Arrestin-2 interacts with ESCRT-0 proteins STAM-1 and HRS to control the amount of chemokine receptor CXCR4 targeted into the degradative pathway by regulating the ubiquitination status of HRS.
Abstract
The chemokine receptor CXCR4, a G protein-coupled receptor, is targeted for lysosomal degradation via a ubiquitin-dependent mechanism that involves the endosomal sorting complex required for transport (ESCRT) machinery. We have reported recently that arrestin-2 also targets CXCR4 for lysosomal degradation; however, the molecular mechanisms by which this occurs remain poorly understood. Here, we show that arrestin-2 interacts with ESCRT-0, a protein complex that recognizes and sorts ubiquitinated cargo into the degradative pathway. Signal-transducing adaptor molecule (STAM)-1, but not related STAM-2, interacts directly with arrestin-2 and colocalizes with CXCR4 on early endosomal antigen 1-positive early endosomes. Depletion of STAM-1 by RNA interference and disruption of the arrestin-2/STAM-1 interaction accelerates agonist promoted degradation of CXCR4, suggesting that STAM-1 via its interaction with arrestin-2 negatively regulates CXCR4 endosomal sorting. Interestingly, disruption of this interaction blocks agonist promoted ubiquitination of hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) but not CXCR4 and STAM-1 ubiquitination. Our data suggest a mechanism whereby arrestin-2 via its interaction with STAM-1 modulates CXCR4 sorting by regulating the ubiquitination status of HRS.
INTRODUCTION
The chemokine receptor CXCR4, a G protein-coupled receptor (GPCR), upon activation by its cognate ligand stromal cell-derived factor (SDF)-1α (CXCL12) is rapidly internalized and targeted into the degradative pathway by a ubiquitin-dependent mechanism (Marchese and Benovic, 2001; Marchese et al., 2003). Activation by CXCL12 induces rapid and transient phosphorylation of serine residues 324 and 325 within the carboxy-terminal tail (C-tail) of CXCR4, thereby promoting binding to the E3 ubiquitin ligase atrophin-1–interacting protein (AIP)4 via a novel WW-domain–mediated interaction culminating in ubiquitination of vicinal lysine residues (Marchese et al., 2003; Bhandari et al., 2009). This is followed by internalization of CXCR4 onto early endosomes where the ubiquitin moiety serves as a sorting signal to direct the receptor to lysosomes for proteolysis (Marchese and Benovic, 2001; Marchese et al., 2003).
In general, the ubiquitin moiety on ubiquitinated receptors interacts with ubiquitin binding domains (UBDs) found in several proteins of the endosomal sorting complex required for transport (ESCRT) machinery (Raiborg and Stenmark, 2009; Shields et al., 2009). The ESCRT machinery consists of four distinct protein complexes (ESCRT 0–III) that act in a sequential and a coordinated manner to target ubiquitinated receptors into multivesicular bodies, which then fuse with lysosomes where degradation occurs. Recruitment into this pathway takes place by the initial recognition of the ubiquitinated receptor by ESCRT-0, which then subsequently recruits ESCRT-I to the endosomal membrane, followed by recruitment of ESCRT II and III, culminating in proper execution of the sorting process (reviewed in Williams and Urbe, 2007; Raiborg and Stenmark, 2009). Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) is a critical element of ESCRT-0 and has been shown to mediate down-regulation of several cell surface signaling receptors (Bache et al., 2003a; Kanazawa et al., 2003; Abella et al., 2005; Hasdemir et al., 2007), including CXCR4 (Marchese et al., 2003). The ubiquitin moiety on CXCR4 is thought to interact with the ubiquitin interacting motif (UIM) found in HRS, thereby targeting CXCR4 into the degradative pathway.
Together with HRS, signal-transducing adaptor molecule (STAM) forms ESCRT-0. STAM was originally identified as an adaptor protein involved in cytokine signaling (Takeshita et al., 1996, 1997). Two STAM isoforms exist, STAM-1 and STAM-2, which share 53% amino acid identity and may be redundant in their function (Lohi et al., 1998; Endo et al., 2000; Pandey et al., 2000; Yamada et al., 2002). Similar to HRS, STAM also binds to ubiquitin and may act in concert with HRS to recruit ubiquitinated receptors for lysosomal sorting (Asao et al., 1997; Takata et al., 2000; Bache et al., 2003b; Kanazawa et al., 2003). STAMs also may modulate endosomal sorting by virtue of their ability to interact with endosomal associated deubiquitinating enzymes AMSH and UBPY, which may modulate the ubiquitination status of both receptors and/or the sorting machinery (McCullough et al., 2004, 2006; Bowers et al., 2006; Row et al., 2006; Kong et al., 2007; Ma et al., 2007). Recently, STAMs have been implicated in endoplasmic reticulum to Golgi trafficking, possibly via their interaction with coat protein II proteins (Rismanchi et al., 2009). However, to our knowledge, their role in GPCR trafficking and signaling remains relatively unknown.
We have recently shown that arrestin-2 mediates endosomal sorting of CXCR4 (Bhandari et al., 2007). Nonvisual arrestins, arrestin-2 and arrestin-3 (a.k.a. β-arrestin1 and β-arrestin2, respectively), are generally known for their ability to regulate GPCR desensitization, internalization, and signaling (Moore et al., 2007), although their role in endosomal sorting remains relatively unexplored. Arrestin-2 interacts with and colocalizes with AIP4 on early endosomes, where it targets CXCR4 for lysosomal sorting (Bhandari et al., 2007). In addition to mediating ubiquitination of CXCR4 at the plasma membrane, AIP4 also interacts with and mediates ubiquitination of HRS, probably on endosomes; however, the function of the ubiquitin moiety remains unknown (Marchese et al., 2003). How arrestin-2 integrates with AIP4 and HRS to carry out CXCR4 sorting into the degradative pathway remains poorly understood.
Here, we show that arrestin-2 interacts with ESCRT-0 via a direct interaction with STAM-1. Disruption of this interaction attenuates CXCR4 mediated ubiquitination of HRS and accelerates CXCR4 degradation, suggesting that the arrestin-2/STAM-1 complex negatively regulates CXCR4 lysosomal sorting via ubiquitination of HRS. This study provides novel mechanistic insight into the role that arrestin-2 has on targeting CXCR4 into the degradative pathway and furthers our understanding of the complex molecular events that mediate endosomal sorting of GPCRs.
MATERIALS AND METHODS
Cell Lines, Antibodies, and Reagents
Human embryonic kidney (HEK) 293 cells (Microbix, Toronto, ON, Canada) and HeLa cells (American Type Culture Collection, Manassas, VA) were maintained in DMEM (HyClone Laboratories, Logan, UT) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories). HRS (M-79) rabbit polyclonal, β-arrestin1/2 rabbit polyclonal (H-290), and mouse monoclonal (21-B1) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-glutathione transferase (GST) monoclonal antibodies (mAb) and glutathione-Sepharose 4B resin were from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). The anti-CXCR4 antibody was described previously (Marchese and Benovic, 2001). The STAM-1 and AMSH polyclonal antibodies were from ProteinTech Group (Chicago, IL). The arrestin-2 and anti-early endosomal antibody (EEA)1 monoclonal antibodies were from BD Biosciences (San Jose, CA). The anti-hemagglutinin (HA) polyclonal and monoclonal antibodies were from Covance Research Products (Berkeley, CA). The anti-FLAG M2, M1, and M2-horseradish peroxidase-conjugated monoclonal antibodies, FLAG polyclonal antibody, alkaline phosphatase-conjugated anti-mouse antibody, and AMD3100 were from Sigma-Aldrich (St. Louis, MO). The alkaline phosphatase substrate kit was from Bio-Rad Laboratories (Hercules, CA). The anti-T7 goat polyclonal antibody was from Abcam (Cambridge, MA). The anti-epidermal growth factor receptor (EGFR) mouse mAb was from Assay Designs (Ann Arbor, MI). The anti-actin mAb was from MP Biomedicals (Aurora, OH). Stromal cell-derived factor-1α (CXCL12) and epidermal growth factor (EGF) were from PeproTech (Rocky Hill, NJ). The Alexa-Fluor 635-conjugated goat anti-mouse, Alexa-Fluor 594-conjugated anti-rat, Alexa-Fluor 488-conjugated goat anti-rabbit, and Alexa-Fluor 568-conjugated goat anti-rabbit antibodies were from Invitrogen (Carlsbad, CA). The small interfering RNA (siRNA) for glyceraldehyde-3-phosphate dehydrogenase (GAPD), STAM-1 (GAACGAAGAUCCGAUGUAU), and associated molecule with the Src homology 3 (SH3) domain of STAM (AMSH) (siGENOME SMARTpool D-012202) were from Dharmacon RNA Technologies (Lafayette, CO).
DNA Constructs
HA-CXCR4, FLAG-ubiquitin, HA-CXCR4-YFP, HA-arrestin-3, and HA-arrestin-2 constructs were described previously (Bhandari et al., 2007). Primers used for generating all constructs are listed in Supplemental Table S1. For STAM-1 truncation mutants (1-195, 1-269, 1-390, 391-540, 337-540, 270-540, 212-540, 144-540), full-length STAM-1 in 3×FLAG-pCMV-10 was amplified by polymerase chain reaction (PCR) using primers flanking various regions of STAM-1 as indicated above and harboring 5′ and 3′ HindIII and XbaI restriction enzyme sites, respectively. PCR fragments were digested and ligated into the HindIII and XbaI sites of 3×FLAG pCMV-10 (Sigma-Aldrich). For STAM-1-ΔGAT, the region encompassing amino acid residues 343-377 was deleted by two-step PCR with mutually annealing overlapping primers and flanking primers based on 3×FLAG-pCMV-10. Amplified product was digested and ligated into HindIII and XbaI sites of 3×FLAG-pCMV-10 and pGEX-4T2 (GE Healthcare). For STAM-1-GAT, amino acid residues 296-380 were amplified by PCR from full-length FLAG-STAM-1 and cloned into the HindIII and XbaI sites of 3×FLAG-pCMV-10 and EcoRI and XhoI sites of pGEX-4T2. For arrestin-2-(25-161) constructs, amino acid residues 25-161 were amplified by PCR from HA-arrestin-2-(1-161) and cloned into HindIII and XbaI sites of 3×FLAG pCMV-10 and SmaI and XhoI sites of pGEX4T2 respectively. For YFP-STAM-1, full-length STAM-1 was amplified from FLAG-STAM-1 and cloned into the HindIII and KpnI sites of pEYFP-C1 vector (Clontech, Mountain View, CA). The sequence of all constructs was verified by sequencing.
GST-Fusion Protein Binding Assays
Escherichia coli BL21 cells transformed with GST-fusion protein constructs or empty vector (pGEX-4T2) were grown overnight in Luria Broth containing 100 μg/ml ampicillin. The next day, cultures were diluted (3.7%) and grown to an OD600 ∼ 0.35–0.40 at 37°C followed by induction with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside for 1 h at 18°C. Cells were then pelleted by centrifugation and resuspended in 1 ml of binding buffer (20 mM Tris-Cl pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 1 mM dithiothreitol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin-A), followed by sonication and centrifugation. Clarified lysates were incubated with glutathione-Sepharose 4B resin for 1 h, washed, and resuspended in binding buffer. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and stained with Gel-Code blue to estimate the protein amounts by comparing the samples to known amounts of purified bovine serum albumin (Fraction V; Roche Diagnostics, Indianapolis, IN). For binding assays, equimolar amounts of purified GST-fusion proteins were incubated with 100 μl of clarified cell lysate of HEK293 cells expressing the desired construct for 2–4 h at 4°C. For binding experiments using purified arrestin-2, GST fusion proteins were incubated with 500 ng of arrestin-2 in 100 μl of binding buffer for 1 h at 4°C. After incubation, samples were washed three times with binding buffer, eluted in 2× sample buffer by boiling for 10 min and bound proteins were detected by SDS-PAGE followed by immunoblotting.
Degradation Assay
HEK293 cells stably expressing HA-CXCR4 or HeLa cells expressing endogenous levels of CXCR4 grown on 10-cm dishes were transfected with 100 nM STAM-1, AMSH, or GAPD siRNA using Lipofectamine 2000 transfection reagent (Invitrogen). To assess the role of STAM-1 and arrestin-2 minigene constructs on CXCR4 degradation, HEK293 cells grown on 10-cm dishes were cotransfected with 1 μg of HA-CXCR4 and 9 μg of FLAG-STAM-1-GAT, FLAG-arrestin-2-(25-161) or empty vector (pCMV-10) using TransIT-LT1 transfection reagent (Mirus, Madison, WI). Twenty-four hours later, cells were passaged onto poly-l-lysine (0.1 mg/ml; Sigma-Aldrich) coated 24-well plates (HEK293 cells) or six-well plates (HeLa cells) and grown for an additional 18–24 h. Cells were washed once and incubated with DMEM containing 10% FBS and 50 μg/ml cyclohexamide to stop protein synthesis for 15 min at 37°C. Cells were then incubated with the same medium containing vehicle (0.5% bovine serum albumin [BSA]) or 30 nM CXCL12 for 1, 2, and 3 h. Cells were washed and collected in 300 μl of 2× sample buffer and then sonicated. Receptor amounts were determined by SDS-PAGE followed by immunoblotting using an anti-HA mAb or anti-CXCR4 antibody, as described previously (Marchese, 2009). To assess EGFR degradation, HeLa cells grown on six-well plates were transfected with 3 μg of FLAG-STAM-1-GAT, FLAG-arrestin-2-(25-161), or empty vector (pCMV-10) using TransIT-LT1 transfection reagent. Forty-eight hours after transfection cells were incubated with DMEM containing 10% FBS and 50 μg/ml cyclohexamide to stop protein synthesis for 15 min at 37°C. Cells were then incubated with the same medium containing vehicle (0.5% BSA) or 100 ng/ml EGF for 1 h. Cells were processed as described above for CXCR4 degradation.
Coimmunoprecipitation Studies
HeLa cells were transiently transfected with HA-Arrestin-2, HA-arrestin-3, or empty vector alone (pcDNA3) using TransIT-LT1 transfection reagent. Forty-eight hours later, cells were collected in 1.5-ml immunoprecipitation buffer [20 mM Na2PO4, pH 6.5, 150 mM NaCl, 1% (vol/vol) Triton-X 100, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin A] and incubated at 4°C for 30 min. Cells were sonicated, centrifuged, and the clarified lysates were incubated with an anti-HA mAb or isotype control antibody to immunoprecipitate HA-tagged arrestin-2/3 followed by immunoblotting to detect bound endogenous STAM-1 and HRS. Endogenous arrestins were immunoprecipitated from HeLa cells using an anti-arrestin2/3 mouse monoclonal or isotype control antibody followed by immunoblotting to detect bound endogenous STAM-1 and HRS. To assess the effect of the STAM-1-GAT minigene on the interaction between STAM-1 and arrestin-2, lysates from HeLa cells transfected with HA-arrestin-2 and FLAG-STAM-1-GAT or pCMV were incubated with an anti-HA or isotype control antibody and immunoprecipitates were analyzed for the presence of endogenous STAM-1. To assess the effect of the arrestin-2-(25-161) minigene on the interaction between STAM-1 and arrestin-2, HeLa cells transfected with T7-STAM-1, HA-arrestin-2, and FLAG-arrestin-2-(25-161) or pCMV were incubated with an anti-T7 polyclonal antibody and immunoprecipitates were analyzed for the presence of HA-arrestin-2 and endogenous HRS.
Confocal Immunofluorescence Microscopy
HEK293 cells transiently transfected with HA-CXCR4-YFP were passaged onto poly-l-lysine–coated coverslips and allowed to grow for 24 h. HeLa cells were used to examine the distribution of endogenous CXCR4. Cells were washed once with warm DMEM containing 20 mM HEPES, pH 7.5, and incubated in the same medium for 3–4 h at 37°C. Cells were treated with 30 nM CXCL12 or vehicle for 30 min, fixed with 3.7% paraformaldehyde, and then permeabilized with 0.05% (wt/vol) saponin for 10 min, similar to a protocol we have described previously (Bhandari et al., 2007). Cells were coincubated with STAM-1, EEA1, or β-arrestin2/3 antibodies. Endogenous CXCR4 in HeLa cells was stained with rat anti-CXCR4 mAb. In brief, after permeabilization and fixation, cells were incubated with 1% BSA in 0.05% saponin-phosphate-buffered saline (PBS) for 30 min at 37°C, followed by incubating with primary antibody for 1 h at 37°C. Primary antibodies for STAM-1 and EEA1 were used at 1:100 dilution and against CXCR4 and β-arrestin2/3 was used at a 1:50 dilution. Cells were washed five times with 0.05% saponin-PBS, followed by incubating with appropriate Alexa-Fluor–conjugated secondary antibodies for 30 min at 37°C. Finally, cells were washed with PBS and fixed again in 3.7% formaldehyde-PBS and mounted onto glass slides using mounting media containing 4,6-diamidino-2-phenylindole. Samples were analyzed using an LSM 510 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) equipped with a Plan-Apo 63×/1.4 oil lens objective. Images were acquired using a 1.4-megapixel cooled extended spectra range RGB digital camera set at 512 × 512 resolution. Acquired images were analyzed using ImageJ, version 1.41o (National Institutes of Health, Bethesda, MD), and the amount of colocalization between proteins was determined using MetaMorph 7.6 (Molecular Devices, Downingtown, PA).
Ubiquitination Assays
For CXCR4 ubiquitination, HEK293 cells stably expressing HA-CXCR4 grown on 10-cm dishes were transfected with 3 μg of FLAG-ubiquitin. Eight hours later, cells were transfected either with 10 μg of FLAG-STAM-1-GAT, FLAG-Arr2-(25-161), or empty vector (pCMV). The next day, cells were passaged onto 6-cm dishes and allowed to grow for an additional 24 h. The next day, cells were serum starved in DMEM containing 20 mM HEPES for 3 h and then treated with 30 nM SDF for 30 min, washed once on ice with cold PBS, and collected in 1 ml of lysis buffer [50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% (wt/vol) sodium deoxycholate, 1% (vol/vol) NP-40, 0.1% (wt/vol) SDS, 20 mM N-ethylmaleimide (NEM), and 10 μg/ml each of leupeptin, aprotinin, and pepstatin A]. Samples were transferred into microcentrifuge tubes and placed at 4°C for 30 min, followed by sonification and centrifugation to pellet cellular debris. Clarified cell lysate was incubated with an anti-HA polyclonal antibody and the immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting using an anti-FLAG antibody conjugated to HRP.
To detect HRS ubiquitination, HEK293 cells stably expressing HA-CXCR4 were transfected with 3 μg of FLAG-ubiquitin. Eight hours later, cells were cotransfected with 8 μg of FLAG-STAM-1-GAT or empty vector (pCMV-10) and 2 μg of T7-tagged HRS. Twenty-four hours later, cells were passaged onto poly-l-lysine–coated 6-cm dishes, and the next day cells were serum starved for 4–5 h in DMEM containing 20 mM HEPES and were treated with 30 nM SDF or vehicle alone for 30 and 60 min. Cells were washed with cold PBS and collected in 1 ml of ubiquitination buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1% Triton-X 100, 5 mM EDTA, 20 mM NEM, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μg/ml pepstatin-A), incubated for 30 min at 4°C, sonicated, and clarified by centrifugation. HRS was immunoprecipitated using an anti-HRS polyclonal antibody and immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting to detect ubiquitinated HRS using an anti-FLAG antibody conjugated to HRP.
For STAM-1 ubiquitination experiments, HeLa cells grown in six-well dishes were cotransfected with 3 μg of T7-STAM-1 and 40 ng of HA-ubiquitin. Eight hours later, cells were transfected with 3 μg of FLAG-STAM-1-GAT or empty vector (pCMV-10). Twenty-four hours later, cells were passed onto poly-l-lysine–coated 6-cm dishes, and the next day cells were serum starved and treated and processed as described above for HRS ubiquitination using a modified ubiquitination buffer (20 mM NaPO4, pH 6.5, 150 mM NaCl, 1% Triton-X 100, 20 mM NEM, and protease inhibitor cocktail). Tagged STAM-1 was immunoprecipitated using an anti-T7 goat polyclonal antibody and immunoprecipitates were analyzed by SDS-PAGE followed by immunoblotting to detect ubiquitinated STAM-1 using an anti-HA mAb.
Internalization and Recycling Assays
For measuring internalization and recycling of CXCR4, HEK293 cells grown on 10-cm dishes were cotransfected with FLAG-CXCR4 (1 μg) and 100 nM STAM-1 or GAPD siRNA using Lipofectamine 2000 transfection reagent. The next day, cells were passaged onto poly-l-lysine–coated 24-well plates and grown for an additional 24 h. Cells were serum starved for 3–4 h; placed on ice; washed once with DMEM containing 0.1% BSA, 20 mM HEPES, and 1 mM Ca2+; and then incubated in the same medium containing the calcium-dependent M1 anti-FLAG antibody for 1 h on ice, which labels cell surface receptors only. Cells were washed and incubated in the same medium containing vehicle or 30 nM CXCL12 for 45 min at 37°C. To remove surface bound antibody, cells were washed three times with Ca2+- and Mg2+-free PBS containing 0.04% EDTA. Cells were incubated in DMEM containing 1 mM Ca2+ and the CXCR4 antagonist AMD3100 (10 μM) to block any further internalization for 30 and 60 min at 37°C. The amount of receptor/antibody that recycled back to the cell surface was quantified by incubating cells with an alkaline-phosphatase conjugated goat anti-mouse immunoglobulin (Ig)G antibody. In brief, cells were washed once with PBS containing 1 mM Ca2+ and then fixed with 3.7% paraformaldehyde for 5 min on ice. After fixation, cells were washed three times and incubated with alkaline phosphatase-conjugated goat anti-mouse antibody diluted in PBS containing 1% BSA for 1 h at room temperature. Cells were then washed with PBS and incubated in p-nitrophenyl phosphate diluted in diethanolamine buffer (Bio-Rad Laboratories) for 5–15 min. Reactions were stopped by adding 0.4 N NaOH and an aliquot was used to measure the absorbance at 405 nm. Percentage of receptor recycling was calculated by dividing the amount of receptor internalized by the amount of receptors recovered after incubation at different time intervals. To calculate the percentage of receptor internalization, the amount of receptor remaining on the cell surface was divided by the total number of receptors present on the cell surface before treatment with agonist.
Statistical Analysis
Statistical analyses were performed using Prism 4.00 for Macintosh (GraphPad Software, San Diego, CA; www.graphpad.com).
RESULTS
Arrestins Interact with ESCRT-0
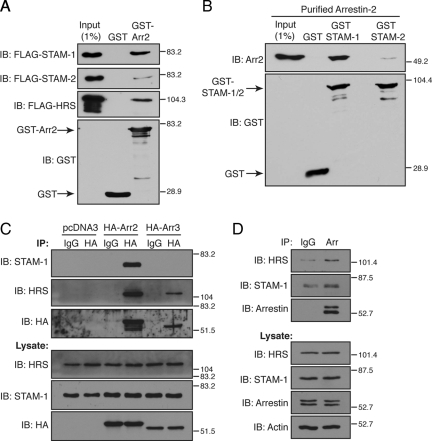
Although we have shown previously that HRS and arrestin-2 mediate endosomal sorting of CXCR4 into the degradative pathway (Marchese et al., 2003; Bhandari et al., 2007), the molecular mechanisms remain poorly understood. To gain mechanistic insight into this process, we initially examined whether arrestin-2 interacts with ESCRT-0 by determining whether it binds to HRS, STAM-1, or STAM-2. To address this, cell lysates prepared from HEK293 cells expressing FLAG-tagged STAM-1, STAM-2, or HRS were incubated with bacterially purified GST-arrestin-2 and GST immobilized on glutathione-Sepharose resin. As shown in Figure 1A, arrestin-2 bound to STAM-1 and HRS but only weakly to STAM-2. To rule out the possibility of an intermediate protein mediating the interaction with STAM-1, similar experiments were performed using purified arrestin-2. As shown in Figure 1B, GST-STAM-1, but not GST-STAM-2 and GST, bound to purified arrestin-2, indicating that the interaction between arrestin-2 and STAM-1 is direct and confirming that arrestin-2 binds poorly to STAM-2. To determine whether arrestin-2 associates with ESCRT-0 in cells, HA-arrestin-2, HA-arrestin-3, or empty vector (pcDNA3) were transfected into HeLa cells followed by immunoprecipitation and immunoblotting to detect the presence of endogenous STAM-1 and HRS. Both STAM-1 and HRS were detected in the immunoprecipitates from cells expressing HA-arrestin-2, suggesting that arrestin-2 associates with HRS and STAM-1 in cells (Figure 1C), whereas HRS, but not STAM-1, was detected in the HA-arrestin-3 immunoprecipitates (Figure 1C). Similarly, endogenous arrestins also coimmunoprecipitated with endogenous STAM-1 and HRS in HeLa cells (Figure 1D). Together, these data show that the interaction between STAM-1 and nonvisual arrestins is limited to arrestin-2 and that HRS interacts with both arrestin-2 and arrestin-3. In addition, our data suggest that arrestin-2 exists in complex with a subpopulation of ESCRT-0 that includes STAM-1 and HRS but not STAM-2.
Figure 1.
Arrestin-2 interacts with ESCRT-0. (A) Equimolar amounts (∼134 nM) of GST-arrestin-2 and GST immobilized on glutathione-Sepharose resin were incubated with lysates from HEK293 cells transiently transfected with FLAG-STAM-1, FLAG-STAM-2, or FLAG-HRS. Bound proteins were detected by immunoblotting using the anti-FLAG M2 antibody. (B) Equimolar amounts (117 nM) of GST-STAM-1, GST-STAM-2, and GST immobilized on glutathione-Sepharose resin were incubated with purified arrestin-2 (∼212 nM). Bound arrestin-2 was detected using an anti-arrestin-2 mAb. (A and B) Blots were stripped and reprobed using an anti-GST antibody to determine the levels of the GST fusion proteins used in the binding assay. (C and D) Lysates from HeLa cells either transiently transfected with HA-arrestin-2, HA-arrestin-3, or empty vector (pcDNA3) (C) or untransfected (D) were incubated with antibodies to immunoprecipitate transfected (C) or endogenous arrestins (D) as described in Materials and Methods. Immunoprecipitates (IP) and lysates were analyzed by SDS-PAGE and immunoblotting as indicated. Shown are representative blots from one of three (A–C) or four (D) independent experiment.
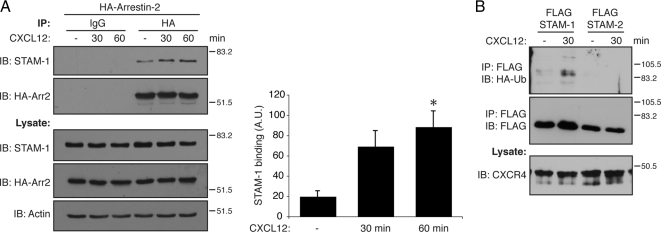
Next, we examined whether the interaction was regulated by activation of CXCR4. HeLa cells, which endogenously express CXCR4, were transfected with HA-arrestin-2 and treated with CXCL12 (30 nM) or vehicle (0.05% BSA-PBS) for various times followed by immunoprecipitation of tagged arrestin-2 and immunoblotting to detect bound endogenous STAM-1. Activation of CXCR4 enhanced the interaction between STAM-1 and arrestin-2 as early as 30 min after agonist treatment that persisted up to 60 min (Figure 2A). Because STAM has been shown to be ubiquitinated (McCullough et al., 2004), we next assessed whether CXCR4 activation promotes ubiquitination of STAM-1. HEK293 cells transfected with FLAG-tagged STAM-1 or STAM-2 and HA-tagged ubiquitin were treated with CXCL12 (100 nM) for 30 min followed by immunoprecipitation of tagged STAM proteins and immunoblotting to detect incorporation of tagged ubiquitin. As shown in Figure 2B, STAM-1 was ubiquitinated by agonist activation of CXCR4, but STAM-2 was not ubiquitinated.
Figure 2.
CXCR4 regulates the arrestin-2/STAM-1 interaction. (A) HeLa cells transiently transfected with HA-arrestin-2 were serum starved as described in Materials and Methods, followed by treatment with 30 nM CXCL12 for 30 and 60 min. Cell lysates were subject to immunoprecipitation using monoclonal anti-HA and isotype control antibodies. Immunoprecipitates and lysates were analyzed by SDS-PAGE and immunoblotting to detect endogenous STAM-1 and HA-arrestin-2. Immunoblots were subject to densitometric analysis, and the bar graph represents the average STAM-1 binding ± SEM normalized to the level of HA-arrestin-2 in the immunoprecipitates. STAM-1 binding to arrestin-2 was significantly increased upon agonist treatment as compared with vehicle. Data were analyzed by one-way ANOVA followed by a Bonferroni's post hoc test (*p < 0.05). (B) STAM-1 is preferentially ubiquitinated upon CXCR4 activation. HEK293 cells cotransfected with HA-CXCR4, FLAG-STAM-1, or FLAG-STAM-2 and HA-ubiquitin were treated with 100 nM CXCL12 for 30 min. FLAG-STAM-1/2 were immunoprecipitated using an anti-FLAG antibody, followed by SDS-PAGE and immunoblotting to detect incorporated HA-ubiquitin. Blots were stripped and reprobed for FLAG-STAM-1/2 to assess loading. Cell lysates were analyzed for the presence of HA-CXCR4. Shown are representative blots from one of three independent experiments.
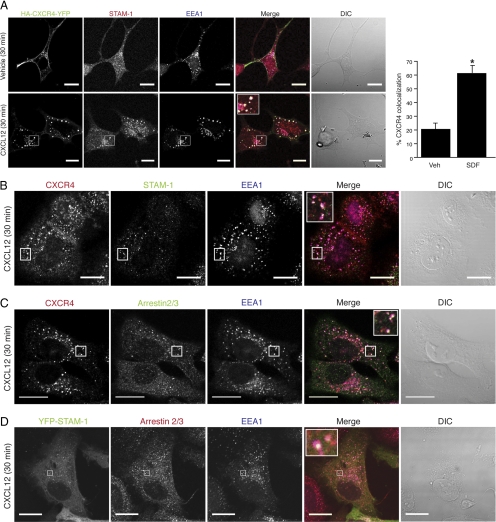
To confirm that arrestin-2 and STAM-1 are found within the same intracellular compartment, we examined their distribution in cells by confocal immunofluorescence microscopy. As shown in Figure 3A, in HEK293 cells transfected with yellow fluorescent protein (YFP)-tagged CXCR4, a construct that we have described previously (Bhandari et al., 2009), CXCR4 was mainly localized to the plasma membrane in vehicle-treated cells, whereas endogenous STAM-1 was mainly localized to punctate vesicles distributed throughout the cytoplasm, many of which also colocalized with EEA1, used here as a marker for early endosomes. In contrast, upon agonist treatment, CXCR4 distributed into an intracellular punctate pattern, indicating that it had internalized into vesicles that also contained STAM-1 and EEA1 (Figure 3A, bottom). We also examined the distribution of endogenous CXCR4 in HeLa cells treated with CXCL12 for 30 min, revealing that CXCR4 colocalized with endogenous STAM-1 (Figure 3B) and arrestin-2/3 (Figure 3C) on EEA1-positive early endosomes. CXCR4 activation also promoted colocalization of arrestin-2/3 and YFP-tagged STAM-1 on early endosomes in HeLa cells (Figure 3D). Together, our data suggest that upon internalization CXCR4 appears on early endosomes together with arrestin-2 and STAM-1.
Figure 3.
Arrestin-2, STAM-1 and CXCR4 colocalize on early endosomes. (A) Serum-starved HEK293 cells expressing HA-CXCR4-YFP were treated with 30 nM CXCL12 or vehicle for 30 min. Cells were fixed, permeabilized and double stained with anti-STAM-1 (red) and anti-EEA1 (blue). White puncta in the merged images represents colocalization between all three proteins. The percentage of colocalization between CXCR4-YFP and STAM-1 was quantified as described in Materials and Methods. Bar graph represents the percent colocalization between CXCR4-YFP and STAM-1 in vehicle and SDF-treated cells ± SEM from 10 cells. Data were analyzed by Student's t test *p < 0.0001. (B–D) Serum-starved HeLa cells were treated with 30 nM CXCL12 or vehicle for 30 min. Cells were fixed, permeabilized, and triple stained with anti-STAM-1 (green), anti-EEA1 (blue), and anti-CXCR4 (red) (B); triple stained with anti CXCR4 (red), anti-arrestin-2/3 (green), and anti-EEA1 (blue) (C); and HeLa cells expressing YFP-STAM-1 were double stained with arrestin-2/3 (red) and EEA1 (blue) (D). White puncta in the merged images represent colocalization between all three proteins. Colocalization between CXCR4 and STAM-1 (B; 20%), CXCR4 and arrestin (C; 30.7%), and YFP-STAM-1 and arrestin-2 (D; 26%). were quantified as described in Materials and Methods. Inset represents 4–8× the size of the boxed region. Differential interference contrast (DIC) images are shown. Shown are representative micrographs from three independent experiments. Bars, 20 μm.
Role of STAM-1 in CXCR4 Trafficking
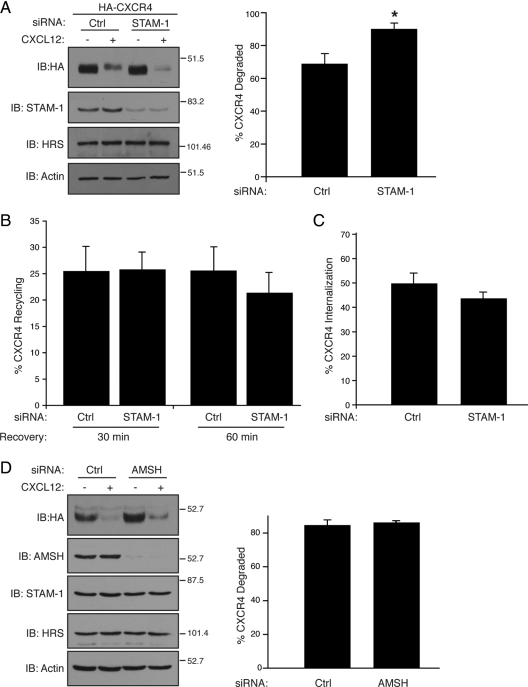
Because these data suggest that STAM-1 has a role in endosomal sorting of CXCR4, we examined agonist promoted degradation of CXCR4 in cells that were depleted of STAM-1 by RNA interference. HEK293 cells stably expressing HA-CXCR4 were transfected with control or STAM-1 siRNA, followed by treatment with CXCL12 (30 nM) for 3 h, and receptor degradation was assessed by immunoblot analysis, as described previously (Marchese et al., 2003). As shown in Figure 4A, siRNA-mediated depletion of STAM-1 lead to a moderate, but statistically significant, increase in CXCR4 degradation, compared with control siRNA-treated cells, suggesting that STAM-1 negatively regulates agonist promoted degradation of CXCR4. As the amount of receptor that is degraded is in part a function of the rate of receptor internalization and recycling, we also examined the effect of depleting STAM-1 on CXCR4 internalization and recycling. Cell surface FLAG-tagged CXCR4 was labeled with the M1 anti-FLAG antibody on ice in the presence of 1 mM Ca2+, because the M1 antibody binds to the FLAG epitope in a calcium-dependent manner. Cells were washed to remove unbound antibody, and the media were replaced with DMEM containing CXCL12 (30 nM) in the continued presence of 1 mM Ca2+ and placed at 37°C for 45 min to allow for internalization of the M1/CXCR4 complexes to take place. Antibody remaining on the surface, mostly representing uninternalized receptor, was removed by incubating cells with PBS containing EDTA (0.04%), a calcium-chelating agent. The amount of antibody (i.e., receptor) that recycled back to the cell surface was quantified by cell surface enzyme-linked immunosorbent assay (ELISA) in parallel wells that were incubated at 37°C for 30 and 60 min. In control siRNA treated cells, ∼20% of internalized CXCR4 recycled back to the cell surface after 30 and 60 min, similar to what we observed in STAM-1–depleted cells, suggesting that STAM-1 depletion had no effect on recycling of CXCR4 (Figure 4B). In addition, agonist-promoted internalization of CXCR4 was similar in STAM-1–depleted cells, compared with control siRNA-treated cells, suggesting that STAM-1 is not involved in CXCR4 internalization (Figure 4C). We also examined the role of AMSH on agonist promoted degradation of CXCR4. AMSH is a deubiquitinating enzyme that interacts with STAM-1 and negatively regulates endosomal sorting of the EGFR (McCullough et al., 2004). As shown in Figure 4D, siRNA-mediated depletion of AMSH did not effect agonist promoted degradation of CXCR4 in HeLa cells, suggesting that AMSH does not regulate endosomal sorting of activated CXCR4. However, CXCR4 levels were elevated in vehicle treated cells transfected with AMSH siRNA (Figure 4D), suggesting that AMSH may regulate degradation of constitutively internalized CXCR4, similar to what has been reported recently (Sierra et al., 2010). Together, our data suggest that STAM-1 negatively regulates CXCR4 degradation likely through a mechanism that directly attenuates endosomal sorting.
Figure 4.
STAM-1 negatively regulates CXCR4 degradation. HEK293 cells stably expressing HA-CXCR4 were transfected with control (GAPD) and STAM-1 siRNA as described in Materials and Methods. Cells were treated with vehicle (PBS containing 0.01% BSA) or 30 nM CXCL12 for 3 h and receptor levels were determined by immunoblotting followed by densitometric analysis. Bars represent the average amount of CXCR4 degraded ± SEM from three independent experiments. *p < 0.05, unpaired t test. (B) CXCR4 recycling was measured in HEK293 cells transfected with FLAG-CXCR4 and siRNA as described in A. Surface receptors were labeled with the M1 anti-FLAG antibody followed by treatment with 30 nM CXCL12 for 45 min in DMEM containing 0.1% BSA, 20 mM HEPES, pH 7.4, and 1 mM Ca2+. Antibody remaining on the cell surface was stripped by two rapid washes with Ca2+/Mg2+ free PBS containing 0.04% EDTA. Cells were then incubated in DMEM containing 1 mM Ca2+ and 10 μM AMD3100 (CXCR4 antagonist) and incubated at 37°C for 30 and 60 min. The amount of antibody reappearing on the cell surface was quantified by ELISA, as described in Materials and Methods, and used as an indicator of receptor recycling. Bars represent the percentage of internalized receptor that recycled ± SEM from three independent experiments. (C) Bars represent the percentage of cell surface receptors internalized in cells treated with CXCL12 compared with vehicle treated cells. The error bars represent SEM from three independent experiments. (D) HeLa cells were transfected with GAPD and AMSH siRNA and treated and analyzed as in A. Data represent the mean ± SEM from three independent experiments.
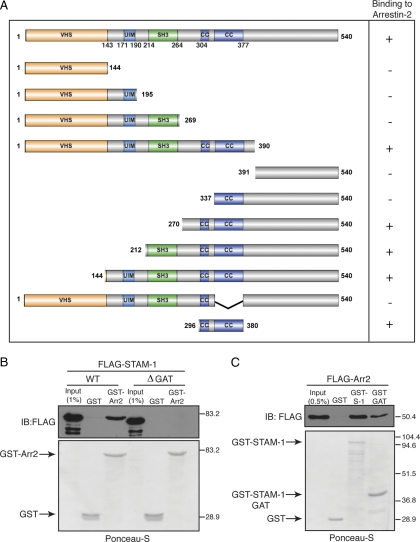
Mapping the Arrestin-2 Binding Site on STAM-1
We recently reported that arrestin-2 positively regulates CXCR4 sorting into the degradative pathway. To gain insight into the function of the arrestin-2/STAM-1 interaction on CXCR4 trafficking, we initially set out to determine the mechanism of the interaction. To accomplish this we mapped the arrestin-2 binding region on STAM-1 by truncation mutagenesis. STAMs contain multiple domains, characterized by the presence of an amino-terminal Vps27, Hrs, STAM homology (VHS) domain, UIM, SH3 (Src homology) domain, immunoreceptor based tyrosine activation motif (ITAM), and a GGA and TOM1 homologous (GAT) domain that partially overlaps with the ITAM (Prag et al., 2007; Ren et al., 2009). We created several STAM-1 N-terminal and C-terminal truncation mutants, according to its domain organization, tagged with the FLAG epitope on the amino-terminal end (Figure 5A). GST-arrestin-2 and GST immobilized on glutathione-Sepharose resin were incubated with lysates expressing the various STAM-1 truncation mutants, and bound proteins were detected by immunoblotting. The results from these experiments are summarized in Figure 5A and the data are shown in Supplemental Figure S1. The arrestin-2 binding region was determined to reside between amino acid residues 296-380 on STAM-1. This region encompasses the GAT domain, which has been shown to form two tandem coiled-coil domains (amino-acid residues 301–377) (Prag et al., 2007; Ren et al., 2009). To further confirm that the GAT domain mediates binding to arrestin-2, deletion of the GAT domain abrogated STAM-1 binding to arrestin-2 (Figure 5B), and the GAT domain alone fused to GST was able to bind to arrestin-2 (Figure 5C).
Figure 5.
The STAM-1 GAT domain is both necessary and sufficient for arrestin-2 binding. (A) STAM-1 truncation mutants are represented schematically. Binding or no binding to GST-arrestin-2 is represented by + and − symbols, respectively, on the right as assessed by data shown in Supplemental Figure S1. (B) Equimolar amounts (∼600 nM) of GST-arrestin-2 and GST were incubated with lysates from HEK293 cells transiently transfected with FLAG-tagged full-length-STAM-1 or STAM-1-ΔGAT. (C) Equimolar amounts (∼117 nM) of (GST-STAM-1,) GST-STAM-1-GAT and GST were incubated with lysates from HEK293 cells transiently transfected with FLAG-tagged arrestin-2. In B and C, bound proteins were detected by immunoblotting, followed by staining blots with Ponceau-S to assess the amount of GST fusion protein used in the binding assay. Shown are representative blots from one of three independent experiments.
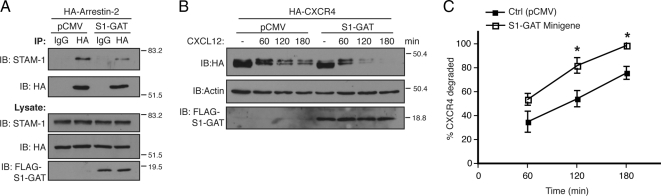
To determine whether the interaction between STAM-1 and arrestin-2 is important for CXCR4 trafficking, we initially expressed the GAT domain as a minigene in cells and assessed whether it disrupted the arrestin-2/STAM-1 interaction. HeLa cells transfected with FLAG-S1-GAT and HA-arrestin-2 were subjected to immunoprecipitation using an anti-HA antibody followed by immunoblotting to detect the presence of endogenous STAM-1 in the immunoprecipitates. As shown in Figure 6A, expression of the GAT domain disrupted the arrestin-2/STAM-1 interaction. To determine the function of the arrestin-2/STAM-1 interaction on lysosomal targeting of CXCR4, we examined the effect of expressing the GAT domain on CXCR4 degradation. Remarkably, expression of the GAT domain significantly accelerated CXCR4 degradation after agonist treatment as compared with empty vector (Figure 6, B and C). Together, these data suggest that the STAM-1/arrestin-2 interaction negatively regulates CXCR4 sorting to lysosomes. Because the STAM-1 GAT domain has been shown recently to bind to HRS and is predicted to be required for the assembly of ESCRT-0 (Ren et al., 2009), it is conceivable that arrestin-2 binding to STAM-1 displaces its interaction with HRS and promotes disassembly of ESCRT-0, which somehow negatively regulates the amount of CXCR4 that is targeted for lysosomal degradation.
Figure 6.
Expression of the GAT domain disrupts the arrestin-2/STAM-1 interaction and accelerates CXCR4 degradation. (A) Lysates from HeLa cells cotransfected with HA-arrestin-2 and FLAG-STAM-1-GAT (S1-GAT) or empty vector (pCMV) were incubated with anti-HA and IgG control antibodies. Immunoprecipitates were analyzed by immunoblotting to detect bound endogenous STAM-1, and lysates were analyzed to assess expression of the various constructs. Shown are representative blots from one of three independent experiments. (B) HA-CXCR4 degradation was assessed in HEK293 cells expressing FLAG-STAM-1-GAT or empty vector (pCMV) as described in Materials and Methods. (C) Graphical representation of percentage of receptor degraded. Error bars represent SEM from three independent experiments. Data were analyzed by two-way ANOVA and followed by a Bonferroni's post hoc test. (*p < 0.0001). Shown are representative blots from one of three independent experiments.
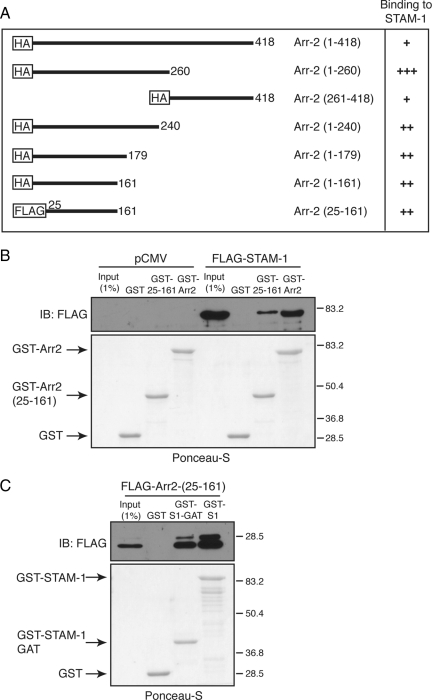
Mapping the STAM-1 Binding Site on Arrestin-2
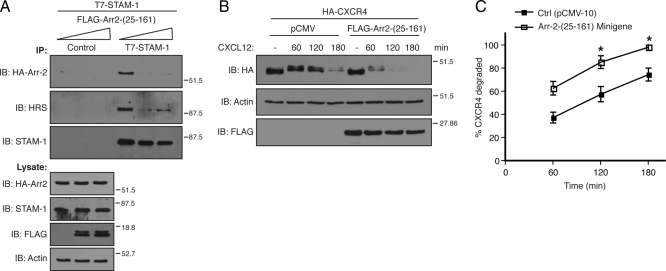
To gain greater insight into this process, we next set out to identify the STAM-1 binding region on arrestin-2 by truncation mutagenesis. Schematic representations of the arrestin-2 truncation mutants used are shown in Figure 7A; most have been described previously (Bhandari et al., 2007). GST-STAM-1 and GST were incubated with lysates prepared from HEK293 cells expressing various HA-tagged arrestin-2 truncation mutants. The results from these binding experiments are summarized in Figure 7A, and the data are shown in Supplemental Figure S2. Both the N- and C-terminal regions of arrestin-2 bound to GST-STAM1, but not GST, although binding to the N-terminal region seemed to be stronger, suggesting that it represented the main binding region. Further deletion of this region revealed that the STAM-1 binding site on arrestin-2 is between amino acid residues 1–161 (Supplemental Figure S2B). We next determined whether expression of this region as a minigene in cells also disrupted the arrestin-2/STAM-1 interaction. However, when expressed in cells the arrestin-2-(1-161) minigene completely blocked CXCR4 degradation (data not shown). The N-terminal lysine residues within arrestin-2 are predicted to serve as phosphosensors and recognize phosphates attached to receptors (Kern et al., 2009), analogous to what has been observed for arrestin-1 (Vishnivetskiy et al., 2000); therefore, the arrestin-2-(1-161) construct may bind to CXCR4 and have a dominant-negative effect on CXCR4 internalization. To rule out any effects at the level of internalization, the first 24 amino acids from the N terminus of arrestin-2 were deleted to create arrestin-2-(25-161), and we initially tested the ability of this mutant to bind to STAM-1. As shown in Figure 7B, GST fused to arrestin-2-(25-161), but not GST alone, efficiently bound to FLAG-STAM-1 expressed in cells. A FLAG-tagged construct of arrestin-2-(25-161) when expressed in HEK293 cells also bound to GST-STAM-1-GAT, suggesting that the STAM-1/GAT domain binding site on arrestin-2 is located between amino acid residues 25-161 (Figure 7C). We next examined whether expression of arrestin-2-(25-161) disrupted the STAM-1/arrestin-2 interaction and modulated CXCR4 degradation. Expression of FLAG-arrestin-2-(25-161) markedly disrupted the interaction between arrestin-2 and STAM-1 (Figure 8A) and significantly accelerated agonist promoted degradation of CXCR4 (Figure 8, B and C), similar to what was observed with the STAM-1 GAT domain (Figure 6). Together these data further indicate that the interaction between STAM-1 and arrestin-2 attenuates CXCR4 trafficking into the degradative pathway.
Figure 7.
Mapping of the STAM-1 binding domain on arrestin-2. (A) Arrestin-2 truncation mutants used in the binding studies are represented schematically. Binding between GST-STAM-1 and HA-tagged arrestin-2 truncation mutants is shown as weak (+), intermediate (++), and strong (+++) on the right. (B) Equimolar amounts (∼234 nM) of GST-arrestin-2, GST-Arr2-(25-161), and GST were incubated with lysates from HEK293 cells transiently transfected with FLAG-tagged STAM-1 and empty vector (pCMV-10). (C) Equimolar amounts (∼276 nM) of GST-STAM-1, GST-STAM-1-GAT, and GST alone were incubated with lysates from HEK293 cells transiently transfected with FLAG-Arr-2-(25-161). In B and C, bound proteins were detected by immunoblotting using an anti-FLAG antibody and blots were stained with Ponceau-S to assess the amount of GST-tagged protein used in the binding assay. Shown are representative blots from one of three independent experiments.
Figure 8.
Expression of Arr2-(25-161) disrupts the STAM-1/arrestin-2 interaction and accelerates CXCR4 degradation. (A) Lysates were prepared from HeLa cells cotransfected with T7-STAM-1, HA-arrestin-2 and increasing amounts (0, 1, and 2.5 μg) of FLAG-Arr2 (25-161). Lysates were divided into equal aliquots and incubated with either an anti-T7 polyclonal antibody or protein G agarose alone (control). Immunoprecipitates were analyzed by immunoblotting to detect bound HA-arrestin-2 and endogenous HRS and lysates were analyzed to assess the expression of the various constructs. Shown are representative blots from one of three independent experiments. (B) HA-CXCR4 degradation was assessed in HEK293 cells expressing FLAG-Arr2-(25-161) or empty vector (pCMV) as described in Materials and Methods. (C) Graphical representation of percent receptor degraded. Error bars represent SEM from three independent experiments. Data were analyzed by two-way ANOVA and followed by a Bonferroni's post hoc test. (*p < 0.0001). Shown are representative blots from one of three independent experiments.
Role of the Arrestin-2/STAM-1 Interaction on the Ubiquitination Status of CXCR4, STAM-1, and HRS
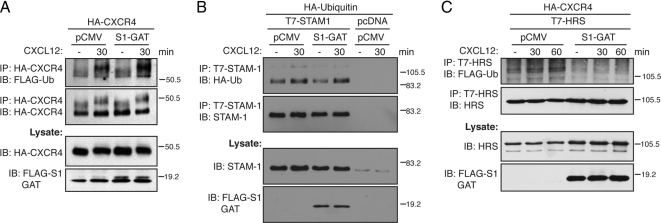
STAM, through its interaction with several deubiquitinating enzymes, may regulate the ubiquitination status of both cargo and of itself (McCullough et al., 2006; Row et al., 2006). Therefore, one possibility is that the STAM-1/arrestin-2 interaction modulates the ubiquitination status of CXCR4 and STAM-1, thereby facilitating CXCR4 trafficking into the degradative pathway. To examine this possibility we examined the effect of expressing the GAT domain on the ubiquitination status of both CXCR4 and STAM-1. Surprisingly, expression of the GAT, compared with empty vector, did not significantly change the ubiquitination status of CXCR4 (Figure 9A) and STAM-1 (Figure 9B), suggesting that the STAM-1/arrestin-2 interaction does not regulate their ubiquitination status. In sharp contrast, expression of the GAT domain blocked CXCR4 mediated ubiquitination of HRS (Figure 9C). Therefore, together our data show that the STAM-1/arrestin-2 interaction is critical for modulating ubiquitination of HRS, which is probably important for regulating sorting of CXCR4 into the degradative pathway.
Figure 9.
Disrupting the STAM-1/arrestin-2 interaction inhibits HRS ubiquitination but does not effect on CXCR4 and STAM-1 ubiquitination. (A) HEK293 cells stably expressing HA-CXCR4 were transfected with FLAG-ubiquitin and STAM-1-GAT domain or pCMV. (B) HeLa cells were transfected with HA-ubiquitin, T7-STAM-1, and STAM-1-GAT or pCMV. (C) Cells were transfected as in A, except T7-HRS was also transfected. Cells were serum starved and treated with 30 nM CXCL12 for 30–60 min, followed by immunoprecipitation and immunoblotting to detect incorporated ubiquitin as described in Materials and Methods. Shown are representative blots from six (A) or three (B and C) independent experiments.
DISCUSSION
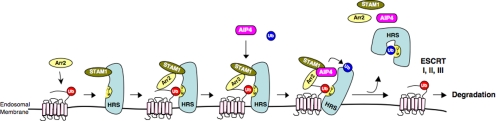
Nonvisual arrestins are known for their ability to mediate GPCR desensitization, trafficking, and signaling (Moore et al., 2007; Kovacs et al., 2009). We have reported recently that arrestin-2 interacts with AIP4 and mediates endosomal sorting of CXCR4 into the degradative pathway (Bhandari et al., 2007). Here, we extend these findings and provide further mechanistic insight into this unprecedented role of arrestin-2. Our data suggest that arrestin-2 mediates multiple interactions with ESCRT-0 on early endosomes, serving to regulate the amount of CXCR4 that is degraded. Arrestin-2 probably links ubiquitinated CXCR4 to ESCRT-0 via an initial interaction with HRS and/or STAM-1. Interestingly, our data reveal that the arrestin-2 interaction with STAM-1 is important for regulating ubiquitination of HRS, which we propose attenuates HRS sorting function, thereby controlling the extent to which CXCR4 is degraded (see model in Figure 10).
Figure 10.
Proposed mechanism for the role of the STAM-1/arrestin-2 complex in endosomal sorting of CXCR4. CXCR4 is ubiquitinated by the E3 ubiquitin ligase AIP4 at the plasma membrane, after which it is internalized onto early endosomes, although ubiquitination is not required for this process. Once on endosomes ubiquitinated CXCR4 is recognized by HRS, likely by an interaction involving the ubiquitin moiety (red) on CXCR4 and the UIM of HRS, and possibly via an interaction with arrestin-2. Arrestin-2 then interacts with STAM-1, which serves to recruit AIP4 culminating in the ubiquitination of HRS. We speculate that this triggers a conformational change in HRS induced by an interaction between the ubiquitin moiety (blue) and the internal UIM. CXCR4 is subsequently committed to downstream interactions with ESCRT-I-III, whereas arrestin-2, STAM-1, AIP4, and autoinhibited HRS are recycled such that another round of sorting can take place.
We used truncation mutagenesis to narrow the arrestin-2 binding region on STAM-1 to the GAT domain and the STAM-1 binding region on arrestin-2 to amino acid residues 25-161. Expression of both of these domains similarly disrupted the arrestin-2/STAM-1 interaction and enhanced agonist promoted degradation of CXCR4. Our data are consistent with the notion that the STAM-1/arrestin-2 interaction negatively regulates sorting of CXCR4 into the degradative pathway. This interaction may be specific to modulating CXCR4 and/or GPCR sorting as EGFR degradation was not altered by expression of the STAM-1 GAT domain and arrestin-2-(25-161) (Supplemental Figure S3). Depletion of STAM-1 by siRNA also enhanced CXCR4 degradation further revealing that STAM-1 negatively regulates CXCR4 endosomal sorting (Figure 4A). In contrast, we have shown previously that arrestin-2 promotes CXCR4 sorting (Bhandari et al., 2007) and when considered with our present data indicate that arrestin-2 has opposing effects on CXCR4 degradation. This suggests that arrestin-2 probably acts at multiple steps in the sorting process and may initially act upstream of STAM-1 to positively regulate sorting of CXCR4 into the degradative pathway. Arrestin-2 interacts with the C-tail of CXCR4 (Busillo et al., 2010); therefore, it is possible that arrestin-2 binds to CXCR4 on endosomes in order to recruit CXCR4 to ESCRT-0, possibly through an interaction with either HRS and/or STAM-1. This is consistent with our data that shows that arrestin-2 colocalizes with CXCR4 and STAM-1 on early endosomes upon agonist activation (Figure 3). Interestingly, a recent study found that Rim8, a Saccharomyces cerevisiae molecule distantly related to mammalian arrestins, may function to directly recruit a putative 7TM receptor to the ESCRT machinery (Herrador et al., 2010). After arrestin-2 initially directs CXCR4 to ESCRT-0, this is probably followed by an interaction with STAM-1 to attenuate CXCR4 degradation. Therefore, our data are consistent with a model in which arrestin-2 influences CXCR4 sorting positively and negatively, and it is a balance of these two activities that dictates the extent to which CXCR4 is degraded.
How does STAM-1 mediate the negative action of arrestin-2 on CXCR4 degradation? Because ubiquitination of HRS is markedly reduced by expression of the GAT domain, it is likely that STAM-1 via its interaction with arrestin-2 regulates the ubiquitination status of HRS to control CXCR4 degradation. This suggests that CXCR4 promoted ubiquitination of HRS (Figure 9C; Marchese et al., 2003) attenuates its sorting activity. HRS contains a UIM that is thought to bind to ubiquitin moieties on cargo to recruit them into the degradative pathway (Hirano et al., 2006). Interestingly, monoubiquitination of UBD containing proteins is thought to induce an intramolecular interaction between the ubiquitin moiety and the internal UBD, which in a protein such as HRS may induce an autoinhibitory conformation such that it can no longer bind to ubiquitin moieties on cargo (Hoeller et al., 2006). Because HRS ubiquitination is reduced by expression of the GAT domain, a loss of autoinhibition probably enhances its sorting function culminating in enhanced degradation of CXCR4. Therefore, CXCR4 promoted ubiquitination of HRS may occur once HRS has completed its sorting function and CXCR4 has been committed to downstream elements of the degradative pathway (Figure 10).
How does arrestin-2/STAM-1 regulate the ubiquitination status of HRS? We have shown previously that arrestin-2 interacts with AIP4 to regulate endosomal sorting of CXCR4 (Bhandari et al., 2007) and that AIP4 mediates agonist promoted ubiquitination of HRS (Marchese et al., 2003). Therefore, it is possible that arrestin-2 together with STAM-1 may serve to bridge AIP4 and HRS to facilitate HRS ubiquitination by AIP4. This is consistent with our data that show that expression of arrestin-2-(25-161) also displaces HRS binding to arrestin-2/STAM-1 (Figure 8A). Alternatively, the arrestin-2/STAM-1 complex may regulate HRS deubiquitination. STAM has been shown to interact with deubiquitinating enzymes, such as AMSH and UBPY, which have been shown to regulate the ubiquitination status of cargo (e.g., EGFR, protease activated receptor 2) and/or of STAM itself (McCullough et al., 2004; Row et al., 2006; Hasdemir et al., 2009). However, the arrestin-2/STAM-1 complex does not seem to modulate the ubiquitination status of CXCR4 and nor of STAM-1 (Figure 9). In addition, depletion of AMSH did not affect agonist promoted degradation of CXCR4 (Figure 4D), suggesting that it may not be linked to this process, although it does not exclude the possibility that other DUBs may be involved (Row et al., 2006; Shenoy et al., 2009). Nevertheless, our data are consistent with the notion that the arrestin-2/STAM-1 complex mediates ubiquitination of HRS probably via AIP4.
Interestingly, STAM-2 seems to be excluded from endosomal sorting of CXCR4 because arrestin-2 binds selectively to STAM-1 (Figure 1A). This suggests that CXCR4 sorting is restricted to ESCRT-0 complexes that contain STAM-1 but not STAM-2. We also observed that activation of CXCR4 selectively enhances STAM-1 ubiquitination over STAM-2 (Figure 2B), further supporting the selectivity of STAM-1 toward CXCR4. However, the arrestin-2/STAM-1 interaction may not be linked to STAM-1 ubiquitination (Figure 9B). Presently, the function of STAM-1 ubiquitination on CXCR4 trafficking remains unknown, although it is possible that it may have a role in some other aspect of CXCR4-related functions. Polyubiquitination of STAM has been linked to its degradation (Row et al., 2006); however, it is doubtful that CXCR4 regulates STAM-1 stability because we did not observe any differences in STAM-1 levels in cells treated with CXCL12 (data not shown).
In summary, our study provides novel mechanistic insight into the role of arrestin-2 in endosomal sorting of CXCR4 via multiple interactions with ESCRT-0. We reveal here that via an interaction with STAM-1, arrestin-2 serves as an adaptor to regulate endosomal ubiquitination events that are critical for regulating sorting of ubiquitinated CXCR4 into the degradative pathway, thereby controlling the amount of CXCR4 that is degraded.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM-75159 (to A. M.) and by American Heart Association predoctoral fellowship 0910098G (to R. M.).
Abbreviations used:
- AIP
atrophin-1–interacting protein
- AMSH
associated molecule with the Src homology 3 domain of STAM
- EGFR
epidermal growth factor receptor
- ESCRT
endosomal sorting complex required for transport
- GPCR
G protein-coupled receptor
- GST
glutathione transferase
- HRS
hepatocyte growth factor-regulated tyrosine kinase substrate
- SDF
stromal cell-derived factor
- STAM
signal transduction adaptor molecule
- UBD
ubiquitin binding domain
- UIM
ubiquitin interacting motif.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0169) on May 26, 2010.
REFERENCES
- Abella J. V., Peschard P., Naujokas M. A., Lin T., Saucier C., Urbe S., Park M. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell. Biol. 2005;25:9632–9645. doi: 10.1128/MCB.25.21.9632-9645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asao H., Sasaki Y., Arita T., Tanaka N., Endo K., Kasai H., Takeshita T., Endo Y., Fujita T., Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 1997;272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- Bache K. G., Brech A., Mehlum A., Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 2003a;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bache K. G., Raiborg C., Mehlum A., Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 2003b;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- Bhandari D., Robia S. L., Marchese A. The E3 ubiquitin ligase atrophin interacting protein 4 binds directly to the chemokine receptor CXCR4 via a novel WW domain-mediated interaction. Mol. Biol. Cell. 2009;20:1324–1339. doi: 10.1091/mbc.E08-03-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D., Trejo J., Benovic J. L., Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J. Biol. Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- Bowers K., Piper S. C., Edeling M. A., Gray S. R., Owen D. J., Lehner P. J., Luzio J. P. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- Busillo J. M., Armando S., Sengupta R., Meucci O., Bouvier M., Benovic J. L. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J. Biol. Chem. 2010;285:7805–7817. doi: 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K., et al. STAM2, a new member of the STAM family, binding to the Janus kinases. FEBS Lett. 2000;477:55–61. doi: 10.1016/s0014-5793(00)01760-9. [DOI] [PubMed] [Google Scholar]
- Hasdemir B., Bunnett N. W., Cottrell G. S. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) mediates post-endocytic trafficking of protease-activated receptor 2 and calcitonin receptor-like receptor. J. Biol. Chem. 2007;282:29646–29657. doi: 10.1074/jbc.M702974200. [DOI] [PubMed] [Google Scholar]
- Hasdemir B., Murphy J. E., Cottrell G. S., Bunnett N. W. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J. Biol. Chem. 2009;284:28453–28466. doi: 10.1074/jbc.M109.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A., Herranz S., Lara D., Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S., Kawasaki M., Ura H., Kato R., Raiborg C., Stenmark H., Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat. Struct. Mol. Biol. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- Hoeller D., et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- Kanazawa C., Morita E., Yamada M., Ishii N., Miura S., Asao H., Yoshimori T., Sugamura K. Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem. Biophys. Res. Commun. 2003;309:848–856. doi: 10.1016/j.bbrc.2003.08.078. [DOI] [PubMed] [Google Scholar]
- Kern R. C., Kang D. S., Benovic J. L. Arrestin2/clathrin interaction is regulated by key N- and C-terminal regions in arrestin2. Biochemistry. 2009;48:7190–7200. doi: 10.1021/bi900369c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong C., Su X., Chen P. I., Stahl P. D. Rin1 interacts with signal-transducing adaptor molecule (STAM) and mediates epidermal growth factor receptor trafficking and degradation. J. Biol. Chem. 2007;282:15294–15301. doi: 10.1074/jbc.M611538200. [DOI] [PubMed] [Google Scholar]
- Kovacs J. J., Hara M. R., Davenport C. L., Kim J., Lefkowitz R. J. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev. Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O., Poussu A., Merilainen J., Kellokumpu S., Wasenius V. M., Lehto V. P. EAST, an epidermal growth factor receptor- and Eps15-associated protein with Src homology 3 and tyrosine-based activation motif domains. J. Biol. Chem. 1998;273:21408–21415. doi: 10.1074/jbc.273.33.21408. [DOI] [PubMed] [Google Scholar]
- Ma Y. M., Boucrot E., Villen J., Affar el B., Gygi S. P., Gottlinger H. G., Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- Marchese A. Ubiquitination of chemokine receptors. Methods Enzymol. 2009;460:413–422. doi: 10.1016/S0076-6879(09)05221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A., Benovic J. L. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J. Biol. Chem. 2001;276:45509–45512. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 2003;5:709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M. J., Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Moore C. A., Milano S. K., Benovic J. L. Regulation of receptor trafficking by GRKs and arrestins. Ann. Rev. Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- Pandey A., Fernandez M. M., Steen H., Blagoev B., Nielsen M. M., Roche S., Mann M., Lodish H. F. Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J. Biol. Chem. 2000;275:38633–38639. doi: 10.1074/jbc.M007849200. [DOI] [PubMed] [Google Scholar]
- Prag G., Watson H., Kim Y. C., Beach B. M., Ghirlando R., Hummer G., Bonifacino J. S., Hurley J. H. The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell. 2007;12:973–986. doi: 10.1016/j.devcel.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Ren X., Kloer D. P., Kim Y. C., Ghirlando R., Saidi L. F., Hummer G., Hurley J. H. Hybrid structural model of the complete human ESCRT-0 complex. Structure. 2009;17:406–416. doi: 10.1016/j.str.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanchi N., Puertollano R., Blackstone C. STAM adaptor proteins interact with COPII complexes and function in ER-to-Golgi trafficking. Traffic. 2009;10:201–217. doi: 10.1111/j.1600-0854.2008.00856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P. E., Prior I. A., McCullough J., Clague M. J., Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- Shenoy S. K., Modi A. S., Shukla A. K., Xiao K., Berthouze M., Ahn S., Wilkinson K. D., Miller W. E., Lefkowitz R. J. Beta-arrestin-dependent signaling and trafficking of 7-transmembrane receptors is reciprocally regulated by the deubiquitinase USP33 and the E3 ligase Mdm2. Proc. Nat. Acad. Sci. USA. 2009;106:6650–6655. doi: 10.1073/pnas.0901083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S. B., Oestreich A. J., Winistorfer S., Nguyen D., Payne J. A., Katzmann D. J., Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra M. I., Wright M. H., Nash P. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J. Biol. Chem. 2010;285:13990–14004. doi: 10.1074/jbc.M109.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H., Kato M., Denda K., Kitamura N. A hrs binding protein having a Src homology 3 domain is involved in intracellular degradation of growth factors and their receptors. Genes Cells. 2000;5:57–69. doi: 10.1046/j.1365-2443.2000.00303.x. [DOI] [PubMed] [Google Scholar]
- Takeshita T., et al. Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem. Biophys. Res. Commun. 1996;225:1035–1039. doi: 10.1006/bbrc.1996.1290. [DOI] [PubMed] [Google Scholar]
- Takeshita T., Arita T., Higuchi M., Asao H., Endo K., Kuroda H., Tanaka N., Murata K., Ishii N., Sugamura K. STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity. 1997;6:449–457. doi: 10.1016/s1074-7613(00)80288-5. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy S. A., Schubert C., Climaco G. C., Gurevich Y. V., Velez M. G., Gurevich V. V. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Yamada M., Ishii N., Asao H., Murata K., Kanazawa C., Sasaki H., Sugamura K. Signal-transducing adaptor molecules STAM1 and STAM2 are required for T-cell development and survival. Mol. Cell. Biol. 2002;22:8648–8658. doi: 10.1128/MCB.22.24.8648-8658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.