During vertebrate development, gene expression is tightly controlled by dynamic regulatory circuits which determine and maintain cell lineages. These regulatory mechanisms, including transcription factor-DNA interactions and epigenetic programming, insure proper spatial and temporal gene expression. In addition to the aforementioned mechanisms, microRNAs (miRNAs) have been shown to play a critical role in controlling a wide range of cellular processes. miRNAs are 18–25 nucleotide long non-coding RNAs that repress mRNA translation or modulate mRNA degradation by binding to the 3’-untranslated region of target mRNAs. The primary miRNA transcripts are transcribed by RNA polymerase II and further processed to mature miRNAs by the Drosha/DGCR8 and Dicer complexes. Individual miRNAs can target hundreds of mRNAs and their expression is often associated with specific cell types or developmental stages.1,2 This suggests that even a single miRNA can have a significant impact on numerous biological processes.
Several studies have shown that microRNA-214 (miR-214) is instrumental in determining cell fate in several cell types.3,4,6 Yet the modalities by which miR-214 modulates cell lineage specification can be quite diverse. In zebrafish, miR-214 is required for muscle cell fate decision during somitogenesis. Inhibition of miR-214 expression decreases slow-muscle cell types in the developing somites. One miR-214 target in zebrafish is suppressor of fused (su(fu)), a negative regulator of Hedgehog signaling. By repressing su(fu) expression in different somite compartments, miR-214 mediates muscle cell fate transition through the Hedgehog pathway.3 In mouse skeletal muscle, miR-214 shows robust expression in differentiating myoblasts. Overexpression of miR-214 in muscle cells results in premature expression of muscle genes and acceleration of muscle differentiation while blockage of its expression promotes myoblast proliferation and dampens myogenesis.4 Genetic ablation of a region containing the murine mir-214 locus leads to several developmental defects, including reduced skeletal muscle mass.5 Unlike zebrafish, in mouse skeletal muscle, miR-214 targets the Polycomb group protein (PcG) Enhancer of zest homologue 2 (Ezh2) to regulate muscle cell differentiation.4 Ezh2 trimethylates lysine 27 of histone H3 (H3K27me3) and represses gene transcription. It has been shown that Ezh2 is developmentally regulated during myogenesis and blocks muscle differentiation by imposing H3K27me3 on muscle specific genes. It is critical to remove Ezh2 binding and its cognate methylation for appropriate muscle gene activation.7 Therefore, repression of Ezh2 by miR-214 is essential for initiating muscle differentiation.4 Interestingly, this miR-214-dependent Ezh2 regulation is also observed in embryonic stem (ES) cells. In ES cells induced to differentiate by retinoic acid, upregulation of miR-214 expression coincides with reduction of Ezh2 protein. Ectopic expression of miR-214 in pluripotent ES cells reduces Ezh2 protein level thus derepressing transcription of developmental regulators leading to loss of ES cell pluripotency.4 In Xenopus, miR-214 is highly expressed in multipotent retinal progenitors at early embryonic stages. By repressing Xotx2 and Xvsx, two key regulators of late retinal neurons, miR-214 controls the developmental timing of these progenitors and determines their fate.6
The importance of miR-214 in cell fate commitment suggests that miR-214 expression also needs to be precisely controlled. The relevance of miRNA dynamic regulation is well illustrated in skeletal muscle where miR-214 transcription is regulated by a double-negative feedback loop in which miR-214 is repressed by Ezh2 and activated by myoD/myogenin.4 In addition of being regulated at the transcriptional level, the processing of primary-214 transcripts to mature miRNAs is controlled by p72 Dead-box RNA helicase subunits in the mouse Drosha complex.8 Of note, p72 itself associates with MyoD to promote muscle differentiation.9 Thus, p72 helicase could promote differentiation by both co-activating MyoD-dependent transcription and favoring miR-214 processing. Figure 1 summarizes a working model of the regulatory network involving miR-214 during muscle differentiation. Muscle cell fate is determined by a series of events including transcription and biogenesis of miR-214 and the feedback loop between miR-214 and its target. The mechanisms discussed here may serve as a potential model for miRNA mediated regulatory networks in other biological systems.
Figure 1.
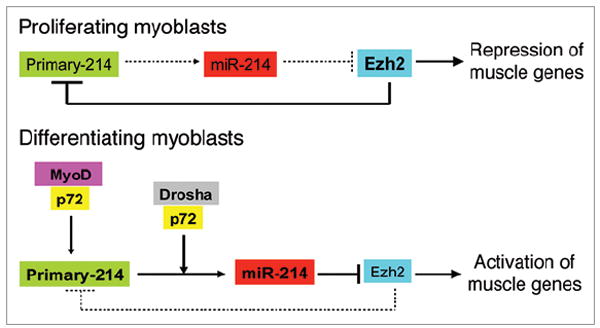
In proliferating myoblasts, where it is highly expressed, Ezh2 represses primary-miR-214 transcription as well as other muscle specific genes to maintain the myoblasts in undifferented state. Upon differentiation, Ezh2 expression is reduced and miR-214 locus is derepressed. Robust production of miR-214 is achieved by MyoD/p72 mediated transcriptional activation and Drosha/p72 processing. miR-214 then negatively feeds back on Ezh2 by reducing its mRNA translation to further accelerate muscle differentiation.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal and Skin Diseases of the National Institutes of Health.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/11472
Comment on: Juan AH, et al. Mol Cell 2009; 36:61–74.
References
- 1.Bartel DP. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, et al. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Flynt AS, et al. Nat Genet. 2007;39:259–63. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juan AH, et al. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, et al. Dev Dyn. 2008;237:3738–48. doi: 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 6.Decembrini S, et al. Proc Natl Acad Sci USA. 2009;106:21179–84. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caretti G, et al. Genes Dev. 2004;18:2627–38. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda T, et al. Nat Cell Biol. 2007;9:604–11. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 9.Caretti G, et al. Dev Cell. 2006;11:547–60. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]


