TANK interacts with PLK1 in vitro and in vivo. Overexpression of PLK1 specially prevents TNFR-induced NF-κB transactivation and the DNA binding activity of NF-κB. The PLK1 reduces the ubiquitination of NEMO activated by TNF-α.
Abstract
TANK/I-TRAF is a TRAF-binding protein that negatively regulates NF-κB activation. The underlying mechanism of this activity remains unclear. Here we show that TANK directly interacts with PLK1, a conserved cell cycle–regulated kinase. PLK1 inhibits NF-κB transcriptional activation induced by TNF-α, IL-1β, or several activators, but not by nuclear transcription factor p65. PLK1 expression reduces the DNA-binding activity of NF-κB induced by TNF-α. Moreover, endogenous activation of PLK1 reduces the TNF-induced phosphorylation of endogenous IκBα. PLK1 is bound to NEMO (IKKγ) through TANK to form a ternary complex in vivo. We describe a new regulatory mechanism for PLK1: PLK1 negatively regulates TNF-induced IKK activation by inhibiting the ubiquitination of NEMO. These findings reveal that the scaffold protein TANK recruits PLK1 to negatively regulate NF-κB activation and provide direct evidence that PLK1 is required for the repression function of TANK.
INTRODUCTION
TRAF-associated NF-κB activator (TANK), also known as I-TRAF (TRAF-interacting protein), was initially identified as a protein associated with TRAF1, TRAF2, and TRAF3 (Cheng and Baltimore, 1996; Kaye et al., 1996; Rothe et al., 1996; see Abbreviations footnote). TANK emerged as a bifunctional adaptor protein, as it was found to function as an inhibitor of TRAF-mediated NF-κB activation and also as a cofactor for the activation of NF-κB induced by TRAF2, TBK1, or IκB kinase epsilon (IKKε) (Pomerantz and Baltimore, 1999; Nomura et al., 2000). This biphasic response is dependent on the bipartite organization of TANK. The N-terminal domain of TANK coactivates NF-κB, whereas the C-terminal domain completely counteracts this stimulation. The C-terminal domain of TANK inhibits signaling from intact upstream cytokines, receptors, or TRAF2, but does not directly affect the subunit of NF-κB. TANK acts in TRAF2-dependent signal transduction pathways that activate NF-κB. TANK may play an inhibitory role in TRAF2 function by preventing the spontaneous aggregation of TRAF2 and competing for binding to upstream receptors (Cheng and Baltimore, 1996; Kaye et al., 1996; Rothe et al., 1996). In addition to being a general regulator of TRAF protein function, TANK is also connected to the downstream IKK complex: TANK binds to the essential regulator subunit IKKγ/NEMO (Chariot et al., 2002). On tumor necrosis factor alpha (TNF-α) stimulation, TANK is recruited to the IKK complex and is required for TNF-α–mediated NF-κB activation and the expression of selected target genes. These results suggest a model in which TANK positively regulates NF-κB activation by connecting upstream signaling molecules, such as TBK1, to the IKK complex and p65 (Bonif et al., 2006). However, the mechanism by which TANK inhibits the activation of NF-κB and its downstream signaling remains elusive.
The NF-κB family of transcription factors is active not only in immune and inflammatory responses, but also in cell cycle regulation, differentiation, and apoptosis (Pahl, 1999). These transcription factors affect the expression of numerous components of the immune system and a variety of proteins that inhibit apoptosis and promote cell survival/proliferation. The proteins of the NF-κB family affect cell cycle regulation through the transcriptional regulation of the CDK (cyclin-dependent kinase)/CDKI (CDK inhibitor) system. Furthermore, specific CDKs have been found to regulate the transcriptional activity of NF-κB, and the corresponding CDKI can stimulate κB-dependent gene expression (Perkins et al., 1997). However, whether cell cycle regulators can modulate the activation of the NF-κB signaling pathway requires further investigation. Recently, PLK1, a cell cycle regulation kinase, was reported to phosphorylate IKKβ to inhibit its activation (Higashimoto et al., 2008). This finding provides direct evidence that a cell cycle regulator affects NF-κB activation. PLK1 is a key regulator of cell cycle processes in eukaryotic cells. It regulates a variety of M phase–specific events, including centrosome maturation, bipolar spindle formation, microtubule motor regulation, and cytokinesis (Golsteyn et al., 1996; Yarm, 2002; van Vugt and Medema, 2005; Li and Li, 2006). PLK1 is a target of the DNA damage checkpoint, and its activity is inhibited by DNA damage signals (Smits et al., 2000; van Vugt et al., 2001).
In this study, TANK was identified as a partner of PLK1 through yeast two-hybrid screening. We then confirmed that TANK interacts with PLK1 in vitro and in vivo. Furthermore, we found that overexpression of PLK1 specifically prevents TNF receptor (TNFR)-induced NF-κB transactivation and the DNA-binding activity of NF-κB. We confirmed that PLK1 is a negative regulator of NF-κB transactivation that is activated by upstream stimulators, including cytokines and activators, but not by p65. We also determined that the negative regulation of PLK1 on NF-κB activation is associated with the interaction of PLK1 and TANK. Moreover, we investigated the roles of PLK1 on modulating the activity of IKK complex. We found that PLK1 specifically interacts with NEMO and is bridged by TANK to form a ternary complex under physiological conditions. PLK1 reduces the ubiquitination of NEMO, which is activated by TNF-α. We identified a new molecular mechanism in which PLK1 participates in the negative regulation of NF-κB in addition to its ability to phosphorylate IKKβ.
MATERIALS AND METHODS
Plasmids and Reagents
The human genes encoding PLK1, TANK, and TRAF2 were amplified from a human liver cDNA library (Invitrogen, Carlsbad, CA). Full-length PLK1 and PLK1 deletion mutants were generated by PCR, followed by subcloning into the pFlag-CMV-2 (Sigma, St. Louis, MO) or pDsRed1-N1 (Clontech, Palo Alto, CA) vectors. The kinase defective mutant pCMV-PLK1-K82R was purchased from Origene (Rockville, MD). Full-length TANK was cloned into the pCMV-Myc (Clontech, CA) and pEGFP-C3 (Clontech, CA) eukaryotic expression vectors. TANK deletion mutants were generated by PCR or recombinant PCR, followed by subcloning into the pCMV-Myc (Clontech) vector. The NEMO, MyD88, p65 TBK1, IKKε, and TLR4 mammalian expression plasmids were constructed in our lab (unpublished results). The Renilla luciferase expression vector (pRL-TK) was purchased from Promega (Madison, WI), and the luciferase reporter plasmid (κB-Luc) was obtained from Stratagene (La Jolla, CA). The human IKKα and IKKβ plasmids were gifts from Z. G. Liu (National Cancer Institute, National Institutes of Health), and the RIP expression vector was from X. Lin (Anderson Cancer Center). TNF-α, interleukin 1 beta (IL-1β), LPS, thymidine nocodozole, the anti-Flag (M2) antibody, and the anti-hemagglutinin (HA; 12CA5) antibody were purchased from Sigma. Protein A/G Plus-agarose, rabbit immunoglobulin G (IgG), mouse IgG, and antibody reagents, including anti-Myc (9E10), anti-Myc (9E10) horseradish peroxidase (HRP), anti-PLK1 (E-2), anti-TANK (C-20), and anti-IKKγ (FL-419) were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-ubiquitin (P4D1), anti-IκBα, anti-phospho-IκBα and anti-phospho-IKKα (Ser180)/IKKβ (Ser181) antibodies were from Cell Signaling (Beverly, MA). The HRP-conjugated anti-glutathione S-transferase (GST; M071–7) antibody was from Medical and Biological Laboratories (Nagoya, Japan). Glutathione Sepharose 4B was from GE Healthcare Bio-Science AB (Uppsala, Sweden). The fluorescein isothiocyanate–conjugated second antibody was from Zhongshan Goldenbridge Biotechnology (Beijing, China). λ phosphatase was purchased from New England BioLabs.
Yeast Two-Hybrid Screening
The gene encoding PLK1 was cloned into the pDBLeu vector and used as bait to screen a human liver cDNA library (Invitrogen) by yeast two-hybrid in Saccharomyces cerevisiae MaV203 according to the manufacturer's protocol. Positive clones were selected as previously described (Zhou et al., 2008).
Cell Culture and Transfection
HEK293 (human embryonic kidney 293) cells were cultured in DMEM supplemented with 10% (vol/vol) fetal calf serum (FBS). HeLa cells were maintained in DMEM supplemented with 10% FBS and 10% penicillin/streptomycin. K562 cells were maintained in RPMI 1640 supplemented with 10% FBS and 10% penicillin/streptomycin. All cells were cultured at 37°C and 5% CO2. Cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
In Vitro Binding Assays
To obtain GST-PLK1 proteins, PLK1 cDNA was cloned into the pGEX-4T-2 (GE Healthcare Bio-Science) vector. To make His-TANK and His-NEMO fusion proteins, TANK and NEMO cDNA sequences were cloned into the pET-28a-c(+) vector (Novagen, Darmstadt, Germany). BL21(DE3) bacteria containing each resultant plasmid were diluted 1:100 from an overnight culture and grown for 4–6 h at 37°C until the OD600 reached 0.4–0.6. Expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for 10–16 h at 20°C and a slow rotation speed. The bacteria were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS) containing a protease inhibitor cocktail and sonicated. For the in vitro binding assays, the GST-PLK1 supernatant was incubated with Glutathione Sepharose 4B (GE Healthcare Bio-Sciences) for 8 h at 4°C. The beads were washed three times with PBS and then incubated with supernatant containing His-TANK or His-NEMO for 12 h at 4°C. After the incubation, the beads were extensively washed with PBS, and the same volume of 2× loading buffer was added to the beads, followed by Western blotting.
Immunoprecipitation Assay
HEK293 cells were transfected with the indicated plasmids for protein overexpression. Cells were harvested 24–48 h after transfection. Cell lysates were prepared in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Tween 20, 0.2% NP-40, 10% glycerol) supplemented with a protease inhibitor cocktail (Roche, Nutley, NJ) and phosphatase inhibitors (10 mM NaF and 1 mM Na3VO4). During λ-phosphatase treatment (5 U/mg protein extract; 30 min at 30°C), the phosphatase inhibitors were omitted. Immunoprecipitations were performed using the appropriate antibodies and protein A/G-agarose (Santa Cruz) at 4°C. Lysates and immunoprecipitates were incubated with the indicated primary antibodies and the appropriate secondary antibody, followed by detection with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL). To detect the endogenous interaction between PLK1 and TANK, an immunoprecipitation was performed with either an anti-PLK1 or an anti-TANK antibody. PLK1 or TANK was then detected in the lysates and immunoprecipitates using the indicated antibodies.
Fluorescence Microscopy and Immunostaining
HEK293 cells were transfected with polymeric enhanced green fluorescent protein (pEGFP)-PLK1 and polymeric red fluorescent protein (pRFP)-TANK. After 24 h of transfection, the cells were fixed with methanol. The nuclei of the cells were stained with 0.1 g/ml DAPI. The cells were observed using a fluorescence microscope. To observe the localization of endogenous proteins, HEK293 cells were rinsed with PBS, fixed with 4% paraformaldehyde for 30 min, permeabilized with 1% Triton in PBS for 10–15 min, blocked with 3% BSA (bovine serum albumin) in PBS for 2 h, stained with various antibodies, and finally visualized with a fluorescein isothiocyanate-conjugated second antibody. The images were acquired by fluorescence microscopy.
Luciferase Reporter Assays
HEK293 cells were transfected with 0.1 μg of the κB-luciferase reporter gene (Stratagene) plus 2 ng of the Renilla luciferase expression vector pRL-TK (Promega), with or without various amounts of the pFlag-CMV-PLK1 expression vector. After treatment for 6–7 h with 10 ng/ml TNF-α or IL-1β, the cells were collected. Luciferase activity was assessed as previously described (Yu et al., 2008). All experiments were repeated at least three times.
RNA Interference
The double-stranded RNA duplexes were synthesized by GeneChem (Shanghai, China). The siRNA oligonucleotide used to target human TANK was UCA CUU CAA CAG ACU AUU ATT (Guo and Cheng, 2007). Duplexes of small interfering RNA (siRNA) were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A nonsilencing duplex was used as a control. 48 h after transfection, the cells were harvested. To examine the effect of silencing TANK on NF-κB reporter gene activity in HEK293 cells, the cells were transfected with 40 nM siRNA. 48 h later, they were transfected with the pNF-κB-Luc reporter and retransfected with the siRNA. After 6–7 h of treatment with TNF-α (10 ng/ml), the cells were lysed for use in a luciferase reporter assay.
Electrophoretic Mobility Shift Assay
Nuclear protein extracts were prepared using the ProteoJETTM Cytoplasmic and Nuclear Protein Extraction Kit K0311 (Fermentas, Canada). The electrophoretic mobility shift assay was performed as described in the instructions of the LightShift Chemiluminescent Electrophoretic Mobility Shift Assay (EMSA) Kit (Pierce, Rockford, IL) using biotin-labeled NF-κB double-stranded oligonucleotides (top strand: 5′-AGT TGA GGG GAC TTT CCC AGG C-3′; bottom strand: 3′-TCA ACT CCC CTG AAA GGG TCC G-5′; Sangon Biotech, Shanghai, China).
RESULTS
Identification of TANK as a PLK1-interacting Protein by Yeast Two-Hybrid Screening
To identify novel proteins that bind to PLK1, full-length human PLK1 was used as bait to screen a human liver cDNA library. Six independent positive clones were found that encoded full-length TANK or portions of TANK (also known as TRAF-interacting protein or I-TRAF). TANK was previously identified as an adaptor protein in the TRAF2-dependent NF-κB-activating pathway and has a potential effect in the regulation of NF-κB, but its precise role is controversial (Cheng and Baltimore, 1996; Kaye et al., 1996; Rothe et al., 1996; Pomerantz and Baltimore, 1999; Nomura et al., 2000; Chariot et al., 2002; Bonif et al., 2006). TANK binds to the TRAF family members TRAF1/2/3 (Cheng and Baltimore, 1996; Kaye et al., 1996; Rothe et al., 1996) and the IκB-related kinases, IKKα/β/γ, TBK1, and IKKε (Pomerantz and Baltimore, 1999; Nomura et al., 2000; Chariot et al., 2002).
TANK Interacts with PLK1 In Vivo
To confirm that PLK1 and TANK interact in mammalian cells, Flag-PLK1 and Myc-TANK were cotransfected in HEK293 cells. The proteins were immunoprecipitated with either an anti-FLAG or an anti-Myc antibody, and the presence of associated proteins was determined by Western blot analysis. The two proteins were shown to coimmunoprecipitate using an anti-Flag antibody to precipitate PLK1, followed by blotting with an anti-Myc antibody to detect TANK, and vice versa (Figure 1A). Next, we examined whether endogenous PLK1 and TANK interact under physiological conditions by immunoprecipitation. TANK was present in immunoprecipitates obtained from K562 cell extracts with an antibody against PLK1 (anti-PLK1), whereas TANK did not coprecipitate when a control antibody, normal mouse IgG, was used. Furthermore, the PLK1 protein was detected when the lysate was immunoprecipitated with an anti-TANK antibody, but not normal goat IgG (Figure 1B).
Figure 1.
PLK1 interacts with TANK in vivo. (A) Coimmunoprecipitation of Flag-PLK1 and Myc-TANK. HEK293 cells were cotransfected with Flag-PLK1 and Myc-TANK. After 24 h, anti-Flag or anti-Myc immunoprecipitates (IP) were subjected to immunoblotting (IB) with the appropriate antibodies. (B) Immunoblot analysis of the interaction between endogenous PLK1 and TANK in K562 cell lysates after immunoprecipitation with mouse IgG, anti-PLK1, goat IgG, or anti-TANK antibodies.
Mapping the Regions Required for thePhosphorylation-independent Interaction Between PLK1 and TANK
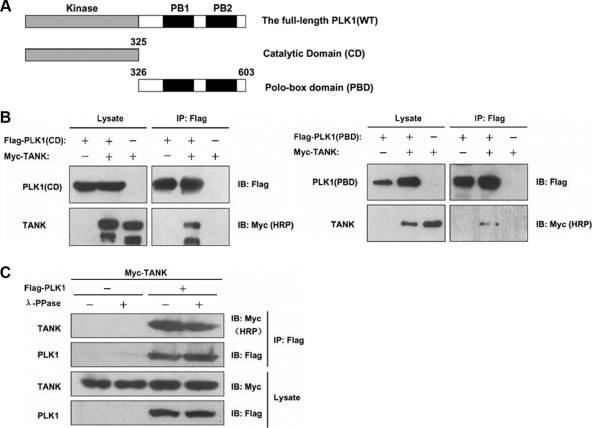
The members of the PLK family have a high degree of homology within their N-terminal catalytic domains (CD) and C-terminal noncatalytic domains, which include several highly conserved polo-box domains (PBD). The PBD is crucial for the subcellular distribution of PLKs to mitotic structures and substrate-specific interactions (Golsteyn et al., 1995; Smits et al., 2000).
To further characterize the TANK-binding region of PLK1, we constructed two PLK1 truncation mutants: one contained the CD domain (aa 1-325) and the other contained the PBD region (aa 326-603), which includes two tandem polo boxes (Figure 2A). HEK293 cells were cotransfected with Flag-PLK1-CD or Flag-PLK1-PBD and Myc-TANK. The lysates were immunoprecipitated with an anti-Flag antibody and subjected to Western blot analysis to detect TANK using an anti-Myc antibody. As shown in Figure 2B, both PLK1-CD and PLK1-PBD interacted with TANK. These results indicate that aa 1-325 and aa 326-603, which include the kinase domain and PBD binding domain of PLK1, respectively, are both essential for the interaction with TANK. The ability of the PBD domain of PLK1 to bind to TANK indicated that the kinase activity of PLK1 is not necessary for its interaction with TANK.
Figure 2.
The interaction between PLK1 and TANK requires the CD or PBD domain of PLK1, but is independent of PLK1 phosphorylation. (A) Domain architecture of full-length PLK1 and two deletion mutant constructs. The two polo boxes of PLK1 are filled in black. (B) Coimmunoprecipitation of Flag-PLK1-CD (left) or Flag-PLK1-PBD (right) and Myc-TANK. Both Flag-PLK1-CD and Flag-PLK1-PBD were cotransfected with Myc-TANK in HEK293 cells. After 24 h, anti-Flag immunoprecipitates were subjected to immunoblotting with the indicated antibodies. (C) HEK293 cells were transfected with Myc-TANK together with Flag-PLK1 or pFlag-CMV-2. After 24 h, cells were lysed in lysis buffer, treated or not with 5U/μg λ phosphatase, and subjected to immunoprecipitation (IP) with an anti-Flag antibody. The presence of TANK and PLK1 in the immunocomplexes and in the total cell extracts was revealed by immunoblot analysis using anti-Myc and anti-Flag antibodies.
To delineate the region in TANK that is required for the interaction with PLK1, we tested the ability of various truncation mutants of TANK to bind to the full-length PLK1 protein using coimmunoprecipitation assays. Among the N-terminal deletion mutants of TANK protein, a fragment that lacked the first 50 aa (ΔN50) was still able to interact with TANK, but another fragment that lacked the first 100 aa (ΔN100) was not (Figure 3, A and B). Among the various C-terminal deletions of TANK, those lacking part or all of the last 100 aa (ΔC100) were still able to bind PLK1, whereas the fragment that lacked the last 150 aa (ΔC150) was not (Figure 3, C and D). On the basis of these findings, we presume that the region containing aa 51-325 of TANK is sufficient to mediate its interaction with PLK1, and the regions containing aa 51-100 and aa 275-325 might contain PLK1 binding sites.
Figure 3.
Mapping of the PLK1-interacting sites on TANK. (A and C) Schematic illustration of the TANK expression constructs tested for their interaction with PLK1. (B and D) Coimmunoprecipitations of Myc-TANK, Myc-ΔN TANK truncations (B), and Myc-ΔC TANK truncations (D) with Flag-PLK1. HEK293 cells were transfected with the indicated plasmids. Total extracts were immunoprecipitated with an anti-FLAG antibody (PLK1), followed by Western blot analysis with an anti-Myc (TANK) antibody (top panels). The presence of the various TANK truncation mutants and Flag-PLK1 are demonstrated in the Western blot (bottom panels). WCL, whole cell lysates. The lanes containing full-length (FL) TANK or mutants of TANK are indicated. (E) Domain architecture of full-length TANK and three TANK mutants: aa 51-325, Δ51-100 and Δ275-325. (F) TANK mutants 51-325, Δ51-100 and Δ275-325 weakly bind to wild-type PLK1 (wt). HEK293 cells were transfected with the indicated plasmids and lysed. Total extracts were immunoprecipitated with an anti-Flag antibody (PLK1), followed by Western blot analysis with an anti-Myc antibody (wild-type and mutant TANK; top panels). The presence of wild-type and mutant Myc-TANK and Flag-PLK1 in the extracts is shown (bottom panels). A representative result of three independent experiments is shown.
We analyzed the protein sequence of TANK and found a PLK1 kinase substrate motif in aa 284-289 and four PLK1 PBD domain binding motifs in aa 67-69, 116-118, 243-245 and 248-250 (Figure 3E). We hypothesized that these motifs might mediate the interaction between PLK1 and TANK. According to aforementioned results, the TANK mutant ΔN100, which lacked the first PLK1 PBD domain-binding motif, and the mutant ΔC150, which lacked the PLK1 kinase substrate motif, failed to interact with PLK1. This finding suggests that these two motifs of TANK may be required for its interaction with PLK1. To prove this hypothesis, three TANK mutants were constructed: aa 51-325, TANKΔ51-100, and TANKΔ275-325 (Figure 3E). We found that these three mutants could bind to PLK1, but their binding activities were remarkably less than full-length TANK (Figure 3F). These results suggest that aa 51-100 and aa 275-325 of TANK have some effect on its interaction with PLK1, but deletion of these two regions cannot completely disturb this interaction. Together with the weak interaction between TANK (51-325) and PLK1, we hypothesize that the interaction between TANK and PLK1 correlates with the protein conformation, but does not depend on the phosphorylation of TANK. These conclusions are consistent with the PLK1 truncation mutant assays.
To corroborate the dependence of TANK phosphorylation on its interaction with PLK1, we expressed TANK with PLK1 in HEK293 cells, and the cell extracts were treated with λ phosphatase before PLK1 immunoprecipitation. As expected, the results show that the association of TANK with PLK1 was unaffected by the phosphatase treatment (Figure 2C).
PLK1 and TANK Colocalizes in the Cytoplasm
PLK1 is localized to the nucleus and cytoplasm during the S and G2 phases (Taniguchi et al., 2002) and concentrates in the nucleus in the M phase (Golsteyn et al., 1995). As an intercellular adaptor molecule, TANK is mainly localized to the cytoplasm (Rothe et al., 1996). To investigate the subcellular localization of PLK1 and TANK, HEK293 cells were transfected with PLK1, TANK, or both (Figure 4A). When coexpressed, PLK1 and TANK colocalized in the cytoplasm. The expression of PLK1 did not affect the localization of TANK, whereas TANK recruited PLK1 to the cytoplasm. We then wondered whether ectopic TANK expression affects the subcellular localization of the endogenous PLK1 protein. HEK293 cells were transfected with GFP-TANK for 24 h, and the localization of endogenous PLK1 was examined by immunostaining fixed cells with an anti-PLK1 antibody (Figure 4B). PLK1 localization was dramatically altered upon ectopic expression of GFP-TANK. It accumulated in the cytoplasm with TANK.
Figure 4.
PLK1 and TANK colocalize in the cytoplasm. (A) Either GFP-PLK1 or GFP-TANK was transfected in HEK293 cells (top and middle panels), and GFP-PLK1 and RFP-TANK were cotransfected in HEK293 cells (bottom panel). Fluorescence was visualized, and images were captured with a fluorescence microscope at 24 h after transfection. Nuclei were stained with DAPI (blue), and the images were merged. Yellow represents colocalized PLK1 and TANK proteins. (B) Ectopic expression of TANK influences the localization of endogenous PLK1. HEK293 cells were transfected with control pEGFP-C3 or GFP-TANK (green). After 24 h, the cells were fixed, permeabilized, and incubated with an anti-PLK1 antibody to show the subcellular localization of PLK1 (red) by indirect immunostaining. DNA was counterstained with DAPI (blue). Merged images show the superimposition of red and blue, or red, blue, and green signals.
PLK1 Inhibits NF-κB Transcriptional Activation through IKK Deactivation
To determine whether PLK1 regulates NF-κB activation through TANK, we examined the effect of PLK1 on the transcriptional activation of NF-κB using a luciferase reporter gene that was under the control of the human κB promoter (Li et al., 2004). We found that at a low level of expression, TANK weakly activated NF-κB and PLK1 inhibited NF-κB activation by TANK in dose-dependent manner (Figure 5A). We also confirmed the inhibitory effect of PLK1 on NF-κB activation induced by TNF-α or IL-1β (Figure 5B). We then determined whether PLK1 especially affected TNFR signal–induced NF-κB activation. In Figure 5C, we compare the effect of PLK1 on NF-κB activation mediated by TNF-α, IL-1β, and lipopolysaccharide (LPS). We found that NF-κB activation induced by LPS was almost unaffected by PLK1. To further confirm the PLK1-mediated inhibition of NF-κB activation, we examined the effect of PLK1 on NF-κB activation mediated by the adaptor proteins TRAF2, RIP, and MyD88. Overexpression of PLK1 blocked the NF-κB activation triggered by any of the three proteins (Figure 5D). NF-κB activation resulting from overexpression of IKKα, IKKβ, or TBK1 was also inhibited by PLK1, but NF-κB activation induced by overexpression of the NF-κB subunit p65 was unaffected by PLK1 (Figure 5D). The adaptor proteins and IKK catalytic units activated NF-κB and required IKK activation, but p65 did not. We conclude that IKK deactivation was responsible for the PLK1-mediated inhibition of NF-κB. We hypothesize that LPS stimuli activate NF-κB through another signaling pathway when the activation of IKKs is inhibited by PLK1.
Figure 5.
PLK1 is a negative regulator of NF-κB activation induced by TNFR, and this inhibition is dependent on PLK1 kinase activity. (A and B) The activation of NF-κB is inhibited by PLK1 and induced by low expression of TANK or treatment with the cytokines TNF-α and IL-1β. HEK293 cells were transiently transfected with κB-responsive luciferase reporter plasmids, pRL-TK control plasmids and increasing amounts of Flag-PLK1. After 24 h, the cell extracts were collected for luciferase activity measurements. The cells were treated for 6–7 h with TNF-α (10 ng/ml) or IL-1β (10 ng/ml) before lysis as indicated. (C) PLK1 does not affect the LPS-induced activation of NF-κB. Luciferase assay of HEK293 cells transiently transfected with κB-responsive luciferase reporter, pRL-TK control and Flag-PLK1. Because HEK293 cells do not express TLRs, we transfected Myc-TLR4 into LPS-stimulated cells. After 24 h, cells were treated for 6–7 h with TNF-α, IL-1β, or LPS (1 μg/ml) before lysis as indicated. (D) PLK1 strongly inhibits NF-κB activation when coexpressed with TRAF2, IKKα, IKKβ, or RIP; weakly inhibits NF-κB when coexpressed with MyD88 or TBK1; but does not inhibit NF-κB when coexpressed with p65. HEK293 cells were transiently transfected with κB-responsive luciferase reporter plasmids, pRL-TK control plasmids, Flag-PLK1 and the additional indicated plasmids. After 24 h, the cell extracts were collected for the luciferase activity measurements. The TANK vector was used as a positive control, and pFlag-CMV-2 was used as a negative control (CON). (E) Wild-type PLK1 dramatically reduces NF-κB activity, but PLK1-K82R does not. HEK293 cells were transfected with wild-type PLK1 or PLK1-K82R, the κB-responsive luciferase reporter, and various constructs. TNF stimulation was performed as described earlier. (F) Cell cycle–blocking reagents influence the activation of NF-κB. HEK293 cells were transfected with the κB-responsive luciferase reporter plasmid, and 24 h after transfection, the cells were treated with DMSO (control), nocodazole (100 ng/ml), or thymidine (2 mM) for 18 h. Values are means ± SD; (n = 3). Data are representative of three experiments.
Expression of a Kinase-defective Mutant of PLK1 Reduces Its Inhibition of NF-κB
To determine whether the inhibition of NF-κB by PLK1 relies on its kinase activity, we expressed a kinase defective mutant, PLK1-K82R, in HEK293 cells and tested the transcriptional activity of NF-κB by reporter gene assay. Wild-type PLK1 dramatically reduced the NF-κB activity induced by TNF-α, IKK (IKKβ), and adaptor proteins (RIP, MyD88), but the PLK1-K82R mutant only slightly inhibited NF-κB (Figure 5E). This experiment suggests that the kinase activity of PLK1 plays a key role in the inhibition of NF-κB.
Endogenous Activity of PLK1 Induced by CellCycle-blocking Reagents Influences the Activation of NF-κB
To discount the possibility that the inhibitory effect of PLK1 on NF-κB activation was a result of overexpression artifacts, we examined the effects of nocodazole and thymidine, which induce and reduce, respectively, the endogenous activity of PLK1 by controlling the cell cycle (Jang et al., 2002), on the transcriptional activation of NF-κB. The κB-luciferase construct and pRL-TK were coexpressed in HEK293 cells. After 24 h of transfection, cells were treated with DMSO (control), nocodazole (100 ng/ml), or thymidine (2 mM) for 18 h. Most cells treated with nocodazole for 16–18 h are known to remain in M phase, in which PLK1 is phosphorylated and activated, and most cells treated with thymidine are known to arrest between G1 and S phases, in which PLK1 is expressed at a low level and is inactive. The transcriptional activation of NF-κB was significantly reduced in nocodazole-treated cells and increased in thymidine-treated cells (Figure 5F).
Interaction of PLK1 with TANK Promotes the Inhibition of NF-κB
To investigate the involvement of TANK in the PLK1-induced repression of NF-κB activation, we examined the effect of TANK knockdown on Flag-PLK1 after treatment with TNF-α. The efficiency of siRNA to reduce TANK protein levels in HEK293 cells is shown in Figure 6A. When endogenous TANK protein was knocked down in HEK293 cells, the NF-κB activation induced by TNF-α was two-fold greater than the control (Figure 6B, left), but the PLK1-induced repression of NF-κB activation by TNF-α was reduced (Figure 6B, right). It has been reported that overexpression of TANK in incremental levels results in a biphasic NF-κB activation response. At a low expression level, TANK slightly activates NF-κB, but at a high expression level, TANK strongly inhibits NF-κB activation in HEK293 cells (Cheng and Baltimore, 1996; Rothe et al., 1996). Knockdown of TANK enhanced NF-κB activation: this finding demonstrated that the role of TANK in the TNF-α–mediated NF-κB signaling pathway is complex (Figure 6B, left , lane 3). We believe that TANK might participate in several branches of various signaling pathways and, thus, has different effects on NF-κB activation. To confirm whether the effect of PLK1 on NF-κB changed after knockdown of endogenous TANK, we calculated the ratio of NF-κB that was repressed by PLK1 (the relative magnitudes of the reporter gene activity of NF-κB before and after overexpression of PLK1). We found that when endogenous TANK was knocked down in HEK293 cells, PLK1 was still able to repress NF-κB, but the ratio of NF-κB that was repressed by PLK1 was remarkably decreased (Figure 6B, right). All of these data demonstrate that PLK1 interacts with TANK to enhance the inhibitory effect of PLK1 on NF-κB.
Figure 6.
Knockdown of endogenous TANK reduces the PLK1-induced repression of NF-κB activation by TNF-α. (A) Exogenously expressed and endogenous TANK protein levels were markedly reduced by siRNA-TANK. Increasing amounts of siRNA-TANK together with Myc-TANK or Myc vectors were cotransfected in HEK293 cells. After 48 h, cells were harvested and subjected to Western blot analysis using the indicated antibodies. (B) SiRNA-TANK (40 nM) enhanced the activation of NF-κB induced by TNF-α in HEK293 cells (upper panel, lane 3). Overexpression of PLK1 also inhibited the activation of NF-κB when cotransfected with siRNA-TANK (upper panel, lane 4). The ratio of NF-κB that was repressed by PLK1 corresponds to the relative magnitudes of the reporter gene activity of NF-κB before and after overexpression of PLK1. These ratios were remarkably depressed when endogenous TANK was knocked down in HEK293 cells (lower). HEK293 cells were transfected with NF-κB-responsive luciferase reporter plasmids, pRL-TK control plasmids and the siRNA as indicated. After 24 h, cells were either left untreated or stimulated with TNF-α (10 ng/ml) for 6–7 h before lysis, as indicated. The cell extracts were collected for luciferase activity measurements. Values are the means ± SD (n = 3). Data are representative of three experiments.
Expression of PLK1 Reduces the DNA-binding Activity of NF-κB Induced by TNF-α
We next examined the effect of PLK1 expression on the TNF-induced DNA binding of NF-κB. For DNA-binding measurements, HEK293 cells were transfected with Flag-PLK1 and control vectors for 24 h. Cells were treated with TNF-α for 0–30 min. The DNA-binding activity of NF-κB was measured by EMSA using nuclear extracts. The DNA-binding activity of NF-κB was increased by TNF-α treatment, and transfection of PLK1 reduced the TNF-induced NF-κB DNA binding (Figure 7A).
Figure 7.
PLK1 reduces the DNA-binding activity of NF-κB and the TNF-induced phosphorylation and degradation of IκBα. (A) Expression of PLK1 reduces the DNA-binding activity of NF-κB. HEK293 cells were transfected with Flag-PLK1 or pCMV-Flag-2. After 24 h, the cells were treated with 20 ng/ml TNF-α for the indicated times. Nuclear extracts were then prepared for EMSA. Protein p65 and PLK1 in the nucleus were analyzed by Western blotting. (B) Endogenous PLK1 activation reduced the TNF-induced phosphorylation and degradation of IκBα. HeLa cells were treated with DMSO (control), nocodazole (100 ng/ml), or thymidine (2 mM) for 18 h and then stimulated or not with 20 ng/ml TNF-α for the indicated times. Cells were harvested, and phospho-IκBα levels were determined by immunoblot (IB) analysis (top). IκBα, PLK1, phospho-PLK1, and tubulin levels were also determined by immunoblotting.
Activation of PLK1 Is Independent of TNF-α Treatment and Reduces the TNF-induced Phosphorylation and Degradation of Endogenous IκBα
NF-κB activation after viral or bacterial infection involves the signal-induced phosphorylation and subsequent degradative polyubiquitylation of the inhibitory κBα protein (IκBα) through the canonical IκB kinase (IKK)-dependent pathway (Perkins, 2007). To assess the effect of PLK1 on the cellular activity of IKK, we determined the TNF-induced phosphorylation and protein levels of IκBα in HeLa cells. HeLa cells were incubated with DMSO, thymidine, or nocodazole for 18 h and then treated with TNF-α for 0, 5, 15, and 30 min. As shown in Figure 7B, the protein level of endogenous PLK1 was reduced in thymidine-induced cells, but was dramatically increased in nocodazole-induced cells. Phosphorylated PLK1, which corresponds to the activated PLK1, was only detected in nocodazole-induced cells. TNF-α treatment did not impact the protein level or phosphorylation status of PLK1. Immunoblot analysis of cell extracts using a phospho-specific anti-IκBα antibody showed that TNF-α treatment increased the level of phosphorylated IκBα in DMSO-induced cells. In thymidine-induced cells, the phosphorylation of IκBα was enhanced, whereas phosphorylated IκBα could not be detected in nocodazole-induced cells. Because IκBα is phosphorylated by IKKs, we can conclude that the activation of PLK1 negatively regulates IKK activity.
PLK1 Interacts with NEMO
Considering the former results, PLK1 might directly inhibit the activation of NF-κB through the IKK complex. Therefore, we tested the interaction of PLK1 with the three subunits of the IKK complex. First, the interaction between PLK1 and IKKα, IKKβ, or NEMO (IKKγ) was tested by coimmunoprecipitation assay in HEK293 cells. Only the regulatory subunit NEMO was able to bind PLK1 (Figure 8A) in HEK293 cells. Because we demonstrated that PLK1 negatively regulates TNF-mediated NF-κB activation, we then analyzed whether the interaction of PLK1 with TANK or NEMO is regulated by TNF-α. As shown in the left of Figure 8C, the protein level of TANK, which binds to PLK1, does not vary upon TNF-α treatment. We obtained similar results for the interaction between NEMO and PLK1 (Figure 8C, right). We concluded that TNF-α does not have a prominent effect on the binding of PLK1 to TANK or NEMO.
Figure 8.
PLK1 interacts with NEMO. (A) PLK1 interacts with NEMO, but not with IKKα/β in HEK293 cells. Immunoassay of lysates of HEK293 cells that were cotransfected with vectors expressing Myc-NEMO, HA-IKKα/β, and Flag-PLK1, immunoprecipitated with an anti-Flag antibody, and detected with anti-Myc (HRP) or anti-HA antibodies. (B) PLK1 directly interacts with TANK, but not with NEMO in vitro. Immunoblot analysis of bound proteins in lysates of bacteria (BL21) that were expressing His-tagged TANK (left) or NEMO (right) and incubated with Sepharose beads coupled to either GST alone or a GST-PLK1 fusion protein. (C) Interactions between PLK1 and TANK or NEMO are affected by TNF-α. Immunoassay of lysates of HEK293 cells that were cotransfected with vectors expressing Myc-TANK, Myc-NEMO and Flag-PLK1, stimulated for 0–15 min with TNF-α (20 ng/ml) before lysis, immunoprecipitated with an anti-Flag antibody, and detected with an anti-Myc (HRP) antibody.
TANK Connects PLK1 with NEMO to Form a Ternary Complex
In vitro assays showed that PLK1 directly interacts with TANK, but not with NEMO (Figure 8B). TANK has been reported to bind to NEMO to cause upstream signaling proteins to link to IKK complexes (Chariot et al., 2002). We propose that the scaffold protein TANK connects PLK1 with NEMO, and PLK1, TANK, and NEMO form a complex. To test these hypotheses, we transfected combinations of Flag-PLK1, Myc-TANK, Myc-NEMO, and Flag-NEMO in HEK293 cells. Either anti-Flag or anti-Myc antibodies were used to immunoprecipitate PLK1 and TANK, respectively. Anti-Myc or anti-Flag Western blot analysis was performed with the immunoprecipitates and revealed the presence of the other two proteins (Figure 9A). Therefore, a ternary complex of PLK1, TANK, and NEMO should have formed in the HEK293 cells. Next we examined whether this complex exists in physiological conditions by immunoprecipitating endogenous proteins. Both PLK1 and TANK were present in immunoprecipitates obtained from K562 cell extracts with an anti-NEMO antibody, whereas neither PLK1 nor TANK coprecipitated when we used a control antibody, normal rabbit IgG (Figure 9B, left). The same results were obtained using an anti-PLK1 antibody to immunoprecipitate TANK and NEMO (Figure 9B, right). To test the effect of TANK on the interaction of PLK1 with NEMO, we transfected combinations of Myc-TANK, siRNA-TANK, Flag-PLK1, and Myc-NEMO in HEK293 cells. An anti-Flag antibody was used to immunoprecipitate the cell extracts. As shown in the left of Figure 9C, ectopically expressed TANK did not affect the protein level of NEMO, which binds to PLK1, but knockdown of TANK using siRNA sharply reduced the level of the NEMO protein. We also performed this experiment under physiological conditions. In the right of Figure 9C, the endogenous interaction between PLK1 and NEMO was also reduced by knockdown of endogenous TANK. These results show that TANK is essential for the interaction between PLK1 and NEMO. The quantity of the endogenous TANK protein is sufficient for mediating the interaction between ectopically expressed PLK1 and NEMO, and an increase in the protein level of TANK did not markedly enhance this interaction (Figure 9C, left, lane 3).
Figure 9.
TANK connects PLK1 with NEMO to form a ternary complex. (A) HEK293 cells were cotransfected with Flag-PLK1 along with Myc-TANK, Myc-NEMO, or Flag-NEMO as indicated. The cell lysates were immunoprecipitated with an anti-Flag antibody and detected with an anti-Myc(HRP) antibody (left), or immunoprecipitated with an anti-Myc antibody and detected with an anti-Flag (HRP) antibody (right). (B) Immunoblot analysis of the interaction between endogenous PLK1, NEMO, and TANK in K562 cell lysates after immunoprecipitation with rabbit IgG or anti-NEMO (left) and mouse IgG or anti-PLK1 (right) antibodies. (C) Immunoassay of lysates of HEK293 cells that were cotransfected with the indicated vectors and siRNA, immunoprecipitated with an anti-Flag antibody and detected with the indicated antibodies (left). Immunoblot analysis of the interaction between endogenous PLK1, NEMO, and TANK in the lysates of HeLa cells that were transfected with siRNA-TANK or not, and after immunoprecipitation with mouse IgG or anti-PLK1 antibodies (right). Data are representative of three experiments.
TANK-interacting Protein TBK1 and IKKε Do Not Interact with PLK1
TANK was previously shown to assist NF-κB activation as part of a complex with the IKK-related kinases TANK-binding kinase 1 (TBK1) and IKKε, two kinases that are distantly related to canonical IKKs (Chariot et al., 2002). We aimed to determine whether PLK1 could bind to these two kinases. As shown in Figure 10A, neither TBK1 nor IKKε interacted with PLK1 in HEK293 cells. To investigate whether IKKε may be involved in regulating the ability of TANK to interact with PLK1, HEK293 cells were transfected with Myc-IKKε, Myc-TANK, or Flag-PLK1 as described above. Flag-PLK1 was immunoprecipitated, and the resulting immunoprecipitates were subjected to anti-Myc Western blot analysis. Expression of IKKε had no effect on the interaction between TANK and PLK1. Coexpression of transfected TANK resulted in slightly detectable coimmunoprecipitation of IKKε and PLK1 (Figure 10C). We also tested whether a TANK construct that lacked the IKKε-interacting domain (aa 111–170) could still interact with PLK1. As shown in Figure 10B, the TANK mutant without the IKKε-binding domain no longer interacted with PLK1. According to former results, the IKKε-interacting domain is part of the N-terminal domain of TANK that is necessary for its interaction with PLK1. This domain is suggested to be a key region in TANK that recruits IKKε and PLK1 to interact with TANK.
Figure 10.
TANK interacting protein TBK1 and IKKε do not interact with PLK1. (A) Immunoassay of lysates of HEK293 cells that were cotransfected with vectors expressing Flag-TBK1 and GFP-PLK1 or Flag-PLK1 and Myc-IKKε, immunoprecipitated (IP) with an anti-Flag antibody, and analyzed by immunoblot (IB) analysis with an anti-GFP (left) or an anti-Myc (right) antibody. (B) Immunoassay of lysates of HEK293 cells that were cotransfected with Myc-TANK or TANKΔIKKε and Flag-PLK1, immunoprecipitated (IP) with an anti-Flag antibody, and analyzed by immunoblot (IB) analysis with a HRP-conjugated anti-Myc antibody. (C) Immunoassay of lysates of HEK293 cells that were transfected with Flag-PLK1, Myc-TANK, and increasing amounts of Myc-IKKε; PLK1 in the lysates was immunoprecipitated with an anti-Flag antibody, followed by immunoblot analysis with the indicated antibodies.
PLK1 Inhibits the Ubiquitination of NEMO
The IKK complex serves as the key regulator for the activation of NF-κB by various stimuli. It contains two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ/NEMO. The activation of the IKK complex is dependent on the ubiquitination of NEMO. We examined the change in the ubiquitination of NEMO in the PLK1-mediated inhibition of NF-κB activation. First, we determined whether overexpression of PLK1 had any effect on NEMO-Ubiquitin (Ub) conjugates in transient transfection experiments using HEK293 cells in which both epitope-tagged Ub and NEMO were expressed (Figure 11A). We found that overexpression of PLK1 weakens the NEMO-Ub conjugates. The same results were obtained under physiological conditions by immunoprecipitating endogenous NEMO proteins in TNF-α–induced HEK293 cells (Figure 11B).
Figure 11.
PLK1 inhibits NEMO ubiquitination. (A) HA-Ub and Myc-NEMO were coexpressed in HEK293 cells with Flag-PLK1 or empty vectors. Extracts were prepared as described previously, and anti-Myc immunoprecipitates were probed with an anti-HA antibody. (B) HeLa cells were transfected with Flag-PLK1 or empty vectors, and 48 h later, the cells were treated with TNF-α (20 ng/ml) for 0–30 min. Cell lysates were immunoprecipitated with an anti-NEMO antibody. The cell lysates and immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-Ub or anti-NEMO antibodies. Data are representative of at least three independent experiments.
DISCUSSION
The NF-κB/Rel family of transcription factors are active in cell cycle regulation. Presently, there are many reports that describe cell cycle regulation through the CDK/CDKI system (Joyce et al., 2001; Prajapati et al., 2006). However, there are few reports that cell cycle regulators also regulate the activity of NF-κB. For example, the adaptor protein TANK regulates TRAF2-induced NF-κB signaling pathway was reported (Cheng and Baltimore, 1996; Rothe et al., 1996). Recently it was been revealed that the TANK binds to downstream IKKs complex and regulates its activation (Chariot et al., 2002; Bonif et al., 2006). They provided evidences to the mechanisms of TANK positive regulate the activation of NF-κB, but how TANK negatively regulates its activity remains to be determined. Especially, how the adaptor protein TANK transmit the negative signal to the downstream cascades to terminate the activation of NF-κB.
We have shown here that TANK inhibits the NF-κB activity by recruiting PLK1 to NEMO/IKKγ. Importantly, the PLK1-mediated inhibition of NF-κB depends on IKK activation and requires interaction with TANK. In addition, we confirmed that PLK1 interacts with the regulatory subunit NEMO of the IKK complex and that PLK1, TANK and NEMO form a ternary complex under physiological conditions. Moreover, knockdown of TANK markedly reduces the interaction of PLK1 and NEMO. The present data suggest that TANK recruits PLK1 to the IKK complex and that both negatively regulate downstream NF-κB signaling.
PLK1 has been recently reported to negatively regulate TNF-induced IKK activation through the phosphorylation of the gamma binding domain (γBD) of IKKβ (Higashimoto et al., 2008). This inhibition effect relies on the kinase activity of PLK1. Using an immune complex kinase assay system to test the activities of the endogenous IKK complex, overexpression of a constitutively active form of PLK1 was shown to down-regulate TNF-α–induced IKK activation, but wild-type PLK1 did not have any effect (Higashimoto et al., 2008). In contrast to these results, we found that overexpression of wild-type PLK1 observably reduced NF-κB activation induced by TNF-α, IL-1β, or several activators in HEK293 cells. In this study, we tested the transcriptional activation of NF-κB with a luciferase reporter assay. Expression of wild-type PLK1 in HEK293 cells observably reduced NF-κB activation induced by TNF-α and several activators, and these results are consistent with a previous report (Higashimoto et al., 2008). We have shown that PLK1 interacts with TANK in a phosphorylation-independent manner. However, the kinase activity of PLK1 should play a key role in its negative regulation of NF-κB. Another molecular mechanism might exist in which PLK1 inhibits NF-κB activation through its interaction with TANK, but does not require its kinase activity.
We analyzed the physical interaction between PLK1 and the three subunits of the IKK complex, IKKα, IKKβ, and IKKγ (NEMO), and found that PLK1 only interacted with NEMO. This result is unexpected due to the fact that PLK1 directly phosphorylates IKKβ (Higashimoto et al., 2008). We hypothesize that the interaction of PLK1 and IKKβ might be transient or mediated by other proteins. Moreover, we confirmed that the interaction between PLK1 and NEMO depends on TANK. To ascertain whether PLK1 affects IKK activation through NEMO, we tested the effect of PLK1 on the ubiquitination of NEMO. We found that overexpression of PLK1 reduced the TNF-α–induced ubiquitination of NEMO. Therefore, it is likely that PLK1 regulates the TNF-α–induced activation of IKK by modifying both the phosphorylation of IKKβ and the ubiquitination of NEMO. PLK1 has been reported to physically interact with the deubiquitinating enzyme cylindromatosis (CYLD) (Stegmeier et al., 2007). CYLD removes Lys-63–linked ubiquitin chains from IKK signaling components, including NEMO, and thereby inhibits NF-κB pathway activation (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003). Taken together with our results, it would be worthwhile to investigate the roles of CYLD in the PLK1-mediated deubiquitination of NEMO.
Because the expression level, activity and localization of PLK1 changes during the cell cycle, we wondered if PLK1 is a general negative regulator of NF-κB or, alternatively, if PLK1 inhibits NF-κB activity only under certain conditions. According to our results from the subcellular localization assays, we found that TANK binds PLK1 in the cytoplasm. During S phase, G2 phase, and prophase of mitosis, PLK1 shows both nuclear and cytoplasmic localization (Taniguchi et al., 2002). We suppose that PLK1 is recruited to TANK and plays a negative regulatory role on NF-κB during these stages of the cell cycle. This hypothesis has been proven in cell cycle block assays in which nocodazole treatment caused cells to remain in G2/M phase and reduced the transcriptional activation of NF-κB and the phosphorylation of IκBα.
It is well known that PLK1 plays an important role in mitosis in eukaryotic cells. According to recent studies, PLK1 also plays unexpected roles during interphase. In vertebrate cells, PLK1 has emerged as a novel player in maintaining genomic stability during DNA replication in S phase and as an important modulator of the DNA damage checkpoint during the G2/M transition. In response to DNA damage signals, PLK1 is inhibited through ATM/ATR (van Vugt et al., 2001). Activation of the NF-κB signaling pathway is one of the cellular responses evoked to maintain homeostasis after DNA damage (Janssens et al., 2005; Habraken and Piette, 2006; Janssens and Tschopp, 2006) as well as the checkpoint pathway. According to previous results and our results, the adaptor protein TANK can separately interact with both NEMO and PLK1. This linkage might synergistically connect the responses of the two signaling pathways to DNA damage. In recent years, great progress has been made with respect to the regulatory networks of NF-κB and the cell cycle. Nevertheless, the detailed molecular mechanisms connecting these two networks have not been clarified. Our research provides a new clue to the cooperative regulation of the NF-κB signaling pathway and cell cycle control, which has potential value in developing new strategies for the treatment of various human diseases, especially chronic autoimmune disorders and cancer.
In conclusion, we demonstrate that the cell cycle regulatory kinase PLK1 is recruited to the IKK complex through TANK and is required for the negative effect of TANK on NF-κB activation. PLK1 inhibits NF-κB activation through the deactivation of IKKs, but the physiological signal that triggers this interaction remains to be uncovered. Previous reports demonstrated that TANK is phosphorylated by IKKε and TBK1 and connects these upstream kinases to the IKK complex (Nomura et al., 2000; Chariot et al., 2002). However, we did not find an interaction between these two IKK-related kinases and PLK1, and the expression of IKKε had no effect on the interaction between TANK and PLK1. In addition to its role in NF-κB signaling, TANK is also involved in IRF3/7 signaling pathway regulation induced by the TLRs (Toll-like receptors) through its interaction with TBK1 and IKKε (Fitzgerald et al., 2003; Kawagoe et al., 2009). Our results suggest that TBK1 and IKKε do not participate in the negative regulation of NF-κB by PLK1.
ACKNOWLEDGMENTS
We greatly appreciate Z. G. Liu. and X. Lin for generously providing plasmids. This work was supported by the National High-Tech Research and Development Program (2006AA02A310), the Special Funds for Major State Basic Research of China (2006CB910802), the Chinese National Natural Science Foundation (Innovation group project, 30621063), and the State Key Laboratory of Proteomics (SKLP-K200801).
Abbreviations used:
- CD
catalytic domain
- IκB
inhibitor κB
- IKK
IκB kinase
- IL-1β
interleukin 1β
- NEMO
NF-κB essential modulator
- NF-κB
nuclear factor κB
- PBD
polo-box domain
- PLK1
polo-like kinase 1
- TANK
TRAF-associated NF-κB activator
- TNF
tumor necrosis factor
- TRAF
TNF receptor–associated factor
- Ub
ubiquitin
- siRNA
small interfering RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0715) on May 19, 2010.
REFERENCES
- Bonif M., et al. TNFalpha- and IKKbeta-mediated TANK/I-TRAF phosphorylation: implications for interaction with NEMO/IKKgamma and NF-kappaB activation. Biochem. J. 2006;394:593–603. doi: 10.1042/BJ20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Chariot A., Leonardi A., Muller J., Bonif M., Brown K., Siebenlist U. Association of the adaptor TANK with the I kappa B kinase (IKK) regulator NEMO connects IKK complexes with IKK epsilon and TBK1 kinases. J. Biol. Chem. 2002;277:37029–37036. doi: 10.1074/jbc.M205069200. [DOI] [PubMed] [Google Scholar]
- Cheng G., Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Golsteyn R. M., Lane H. A., Mundt K. E., Arnaud L., Nigg E. A. The family of polo-like kinases. Prog. Cell Cycle Res. 1996;2:107–114. doi: 10.1007/978-1-4615-5873-6_11. [DOI] [PubMed] [Google Scholar]
- Golsteyn R. M., Mundt K. E., Fry A. M., Nigg E. A. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Cheng G. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J. Biol. Chem. 2007;282:11817–11826. doi: 10.1074/jbc.M700017200. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Piette J. NF-kappaB activation by double-strand breaks. Biochem. Pharmacol. 2006;72:1132–1141. doi: 10.1016/j.bcp.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Higashimoto T., Chan N., Lee Y. K., Zandi E. Regulation of I(kappa)B kinase complex by phosphorylation of (gamma)-binding domain of I(kappa)B kinase (beta) by Polo-like kinase 1. J. Biol. Chem. 2008;283:35354–35367. doi: 10.1074/jbc.M806258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y. J., Ma S., Terada Y., Erikson R. L. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 2002;277:44115–44120. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- Janssens S., Tinel A., Lippens S., Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Janssens S., Tschopp J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 2006;13:773–784. doi: 10.1038/sj.cdd.4401843. [DOI] [PubMed] [Google Scholar]
- Joyce D., Albanese C., Steer J., Fu M., Bouzahzah B., Pestell R. G. NF-kappaB and cell-cycle regulation: the cyclin connection. Cytokine Growth Factor Rev. 2001;12:73–90. doi: 10.1016/s1359-6101(00)00018-6. [DOI] [PubMed] [Google Scholar]
- Kawagoe T., Takeuchi O., Takabatake Y., Kato H., Isaka Y., Tsujimura T., Akira S. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat. Immunol. 2009;10:965–972. doi: 10.1038/ni.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye K. M., Devergne O., Harada J. N., Izumi K. M., Yalamanchili R., Kieff E., Mosialos G. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc. Natl. Acad. Sci. USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A., Chable-Bessia C., Cantarella G., Israel A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Li C. Y., Zhan Y. Q., Xu C. W., Xu W. X., Wang S. Y., Lv J., Zhou Y., Yue P. B., Chen B., Yang X. M. EDAG regulates the proliferation and differentiation of hematopoietic cells and resists cell apoptosis through the activation of nuclear factor-kappa B. Cell Death Differ. 2004;11:1299–1308. doi: 10.1038/sj.cdd.4401490. [DOI] [PubMed] [Google Scholar]
- Li J. J., Li S. A. Mitotic kinases: the key to duplication, segregation, and cytokinesis errors, chromosomal instability, and oncogenesis. Pharmacol. Ther. 2006;111:974–984. doi: 10.1016/j.pharmthera.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Nomura F., Kawai T., Nakanishi K., Akira S. NF-kappaB activation through IKK-i-dependent I-TRAF/TANK phosphorylation. Genes Cells. 2000;5:191–202. doi: 10.1046/j.1365-2443.2000.00315.x. [DOI] [PubMed] [Google Scholar]
- Pahl H. L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Perkins N. D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- Perkins N. D., Felzien L. K., Betts J. C., Leung K., Beach D. H., Nabel G. J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- Pomerantz J. L., Baltimore D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18:6694–6704. doi: 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati S., Tu Z., Yamamoto Y., Gaynor R. B. IKKalpha regulates the mitotic phase of the cell cycle by modulating Aurora A phosphorylation. Cell Cycle. 2006;5:2371–2380. doi: 10.4161/cc.5.20.3359. [DOI] [PubMed] [Google Scholar]
- Rothe M., Xiong J., Shu H. B., Williamson K., Goddard A., Goeddel D. V. I-TRAF is a novel TRAF-interacting protein that regulates TRAF-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 1996;93:8241–8246. doi: 10.1073/pnas.93.16.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits V. A., Klompmaker R., Arnaud L., Rijksen G., Nigg E. A., Medema R. H. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Sowa M. E., Nalepa G., Gygi S. P., Harper J. W., Elledge S. J. The tumor suppressor CYLD regulates entry into mitosis. Proc. Natl. Acad. Sci. USA. 2007;104:8869–8874. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi E., Toyoshima-Morimoto F., Nishida E. Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J. Biol. Chem. 2002;277:48884–48888. doi: 10.1074/jbc.M206307200. [DOI] [PubMed] [Google Scholar]
- Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- van Vugt M. A., Medema R. H. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- van Vugt M. A., Smits V. A., Klompmaker R., Medema R. H. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J. Biol. Chem. 2001;276:41656–41660. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- Yarm F. R. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wang J., Li W., Yuan Y. Z., Li C. Y., Qian X. H., Xu W. X., Zhan Y. Q., Yang X. M. Proteomic screen defines the hepatocyte nuclear factor 1alpha-binding partners and identifies HMGB1 as a new cofactor of HNF1alpha. Nucleic Acids Res. 2008;36:1209–1219. doi: 10.1093/nar/gkm1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Li L., Liu Q., Xing G., Kuai X., Sun J., Yin X., Wang J., Zhang L., He F. E3 ubiquitin ligase SIAH1 mediates ubiquitination and degradation of TRB3. Cell Signal. 2008;20:942–948. doi: 10.1016/j.cellsig.2008.01.010. [DOI] [PubMed] [Google Scholar]