Lens fiber cell differentiation, denucleation, and abnormal apoptosis were examined through lens-specific expression of the N-terminal nuclear receptor box of NCOA6 in combination with lens-specific deletion of Ncoa6 in mice. The present data show that early apoptosis in lens fiber cells inhibits their subsequent denucleation.
Abstract
Nuclear receptor coactivator 6 (NCOA6) is a multifunctional protein implicated in embryonic development, cell survival, and homeostasis. An 81-amino acid fragment, dnNCOA6, containing the N-terminal nuclear receptor box (LXXLL motif) of NCOA6, acts as a dominant-negative (dn) inhibitor of NCOA6. Here, we expressed dnNCOA6 in postmitotic transgenic mouse lens fiber cells. The transgenic lenses showed reduced growth; a wide spectrum of lens fiber cell differentiation defects, including reduced expression of γ-crystallins; and cataract formation. Those lens fiber cells entered an alternate proapoptotic pathway, and the denucleation (karyolysis) process was stalled. Activation of caspase-3 at embryonic day (E)13.5 was followed by double-strand breaks (DSBs) formation monitored via a biomarker, γ-H2AX. Intense terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) signals were found at E16.5. Thus, a window of ∼72 h between these events suggested prolonged though incomplete apoptosis in the lens fiber cell compartment that preserved nuclei in its cells. Genetic experiments showed that the apoptotic-like processes in the transgenic lens were both p53-dependent and p53-independent. Lens-specific deletion of Ncoa6 also resulted in disrupted lens fiber cell differentiation. Our data demonstrate a cell-autonomous role of Ncoa6 in lens fiber cell differentiation and suggest novel insights into the process of lens fiber cell denucleation and apoptosis.
INTRODUCTION
Early stages of embryonic lens development culminate with the formation of the lens vesicle, made up of undifferentiated lens precursor cells (Cvekl and Duncan, 2007). Under the influence of fibroblast growth factors and bone morphogenetic proteins produced by the neuroretina, cells from the posterior part of the lens vesicle initiate the differentiation processes, including cell cycle withdrawal, cell growth, and elongation after embryonic day (E)10.5 of mouse embryonic development (Lovicu and McAvoy, 2005; Griep, 2006; Robinson, 2006). Within ∼48 h, these cells reach the anterior part of the lens vesicle as primary lens fiber cells. In contrast, cells located in the anterior part of the lens vesicle retain their proliferative capacity and organize into a sheet of cuboidal anterior lens epithelium (Lovicu and McAvoy, 2005). As those cells reach the lens equator (transitional zone), their differentiation is induced to form secondary fibers, which wrap around the previously formed fiber cells. Lens fiber cell differentiation is characterized by cell elongation, and temporally and spatially controlled expression of crystallins and other lens-specific proteins (Piatigorsky, 1981; Graw, 2003; Cvekl and Duncan, 2007). Lens fiber cell differentiation also includes coordinated degradation of organelles, including the nucleus. Destruction of subcellular organelles is required to eliminate sources of light scattering (Yan et al., 2006; Bassnett, 2009).
Degradation of nuclei during mammalian development and differentiation is a special process limited to lens fiber cells, erythrocytes, and keratinocytes. These enucleated cells can be viewed as “storage” cells containing large amounts of specific proteins. In other words, crystallins, hemoglobins, and keratins are required to generate a transparent and refractive tissue (lens), transport oxygen (erythrocytes), and provide a protective barrier for the skin (keratinocytes), respectively. Lens fiber cell-specific degradation of nuclei (denucleation) is morphologically characterized by differently shaped nuclei and the formation of an organelle-free zone (OFZ) in the center of lens cortex (Bassnett, 2009). The denucleation process depends on lens-preferred lysosomal DNase IIβ, an acid endonuclease, to degrade chromatin in lens fiber cells (Nishimoto et al., 2003). Mammalian erythrocytes are enucleated via nuclear engulfment by macrophages (Yoshida et al., 2005). The skin keratinocytes lose their nuclei through an apoptotic caspase-independent process, termed cornification (Lippens et al., 2009). Thus, lens fiber cells, erythrocytes, and keratinocytes degrade their nuclei through distinct cellular and molecular mechanisms.
Before executing the default denucleation process, lens fiber cells are prone to enter an alternate, proapoptotic pathway. Lens-specific expression of cell cycle regulators, including viral oncogenes in transgenic lens fiber cells (Griep et al., 1993; Fromm et al., 1994; Nakamura et al., 1995; Pan and Griep, 1995; Gomez Lahoz et al., 1999; Chen et al., 2000, 2002, 2004), induced ectopic apoptosis in the lens fiber cell compartment. Although the majority of these studies used terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay to identify apoptotic cells, two recent reports, which focused on abnormal lens formation (Wang et al., 2005; Cang et al., 2006), have identified DNA double-strand breaks (DSBs) formation via a biomarker, γ-H2AX, to monitor DNA damage. In mammalian cells, generation of DSBs is either followed by successful DNA repair and cell survival, or programmed cell death is executed (Canman, 2003; Petrini and Stracker, 2003). However, it is possible that normal denucleation process in lens fiber cells can also generate this type of DNA damage. Thus, temporospatial characterization of DSBs formation inside and outside of the presumptive OFZ during normal and abnormal lens development should provide novel insight into lens fiber cell denucleation.
Here, we considered transgenic expression of proteins that could abrogate both antiapoptotic (protective) mechanisms of the lens (Andley, 2007) and simultaneously perturb lens fiber cell karyolysis (Bassnett, 2009). Initially, we focused on three genes, Brg1 (Liu et al., 2004), Ncoa6 (Mahajan et al., 2004), and Rybp (Stanton et al., 2007), encoding proteins with diverse roles in preventing apoptosis as well as regulating chromatin structure. Previously, these proteins were implicated as regulators of lens development (Kim et al., 2002; Yang et al., 2006a; Pirity et al., 2007). Our pilot experiments suggested that Ncoa6 was a good model gene to study lens fiber cell differentiation and denucleation.
Nuclear receptor coactivator 6 (Ncoa6; other aliases are Asc-2, Aib3, TRBP, RAP250, NRC, and PRIP) acts as an ubiquitously expressed coactivator of specific nuclear receptors and several other transcription factors (Mahajan and Samuels, 2008). NCOA6 encodes a large protein composed of 2063 amino acids with two nuclear receptor (NR) boxes (featuring the LXXLL sequence), two transactivation domains, and a C-terminal regulatory domain (see Figure 1A) (Mahajan et al., 2007). Several previous studies revealed antiapoptotic function of NCOA6 and its up-regulation in tumor cells (Anzick et al., 1997; Mahajan et al., 2004; Yeom et al., 2006; Zhu et al., 2009). Targeted knockout of Ncoa6 resulted in embryonic lethality between E8.5 and E12.5 (Kuang et al., 2002; Antonson et al., 2003; Zhu et al., 2003; Mahajan et al., 2004). This early lethality prevented studies of lens fiber cell differentiation. An 81-amino acid fragment of NCOA6 (dnNCOA6) including the N-terminal NR box (LXXLL-1 motif; see Figure 1) has been shown to act as a dominant inhibitor of NCOA6 that blocks the interaction of retinoic acid (RA) receptors and NCOA6 (Kim et al., 2002). Transgenic mice that express dnNCOA6 under the ubiquitous β-actin promoter revealed a spectrum of ocular abnormalities including microphthalmia and cataract (Kim et al., 2002) as well as a plethora of defects in other tissues (Kim et al., 2002). However, abnormal lens development in this system could be caused by indirect effects (e.g., perturbed optic cup and early development of the neuroretina) (Mathers et al., 1997; Porter et al., 1997; Lee et al., 2005). It was also possible that the damage to the lens was initiated in the lens progenitor/precursor cells before their differentiation (Yamada et al., 2003; Liu et al., 2006).
Figure 1.
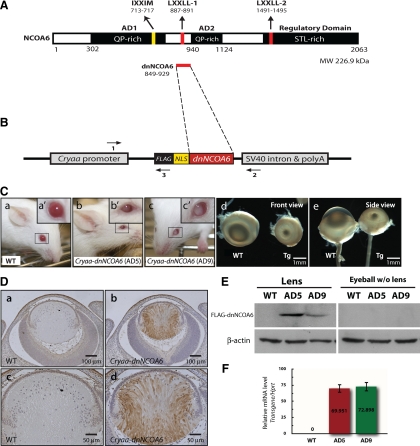
Generation and phenotype analysis of the Cryaa-dnNCOA6 transgenic mouse model. (A) Schematic diagram of the NCOA6 protein structure (2063 amino acid residues). NCOA6 contains two QP-rich ADs, two canonical NR boxes (LXXLL-1 and LXXLL-2), which are important for interaction with ligand-bound NRs, one noncanonical NR box (IXXMM), and the C-terminal STL-rich regulatory domain. (B) Schematic diagram of the Cryaa-dnNCOA6 transgenic construct. cDNA of dnNCOA6 including amino acid 849–929 of NCOA6 with a NLS and 3xFLAG tag in the 5′ end was inserted between the Cryaa (αA-crystallin) promoter and the intron-polyadenylation sequences of the small t antigen from SV40 (SV40 intron & polyA). The three primers used for genotyping are shown as horizontal arrows. (C) Two founders of Cryaa-dnNCOA6, AD5 (b) and AD9 (c), were generated and showed microphthalmia and cataract compared with WT (a) mice. Eyeballs from WT and AD5 animals are aligned in front view (d) and side view (e) to compare phenotypes. Transgenic lenses displayed a reduction of ∼3.84- to 4.75-fold in size compared with WT (see Materials and Methods). Transgene copy number of line AD5 and AD9 is 12∼16 and 12∼19 copies per genome, respectively. (D) Lens-specific expression of the FLAG-dnNCOA6 fragment was evaluated by immunohistochemistry using an anti-FLAG antibody (brown) in E14.5 WT (a and c) and transgenic (b and d) eyes. Nuclei (blue) were counterstained with hematoxylin. Scale bar is shown in each panel. (E) Western blot analysis of lens-specific FLAG-dnNCOA6 fragments expression in the lens and eyeball (without lens) tissues from WT, AD5, and AD9. β-Actin was used for loading control. (F) Relative expression levels of transcripts encoding dnNCOA6 in AD5 and AD9 transgenic lines were analyzed by qRT-PCR. Primers used (see Materials and Methods) are specific to the transgene (human sequence) and did not cross-react with the endogenous mouse Ncoa6 transcripts.
In this report, multiple roles of Ncoa6 in lens fiber differentiation were examined through the expression of lens-specific dnNCOA6 transgene and the deletion of Ncoa6 in the lens lineage. The specificity of dnNCOA6 action was examined in Ncoa6 heterozygous mutant background. Next, the lens-specific dnNCOA6 transgenic mouse model was used to examine the interference between the proapoptotic program initiated in transgenic lenses and the denucleation process. Together, these studies reveal that dnNCOA6-induced apoptosis was incomplete in the lens fiber cell compartment while the normal denucleation process was arrested.
MATERIALS AND METHODS
Generation and Analysis of Cryaa-dnNCOA6 Transgenic Mice
Nuclear localization sequence (NLS) in frame fusion with dnNCOA6 (amino acid 849–929 of NCOA6 protein), NLS-dnNCOA6, was amplified by polymerase chain reaction (PCR) from pCAGGS-ASC2co2CN (a gift from Dr. Jae Woon Lee, Pohang University of Science and Technology) and subcloned downstream of 3xFLAG-tag in the 3XFLAG-CMV-10 vector (Sigma-Aldrich, St. Louis, MO). 3xFLAG-NLS-dnNCOA6 was then amplified by PCR and inserted into the BglII site between the Cryaa promoter (−366 to + 46) (Overbeek et al., 1985) and simian virus 40 (SV40) intron-polyadenylation sequences of pACP3 vector (Duncan et al., 2000; see Figure 1B). PstI digestion generated a 687-base pair fragment between the cDNA sequence and the PstI site before the Cryaa promoter in pACP3 vector confirmed the orientation. The construct was then linearized with NotI and sent to the Albert Einstein College of Medicine Transgenic Facility (Bronx, NY) for microinjection to generate Tg (Cryaa-dnNCOA6) mice. We will, from here on, refer to these mice as Cryaa-dnNCOA6 mice. Transgenic mice were generated and maintained in FVB/N background. All mice used in this study were maintained at the Albert Einstein College of Medicine Institute for Animal Studies under specific pathogen-free conditions in accordance with institutional guidelines. For staging of embryos, vaginal plug formed around noon of the day was considered as E0.5. Animals were staged by denoting the day of birth as neonate and subsequent days as postnatal day (P)1, P2, etc. Two primer sets were used for genotyping (primer 1: 5′-GCTCCTGTCTGACTCACTGC-3′; primer 2: 5′-GCTTTAAATCTCTGTAGGTAGTTTGTC-3′; primer 3: 5′-CATCGTCATCCTTGTAATC-3′). Primer 1 and 2 generate a 509-base pair PCR product across the Cryaa promoter and the SV40 intron. Primer 1 and 3 generate a 144-base pair PCR product across the Cryaa promoter and the FLAG-tag sequence (see Figure 1B).
Primers spanning the intron and exon of Ccni (5′-TCTTCTCCCTCCTCAGACG-3′ and 5′-CCGTTACCACCTCATGATCC-3′), B2m (5′-CCCTGGCTGGCTCTCATT-3′ and 5′-ACTGAAGCGACCGCGACT-3′), and Cryaa gene (5′-CCTTCCTGTCTTCCACCATC-3′ and 5′-GCAGCTAGGAGGAACCAGTG-3′), and primers including part of the transgenic Cryaa promoter, Cryaa-tg (5′-CCCGAGCTGAGCATAGACAT-3′ and 5′-AGTCAGACAGGAGCCTCTGG-3′), were used to determine transgenic copy number from three biological replicates in a quantitative (real-time) PCR (qRT-PCR) analysis using a 7900HT fast real-time PCR system with Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). Primers of Ccni, B2m, and Cryaa genes were used as internal controls and Cryaa-tg primers were used to determine transgene copy number.
Primers (5′-CCAGAGGCTCCTGTCTGACT-3′ and 5′-CCGTCATGGTCTTTGTAGTCC-3′) specific to dnNCOA6 cDNA sequence were used to compare the relative expression levels of dnNCOA6 transgene in wild-type (WT) and transgenic newborn lenses using qRT-PCR. Total RNA was prepared using RNeasy Microkit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. cDNA was generated with oligo(dT20) primers (Invitrogen, Carlsbad, CA) and SuperScript II Reverse Transcriptase (Invitrogen), according to the manufacturer's protocol. Each 20-μl reaction was diluted 10 times, and 2 μl was used for each qRT-PCR reaction. The relative gene expression levels from three biological replicates were analyzed. For data normalization, expression of three reference genes, B2m, Hprt, and Sdha, was examined (Wolf et al., 2009).
Histological Examination and Microscope
Age-matched embryos or postnatal eyeballs were fixed in 10% neutral buffered Formalin overnight. Paraffin embedding, sectioning (5-μm sagittal sections at the midline of the eye) and hematoxylin and eosin (H&E) staining were performed by the Albert Einstein College of Medicine Histotechnology and Comparative Pathology Facility. Routine light imaging was performed on an Axioskop II microscope with Axiovision software (Carl Zeiss, Thornwood, NY). Fluorescence imaging was performed on an upright BX61 microscope (CoolSNAP HQ camera; Olympus, Melville, NY) and processed with IPLab acquisition software (BD Biosciences, San Jose, CA). Pictures from serial eye sections were taken, and lens size was measured using ImageJ software (National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/) according to the scale. Alternatively, lenses were dissected from Formalin-fixed eyeballs and pictures were taken with an MZ FLIII stereomicroscope (Leica Microsystems, Deerfield, IL) with bright-field transmitted light. All pictures were processed in ImageJ to measure the surface area and height of each lens for comparison. Every set of experiment compared littermates and was repeated at least three times.
Western Blot Analysis
Lens and eyeball (without lens) tissues were dissected from newborn mice and homogenized in mammalian lysis buffer (150 mM NaCl, 1 mM NaF, 50 mM Tris, pH 8.0, 0.5% NP-40, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and proteinase inhibitors [Roche Diagnostics, Indianapolis, IN]) to obtain protein lysates. Ten to 100 μg of each protein sample was run on a 12 or 15% acrylamide gel. Proteins were then transferred to a nitrocellulose membrane. The membrane was blocked with 5% skim milk in Tris-buffered saline (TBS) for 1 h and incubated with primary antibody using the recommended dilution overnight at 4°C. The membrane was then washed with TBS, incubated with horseradish peroxidase-conjugated secondary antibody (1:2000; Promega, Madison, WI), and visualized with ECL Plus western blotting detection reagents (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) according to manufacturer's instructions. The dilution used for each antibody is listed as follows: mouse monoclonal anti-FLAG antibody (mAb) (1:1000; Sigma-Aldrich), rabbit polyclonal anti-c-Maf antibody (1:1000; Bethyl Laboratories, Montgomery, TX), mouse monoclonal anti-p53 antibody (1:250; Merck, Whitehouse Station, NJ), rabbit polyclonal anti-αA-crystallin antibody (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-αB-crystallin antibody (1:2000; Assay Designs, Ann Arbor, MI), rabbit polyclonal anti-β-crystallin antibody (1:500; Santa Cruz Biotechnology), rabbit polyclonal anti-γ-crystallin antibody (1:500; Santa Cruz Biotechnology), monoclonal anti-β-actin antibody (1:2000; Sigma-Aldrich), and mouse monoclonal anti-NCOA6 antibody (1:1000; produced in Dr. Jianming Xu's laboratory). The protein bands from Western blots were quantified with ImageJ software and normalized using β-actin as a loading control.
Immunohistochemistry and Immunofluorescence
Tissue sections were deparaffinized in 60°C oven and xylene and rehydrated through an alcohol gradient series. Sections were heated and cooled to room temperature in sodium citrate buffer for 30 and 20 min, respectively. Sections were then treated with 0.3% H2O2 and incubated with blocking buffer (5% donkey serum and 2% bovine serum albumin [BSA] in TBS). Rabbit polyclonal anti-FLAG (1:500; Sigma-Aldrich) or rat monoclonal anti-Ki-67 (1:500; Dako North America, Carpinteria, CA) antibody was diluted in blocking solution with 1-h incubation at room temperature. After washing with TBS, slides were incubated with biotinylated goat anti-rabbit immunoglobulin (Ig)G (1:500) for 1 h at room temperature. The signal was detected using streptavidin-conjugated horseradish peroxidase and peroxidase activity was visualized with diaminobenzidine/H2O2 (Vector Laboratories, Burlingame, CA). Slides were then counterstained with hematoxylin. For immunofluorescence, treatment with H2O2 was omitted and tissue sections were incubated with blocking solution (5% goat serum and 2% BSA in TBS) for 30 min. Rabbit polyclonal anti-cleaved caspase-3 Asp175 (1:200; Cell Signaling Technology, Danvers, MA); rabbit polyclonal anti-c-Maf (1:500; Bethyl Laboratories); rabbit polyclonal anti-γ-crystallin (1:100; Santa Cruz Biotechnology); rabbit polyclonal anti-β-crystallin (1:100; Santa Cruz Biotechnology); mouse monoclonal anti-FLAG (1:1000; Sigma-Aldrich); rabbit polyclonal anti-phospho-H2AX (Ser139), γ-H2AX (1:200; Millipore, Billerica, MA); rabbit polyclonal anti-protein disulfide isomerase (PDI) (1:500; Assay Designs, Ann Arbor, MI); and rabbit polyclonal anti-phospho-histone H3 (1:200; Millipore) were diluted in blocking solution and incubated overnight at 4°C. Slides were then washed with TBS and incubated with the secondary antibody (1:500; Alexa Fluorescence 568 or 488 goat anti-rabbit/mouse IgG [Invitrogen]) and 4′,6-diamidino-2-phenylindole (DAPI) in blocking solution for 45 min at room temperature, followed by TBS washing. Slides were mounted with VECTASHIELD mounting medium (Vector Laboratories).
Scanning Electron Microscope (SEM)
Three-month-old mouse lenses were fixed in 0.08 M sodium cacodylate, 1.25% glutaraldehyde, and 1% paraformaldehyde, pH 7.4, at room temperature overnight. After fixation, the lens capsule and several outermost layers of fiber cells were peeled off to show the fiber pattern. Lens samples were then dehydrated through a graded series of ethanol and processed by critical point drying using liquid carbon dioxide in a 795 critical point drier (Tousimis Samdri, Rockville, MD). The lenses were subsequently transferred to a filter paper, placed in a vacuum desiccator, mounted on a stub, and sputter coated for 2 min with gold-palladium in a Vacuum Desk-2 sputter coater (Denton, Cherry Hill, NJ). Specimens were examined with a JSM6400 scanning electron microscope (JEOL, Peabody, MA), using an accelerating voltage of 10 kV.
Detection of Apoptosis
Apoptosis was analyzed using DeadEnd Fluorometric TUNEL System (Promega). Tissue sections were deparaffinized and rehydrated as described above, followed by TUNEL staining according to the manufacturer's manual. Three sections from each eye/genotype were stained and TUNEL-positive nuclei in lens fiber cells were quantified. Every set of experiment compared littermates and was repeated at least three times.
Role of p53 in Suppressing Apoptosis in Cryaa-dnNCOA6 Lenses
To examine the role of p53 in the ectopic apoptosis induced in Cryaa-dnNCOA6 lenses, p53+/null mice were crossed with p53+/null; Cryaa-dnNCOA6 mice to generate p53null/null; Cryaa-dnNCOA6 and p53+/+; Cryaa-dnNCOA6 E14.5 embryos. At least four midline eye sections were analyzed for DSBs from each embryo. Nine embryos from p53null/null; Cryaa-dnNCOA6 and 10 embryos from p53+/+; Cryaa-dnNCOA6 were analyzed. For the rescue effect in lens size, E16.5 embryos were generated from the crossing described above. Serial midline eye sections were subjected to H&E staining and routine light imaging was performed. Lenses from neonate littermates were used to quantify the rescue effect in lens size. Every set of experiment was repeated at least three times.
Lens-specific Deletion of Ncoa6 and Tissue-specific Genomic DNA Extraction for Genotyping
Generation of the Ncoa6 conditional knockout mice will be described elsewhere (Li and Xu, unpublished data). In brief, the seventh exon of Ncoa6 gene was flanked by a pair of loxP sites (Ncoa6flox/flox). The deleted Ncoa6 allele did not contain exon 7 and caused a reading frame shift between exons 6 and 8. Ncoa6 floxed mice were crossed with EIIa-Cre transgenic line (The Jackson Laboratory, Bar Harbor, ME) to obtain Ncoa6 null allele. To generate lens-specific Ncoa6 knockout mice, Ncoa6flox/null and Ncoa6flox/flox mice were crossed with Le-Cre transgenic line to delete the floxed allele in the lens lineage. Transmission of the targeted allele and knockout allele were confirmed by PCR. Le-Cre mice were maintained in FVB/N background and genotyped as described previously (Ashery-Padan et al., 2000). Lens, eyeball (without lens) and ear tissues were dissected from 2- to 3-mo-old mice. DNeasy blood & tissue kit (QIAGEN) was used to extract genomic DNA. Double-distilled H2O (200 μl) was used to elute the genomic DNA, and 1 μl of elution product was used for each genotyping reaction.
RESULTS
Initial Characterization of Cryaa-dnNCOA6 Transgenic Mouse Model
The panocular defects shown in the earlier dnNCOA6 transgenic mouse model under the control of CMV enhancer/β-actin promoter (Kim et al., 2002) prompted us to ask whether Ncoa6 plays a cell-autonomous or a nonautonomous role in lens development. To address this issue, we used a lens-specific Cryaa (αA-crystallin) promoter as a tool to express a dnNCOA6 fragment in postmitotic lens fiber cells (Overbeek et al., 1985; Westphal et al., 1985; Wawrousek et al., 1990). A FLAG epitope and an NLS were added to the N terminus of the dnNCOA6 fragment (Figure 1B). Two transgenic lines, AD5 and AD9, were generated and gross morphological and histological analyses revealed specific eye phenotypes, microphthalmia, and cataract (Figure 1C) in both lines. Because line AD5 exhibited more severe lens phenotypes, it was used throughout this study. To confirm the lens-specific expression of the dnNCOA6 fragment, E14.5 eye sections from WT and Cryaa-dnNCOA6 mouse embryos were stained with an anti-FLAG antibody to detect transgenic protein expression. The data showed lens-specific expression of the dnNCOA6 fragment in lens fiber cells (Figure 1D). In addition, no FLAG staining was detected in the lens epithelium (Figure 1D and Supplemental Figure 1). The lens-specific expression of dnNCOA6 fragments was further confirmed by Western blot in which lens and eyeball (without lens) whole tissue lysates from WT and both transgenic lines were analyzed with an anti-FLAG antibody (Figure 1E). The same antibody was used to examine the onset of transgenic protein expression. Our results showed lens-specific transgenic protein expression at E12.5 and E13.5 mouse lens fiber cells (Supplemental Figure S1). The relative expression level of the transgene was examined by qRT-PCR using primers specific to the transgenic construct. The result showed comparable levels of transgenic transcripts in both lines using cDNAs prepared from neonatal lenses (Figure 1F). Next, NCOA6-specific mAb was used in Western blotting to determine expression levels of the endogenous NCOA6 protein in transgenic lenses. Our data (Supplemental Figure S2) showed no significant change in NCOA6 protein expression levels in WT and transgenic lens protein extracts. Thus, the initial studies of the Cryaa-dnNCOA6 mouse model support the cell-autonomous role of Ncoa6 in lens fiber cell differentiation.
Lens-specific Expression of dnNCOA6 Caused Multiple Defects in Lens Growth, Lens Fiber Cell Elongation, and Their Terminal Differentiation
To further examine the lens abnormalities in Cryaa-dnNCOA6 mice, histological analysis of eye tissues from different embryonic and postnatal stages were conducted. We found minor differences including a posterior shift of some nuclei in E12.5 transgenic lens, whereas E12.5 WT lens seemed normal (compare Figure 2, A and B). At ∼E15.5, some irregular lens fiber cells containing nuclei with abnormal morphology were found in the transgenic lens (compare Figure 2, C and D). Higher magnification images revealed pyknotic nuclei in the transgenic lens (Figure 2D′), whereas the WT lens appeared normal (Figure 2C′). Pyknotic nuclei denote chromatin shrinkage and hyperchromasia (Bassnett, 2009). The size difference between the WT and transgenic lens is apparent in neonatal lenses and more pronounced in P21 lenses (Figure 2, E–H).
Figure 2.
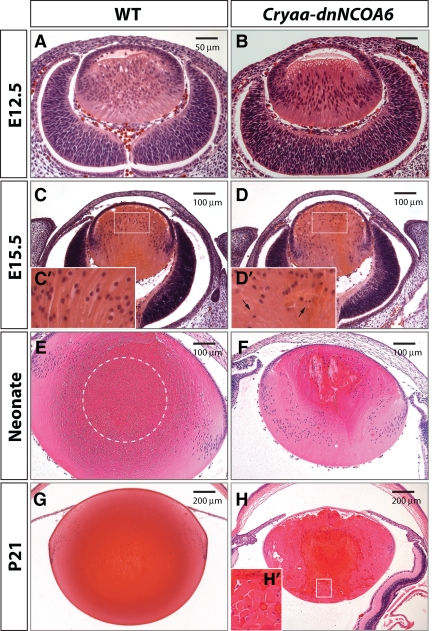
Histological analysis of lens development revealed lens fiber cell differentiation defects in Cryaa-dnNCOA6 lenses. H&E staining was performed with WT (A, C, E, and G) and transgenic (B, D, F, and H) mouse eye sections at E12.5, E15.5, neonatal stage, and P21, respectively, to show lens morphology. At E15.5, the transgenic lens appeared smaller (D) compared with WT (C) and exhibits pyknotic nuclei. Higher magnification images of E15.5 lens fiber cell nuclei are shown as insets (C′ and D′) and pyknotic nuclei are indicated by arrows. The OFZ is located in the center of the WT lens (E and G) and is indicated by dashed circle in E. Transgenic lenses display irregular fiber pattern and pyknotic staining (F and H). The rounded end fragment and cortical liquefaction in dark red are revealed in the inset (H′). Scale bar is shown in each panel.
In the WT neonatal lens, an OFZ exists in the center of the lens (Figure 2E). In contrast, it is absent in the transgenic lens (Figure 2F). The center of the Cryaa-dnNCOA6 lens is filled with fibers containing nuclei, suggesting lens fiber cell denucleation defects. In addition, the deteriorating lens exhibits further abnormalities such as posterior shift of the transitional zone, swollen and shortened fiber cell length, rounded end fragments and cortical liquefaction (Figure 2, F, H, and H′). Fiber cell degeneration and liquefaction suggest abnormal cell–cell adhesion between lens fiber cells. These are common manifestations of cataract formation (Smith, 2002). The entire structure of the lens fiber cell compartment further deteriorates in the P21 transgenic lens as evidenced by the more pronounced structural defects of lens fibers, including an empty space between the epithelium and fiber cell mass (Figure 2H). The pyknotic nuclei persist in the presumptive OFZ, suggesting a permanent block of the lens fiber cell denucleation process.
Next, SEM was used to analyze the lens microstructure, including the suture formation, fiber cell morphology and cell-to-cell interactions. As the fiber cells grow, they elongate and eventually meet other fibers in the anterior and posterior part of the lens in a linear manner forming suture lines, which become the anterior and posterior Y-shaped sutures in opposite orientation (Rafferty and Esson, 1974). The results showed that the Y suture was not observed in Cryaa-dnNCOA6 lens (compare Figure 3, A and D). In addition, transgenic fiber cells were expanded and skewed (compare Figure 3, B and E). In the WT lens, lens fiber cells were regularly arranged with a “ball-and-socket” structure to hold the adjacent lens fibers together (Figure 3, C and C′). In contrast, no ball-and-socket structures were apparent in transgenic lenses (Figure 3, F and F′).
Figure 3.
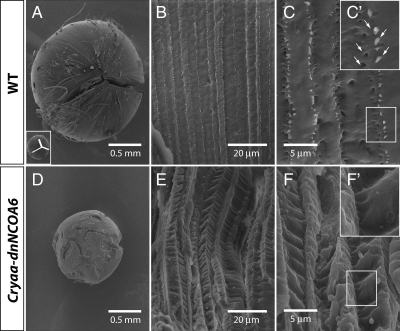
Loss of Y-suture and deformed lens fiber cells in Cryaa-dnNCOA6 lenses. SEM analysis was carried out on 3-mo-old lenses from WT (A–C) and transgenic mice (D–F). Normal Y-suture found in the WT lens (A) is absent in the transgenic lens (D). The Y-suture is illustrated as inset in A. Higher magnification reveals the enlarged and disrupted fiber cell pattern in the transgenic lens (E), whereas the WT lens showed aligned fiber cell pattern (B). Ball-and-socket structure (indicated by white arrows in C′), which is important to hold fiber cells together, is not apparent in the transgenic lens (F and F′) compared with the WT lens (C and C′). Scale bar is given in each panel.
To further examine the terminal differentiation defects in lens fiber cells, an antibody against PDI, an endoplasmic reticulum (ER) marker (Denecke et al., 1992), was used to mark the presence of organelles (Bassnett, 2002). In normal lens, an OFZ, composed of mature fiber cells, forms in the center of the lens cortex after denucleation. Our data showed that PDI expression increased as lens fiber cells differentiate and gradually decreased in fiber cells close to the OFZ in the neonatal WT lens (Figure 4, A–C). As the OFZ formed in the center of the WT lens, no nuclear and PDI staining was observed. In contrast, both nuclei and PDI staining was detected in the center of the transgenic lens (Figure 4, D–F). Some aggregates of ER were even detected in the frontal part of the Cryaa-dnNCOA6 lens (Figure 4F). We conclude that the terminal differentiation process in transgenic lenses was not properly executed; subcellular organelles, including nuclei, were not eliminated and early appearance of pyknotic nuclei (Figure 2) suggested the initiation of apoptosis.
Figure 4.
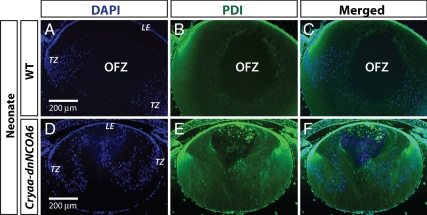
OFZ is absent in the Cryaa-dnNCOA6 lens. An anti-PDI antibody was used to label ER, indicating the existence of organelles, in WT (A–C) and transgenic lenses (D–F) at neonatal stage. Nuclei were stained (blue signal) with DAPI. The center of WT lens cortex is negative with DAPI and anti-PDI antibody staining, which is labeled as OFZ. Lens epithelium, LE; transitional zone (bow region), TZ. Scale bar is shown in A and D.
Initiation of Prolonged and Incomplete Apoptotic-like Processes in the Cryaa-dnNCOA6 Lens
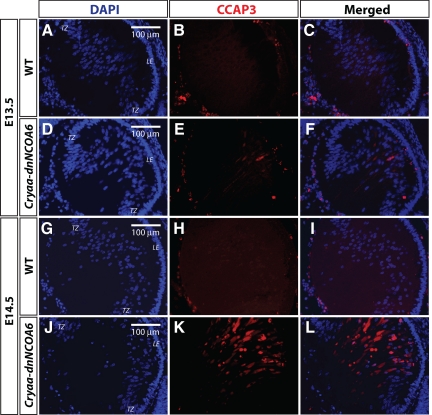
Considering the important role of Ncoa6 gene in cell survival (Mahajan et al., 2004) and the presence of pyknotic nuclei in the Cryaa-dnNCOA6 lens, an indicator of apoptosis (Figure 2), we next examined the transgenic lens for evidence of apoptosis. The early stages of apoptosis are associated with the activation of caspase-3, a death protease that is activated by initiator caspases (Siegel, 2006). We monitored activated caspase-3 (cleaved caspase-3, CCAP3) in Cryaa-dnNCOA6 lenses and identified CCAP3-positive cells in both E13.5 and E14.5 transgenic lenses in lens fiber cell compartment but not in the WT control (Figure 5). These data suggest that activated caspase-3 was induced in lens fiber cells within ∼24 h after the onset of dnNCOA6 expression in elongating primary lens fiber cells.
Figure 5.
Caspase-3 activation in differentiating transgenic lens fiber cells. E13.5 and E14.5 WT and transgenic lenses were subjected to immunofluorescence analysis with CCAP3 antibody. The E13.5 transgenic lens showed staining in the lens fiber cell compartment (D–F) compared with WT (A–C). The amount of CCAP3 staining increased in the E14.5 transgenic lens (J–L) and no staining was detected in the WT lens (G–I). Nuclei (blue signal) were stained with DAPI. Lens epithelium, LE; transitional zone, TZ. Scale bar is shown in panels A, D, G, and J.
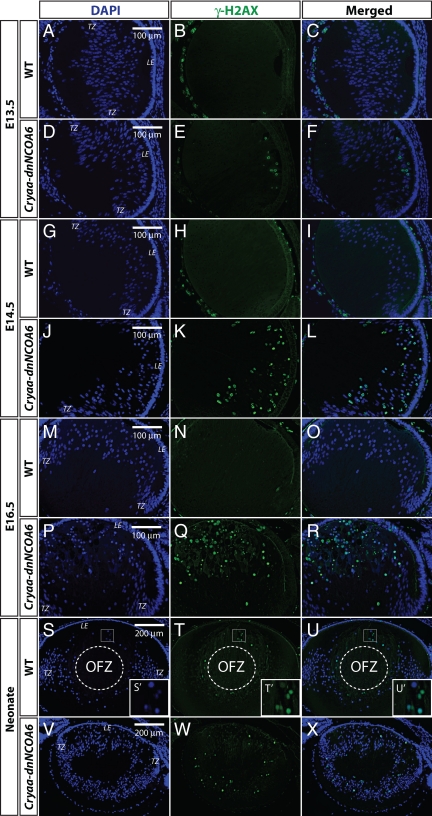
In general, DNA lesion in mammalian cells can create DSBs, which trigger a signaling cascade to form a repair complex at individual break points (Petrini and Stracker, 2003). At the very early step, upstream kinases phosphorylate histone H2A variant H2AX at Ser 139 (γ-H2AX) surrounding DSBs and γ-H2AX facilitates the recruitment of proteins to repair foci (Fernandez-Capetillo et al., 2004; Thiriet and Hayes, 2005). Thus, a γ-H2AX antibody was used to examine embryonic and neonatal lenses. The DSBs were detected in E13.5 transgenic lenses and became more abundant in E14.5, E16.5, and neonatal transgenic lenses (Figure 6), indicating the presence of severe DNA damage. In addition, we found the presence of T2609 phosphorylated DNA protein kinase catalytic subunit (DNA-PKcs) (Supplemental Figure S3), which is activated after DSBs in nonhomologous end-joining DNA repair pathway (Chan et al., 2002).
Figure 6.
Identification of DNA double-strand breaks (DSBs) through γ-H2AX immunofluorescence in WT and Cryaa-dnNCOA6 lens fiber cells. γ-H2AX antibody was used to label DSBs generated through DNA damage or normal denucleation process. DSBs are detected as early as E13.5 in the transgenic lens fiber cell compartment (D–F) and are absent in the WT lens (A–C). The number of DSBs increased in E14.5 and E16.5 transgenic lens fiber cells (J–L and P–R), whereas WT controls are negative (G–I and M–O). In the neonatal WT lens, nuclei of lens fiber cells at the margin of the OFZ display strong γ-H2AX staining (S–U and S′–U′). The neonatal transgenic lens exhibits dispersed DSBs staining in fiber cells and the absence of OFZ (V–X). The OFZ is indicated by dashed circle. Nuclei (blue signal) were stained with DAPI. Mesenchymal cells and hyaloid vascular structure surrounding the lens often generate nonspecific signals, which do not interfere with our observations in the lens fiber cell compartment. Lens epithelium, LE; transitional zone, TZ. Scale bar is shown in A, D, G, J, M, P, S, and V.
At the morphological level, lens fiber cells start the denucleation process at ∼E17 (Vrensen et al., 1991; Bassnett, 2009). No DSBs were detected in WT embryonic lenses younger than or equal to E16.5 (Figure 6, A–C, G–I, and M–O). In WT E17.5 lenses, primary lens fiber cells close to the presumptive OFZ showed positive γ-H2AX staining (data not shown). In contrast, newborn WT lenses stained with γ-H2AX in the margin of OFZ in secondary lens fiber cells (Figure 6, S–U and S′–U′).
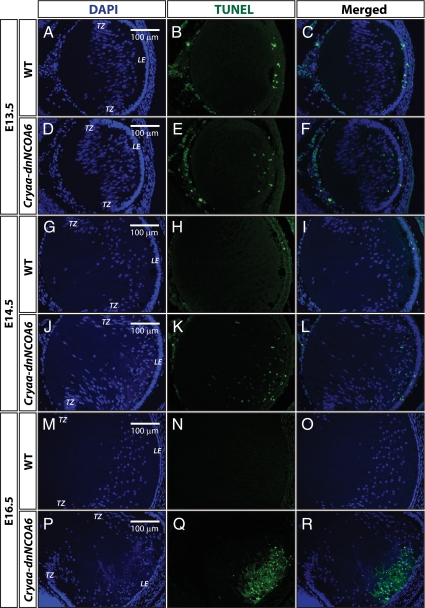
Next, embryonic lens tissues were examined for apoptosis by TUNEL assay. Some TUNEL-positive cells were detected in the proliferating lens epithelium and the fiber cell compartment in both E13.5 and E14.5 transgenic lenses (Figure 7, D–F and J–L). In E16.5 transgenic lenses, the apoptotic signals increased dramatically in lens fiber cell compartment (Figure 7, P–R). Some sporadic apoptotic nuclei are observed in WT lens epithelium cells; however, no apoptotic cells were found in differentiating WT lens fiber cells (Figure 7, A–C, G–I, and M–O). Evidence for massive DNA damage or cell death was suggested by TUNEL signals in E16.5 transgenic lenses (Figure 7, P–R). However, completion of apoptosis is uncertain, because abnormal lens fiber cells with pyknotic nuclei are found in both neonatal and P21 lenses (Figures 2 and 4).
Figure 7.
TUNEL assays in WT and Cryaa-dnNCOA6 lens fiber cells. (A–C) At E13.5, sporadic apoptotic nuclei were detected only in the epithelium cells of WT lenses. (D–F) In contrast, apoptotic nuclei were detected in both epithelium and lens fiber cells of transgenic lenses. TUNEL-positive nuclei: 5.47 ± 2.00 nuclei/section. (J–L) Similar results were observed in E14.5 lenses with slightly elevated number of apoptotic nuclei in transgenic lens fiber cells. TUNEL-positive nuclei: 12.88 ± 7.59 nuclei/section. (P–R) At E16.5, strong TUNEL signals were detected in the frontal part of lens fiber cell compartment in the transgenic lens. No TUNEL-positive nuclei were observed in WT lens fiber cells (A–C, G–I, and M–O). Nonspecific signals outside of the lens are explained in Figure 6. Lens epithelium, LE; transitional zone, TZ. Scale bar is shown in A, D, G, J, M, and P.
In summary, no DSBs were detected in E16.5 WT lenses (beginning of primary lens fiber cell denucleation process), but DSBs were found in both denucleating primary (E17.5) and secondary fiber cells (neonate). In transgenic lenses, DSBs and the apoptosis signals appeared before the normal denucleation process (∼E17). Nevertheless, the apoptotic-like process here differs from the typical apoptosis because the onset of caspase-3 activation and formation of intense TUNEL signals are separated by ∼72 h. Both lens fiber cell elimination and denucleation processes were arrested suggesting that lens fiber cells operate a specific mechanism to preserve cells, even with nuclei exhibiting both morphological and molecular abnormalities.
Abnormal Lens Fiber Cell Differentiation Is Suppressed in the Absence of p53
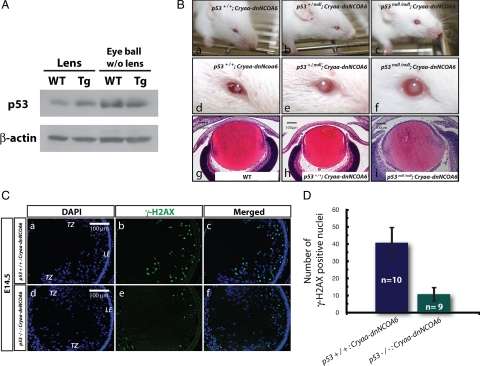
It has been reported previously that the overexpression of human p53 gene induced apoptosis in the transgenic mouse lens and that the apoptosis was rescued by a p53 mutant transgene, which can interfere with p53 normal function (Nakamura et al., 1995). E7, an oncoprotein encoded by the human papillomavirus HPV-16, induced cell cycle reentry in transgenic lens fiber cells and p53-dependent and p53-independent apoptosis (Pan and Griep, 1994, 1995). Here, we found that p53 protein expression was elevated in Cryaa-dnNCOA6 lenses in the range of 1.42- to 2.12-fold compared with the WT but not in other ocular tissues (Figure 8A). We also found three p53 target genes, i.e., p21, Bim, and Perp, up-regulated in transgenic lenses at the RNA level (data not shown).
Figure 8.
Abnormal lens fiber cell apoptosis is both p53-dependent and p53-independent. (A) Western blot was performed to analyze p53 protein expression in the lens and eyeball (without lens) of WT and transgenic mice at neonatal stage. β-Actin was used for loading control. (B) External observation of adult mouse eyes (a–f) and histological analysis of E16.5 mouse eyes (g–i) in p53+/+; Cryaa-dnNCOA6, p53+/null; Cryaa-dnNCOA6, p53null/null; Cryaa-dnNCOA6, and WT mice. (C) Reduced number of DSBs in p53null/null; Cryaa-dnNCOA6 lenses (d–f) compared with p53+/+; Cryaa-dnNCOA6 lenses at E14.5 (a–c). Nuclei (blue signal) were stained with DAPI. Scale bar is shown in a and d. (D) To quantify the amounts of DSBs in Cryaa-dnNCOA6 lenses in WT or p53 null background, the number of γ-H2AX-positive nuclei was compared between E14.5 p53+/+; Cryaa-dnNCOA6 (n = 10) and p53null/null; Cryaa-dnNCOA6 (n = 9) lenses. Nonspecific signals outside of the lens are explained in Figure 6. Lens epithelium, LE; transitional zone, TZ.
Next, to examine whether defects in Cryaa-dnNCOA6 lenses were p53-dependent in lens fiber cells, we crossed the Cryaa-dnNCOA6 mice to mice carrying a null mutation in the p53 gene (Donehower et al., 1992) by using a strategy reported previously (Hettmann et al., 2000; Wang et al., 2005; Cang et al., 2006; Yang et al., 2006b). As shown in Figure 8B, increased p53+/null; Cryaa-dnNCOA6 mice eye (lens) size compared with p53+/+; Cryaa-dnNCOA6 was found. In p53null/null; Cryaa-dnNCOA6 mice, eye size improvement was even more pronounced. Quantitative analysis using neonate lenses revealed 1.35- to 1.65-fold increase of lens size in p53null/null compared with WT background. Histological data from E16.5 WT control, p53+/+; Cryaa-dnNCOA6 and p53null/null; Cryaa-dnNCOA6 are shown in Figure 8B for comparison. To further evaluate the p53 rescue effect, embryonic tissue sections from E14.5 p53+/+; Cryaa-dnNCOA6 and p53null/null; Cryaa-dnNCOA6 were examined with a γ-H2AX antibody and the amounts of DSBs in the lens were quantified. The deletion of p53 significantly reduced the amount of DSBs induced by Cryaa-dnNCOA6 transgene (Figure 8C). There was ∼60% reduction in DSB-positive nuclei in p53null/null; Cryaa-dnNCOA6 lenses compared with the normal p53 background (Figure 8D). These findings support the idea that defects in the Cryaa-dnNCOA6 differentiating lens fiber cells can be alleviated in the p53 null background and that the formation of apoptotic-like cells was both p53-dependent and p53-independent.
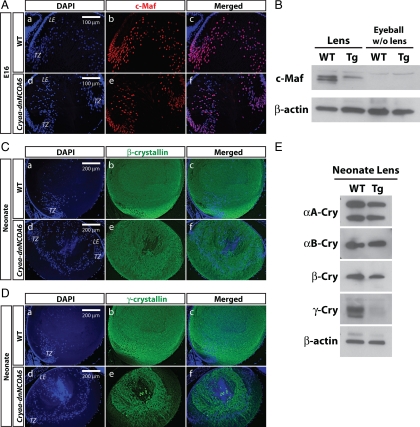
Decrease of c-Maf Protein Expression in Cryaa-dnNCOA6 Lens Fiber Cells Is Accompanied by Reduced γ-Crystallin Proteins Expression
Previous studies of ubiquitously expressed dnNCOA6 identified reduced expression of αB- and γ-crystallins and linked these results with the presence of retinoic acid responsive elements (RAREs) in their 5′-regulatory regions (Kim et al., 2002). Here, we first examined the expression levels of two lens fiber cell differentiation factors, c-Maf and Pax6 (Shaham et al., 2009). Transcription factor c-Maf is crucial for the high levels of crystallin gene expression (Kawauchi et al., 1999; Kim et al., 1999; Ring et al., 2000; Rajaram and Kerppola, 2004; Yang et al., 2004). Our results showed that the expression of c-Maf protein was decreased in the cortex region of the Cryaa-dnNCOA6 lens (Figure 9A). In addition, whole cell lysates from WT and transgenic lens and eyeball (without lens) were analyzed by protein immunoblot with an anti-c-Maf antibody. We found 2.37- to 2.64-fold reduction of c-Maf protein in the transgenic lens compared with WT (Figure 9B). Pax6 regulates lens development and crystallin gene expression at multiple levels (Cvekl and Duncan, 2007). It also regulates c-Maf gene expression (Sakai et al., 2001). However, no difference was observed in the expression level of Pax6 protein between the WT and transgenic lens (data not shown). Thus, down-regulation of the c-Maf protein level in the Cryaa-dnNCOA6 lens was independent of Pax6. Next, we used crystallin-specific antibodies to examine lens fiber cell differentiation. Down-regulation of both β- and γ-crystallins was observed by immunofluorescence (Figure 9, C and D). Strong down-regulation of γ-crystallin proteins was confirmed by Western blot (Figure 9E). From these results we conclude that expression of γ-crystallins was more affected in transgenic lenses compared with other crystallins examined (αB- and β-crystallins).
Figure 9.
Down-regulation of c-Maf and γ-crystallins in the Cryaa-dnNCOA6 transgenic lenses. (A) Immunofluorescence staining was carried out in E16 WT and transgenic lenses. In WT, c-Maf protein expression was up-regulated in the transitional zone and persisted in lens fiber nuclei (A, a–c). However, many lens fiber nuclei lost c-Maf protein expression in the transgenic lens (A, d–f). Nuclei (blue signal) were stained with DAPI. (B) Western blot analysis of the c-Maf protein expression level in WT and transgenic neonatal lenses (2.37- to 2.64-fold reduction in transgenic lenses) and eyeballs (without lens). β-Actin was used as loading control. (C) Some reduction of β-crystallins in transgenic lenses was observed compared with WT lenses. Lens epithelium, LE; transitional zone, TZ. (D) γ-Crystallin proteins expression is down-regulated in transgenic lenses compared with WT lenses. (E) Western blot analysis of αA-crystallin (αA-Cry), αB-crystallin (αB-Cry), β-crystallin (β-Cry), and γ-crystallin (γ-Cry) protein expression levels in neonatal WT and transgenic lenses. β-Actin served as loading control. The protein expression ratios of WT to transgenic lenses are indicated as follows: αA-Cry (1.08–1.18), αB-Cry (1.18–1.23), β-Cry (1.46–1.56), and γ-Cry (1.56–2.95).
Lens-specific Deletion of Ncoa6 Caused Microphthalmia and Cataract Formation
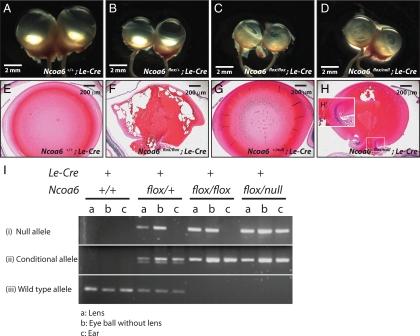
Although a cell-autonomous role of Ncoa6 in lens development is suggested by experiments described above, examination of lens-specific Ncoa6 knockout offers an opportunity to test this mechanism independently. To delete Ncoa6 in the lens, Le-Cre mice (Ashery-Padan et al., 2000) were crossed with floxed Ncoa6 mice for specific deletion in the surface ectoderm/lens lineage. We observed smaller and cloudy lenses (Figure 10, C, D, F, and H) with persisting nuclei (Figure 10H′). In contrast, Ncoa6 heterozygous mutants did not reveal any significant lens phenotype (Figure 10, B and G). Although phenotypes exhibited by Ncoa6flox/flox; Le-Cre and Ncoa6flox/null; Le-Cre mice were milder than those in Cryaa-dnNCOA6 mice, the penetrance was 100% (Table 1). Genotyping to detect WT, conditional, and null alleles (see Materials and Methods) revealed incomplete deletion of the floxed Ncoa6 allele in the lens (Figure 10I).
Figure 10.
Deletion of Ncoa6 in prospective lens ectoderm led to microphthalmia and cataract. (A–D) Eyeball morphology of four genotypes: Ncoa6+/+; Le-Cre, Ncoa6flox/+; Le-Cre, Ncoa6flox/flox; Le-Cre, and Ncoa6flox/null; Le-Cre at ∼3 mo old. (E–H) H&E staining of 3-mo-old lenses from Ncoa6+/+; Le-Cre and Ncoa6flox/flox; Le-Cre mice, and 1-mo-old lenses from Ncoa6+/null; Le-Cre and Ncoa6flox/null; Le-Cre mice. Scale bar is shown in each panel. (I) To analyze the deletion efficiency of the floxed allele, lens, eyeball (without lens), and ear tissues from Ncoa6+/+; Le-Cre, Ncoa6flox/+; Le-Cre, Ncoa6flox/flox; Le-Cre, and Ncoa6flox/null; Le-Cre mice were used to extract genomic DNA for genotyping. Analysis of ear tissue confirmed the tissue-specific expression of Le-Cre. In Ncoa6+/+; Le-Cre tissues, only the WT allele was detected. Most Ncoa6flox/+; Le-Cre tissues showed the WT, conditional and null alleles except ear tissues in which no null allele is detected. In Ncoa6flox/flox; Le-Cre mice, the null allele is detected in the lens; however, the deletion is incomplete as in the Ncoa6flox/null; Le-Cre lens.
Table 1.
Eye phenotypes observed in the lens-specific Ncoa6 conditional knockout mice
| Genotype | Phenotype | Onset age |
|---|---|---|
| Ncoa6+/+; Le-Cre | None | |
| Ncoa6+/null; Le-Cre | None | |
| Ncoa6flox/flox; Le-Cre | Small eye (17/17) | 1∼5 mo |
| Severe cataract (13/17) | 1∼5 mo | |
| Mild cataract (4/17) | 1∼2.5 mo | |
| Ncoa6flox/null; Le-Cre | Small eye and cataract (14/14) | 21 d ∼2.5 mo |
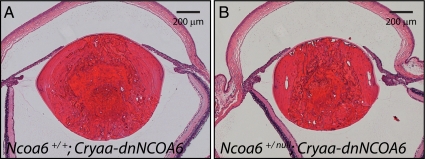
The dnNCOA6 fragment was shown to interfere with retinoid-dependent recruitment of NCOA6 to RAREs, which could not be rescued by TRAP220 and SRC-2, two other coactivators involved in RA-dependent gene regulation (Kim et al., 2002). However, no genetic experiment has been performed to test the role of dnNCOA6 as a dominant inhibitor in vivo. To prove the loss-of-function effect of dnNCOA6 in the lens, Ncoa6+/null mice were crossed with Cryaa-dnNCOA6 mice to generate Ncoa6+/null; Cryaa-dnNCOA6, and Ncoa6+/+; Cryaa-dnNCOA6 mice. Both Ncoa6 +/null; Cryaa-dnNCOA6 and Ncoa6 +/+; Cryaa-dnNCOA6 mice have cataract phenotype and the lenses of Ncoa6+/null; Cryaa-dnNCOA6 mice were 23 ± 11% smaller than Ncoa6+/+; Cryaa-dnNCOA6 mice (Figure 11).
Figure 11.
Increased lens abnormalities in Ncoa6+/null; Cryaa-dnNCOA6 mice. Heterozygous Ncoa6 mice were crossed with Cryaa-dnNCOA6 mice to generate Ncoa6+/+; Cryaa-dnNCOA6 and Ncoa6 +/null; Cryaa-dnNCOA6 mice. Serial eye sections were used to compare the lens size of each genotype. The result indicates that the Ncoa6+/null; Cryaa-dnNCOA6 lens (B) is smaller than the Ncoa6+/+; Cryaa-dnNCOA6 lens (A). Quantitative analysis using P21 lenses revealed that the Ncoa6 +/null; Cryaa-dnNCOA6 lens is 23 ± 11% smaller than the Ncoa6+/+; Cryaa-dnNCOA6 lens. Eyes from three mice of each genotype were analyzed. Scale bar is shown in each panel.
DISCUSSION
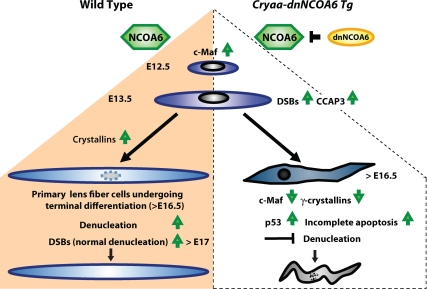
The present data show multiple novel functions of Ncoa6 in lens fiber cell differentiation. Lens-specific expression of a dominant-negative fragment that abrogates the function of NCOA6 elicited a series of lens fiber cell-specific differentiation defects, including disruption of crystallin protein expression, initiation of prolonged yet incomplete apoptosis in the lens fiber cell compartment, and inhibition of lens fiber cell denucleation (Figure 12). In parallel, the lens-specific knockout of Ncoa6 resulted in lens fiber cell differentiation defects including the persisting nuclei, which indicates inhibition of lens fiber cell karyolysis. The lens vesicle in E11.5 Ncoa6 knockout mice was normal (data not shown). These findings reveal that Ncoa6 is not required for the formation of lens lineage/precursor cells. The incomplete deletion of the floxed Ncoa6 allele by the Le-Cre system found here and reported previously (Swamynathan et al., 2007) could explain the milder defects in the Ncoa6 conditional knockout compared with the Cryaa-dnNCoA6 transgenic mouse model. Furthermore, the lens-specific deletion of Ncoa6 did not exhibit morphologically detectable phenotypes until 3–4 wk of age (Table 1), and these defects were limited to lens fiber cells. We found that a single copy of Ncoa6 was sufficient to support normal lens development. In addition, the aggravation of the lens phenotype in Ncoa6+/null; Cryaa-dnNCOA6 mice confirmed genetically the specificity of the dnNCOA6 action. Collectively, our data revealed an essential cell-autonomous role of Ncoa6 in lens fiber differentiation and denucleation.
Figure 12.
Summary diagram of lens fiber cell differentiation and the role of NCOA6 in the denucleation process. In WT lens (left side of the diagram), up-regulation of c-Maf and crystallins is essential for lens fiber cell differentiation. Their terminal differentiation requires orchestrated degradation of all subcellular organelles to avoid light scattering. Nuclear degradation is a relatively lengthy process (3–4 d in mouse; Bassnett, 2009). In Cryaa-dnNCOA6 transgenic lenses (right side of the diagram), p53-dependent and p53-independent proapoptotic processes become evident as early as at E13.5 due to the generation of CCAP3 followed by the appearance of DSBs monitored via γ-H2AX–specific antibodies. Although abundant TUNEL signals are found at E16.5, the nuclear degradation is not properly executed and abnormally differentiated lens fibers, with significantly reduced expression of c-Maf and γ-crystallins and retained nuclei, are preserved in the transgenic lenses.
At the molecular level, our data showed that lens-specific expression of dnNCOA6 preferentially reduced γ-crystallin protein expression. Expression of αB- and β-crystallins was less affected. In contrast, ubiquitously expressed dnNCOA6 inhibited expression of αB-, γC-, γD-, and γF-, but not αA-, βA3/A1-, γA-, and γB-crystallins at RNA level (Kim et al., 2002). Our data showed reduced but not abolished expression of c-Maf, an upstream regulatory protein of crystallins, in the Cryaa-dnNCOA6 lens. These findings indicate that expression of γ-crystallins seems to be most sensitive to reduced expression levels of c-Maf. In c-Maf+/− lenses, no change in crystallin gene expression profiles was found (Ring et al., 2000). Reduced expression of αB- and γF-crystallin genes in CMV/β-actin-dnNCOA6 transgenic mice (Kim et al., 2002) was explained via the inhibition of RA signaling by dnNCOA6. Previously, RA signaling and retinoic acid receptor/retinoid X receptor nuclear receptors were shown to regulate these crystallin promoters (Tini et al., 1993; Gopal-Srivastava et al., 1998; Chauhan et al., 2004). However, RA signaling is attenuated in E13.5 differentiating lens fiber cells as measured by the activity of the RARE-lacZ reporter in the embryonic eye (Rossant et al., 1991; Balkan et al., 1992; Enwright and Grainger, 2000). Thus, down-regulation of c-Maf might contribute to reduced expression of γ-crystallins after E13.5 in the lens, whereas both RA signaling and c-Maf could play roles in the regulation of αB-, γC-, γD-, and γF-crystallins during E12.5–E13.5 of lens differentiation.
Our study elucidates the apoptotic-like process in the lens fiber cell compartment and their relationship to karyolysis. Programmed cell death takes place in a time frame of 2–12 h (Potten, 1996; Feinstein-Rotkopf and Arama, 2009), and the denucleation process takes over a course of 3–4 d (Bassnett, 2009). The final stage of apoptosis is the formation of apoptotic bodies followed by engulfment of cell corpses by nearby cells. In contrast, denucleated lens fiber cells are preserved. Several other models examined apoptosis induced by lens-specific transgenes. These studies identified abrupt differentiation and apoptosis in lens fiber cells (Stolen and Griep, 2000; de Iongh et al., 2001) or showed that lens fiber cells reentered cell cycle and displayed apoptosis (Pan and Griep, 1995; Chen et al., 2002, 2004; Xie et al., 2006). No evidence for cell cycle reentry in the Cryaa-dnNCOA6 lens was detected (Supplemental Figure S4). The first indication of apoptosis in transgenic lenses was found at E13.5 with the activation of caspase-3. However, abundant TUNEL-positive cells were detected at E16.5 in transgenic lenses. Caspase-3 is required for cytoplasmic (membrane blebbing and cell shrinkage) and nuclear events (DNA fragmentation) associated with apoptosis (Janicke et al., 1998; Zheng et al., 1998; Porter and Janicke, 1999). The 3-d gap between the earliest sign of apoptosis and abundant TUNEL-positive cells is unusual (Feinstein-Rotkopf and Arama, 2009). We speculate that the programmed cell death induced in Cryaa-dnNCOA6 lenses could be desensitized like in certain cancer cells (Kawabata et al., 1999) as the lens fiber cells are programmed for denucleation (Cvekl and Mitton, 2010). In addition, we did not find evidence for the generation of apoptotic bodies and their engulfment by surrounding lens fiber cells. Transgenic lens fiber cells were preserved in the fiber cell compartment and exhibited denucleation defects.
The present data showed the formation of DSBs and activation of DNA-PKcs in normal lens fiber cells in the margin of OFZ. Both of these processes also occurred in E13.5 Cryaa-dnNCOA6 lenses. In WT lenses, DSBs were detected from E17.5 in the region of prospective OFZ. In neonatal lenses, DSBs were found in the margin of the OFZ marking the denucleating secondary lens fibers. Interestingly, transgenic lens fiber cells showed denucleation defects. Thus, it is intriguing that although both processes, i.e., apoptosis and denucleation, are aimed to eliminate nuclei; the outcome of their interaction is the retention of nuclei. We propose that apoptosis activated in Cryaa-dnNCOA6 elongating lens fibers was arrested and prevented the completion of karyolysis. Alternatively, the normal lens fiber cell differentiation program was disrupted early (∼E13.5–E17) and the denucleation process was never properly activated.
p53 plays a central role in genome integrity and apoptosis regulation (Zhivotovsky and Kroemer, 2004). We found up-regulation of p53 in Cryaa-dnNCOA6 lenses, which is consistent with the initiation of proapoptotic process in the lens fiber cell compartment. In the WT lens, p53 is not required for lens fiber cell differentiation because p53 null lenses seem normal (Donehower et al., 1992). The p53-dependent pathway becomes important only if cell cycle reentry (Pan and Griep, 1995) or proapoptotic processes are initiated in differentiating lens fiber cells such as in the present study. Our data showed that apoptosis in Cryaa-dnNCOA6 lens fiber cells was both p53-dependent and p53-independent, in agreement with studies of other genes in the lens (Morgenbesser et al., 1994; Pan and Griep, 1995; Li et al., 2005).
In this study, early-induced proapoptotic processes are significantly delayed in the lens fiber cell compartment. Both αA- and αB-crystallins are important to prevent apoptosis in the lens (Li et al., 2005; Morozov and Wawrousek, 2006; Xi et al., 2008), tumor cells (Stegh et al., 2008), and in other cell types (Alge et al., 2002; Kamradt et al., 2002), Here, we did not identify reduction of αA- and αB-crystallins protein expression in the Cryaa-dnNCOA6 lens. These findings suggest that anti-apoptotic role(s) of Ncoa6 in the lens are not mediated via anti-apoptotic activities of α-crystallins. Between E17 and 18, apoptotic-like lens fiber cell nuclei should receive additional destructive signals via the denucleation (karyolysis) pathway as described above. It is tempting to speculate that antiapoptotic proteins such as NCOA6 and crystallins regulate the antiapoptotic and denucleation pathways. Their existence preserves and ensures that lens fiber cells survive as “frozen” bags to generate a transparent and refractive lens. It is also interesting to note that specific mouse γ-crystallin mutant lenses displayed abnormalities in the denucleation process (Graw, 2004, 2009; Graw et al., 2004; Sandilands et al., 2002). Identification and functional characterization of novel proteins and enzymes that regulate lens fiber cell denucleation is needed for better understanding how lens fiber cells degrade genomic DNA and protect themselves from entering apoptosis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jae Woon Lee for providing pCAGGS-ASC2co2CN plasmid. We are grateful to Drs. Melinda Duncan, Anne Griep, Paul Overbeek, Louise Wolf, and Uwe Werling for critical reading of this manuscript. Histology and analytical imaging facilities at the Albert Einstein College of Medicine are acknowledged for their support, special thanks to Drs. Ruth Ashery-Padan and Judy West-May for providing us Le-Cre mice, and Dr. Winfried Edelmann for EIIa-Cre and p53 knockout mice. Data in this article are from a thesis to be submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University. This study was supported by National Institutes of Health grants R01EY-012200 (to A. C.), EY-014237 (to A. C.), and CA-119689 (to J. X.). A. C. is a recipient of the Irma T. Hirschl Career Scientist Award.
Abbreviations used:
- dn
dominant-negative
- DSB
double-strand break
- ER
endoplasmic reticulum
- H&E
hematoxylin and eosin
- NR
nuclear receptor
- OFZ
organelle-free zone
- RARE
retinoic acid-responsive elements
- SEM
scanning electron microscope
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1031) on May 19, 2010.
REFERENCES
- Alge C. S., Priglinger S. G., Neubauer A. S., Kampik A., Zillig M., Bloemendal H., Welge-Lussen U. Retinal pigment epithelium is protected against apoptosis by alphaB-crystallin. Invest. Ophthalmol. Vis. Sci. 2002;43:3575–3582. [PubMed] [Google Scholar]
- Andley U. P. Crystallins in the eye: function and pathology. Prog. Retin. Eye Res. 2007;26:78–98. doi: 10.1016/j.preteyeres.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Antonson P., Schuster G. U., Wang L., Rozell B., Holter E., Flodby P., Treuter E., Holmgren L., Gustafsson J. A. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol. Cell Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R., Marquardt T., Zhou X., Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkan W., Colbert M., Bock C., Linney E. Transgenic indicator mice for studying activated retinoic acid receptors during development. Proc. Natl. Acad. Sci. USA. 1992;89:3347–3351. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. Lens organelle degradation. Exp. Eye Res. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp. Eye Res. 2009;88:133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang Y., Zhang J., Nicholas S. A., Bastien J., Li B., Zhou P., Goff S. P. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell. 2006;127:929–940. doi: 10.1016/j.cell.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Canman C. E. Checkpoint mediators: relaying signals from DNA strand breaks. Curr. Biol. 2003;13:R488–R490. doi: 10.1016/s0960-9822(03)00410-x. [DOI] [PubMed] [Google Scholar]
- Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan B. K., Yang Y., Cveklova K., Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 2004;32:1696–1709. doi: 10.1093/nar/gkh334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ash J. D., Branton P., Fromm L., Overbeek P. A. Inhibition of crystallin expression and induction of apoptosis by lens-specific E1A expression in transgenic mice. Oncogene. 2002;21:1028–1037. doi: 10.1038/sj.onc.1205050. [DOI] [PubMed] [Google Scholar]
- Chen Q., Hung F. C., Fromm L., Overbeek P. A. Induction of cell cycle entry and cell death in postmitotic lens fiber cells by overexpression of E2F1 or E2F2. Invest. Ophthalmol. Vis. Sci. 2000;41:4223–4231. [PubMed] [Google Scholar]
- Chen Q., Liang D., Yang T., Leone G., Overbeek P. A. Distinct capacities of individual E2Fs to induce cell cycle re-entry in postmitotic lens fiber cells of transgenic mice. Dev. Neurocci. 2004;26:435–445. doi: 10.1159/000082285. [DOI] [PubMed] [Google Scholar]
- Cvekl A., Duncan M. K. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog. Retin. Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A., Mitton K. P. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105 doi: 10.1038/hdy.2010.16. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Iongh R. U., Gordon-Thomson C., Chamberlain C. G., Hales A. M., McAvoy J. W. Tgfbeta receptor expression in lens: implications for differentiation and cataractogenesis. Exp. Eye Res. 2001;72:649–659. doi: 10.1006/exer.2001.1001. [DOI] [PubMed] [Google Scholar]
- Denecke J., De Rycke R., Botterman J. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 1992;11:2345–2355. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr, Butel J. S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Duncan M. K., Cvekl A., Li X., Piatigorsky J. Truncated forms of Pax-6 disrupt lens morphology in transgenic mice. Invest. Ophthalmol. Vis. Sci. 2000;41:464–473. [PubMed] [Google Scholar]
- Enwright J. F., 3rd, Grainger R. M. Altered retinoid signaling in the heads of small eye mouse embryos. Dev. Biol. 2000;221:10–22. doi: 10.1006/dbio.2000.9652. [DOI] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y., Arama E. Can't live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis. 2009;14:980–995. doi: 10.1007/s10495-009-0346-6. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. H2AX: the histone guardian of the genome. DNA Rep. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Fromm L., Shawlot W., Gunning K., Butel J. S., Overbeek P. A. The retinoblastoma protein-binding region of simian virus 40 large T antigen alters cell cycle regulation in lenses of transgenic mice. Mol. Cell Biol. 1994;14:6743–6754. doi: 10.1128/mcb.14.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Lahoz E., et al. Cyclin D- and E-dependent kinases and the p57(KIP2) inhibitor: cooperative interactions in vivo. Mol. Cell Biol. 1999;19:353–363. doi: 10.1128/mcb.19.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal-Srivastava R., Cvekl A., Piatigorsky J. Involvement of retinoic acid/retinoid receptors in the regulation of murine alphaB-crystallin/small heat shock protein gene expression in the lens. J. Biol. Chem. 1998;273:17954–17961. doi: 10.1074/jbc.273.28.17954. [DOI] [PubMed] [Google Scholar]
- Graw J. The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- Graw J. Congenital hereditary cataracts. Int. J. Dev. Biol. 2004;48:1031–1044. doi: 10.1387/ijdb.041854jg. [DOI] [PubMed] [Google Scholar]
- Graw J. Genetics of crystallins: cataract and beyond. Exp. Eye Res. 2009;88:173–189. doi: 10.1016/j.exer.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Graw J., Neuhauser-Klaus A., Klopp N., Selby P. B., Loster J., Favor J. Genetic and allelic heterogeneity of Cryg mutations in eight distinct forms of dominant cataract in the mouse. Invest. Ophthalmol. Vis. Sci. 2004;45:1202–1213. doi: 10.1167/iovs.03-0811. [DOI] [PubMed] [Google Scholar]
- Griep A. E. Cell cycle regulation in the developing lens. Semin. Cell Dev. Biol. 2006;17:686–697. doi: 10.1016/j.semcdb.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep A. E., Herber R., Jeon S., Lohse J. K., Dubielzig R. R., Lambert P. F. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol. 1993;67:1373–1384. doi: 10.1128/jvi.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmann T., Barton K., Leiden J. M. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev. Biol. 2000;222:110–123. doi: 10.1006/dbio.2000.9699. [DOI] [PubMed] [Google Scholar]
- Janicke R. U., Sprengart M. L., Wati M. R., Porter A. G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- Kamradt M. C., Chen F., Sam S., Cryns V. L. The small heat shock protein alpha B-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J. Biol. Chem. 2002;277:38731–38736. doi: 10.1074/jbc.M201770200. [DOI] [PubMed] [Google Scholar]
- Kawabata Y., Hirokawa M., Kitabayashi A., Horiuchi T., Kuroki J., Miura A. B. Defective apoptotic signal transduction pathway downstream of caspase-3 in human B-lymphoma cells: a novel mechanism of nuclear apoptosis resistance. Blood. 1999;94:3523–3530. [PubMed] [Google Scholar]
- Kawauchi S., Takahashi S., Nakajima O., Ogino H., Morita M., Nishizawa M., Yasuda K., Yamamoto M. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 1999;274:19254–19260. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- Kim J. I., Li T., Ho I. C., Grusby M. J., Glimcher L. H. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl. Acad. Sci. USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol. Cell Biol. 2002;22:8409–8414. doi: 10.1128/MCB.22.24.8409-8414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S. Q., Liao L., Zhang H., Pereira F. A., Yuan Y., DeMayo F. J., Ko L., Xu J. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J. Biol. Chem. 2002;277:45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Wroblewski E., Philips G. T., Stair C. N., Conley K., Reedy M., Mastick G. S., Brown N. L. Multiple requirements for Hes 1 during early eye formation. Dev. Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. W., Liu J. P., Mao Y. W., Xiang H., Wang J., Ma W. Y., Dong Z., Pike H. M., Brown R. E., Reed J. C. Calcium-activated RAF/MEK/ERK signaling pathway mediates p53-dependent apoptosis and is abrogated by alpha B-crystallin through inhibition of RAS activation. Mol. Biol. Cell. 2005;16:4437–4453. doi: 10.1091/mbc.E05-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippens S., Hoste E., Vandenabeele P., Agostinis P., Declercq W. Cell death in the skin. Apoptosis. 2009;14:549–569. doi: 10.1007/s10495-009-0324-z. [DOI] [PubMed] [Google Scholar]
- Liu K., Luo Y., Lin F. T., Lin W. C. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Lagutin O. V., Mende M., Streit A., Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu F. J., McAvoy J. W. Growth factor regulation of lens development. Dev. Biol. 2005;280:1–14. doi: 10.1016/j.ydbio.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Mahajan M. A., Das S., Zhu H., Tomic-Canic M., Samuels H. H. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol. Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M. A., Murray A., Levy D., Samuels H. H. Nuclear receptor coregulator (NRC): mapping of the dimerization domain, activation of p53 and STAT-2, and identification of the activation domain AD2 necessary for nuclear receptor signaling. Mol. Endocrinol. 2007;21:1822–1834. doi: 10.1210/me.2005-0529. [DOI] [PubMed] [Google Scholar]
- Mahajan M. A., Samuels H. H. Nuclear receptor coactivator/coregulator NCoA6(NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Nucl. Recept. Signal. 2008;6:e002. doi: 10.1621/nrs.06002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers P. H., Grinberg A., Mahon K. A., Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Morgenbesser S. D., Williams B. O., Jacks T., DePinho R. A. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Morozov V., Wawrousek E. F. Caspase-dependent secondary lens fiber cell disintegration in alphaA-/alphaB-crystallin double-knockout mice. Development. 2006;133:813–821. doi: 10.1242/dev.02262. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Pichel J. G., Williams-Simons L., Westphal H. An apoptotic defect in lens differentiation caused by human p53 is rescued by a mutant allele. Proc. Natl. Acad. Sci. USA. 1995;92:6142–6146. doi: 10.1073/pnas.92.13.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S., Kawane K., Watanabe-Fukunaga R., Fukuyama H., Ohsawa Y., Uchiyama Y., Hashida N., Ohguro N., Tano Y., Morimoto T., Fukuda Y., Nagata S. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–1074. doi: 10.1038/nature01895. [DOI] [PubMed] [Google Scholar]
- Overbeek P. A., Chepelinsky A. B., Khillan J. S., Piatigorsky J., Westphal H. Lens-specific expression and developmental regulation of the bacterial chloramphenicol acetyltransferase gene driven by the murine alpha A-crystallin promoter in transgenic mice. Proc. Natl. Acad. Sci. USA. 1985;82:7815–7819. doi: 10.1073/pnas.82.23.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Griep A. E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- Pan H., Griep A. E. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- Petrini J. H., Stracker T. H. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981;19:134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- Pirity M. K., Wang W. L., Wolf L. V., Tamm E. R., Schreiber-Agus N., Cvekl A. Rybp, a polycomb complex-associated protein, is required for mouse eye development. BMC Dev. Biol. 2007;7:39. doi: 10.1186/1471-213X-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A. G., Janicke R. U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- Porter F. D., et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Potten C. S. What is an apoptotic index measuring? A commentary. Br. J. Cancer. 1996;74:1743–1748. doi: 10.1038/bjc.1996.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty N. S., Esson E. A. An electron-microscope study of adult mouse lens: some ultrastructural specializations. J. Ultrastruct. Res. 1974;46:239–253. doi: 10.1016/s0022-5320(74)80059-6. [DOI] [PubMed] [Google Scholar]
- Rajaram N., Kerppola T. K. Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract. Mol. Cell Biol. 2004;24:5694–5709. doi: 10.1128/MCB.24.13.5694-5709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring B. Z., Cordes S. P., Overbeek P. A., Barsh G. S. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Robinson M. L. An essential role for FGF receptor signaling in lens development. Semin. Cell Dev. Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Zirngibl R., Cado D., Shago M., Giguere V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Sakai M., Serria M. S., Ikeda H., Yoshida K., Imaki J., Nishi S. Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res. 2001;29:1228–1237. doi: 10.1093/nar/29.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands A., et al. Altered aggregation properties of mutant gamma-crystallins cause inherited cataract. EMBO J. 2002;21:6005–6014. doi: 10.1093/emboj/cdf609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O., Smith A. N., Robinson M. L., Taketo M. M., Lang R. A., Ashery-Padan R. Pax6 is essential for lens fiber cell differentiation. Development. 2009;136:2567–2578. doi: 10.1242/dev.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. M. Caspases at the crossroads of immune-cell life and death. Nat. Rev. Immunol. 2006;6:308–317. doi: 10.1038/nri1809. [DOI] [PubMed] [Google Scholar]
- Smith R. S. Systematic Evaluation of the Mouse Eye: Anatomy, Pathology and Biomethods. Boca Raton, FL: CRC Press; 2002. [Google Scholar]
- Stanton S. E., Blanck J. K., Locker J., Schreiber-Agus N. Rybp interacts with Hippi and enhances Hippi-mediated apoptosis. Apoptosis. 2007;12:2197–2206. doi: 10.1007/s10495-007-0131-3. [DOI] [PubMed] [Google Scholar]
- Stegh A. H., Kesari S., Mahoney J. E., Jenq H. T., Forloney K. L., Protopopov A., Louis D. N., Chin L., DePinho R. A. Bcl2L12-mediated inhibition of effector caspase-3 and caspase-7 via distinct mechanisms in glioblastoma. Proc. Natl. Acad. Sci. USA. 2008;105:10703–10708. doi: 10.1073/pnas.0712034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolen C. M., Griep A. E. Disruption of lens fiber cell differentiation and survival at multiple stages by region-specific expression of truncated FGF receptors. Dev. Biol. 2000;217:205–220. doi: 10.1006/dbio.1999.9557. [DOI] [PubMed] [Google Scholar]
- Swamynathan S. K., Katz J. P., Kaestner K. H., Ashery-Padan R., Crawford M. A., Piatigorsky J. Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol. Cell Biol. 2007;27:182–194. doi: 10.1128/MCB.00846-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiriet C., Hayes J. J. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Tini M., Otulakowski G., Breitman M. L., Tsui L. C., Giguere V. An everted repeat mediates retinoic acid induction of the gamma F-crystallin gene: evidence of a direct role for retinoids in lens development. Genes Dev. 1993;7:295–307. doi: 10.1101/gad.7.2.295. [DOI] [PubMed] [Google Scholar]
- Vrensen G. F., Graw J., De Wolf A. Nuclear breakdown during terminal differentiation of primary lens fibres in mice: a transmission electron microscopic study. Exp. Eye Res. 1991;52:647–659. doi: 10.1016/0014-4835(91)90017-9. [DOI] [PubMed] [Google Scholar]
- Wang Y., Ko B. C., Yang J. Y., Lam T. T., Jiang Z., Zhang J., Chung S. K., Chung S. S. Transgenic mice expressing dominant-negative osmotic-response element-binding protein (OREBP) in lens exhibit fiber cell elongation defect associated with increased DNA breaks. J. Biol. Chem. 2005;280:19986–19991. doi: 10.1074/jbc.M501689200. [DOI] [PubMed] [Google Scholar]
- Wawrousek E. F., Chepelinsky A. B., McDermott J. B., Piatigorsky J. Regulation of the murine alpha A-crystallin promoter in transgenic mice. Dev. Biol. 1990;137:68–76. doi: 10.1016/0012-1606(90)90008-7. [DOI] [PubMed] [Google Scholar]
- Westphal H., Overbeek P. A., Khillan J. S., Chepelinsky A. B., Schmidt A., Mahon K. A., Bernstein K. E., Piatigorsky J., de Crombrugghe B. Promoter sequences of murine alpha A crystallin, murine alpha 2(I) collagen or of avian sarcoma virus genes linked to the bacterial chloramphenicol acetyl transferase gene direct tissue-specific patterns of chloramphenicol acetyl transferase expression in transgenic mice. Cold Spring Harb. Symp. Quant. Biol. 1985;50:411–416. doi: 10.1101/sqb.1985.050.01.051. [DOI] [PubMed] [Google Scholar]
- Wolf L. V., Yang Y., Wang J., Xie Q., Braunger B., Tamm E. R., Zavadil J., Cvekl A. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS One. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J. H., Bai F., Gross J., Townsend R. R., Menko A. S., Andley U. P. Mechanism of small heat shock protein function in vivo: a knock-in mouse model demonstrates that the R49C mutation in alpha A-crystallin enhances protein insolubility and cell death. J. Biol. Chem. 2008;283:5801–5814. doi: 10.1074/jbc.M708704200. [DOI] [PubMed] [Google Scholar]
- Xie L., Overbeek P. A., Reneker L. W. Ras signaling is essential for lens cell proliferation and lens growth during development. Dev. Biol. 2006;298:403–414. doi: 10.1016/j.ydbio.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Yamada R., Mizutani-Koseki Y., Hasegawa T., Osumi N., Koseki H., Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–1770. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- Yan Q., Liu J. P., Li D. W. Apoptosis in lens development and pathology. Differentiation. 2006;74:195–211. doi: 10.1111/j.1432-0436.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chauhan B. K., Cveklova K., Cvekl A. Transcriptional regulation of mouse alphaB- and gammaF-crystallin genes in lens: opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J. Mol. Biol. 2004;344:351–368. doi: 10.1016/j.jmb.2004.07.102. [DOI] [PubMed] [Google Scholar]
- Yang Y., et al. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006a;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. G., Frappart P. O., Frappart L., Wang Z. Q., Tong W. M. A novel function of DNA repair molecule Nbs1 in terminal differentiation of the lens fibre cells and cataractogenesis. DNA Rep. 2006b;5:885–893. doi: 10.1016/j.dnarep.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Yeom S. Y., et al. Regulation of insulin secretion and beta-cell mass by activating signal cointegrator 2. Mol. Cell Biol. 2006;26:4553–4563. doi: 10.1128/MCB.01412-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Kawane K., Koike M., Mori Y., Uchiyama Y., Nagata S. Phosphatidylserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature. 2005;437:754–758. doi: 10.1038/nature03964. [DOI] [PubMed] [Google Scholar]
- Zheng T. S., Schlosser S. F., Dao T., Hingorani R., Crispe I. N., Boyer J. L., Flavell R. A. Caspase-3 controls both cytoplasmic and nuclear events associated with Fas-mediated apoptosis in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:13618–13623. doi: 10.1073/pnas.95.23.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky B., Kroemer G. Apoptosis and genomic instability. Nat. Rev. Mol. Cell Biol. 2004;5:752–762. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- Zhu Y. J., Crawford S. E., Stellmach V., Dwivedi R. S., Rao M. S., Gonzalez F. J., Qi C., Reddy J. K. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J. Biol. Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- Zhu Y. T., Hu L., Qi C., Zhu Y. J. PRIP promotes tumor formation through enhancing serum-responsive factor-mediated FOS expression. J. Biol. Chem. 2009;284:14485–14492. doi: 10.1074/jbc.M900935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.