Abstract
Apicomplexa is a phylum within the kingdom Protista that contains some of the most significant threats to public health. One of the members of this phylum, Toxoplasma gondii, is amenable to molecular genetic analyses allowing for the identification of factors critical for colonization and disease. A pathway found to be important for T. gondii pathogenesis is the Ran network of nuclear trafficking. Bioinformatics analysis of apicomplexan genomes shows that while Ran is well conserved, the key regulators of Ran—Regulator of Chromosome Condensation 1 and Ran GTPase activating protein—are either highly divergent or absent. Likewise, several import and export receptor molecules that are crucial for nuclear transport are either not present or have experienced genetic drift such that they are no longer recognizable by bioinformatics tools. In this minireview we describe the basics of nuclear trafficking and compare components within apicomplexans to defined systems in humans and yeast. A detailed analysis of the nuclear trafficking network in these eukaryotes is required to understand how this potentially unique cellular biological pathway contributes to host–parasite interactions.
Introduction
Apicomplexan parasites are a major health concern, causing disease to humans and animals worldwide. In addition to their medical relevance, apicomplexans should also be appreciated as divergent eukaryotes with unique cellular biology. Members of this phylum that are medically important to humans and of common research interest include Plasmodium spp., Cryptosporidium spp., and Toxoplasma gondii. Between 350 and 500 million people worldwide become infected by Plasmodium annually. Most of the over one million people who succumb to the Plasmodium infection each year are children in sub–Saharan Africa. Cryptosporidium causes severe diarrhea in children of developing countries and immunocompromised individuals worldwide. T. gondii is a very successful colonizer of humans, with an estimated one third of the world's human population infected (Tenter et al., 2000). While normally dormant for the duration of the host's lifetime, T. gondii can become an opportunistic pathogen that causes encephalitis in immunocompromised patients, and can be passed vertically to developing fetuses during gestation (Dubey, 1994; Dubey, 1998).
T. gondii, like other Apicomplexa, is a single-cell eukaryote with many unique organelles, including micronemes, rhoptries, dense granules, and the apicoplast. Along with these distinct organelles, T. gondii contains the typical organelles found in other eukaryotes such as the Golgi, endoplasmic reticulum, a single mitochondrion, and a nucleus. However, the typical organelles do not always contain standard components as defined by model eukaryotes. In particular, recent studies have identified a highly divergent ortholog of the Regulator of Chromosome Condensation 1 (RCC1) that is critical for parasite pathogenesis (Frankel et al., 2007) and functions as a guanine nucleotide exchange factor (GEF) for the GTPase Ran (Frankel and Knoll, 2008). These studies have opened questions about the sequence divergence and apparent absence of key components of nuclear transport in apicomplexans.
Here we review the basics of nucleocytoplasmic transport, highlighting the potential differences in transport machinery present in apicomplexan parasites. We have performed bioinformatics analyses and compiled a comprehensive table that includes the orthologs present or absent in the three human pathogens, T. gondii, Plasmodium falciparum, and Cryptosporidium parvum, compared to human nuclear trafficking components. The apparent absence of sequence similarity to many factors of nuclear transport requires an investigation to determine whether unique nucleocytoplasmic transport machinery exists in apicomplexans or if there are simply fewer components. If a simplified system does exist in apicomplexans, due to the increasing genetic and proteomics tools available for T. gondii, we propose the possible advantages of using T. gondii as a model system to study the complex mechanisms of nuclear transport.
The Master Switch, Ran
The first step of understanding nuclear transport is to identify the main protein driving it, Ran, a member of the Ras GTPase superfamily. All Ras family members are bound to GTP or GDP, serving as an “on/off ” switch. Structural changes occur in Ran depending on its nucleotide-bound state, which results in different substrate specificity. Ran has a high level of sequence identity between eukaryotic organisms, including the protozoa, representing its well-conserved function among species. Along with driving nuclear transport, there are two other described functions for Ran: reassembly of the nuclear envelope during mitosis and spindle formation (Di Fiore et al., 2004). The transport of cargo between the nucleus and cytosol is tightly controlled by Ran and its nucleotide-bound state. Using nonhydrolyzable analogs of GTP and GDP, as well as dominant-negative Ran mutants, it was determined that the GTP-bound form of Ran is the sole energy source for nucleocytoplasmic transport (Melchior et al., 1993; Carey et al., 1996; Palacios et al., 1996; Murphy et al., 1997; Richards et al., 1997). There is a clear asymmetry of the two bound nucleotide states of Ran, with Ran-GTP being primarily located in the nucleus and Ran-GDP in the cytoplasm. This differential localization of Ran requires the spatial separation of its two opposing regulators, RCC1 and Ran GTPase activating protein (RanGAP) (reviewed in Moore, 1998).
RCC1 is an essential nuclear protein bound to chromatin that serves as a GEF for Ran, replacing RanGDP with GTP (Fig. 1) (Bischoff and Postingl, 1991). RCC1 has been shown to bind to histones H2A and H2B, which enhances its GEF activity (Nemergut et al., 2001). RCC1 proteins contain seven tandem repeats of 50–60 amino acids termed RCC1 repeats. These tandem repeats form a seven blade propeller structure with one side binding to Ran for exchange activity and the other bound to chromatin (Azuma et al., 1996, 1999; Renault et al., 1998). Surprisingly, none of the sequenced protozoa contain typical RCC1 proteins with seven tandem RCC1 domains.
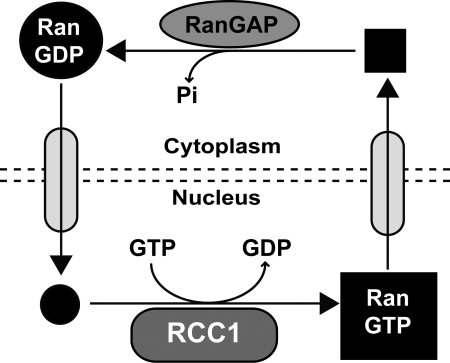
FIG. 1.
The Ran cycle. Ran cycles between the nucleus and cytoplasm bound to either GDP or GTP. RanGDP enters the nucleus and encounters RCC1. GDP is then replaced with GTP by RCC1, which creates a predominance of RanGTP. In the cytoplasm, RanGTP quickly interacts with RanGAP, and the GTP is hydrolyzed to GDP. The actions of RCC1 and RanGAP create a steep gradient of RanGDP in the cytosol and RanGTP in the nucleus (represented by a large circle or box, respectively).
Due to the low intrinsic GTPase activity of Ran alone, RanGAP is located in the cytosol and enhances the hydrolysis of RanGTP to RanGDP approximately 1000-fold. This GTPase activity is aided by Ran binding proteins 1 and 2 in mammalian systems; however, yeast encodes only Ran binding protein 1 (Bischoff and Gorlich, 1997). RanGAP possesses structural symmetry, consisting of 11 leucine-rich repeats that form a crescent shape (Hillig et al., 1999). Mammalian RanGAP functions as a dimer, whereas yeast forms do not. RanGAP also contains nuclear export signals (NES) to ensure that if any protein enters the nucleus, it is quickly exported. Similar to the divergence seen with RCC1, bioinformatics analyses show that none of the sequenced protozoa contain any protein with similarity to RanGAP. With RCC1 exclusively nuclear and RanGAP cytosolic, it creates an essential gradient of RanGDP in the cytoplasm and RanGTP in the nucleus. Perturbations to the Ran system, either by altering the cellular localization of RCC1 (Akhtar et al., 2001) or by introducing dominant negative Ran mutants (Carey et al., 1996), are lethal. It is perplexing how protozoa regulate such an essential process as nuclear trafficking with divergent Ran regulators.
The Ins and Outs of Nuclear Trafficking
In eukaryotes, the nuclear and cytoplasmic compartments are divided by the nuclear envelope. Cells use this division to regulate numerous functions, such as transcription and translation, by controlling the movement of molecules between the nucleus and the cytosol. Proteins smaller than approximately 40 kDa are able to passively diffuse through the nuclear pore complex (NPC), whereas larger proteins require active transport through the assistance of karyopherins, specific transport receptors that shuttle between the nucleus and cytosol (reviewed in Macara, 2001). Karyopherins can be subdivided into those that transport molecules into the nucleus (importins) and those that transport molecules out of the nucleus (exportins). Karyopherins are able to distinguish between the diverse proteome to target specific cargo molecules for transport. In yeast, it is estimated that over 2000 proteins are shuttled between the nucleus and the cytoplasm, yet the receptors and signals are known for only a few.
Characterized Import Components
Importin β is responsible for transporting a large number of cargo molecules into the nucleus. The importin β superfamily consists of 14 members in yeast and more than 20 in mammals. Most substrates are identified by their nuclear localization signal (NLS), which is encoded in either a mono- or a bi-partite sequence characterized as stretches of basic amino acids (Hodel et al., 2001). The identification of NLS sequences using bioinformatics methods can be difficult because nonfunctional NLSs can be buried within the tertiary structure, or functional NLS sequences can be missed if they are short or if protein folding brings together basic amino acids. Additionally, with growing knowledge of import and export receptors, new binding sites and sequences required for nuclear transport will likely be discovered. Imported proteins will most often bind to an importin α molecule, although direct binding to importin β does occur. Importin α serves as a middleman, to identify cargo destined for the nucleus and then interact with importin β, which is needed for binding and movement through the NPC (Fig. 2) (reviewed in Fried and Kutay, 2003). There are at least six identified importin α isoforms in humans, two in Schizosaccharomyces pombe, and only one in Saccharomyces cerevisiae (SRP1). In addition, transportin is another importin that does not require an adaptor (importin α) to serve as a receptor for nuclear import. Transportin recognizes a 38-residue signal termed the M9 domain, which does not contain basic amino acids and is best characterized for hnRNPA1 (Bogerd et al., 1999).
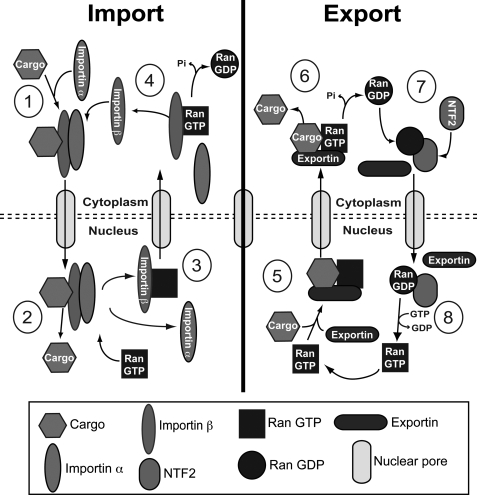
FIG. 2.
Nucleocytoplasmic transport. For import of molecules, cytoplasmic cargo is identified by importin α, which then binds to importin β (1). This ternary complex translocates through the nuclear membrane and into the nucleus. Once there, RanGTP binds to importin β and causes a dissociation of the complex, which releases cargo to the nucleus (2). Import receptors are then recycled back to the nucleus (3) through binding of RanGTP and export to the cytosol. RanGTP is then hydrolyzed to the GDP-bound state and causes the release of the import receptors (4) and the cycle starts over again. Export of cargo undergoes a similar mechanism. Exported molecules will bind to the export receptor with RanGTP and exit the nucleus (5). Next RanGTP is hydrolyzed to cause release of cargo into the cytoplasm (6). NTF2 specifically identifies RanGDP and returns it to the nucleus (7) for RCC1 to then exchange it to RanGTP (8).
Once the cargo–receptor complex makes its way into the nucleus, it encounters Ran bound to GTP. RanGTP binds to importin β, which causes a conformational change and subsequent dissociation of the complex that releases the nuclear cargo (reviewed in Cook et al., 2007). Importin β, coupled to RanGTP, shuttles back to the cytoplasm where it quickly encounters RanGAP. This leads to the hydrolysis of RanGTP to GDP to release Ran from importin β. For cargo requiring importin α, a specific transport receptor termed CAS binds with a second RanGTP, and then the ternary complex shuttles back to the cytosol to interact with RanGAP and start the cycle over. RanGDP in the cytosol is transferred back to the nucleus by its exclusive transporter, NTF2, to be exchanged to its GTP-bound state by RCC1 (Ribbeck et al., 1998). The energy required to fulfill a complete cycle is the hydrolysis of GTP on Ran.
Characterized Export Components
Protein export from the nucleus takes a similar route as import. The most prominent exportin is CRM1, named Xpo1p in yeast. It recognizes proteins with a short leucine-rich region, which acts as a NES. To exit, cargo forms a ternary complex with an exportin receptor and RanGTP. This complex traverses the NPC to the cytosol where RanGAP induces hydrolyses of GTP on Ran. The conformational change induced by the nucleotide change causes the exported cargo to be released. Empty exportin and RanGDP are then recycled back to the nucleus. There are additional cofactors that aid in either the binding of cargo to CRM1 or the release of cargo from its receptor. Examples include RanBP3, Nxt1, RanBP1, and, possibly, eIF-5A. RanBP3 binds directly to CRM1 and enhances its affinity for RanGTP- and NES-containing cargo (Lindsay et al., 2001). Nxt1 binds to RanGTP to aid in cargo–receptor complex dissociation at the NPC (Black et al., 2001).
In addition to CRM1, there are at least six other export receptors in humans. The yeast exportin Msn5 (closest ortholog is exportin 5 in humans) is interesting as it also serves as an importin for at least one cargo (RPA) (Yoshida and Blobel, 2001). Exportin 6 is responsible for the export of profilin–actin complexes (Stuven et al., 2003), and exportin 7 performs general export using a CRM1-independent export signal (Mingot et al., 2004). Exportin-t (called Los1p in yeast) is responsible for tRNA export (Kutay et al., 1998), while TAP (called Mex67 in yeast) performs mRNA export (Segref et al., 1997). There are likely other import and export receptors yet to be elucidated.
Components of Nuclear Trafficking in Apicomplexans
No predicted proteins in any sequenced apicomplexan encode seven tandem RCC1 repeats and are predicted to be nuclear. MAL7P1.38, the P. falciparum ortholog with the highest E-value against human RCC1, is 688 amino acids, contains only three repeats, and is predicted to be nuclear (Table 1). The reported RCC1 ortholog in P. falciparum is 1327 amino acids, much larger than the conventional RCC1 proteins of ∼420 amino acids (Ji et al., 1998). This P. falciparum protein contains six RCC1 repeats and is localized to the nucleus, but has not yet been confirmed to be a functional GEF. These RCC1 repeat–containing proteins may have redundant GEF activity or may be active during different stages of the lifecycle. T. gondii encodes 13 proteins that contain RCC1 repeats. The T. gondii RCC1 ortholog (TgRCC1) has the highest similarity to human RCC1 and localizes to the parasite nucleus (Frankel et al., 2007). The C. parvum ortholog with the highest sequence identity is cgd6_2770. This 487 amino acid protein has seven tandem RCC1 repeats, but no obvious NLS to direct the protein to the nucleus. In S. cerevisiae, the Ats1p protein contains four RCC1 repeats and is localized to the cytosol, yet this protein does not appear to function as a GEF and is not an essential protein (Kirkpatrick and Solomon, 1994). It is possible that the apicomplexan parasites encode a protein similar to Ats1p, whose function(s) is yet to be determined.
Table 1.
Comparison of Nuclear Transport Components
| Protein name | Accession no. | S. cerevisiae | T. gondii (E-value) | P. falciparum (E-value) | C. parvum (E-value) |
|---|---|---|---|---|---|
| RCC1 | BAA00469 | Prp20 | 31.m00904 (7.5e-27) | MAL7P1.38 (9.4e-24) | cgd6_2770 (2.1e-23) |
| RanGAP | CAA57714 | RNA1 | None | None | None |
| Ran | P62826 | GSP1 | 50.m00042 (4.8e-84) | PF11_0183 (4.6e-79) | cgd7_220 (3.0e-84) |
| Importin alpha | NP_002255 | Srp1p | 52.m00001 (5.9e-127) | PF08_0087 (1.2e-110) | cgd8_3260 (8.4e-116) |
| Importin beta | NP_002256 | Kap95 | 83.m01307 (e-100) | PF08_0069 (4.1e-77) | cgd7_3030 (3.1e-106) |
| Transportin 1 | NP_002261 | Kap104 | 35.m00891 (e-118) | PFF1345w (3.9e-88) | cgd8_3440 (5.3e-98) |
| Transportin 2 | NP_038461 | Kap104 | 35.m00891 (e-121) | PFF1345w (4.1e-83) | cgd8_3440 (1.2e-88) |
| Transportin SR1 | AAD38537 | Mtr10 | 57.m01705 (6.0e-09) | None | None |
| Transportin SR2 | NP_036602 | Kap122p | 57.m01705 (7.0e-09) | None | None |
| Importin 4 | NP_078934 | Yrb4p | 44.m02667 (2.0e-24) | PFE1195w (4.0e-07) | cgd7_3270 (2.1e-05) |
| Importin 5/RanBP5 | CAA70103 | Pse1p | 44.m02525 (4.0e-83) | PFE1195w (3.3e-98) | cgd7_3270 (1.8e-97) |
| Importin 7 | NP_006382 | Nmd5p | 42.m03288 (9.0e-32) | MAL7P1.202 (2.0e-17) | cgd7_790 (4.1e-07) |
| Importin 8 | NP_006381 | Kap119p | 42.m03288 (2.0e-25) | MAL7P1.202 (3.4e-16) | cgd3_1790 (3.4e-06) |
| Importin 9 | NP_060555 | Kap114p | 44.m02525 (0.003) | None | None |
| Importin 11/RanBP11 | AAF21936 | Lph2p | 52.m01606 (4.0e-04) | PF14_0304 (5.4e-08) | cgd3_1790 (5.4e-05) |
| Importin 13 | NP_055467 | Pdr6p | 50.m03311 (5.0e-05) | PFC0135c (0.0045) | None |
| NTF2 | NP_005787 | Ntf2 | 44.m00054 (7.0e-20) | PF14_0122 (8.0e-17) | cgd4_590 (1.4e-23) |
| Nxt1 | AAD54942 | Nxt1 | 44.m00054 (8.6e-07) | PF14_0122 (1.8e-06) | cgd4_590 (1.4e-08) |
| CRM1/Exportin 1 | NP_003391 | Xpo1p | 50.m03311 (6.0e-189) | PFC0135c (1.8e-123) | cgd3_3060 (7.4e-117) |
| TAP | NP_006353 | Mex67p | None | None | None |
| CAS | NP_001307 | Cse1p | 52.m01606 (6.0e-88) | PFI1590c (3.0e-33) | cgd3_1790 (3.4e-35) |
| Exportin-t | NP_009166 | Los1p | 50.m03311 (3.0e-06) | MAL13P1.83 (1.6e-23) | cgd7_1780 (2.0e-16) |
| Exportin 4 | NP_071904 | None | 41.m00005 (1.6e-07) | PFI0490c (2.6e-08) | cgd7_1670 (6.2e-08) |
| Exportin 5 | NP_065801 | Msn5p | 50.m03311 (7.0e-12) | PFC0135c (9.1e-13) | cgd3_3060 (2.7e-08) |
| Exportin 6 | NP_055986 | None | 50.m03311 (3.7e-10) | PFC0135c (1.8e-07) | cgd3_3060 (7.2e-05) |
| Exportin 7 | NP_055839 | None | 41.m00005 (e-161) | PFI0490c (4.9e-112) | cgd7_1670 (1.7e-134) |
| RanBP1 | CAG30442 | Yrb1p | 83.m01206 (7.0e-24) | PFD0950w (8.1e-24) | cgd5_3950 (4.2e-31) |
| RanBP3 | Q9H6Z4 | Ranbp3 | None | PFD0950w (0.025) | cgd5_3950 (0.0042) |
| RanBP6 | AAC14260 | Kap121p | 44.m02525 (6.0e-76) | PFE1195w (8.3e-89) | cgd7_3270 (7.6e-73) |
| RanBP17 | NP_075048 | None | 41.m00005 (e-121) | PFI0490c (5.3e-105) | cgd7_1670 (2.7e-131) |
Human nuclear transport constituents (accession numbers listed in the second column) were used to BLAST search the annotated genomes of T. gondii, P. falciparum, and C. parvum. The S. cerevisiae orthologs are listed in the third column. The annotated parasite protein is listed with the E-value given in parentheses. To ascertain that the absence of protein orthologs was not due to inaccurate protein annotations, we used tblastn and blastn searches of the parasite genomes with the mRNA nucleotide sequence of human genes as the query. These searches did not uncover any additional orthologs. For E-values in italics, the annotated protein appeared in the BLAST search but has a higher E-value to another protein.
At 1155 amino acids, TgRCC1 is unusually large and has been proven to be involved in parasite pathogenesis (Frankel et al., 2007). It was shown that TgRCC1 mutant parasites were growth defective in serum-starved host cells, and thus it was speculated that the rate of nuclear trafficking is important for eukaryotic pathogens under the stress of nutrient limitation in vivo (Frankel et al., 2007). Even though TgRCC1 contains only five RCC1 domains, it is able to complement a temperature-sensitive mammalian RCC1 mutant cell line (TsBN2) (Frankel and Knoll, 2008). The ability of TgRCC1 to complement the TsBN2 cell line creates a potential model for the functional analysis of parasite RCC1 proteins. TgRCC1 contains a zinc finger motif that is found in the nucleoporins Nup153 and Nup358 (RanBP2). This zinc-finger motif from RanBP2 has been shown to interact directly with Ran (Yaseen and Blobel, 1999). While we speculated that the zinc-finger aids in Ran binding in the absence of the typical seven RCC1 repeats, alteration of the TgRCC1 zinc-finger motif did not ablate function (Frankel and Knoll, 2008). C. parvum also contains cgd4_1350, a 958 amino acid protein that is predicted to be nuclear with four RCC1 repeats and the same zinc-finger motif present in TgRCC1. It is likely that a function for this conserved zinc-finger motif will be uncovered with further analysis of these RCC1 orthologs.
Sequence similarity searches have been unable to identify a RanGAP ortholog in any protozoan. Keyword searches among annotated proteins in the T. gondii genome database identified one candidate, but sequence analysis shows a strong similarity to RanBP1. Perhaps the RanGAP function in apicomplexans is performed by a single protein with multiple cellular responsibilities (i.e., a fusion of Ran binding protein 1 and RanGAP). It is also possible that a completely unique protein possesses the RanGAP function. Another possibility is that T. gondii Ran has a higher intrinsic GTPase activity than other homologs, removing the need for RanGAP. Higher intrinsic GTPase activity would require additional cofactors in the nucleus to prevent this activity, and would reveal a novel Ran network. We have observed that TgRan displays similar kinetics of GTPase activity to human Ran (Frankel and Knoll, 2008), suggesting the presence of an unidentified TgRanGAP. It will be critical to determine what protein is performing the RanGAP function.
Apicomplexans encode Ran and NTF2 orthologs to recycle Ran from the cytosol to the nucleus. While T. gondii Ran shares 71% amino acid identity to human, there are clear differences in Ran properties from described systems. For example, TgRan is found throughout the parasite cell, as opposed to the predominantly nuclear localization in all other eukaryotes examined (Frankel and Knoll, 2008). In addition, T. gondii is able to stably express dominant negative Ran mutants that are lethal in other systems. Purified human Ran T24N, which has been shown to have essentially no affinity for GTP and bound to either GDP or no nucleotide at all, binds to RCC1 and inactivates it, thus acting as a dominant negative (Kornbluth et al., 1994). Ran G19V is a gain-of-function lethal mutant because Ran becomes insensitive to RanGAP activation and is locked in the GTP-bound state (Lounsbury et al., 1996). Expression of TgRan T24N recapitulated phenotypes observed for TgRCC1 mutant parasites, specifically attenuation in mice and a growth defect in serum-starved host cells (Frankel and Knoll, 2008). An elucidation of the Ran network in T. gondii will help us to understand why these mutant forms of Ran can be expressed and which enzyme is capable of RanGAP activity.
Import Components in Apicomplexans
An importin α ortholog has previously been reported in T. gondii (Bhatti and Sullivan, 2005). While BLAST searches of the T. gondii database with both human and the T. gondii importin α identified multiple proteins with similarity, only the annotated protein identified by Bhatti et al. contains the characteristic importin β binding domain (Bhatti and Sullivan, 2005). This importin β binding domain is sufficient and essential for binding to importin β. It will be important to determine if there are more importin α family members in T. gondii. Importin β is conserved in all sequenced apicomplexans, with relatively higher similarity (Table 1). While orthologs of both importin α and β have been reported for P. falciparum, functional analysis has not yet been performed (Mohmmed et al., 2003).
In addition to importin β, apicomplexans contain orthologs of importin β family members, including transportins and importins 4, 5, and 7. However, we were unable to identify orthologs for either human or yeast importin 4b, 8, 9, 11, and 13. Interestingly, while T. gondii encodes both importin 4 and 5 orthologs, P. falciparum and C. parvum each encode only one, which has a higher sequence identity to importin 5 (the E-values compared to either importin 4 or 5 are shown on Table 1). While humans contain 20 members of the importin β family and S. cerevisiae contains 13, T. gondii appears to contain only 5. If this is true, it will be the most minimal system for transport reported. It will be interesting to understand how T. gondii is able to perform the tremendous task of shuttling proteins between the nucleus and cytoplasm using so few receptors. Without the redundancy and complexity in other systems such as yeast or Xenopus, the apicomplexans may serve as an excellent model to determine the mechanism of protein import into the nucleus. No studies have looked experimentally at the number of karyopherin members in protozoa. Parasites may contain unique means of receptor-mediated transport or simply encode a small number of proteins to transport a large number of cargos.
Export Components in Apicomplexans
The majority of nuclear export is supported by CRM1. All of the apicomplexans contain a CRM1 ortholog with high sequence conservation (Table 1). CAS, which is required for cycling importin α back to the nucleus, and exportin 7 (RanBP16) are also well conserved. The NES for exportin 7 does not use the typical leucine-rich region, but instead includes folded motifs with basic residues (Mingot et al., 2004). Exportin 7 has broad substrate specificity, which may be used widely in apicomplexans in the absence of other exportins. There are no orthologs for other reported export receptors such as exportin 4, 5 (discussed below), and 6 (RanBP20). In addition, there are no identifiable orthologs for the CRM1 cofactors RanBP3 and Nxt1 in apicomplexan parasites. While P. falciparum and C. parvum contain orthologs of eIF-5A, it appears that T. gondii does not. The karyopherin Msn5, which functions in both import and export in yeast, is not conserved in apicomplexans.
Apicomplexans appear to be missing the receptor for mRNA transport, TAP (Mex67 in yeast). Interestingly, P. falciparum encodes a protein with weak similarity to the yeast Mex67 (annotated protein PF14_0305; e-value of 4.4e−05). How do apicomplexans transport mRNAs? Cells may survive by passive diffusion of the RNA molecules, but given the large size of many of the T. gondii mRNAs, it is unlikely that these RNAs merely diffuse into the cytosol. Perhaps the T. gondii export receptors that share similarity to human and yeast equivalents share additional functions beyond what is described in other systems. In Trypanosoma brucei and T. cruzi, CRM1 functions to also export mRNAs (Cuevas et al., 2005). Thus, CRM1 in apicomplexans may be shuttling RNAs out to the cytosol in addition to other cargo.
T. gondii does not encode a protein with similarity to exportin-t, which is responsible for export of tRNAs, while both P. falciparum and C. parvum contain orthologs. Studies using Xenopus oocysts show that inhibition of exportin-t reduces the trafficking of tRNAs by 80% (Arts et al., 1998). This study illustrated that there may be additional mechanisms responsible for the transport of tRNAs. Exportin 5 is able to bind to tRNAs in a Ran-dependant fashion to transport them (Bohnsack et al., 2002); however, apicomplexans do not contain an exportin 5 ortholog. T. gondii may possess an unknown export receptor for tRNA, as the annotation of the T. gondii genome has not been confirmed for many proteins. Unraveling the mechanisms of how these parasites are able to perform all of these critical functions with proteins of unknown sequence similarity will undoubtedly bring forth new knowledge to the fields of parasitology and cell biology. These studies may reveal novel machinery and transport mechanisms.
Conclusions and Future Directions
The study of nuclear transport in apicomplexans is an understudied area of research; however, the availability of tools for the manipulation of apicomplexans makes them excellent models (Kim and Weiss, 2004). Multiple selectable markers are available to perform targeted disruption of various genes of interest. Fluorescent tags (e.g., GFP) have been used for photobleaching to monitor the movement of parasite proteins in viable cells (Hu et al., 2002; Klonis et al., 2002). The tet-inducible system allows induction or repression of genes of interest (Meissner et al., 2001, 2002). For instance, the tet-inducible expression system was used to evaluate the key role of a microneme protein (MIC2) during the invasion process of T. gondii (Huynh and Carruthers, 2006). It was recently reported that fusion of a ligand-controlled destabilization domain (ddFKBP) to a protein of interest would allow for rapid degradation (Herm-Gotz et al., 2007). Importantly, this action is as quick as 20 min and reversible. These tools will enable researchers to replace native copies of components of nuclear trafficking with inducible genes to determine their role in transport and their kinetics of movement, as well as to identify additional transport machinery.
Sequence analysis with human and yeast components of nuclear trafficking to the sequenced genomes of T. gondii, P. falciparum, and C. parvum has revealed several unexpected findings. Apicomplexans appear to be either missing several key nuclear transport proteins compared to other model organisms, or significant genetic drift has occurred in these nuclear transport proteins such that they are no longer recognized by BLAST searches. It is possible that these parasites encode minimal transport components that perform multiple, diverse functions. These unique parasites may also have mechanisms for nucleocytoplasmic transport that have not been previously described. In particular, questions arise about how these parasites transport RNA molecules out to the cytoplasm, and what protein functions as a RanGAP. For such a critical process as nuclear transport, late branching eukaryotes have multiple transport mechanisms, which apicomplexans appear to lack. This dearth of functional redundancy may enable a simplified analysis of this complex process. While the study of nuclear trafficking mechanisms in apicomplexans will answer many important cellular biological questions, these studies can potentially unveil new drug targets in these medically relevant pathogens.
Acknowledgments
We thank Angela Pollard for her editorial assistance. The work studying how nuclear trafficking defects affect virulence is supported by NIH Award A1054603.
Disclosure Statement
No competing financial interests exist.
References
- Akhtar N. Hagan H. Lopilato J.E. Corbett A.H. Functional analysis of the yeast Ran exchange factor Prp20p: in vivo evidence for the RanGTP gradient model. Mol Genet Genomics. 2001;265:851–864. doi: 10.1007/s004380100480. [DOI] [PubMed] [Google Scholar]
- Arts G.J. Kuersten S. Romby P. Ehresmann B. Mattaj I.W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y. Renault L. Garcia-Ranea J.A. Valencia A. Nishimoto T. Wittinghofer A. Model of the ran-RCC1 interaction using biochemical and docking experiments. J Mol Biol. 1999;289:1119–1130. doi: 10.1006/jmbi.1999.2820. [DOI] [PubMed] [Google Scholar]
- Azuma Y. Seino H. Seki T. Uzawa S. Klebe C. Ohba T. Wittinghofer A. Hayashi N. Nishimoto T. Conserved histidine residues of RCC1 are essential for nucleotide exchange on Ran. J Biochem (Tokyo) 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- Bhatti M.M. Sullivan W.J., Jr Histone acetylase GCN5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J Biol Chem. 2005;280:5902–5908. doi: 10.1074/jbc.M410656200. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Black B.E. Holaska J.M. Levesque L. Ossareh-Nazari B. Gwizdek C. Dargemont C. Paschal B.M. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J Cell Biol. 2001;152:141–155. doi: 10.1083/jcb.152.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P. Benson R.E. Truant R. Herold A. Phingbodhipakkiya M. Cullen B.R. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J Biol Chem. 1999;274:9771–9777. doi: 10.1074/jbc.274.14.9771. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T. Regener K. Schwappach B. Saffrich R. Paraskeva E. Hartmann E. Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey K.L. Richards S.A. Lounsbury K.M. Macara I.G. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. Bono F. Jinek M. Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Cuevas I.C. Frasch A.C. D'Orso I. Insights into a CRM1-mediated RNA-nuclear export pathway in Trypanosoma cruzi. Mol Biochem Parasitol. 2005;139:15–24. doi: 10.1016/j.molbiopara.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Di Fiore B. Ciciarello M. Lavia P. Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle. 2004;3:305–313. [PubMed] [Google Scholar]
- Dubey J.P. Toxoplasmosis. J Am Vet Med Assoc. 1994;205:1593–1598. [PubMed] [Google Scholar]
- Dubey J.P. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- Frankel M.B. Knoll L.J. Functional analysis of key nuclear trafficking components reveals an atypical Ran network required for parasite pathogenesis. Mol Microbiol. 2008;70:410–420. doi: 10.1111/j.1365-2958.2008.06419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel M.B. Mordue D.G. Knoll L.J. Discovery of parasite virulence genes reveals a unique regulator of chromosome condensation 1 ortholog critical for efficient nuclear trafficking. Proc Natl Acad Sci USA. 2007;104:10181–10186. doi: 10.1073/pnas.0701893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herm-Gotz A. Agop-Nersesian C. Munter S. Grimley J.S. Wandless T.J. Frischknecht F. Meissner M. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nat Methods. 2007;4:1003–1005. doi: 10.1038/nmeth1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig R.C. Renault L. Vetter I.R. Drell T.t. Wittinghofer A. Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- Hodel M.R. Corbett A.H. Hodel A.E. Dissection of a nuclear localization signal. J Biol Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Hu K. Roos D.S. Murray J.M. A novel polymer of tubulin forms the conoid of Toxoplasma gondii. J Cell Biol. 2002;156:1039–1050. doi: 10.1083/jcb.200112086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh M.H. Carruthers V.B. Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2006;2(e84):0753–0762. doi: 10.1371/journal.ppat.0020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D.D. Sultan A.A. Chakrabarti D. Horrocks P. Doerig C. Arnot D.E. An RCC1-type guanidine exchange factor for the Ran G protein is found in the Plasmodium falciparum nucleus. Mol Biochem Parasitol. 1998;95:165–170. doi: 10.1016/s0166-6851(98)00107-8. [DOI] [PubMed] [Google Scholar]
- Kim K. Weiss L.M. Toxoplasma gondii: the model apicomplexan. Int J Parasitol. 2004;34:423–432. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D. Solomon F. Overexpression of yeast homologs of the mammalian checkpoint gene RCC1 suppresses the class of alpha-tubulin mutations that arrest with excess microtubules. Genetics. 1994;137:381–392. doi: 10.1093/genetics/137.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonis N. Rug M. Harper I. Wickham M. Cowman A. Tilley L. Fluorescence photobleaching analysis for the study of cellular dynamics. Eur Biophys J. 2002;31:36–51. doi: 10.1007/s00249-001-0202-2. [DOI] [PubMed] [Google Scholar]
- Kornbluth S. Dasso M. Newport J. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J Cell Biol. 1994;125:705–719. doi: 10.1083/jcb.125.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U. Lipowsky G. Izaurralde E. Bischoff F.R. Schwarzmaier P. Hartmann E. Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lindsay M.E. Holaska J.M. Welch K. Paschal B.M. Macara I.G. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol. 2001;153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury K.M. Richards S.A. Carey K.L. Macara I.G. Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J Biol Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- Macara I.G. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M. Brecht S. Bujard H. Soldati D. Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res. 2001;29(E115):1–10. doi: 10.1093/nar/29.22.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M. Schluter D. Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Melchior F. Paschal B. Evans J. Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot J.M. Bohnsack M.T. Jakle U. Gorlich D. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 2004;23:3227–3236. doi: 10.1038/sj.emboj.7600338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohmmed A. Kishore S. Dasaradhi P.V. Patra K. Malhotra P. Chauhan V.S. Cloning and characterization of Plasmodium falciparum homologs of nuclear import factors, karyopherin alpha and karyopherin beta. Mol Biochem Parasitol. 2003;127:199–203. doi: 10.1016/s0166-6851(02)00331-6. [DOI] [PubMed] [Google Scholar]
- Moore M.S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Murphy G.A. Moore M.S. Drivas G. Perez de la Ossa P. Villamarin A. D'Eustachio P. Rush M.G. A T42A Ran mutation: differential interactions with effectors and regulators, and defect in nuclear protein import. Mol Biol Cell. 1997;8:2591–2604. doi: 10.1091/mbc.8.12.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut M.E. Mizzen C.A. Stukenberg T. Allis C.D. Macara I.G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- Palacios I. Weis K. Klebe C. Mattaj I.W. Dingwall C. RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L. Nassar N. Vetter I. Becker J. Klebe C. Roth M. Wittinghofer A. The 1.7 a crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Ribbeck K. Lipowsky G. Kent H.M. Stewart M. Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S.A. Carey K.L. Macara I.G. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Segref A. Sharma K. Doye V. Hellwig A. Huber J. Luhrmann R. Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuven T. Hartmann E. Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profiling-actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenter A.M. Heckeroth A.R. Weiss L.M. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen N.R. Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]