Abstract
F-9775A and F-9775B are cathepsin K inhibitors that arise from a chromatin remodelling deletant strain of Aspergillus nidulans. A polyketide synthase gene has been determined to be responsible for their formation and for the simpler, archetypical polyketide orsellinic acid. We have discovered simple culture conditions that result in the production of the three compounds, and this facilitates analysis of the genes responsible for their synthesis. We have now analysed the F9775/orsellinic acid gene cluster using a set of targeted deletions. We find that the polyketide synthase alone is required for orsellinic acid biosynthesis and only two additional genes in the cluster are required for F9775 A and B synthesis. Our deletions also yielded the bioactive metabolites gerfelin and diorcinol.
Introduction
Fungal genera such as Aspergillus are recognized to produce a variety of structurally complex secondary metabolites, many of which have significant relevance to human health. Examples of such metabolites include lovastatin, a well-known antihypercholesterolemic agent produced by A. terreus, and aflatoxin, a carcinogen produced by A. flavus.1,2 The recent sequencing of a number of fungal genomes reveals that fungi have many more secondary metabolism pathways than previously thought.3-6 For example, A. nidulans contains 27 polyketide synthase (PKS) and 14 nonribosomal peptide synthetase (NRPS) genes.3 Since far fewer secondary metabolites have been identified from this heavily studied species, the products of many of these biosynthetic pathways are still unknown.
Fungal polyketides are produced by multidomain iterative PKSs. Fungal PKSs can be further grouped into nonreduced (NR), partially reduced (PR), and highly reduced (HR) PKSs by examining the domains encoded in the gene.7-9 NR-PKSs usually contain a starter unit ACP transacylase (SAT) domain responsible for starter unit selection, as well as domains responsible for chain extension: β-ketoacyl synthase (KS), acyl transferase (AT), product template (PT), and acyl carrier protein (ACP). The domains found in NR-PKSs after the chain extension domains are highly varied. They can include thioesterase/Claisen-cyclase (TE/CLC), methyltransferase (CMeT) and reductase (R) domains. Of the nonreduced polyketides, orsellinic acid (1, Fig. 1) is potentially the simplest tetraketide produced from the condensation of a starter acetate group and three malonyl extender units.
Fig. 1.
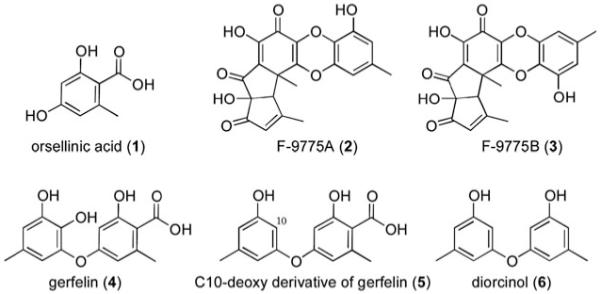
Chemical structures of orsellinic acid (1) and F-9775A (2) and B (3) gerfelin (4), C-10-deoxy gerfelin (5), diorcinol (6).
F-9775A (2) and F-9775 B (3) are yellow polyketides initially observed from Paecilomyces carneus and demonstrated to inhibit the cysteine protease cathepsin K.10 We had shown that deletion of cclA, a bre2 ortholog involved in histone H3 lysine 4 methylation, led to the generation of F-9775 A and B under culture conditions in which these metabolites were normally not detected.11 The deletion led to increased mRNA transcripts of genes AN7909.4 (using the widely accepted gene designations for this species) through AN7915.4, suggesting that at least some of these genes were required for F9775 biosynthesis. Indeed, deletion of AN7909.4 resulted in the disappearance of the metabolites.
While this manuscript was in preparation, it was reported by Schroeckh et al. that AN7909.4 is responsible not only for F-9775 A and B but for the simple polyketide orsellinic acid.12 Orsellinic acid synthase (OSAS) was one of the first discovered fungal PKSs, isolated from Penicillium madriti and reported in 1968.13 A similar, methyltransferase (CMeT) containing enzyme, methylorsellinaldehyde synthase, which produced 3-methylorcinaldehyde when heterologously expressed, has been described in the fungus, Acremonium strictum.14 Schroeckh et al. had detected orsellinic acid and F-9775 A and B through cocultivating A. nidulans with a soil-dwelling actinomycete, with the concurrent upregulation of the five genes beginning with AN7909.4 (and termed orsA through orsE).12
We have tried a variety of growth conditions and media to obtain enhanced expression of A. nidulans secondary metabolites and have found conditions that give expression of orsellinic acid as well as F-9775 A and B in high titers from strains that do not carry the cclA deletion. This has facilitated our analysis of the orsellinic acid/F9775 cluster. Through a series of targeted deletions, we report that AN7909.4 is not only necessary but evidently sufficient for orsellinic acid production, without the requirement of a regulatory or tailoring gene. In contrast, three contiguous genes including AN7909.4 are essential for F-9775 A and B synthesis, but deletion of other genes in the putative orsellinic acid/F9775 cluster did not prevent production of the three compounds. The deletions also resulted in accumulation of the bioactive compounds gerfelin and diorcinol.
Results and discussion
Isolation of orsellinic acid and F-9775 A and B in A. nidulans through a culture condition variant
In our previous studies, a wild type A. nidulans strain cultured under liquid or solid glucose minimal media and yeast extract media produced copious amounts of the aromatic compound sterigmatocystin, the penultimate product of aflatoxin, together with additional secondary metabolites, terrequinone, emericellamides, dehydroaustinol, and austinol.15-17 As mentioned, F-9775 A and B were undetected under normal conditions but emerged in a cclA deletant strain11 or from co-cultivation with an actinomycete.12 A third approach to turn on the production of different metabolites is to alter the conditions in which the organism is cultivated, a strategy that has been called one strain many compounds (OSMAC).18 In order to discover additional secondary metabolites produced by this organism, wild type R153 A. nidulans strain was subjected to 20 different conditions (various media, different cultivation times, stationary or submerged; see Experimental section for details) and screened by HPLC-DAD-MS. In stationary liquid Czapek media we identified three aromatic compounds not substantially found in any other media cultures that we studied. A larger fermentation (12 × 150 mL) resulted in the successful isolation of the three metabolites. One of the products is the aromatic polyketide orsellinic acid, a metabolite we have not observed in A. nidulans using any other approach. Orsellinic acid was identified by acquired spectroscopic data (1H NMR, 13C NMR, UV, and mass spectrometry) and by comparison to commercially available orsellinic acid (Alfa Aesar, Fig. S4–S7). The other two metabolites were the cathepsin K inhibitors F-9775A and F-9775B. As previously reported, F-9775A and F-9775B were characterized by both 1H NMR and 13C NMR (Fig. S8–S11 in the supplementary data†).11 Given the co-expression of orsellinic acid with the cathepsin K inhibitors, we suspect that orsellinic acid is also produced in the cclA mutant strain, but, as we have found that it is a poor substrate in our electrospray ionization (ESI) condition for initial screening, the amount was below the detection threshold. Stationary liquid Czapek’s media, by comparison, affords high titers of orselinic acid and all related metabolites mentioned herein.
Linking orsellinic acid synthase to one of the NR-PKSs in A. nidulans
The isolation and characterization of orsellinic acid from A. nidulans cultured in Czapek media suggested that it was now possible to locate orsellinic acid synthase by investigating every candidate NR-PKS in the genome. As two of the 12 NR-PKSs have known function, generating sterigmatocystin and the wA spore pigment, we focused on the ten unannotated genes. Previously, we had deleted these ten genes to determine which were responsible for the metabolites that emerged from cclAΔ, including F-9775 A and B.11 The deletions were accomplished using our previously reported strategy involving a nkuA strain and fusion PCR.19 The targeted genes were replaced with the A. fumigatus pyrG gene.20 All deletants were verified using diagnostic PCR (data not shown). LC-DAD-MS analysis of the ten NR-PKS deletants showed that only AN7909.4Δ, the same deletant that eliminated production of F-9775 A and B (2 and 3), failed to produce orsellinic acid (1, Fig. 2 and S1), in agreement with Schroeckh et al.12 None of the other NR-PKS genes studied had any noticeable effect on 1, 2, or 3. This reveals that orsellinic acid in A. nidulans is not a shunt product that can be issued from various NR-PKSs.
Fig. 2.
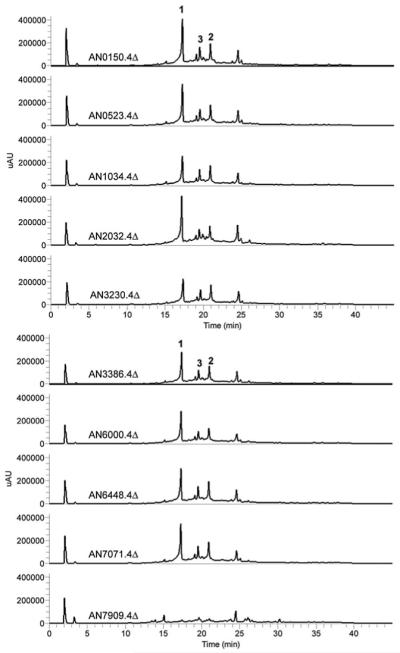
(A) HPLC profiles of extracts from the ten NR-PKS knockout strains as detected by UV absorption at 254 nm.
The orsellinic acid synthase (AN7909.4) is a 2104 amino acid multidomain iterative type I PKS that shares similarities with other nonreduced PKSs identified from fungal genome sequences. The sequence suggests the domain organization is SAT, KS, AT, PT, ACP, ACP, and TE/CYC. This arrangement, including the two ACP domains, is common in nonreduced fungal PKSs. The reason for the presence of more than one ACP is unclear, but notably Fujii, Ebizuka, and coworkers have shown that the A. nidulans wA gene continued to yield the polyketide product if either of the two ACP domains was inactivated, whereas inactivation of both domains abrogated activity.21 The bacterial OSAS PKS, also iterative type I, from Streptomyces has been reported as part of the calicheamicin and the avilamycin A gene clusters.22,23 The domain structure of the bacterial OSAS is KS, AT, DH, and ACP and differs from the fungal OSAS we have discovered. Specifically, the bacterial OSAS contains an additional dehydratase domain (DH) and lacks the PT and off loading TE/CYC domains. The bacterial OSAS appears to be homologous to the fungal partially reduced PKSs and not the nonreduced PKSs.9
Genes surrounding the orsellinic acid synthase
Since secondary metabolism genes in A. nidulans are usually clustered, we were interested in identifying additional genes that could possibly help explain the relation between orsellinic acid and F9775A (2) and B (3). Our previous gene expression analyses had indicated that AN7911.4-AN7915.4 were upregulated together,11 and Schroeckh et al. found increased gene expression for AN7911.4-AN7914.4,12 suggesting that at least some of these genes might be involved in production of orsellinic acid and F9775 A and B. In the same method described earlier, we individually deleted the eight genes preceding AN7909.4 (AN7901.4-AN7908.4), and the five genes following AN7909.4 (AN7911.4-AN7915.4). Because the annotation of the A. nidulans genome has been revised three times, the order of genes is not strictly sequential, accounting for the numerical gap between AN7909.4 and AN7911.4 (Fig. 3A). All transformants were verified using diagnostic PCR, and the deletions of AN7909.4, AN7911.4, and AN7912.4 were further verified by Southern blot analyses (data not shown). LC-DAD-MS analysis of three separate deletant strains per gene showed that only two mutants, AN7911.4 and AN7912.4, failed to produce F9775 A (2) and B(3) although continued to yield orsellinic acid, whereas the others still produce all three compounds (Fig. 3B). The AN7911.4 gene product has high homology to 2,6-dihydroxybenzoic acid decarboxylase isolated from Rhizobium radiobacter (35% identity/50% similarity).24 The AN7912.4 gene product has high homology to several putative tyrosinases and is homologous to a monooxygenase involved in trichothecene synthesis in Fusarium sporotrichioides (30% identity/41% similarity),25 as revealed by a BLAST search of the NCBI non-redundant database. AN7913.4 and AN7914.4 appear to code for a hypothetical protein and an alcohol dehydrogenase, respectively. The chemical complexity of F9775 A (2) and B (3) would suggest that additional post-PKS genes located elsewhere in the genome might be necessary to complete their biosynthesis.
Fig. 3.
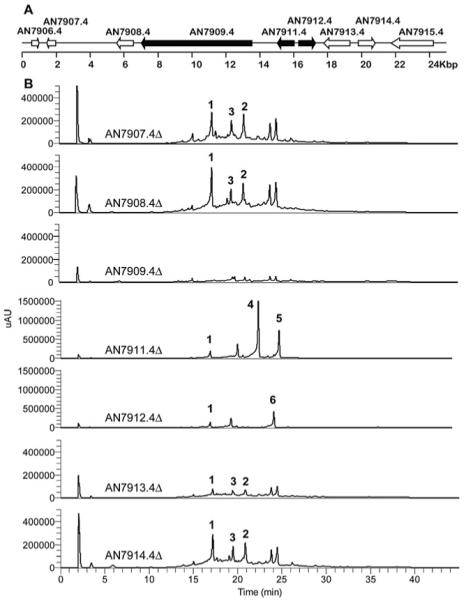
(A) Organization of the gene cluster surrounding the orsellinic acid synthase PKS (AN7907.4) in A. nidulans. Each arrow indicates the direction of transcription and the relative sizes of the ORFs deduced from analysis of the nucleotide sequences. ORFs AN7909.4 (PKS), AN7911.4 (decarboxylase) and AN7912.4 (tyrosinase) are involved in F-9775 A and B biosynthesis. (B) HPLC profile of extracts as detected by UV absorption at 254 nm.
The UV traces also showed that both AN7911.4Δ and AN7912.4Δ deletion strains produce new aromatic compounds. AN7911.4Δ and AN7912.4Δ strains were grown in large scale, and the metabolites were purified from both strains. From the AN7911.4Δ strain we were able to isolate, and characterize by NMR, gerfelin (4, Fig. S12 and S13 in the supplementary data†), the C10-deoxy derivative of gerfelin (5, Fig. S14 and S15 in the supplementary data†), and orsellinic acid (1). Gerfelin has been previously isolated from the fungus Beauveria felina and has been shown to be an inhibitor of human geranylgeranyl diphosphate synthase.26 The methyl ester of gerfelin also suppresses osteoclastogenesis by inhibiting glyoxalase I.27
From the AN7912.4Δ deletant strain we were able to isolate diorcinol (6) and orsellinic acid (1) (Fig. S16 to S19 in the supplementary data†). The metabolite diorcinol has been previously isolated from a marine-derived isolate of A. versicolor and was shown to have antibiotic activities.28-30 It is possible that the metabolites we observed from the AN7911.4 and AN7912.4 knockout strains are not on the direct pathway of F-9775 A (2) and B (3) production but are shunt products orchestrated by the same enzymes involved in F-9775 A (2) and B(3) generation. Nevertheless, gerfelin (5) and diorcinol (6), not typically detected in A. nidulans, are of interest as natural products due to their own biological activities. In accord with the recent work on orsellinic acid12 we identify AN7909.4, AN7911.4, and AN7912.4 as orsA, orsB, and orsC, respectively. However, we find that only orsA appears to be necessary for orsellinic acid generation, without the need of any regulatory or tailoring gene. Interestingly, deletion of cclA or co-cultivating with an actinomycete led to the upregulation of three and two, respectively, contiguous genes after AN7912.4 that we have found are not required for synthesis of orsellinic acid or F-9775 A and B. At this point it is not clear whether the upregulation of these genes serves any function, whether the corresponding proteins are formed, and whether such proteins are functional. Our data do make the important point, however, that upregulation of genes in clusters does not necessarily imply that the products of the genes are essential components of the biosynthetic pathway encoded by the cluster.
Conclusion
The orsellinic acid synthase is possibly the simplest tetraketide synthase known in fungi and thus represents an ideal platform for future experiments on this important class of enzymes. We find that orsA, the gene that is necessary for orsellinic acid, is apparently sufficient. The identification of the orsellinic acid synthase gene in A. nidulans is particularly significant considering the wealth of tools available for this model fungal organism. We were able to detect the production of orsellinic acid in A. nidulans when cultivated in stationary Czapek media, and this approach allowed us to obtain it and the cathepsin K inhibitors F-9775 A and B, as well as gerfelin, and diorcinol, in high titers. We have been able to identify two genes located next to the orsellinic acid synthase gene that are involved in the biosynthesis of the cathepsin K inhibitors, the deletion of which leads to metabolites of biological interest. Additional co-regulated OSAS cluster genes appear to have no effect on the biosynthesis of any of these metabolites.
Experimental
Generation of fusion PCR fragments, A. nidulans protoplasting and transformation
The gene targeting procedures of Szewczyk et al.31 were used for all gene deletions. Two ~1000 base pair fragments upstream and downstream of each targeted gene were amplified from genomic A. nidulans DNA by PCR. Primers used in this study are listed in Table S1 in the supplementary information.† The two amplified flanking sequences and an A. fumigatus pyrG selectable marker cassette were fused together by PCR using two nested primers. A. nidulans strains used in this study are listed in Table S2.† Protoplast generation and transformation were carried out as described.20,31 A strain LO2026 (Table S2†) carrying a deletion of the stcJ gene (stcJΔ) that prevents sterigmatocystin production was used as a recipient strain. The rationale for using an stcJΔ strain is that acetyl and malonyl CoA precursors may be freed up for other products and that the absence of sterigmatocystin may facilitate purification. The transformations were verified by diagnostic PCR. Diagnostic PCR of the deletant strains was performed using the external primers used in the first round of PCR. The difference in the size between the gene replaced by the selective marker and the native gene allowed us to determine if transformants carried correct gene replacements. In cases in which the sizes of both the WT and deletant products were similar, the diagnostic PCR was performed using one of the external primers and the primer located inside the marker gene. In those cases, the deletants gave the PCR product of the expected size while no product was present in non-deletants.
Deletions of AN7909.4, AN7911.4, and AN7912.4 were further verified by Southern hybridizations using the method of Oakley et al.32 DNA from transformants was prepared using DNeasy Plant Mini Kits (Qiagen). Hybridizations were with radioactively-labeled transforming fragments. In each case at least two transformants carrying the correct gene replacement and no additonal insertions of transforming DNA were identified and used for further study.
Fermentation and LC/MS analysis
Aspergillus nidulans wild type strain R153 was cultivated in 20 different conditions. Unless otherwise noted, all conditions were at 37 °C and 250 rpm, for 4 days: #1 glucose minimal media (10 g glucose, 6 g NaNO3, 0.52 g KCl, 0.52 g MgSO4, 1.52 g KH2PO4, 1 mL trace element solution, 1 L H2O); #2 lactose minimal media (as #1, except for lactose as the sugar source); #3 arabinose minimal media (as #1, except for arabinose as the sugar source); #4 xylose minimal media (as #1, except for xylose as the sugar source); #5 ammonium-based media (as #2, except for ammonium sulfate as the nitrogen source); #6 RPMI media (RPMI minimal media purchased from GIBCO); #7 rice media (300 g brown rice boiled and filtered, 1 L H2O); #8 potato glucose media (300 g dried potatoes boiled and filtered, 20 g glucose, 1 mL trace element solution, 1 L H2O); #9 YEP media (10 g yeast extract, 20 g peptone, 20 g glucose, 1 mL trace element solution, 1 L H2O); #10 lactose minimal media with 5% DMSO; #11 lactose minimal media, no shaking; #12 YAG plates (5 g yeast extract, 20 g glucose, 15 g agar, 1 mL trace element solution, 1 L H2O); #13 YAG plates adjusted to pH = 4 with HCl; #14 YAG plates adjusted to pH = 9 with NaOH; #15 YES plates (20 g yeast extract, 120 g sucrose, 20 g agar, 1 L H2O); #16 glucose media without trace element solution; #17 Czapek’s media (3.0 g L−1 NaNO3, 0.50 g L−1 KCl, 0.50 g L−1 MgSO4 7H2O, 1.0 g L−1 K2HPO4, 30 g L−1 sucrose, 1 ml L−1 trace element solution); #18 Czapek’s yeast agar (Czapek’s media with 15 g agar); #19 Czapek’s media, no shaking; #20 Czapek’s media, no shaking, room temperature, two weeks.
For LC-DAD-MS analysis culture medium was collected and extracted twice with a volume of EtOAc equal to the culture volume. The combined EtOAc layers were evaporated in vacuo, redissolved in 1 mL of MeOH and 10 μL was injected for HPLC-DAD-MS analysis. LC/MS was carried out using a ThermoFinnigan LCQ Advantage ion trap mass spectrometer with a RP C18 column (Alltech Prevail; 2.1 × 100 mm with a 3 μm particle size) at a flow rate of 125 μl min−1. The HPLC solvent gradient was 95% MeCN–H2O (solvent B) in 5% MeCN–H2O (solvent A) both containing 0.05% formic acid: 0% B from 0 to 5 min, 0 to 100% B from 5 to 35 min, 100% B from 35 to 40 min, 100 to 0% B from 40 to 45 min, and re-equilibration with 0% B from 45 to 50 min. Spores of LO2026 (the control strain) or three strains of each gene deletant AN7901.4Δ through AN7915.4Δ (for individual strain numbers, see Table S2†) were individually inoculated (5 × 106/mL) into 150 mL Czapek minimal medium (3.0 g L−1 NaNO3, 0.50 g L−1 KCl, 0.50 g L−1 MgSO4·7H2O, 1.0 g L−1 K2HPO4, 30 g L−1 sucrose, 1 ml L−1 trace element solution) supplemented when necessary with pyridoxine (0.5 μg ml−1) or uracil (1 mg ml−1) and uridine (10 mM) and cultivated at room temperature with no shaking. (To prepare the trace metal solution, 2.2 g ZnSO4·H2O, 1.1 g H3BO3, 0.5 g MnCl2·H2O, 0.5 g FeSO4·H2O, 94.5 mg CoCl2, 0.16 g CuSO4·5H2O, 0.11 g (NH4)6Mo7O24·4H2O, and 5.0 g Na4EDTA were added in that order to 80 mL H2O, dissolving each completely before adding the next. The solution was heated to boiling, cooled to 60 °C, and the pH was adjusted to 6.5 using KOH pellets. The solution was allowed to cool to room temperature, and the volume was adjusted to 100 mL.) After 2 weeks, culture medium was collected by filtration, acidified with concentrated HCl to a pH of 2, and extracted with the same volume of EtOAc two times. The combined EtOAc layers were evaporated in vacuo and re-dissolved in MeOH (1 mg ml−1) for HPLC-DAD-MS analysis.
For structural elucidation, the LO2026 (stcJΔ), LO2517 (stcJΔ/AN7911.4Δ), and LO2525 (stcJΔ/AN7912.4Δ) strains were each cultivated in 12 aliquots of 150 mL Czapek media, acidified with concentrated HCl to a pH of 2, extracted twice with EtOAc, and evaporated, then applied to a Sep Pak cartridge to remove residual salts. The crude materials were separated by preparative HPLC [Phenomenex Luna 5 μm C18 (2), 250 × 21.2 mm] with a flow rate of 5.0 ml min−1 and measured by a UV detector at 250 nm. The gradient system for orsellinic acid (1), F-9775A (2), and B (3) was MeCN (solvent B) in 5% MeCN–H2O (solvent A), both containing 0.05% TFA: 10 to 40% B from 0 to 40 min, 40 to 100% B from 40 to 41 min, 100% B from 41 to 46 min, 100% B to 10% B from 46 to 47 min, and re-equilibration with 10% B from 47 to 52 min (tR = 15.3, 30.1, and 24.4, min, respectively). The gradient system for gerfelin (4), the C10-deoxy derivative of gerfelin (5), and diorcinol (6) was 25 to 55% B from 0 to 40 min, 55 to 100% B from 40 to 41 min, 100% B from 41 to 46 min, 100% B to 25% B from 46 to 47 min, and re-equilibration with 25% B from 47 to 52 min (tR = 18.6, 26.3, and 24.3 min, respectively).
Orsellinic acid (1) and gerfelin (4) were further eluted (9 : 1 : 0.1 dichloromethane : methanol : glacial acetic acid) through a small plug of silica gel to yield 6.7 and 19.5 mg, respectively, of isolated product. The C10-deoxy derivative of gerfelin (5) was further purified using preparatory TLC (9 : 1 : 0.1 dichloromethane : methanol : glacial acetic acid; 8.0 mg). 2 (4.7 mg), 3 (3.9 mg), and diorcinol (29 mg) required no further isolation after HPLC. The amounts represent the following titers: 1, 3.7 mg L−1; 2, 2.6 mg L−1; 3, 2.2 mg L−1; 4, 10.8 mg L−1; 5, 4.4 mg L−1; 6, 16.1 mg L−1)
Orsellinic acid (1)
Colorless needles; mp 188–189 °C (lit.33 mp 186–189 °C; IR 3291, 1607, 1599, 1305, 1196, 1164 cm−1; For UV-Vis and ESIMS spectra, see Fig. S3†; 1H NMR (CD3OD, Fig. S4†): δ = 2.48 (3H, br s), 6.13 (1H, d, J = 2.4 Hz), 6.18 (1H, br d, J = 2.4 Hz); 13C NMR (CD3OD, Fig. S5†): δ = 24.5, 101.7, 105.9, 112.3, 145.4, 163.7, 167.1, 175.3. 1H and 13C NMR data (CDOD3), in good agreement with the commercial source (Alfa Aesar).
F-9775A (2)
Yellow powder; IR 3306, 1733, 1637, 1530, 1512, 1322, 1230, 1084; 1H NMR (DMSO-d6): δ = 1.74 (3H, s), 2.14 (3H, s), 2.24 (3H, s), 3.36 (1H, s), 6.07 (1H, br s), 6.39 (1H, br s), 6.46 (1H, br s), 6.84 (1H, s), 10.0 (1H, s); 13C NMR (DMSO-d6): δ = 19.6, 20.5, 28.6, 43.9, 59.4, 87.2, 107.4, 114.6, 119.2, 127.4, 131.3, 131.6, 133.8, 139.8, 145.6, 148.1, 154.4, 174.4, 174.9, 194.6, 199.4.
F-9775B (3)
Yellow powder; IR 3306, 1732, 1638, 1527, 1512, 1322, 1230, 1084; 1H NMR (DMSO-d6): δ = 1.76 (3H, s), 2.13 (3H, s), 2.23 (3H, s), 3.46 (1H, s), 6.05 (1H, br s), 6.29 (1H, br s), 6.42 (1H, br s), 6.75 (1H, s), 9.92 (1H, s); 13C NMR (DMSO-d6): δ = 19.4, 20.7, 29.0, 44.1, 59.3, 87.3, 107.4, 113.5, 118.9, 125.8, 130.9, 131.4, 135.4, 141.5, 145.4, 148.8, 155.1, 174.6, 174.9, 194.2, 199.7.
Gerfelin (4)
White powder; IR 3189, 1666, 1599, 1446, 1257, 1204, 1150 cm−1; For UV-Vis and ESIMS spectra, see Fig. S3†; 1H NMR (DMSO-d6, Fig. S12†): δ = 2.13 (3H, br s), 2.39 (3H, br s), 6.02 (1H, d, J = 2.4 Hz), 6.24 (1H, br d, J = 2.4 Hz), 6.27 (1H, br d, J = 1.6 Hz), 6.53 (1H, br d, J = 1.6 Hz), 8.47 (1H,s), 9.44 (1H, s); 13C NMR (DMSO-d6, Fig. S13†): δ = 20.4, 22.8, 100.7, 109.2, 110.3, 113.1, 113.5, 128.0, 135.4, 141.8, 141.8, 147.0, 161.4, 162.6, 172.4. 1H and 13C NMR data (DMSO), in good agreement with the published data.34
2-Hydroxy-4-(3-hydroxy-5-methylphenoxy)-6-methylbenzoic acid (5)
White powder; IR 3392, 1698, 1687, 1208, 1142 cm−1; For UV-Vis and ESIMS spectra, see Fig. S3†; 1H NMR (DMSO-d6, Fig. S14†): δ = 2.21 (3H, br s), 2.38 (3H, br s), 6.20 (1H, d, J = 2.4 Hz), 6.23 (1H, br s), 6.32 (1H, br s), 6.33 (1H, d, J = 2.4 Hz), 6.43 (1H, br s), 9.59 (1H, s); 13C NMR (DMSO-d6, Fig. S15†): δ = 21.1, 22.3, 102.7, 104.0, 111.0, 111.4, 111.4, 112.3, 140.5, 141.7, 156.0, 158.6, 160.0, 161.9, 171.8. 1H and 13C NMR data (DMSO-d6), were similar with the published data, but there were discrepancies of as much as 0.25 ppm for 1H NMR and 4.8 ppm for 13C NMR.35 We performed additional NMR experiments (DEPT, HMQC, HMBC, COSY), which corroborated our assignment of 5 as 2-hydroxy-4-(3-hydroxy-5-methylphenoxy)-6-methylbenzoic acid.
Diorcinol (6)
Light orange oil; IR 3365, 1616, 1601, 1320, 1149, 1039, 835 cm−1; For UV-Vis and ESIMS spectra, see Fig. S3†; 1H NMR (CDCl3, Fig. S16†): δ = 2.24 (3H, br s), 6.29 (1H, br s), 6.40 (2H, br s); 13C NMR (CDCl3, Fig. S17†): δ = 21.4, 103.4, 111.2, 112.2, 141.0, 156.3, 157.9; 1H NMR (CDOD3, Fig. S18†): δ = 2.22 (3H, br s), 6.19 (1H, br s), 6.26 (1H, br s), 6.35 (1H, br s); 13C NMR (CDOD3, Fig. S19†): δ = 21.7, 104.3, 111.9, 112.1, 141.8, 159.7, 159.8. 1H and 13C NMR data (CDCl3 and CDOD3), in good agreement with the published data.29,36
Supplementary Material
Acknowledgements
The project described was supported in part by NSF grant MCB-0236393 to N.P.K., and Grants R01GM031837 to B.R.O and PO1GM084077 from the National Institute of General Medical Sciences to N. P. K., B. R. O., and C. C. C. W.
Biography

Clay C. C. Wang
Clay Wang was born in Tokyo, Japan, in 1972 and grew up in Taipei, Taiwan. He completed his undergraduate degree in Chemistry from Harvard University in 1996 working in the laboratory of Professor George M. Whitesides and his PhD in Chemistry from California Institute of Technology in 2001 in the laboratory of Professor Peter B. Dervan. He was an NIH-NRSA fellow with Professor Chaitan Khosla at Stanford University. He started his own independent research program at the University of Southern California in 2003. He is currently an Associate Professor at the Department of Pharmacology and Pharmaceutical Sciences and at the Department of Chemistry. His research interest is in natural product biosynthesis, metabolic engineering, and secondary metabolism regulation. He is a recipient of an American Cancer Society Research Scholarship.
Footnotes
Electronic supplementary information (ESI) available: Primers and A. nidulans strains used in this study; NMR spectra; EIC data. See DOI: 10.1039/b904541d
This article is part of the 2009 Molecular BioSystems ‘Emerging Investigators’ issue: highlighting the work of outstanding young scientists at the chemical- and systems-biology interfaces.
References
- 1.Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 2.Payne GA, Nystrom GJ, Bhatnagar D, Cleveland TE, Woloshuk CP. Appl. Environ. Microbiol. 1993;59:156–162. doi: 10.1128/aem.59.1.156-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, Df’Enert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 4.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 5.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O’Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 6.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hom-bergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 7.Bingle LE, Simpson TJ, Lazarus CM. Fungal Genet. Biol. 1999;26:209–223. doi: 10.1006/fgbi.1999.1115. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson TP, Rudd BA, Dawson M, Lazarus CM, Simpson TJ, Cox RJ. Chem. Biol. 2001;8:157–178. doi: 10.1016/s1074-5521(00)90064-4. [DOI] [PubMed] [Google Scholar]
- 9.Cox RJ. Org. Biomol. Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 10.Sato S, Morishita T, Hosoya T, Ishikawa Y. JP 11001480A Japan Pat. 1999
- 11.Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaucher GM, Shepherd MG. Biochem. Biophys. Res. Commun. 1968;32:664–671. doi: 10.1016/0006-291x(68)90290-8. [DOI] [PubMed] [Google Scholar]
- 14.Bailey AM, Cox RJ, Harley K, Lazarus CM, Simpson TJ, Skellam E. Chem. Commun. 2007:4053–4055. doi: 10.1039/b708614h. [DOI] [PubMed] [Google Scholar]
- 15.He J, Wijeratne EM, Bashyal BP, Zhan J, Seliga CJ, Liu MX, Pierson EE, Pierson LS, 3rd, VanEtten HD, Gunatilaka AA. J. Nat. Prod. 2004;67:1985–1991. doi: 10.1021/np040139d. [DOI] [PubMed] [Google Scholar]
- 16.Oh DC, Kauffman CA, Jensen PR, Fenical W. J. Nat. Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 17.Marquez-Fernandez O, Trigos A, Ramos-Balderas JL, Viniegra-Gonzalez G, Deising HB, Aguirre J. Eukaryotic Cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bode HB, Bethe B, Hofs R, Zeeck A. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 19.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CC. Chem. Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Chem. Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 22.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 23.Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. J. Bacteriol. 1997;179:6271–6278. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii Y, Narimatsu Y, Iwasaki Y, Arai N, Kino K, Kirimura K. Biochem. Biophys. Res. Commun. 2004;324:611–620. doi: 10.1016/j.bbrc.2004.09.091. [DOI] [PubMed] [Google Scholar]
- 25.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. Fungal Genet. Biol. 2001;32:121–133. doi: 10.1006/fgbi.2001.1256. [DOI] [PubMed] [Google Scholar]
- 26.Zenitani S, Tashiro S, Shindo K, Nagai K, Suzuki K, Imoto M. J. Antibiot. 2003;56:617–621. doi: 10.7164/antibiotics.56.617. [DOI] [PubMed] [Google Scholar]
- 27.Kawatani M, Okumura H, Honda K, Kanoh N, Muroi M, Dohmae N, Takami M, Kitagawa M, Futamura Y, Imoto M, Osada H. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11691–11696. doi: 10.1073/pnas.0712239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fremlin LJ, Piggott AM, Lacey E, Capon RJ. J. Nat. Prod. 2009;72:666–670. doi: 10.1021/np800777f. [DOI] [PubMed] [Google Scholar]
- 29.Itabashi T, Nozawa K, Nakajima S, Kawai K. Chem. Pharm. Bull. 1993;41:2040–2041. [Google Scholar]
- 30.Bunyapaiboonsri T, Yoiprommarat S, Intereya K, Kocharin K. Chem. Pharm. Bull. 2007;55:304–307. doi: 10.1248/cpb.55.304. [DOI] [PubMed] [Google Scholar]
- 31.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Nat. Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- 32.Oakley CE, Weil CF, Kretz PL, Oakley BR. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- 33.Harris TM, Murphy GP, Poje AJ. J. Am. Chem. Soc. 1976;98:7733–7741. [Google Scholar]
- 34.Islam MS, Kitagawa M, Imoto M, Kitahara T, Watanabe H. Biosci., Biotechnol., Biochem. 2006;70:2523–2528. doi: 10.1271/bbb.60287. [DOI] [PubMed] [Google Scholar]
- 35.Ivanova V, Kolarova M, Aleksieva K, Graefe U, Schlegel B. Prep. Biochem. Biotechnol. 2007;37:39–45. doi: 10.1080/10826060601039436. [DOI] [PubMed] [Google Scholar]
- 36.Takenaka Y, Tanahashi T, Nagakura N, Hamada N. Chem. Pharm. Bull. 2003;51:794–797. doi: 10.1248/cpb.51.794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


