Abstract
Morphological diversity within closely related species is an essential aspect of evolution and adaptation. Mutations in the Melanocortin 1 receptor (Mc1r) gene contribute to pigmentary diversity in natural populations of fish, birds, and many mammals. However, melanism in the gray wolf, Canis lupus, is caused by a different melanocortin pathway component, the K locus, that encodes a beta-defensin protein that acts as an alternative ligand for Mc1r. We show that the melanistic K locus mutation in North American wolves derives from past hybridization with domestic dogs, has risen to high frequency in forested habitats, and exhibits a molecular signature of positive selection. The same mutation also causes melanism in the coyote, Canis latrans, and in Italian gray wolves, and hence our results demonstrate how traits selected in domesticated species can influence the morphological diversity of their wild relatives.
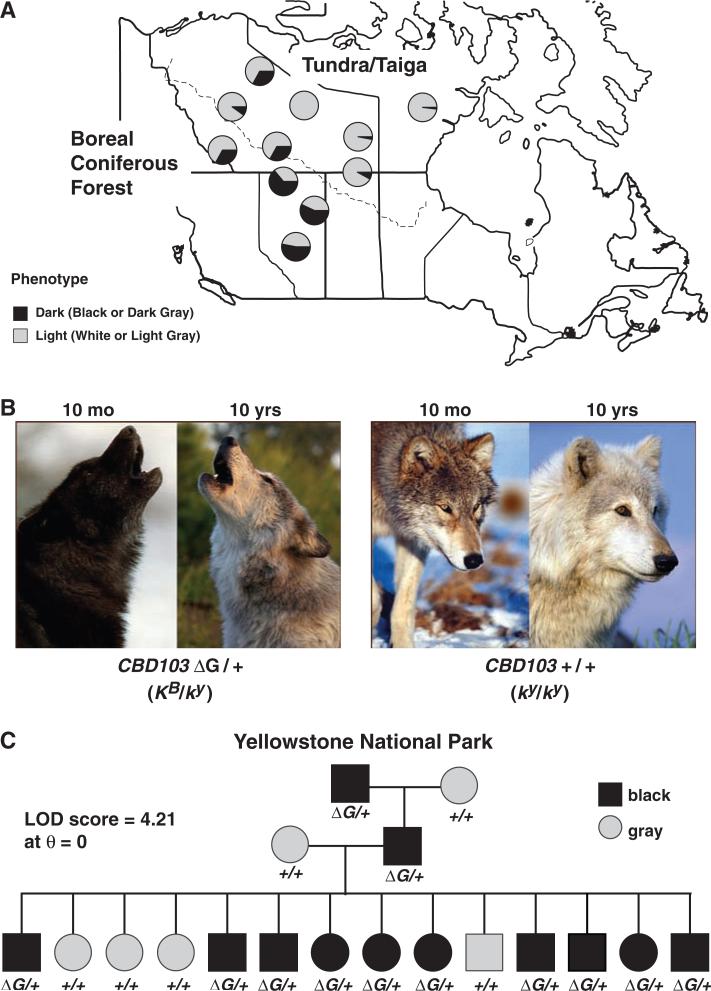
The correspondence between coat color and habitat is often attributed to natural selection, but rarely is supporting evidence provided at the molecular level. In North American gray wolves, coat color frequencies differ between wolves of forested and open habitats throughout western North America (1), including Denali National Park (2) and the Kenai Peninsula in Alaska (3), and much of the Canadian Arctic (4, 5). These differences are especially dramatic between wolves of the high tundra that are migratory and follow barren-ground caribou to their breeding areas, and wolves that are year-round residents in the neighboring boreal forest and hunt nonmigratory prey. Dark-colored wolves are extremely rare in the tundra but increase in frequency along a southwest cline toward forested areas (Fig. 1A). The potential selective value of dark versus light coat color has been suggested to include concealment during predation and/or indirect effects due to pleiotropy, but remains unresolved because the underlying gene(s) have not been identified (5–7).
Fig. 1.
Distribution of melanism and K locus genotypes in North American gray wolves. (A) Location and coat color phenotype of Canadian samples used here and as described (4). (B) Age-related graying and the associated difficulty of inferring genotype from phenotype in gray animals. Each pair of photos shows the same individual at different ages (10 months and 10 years) and documents an increasingly gray appearance at 10 years, reflecting the dilution of eumelanin in the KB/ky individual (left pair of images) and dilution of both eumelanin and pheomelanin in the ky/ky individual (right pair of images). [Images courtesy of Monty Sloan, Wolf Park, Battle Ground, Indiana] (C) Co-segregation of KB and black coat color in a three-generation pedigree from the Leopold pack in Yellowstone National Park (17). ΔG indicates the dominant KB allele, whereas + indicates the wild-type allele, ky.
In many vertebrates, natural pigmentary variation is controlled by the agouti– melanocortin 1 receptor (Mc1r) pathway, a ligand receptor pair that modulates the amount and type of pigment—red/yellow pheomelanin or brown/black eumelanin—produced by melanocytes in skin, hair, or feathers. Gain-of-function Mc1r mutations are well-recognized causes of melanism in many domestic and laboratory animal species (8, 9), as well as in several natural populations of birds (10), rodents (11, 12), and canids (13). Recently, we found that pigment type-switching in domestic dogs involves an additional component of the melanocortin pathway, the K locus, which encodes a beta-defensin protein, CBD103 (14, 15).
Coat color in Canadian wolves is genetically complex, with phenotypes ranging from white to gray to black, and is also confounded by an independent effect of graying with age (Fig. 1B). However, in Yellowstone National Park, where a small number of founder animals from Canada were recently reintroduced (16, 17), gray and black coat colors segregate as a Mendelian trait. We surveyed molecular variation in Agouti, Mc1r, and CBD103 in wolves from North America and identified several Mc1r and Agouti polymorphisms. However, none of these were predicted to affect gene function and did not associate with black coat color (table S1). In contrast, in a 14-member, three-generation kindred from Yellowstone, we observed complete co-segregation between black coat color and markers at the K locus [logarithm of the odds ratio for linkage (lod) score = 4.21 at the maximum likelihood estimate of recombination fraction (θ) = 0, Fig. 1C], which is unlinked and lies on a different chromosome from Agouti and Mc1r.
In dogs, the ancestral CBD103 allele (ky) confers normal Agouti and Mc1r gene action, whereas a 3–base pair (bp) deletion (CBD103ΔG23 or KB) suppresses Agouti gene action, leading to dominant inheritance of a black coat (14, 15). We observed the same 3-bp deletion in 102 out of 104 black-colored wolves from Yellowstone and 9 out of 9 from the Canadian Arctic. Conversely, CBD103ΔG23 was absent from 120 of 120 gray-colored wolves from Yellowstone and from 22 of 22 white-colored wolves from the Canadian Arctic (Table 1). We also found CBD103ΔG23 in 6 of 10 gray-colored wolves from the Canadian Arctic, suggesting that gray coat color can result either from the absence of CBD103ΔG23 and a modified agouti phenotype (in which individual hairs contain both cream-colored pheomelanin and dark eumelanin) or from secondary factors such as age that dilute the pigmentation of hairs that contain only eumelanin. [Additional genealogy studies of the Yellowstone population (17) together with the paucity of Mc1r variation in wolves (table S1) suggest that black coat color reported for the two ky/ky Yellowstone wolves is likely to reflect phenotypic ambiguity or misclassification at the time of sampling.] Allele frequencies for CBD103ΔG23 in tundra and forest wolves overall were estimated at 0.02 and 0.19, corresponding to phenotype frequencies of 2 to 33% and 33 to 64% for dark wolves in tundra and forest populations, respectively (Fig. 1A) (4).
Table 1.
Distribution of CBD103 alleles in wolves and coyotes. N/A, not applicable.
| Animal and location | Phenotype† |
|||
|---|---|---|---|---|
| White | Gray | Black | ||
| Forest wolves* | Total no. | 12 | 2 | 7 |
| No. carrying KB | 0 | 1 | 7 | |
| Tundra/taiga wolves* | Total no. | 10 | 8 | 2 |
| No. carrying KB | 0 | 5 | 2 | |
| Yellowstone wolves | Total no. | 0 | 120 | 104 |
| No. carrying KB | N/A | 0 | 102 | |
| Coyotes‡ | Total no. | 0 | 61 | 6 |
| No. carrying KB | N/A | 0 | 6 | |
Forest and tundra/taiga wolves are from the Canadian Arctic (Fig. 1A). The overall frequency of dark (gray or black) wolves is 62 and 7% in the forest and tundra/taiga, respectively (4), and the genotype distributions shown do not represent population-based frequencies. All forest and tundra/taiga wolves carrying KB were KB/ky; in the Yellowstone population, 10 were KB/KBand 92 were KB/ky.
This categorical designation of phenotypes, as defined at sample collection, does not fully capture the spectrum of normal coat color variation as indicated in Fig. 1B.
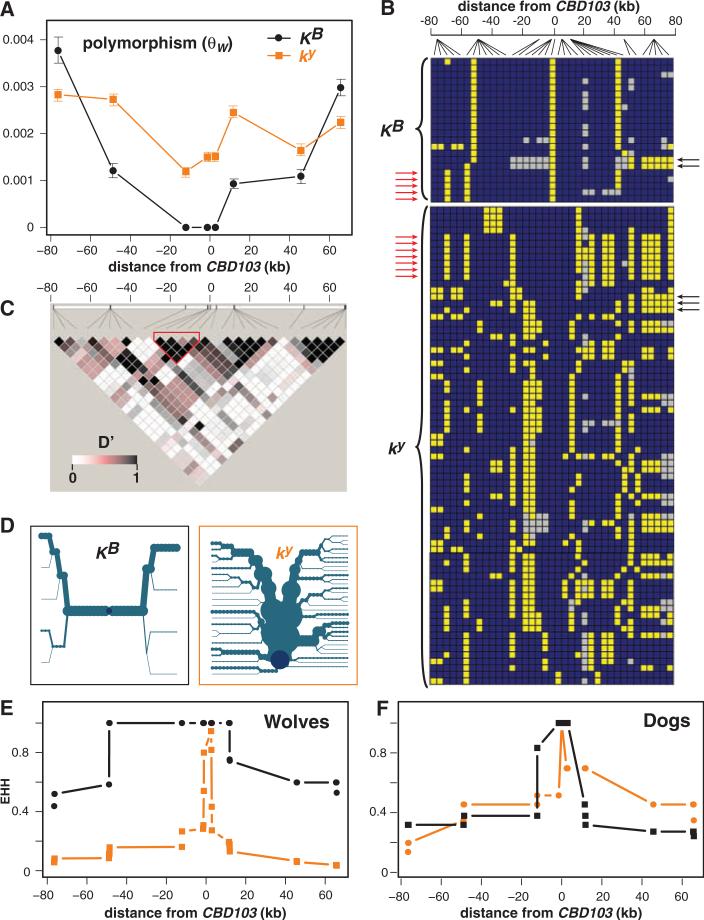
To investigate the evolutionary history of the melanistic K allele, we sequenced eight single-copy noncoding segments distributed across an ~150-kb region centered on CBD103 in 32 Arctic and 15 unrelated Yellowstone wolves, as well as in 12 domestic dogs: 6 ky/ky (akita, basenji, boxer, bulldog, Doberman pinscher, and great dane) and 6 KB/KB (curly-coated retriever, Dalmatian, great dane, Labrador retriever, poodle, and Portuguese water dog). We identified 52 biallelic polymorphisms across all canids (36 in wolves) and estimated haplotype structure (tables S3 and S4, Fig. 2B, and fig. S2). The rate of polymorphism among all wolf amplicons was one single-nucleotide polymorphism (SNP) per 510 bp (Watterson's estimator, θW = 1.96 × 10−3), which is similar to genome-wide measurements of polymorphism between the boxer and the gray wolf (1 out of 580 bp) and the coyote (1 out of 420 bp) (18). However, partitioning our data according to K locus genotype and proximity to CBD103 revealed little or no polymorphism among KB-bearing chromosomes close to CBD103, rising to levels at or above those observed in ky-bearing chromosomes in the 75 kb spanning either side of the locus (Fig. 2A). This pattern, and the analogous one for nucleotide diversity (π, fig. S1), is also reflected in a significant difference in haplotype diversity between KB (8 unique of 22 total) and ky (59 unique of 72 total) chromosomes (χ2 = 14.2, P < 0.001). Together with the correlations between coat color and habitat (2–5), the combination of low diversity and high frequency suggests that KB has been under positive selection in North American forest wolves.
Fig. 2.
Polymorphism and haplotype structure of the K locus in North American gray wolves [(A) to (E), 1 KB/KB, 20 KB/ky, and 26 ky/ky] and domestic dogs [(F), 6 KB/KB and 6 ky/ky]. (A) Polymorphism (θW, ±SD) as a function of distance from CBD103. (B) Wolf haplotype structure was inferred on the basis of 36 SNPs; each row represents a KB- or ky- bearing chromosome; blue and yellow squares represent the major and minor alleles, respectively; and the gray squares represent missing data. Red and black arrows indicate examples of haplotypes likely to represent historical recombination between KB- and ky-bearing chromosomes at the 5′ and 3′ ends of the locus, respectively. (C) Pairwise LD values (expressed as D') for all wolf chromosomes; the red outline indicates a core region (as in Fig. 3) unlikely to have undergone historical recombination. (D) Haplotype bifurcation diagrams for KB- or ky-bearing chromosomes, in which the central dark blue dot represents CBD103, branches represent haplotype divergence, and the thickness of the lines is proportional to the number of chromosomes. (E and F) EHH for KB- or ky-bearing chromosomes in wolves (E) and dogs (F) as a function of distance from CBD103ΔG23.
Overall, the patterns of linkage disequilibrium (LD) across 150 kb surrounding the K locus were similar to comparisons between different breeds of domestic dogs (18), with relatively small haplotype blocks, including an ~4-kb CBD103 core region within which there is no evidence for historical recombination (Fig. 2C). However, different evolutionary histories for the Arctic wolf KB and ky alleles were apparent when the SNP patterns (Fig. 2B) were depicted as haplotype bifurcation diagrams (Fig. 2D), which highlight a central region of ~60 kb devoid of polymorphism among wolf KB haplotypes. This characteristic, and the corresponding difference between KB and ky chromosomes, were represented quantitatively by the extended haplotype homozygosity (EHH) statistic (19), which is the empirical probability that two chromosomes chosen at random remain identical at progressively increasing distances from CBD103. As depicted in Fig. 2, E and F, the distribution of EHH was considerably broader for KB as compared to ky chromosomes in wolves, whereas the distributions were nearly identical for KB as compared to ky chromosomes in dogs. Together with additional analyses of genome-wide SNP data [supporting online material (SOM) text and fig. S3], these observations suggest that KB has risen to high frequency by a selective sweep.
As in black dogs and melanistic wolves, CBD103ΔG23 was associated with coat color in 67 coyotes (6 black and 61 gray, Table 1 and table S2). These findings suggest three possible evolutionary histories. First, the 3-bp deletion may be relatively old, having occurred in a canid ancestor more than 1 million years ago before the divergence of coyotes from wolves. Second, the 3-bp deletion may have occurred more recently in one of the species, followed by introgression into the others. Finally, the 3-bp deletion may represent a mutational hotspot, having recurred independently in coyotes, wolves, and dogs. To distinguish among these possibilities, we ascertained and compared coyote haplotypes (6 KB and 18 ky) with those from the North American wolf and dog.
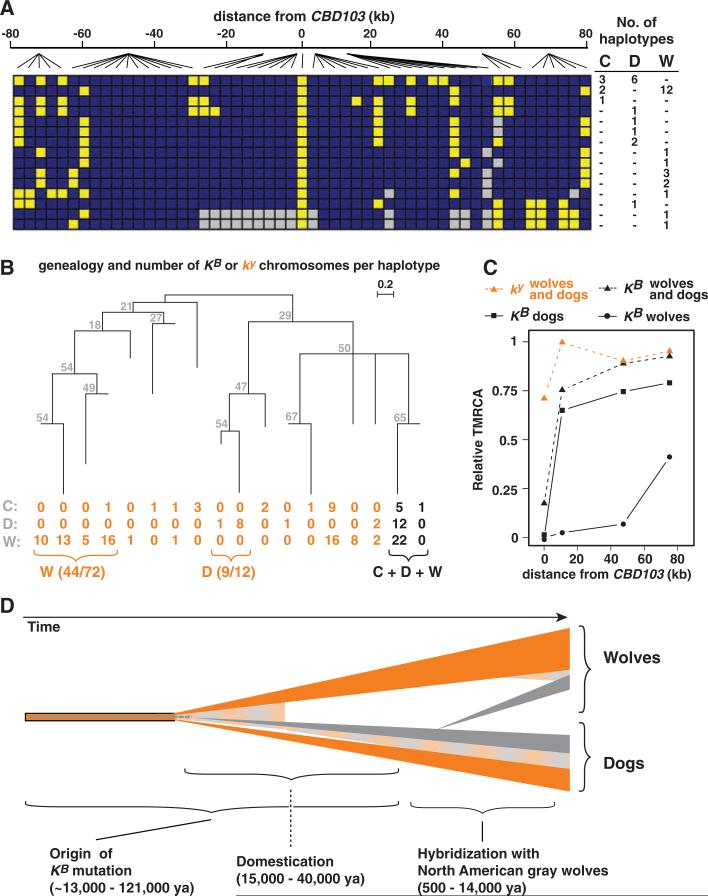
The pattern of haplotype diversity for all three canids was similar to that observed in wolves alone and showed significantly less diversity among KB (15 unique of 40 total) relative to ky (66 unique of 102 total) chromosomes (χ2 = 9.7, P = 0.003). Of the 15 unique KB haplotypes, 1 haplotype was observed in three coyotes and six dogs, and a second haplotype was observed in two coyotes and 12 wolves (Fig. 3A). However, none of the 66 unique ky haplotypes were observed in more than one species (fig. S2).
Fig. 3.
Evolutionary relationships and history of the K locus in canids. (A) KB haplotype structure in wolflike canids based on genotypes defined by 52 SNPs. Each row represents a KB-bearing haplotype found in coyotes (C), dogs (D), or wolves (W) listed with their respective frequencies on the right and colored as in Fig. 2B. (B) Inferred genealogical relationships of the core region (Fig. 2C) haplo-types (with bootstrap values from 500 replicates shown next to branches).Each branch represents 1 of 18 different haplotypes, with the number of chromosomes for each haplotype indicated underneath according to species. (C) TMRCA estimates for indicated chromosome subsets calculated according to a molecular clock (22) and expressed as a fraction of the divergence time for all wolflike canids. Individual points represent sets of chromosome segments whose relative TMRCA increases as a function of distance from CBD103, presumably due to ancient hybridization and recombination. (D) Timeline scenario for K locus evolution in dogs and wolves, in which ancestral ky chromosomes are indicated in orange, derivative KB chromosomes in gray, and recombinant chromosomes as an orange-gray checkered pattern. The ky-to-KB mutation may have overlapped or even predated domestication, but the introgression of KB into North American gray wolves is more recent.
Reconstruction of a phylogenetic network for the entire 150-kb region is complicated by historical recombination between extant KB and ky chromosomes (arrows in Fig. 2B) and the lack of a suitable approach for inferring accurate gene genealogies in the presence of recombination (20). However, by focusing on the 4-kb CBD103 core region (Fig. 2C), a simple neighbor-joining tree was constructed for 18 core region haplotypes representing 142 (94 wolf, 24 dog, and 24 coyote) chromosomes (Fig. 3B). In this tree, all the KB chromosomes define a 2-haplotype cluster, whereas the remaining 16 haplotypes (which represent all the ky chromosomes) are more dispersed. Furthermore, many of the ky chromosomes cluster by species (9 out of 12 of the dogs and 44 out of 72 of the wolves), unlike the KB chromosomes. This contrasting phylogenetic pattern suggests that the KB mutation occurred in a single species and was later distributed among dogs, wolves, and coyotes by interspecific hybridization. [The 24 ky haplotypes from coyotes are no closer to each other than to ky haplotypes from wolves or dogs (Fig. 3B), which is consistent with their history of hybridization with other canids (21)].
To gain additional insight into how K locus variation in dogs and wolves arose, we estimated coalescent time to the most recent common ancestor (TMRCA) as a function of cumulative distance from CBD103 for ky and KB chromosomes from wolves, dogs, and both groups together. We applied a molecular clock approach to sequencing data from individual amplicons across the entire 150-kb region (Fig. 2), which assumes that mutations occur at the same constant rate at all sites in wolves and dogs and integrates the effects of both recombination and demography (22). Close to CBD103, TMRCA estimates were near zero for all KB subsets (Fig. 3C) because there is little or no polymorphism in this region (Fig. 3A). However, at greater distances from CBD103 (10 to 50 kb),estimates for dog chromosomes are similar to those of dog and wolf chromosomes considered together, regardless of genotype. This suggests that KB in dogs is sufficiently old to have undergone extensive recombination with ky chromosomes, and that the recombination history includes hybridization between dogs and wolves. However, in the same 10- to 50-kb range, TMRCA estimates for wolf KB chromosomes were considerably less than those from dog KB chromosomes (or from dog and wolf KB chromosomes considered together), suggesting that KB was introduced into North American wolves from dogs, not vice versa.
Introgression of KB from dogs into North American wolves is also supported by geographical and ecological considerations. KB is widely distributed among domestic dogs, including ancient breeds originating in Asia and Africa. In wolves, however, melanism has been reported outside North America only in Italy, where it is associated with molecular and/or morphologic evidence of recent hybridization with free-ranging dogs (23). Indeed, we also examined 22 samples from the Italian Apennines and observed KB in six of seven black “wolves” (including one previously classified to be a dog-wolf hybrid) but 0 of 15 gray wolves. In contrast, genome-wide SNP analysis of 10 KB/ky and 10 ky/ky North American wolves showed no evidence for recent dog-wolf hybridization (SOM text and fig. S3B).
The dog was domesticated between 15,000 and 40,000 years ago in East Asia from gray wolves (24, 25), and we estimate that KB is at least 46,886 years old (95% confidence limit: 12,779 to 121,182 years); therefore, we cannot distinguish whether KB arose before or after domestication. However, if KB arose in Old World wolves before domestication, our data indicate that it must have been lost from the gene pool and reacquired in North America, perhaps from Native American dogs that accompanied humans across the Bering Strait 12,000 to 14,000 years ago (26) (Fig. 3D).
The wolf in the United States faces grave threats, in some cases by eradication, and in others by hybridization, such as in the Great Lakes region (27). However, apparent selection for the KB locus in North American gray wolves shows how genetic diversity—preserved by humans in domestic dogs—may flourish in wild wolf populations. As the available tundra habitat declines because of development and/or global warming, the frequency of the KB mutation may increase further in northern latitudes. Thus, the introduction of genetic diversity into a natural population from a mutation originally selected in domesticated animals may, ironically, provide a mechanism for that population to adapt to a changing environment. Interspecific hybridization has been widely observed between other domesticated species of animals and plants (28–30). Our results imply that variants that appear under domestication can be viable in the wild and enrich the genetic legacy of natural populations.
Supplementary Material
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/1165448/DC1 Materials and Methods
References and Notes
- 1.Gipson PS, et al. Wildl. Soc. Bull. 2002;30:821. [Google Scholar]
- 2.Mech LD, Adams LG, Meier TJ, Burch JW, Dale BW. The Wolves of Denali. Univ. of Minnesota Press; Minneapolis, MN: 1998. [Google Scholar]
- 3.Peterson RO, Wollington JD, Bailey TN. Wildl. Monogr. 1984;88:3. [Google Scholar]
- 4.Musiani M, et al. Mol. Ecol. 2007;16:4149. doi: 10.1111/j.1365-294X.2007.03458.x. [DOI] [PubMed] [Google Scholar]
- 5.Jolicoeur P. Evolution. 1959;13:283. [Google Scholar]
- 6.Majerus MEN. Melanism: Evolution in Action. Oxford Univ. Press; Oxford: 1998. [Google Scholar]
- 7.Ducrest AL, Keller L, Roulin A. Trends Ecol. Evol. 2008;23:502. doi: 10.1016/j.tree.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Klungland H, Vage DI. Ann. N. Y. Acad. Sci. 2003;994:331. doi: 10.1111/j.1749-6632.2003.tb03197.x. [DOI] [PubMed] [Google Scholar]
- 9.Andersson L. Ann. N. Y. Acad. Sci. 2003;994:313. doi: 10.1111/j.1749-6632.2003.tb03195.x. [DOI] [PubMed] [Google Scholar]
- 10.Mundy NI, et al. Science. 2004;303:1870. doi: 10.1126/science.1093834. [DOI] [PubMed] [Google Scholar]
- 11.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. Science. 2006;313:101. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 12.Nachman MW, Hoekstra HE, D'Agostino SL. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5268. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vage DI, et al. Nat. Genet. 1997;15:311. doi: 10.1038/ng0397-311. [DOI] [PubMed] [Google Scholar]
- 14.Kerns JA, et al. Genetics. 2007;176:1679. doi: 10.1534/genetics.107.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candille SI, et al. Science. 2007;318:1418. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bangs EE, Fritts S. Wildl. Soc. Bull. 1996;24:402. [Google Scholar]
- 17.vonHoldt BM, et al. Mol. Ecol. 2008;17:252. doi: 10.1111/j.1365-294X.2007.03468.x. [DOI] [PubMed] [Google Scholar]
- 18.Lindblad-Toh K, et al. Nature. 2005;438:803. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 19.Sabeti PC, et al. Nature. 2002;419:832. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 20.Woolley SM, Posada D, Crandall KA. PLoS ONE. 2008;3:e1913. doi: 10.1371/journal.pone.0001913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy MS, Geffen E, Smith D, Ostrander EA, Wayne RK. Mol. Biol. Evol. 1994;11:553. doi: 10.1093/oxfordjournals.molbev.a040137. [DOI] [PubMed] [Google Scholar]
- 22.Tang H, Siegmund DO, Shen P, Oefner PJ, Feldman MW. Genetics. 2002;161:447. doi: 10.1093/genetics/161.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randi E, Lucchini V. Conserv. Genet. 2002;3:29. [Google Scholar]
- 24.Vila C, et al. Science. 1997;276:1687. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 25.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Science. 2002;298:1610. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 26.Leonard JA, et al. Science. 2002;298:1613. doi: 10.1126/science.1076980. [DOI] [PubMed] [Google Scholar]
- 27.Leonard JA, Wayne RK. Biol. Lett. 2008;4:95. doi: 10.1098/rsbl.2007.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecis R, et al. Mol. Ecol. 2006;15:119. doi: 10.1111/j.1365-294X.2005.02812.x. [DOI] [PubMed] [Google Scholar]
- 29.Halbert N, Derr J. J. Hered. 2007;98:1. doi: 10.1093/jhered/esl051. [DOI] [PubMed] [Google Scholar]
- 30.Ellstrand N, Prentice H, Hancock J. Annu. Rev. Ecol. Syst. 1999;30:539. [Google Scholar]
- 31.Supported by grants from NIH (G.S.B.), NSF (R.K.W., D.R.S., and D.W.S.), and the Swedish Research Council (J.A.L.). We are grateful to H. Chen and S. Schmutz for advice, to H. Manuel for technical assistance, and to members of the U.S. Department of Agriculture Wildlife Services and private citizens for assistance with sample collection. Sequences generated in this study are deposited in GenBank under accession numbers FJ609634 to FJ609641.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.