Abstract
The epidermal growth factor receptor (EGFR) is a central regulator of proliferation and progression in human cancers. Five EGFR inhibitors, two monoclonal antibodies and three TKIs, have recently gained FDA approval in oncology (cetuximab, panitumumab, erlotinib, gefitinib and lapatinib). These strategies of EGFR inhibition demonstrate major tumor regressions in approximately 10–20% of advanced cancer patients. However, many tumors eventually manifest acquired resistance to treatment. In this study we established and characterized a model to study molecular mechanisms of acquired resistance to the EGFR monoclonal antibody cetuximab. Using high-throughput screening we examined the activity of 42 receptor tyrosine kinases in resistant tumor cells following chronic exposure to cetuximab. Cells developing acquired resistance to cetuximab exhibited increased steady-state EGFR expression secondary to alterations in trafficking and degradation. In addition, cetuximab-resistant cells manifested strong activation of HER2, HER3 and cMET. EGFR upregulation promoted increased dimerization with HER2 and HER3 leading to their transactivation. Blockade of EGFR and HER2 led to loss of HER3 and PI(3)K/Akt activity. These data suggest that acquired-resistance to cetuximab is accompanied by dysregulation of EGFR internalization/degradation and subsequent EGFR-dependent activation of HER3. Taken together these findings suggest a rationale for the clinical evaluation of combinatorial anti-HER targeting approaches in tumors manifesting acquired resistance to cetuximab.
Keywords: EGFR, cetuximab, acquired-resistance
INTRODUCTION
The epidermal growth factor receptor (EGFR) is a member of the HER family of receptor tyrosine kinases and consists of four members; EGFR (ErbB1/HER1), HER2/neu (ErbB2), HER3 (ErbB3) and HER4 (ErbB4). Stimulation of the receptor through ligand binding activates the intrinsic receptor tyrosine kinase and promotes receptor homo- or heterodimerization with HER family members. EGFR activation leads to downstream stimulation of several signaling cascades, including MAPK and PI(3)K/Akt that influence cell proliferation, angiogenesis, invasion and metastasis(Citri and Yarden, 2006). Aberrant expression or activity of EGFR is identified in many human epithelial cancers.
Targeting EGFR has been intensely pursued in the last decade as a cancer treatment strategy. One approach uses monoclonal antibodies (mAbs) to target the extracellular domain of the EGFR to block natural ligand binding (Mendelsohn, 2003). Cetuximab (IMC-C225, Erbitux) prevents receptor activation and dimerization and ultimately induces receptor internalization and downregulation (Sunada et al., 1986). Cetuximab exhibits promising anti-tumor activity as monotherapy or in combination with chemotherapy and/or radiation in head and neck cancer and metastatic colorectal cancer (Baselga and Arteaga, 2005; Bonner et al., 2006; Cunningham et al., 2004; Mendelsohn and Baselga, 2006). A second approach involves the use of small molecule tyrosine kinase inhibitors (TKIs) that bind to the ATP-binding site in the tyrosine kinase domain (TKD) of the EGFR and HER2. Three anti-EGFR TKIs, erlotinib (OSI-774, Tarceva), gefitinib (ZD1839, Iressa) and lapatinib (GW572016, Tykerb) are approved by the FDA for use in oncology.
Both approaches to EGFR inhibition show considerable clinical promise. However, increasing evidence suggests that patients who initially respond to EGFR inhibitors may subsequently become refractory (Pao et al., 2005). Therefore, an improved understanding of molecular mechanisms of acquired resistance to EGFR inhibitors may provide valuable leads to enhance the efficacy of this class of agents. The identification of catalytic domain EGFR mutations that predict response to EGFR-TKIs in selected lung cancer patients represents a landmark development in the EGFR field (Sequist et al., 2007). Although EGFR TKD mutations appear to correlate with response to the TKIs erlotinib and gefitinib, no such correlation exists for cetuximab response (Mukohara et al., 2005).
To study mechanisms of acquired resistance to cetuximab, we established a series of cetuximab-resistant clones in vitro following long-term exposure to cetuximab in NSCLC (H226) and HNSCC (SCC-1) cell lines. Following establishment of stable clones, we performed high-throughput screening to examine the activity of 42 membrane receptor tyrosine kinases (RTKs). Through comparative analysis of cetuximab-resistant versus parental lines, we identified that EGFR along with HER2, HER3 and cMET are all highly activated in the resistant clones. Further studies suggest that acquired resistance to cetuximab reflects dysregulation of EGFR internalization/degradation and subsequent EGFR-dependent activation of HER3.
RESULTS
Establishment of cetuximab-resistant lines
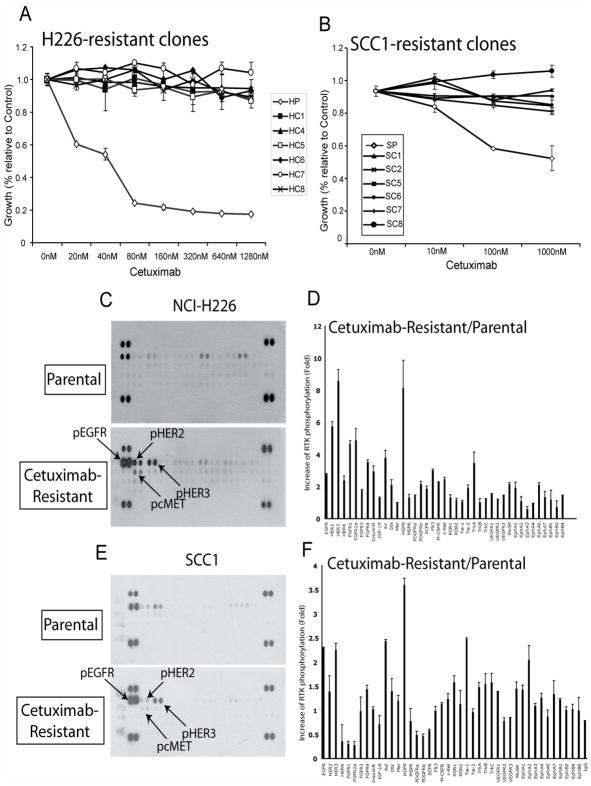
We established cetuximab resistant tumor cell lines using the human NSCLC line NCI-H226 (H226) and the HNSCC line UMSCC-1 (SCC1) to use as a model system to elucidate molecular mechanisms of acquired-resistance to cetuximab. These lines were chosen based on three primary criteria; 1) Cetuximab is used in therapy for both tumor types, 2) the cell lines are sensitive to cetuximab and 3) the cell lines have no TKD mutations. To generate resistant lines, H226 and SCC1 cells were continuously exposed to increasing concentrations of cetuximab over six months. Following the development of heterogeneous populations of cetuximab-resistant cells we isolated individual subclones of cetuximab-resistant lines. This process resulted in six stable resistant clones for the H226 NSCLC line designated HC1, HC4, HC5, HC6, HC7 and HC8. The sensitive parental line was designated HP. For the SCC1 HNSCC line six stable resistant clones were generated (SC1, SC2, SC5, SC6, SC7, SC8). As shown in Figure 1A, all HC clones displayed a robust cetuximab-resistant phenotype when challenged with increasing concentrations of cetuximab as compared to parental controls. Similar results were observed with the SCC1 cetuximab-resistant clones (Figure 1B). Sequence analysis of the EGFR TKD in H226 cells after the establishment of resistant clones indicated no mutations developed during the selection process in either the resistant or parental cells (data not shown).
Figure 1. phospho-receptor tyrosine kinase (RTK) array in NSCLC H226 and HNSCC SCC1 cells demonstrate upregulation of EGFR, HER2, HER3 and cMET.
A: Cetuximab dose response curve using NSCLC line NCI-H226 cetuximab-resistant clones. Cells were treated with the indicated concentration of cetuximab and growth was measured using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Data points are represented as mean ±SEM (n=3).
B: Cetuximab dose response curve using HNSCC line SCC-1 cetuximab-resistant clones. Cells were treated with the indicated concentration of cetuximab and growth was measured using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Data points are represented as mean ±SEM (n=3).
C-F: HER family members have increased activity in HNSCC and NSCLC cetuximab-resistant cells. Cetuximab-sensitive (Parental) and cetuximab-resistant cells were grown to confluence followed by protein extraction. The cell extracts were incubated with membranes containing antibodies to 42 different RTKs. The membranes were washed and incubated with a pan anti-phosphotyrosine antibody to measure the levels of active receptor. Quantitation of phospho-RTK was completed using scanned images from ImageQuant software. Data points are represented as mean ±SEM (n=3).
Upregulation of EGFR and activation of HER2, HER3 and cMet
After successful establishment of cetuximab-resistant clones, we performed high-throughput comparative analyses measuring phosphorylated RTKs in the resistant vs. parental lines to test the hypothesis that acquired resistance to EGFR inhibition results from the activation of alternative RTKs that share overlapping signal transduction elements with the EGFR. To test this hypothesis, we screened the activity of a panel of activated RTKs using an antibody-based array from R&D Systems (Minneapolis, MN) as shown in Figure 1C. Following quantification of scanned images using ImageQuant software, the relative expression of specific phosphorylated RTKs between cetuximab-resistant and parental cells was determined (Figure 1D). The identical experimental approach was performed using the SCC1 cetuximab-resistant lines and parental control (Figure 1E and F). From this high-throughput screen, several phosphorylated RTKs were notably up-regulated in both cetuximab-resistant NSCLC and HNSCC tumor lines including HER family members (EGFR, HER2 and HER3) and the hepatocyte growth factor receptor (HGFR, c-MET). These results indicated that these independent tumor cell lines, chronically challenged with cetuximab, manifested highly similar patterns of altered RTK expression and or activation.
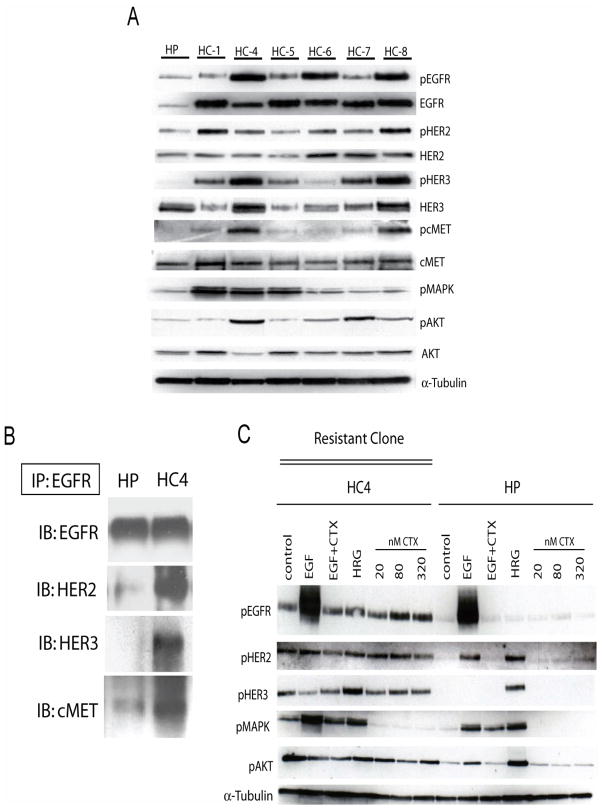
To validate results of the phospho-RTK array in individual cetuximab-resistant clones we performed standard Western blot analysis on the parental and cetuximab-resistant clones of H226 to measure levels of EGFR, HER2, HER3, cMET, and members of their downstream signaling cascades, including the phosphorylated forms of MAPK and Akt. The results demonstrated that findings from the phospho-RTK array were consistent in all of the cetuximab-resistant clones (Figure 2A). Although the activity of EGFR, HER2, HER3 and cMET was increased relative to the parental line, only EGFR steady-state expression was dramatically increased in cetuximab-resistant clones. Furthermore, analysis of EGFR binding partners using immunoprecipitation techniques indicated that EGFR displayed increased binding with HER2, HER3 and cMET as compared to the cetuximab-sensitive parental cells (Figure 2B). Taken together these data suggest that prolonged exposure to cetuximab induces upregulation of EGFR with resultant enhanced heterodimerization of EGFR with HER2, HER3 and cMET.
Figure 2. EGFR is upregulated in cetuximab-resistant cells.
A: Characterization of expression of HER family members and down stream Akt and MAPK in cetuximab-resistant clones (HC1, HC4-HC8). Protein was collected and fractionated by SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as loading control. HP; cetuximab-sensitive parental line, HC; cetuximab-resistant clones.
B: EGFR has increased association with HER2, HER3, and cMET in cetuximab-resistant cells. Cetuximab-resistant cells were harvested and EGFR was immunoprecipitated from the cetuximab-resistant clone HC4 and the parental HP cells with an anti-EGFR antibody. The immunoprecipitates were fractionated on SDS-PAGE followed by immunoblotting for the indicated proteins. HP; cetuximab-sensitive parental line, HC4; cetuximab-resistant clone.
C: Cetuximab treatment does not modulate EGFR phosphorylation in cetuximab-resistant clone (HC4). Parental cells (HP) and cetuximab-resistant clone (HC4) were treated with vehicle (control) or increasing concentrations of cetuximab (CTX) for 24 hours. Stimulation was with EGF (10ng/ml) and HRG (5μM) for 45 minutes. Protein was collected and fractionated by SDS-PAGE followed by immunoblotting for the indicated proteins. α-tubulin was used as a loading control.
The increased expression and heterodimerization activity of EGFR correlates with increased baseline activity of HER2, HER3, MAPK and Akt relative to the parental line (HP) (Figure 2C, control lanes). To examine if cetuximab treatment could inhibit the activity of EGFR and subsequent HER2 and HER3 activity we challenged cetuximab-resistant cells (clone HC4) with increasing concentrations of cetuximab (20–320nM) and measured the phosphorylation of EGFR at tyrosine 1173, relative to parental cells (HP). The results indicated that cetuximab had no effect on the autophosphorylation of EGFR or the activity of HER2, HER3 or Akt. In addition, cetuximab was able to block EGF induced activation of EGFR (Figure 2C, EGF+CTX lanes) indicating that cetuximab retains the capacity to bind the receptor and inhibit EGF binding in cetuximab-resistant cells. Furthermore, cetuximab blocked MAPK activity in cetuximab-resistant cells indicating that MAPK may play a minor role in resistance.
Impaired endocytosis of EGFR
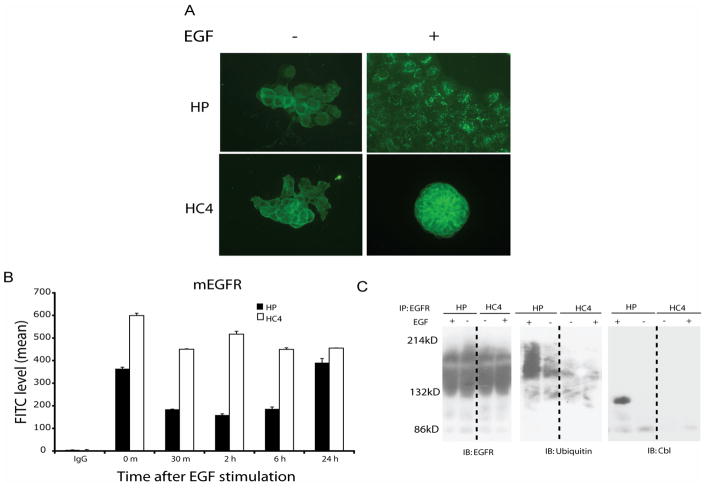
To examine underlying mechanisms of elevated EGFR expression and activation in cetuximab-resistant cells we hypothesized that alterations in receptor trafficking may be involved. To test this hypothesis we analyzed patterns of ligand-stimulated EGFR internalization in resistant cells relative to parental controls. Using immunofluorescent staining, we found an accumulation of intracellular endocytic vesicles staining for EGFR in parental cells following 45 min of EGF stimulation. However, EGFR remained in the extracellular membrane and was not internalized in cetuximab-resistant cells, as shown in Figure 3A. Furthermore, quantifying the surface EGFR by flow cytometry confirmed these findings and showed that parental lines manifested rapid internalization of the EGFR following EGF stimulation with return to baseline within 24 hours (Figure 3B). However, cetuximab-resistant clones showed no receptor internalization in response to EGF stimulation. These results suggest that cetuximab-resistant cells have impaired ability to internalize the EGFR and this mechanism may contribute to the increased expression and activity of EGFR in resistant cells.
Figure 3. Cetuximab-resistant cells have increased EGFR plasma membrane expression.
A: EGFR is internalized following EGF stimulation in parental (HP), but not in cetuximab-resistant cells (HC4). Internalization of EGFR was determined by immunofluorescent staining as described in “Experimental Procedures” using FITC-conjugated anti-EGFR antibody 45 min following EGF (25 ng/ml) stimulation. Internalized EGFR was indicated by the accumulation of intracellular endocytic vesicles staining for EGFR.
B: Cell surface EGFR is decreased following EGF stimulation in parental (HP), but not in cetuximab-resistant cells (HC4) The levels of EGFR on the cell surface were monitored following 0~24 hr of EGF (25 ng/ml) stimulation by flow cytometry as described in “Experimental Procedures”. The numbers represent mean fluorescence intensity of anti-EGFR staining after subtracting IgG background fluorescence.
C: Lack of c-Cbl recruitment and EGFR ubiquitination in cetuximab-resistant cells following EGF stimulation. Cetuximab-resistant (HC4) and parental (HP) cells were either untreated (-) or treated with EGF (25 ng/ml) for 45 min. Thereafter, cell lysates were prepared and analyzed by immunoprecipitation (IP) and immunoblotting (IB) with the indicated antibodies.
To further investigate if cetuximab-resistant cells have impaired ability to degrade EGFR, we examined Cbl-mediated ubiquitinylation of EGFR that is required for lysosomal degradation of the receptor. Upon EGF stimulation, the active EGFR recruits c-Cbl, an E3 ubiquitin ligase, to tyrosine 1045. This interaction enhances receptor ubiquitination and thus directs the receptor to lysosomal degradation (Grovdal et al., 2004). We examined this hypothesis by immunoprecipitating EGFR and found no detectable levels of c-Cbl associated with EGFR in cetuximab-resistant cells whereas the parental lines showed strong association of c-Cbl after EGF stimulation (Figure 3C). Loss of c-Cbl association with the EGFR led to significantly less EGFR ubiquitination after EGF stimulation in the cetuximab-resistant cells versus parental. Sequence analysis of the c-Cbl binding site (Y1045) indicated no mutations of it or surrounding nucleotides. However, Y1045 exhibited increased baseline and EGF stimulated phosphorylation relative to the parental lines (data not shown). Therefore loss of c-Cbl binding to the EGFR in cetuximab resistant cells must occur by other mechanisms. Together these data suggest that impaired degradation may contribute to the increased expression and activity of EGFR in cetuximab-resistant cells and may be a potential mechanism of cetuximab-resistance.
EGFR-dependent activation of HER2, HER3 and cMet
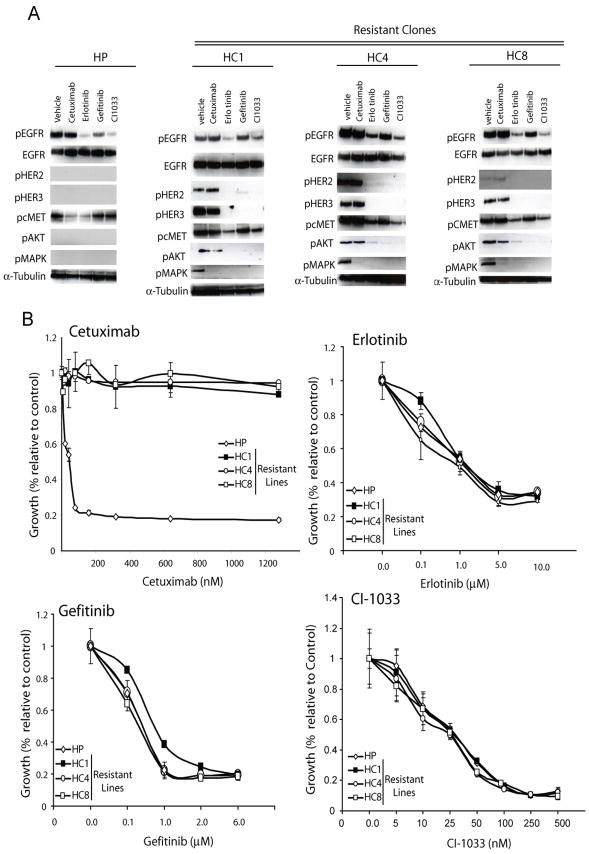
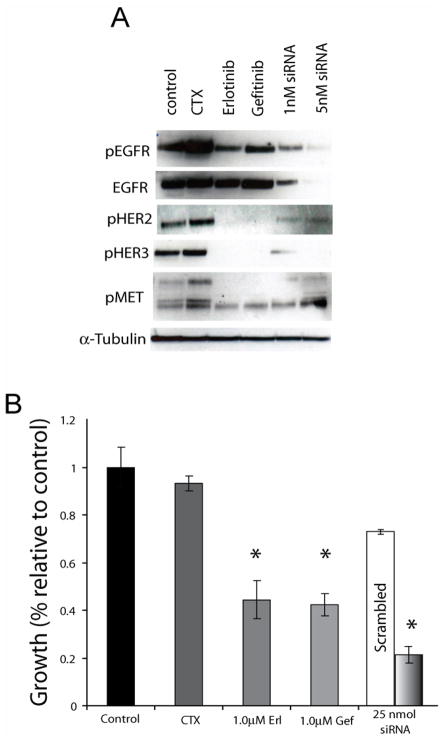
We hypothesized that impaired endocytosis of EGFR was, in part, responsible for the increased levels and subsequent activity of EGFR ultimately sending signals to proliferative and anti-apoptotic pathways. To examine this hypothesis we challenged cetuximab-resistant cells (HC1, HC4, HC8) with a battery of small molecule TKIs (Figure 4A), including erlotinib, gefitinib and the pan-HER irreversible inhibitor CI-1033. All TKIs decreased EGFR phosphorylation at tyrosine 1173. This down regulation of active EGFR correlated with a robust decrease in activity of HER2, HER3, cMET, Akt, and MAPK. Active HER2, HER3 and Akt were undetectable by Western blot in the parental lines. To determine if TKIs had an impact on proliferative potential we performed growth proliferation assays comparing parental and cetuximab-resistant clones challenged with increasing concentrations of each TKI. This treatment led to profound growth inhibitory effects in cetuximab-resistant clones, responding in parallel to their parental counterparts (Figure 4B). Recognizing the relative non-specificity of EGFR TKIs, and to examine if EGFR alone was responsible for transphosphorylation of HER2, HER3, and cMET, we used siRNAs directed against the EGFR. Cetuximab-resistant cells treated with EGFR siRNA showed a dose dependent down-regulation of HER2, HER3, and cMET activity (Figure 5A) and this correlated with a strong inhibitory impact on the proliferative potential of the cetuximab-resistant clones (Figure 5B). Taken together these data suggest that EGFR is responsible for transphosphorylation of HER2, HER3, and cMET. Furthermore, a loss of the activity of these molecules results in decreased proliferative potential in cetuximab-resistant cells.
Figure 4. TKIs retain capacity to inhibit HER signaling in cetuximab-resistant clones.
A: HER family members and downstream signaling molecules are attenuated after treatment with TKIs in cetuximab-resistant clones. Parental cells (HP) and cetuximab-resistant clones (HC1, HC4 and HC8) were treated with vehicle (DMSO), cetuximab (100nM), erlotinib (10uM), gefitinib (1uM) or CI-1033(500nM) for 24 hours. Protein lysates were fractionated on SDS-PAGE followed by Western blotting the indicated proteins. α-Tubulin was used as a loading control.
B: TKIs inhibit proliferation in cetuximab-resistant clones Parental cells (HP) and cetuximab-resistant clones (HC1, HC4, HC8) were plated and allowed to adhere overnight. Growth was measured at 72 hours after drug treatment using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Treatment groups were cetuximab, erlotinib, gefitinib, CI-1033. Data points are represented as mean ±SEM (n=6).
Figure 5. HER2 and HER3 activity in cetuximab-resistant cells is EGFR-dependent.
A: Effects of EGFR knockdown on HER2 and HER3 activity. HC4 cells were plated and treated with the vehicle DMSO (control), 1μM erlotinib, 1μM gefitinib, 1nM of EGFR siRNA, 1 or 5 nM EGFR siRNA. 48 hours later protein was colleted and fractionated by SDS-PAGE and immunoblotted for the indicated proteins.
B: Effects of EGFR knockdown on proliferation of cetuximab-resistant cells. Cetuximab-resistant clone HC4 was plated and treated with control (DMSO), 100nM cetuximab, 1 μM erlotinib, 1 μM gefitinib, or 25 nM of EGFR siRNA. 72 hours later growth proliferation was measured as outlined in the materials and methods. *, P<0.05
Inhibition of cMET activity in cetuximab-resistant cells
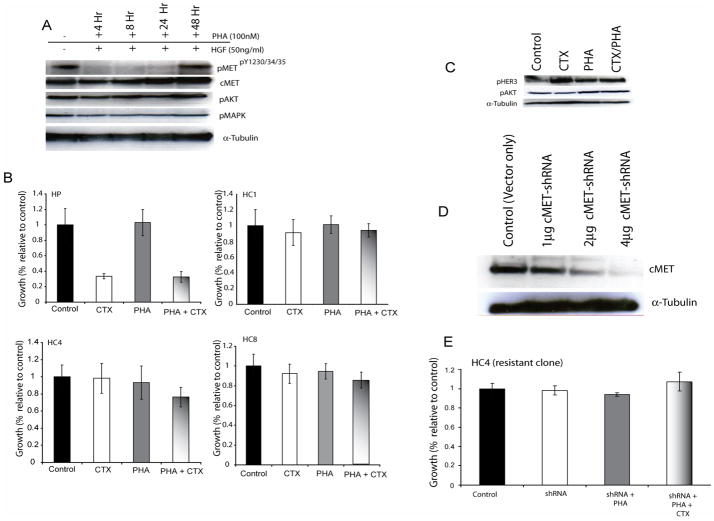
Active cMET was detected in our preliminary studies examining alternative RTK activation in cells with acquired resistance to cetuximab (Figure 1C) and these results were further verified by analysis of individual cetuximab-resistant clones for cMET activity (Figure 2A). To examine if increased cMET activity resulted in cetuximab resistance we used a selective small molecule cMET inhibitor, PHA665752 to inhibit cMET activity (figure 6A) (Christensen et al., 2003). Cetuximab-resistant cells (clone HC4) were treated with PHA and stimulated HGF prior to protein collection. PHA665752 exhibited potent inhibition of cMET autophosphorylation (Y1230/1234/1235) but had no effect on downstream signaling molecules Akt and MAPK activity. To determine the effects of PHA665752 on proliferation we performed growth proliferation assays on cetuximab-resistant clones using PHA665752 alone or in combination with cetuximab (Figure 6B). The parental cells responded briskly to cetuximab challenge but had no response to PHA665752. Furthermore, all cetuximab-resistant clones showed cetuximab resistance and no response to either PHA665752 alone or in combination with cetuximab. This combination treatment had no effect on the activity of HER3 or Akt (Figure 6C). To expand upon these series of tests we designed small hairpin RNA (shRNA) vectors directed against the cMET open reading frame to knockdown the expression levels of cMET in cetuximab-resistant cells (Figure 6D). Growth proliferation assays were performed after transfection of the cMET-shRNA expression vectors (Figure 6E). Cells were transfected with the expression vector and treated with PHA665752 or in combination with cetuximab. The loss of cMET expression did not affect the proliferative potential of cetuximab-resistant cells, nor re-sensitize cells to the anti-proliferative effect of cetuximab. Taken together these results suggest that despite upregulation, cMET activity does not significantly contribute to the ability of cetuximab-resistant clones to proliferate in the presence of cetuximab.
Figure 6. Inhibition of cMET activity does not re-sensitize cetuximab-resistant cells to cetuximab.
A: PHA665752 inhibits cMET activity without affecting Akt or MAPK signaling. Cetuximab-resistant clone HC4 was treated with PHA665752 for 4, 8, 24 or 48 hour. Cells were then stimulated with HGF for 45 minutes. Western blot shows inhibition of cMET activity up to 24 hours when treated with PHA665752.
B: PHA665752 alone or in combination with cetuximab does not inhibit growth of cetuximab- resistant cells. Growth was measured at 72 hours after drug treatment using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Treatment groups were cetuximab (CTX), PHA665752 (PHA) or the combination of the two drugs. HP; cetuximab-sensitive parental line, HC; cetuximab-resistant clones. Data points are represented as mean ±SEM (n=3).
C: Western blotting of cetuximab-resistant clone (HC4) cells after treatment with cetuximab, PHA665752 or the combination. Cetuximab-resistant clone HC4 was treated with cetuximab (100nM), PHA665752 (100nM) or the combination. 24 hours later protein was collected and fractionated by SDS-PAGE and immunoblotted for the indicated proteins.
D: Western blotting of cetuximab-resistant clone (HC4) cells after transient transfection with cMET shRNA vectors. Control is empty vector control. 1, 2, and 4 micrograms of vector were transfected. Cells were collected 48 hours after treatment. 48 hours after transfection protein was collected and fractionated by SDS-PAGE and immunoblotted for the indicated proteins.
E: Loss of cMET does not affect growth of cetuximab-resistant cells (HC4). Cells were transfected with cMET shRNA vectors followed by treatment with vehicle (shRNA only), PHA665752 (shRNA+PHA group) or PHA665752 or cetuximab (shRNA+PHA+CTX group). Growth was measured at 72 hours after treatment using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Data points are represented as mean ±SEM (n=6).
Inhibition of HER2 or HER3 activity
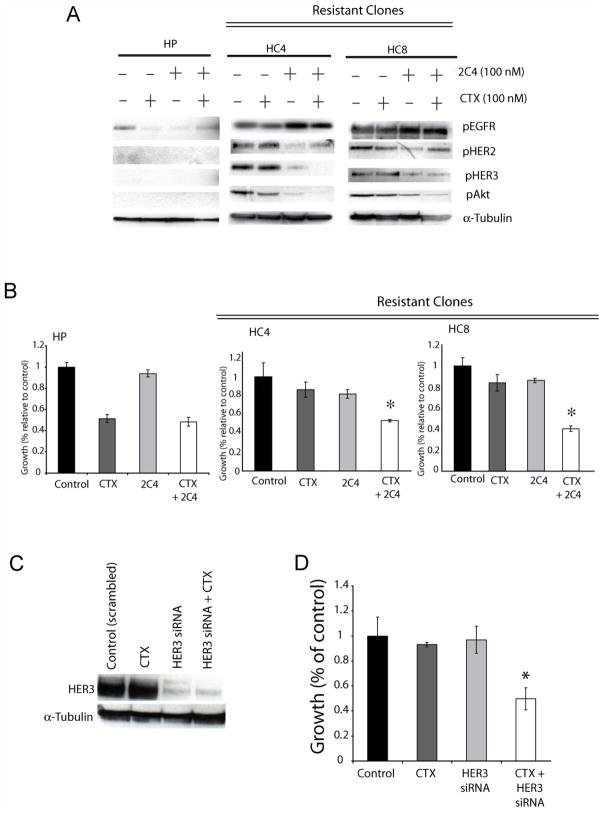
To further elucidate the roles of HER family members in acquired resistance to cetuximab we examined combinations of HER targeting agents. HER2 and HER3 signal primarily when heterodimerized to other HER family members, and HER2 is a preferred heterodimerization partner for HER3 (Citri et al., 2003). Therefore, we hypothesized that HER2/HER3 heterodimers may contribute to the cetuximab-resistant phenotype. To test this hypothesis we used 2C4, a monoclonal antibody that inhibits HER2 heterodimerization (Franklin et al., 2004). Parental and cetuximab-resistant cells (clone HC4 and HC8) were treated with cetuximab, 2C4 or the combination. Cetuximab alone had modest effects on HER2, HER3 or Akt activity whereas 2C4 treatment decreased activity of HER2, HER3, and Akt in resistant cells (Figure 7A). However, combining cetuximab with 2C4 a more potent suppression of HER3 and Akt was observed and this correlated with strong antiproliferative effects (Figure 7B). These data suggest that HER3 activity, dependent on EGFR and HER2, promotes survival in cetuximab-resistant cells.
Figure 7. Inhibition of HER2 or HER3 activity sensitizes cetuximab-resistant cells.
A: Combinatorial treatment of cetuximab-resistant clones HC4 and HC8 with cetuximab and 2C4 leads to loss of HER3 and AKT activity. Parental cells (HP) and cetuximab-resistant clones HC4 and HC8 were treated with cetuximab (CTX), 2C4 (2C4) or the combination. Protein lysates were fractionated on SDS-PAGE followed by Western blotting for the indicated proteins. α-Tubulin was used as a loading control.
B: Combinatorial treatment of cetuximab-resistant clones with cetuximab and 2C4 leads to growth inhibition. Parental cells (HP) and cetuximab-resistant clones (HC4, HC8) were plated and allowed to adhere overnight. Growth was measured at 72 hours after drug treatment using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Treatment groups were cetuximab (100nM), 2C4 (100nM) or the combination. Data points are represented as mean ±SEM (n=6). *, P<0.05
C: Western blotting of cetuximab-resistant clone (HC4) cells after transient transfection with HER3 siRNA. Treatments are: Scrambled siRNA (50nM), Cetuximab (100nM), HER3 siRNA (50nM) or the combination. 48 hours after treatment protein was collected and fractionated by SDS-PAGE and immunoblotted for the indicated proteins.
D: HER3 knockdown re-sensitizes cetuximab-resistant cells to cetuximab. Cells were transfected with scrambled (50nM, control), 100nM cetuximab, HER3 (50nM) siRNA in combination with 100nM cetuximab. The cells were collected 48 hours later for Western blot analysis of the indicated proteins. Growth was measured at 72 hours after treatment using the growth proliferation assay as described in the experimental procedures and plotted as a percentage of growth relative to the untreated control cells. Data points are represented as mean ±SEM (n=6). *, P<0.05
We find HER3 to be highly activated in our RTK array screening evaluation of cetuximab resistant cells (Figure 1C). Indeed, recent reports suggest that transactivation of HER3 is important in conferring resistance to EGFR inhibitors (Engelman et al., 2007; Sergina et al., 2007). To examine whether the loss of HER3 could re-sensitize cetuximab-resistant cells to cetuximab we used siRNAs against HER3 (Figures 7C and 7D). These findings indicate that the loss of HER3 can re-sensitize cetuximab-resistant cells to cetuximab therapy. Taken together these results indicate that cells may overcome EGFR inhibition by cetuximab through transactivation of HER2 and HER3 in an EGFR-dependent matter, and that HER3 signaling may represent a critical signaling mediator in acquired resistance to cetuximab therapy.
DISCUSSION
The development of acquired resistance to cancer therapeutic drugs represents a major obstacle to successful treatment. Tempering promise for the EGFR molecular targeting agents is the emergence of reports that illustrate acquired resistance. It is therefore important to understand mechanisms of acquired resistance to anti-EGFR therapy in an effort to avoid, overcome or diminish this phenomenon. In the current study, we investigate mechanisms of acquired resistance to cetuximab using newly established cetuximab-resistant tumor clones following long-term exposure to cetuximab. Using high-throughput screening to examine the activity of 42 RTKs, we found that EGFR, HER2, HER3 and cMET were highly activated in cetuximab-resistant clones derived from either HNSCC or NSCLC lines (Benavente et al., 2004; Gondi et al., 2006). We present data to suggest that acquired resistance to cetuximab reflects dysregulation of EGFR internalization/degradation and subsequent EGFR-dependent activation of HER2 and HER3. Furthermore, it appears that HER3 activity, dependent on EGFR and HER2, represents a critical step for cells to escape from cetuximab inhibition.
Although acquired resistance to TKI therapy has been attributed to mutations in the cytoplasmic tail of the EGFR, no such correlation between EGFR mutation and cetuximab resistance exists. Our results implicate a novel EGFR-dependent mechanism in acquired resistance to EGFR-targeted monoclonal antibody therapy. Evidence suggests that loss of c-Cbl-regulated EGFR degradation contributes to EGFR overexpression in cancer cells. Several recent reports directly implicate c-Cbl in regulation of the EGFRvIII mutant (Grandal et al., 2007; Han et al., 2006). These reports demonstrate that hypophosphorylation of residue Y1045 of the EGFRvIII leads to defective downregulation mediated by c-Cbl. Other studies examining EGFR TKD mutants in NSCLC indicate that the TKI-sensitizing mutation EGFRΔ746–750 reduces phosphorylation at Y1045 and may in part be responsible for increased signaling from this mutant receptor (Furukawa et al., 2007). Consistent with these findings our data suggests that cells chronically exposed to cetuximab have dysregulated c-Cbl-mediated EGFR degradation (Figure 3C). These results suggest that increased expression and activity of the EGFR in cetuximab-resistant cells may be caused, in part, by dysregulated receptor internalization and sorting.
Increasing evidence identifies cMET as an attractive target for molecular-targeted cancer therapy (Peruzzi and Bottaro, 2006). In addition, cMET has recently been implicated in acquired resistance to gefitinib. Engelman et al reported that NSCLC HCC827 cells chronically exposed to gefitinib in vitro led to the amplification of cMET resulting in the constitutive activation of the HER3-Akt signaling pathway (Engelman et al., 2007). Unlike the above findings, we did not observe protein amplification of cMET expression (Figure 2A). We found that cetuximab-resistant clones had constitutively phosphorylated cMET in the absence of its ligand, HGF. This over-activity of cMET appeared to be dependent on dimerization with EGFR and an increased association of cMET with the EGFR was demonstrated by co-immunoprecipitation (Figure 2B). TKIs and siRNA directed against the EGFR led to a dramatic decrease in cMET phosphorylation (Figure 4A and 5A). Although EFGR TKIs lead to the decrease of cMET, MAPK and Akt activity, small molecule inhibitors targeting cMET directly (PHA665752) did not lead to the loss of MAPK or Akt, two critical downstream molecules of the EGFR and cMET signaling pathways. Furthermore, PHA could not re-sensitize cetuximab-resistant cells to cetuximab therapy. These data indicate that although EGFR heterodimerizes with and activates cMET, this may not represent a critical determinant of acquired-resistance to cetuximab. The specific consequences of the EGFR/cMET interaction in acquired-resistance to cetuximab remain to be defined.
Another major consequence of the increased EGFR activity in cetuximab-resistant cells is the binding and activation of HER2 and HER3 (Figure 2). HER2 activity is EGFR dependent in these cells, as siRNA against EGFR can knock down HER2 activity (Figure 5A). However, it appears that once active, HER2 can transactivate HER3, as the HER2 heterodimerization inhibitor 2C4 partially inhibits HER3 signaling (Figure 7A). Furthermore, it appears that HER3 signaling is more crucial, as our results (Figure 7D) demonstrate that direct targeting of HER3 by siRNA in cetuximab-resistant cells can restore cetuximab-sensitivity. Although increased EGFR activity in cetuximab-resistant cells results in increased dimerization and activation of HER2 and HER3, the role in which HER family ligands plays a role in resistance remains to be investigated. These findings are consistent with several recent reports that suggest HER3 plays an important role in regulating tumor response to EGFR or HER2 targeting agents. Erjala et al demonstrated a correlation between HER3 expression and gefitinib resistance (Erjala et al., 2006). Similarly, Fujimoto et al pointed to HER3 as a potentially important mediator of EGFR inhibition by gefitinib in NSCLC cells (Fujimoto et al., 2005). In HER2-driven breast cancer cells, Sergina et al demonstrated that activation of HER3 and downstream PI3K/AKT signaling can evade the inhibition by gefitinib both in vitro and in vivo. Furthermore, knocking down HER3 signaling by siRNA restores potent pro-apoptotic activity of gefitinib in breast cancer cells (Sergina et al., 2007). Zhou et al reported that heregulin-dependent HER3 activation contributes to gefitinib resistance in NSCLC A549 cells. Inhibiting heregulin release by targeting ADAM proteases restored gefitinib sensitivity (Zhou et al., 2006). Collectively, these studies indicate that HER3 mediates tumor response to EGFR TKIs and activation of HER3 signaling help cells to escape the antitumor effects of these agents. Our data indicate that cetuximab resistance appears to be dependent on HER3 signaling. Taken together these data suggest that HER3 signaling may be a critical shared pathway for acquired resistance to cetuximab or TKIs directed against the EGFR.
A potential limitation of the current study is that cetuximab-resistant cells were developed in vitro. Previous studies suggest that altered control of angiogenesis may play a role in acquired resistance to cetuximab in vivo (Ciardiello et al., 2004). Nevertheless, the current findings are highly consistent with recent reports of in vivo acquired resistance to the HER2 targeting antibody herceptin. Ritter et al indicate that cells chronically challenged with herceptin exhibit elevated levels of activated EGFR as well as increased EGFR/HER2 dimers. Further, their results imply that cells resistant to herceptin retain a brisk anti-proliferative response to EGFR TKIs (Ritter et al., 2007). These corroborative findings suggest that the development of acquired resistance to HER family targeting antibodies, in vitro or in vivo, result in a dysregulation and increased dependence on HER family signaling.
The findings herein suggest potential clinical implications when considering optimal cancer therapeutic strategies. In this model we demonstrate that acquired resistance to the mAb cetuximab occurs via an EGFR-dependent mechanism. Therefore, targeting the EGFR by another mechanism, for example with EGFR TKI’s such as gefitinib and erlotinib, may retain effectiveness for tumors manifesting cetuximab resistance (Figure 4) (Huang et al., 2004; Matar et al., 2004). Furthermore, EGFR-dependent transactivation of HER2 and HER3 appears to play an important role in resistance to cetuximab since the HER2 heterodimerization inhibiting antibody 2C4 can re-sensitize resistant cells to cetuximab (Figure 7A, B). Several, clinical trials utilizing combination therapy approaches directed against HER family members are in progress (Reid et al., 2007), and our results suggest this may be an effective strategy to prevent or overcome acquired resistance to cetuximab.
In conclusion this model system of acquired resistance to cetuximab identifies candidate RTKs that contribute to the underlying molecular mechanisms of acquired resistance. The findings presented suggest that chronic therapy with cetuximab results in dysregulation of EGFR processing and subsequent activation of HER2, HER3, cMET, MAPK, and Akt. Acquired-resistance to cetuximab may result from the activation of RTKs, such as HER2 and HER3, which share overlapping proliferative and survival signaling pathways. These findings carry clinical implications and suggest that novel combinations of molecularly-targeted therapies blocking both EGFR and other HER family members may provide a strategy to overcome acquired resistance and enhance the effectiveness of EGFR-targeted therapy.
EXPERIMENTAL PROCEDURES
Cell Lines
The human non-small cell lung cancer line NCI-H226 was obtained from ATCC and maintained in 10% fetal bovine serum in RPMI. UM-SCC1 cells were obtained from University of Michigan and maintained in 10% fetal bovine serum in DMEM supplemented with 1% hydrocortisone. Cell culture media and supplements were obtained from Life Technologies, Inc. (Gaithersburg, MD).
Compounds
Gefitinib (ZD1839, Iressa™) was provided by AstraZeneca (Macclesfield, UK). Erlotinib (OSI-774, Tarceva™) was provided by Genentech, Inc (San Francisco, CA). Cetuximab (C225, Erbitux™) was provided by ImClone Systems Inc. (New York, NY). 2C4 was provided by Genentech, Inc (San Francisco, CA). CI-1033 and PHA665752 were provided by Pfizer Global Research Inc. (Ann Arbor, MI).
Antibodies and proteins
All antibodies were purchased from commercial sources as indicated below: AKT, EGFR, p-EGFR(Tyr1173), HER3, MET and HRP-conjugated goat-anti-rabbit IgG, goat-anti-mouse IgG and donkey-anti-goat IgG antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). pHER2(Y1221/1222), pHER3(Y1289), pAKT(S473), p-MAPK (Thr202/Tyr204), c-Cbl, were obtained from Cell Signaling Technology (Beverly, MA). HER2 was purchased from Neomarker (Fremont, CA), pCMET (Y1230/1234/1235) from Biosource International (Camarillo, California), tubulin from Calbiochem (San Diego, CA). Purified recombinant EGF and HRG were purchased from R&D Systems (Minneapolis, MN). Annexin V-FITC apoptosis detection kit was obtained from BD Biosciences Pharmingen (San Diego, CA). Anti-ubiquitin and all other chemicals were purchased from Sigma (St. Louis, MO).
Establishment of acquired resistance to EGFR inhibitors
Over a period of six months, tumor cells in culture were continuously exposed to increasing concentrations of cetuximab. Commencing with the IC50 of cetuximab, the exposure dose was progressively doubled every 10–14 days until 8 dose doublings had been successfully achieved. In parallel, controlled parental cells were exposed to the PBS vehicle. The established resistant cell lines were then maintained in continuous culture with the maximally achieved dose of cetuximab.
Phospho-Receptor Tyrosine Kinase (Phospho-RTK) Array
We screened the activity of a panel of receptor tyrosine kinases (RTKs) using an antibody-based array from R&D Systems (Minneapolis, MN). This phospho-RTK array specifically screens for membrane-associated proteins/receptors. Antibodies against 42 different RTKs were pre-spotted in duplicate on nitrocellulose membranes, and cell lysates from our EGFR-inhibitor resistant or parental cells were incubated with the membrane. Thereafter, a pan anti-phosphotyrosine antibody was used to detect the phosphorylated tyrosine on activated RTKs. Following quantification of scanned images using ImageQuant software, the relative expression of specific phosphorylated RTKs between cetuximab-resistant and parental H226 cells was determined.
Cell proliferation assay
Exponentially growing cells were seeded in 6 well plates. Following treatment, monolayers were washed with PBS and fixed/stained with 0.5% crystal violet. Plates were air dried overnight and dye was eluted with 0.1 M sodium citrate (pH 4.2) in ethanol (1:1). Elution was transferred to 96 well plates, and the absorbance was read at 540 nm to determine cell growth.
Immunoblotting analysis
Following treatment, cells were lysed with Tween-20 lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Tween-20, 10% glycerol, 2.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and 10 μg/ml of leupeptin and aprotinin), sonicated and protein was quantitated using a standard Bradford absorbance assay. Equal amounts of protein were fractionated by SDS-PAGE. Thereafter, proteins were transferred to PVDF membrane and analyzed by incubation with the appropriate primary antibody. Proteins were detected via incubation with HRP-conjugated secondary antibodies and ECL chemiluminescence detection system.
Immunoprecipitation
Cells were lysed with NP-40 lysis buffer [50 mM HEPES (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% Deoxycholic acid 10% glycerol, 2.5 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 10 μg/ml of leupeptin and aprotinin]. Cell lysates containing 0.5 mg of protein were incubated overnight with 2 μg of anti-EGFR antibody followed by 30 μl of proteinA/G agarose beads for 2 hr. The immunoprecipitates were pelleted and washed three times with lysis buffer and 3 times with PBS. The captured immunocomplexes were then boiled in 2× SDS sample buffer for 5 min and subjected to immunoblot analysis as described above.
Detection of surface EGFR level by flow cytometry
Following treatment, cells were washed in PBS and harvested by trypsinization. A cell suspension containing 1 × 105 cells in 100 μl PBS containing 1% BSA was incubated with 2 μg of either control IgG or anti-EGFR antibodies for 1 hr at 4°C. Following washes with PBS, cells were incubated with FITC-conjugated antibody for 30 min followed by flow cytometry analysis.
Immunofluorescent staining of EGFR
Cells (5000 cells/well) were seeded on a 4-well glass chamber slide (Nalgene Nunc, Naperville, IL). Following EGF stimulation, cells were washed with PBS and fixed with 4% paraformaldehyde for 10 min. Cells were permeabilized with PBS containing 0.1% Triton-X 100 and 1% BSA for 20 min and blocked with 3% BSA in PBS fro 15 min. Cells were incubated with anti-EGFR antibody (1:100) for 2 hrs followed by 30 min FITC-conjugated antibody. Following washes with PBS, slides were mounted using ProLong antifade mounting solution (Molecular Probes). Fluorescence microscopy and photography were carried out using FITC filter array on an Olympus fluorescent microscope (Scope BX51).
RNA interference studies
All siRNAs were obtained from Dharmacon (Lafayette, CO). Transfection of Dharmacon siRNA into cell lines was achieved by using Dharmafect 1 reagent.
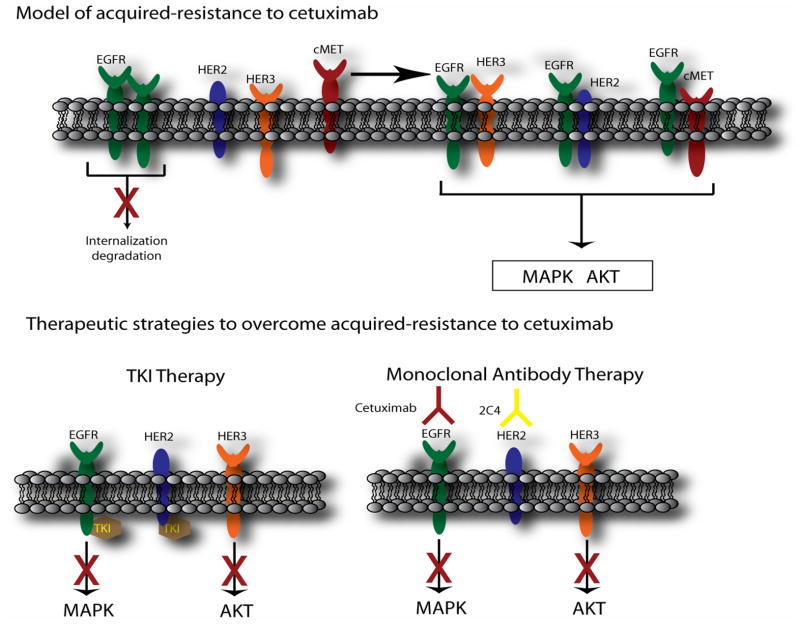
Figure 8. Model of acquired-resistance to cetuximab and therapeutic strategies to overcome resistance.
Chronic exposure to cetuximab leads to dysregulation of c-Cbl mediated internalization and degradation of the EGFR. This increased steady-state expression of EGFR serves to bind and activate HER2 or HER3 and thereby maintain signaling to MAPK and Akt pathways in the presence of cetuximab. TKIs targeting EGFR and HER2 lead to blockade of HER3 transactivation and subsequent pro-survival signals to Akt. In addition, combinatorial monoclonal antibody therapy using cetuximab and 2C4 leads to inactivation of HER3 and down regulation of its signaling pathways.
Acknowledgments
We would like to thank AstraZeneca (gefitinib), Genentech (erlotinib, 2C4), ImClone (cetuximab), OSI (erlotinib), and Pfizer Global Research (PHA665752 and CI-1033) for supplying EGFR- and cMET- targeting agents. This work was supported in part by NIH R01 CA 113448-01 (PMH) and American Cancer Society Postdoctoral Fellowship (DLW).
The abbreviations used are
- DMSO
dimethyl sulfoxide
- EGFR
epidermal growth factor receptor
- ELISA
enzyme-linked immunosorbent assay
- HGF
hepatocyte growth factor
- MAPK
mitogen-activated protein kinase
- mAb
monoclonal antibody
- NSCLC
non-small cell lung cancer
- PI(3)K
phosphatidylinositol 3′-kinase
- RTK
receptor tyrosine kinase
- SCC
squamous cell carcinoma
- TKD
tyrosine kinase domain
- TKI
tyrosine kinase inhibitor
References
- Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol. 2005;23:2445–59. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- Benavente S, Huang S, Armstrong E, Chi A, Chinnaiyan P, Harari PM. Establishment of acquired resistance to epidermal growth factor receptor (EGFR) inhibitors in human tumor cell lines. AACR Meeting Abstracts. 2004:1230-a. [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–55. [PubMed] [Google Scholar]
- Ciardiello F, Bianco R, Caputo R, Caputo R, Damiano V, Troiani T, et al. Antitumor activity of ZD6474, a vascular endothelial growth factor receptor tyrosine kinase inhibitor, in human cancer cells with acquired resistance to antiepidermal growth factor receptor therapy. Clin Cancer Res. 2004;10:784–93. doi: 10.1158/1078-0432.ccr-1100-03. [DOI] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science. 2007 doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Erjala K, Sundvall M, Junttila TT, Zhang N, Savisalo M, Mali P, et al. Signaling via ErbB2 and ErbB3 Associates with Resistance and Epidermal Growth Factor Receptor (EGFR) Amplification with Sensitivity to EGFR Inhibitor Gefitinib in Head and Neck Squamous Cell Carcinoma Cells. Clin Cancer Res. 2006;12:4103–4111. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Wislez M, Zhang J, Iwanaga K, Dackor J, Hanna AE, et al. High Expression of ErbB Family Members and Their Ligands in Lung Adenocarcinomas That Are Sensitive to Inhibition of Epidermal Growth Factor Receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]
- Furukawa M, Nagatomo I, Kumagai T, Yamadori T, Takahashi R, Yoshimura M, et al. Gefitinib-sensitive EGFR lacking residues 746–750 exhibits hypophosphorylation at tyrosine residue 1045, hypoubiquitination, and impaired endocytosis. DNA Cell Biol. 2007;26:178–85. doi: 10.1089/dna.2006.0573. [DOI] [PubMed] [Google Scholar]
- Gondi V, Huang S, Benavente S, Armstrong E, Harari PM. Potential mechanisms of acquiredrResistance to EGFR inhibitors. AACR Meeting Abstracts. 2006:294-b. [Google Scholar]
- Grandal MV, Zandi R, Pedersen MW, Willumsen BM, van Deurs B, Poulsen HS. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis. 2007;28:1408–17. doi: 10.1093/carcin/bgm058. [DOI] [PubMed] [Google Scholar]
- Grovdal LM, Stang E, Sorkin A, Madshus IH. Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp Cell Res. 2004;300:388–95. doi: 10.1016/j.yexcr.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Han W, Zhang T, Yu H, Foulke JG, Tang CK. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol Ther. 2006;5:1361–8. doi: 10.4161/cbt.5.10.3226. [DOI] [PubMed] [Google Scholar]
- Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–62. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- Matar P, Rojo F, Cassia R, Moreno-Bueno G, Di Cosimo S, Tabernero J, et al. Combined epidermal growth factor receptor targeting with the tyrosine kinase inhibitor gefitinib (ZD1839) and the monoclonal antibody cetuximab (IMC-C225): superiority over single-agent receptor targeting. Clin Cancer Res. 2004;10:6487–501. doi: 10.1158/1078-0432.CCR-04-0870. [DOI] [PubMed] [Google Scholar]
- Mendelsohn J. Antibody-mediated EGF receptor blockade as an anticancer therapy: from the laboratory to the clinic. Cancer Immunology & Immunotherapy. 2003;52:342–6. doi: 10.1007/s00262-002-0354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33:369–85. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst. 2005;97:1185–94. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005;2:1–11. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi B, Bottaro DP. Targeting the c-Met Signaling Pathway in Cancer. Clin Cancer Res. 2006;12:3657–3660. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- Reid A, Vidal L, Shaw H, de Bono J. Dual inhibition of ErbB1 (EGFR/HER1) and ErbB2 (HER2/neu) Eur J Cancer. 2007;43:481–9. doi: 10.1016/j.ejca.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, et al. Human Breast Cancer Cells Selected for Resistance to Trastuzumab In vivo Overexpress Epidermal Growth Factor Receptor and ErbB Ligands and Remain Dependent on the ErbB Receptor Network. Clin Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–41. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunada H, Magun BE, Mendelsohn J, MacLeod CL. Monoclonal antibody against epidermal growth factor receptor is internalized without stimulating receptor phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:3825–9. doi: 10.1073/pnas.83.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]