financial interests exist.
References
- Basnakian A.G. Apostolov E.O. Yin X. Abiri S.O. Stewart A.G. Singh A.B. Shah S.V. Endonuclease G promotes cell death of non-invasive human breast cancer cells. Exp Cell Res. 2006;312:4139–4149. doi: 10.1016/j.yexcr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basnakian A.G. Apostolov E.O. Yin X. Napirei M. Mannherz H.G. Shah S.V. Cisplatin nephrotoxicity is mediated by deoxyribonuclease I. J Am Soc Nephrol. 2005;16:697–702. doi: 10.1681/ASN.2004060494. [DOI] [PubMed] [Google Scholar]
- Basnakian A.G. Ueda N. Kaushal G.P. Mikhailova M.V. Shah S.V. DNase I-like endonuclease in rat kidney cortex that is activated during ischemia/reperfusion injury. J Am Soc Nephrol. 2002;13:1000–1007. doi: 10.1681/ASN.V1341000. [DOI] [PubMed] [Google Scholar]
- Bergsmedh A. Ehnfors J. Kawane K. Motoyama N. Nagata S. Holmgren L. DNase II and the Chk2 DNA damage pathway form a genetic barrier blocking replication of horizontally transferred DNA. Mol Cancer Res. 2006;4:187–195. doi: 10.1158/1541-7786.MCR-05-0262. [DOI] [PubMed] [Google Scholar]
- Chu D. Rowe J. Lee H.C. Evaluation of the current models for the evolution of bacterial DNA uptake signal sequences. J Theor Biol. 2006;238:157–166. doi: 10.1016/j.jtbi.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Djurovic S. Iversen N. Jeansson S. Hoover F. Christensen G. Comparison of nonviral transfection and adeno-associated viral transduction on cardiomyocytes. Mol Biotechnol. 2004;28:21–32. doi: 10.1385/MB:28:1:21. [DOI] [PubMed] [Google Scholar]
- Ernest S. Bello-Reuss E. Expression and function of P-glycoprotein in a mouse kidney cell line. Am J Physiol. 1995;269:C323–C333. doi: 10.1152/ajpcell.1995.269.2.C323. [DOI] [PubMed] [Google Scholar]
- Freitas S.S. Azzoni A.R. Santos J.A. Monteiro G.A. Prazeres D.M. On the stability of plasmid DNA vectors during cell culture and purification. Mol Biotechnol. 2007;36:151–158. doi: 10.1007/s12033-007-0028-y. [DOI] [PubMed] [Google Scholar]
- Glasspool-Malone J. Steenland P.R. McDonald R.J. Sanchez R.A. Watts T.L. Zabner J. Malone R.W. DNA transfection of macaque and murine respiratory tissue is greatly enhanced by use of a nuclease inhibitor. J Gene Med. 2002;4:323–332. doi: 10.1002/jgm.259. [DOI] [PubMed] [Google Scholar]
- Huang K.J. Ku C.C. Lehman I.R. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci USA. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S. Kawasaki N. Isolation and characterization of the Schizosaccharomyces pombe cDNA encoding the mitochondrial endonuclease(1) Biochim Biophys Acta. 2001;1519:111–116. doi: 10.1016/s0167-4781(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Irvine R.A. Adachi N. Shibata D.K. Cassell G.D. Yu K. Karanjawala Z.E. Hsieh C.L. Lieber M.R. Generation and characterization of endonuclease G null mice. Mol Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowska M. Garncarz W. Pietrowska M. Garrard W.T. Widlak P. Regulation of the human apoptotic DNase/RNase endonuclease G: involvement of Hsp70 and ATP. Apoptosis. 2005;10:821–830. doi: 10.1007/s10495-005-0410-9. [DOI] [PubMed] [Google Scholar]
- Lacks S.A. Deoxyribonuclease I in mammalian tissues. Specificity of inhibition by actin. J Biol Chem. 1981;256:2644–2648. [PubMed] [Google Scholar]
- Li L.H. Sen A. Murphy S.P. Jahreis G.P. Fuji H. Hui S.W. Apoptosis induced by DNA uptake limits transfection efficiency. Exp Cell Res. 1999;253:541–550. doi: 10.1006/excr.1999.4666. [DOI] [PubMed] [Google Scholar]
- Metifiot M. Faure A. Guyonnet-Duperat V. Bellecave P. Litvak S. Ventura M. Andreola M.L. Cellular uptake of ODNs in HIV-1 human-infected cells: a role for viral particles in DNA delivery? Oligonucleotides. 2007;17:151–165. doi: 10.1089/oli.2006.0061. [DOI] [PubMed] [Google Scholar]
- Napirei M. Gultekin A. Kloeckl T. Moroy T. Frostegard J. Mannherz H.G. Systemic lupus-erythematosus: deoxyribonuclease 1 in necrotic chromatin disposal. Int J Biochem Cell Biol. 2006;38:297–306. doi: 10.1016/j.biocel.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Napirei M. Karsunky H. Zevnik B. Stephan H. Mannherz H.G. Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- Nowak G. Price P.M. Schnellmann R.G. Lack of a functional p21WAF1/CIP1 gene accelerates caspase-independent apoptosis induced by cisplatin in renal cells. Am J Physiol Ren Physiol. 2003;285:F440–F450. doi: 10.1152/ajprenal.00233.2002. [DOI] [PubMed] [Google Scholar]
- Nur E.K.A. Li T.K. Zhang A. Qi H. Hars E.S. Liu L.F. Single-stranded DNA induces ataxia telangiectasia mutant (ATM)/p53-dependent DNA damage and apoptotic signals. J Biol Chem. 2003;278:12475–12481. doi: 10.1074/jbc.M212915200. [DOI] [PubMed] [Google Scholar]
- Peitsch M.C. Irmler M. French L.E. Tschopp J. Genomic organisation and expression of mouse deoxyribonuclease I. Biochem Biophys Res Commun. 1995;207:62–68. doi: 10.1006/bbrc.1995.1153. [DOI] [PubMed] [Google Scholar]
- Shimokawa T. Okumura K. Ra C. DNA induces apoptosis in electroporated human promonocytic cell line U937. Biochem Biophys Res Commun. 2000;270:94–99. doi: 10.1006/bbrc.2000.2388. [DOI] [PubMed] [Google Scholar]
- Stacey K.J. Ross I.L. Hume D.A. Electroporation and DNA-dependent cell death in murine macrophages. Immunol Cell Biol. 1993;71(Pt 2):75–85. doi: 10.1038/icb.1993.8. [DOI] [PubMed] [Google Scholar]
- Tanswell A.K. Staub O. Iles R. Belcastro R. Cabacungan J. Sedlackova L. Steer B. Wen Y. Hu J. O'Brodovich H. Liposome-mediated transfection of fetal lung epithelial cells: DNA degradation and enhanced superoxide toxicity. Am J Physiol. 1998;275:L452–L460. doi: 10.1152/ajplung.1998.275.3.L452. [DOI] [PubMed] [Google Scholar]
- Thomas K.R. Capecchi M.R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986;324:34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]
- Torchilin V.P. Recent approaches to intracellular delivery of drugs and DNA and organelle targeting. Annu Rev Biomed Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- Walport M.J. Lupus, DNase and defective disposal of cellular debris. Nat Genet. 2000;25:177–181. doi: 10.1038/75963. [DOI] [PubMed] [Google Scholar]
- Welter J.F. Solchaga L.A. Stewart M.C. High-efficiency nonviral transfection of primary chondrocytes. Methods Mol Med. 2004;100:129–146. doi: 10.1385/1-59259-810-2:129. [DOI] [PubMed] [Google Scholar]
- Widlak P. Li L.Y. Wang X. Garrard W.T. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J Biol Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- Wilber A. Lu M. Schneider M.C. Deoxyribonuclease I-like III is an inducible macrophage barrier to liposomal transfection. Mol Ther. 2002;6:35–42. doi: 10.1006/mthe.2002.0625. [DOI] [PubMed] [Google Scholar]
- Yan B. Wang H. Li F. Li C.Y. Regulation of mammalian horizontal gene transfer by apoptotic DNA fragmentation. Br J Cancer. 2006;95:1696–1700. doi: 10.1038/sj.bjc.6603484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X. Apostolov E.O. Shah S.V. Wang X. Bogdanov K.V. Buzder T. Stewart A.G. Basnakian A.G. Induction of renal endonuclease G by cisplatin is reduced in DNase I-deficient mice. J Am Soc Nephrol. 2007;18:2544–2553. doi: 10.1681/ASN.2006080896. [DOI] [PubMed] [Google Scholar]
- Zhang J. Dong M. Li L. Fan Y. Pathre P. Dong J. Lou D. Wells J.M. Olivares-Villagomez D. Van Kaer L. Wang X. Xu M. Endonuclease G is required for early embryogenesis and normal apoptosis in mice. Proc Natl Acad Sci USA. 2003;100:15782–15787. doi: 10.1073/pnas.2636393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z. Feijen J. Lok M.C. Hennink W.E. Christensen L.V. Yockman J.W. Kim Y.H. Kim S.W. Low molecular weight linear polyethylenimine-b-poly(ethylene glycol)-b-polyethylenimine triblock copolymers: synthesis, characterization, and in vitro gene transfer properties. Biomacromolecules. 2005;6:3440–3448. doi: 10.1021/bm050505n. [DOI] [PubMed] [Google Scholar]